Abstract
Objectives: The receptor for advanced glycation end products (RAGE) is an oncogenic trans-membranous receptor expressed in many cells. The aim of this study was to clarify the association between RAGE gene 4 single nucleotide polymorphisms (SNPs) and the risk, invasion, metastasis and overall survival of gastric cancer.
Methods and Results: We performed a hospital-based case-control study involving 369 gastric cancer patients and 493 cancer free controls. Four widely-studied SNPs, rs1800625 (T-429C), rs1800624 (T-374A), rs2070600 (Gly82Ser) and rs184003 (G1704T) in RAGE gene, were genotyped by the polymerase chain reaction - ligase detection reaction method. The RAGE gene rs1800625 TT genotype and T allele were significantly associated with a reduced risk of gastric cancer (TT vs. CC: adjusted odds ratio [OR]: 0.72, 95% CI: 0.55-0.95, p=0.021; T vs. C: adjusted OR: 0.67, 95% CI: 0.46-0.97, p=0.032). No hints of significance were detected for the other three SNPs in association with gastric cancer risk. Moreover, rs1800625 and rs184003 were significantly associated with tumor clinical stage (p=0.010 and 0.032, respectively). Survival curves differed significantly between the genotypes of rs1800625.
Conclusions: RAGE gene SNP rs1800625 was significantly associated with gastric cancer risk, and rs1800625 and rs184003 were related to tumor clinical stage, indicating that RAGE gene may be a gastric cancer-susceptibility gene.
Keywords: Gastric cancer, RAGE, Single nucleotide polymorphism, Risk, Survival
Introduction
Gastric cancer is a major cause of cancer-related deaths worldwide, and it is the fifth most common malignancy 1-3. Gastric cancer has a high incidence rate in East Asia, and in particular nearly 42% of global gastric cancer cases occur in China 4. Despite advances in diagnosis and treatment, the prognosis for patients with advanced gastric cancer remains dismal.5 Gastric carcinogenesis is a complex, multistep and sequential process 6. Like other malignancies, many factors may contribute to the development of gastric cancer, such as nutrition, dietary habits, salt consumption, excessive alcohol intake, tobacco smoking, Helicobacter pylori (H.pylori) infection, chronic gastritis, atrophyintestinal metaplasia and dysplasia 7-12. Besides these factors, a large number of studies have demonstrated that the development of gastric cancer is partly under genetic control 13-15. Recently, several promising genes have been identified as potential gastric cancer susceptibility genes. An important one is the receptor for advanced glycation end products (RAGE) gene.
RAGE is an oncogenic multidisciplinary trans-membranous receptor expressed in a number of cells. As a member of immunoglobulin superfamily of receptors, RAGE carries unique properties that make it a key component in the pathogenesis of many diseases and their impediments, such as inflammation, vascular diseases and different malignancies 16-18. In addition, numerous clinical studies confirmed that the expression of RAGE was associated with prostate cancer 19, gastric cancer 20, gallbladder cancer 21, pancreatic cancer 22, lung cancer 23, colorectal cancer 24 and oral squamous cell carcinoma 25. Besides, growing evidence suggests that some single nucleotide polymorphisms (SNPs) in RAGE gene were related to the angiogenesis and tumor metastasis 25, and blocking RAGE signal can inhibit the growth and metastasis of glioma cells 26. Gu et al. reported that mutation of RAGE gene rs2070600 was associated with a significantly higher risk of gastric cancer 15. Based on above observations, it is reasonable to hypothesize that RAGE gene is a promising candidate of gastric cancer. To test this hypothesis, we here genotyped four well-characterized SNPs in RAGE gene, rs1800625 (T-429C), rs1800624 (T-374A), rs2070600 (Gly82Ser) and rs184003 (G1704T), to see whether their genetic variation can alter the risk of gastric cancer and associate with its invasion, metastasis and overall survival.
Materials and Methods
Ethics Statement
This study was approved by the Research Ethics Committee of Fujian Provincial Cancer Hospital.
