Abstract
Obstructive sleep apnea (OSA) is a common disorder associated with increased risk of cardiovascular disease and mortality. Its prevalence and severity vary across ancestral background. Although OSA traits are heritable, few genetic associations have been identified. To identify genetic regions associated with OSA and improve statistical power, we applied admixture mapping on three primary OSA traits [the apnea hypopnea index (AHI), overnight average oxyhemoglobin saturation (SaO2) and percentage time SaO2 < 90%] and a secondary trait (respiratory event duration) in a Hispanic/Latino American population study of 11 575 individuals with significant variation in ancestral background. Linear mixed models were performed using previously inferred African, European and Amerindian local genetic ancestry markers. Global African ancestry was associated with a lower AHI, higher SaO2 and shorter event duration. Admixture mapping analysis of the primary OSA traits identified local African ancestry at the chromosomal region 2q37 as genome-wide significantly associated with AHI (P < 5.7 × 10−5), and European and Amerindian ancestries at 18q21 suggestively associated with both AHI and percentage time SaO2 < 90% (P < 10−3). Follow-up joint ancestry-SNP association analyses identified novel variants in ferrochelatase (FECH), significantly associated with AHI and percentage time SaO2 < 90% after adjusting for multiple tests (P < 8 × 10−6). These signals contributed to the admixture mapping associations and were replicated in independent cohorts. In this first admixture mapping study of OSA, novel associations with variants in the iron/heme metabolism pathway suggest a role for iron in influencing respiratory traits underlying OSA.
Introduction
Obstructive sleep apnea (OSA) is characterized by recurrent episodes of upper airway obstruction during sleep resulting in oxygen desaturation and sleep fragmentation (1). OSA affects more than 10% of adults (2) and increases risk of adverse health outcomes, including hypertension and cardiovascular disease as well as increased mortality (3–5). Candidate gene studies have identified associations between the apnea hypopnea index (AHI), the chief disease defining metric, measured continuously or dichotomized, with variants in genes associated with inflammation, serotoninergic pathways and ventilatory control (6–8). We have recently identified the first genome-wide significant associations with objectively measured OSA traits (9,10). However, the findings are limited by the statistical power with the modest sample size of the available datasets.
One approach for increasing power while being robust to allelic heterogeneity is admixture mapping (11,12), a powerful analytic tool that can be applied to recently admixed populations whose ancestral populations were from isolated continents. This method assumes that the ancestral populations have different disease prevalence/severity and different allele frequencies (AFs), reflecting adaptations to different environments. Under this assumption, local ancestry can be correlated with a phenotype and such association can be detected using regression analysis. With the ability to accurately infer local ancestries, admixture mapping has successfully identified risk genetic variants associated with several complex disorders (11,13–15).
The potential for admixture mapping to identify genetic association with OSA is supported by epidemiological studies of population differences: compared to European-Americans (EAs), OSA prevalence is higher in Hispanic/Latinos (16), African-American (AA) children and young adults (17–19) and Asians (20,21). A genetic linkage study revealed differences in estimated heritability and genomic regions associated with OSA traits in AAs compared to EAs (22).
To explore these differences, we conducted admixture analyses of three primary OSA phenotypes [AHI, overnight average oxyhemoglobin saturation (SaO2) and percentage time SaO2 < 90%], and a secondary heritable OSA measure (average respiratory event duration) (22) in a large Hispanic/Latino sample. To our knowledge, this is the first admixture mapping study of OSA phenotypes.
Results
Sample characteristics
This study included 11 575 individuals from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) with self-identified Hispanic/Latino background (including Central American, Cuban, Dominican, Mexican, Puerto Rican and South American) (23,24) (Materials and Methods). The sample characteristics varied across background groups (Table 1). The Caribbean Islanders (Cubans, Dominicans and Puerto Ricans) had higher average African ancestry proportion and lower average Amerindian ancestry proportion compared to the mainlanders (Mexicans, Central Americans and South Americans) (25). OSA (defined as an AHI > 5) was present in 31% of the sample. Cubans had the highest prevalence of OSA.
Table 1.