Patients and Samples
This study included 387 patients who had gastric cancer confirmed by histopathology at Fujian Provincial Cancer Hospital between January 2008 and December 2010, and all patients received no chemotherapy or radiotherapy. Of the total 387 patients recruited in this study, 18 patients were lost to follow-up, 369 patients with gastric cancer were assessed in final analysis. Another 493 cases of the same period who took physical examinations at this hospital were selected by a random number table method as control group, and they were matched with patients on age (± 5 years old), sex and place of residence. All study subjects were Han Chinese who lived in Fujian Province for more than 10 years, and had no blood relationship with each other. Tumor histological grade was determined according to the American Joint Committee on Cancer (AJCC) system (2010) and the International Union against Cancer (UICC). Written informed consents were obtained from all patients and controls.
Biospecimen and DNA Extraction
The adjacent tissues of gastric cancer specimens from patients were embedded in paraffin using standard methods. Then, genomic DNA was extracted according to the Tiagen DNA kit instructions. Controls provided 5 ml venous EDTA anticoagulant blood according to Beijing BioTeke of the whole blood extraction DNA kit operating instructions. The amount of DNA was detected by UV spectrophotometer, and absorbance values (A values), A260 and A280 were measured with the ratio ranging from 1.8 to 2.0.
SNP Selection and Genotyping
Four SNPs in RAGE gene, rs1800625 C>T, rs1800624 T>A, rs2070600 G>A and rs184003 G>T, that were widely evaluated with association diabetes and cancer were selected for analysis. SNP genotype information was retrieved from the dbSNP database (http://www.ncbi.nlm.gov/projects/SNP/). Genotyping was performed using the polymerase chain reaction - ligase detection reaction (PCR-LDR) method. The DNA fragment containing the SNP loci was amplified by PCR using an ABI9600 PCR instrument (Applied Biosystems, Foster City, CA, USA). The characteristics of sequences in this study are summarized in Table 1.
Table 1.
Primer sequences of RAGE gene four SNPs and the test of Hardy-Weinberg equilibrium
| SNPs | Upstream sequence | Downstream sequence | Case group* | Control group* |
|---|---|---|---|---|
| rs1800625 | 5'-AAACATGAGAAACCCCAGAA-3' | 5'-TGTACCCTCTAGGGTCTCATTC-3' | 0.699a | 0.277b |
| rs1800624 | 5'-AAACATGAGAAACCCCAGAA-3' | 5'-TGTACCCTCTAGGGTCTCATTC-3' | 0.075 | 0.061 |
| rs2070600 | 5'-CGTCACTCTGCCTCACAGTC-3' | 5'-GTCTCCTTTCCATTCCTGTTC-3' | 0.251 | 0.729 |
| rs184003 | 5'-AATTGGTGGTGGAGCCAGAAG-3' | 5'-TTCAGATACCCTCTCTTCCTC-3' | 0.440 | 0.799 |
*The p values of Hardy-Weinberg equilibrium test.
PCR reaction was carried out in a total reaction volume of 15 μl, containing DNA 1 μl, 1.5 μl 10×buffer, 1.5 μl MgCL2, 0.3 μl DNTP (Fermentas, United States), 0.15 μl of each primer, 0.3 μl Taq enzyme (Fermentas, United States) and 10.25 μl H2O. The steps were as follows: initial denaturation step at 94℃ for 3 min, amplification was carried out by 35 cycles at 94℃ for 15 s, 54℃ for 15 s, and at 72℃ for 30 s, followed by a final elongation cycle at 72℃ for 3 min. The connection reaction was performed at adding the PCR product 3 μl, 1 U 10×Taq DNA ligase buffer (New England Biolabs, United States), 0.125 μl Taq DNA ligase (40 U/μl, New England Biolabs, United States ), probe (10p) 0.01 μl/strip and with H2O added to 10 μl under the condition of by cycles at 94℃ for 30 s, 56℃ for 3 min. LDR fluorescent products were analyzed by ABI 377 sequence analyzer (Perkin-Elmer, Foster City, CA, USA), and the results were analyzed by the fragment analysis software GeneMapper 3.7 (Applied Biosystems, Foster city, CA, USA).