Sample characteristics of the analyzed samples from HCHS/SOL
| All | Cuban | Dominican | Puerto Rican | Mexican | Central American | South American | |
|---|---|---|---|---|---|---|---|
| N | 11 575 | 2105 | 1066 | 2031 | 4298 | 1246 | 829 |
| African Ancestry%, mean (SD) | 13.8 (16.1) | 15.2 (0.7) | 44 (1.5) | 21.8 (1.6) | 4.3 (0.5) | 10.6 (0.9) | 6.4 (0.7) |
| Amerindian Ancestry%, mean(SD) | 30.5 (23.7) | 5.8 (0.5) | 6.5 (0.9) | 13.7 (1.6) | 49.1 (1.1) | 45.4 (1.3) | 45.3 (1.4) |
| European Ancestry%, mean (SD) | 55.7 (20.9) | 79 (0.9) | 49.5 (1.6) | 64.5 (2) | 46.5 (1.1) | 44 (1.3) | 48.3 (1.5) |
| Male% | 41 | 46.9 | 34.6 | 42.4 | 39.5 | 40.4 | 39.8 |
| Age, years, mean (SD) | 46.09 (13.86) | 48.68 (13.33) | 45.25 (14.31) | 47.57 (14.26) | 44.66 (13.81) | 44.71 (13.37) | 46.46 (13.28) |
| BMI, kg/m2, mean (SD) | 29.82 (6.06) |
29.32 (5.89) | 29.34 (5.64) | 30.81 (6.96) | 29.86 (5.91) | 30.06 (5.83) | 28.66 (5.24) |
| AHI, events/h, median (IQR) | 2.00 (0.41–6.62) |
2.42 (0.53–8.33) |
1.46 (0.32–5.35) |
2.34 (0.51–7.37) |
1.86 (0.38–6.35) |
1.87 (0.39–6.31) |
1.74 (0.42–5.69) |
| Average SaO2, %, mean (SD) | 96.45 (0.94) |
96.37 (0.97) |
96.56 (0.78) |
96.33 (1.07) |
96.51 (0.9) |
96.49 (0.98) |
96.44 (0.88) |
| Percentage time SaO2 < 90%, mean (SD) | 0.82 (3.05) |
1.05 (3.45) |
0.65 (2.45) |
0.86 (3.37) |
0.74 (2.73) |
0.96 (3.64) |
0.69 (2.62) |
| Average event duration, s, mean (SD) | 23.65 (7.97) |
23.92 (8.03) |
23.3 (8.21) |
22.76 (7.86) |
23.89 (7.95) |
23.31 (7.48) |
24.78 (8.33) |
The correlations among the traits are provided in Supplementary Material, Table S1. As expected, AHI increased with age, male sex and body mass index (BMI), and was negatively correlated with average SaO2 (ρ = −0.69) and positively correlated with time% SaO2 < 90% (ρ = 0.77). Event duration was weakly correlated with the other OSA traits (Supplementary Material, Table S1).
Since the four OSA traits were skewed, they were rank normalized and adjusted for age, sex, BMI, population structure and relatedness in this study (Materials and Methods). The overall estimated heritability for the OSA traits varied from 11 to 17%.
Global ancestry associations
The associations between rank-normalized OSA traits and global African, Amerindian and European ancestries adjusted for demographic variables and BMI are shown in Table 2. Global African ancestry percentage was associated with lower AHI (β = −0.0005, P = 0.023) and higher average SaO2 (β = 0.0006, P = 0.015), suggesting a protective effect for OSA. European ancestry percentage was associated with lower average SaO2 (β = −0.0007, P = 8.50 × 10−4). Respiratory event duration was shorter in African and European ancestries (β = −0.0019 and − 0.0008, P = 7.52 × 10−9 and 6.20 × 10−4), and longer in Amerindian ancestry (β = 0.0015, P = 6.20 × 10−4). Secondary analyses using European ancestry as a reference group present consistent estimated effects for African and Amerindian ancestries (Supplementary Material, Table S2).
Table 2.