To test the accuracy of PCR-LDR method, a total of 96 DNA samples were randomly selected from the total sample, and they were genotyped using the same method, with exactly the sample findings for duplicated samples.
Statistical Analysis
Statistical analysis was performed using the SPSS software version 19.0 (SPSS Inc). Continuous variables were expressed as mean ± standard deviation (SD). Statistical significance was tested by a chi-square (χ2) or the Fisher's exact test. Satisfaction of all genotypes with the Hardy-Weinberg proportions was calculated in control groups. Linkage disequilibrium was depicted using the Haploview software version 4.2. The genotypes of four examined SNPs in RAGE gene were explored by unconditional logistic regression analyses to obtain odds ratio (OR) and its 95% confidence interval (CI). Survival curves were calculated according to the Kaplan-Meier method and compared using the log-rank test. A two-sided p-value < 0.05 were considered statistically significant.
Results
Clinical Characteristics
The final analysis included 369 cases and 493 controls who were frequency matched as previously described. About 70.7% of gastric cancer patients were male, and patients aged 60 years or over accounted for 48.5%. 69.0% of controls were male, and controls aged 60 years or over accounted for 48.1% (Table 2). There were no significant differences in age and sex between patients and controls (p>0.05). The genotype distributions of four SNPs in controls were checked by the Hardy-Weinberg equilibrium test (Table 1), and no deviations were noticed (all p>0.05).
Table 2.
Demographic characteristics of the study subjects
| Characteristics | Total subjects (n=862) | Case group (n=369) | Control group (n=493) |
|---|---|---|---|
| Age (years) | |||
| ≥ 60 | 416 | 179 (48.5%) | 237 (48.1%) |
| < 60 | 446 | 190 (51.5%) | 256 (51.9%) |
| Sex | |||
| Male | 601 | 261 (70.7%) | 340 (69.0%) |
| Female | 261 | 108 (29.3%) | 153 (31.0%) |
| TNM stage | |||
| I | 46 | ||
| II | 72 | ||
| III | 210 | ||
| IV | 41 | ||
| Lymph node metastasis | |||
| N0 | 88 | ||
| N1 | 86 | ||
| N2 | 81 | ||
| N3 | 114 |
Association between RAGE Gene SNPs and Gastric Cancer Risk
The linkage disequilibrium of four studied SNPs is presented in Supplementary Figure 1. In logistic regression analyses, RAGE gene rs1800625 TT genotype was associated with a significantly reduced risk of gastric cancer (OR: 0.72; 95% CI 0.55-0.95; p=0.021) after adjusting for age and sex, and the power to reject the null hypothesis that this OR equals 1 was 66.1% (Table 3). Additionally, the T allele also reduces the risk of gastric cancer (OR: 0.67; 95% CI 0.46-0.97; p=0.032), with study power of 82.4%. In contrast, there were no significant differences in the genotype and allele distributions of the other three polymorphisms between the cases and controls with the increase risk for the development of gastric cancer (all p>0.05).
Table 3.