Associations between OSA traits and individual global ancestry percentage, adjusting for sex, age, age2, age × sex, BMI and BMI2
| African Ancestry | Amerindian Ancestry | European Ancestry | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Estimate per 1% change of ancestry | SE | P | Estimate per 1% change of ancestry | SE | P | Estimate per 1% change of ancestry | SE | P | |
| AHI | −0.0005 | 0.0002 | 0.023 | 0.0003 | 0.0002 | 0.09 | −0.00004 | 0.0002 | 0.847 |
| Average SaO2 | 0.0006 | 0.0003 | 0.015 | 0.0002 | 0.0002 | 0.204 | −0.0007 | 0.0002 | 8.50 × 10−4 |
| Percentage time SaO2 < 90% | 0.0001 | 0.0003 | 0.658 | −0.0001 | 0.0002 | 0.607 | 0.0005 | 0.0002 | 0.805 |
| Average event duration | −0.0019 | 0.0003 | 7.52 × 10−9 | 0.0015 | 0.0002 | 5.96 × 10−12 | −0.0008 | 0.0003 | 6.20 × 10−4 |
Admixture mapping
We performed admixture mapping analyses for each ancestry group separately across the genome using linear mixed regression models, excluding the regions within 2Mb around the centromere and chromosomal boundary (Materials and Methods) (26). OSA traits were rank normalized, adjusted for age, sex, BMI (and interactions and non-linear terms as appropriate), top five principal components (PCs) and kinship coefficient matrix (Materials and Methods). The admixture mapping results are presented in Supplementary Material, Figs S1–4. Previous permutation analyses suggested the genome-wide significance level was 5.7 × 10−5 for admixture mapping analysis in HCHS/SOL data (27) (Materials and Methods). Our primary analysis of AHI and hypoxemia traits identified African ancestry at chromosome 2q37 genome-wide significantly associated with AHI (β = −0.079, P = 3.7 × 10−5) and 15 additional regions with suggestive admixture mapping evidence including chromosome 18q21, with similar ancestry associations for both AHI and percentage time SaO2 < 90% (P < 10−3; Supplementary Material, Table S3). European ancestry was positively associated with both traits (β = 0.048 and 0.049, P = 2.74 × 10−4 and 2.01 × 10−4, for AHI and percentage time SaO2 < 90%, respectively), whereas Amerindian ancestry was negatively associated with both traits (β = −0.052 and − 0.058, P = 4.46 × 10−4 and 9.15 × 10−5, for AHI and percentage time SaO2 < 90%, respectively). Secondary analysis using respiratory event duration additionally identified one genome-wide significant region ([chromosome 16q12-21], P = 6.4 × 10−6) associated with African ancestry and seven suggestive regions (P < 10−3; Supplementary Material, Table S3).
Fine-mapping analyses
We then followed up 2q37, 18q21 and 16q12–21 by single SNP association and joint SNP-ancestry analyses (Materials and Methods). Since 18q21 was associated with two correlated OSA traits, we further performed a combined trait analysis incorporating their phenotypic correlation (Materials and Methods). Significance level for this analysis was determined as P < 8 × 10−6 accounting for multiple comparisons in multiple traits (13,28) (Materials and Methods).
At chromosome 2q37, single SNP association analysis identified two SNPs in linkage disequilibrium (LD) associated with AHI, led by rs6750391[T] [AC011286.1, effect AF (EAF) = 0.898, β = 0.09, P = 1.78 × 10−6] (Table 3 and Supplementary Material, Table S4, Supplementary Material, Fig. S5). The association effect directions were positive across background groups (Fig. 1). The T allele had a lower frequency in African ancestry populations (AF = 0.49) relative to Amerindian (AF = 0.93) and European ancestry populations (AF = 0.95) (Supplementary Material, Table S5). The association P-value between local African ancestry and AHI was more significant when only local African ancestry was included in the regression model (P = 3.7 × 10−5) than when both rs6750391 and local African ancestry were included (P = 2.8 × 10−2) (Table 4, Fig. 2A), suggesting that rs6750391 partially accounted for the admixture mapping evidence.
Table 3.
Significant independent SNPs (P < 8 × 10−6) under the candidate regions associated with OSA traits in HCHS/SOL
| SNP | B37 position | Nearest gene(s) | Alleles (E/A) | EAF | Info | Trait | β (se) | P |
|---|---|---|---|---|---|---|---|---|
| rs6750391 | 2:237635300 | AC011286.1 | T/C | 0.898 | 0.998 | AHI | 0.090 (0.019) | 1.78 × 10−6 |
| rs74021948 | 16:63065908 | RP11-96H17.1 | A/C | 0.984 | 0.956 | Event duration | 0.302 (0.059) | 3.07 × 10−7 |
| rs12935339 | 16:63132772 | RP11-96H17.1 | G/A | 0.308 | 0.999 | Event duration | −0.074 (0.016) | 3.44 × 10−6 |
| rs57403733 | 18:55281709 | NARS, FECH | G/A | 0.803 | 0.999 | AHI | 0.066 (0.015) | 1.47 × 10−5 |
| Percentage time SaO2 < 90% | 0.078 (0.015) | 3.98 × 10−7 | ||||||
| Combineda | 3.20 × 10−7 |
CPASSOC was performed to combine the effect of AHI and percentage time SaO2 < 90%.
Figure 1.
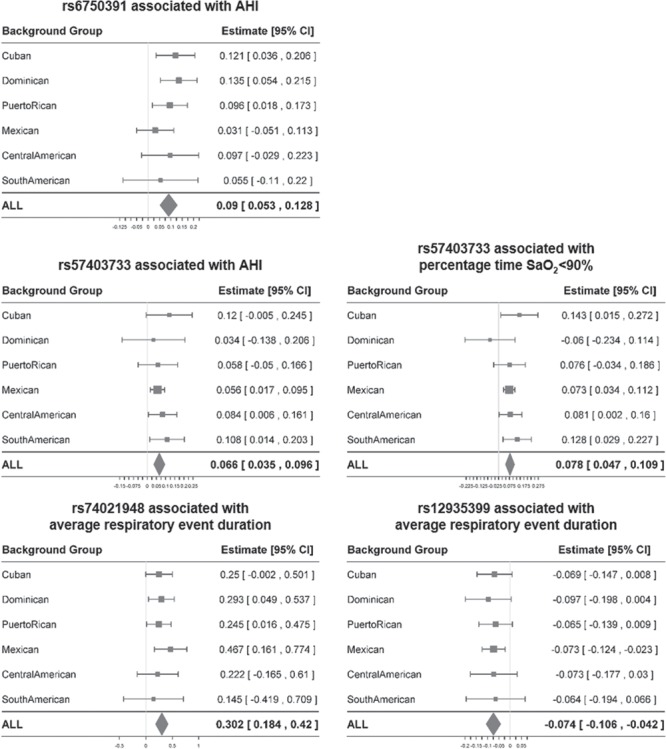
Associations between identified independent variants and OSA traits among background groups.
Table 4.
Comparison between ancestry-OSA association model with and without adjusting for lead SNPs in the three significant regions
| Genomic region (Mb) | Ancestry | Trait | SNP covariate | Est | SE | P |
|---|---|---|---|---|---|---|
| 2:237.54–238.39 | African | AHI | No SNP (baseline) | −0.079 | 0.019 | 3.70 × 10−5 |
| rs6750391 | −0.047 | 0.021 | 2.80 × 10−2 | |||
| 18:51.81–57.12 | Amerindian | AHI | No SNP (baseline) | −0.052 | 0.015 | 4.46 × 10−4 |
| rs57403733 | −0.032 | 0.016 | 4.27 × 10−2 | |||
| European | AHI | No SNP (baseline) | 0.048 | 0.013 | 2.74 × 10−4 | |
| rs57403733 | 0.032 | 0.014 | 2.02 × 10−2 | |||
| Amerindian | Percentage time SaO2 < 90% | No SNP (baseline) | −0.058 | 0.015 | 9.15 × 10−5 | |
| rs57403733 | −0.024 | 0.018 | 1.63 × 10−1 | |||
| European | Percentage time SaO2 < 90% | No SNP (baseline) | 0.049 | 0.013 | 2.01 × 10−4 | |
| rs57403733 | 0.027 | 0.014 | 5.46 × 10−2 | |||
| 16:51.14–64.12 | African | Average event duration | No SNP (baseline) | −0.113 | 0.025 | 6.42 × 10−6 |
| rs74021948 | −0.094 | 0.025 | 2.11 × 10−4 | |||
| rs12935339 | −0.095 | 0.025 | 2.00 × 10−4 | |||
| rs74021948 and rs12935339 | −0.080 | 0.026 | 1.82 × 10−3 |
Figure 2.