Genotype distributions and allele frequencies of RAGE gene four SNPs between cases and controls
| SNPs | Controls (n=493) | Cases (n=369) | OR (95% CI)a | p |
|---|---|---|---|---|
| rs1800625 | ||||
| CC | 6 (1.2%) | 1 (0.3%) | 1.00ref | |
| CT | 77 (15.6%) | 44 (11.9%) | 0.21 (0.03-1.76) | 0.150 |
| TT | 410 (83.2%) | 324 (87.8%) | 0.72 (0.55-0.95) | 0.021 |
| C allele | 89 (9%) | 46 (6.2%) | 1.00ref | |
| T allele | 897 (91%) | 692 (93.8%) | 0.67 (0.46-0.97) | 0.032 |
| TT+CT vs. CC | 4.53 (0.54-37.82) | 0.249 | ||
| rs1800624 | ||||
| TT | 378 (76.7%) | 274 (74.3%) | 1.00ref | |
| AT | 102 (20.7%) | 83 (22.5%) | 0.79 (0.49-1.27) | 0.323 |
| AA | 13 (2.6%) | 12 (3.3%) | 0.88 (0.54-1.45) | 0.619 |
| T allele | 858 (87%) | 631 (85.5%) | 1.00ref | |
| A allele | 128 (13%) | 107 (14.5%) | 1.14 (0.86-1.50) | 0.360 |
| AA+AT vs. TT | 1.14 (0.83-1.56) | 0.236 | ||
| rs2070600 | ||||
| GG | 325 (65.9%) | 221 (59.9%) | 1.00ref | |
| GA | 152 (30.8%) | 134 (36.3%) | 0.78 (0.50-1.22) | 0.270 |
| AA | 16 (3.2%) | 14 (3.8%) | 1.01 (0.65-1.57) | 0.974 |
| G allele | 802 (81.3%) | 576 (78%) | 1.00ref | |
| A allele | 184 (18.7%) | 162 (22%) | 1.22 (0.97-1.55) | 0.092 |
| AA+AG vs. GG | 1.30 (0.98-1.71) | 0.074 | ||
| rs184003 | ||||
| GG | 318 (64.5%) | 240 (65%) | 1.00ref | |
| GT | 157 (31.8%) | 118 (32%) | 1.24 (0.78-1.97) | 0.373 |
| TT | 18 (3.7%) | 11 (3%) | 1.23 (0.77-1.96) | 0.383 |
| G allele | 793 (80.4%) | 598 (81%) | 1.00ref | |
| T allele | 193 (19.6%) | 140 (19%) | 1.04 (0.81-1.32) | 0.75 |
| TT+GT vs. GG | 0.98 (0.74-1.30) | 0.886 |
Abbreviations: OR: odds ratio; CI: confidence interval.
aThe p was calculated after adjusting for age and sex under logistic regression model.
Association between RAGE Gene SNPs and Clinical Features
Considering the association between SNPs and clinical features of patients with gastric cancer, we analyzed whether the variation of SNPs were linked to TNM stage and lymph node metastasis. In our study we found that the polymorphisms of RAGE gene rs1800625 and rs184003 were associated with tumor clinical stage (p=0.010, p=0.032, respectively) (Table 4). However, no evidence demonstrated that the SNPs of RAGE gene was related to Lymph node metastasis of gastric cancer (all p>0.05). As shown in Table 4.
Table 4.
Association between RAGE gene four SNPs and clinical characteristics of gastric cancer
| Clinical characteristics | rs1800625 | rs1800624 | |||||||||||
| CC | CT | TT | p | CT+TT | p | TT | AT | AA | p | AT+AA | p | ||
| Lymph node metastasis | |||||||||||||
| N0 | 0 | 9 | 79 | 0.532 | 88 | 0.303 | 69 | 16 | 3 | 0.562 | 19 | 0.329 | |
| N1-N3 | 1 | 32 | 231 | 263 | 192 | 64 | 8 | 72 | |||||
| TNM stage | |||||||||||||
| I-II | 0 | 15 | 92 | 0.01 | 107 | 0.002 | 84 | 22 | 1 | 0.364 | 23 | 0.233 | |
| III-IV | 1 | 26 | 215 | 241 | 175 | 57 | 10 | 67 | |||||
| rs2070600 | rs184003 | ||||||||||||
| GG | GA | AA | p | GA+AA | p | GG | GT | TT | p | GT+TT | p | ||
| Lymph node metastasis | |||||||||||||
| N0 | 54 | 30 | 4 | 0.984 | 34 | 0.95 | 56 | 28 | 4 | 0.73 | 32 | 0.86 | |
| N1-N3 | 158 | 97 | 9 | 106 | 173 | 84 | 7 | 91 | |||||
| TNM stage | |||||||||||||
| I-II | 63 | 37 | 7 | 0.793 | 44 | 0.908 | 70 | 34 | 3 | 0.032 | 37 | 0.427 | |
| III-IV | 147 | 89 | 6 | 95 | 159 | 76 | 7 | 83 | |||||
Abbreviations: TNM, tumor node metastasis.