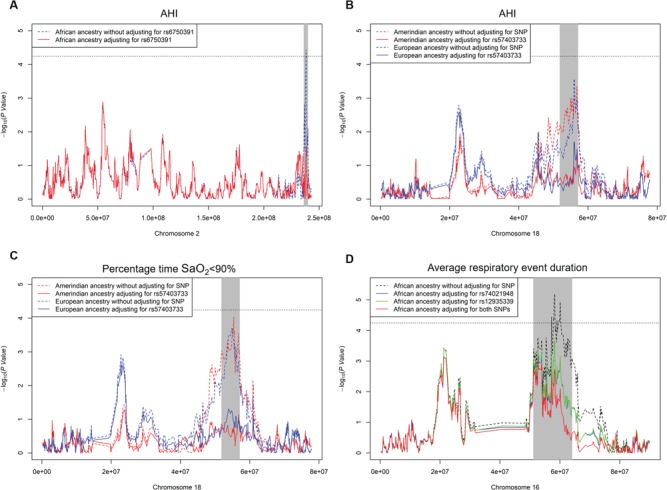
Admixture mapping results of significant ancestry and OSA traits with and without adjusting for target SNPs.
We identified 20 significant SNPs in strong LD overlapping NARS (asparaginyl-tRNA synthetase) and FECH (ferrochelatase) in the 18q21 region, led by rs57403733[G] (EAF = 0.803, associated with the combined AHI and percentage time SaO2 < 90% traits (P = 3.2 × 10−7) (Table 3 and Supplementary Material, Table S4, Supplementary Material, Fig. S6). Rs57403733[G] is positively associated both with AHI and percentage time SaO2 < 90% (β = 0.066 and 0.078, P = 1.47 × 10−5 and 3.98 × 10−7). The association effect directions were highly consistent across background groups (Fig. 1). The G allele had a lower frequency in Amerindian ancestry populations (EAF = 0.79) relative to African (EAF = 0.99) and European ancestry populations (EAF = 0.96) (Supplementary Material, Table S5). Conditional on rs57403733, the association P-values between Amerindian/European ancestries and the AHI and percentage time SaO2 < 90% became less significant (P > 10−2) (Table 4, Fig. 2B and C).
At chromosome 16q12-21, single SNP association analysis identified four SNPs in RP11-96H17.1 associated with our secondary trait, event duration (Table 3 and Supplementary Material, Table S4). Rs74021948[A] (EAF = 0.984, β = 0.302, P = 3.07 × 10−7) was in weak LD with rs12935339[G] (EAF = 0.308, β = −0.074, P = 3.44 × 10−6) and two other SNPs (r2 < 0.2) (Supplementary Material, Fig. S7). Therefore, we investigated rs74021948 and rs12935339 separately. The association directions were consistent among background groups for both SNPs (Fig. 1). Conditioning on rs74021948 or rs12935339, the significance of association between local African ancestry and event duration decreased from P = 6.4 × 10−6 to P = 2 × 10−4. When conditioning on both SNPs, the significance level further decreased (P = 1.82 × 10−3) (Table 4, Fig. 2D).
SNP level replication generalization and bioinformatics analysis
SNPs detected by admixture mapping and follow-up association analyses were tested in 1935 AAs, 6490 EAs and 1240 Hispanic/Latino Americans (HAs) from the replication cohorts (described in Supplementary Table S6). In the NARS/FECH region on chromosome 18, 12 of the 20 significant SNPs had replication P-value <0.05 for percentage time SaO2 < 90% in EAs. The smallest replication P-value was 1.6 × 10−3 for rs3745063 (Supplementary Material, Table S7).
Rs57403733 and rs3745063 are intronic variants of NARS. Rs57403733 is associated with the expression of the gene FECH in transformed fibroblasts cells. Rs3745063 is an expression quantitative trait locus (eQTL) for FECH in lymphoblastoid (29), monocytes and whole blood cells and overlaps promoter histone marks in five tissues (eight cell lines) and enhancer histone marks in 14 tissues (51 cell lines).
We further observed a significant negative association between FECH expression and average SaO2 in 203 HAs from MESA (β = −0.552, P = 0.003), adjusting for age, sex and BMI.
Associations of SNPs at AC011286.1 or RP11-96H17.1 were not replicated.
Discussion
In this first admixture mapping analysis of sleep apnea traits, we leveraged information on diverse ancestral backgrounds available in the largest cohort with objectively measured sleep apnea phenotypes and genome-wide data, identifying several novel signals in regions that differ in local ancestry. Primary analyses identified a genome-wide significant region at chromosome 2q37 that showed a negative association between African ancestry and AHI. Amerindian ancestry and European ancestry at chromosome 18q21 were also suggestively associated with both AHI and degree of hypoxemia during sleep. Secondary analysis identified that African ancestry at chromosome 16q12-21 was also negatively and significantly associated with respiratory event duration. Follow-up joint association analyses in three of the regions which were associated with OSA traits identified variants in AC011286.1, NARS/FECH and RP11-96H17.1(P < 8 × 10−6) that explained part of the admixture mapping evidence. The association of NARS/FECH for percentage time SaO2 < 90% was replicated in independent European cohorts. Notably, bioinformatics and expression data indicated that the variants in chromosome 18 associated with AHI and hypoxemia are also associated with expression of FECH. Furthermore, analysis of available expression data showed that increased FECH expression is associated with lower nocturnal SaO2. Since FECH catalyzes the insertion of iron as part of heme B biosynthesis, these data suggest a novel biological pathway linking OSA with pathways associated with iron/heme metabolism.
We used local ancestry information sampled across the genome to estimate global ancestry as well as to detect risk genomic regions using admixture mapping. Two previous studies have used information on genetic ancestry to study sleep-related traits in recently admixed populations. A study from an admixed Brazilian population identified a negative association between African genetic ancestry and OSA (30), consistent with our results. Halder et al (31) also reported an association between African ancestry and fragmented sleep. This finding also is consistent with our findings that shorter event duration—a marker of low arousal threshold—associated with African ancestry. However, these studies only investigated the association with average individual ancestry proportions (global ancestries) calculated from ancestral informative markers and did not perform admixture mapping or fine mapping to identify specific genetic variants.
Studies of OSA genetics have lagged behind those of other diseases. This partly reflects a need for objectively measuring sleep traits in large samples that are genotyped. Studies have been relatively small and have mostly examined candidate genes and focused on the AHI or the dichotomous trait defined by AHI thresholds, reporting associations with a variety of genes in inflammatory, metabolic and other pathways, that largely were not replicated (6,7,32–39). A genome-wide association analysis including HCHS/SOL identified associations between GPR83 and AHI, and between C6ORF183/CCDC162P and event duration (9); however, independent samples were limited and did not permit replication.
Admixture mapping analysis provided a strategy for improving the power to detect genetic variants associated with OSA, including those that may underlie population differences. The novel association for AHI on chromosome 2q37 maps to an area implicated in a rare disorder, the 2q37 deletion syndrome. This disorder is characterized by hypotonia, facial dysmorphisms and obesity, traits often observed with OSA (40). About half of the patients with this syndrome have sleep disturbances, including one child with sleep apnea (40,41). Several regulatory genes reside in this area, including ones that influence development, such as HDAC4 and TWIST2. In our analysis, two variants within AC011286.1, a linc RNA gene, in this region were associated with AHI. AC011286.1 is expressed in esophageal mucosa (Supplementary Material, Fig. S8), a tissue that shares development origins with the pharynx, which may differentially impact airway collapsibility.
In addition to analyzing the AHI, we also examined two measures of overnight oxygen saturation, which are key features of OSA, that are heritable and predict, and likely mediate, its associated cardiovascular risk and mortality (42–44). We combined information on two highly correlated traits, AHI and percentage time SaO2 < 90%, to further improve power in a region associated with both traits. We also explored associations with average event duration, another OSA trait that varies across ancestral backgrounds, is highly heritable, but not strongly correlated with the AHI (22).