Association between RAGE Gene SNPs and Gastric Cancer Survival
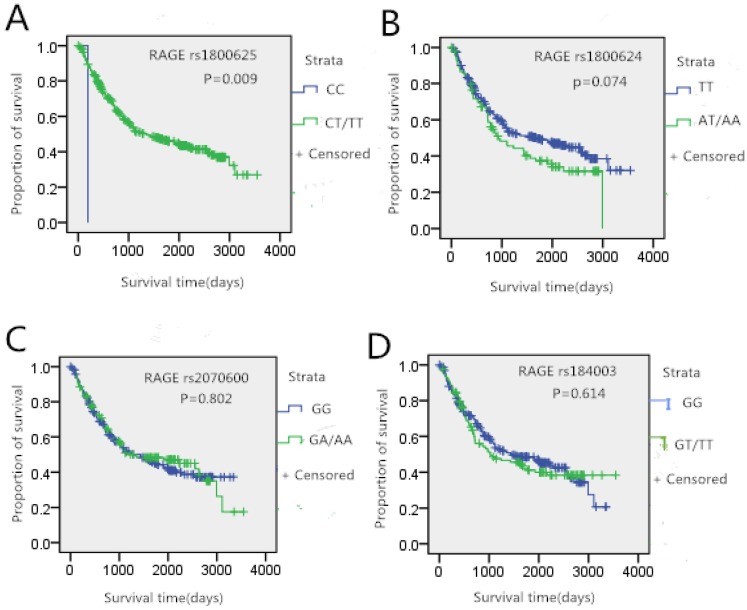
In this study, we further analyzed whether the genotypes of studied SNPs had an association with gastric cancer survival. The Kaplan-Meier curves of survival in gastric cancer patients were shown in Figure 1. There was no significant difference in gender, age, height and weight between gastric cancer survivors and death patients (p>0.05). A difference in survival curves was noted between rs1800625 genotypes. Patients who carried the T gene of rs1800625 had significantly longer survival time (p<0.05, Figure 1A). However, no significantly association between the other three SNPs and gastric cancer survival time was observed.
Figure 1.
Tumor site-specific Kaplan-Meier survival curves according the genotypes of RAGE gene four SNPs in gastric cancer patients
Discussion
The aim of this study was to test the hypothesis that RAGE gene was a promising candidate of gastric cancer. Our findings indicated that in RAGE gene, rs1800625 was associated with gastric cancer risk, and rs1800625 and rs184003 were related to tumor clinical stage, indicating that RAGE gene may be a gastric cancer-susceptibility gene.
It is well known that accumulating genetic variation plays crucial function roles within the tumorigenesis of gastric cancer 27. In order to clarify potential biomarkers of gastric carcinogenesis, we selected four SNPs of RAGE gene and investigated their associations with the risk, metastasis and survival of gastric cancer in a population of Fujian, and our findings proposed that potentially functional SNPs of RAGE gene may play a role in the etiology of gastric cancer.
RAGE activates central cell pathways including mitogen-activated protein kinase, Cdc42 / Rac and nuclear factor-nB signaling by binding to ligands 17,28,29. Previous studies demonstrated that the association between RAGE genetic polymorphisms and gastric cancer risk 15. In this study, we found that RAGE gene rs1800625 TT genotype and T allele were significantly associated with a reduced risk of gastric cancer. In addition, accumulating evidence shows that the expression of RAGE gene may be strongly related to the invasive and metastatic activity of gastric cancer 19,20. Our findings indicated that two SNPs in RAGE gene, rs1800625 and rs184003, were associated with tumor clinical stage. Besides, the patients who carried the T gene of rs1800625 had significantly longer survival time.
Gu et al. found that this SNP was associated with the increased risk of gastric cancer among subgroups of younger subjects aged ≤ 58 years 15. On the other hand, Li et al. reported that the rs2070600 AG genotype was significantly associated with an increased risk of gastric cancer among nonsmokers, nondrinkers and gastric cancer patients 1. However, our results did not show similar results, and may need a large sample size and more detailed risk factor information to validate.
Several limitations for this study should be acknowledged. First, we only genotyped four well-defined SNPs in RAGE gene, and other SNPs were of additional interest in association with the risk and prognosis of gastric cancer. Second, the sample size involved was not large enough to derive a reliable estimate, and so our findings needed to be confirmed in large, well-designed studies. Third, only Chinese subjects were enrolled for analysis, and the extrapolation of our findings to the other ethnic groups was limited.