In the region on chromosome 18q21, we detected 20 variants on NARS/FECH associated with both AHI and percentage time SaO2 < 90%, with 12 also associated in independent cohorts. Many variants are eQTLs for FECH in multiple tissues, suggesting that this might be the relevant gene under this admixture mapping and association peak. Furthermore, using available expression data from monocytes, we found that increased FECH expression was associated with lower nocturnal SaO2. FECH, the last enzyme of the heme biosynthesis pathway which inserts the iron atom into the porphyrin IX ring and generates heme B, is largely expressed in whole blood (Supplementary Material, Fig. S8) (45). Hemoproteins such as hemoglobin myoglobin have diverse biological functions involved in oxygen transport and oxygen storage (46,47). Intracellular free heme is toxic and is degraded to carbon monoxide, biliverdin and iron via heme oxygenase. Heme degradation and its products (e.g. carbon monoxide) play key roles in many biological systems, including glucose metabolism and carotid body oxygen sensing, a mechanism implicated in ventilatory control and sleep apnea (48–50). Iron homeostasis influences redox balance, inflammatory response and energy metabolism (51). Iron chelation influences HIF1-α (Hypoxia Inducible Factor) accumulation and may influence oxygen sensing in the carotid body (52,53), where chemoreceptors play a key role in ventilatory stability and the pathogenesis of sleep apnea (54). Together, our admixture/association findings, supported by bioinformatics and our own expression data, suggest a role for iron/heme in influencing clinically important OSA traits. Future research is needed to determine if our associations reflect variations in hemoglobin and oxygen carrying capacity or effects of iron on ventilatory control or metabolism.
In secondary analyses, we also examined respiratory event duration, which is shorter in AAs than EAs, and partially reflects respiratory arousability and chemoreflex sensitivity, which when abnormal can lead to sleep fragmentation (55–57). In the region of chromosome 16q12-21, we detected four variants on RP11-96H17.1 associated with event duration. RP11-96H17.1 is largely expressed in the brain (Supplementary Material, Fig. S8).
This analysis provides insight into differences in OSA susceptibility across ancestral groups. Notably, African ancestry was associated with a lower AHI and shorter respiratory disturbances. Shorter events, and the reduced physiological tolerance of ventilatory disturbances it reflects, may contribute to sleep fragmentation and sleepiness for any given AHI level, consistent with epidemiological studies reporting shorter sleep duration and sleepiness in AAs (21). Although BMI-adjusted AHI levels are higher in AA children and young adults compared to EAs, this difference dissipates with advancing age (17).
Our study has several strengths. It was conducted on the largest cohort of individuals with Hispanic/Latino background, a highly admixed sample, with objective sleep recording. We focused our analyses on clinically important phenotypes that vary across populations. To gain power we also analyzed two highly correlated traits (AHI and percentage time SaO2 < 90%) by combining their summary statistics (58). Admixture mapping can be more powerful than traditional genome-wide association analyses when trait distributions vary by ancestry. High resolution local ancestries were used, enhancing the accuracy of ancestral estimation. Follow-up two-step fine-mapping analyses (single SNP association and joint SNP-ancestry association analyses) efficiently targeted the variants both associated with the trait and contributing to admixture mapping. Although our replication samples were only modest in size, we were able to replicate 12 of 26 variants in European cohorts, providing the first replicated genetic association finding for OSA traits in a multi-ethnic population.
This study also has several limitations. In-laboratory polysomnography was not performed in the HCHS/SOL, precluding our assessment of sleep architecture. The overall ancestry-specific sample sizes were only modest. Despite this, several top findings replicated in cohorts studied with full polysomnography. Although multiple variants in NARS/FECH were replicated in EAs, the SNP-level replication power was limited by modest sample sizes in other replication cohorts. The complex admixture history and genetic diversity among Hispanic/Latinos present challenges in identifying ideal replication cohorts.
In summary, we identified three novel genomic regions associated with OSA traits in a large Hispanic/Latino cohort using admixture mapping methods. Novel variants and genes detected in those regions were identified that likely explain some of the difference in OSA across ancestral groups. Further investigation of the role of iron/heme pathways, a topic of growing interest due to associations with diabetes, obesity, liver disease and metabolism, and gene regulatory areas on chromosomes 2 and 16 may provide further insight into mechanisms underlying OSA.
Materials and Methods
Sample
The primary sample was the HCHS/SOL, a community-based study in the US, which includes 16 415 adults aged 18–74 with self-identified Hispanic/Latino background (23,24). Individuals were recruited from randomly selected households near four centers in Miami, San Diego, Chicago and the Bronx area of New York. Most participants identified themselves from one of six ethnic background groups, including 1730 Central Americans, 2348 Cubans, 1460 Dominicans, 6471 Mexicans, 2728 Puerto Ricans and 1,068 South Americans. A total of 14 400 individuals had overnight home sleep apnea tests (HSATs) that met study quality criteria and reported no regular use of overnight oxygen or sleep apnea treatment devices. Of these, 11 575 had available genotyping and comprised the primary study sample. The study was approved by the Institutional Review Boards at each participating institution and participants provided written informed consent.
Phenotypes and covariates
During the baseline HCHS/SOL examination (2008–2011), physical measures and questionnaires, blood and HSATs were collected as previously described (24). Age, sex and background were self-reported. BMI was derived from measured weight (kg) and height (cm). Sleep apnea assessments were obtained with the ARES Unicorder 5.2 (B-Alert, Carlsbad, CA) as described before (59). The monitor measures airflow using a nasal cannula and pressure transducer; blood hemoglobin oxygen saturation (SaO2) and pulse rate using a forehead-based reflectance oximeter; body position and movement by an actigraph and snoring by a microphone. Sleep records were transmitted to a central Sleep Reading Center where they were manually scored as described before (60). Respiratory events (apneas or hypopneas) were identified when airflow declined by ≥50% for ≥10 s. Since thermistry was not collected, apneas were not distinguished from hypopneas. In this paper, only respiratory events associated with ≥3% oxyhemoglobin desaturation were analyzed. The AHI was computed as the number of respiratory events divided by the estimated sleep time; this measure highly correlates with polysomnography-derived AHI (61). The average SaO2 level and percent time when SaO2 was <90% used oximetry-based SaO2 values measured continuously during sleep. The average respiratory disturbance event duration was derived by measuring the time between the first breath that was reduced by ≥50% to the termination of the respiratory disturbance event (in seconds).
Genotypes
Genotyping was conducted using an Illumina Omni2.5M SNP array with additional customized content, including 2 536 661 SNPs. Local African, European and Amerindian ancestries were inferred from quality controlled SNPs across the genome using RFMix software (62,63). The global African, Amerindian and European ancestries for each individual, defined as the proportion of each genome inherited from each ancestry, were calculated by the average values of local ancestries at 14 815 uniformly distributed loci on autosomal chromosomes. OSA trait heritability was estimated for the pooled sample and individual background groups using the unbiased Haseman–Elston regression (64).
Global ancestry association analysis
We performed association analyses between OSA traits with each of the global African, Amerindian and European ancestry percentage using linear regression model. Since the OSA traits were skewed, they were rank normalized. Sex, age, age2, age × sex, BMI and BMI2 were adjusted as fixed-effect covariates. We further used global European ancestry as a reference and tested the association between each OSA trait with global African and Amerindian ancestry percentage in one regression model as a secondary analysis. This analysis was less powerful because of larger standard errors introduced by multicollinearity (high correlation between African and Amerindian ancestries).
Admixture mapping analysis
We performed admixture mapping analyses for each ancestry group separately across the genome using a linear mixed regression model (GENESIS package in R). Regions within 2 Mb around the centromere and chromosomal boundary were excluded because of lower local ancestry inference accuracy (26). OSA traits were rank normalized and then adjusted for sex, age, age2, age × sex, BMI and BMI2 as fixed-effect covariates. PCs and kinship coefficients were estimated iteratively, robust to ancestry and population and family structure and admixture, as described before (25,65,66). The top five PCs separated the background group genetic structure well in this sample (25). Population stratification was controlled for by adjusting for fixed effects of five PCs, and family structure was allowed for by adding a variance component proportional to the kinship coefficient matrix. We performed another set of analyses additionally adjusting for sampling weight, households and census blocks. We did not observe significant change from our primary model (Supplementary Material, Figs. S1–4). Admixture mapping reduces the number of comparisons across the genome; therefore, a significance level of 