Taken together, our findings indicated that RAGE gene rs1800625 was associated with gastric cancer risk, and rs1800625 and rs184003 were related to tumor clinical stage, indicating that RAGE gene may be a gastric cancer-susceptibility gene.
Supplementary Material
Supplementary figure.
Acknowledgments
Funding
This study was supported by the Natural Science Foundation of Fujian Province (Grant nos. 2016J01508), Joint Funds for the innovation of science and Technology, Fujian province (Grant number: 2017Y9090) and the Ministry of Health P.R. China (Grant no. WKJ2016-2-05).
Author Contributions
FP, XZ planned and designed the study, and directed its implementation; DH, FP, JL drafted the protocol; QL, XL, HZ obtained statutory and ethics approvals; DH, XL, HZ contributed to data acquisition; FP, DH, XZ had access to all raw data; DH, QL, FP did the data preparation, quality control and data analysis; FP, DH wrote the manuscript.
References
- 1.Li T, Qin W, Liu Y, Li S, Qin X, Liu Z. Effect of RAGE gene polymorphisms and circulating sRAGE levels on susceptibility to gastric cancer: a case-control study. Cancer Cell Int. 2017;17:19. doi: 10.1186/s12935-017-0391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Fidler MM, Gupta S, Soerjomataram I, Ferlay J, Steliarova-Foucher E, Bray F. Cancer incidence and mortality among young adults aged 20-39 years worldwide in 2012: a population-based study. Lancet Oncol. 2017;18:1579–89. doi: 10.1016/S1470-2045(17)30677-0. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM. International variation. Oncogene. 2004;23:6329–40. doi: 10.1038/sj.onc.1207726. [DOI] [PubMed] [Google Scholar]
- 5.Yue Z, Song Y, Wang Z, Luo Y, Jiang L, Xing L. et al. Association of heparanase gene (HPSE-1) single nucleotide polymorphisms with gastric cancer. J Surg Oncol. 2010;102:68–72. doi: 10.1002/jso.21584. [DOI] [PubMed] [Google Scholar]
- 6.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process-First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–40. [PubMed] [Google Scholar]
- 7.Yuzhalin A. The role of interleukin DNA polymorphisms in gastric cancer. Hum Immunol. 2011;72:1128–36. doi: 10.1016/j.humimm.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Russell RM. Nutrition and gastric cancer risk: an update. Nutr Rev. 2008;66:237–49. doi: 10.1111/j.1753-4887.2008.00029.x. [DOI] [PubMed] [Google Scholar]
- 9.Tredaniel J, Boffetta P, Buiatti E, Saracci R, Hirsch A. Tobacco smoking and gastric cancer: review and meta-analysis. Int J Cancer. 1997;72:565–73. doi: 10.1002/(sici)1097-0215(19970807)72:4<565::aid-ijc3>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 10.Xu Z, Zhu H, Luk JM, Wu D, Gu D, Gong W. et al. Clinical significance of SOD2 and GSTP1 gene polymorphisms in Chinese patients with gastric cancer. Cancer. 2012;118:5489–96. doi: 10.1002/cncr.27599. [DOI] [PubMed] [Google Scholar]
- 11.Correa P, Haenszel W, Cuello C, Zavala D, Fontham E, Zarama G. et al. Gastric precancerous process in a high risk population: cross-sectional studies. Cancer Res. 1990;50:4731–6. [PubMed] [Google Scholar]
- 12.Larsson SC, Bergkvist L, Wolk A. Processed meat consumption, dietary nitrosamines and stomach cancer risk in a cohort of Swedish women. Int J Cancer. 2006;119:915–9. doi: 10.1002/ijc.21925. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Yang L, Zhou B, Yu R, Tang N, Wang B. Myeloperoxidase G-463A polymorphism and the risk of gastric cancer: a case-control study. Carcinogenesis. 2006;27:2491–6. doi: 10.1093/carcin/bgl121. [DOI] [PubMed] [Google Scholar]
- 14.Gu H, Yang L, Tang N, Zhou B, Zhu H, Sun Q. et al. Association of endothelin-converting enzyme-1b C-338A polymorphism with gastric cancer risk: a case-control study. Eur J Cancer. 2008;44:1253–8. doi: 10.1016/j.ejca.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Gu H, Yang L, Sun Q, Zhou B, Tang N, Cong R. et al. Gly82Ser polymorphism of the receptor for advanced glycation end products is associated with an increased risk of gastric cancer in a Chinese population. Clin Cancer Res. 2008;14:3627–32. doi: 10.1158/1078-0432.CCR-07-4808. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt AM, Yan SD, Yan SF, Stern DM. The multiligand receptor RAGE as a progression factor amplifying immune and inflammatory responses. J Clin Invest. 2001;108:949–55. doi: 10.1172/JCI14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W. et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–60. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- 18.Xia W, Xu Y, Mao Q, Dong G, Shi R, Wang J. et al. Association of RAGE polymorphisms and cancer risk: a meta-analysis of 27 studies. Med Oncol. 2015;32:442. doi: 10.1007/s12032-014-0442-5. [DOI] [PubMed] [Google Scholar]
- 19.Ishiguro H, Nakaigawa N, Miyoshi Y, Fujinami K, Kubota Y, Uemura H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate. 2005;64:92–100. doi: 10.1002/pros.20219. [DOI] [PubMed] [Google Scholar]
- 20.Kuniyasu H, Oue N, Wakikawa A, Shigeishi H, Matsutani N, Kuraoka K. et al. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J Pathol. 2002;196:163–70. doi: 10.1002/path.1031. [DOI] [PubMed] [Google Scholar]
- 21.Hirata K, Takada M, Suzuki Y, Kuroda Y. Expression of receptor for advanced glycation end products (RAGE) in human biliary cancer cells. Hepatogastroenterology. 2003;50:1205–7. [PubMed] [Google Scholar]
- 22.Takada M, Hirata K, Ajiki T, Suzuki Y, Kuroda Y. Expression of receptor for advanced glycation end products (RAGE) and MMP-9 in human pancreatic cancer cells. Hepatogastroenterology. 2004;51:928–30. [PubMed] [Google Scholar]
- 23.Schraml P, Bendik I, Ludwig CU. Differential messenger RNA and protein expression of the receptor for advanced glycosylated end products in normal lung and non-small cell lung carcinoma. Cancer Res. 1997;57:3669–71. [PubMed] [Google Scholar]
- 24.Jiao L, Taylor PR, Weinstein SJ, Graubard BI, Virtamo J, Albanes D. et al. Advanced glycation end products, soluble receptor for advanced glycation end products, and risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:1430–8. doi: 10.1158/1055-9965.EPI-11-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu XC, Abuduhadeer X, Zhang WB, Li T, Gao H, Wang YH. Knockdown of RAGE inhibits growth and invasion of gastric cancer cells. Eur J Histochem. 2013;57:e36. doi: 10.4081/ejh.2013.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka N, Yonekura H, Yamagishi S, Fujimori H, Yamamoto Y, Yamamoto H. The receptor for advanced glycation end products is induced by the glycation products themselves and tumor necrosis factor-alpha through nuclear factor-kappa B, and by 17beta-estradiol through Sp-1 in human vascular endothelial cells. J Biol Chem. 2000;275:25781–90. doi: 10.1074/jbc.M001235200. [DOI] [PubMed] [Google Scholar]
- 27.Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB. et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–84. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermani A, Hess J, De Servi B, Medunjanin S, Grobholz R, Trojan L. et al. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005;11:5146–52. doi: 10.1158/1078-0432.CCR-05-0352. [DOI] [PubMed] [Google Scholar]
- 29.Sasahira T, Kirita T, Bhawal UK, Yamamoto K, Ohmori H, Fujii K. et al. Receptor for advanced glycation end products (RAGE) is important in the prediction of recurrence in human oral squamous cell carcinoma. Histopathology. 2007;51:166–72. doi: 10.1111/j.1365-2559.2007.02739.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure.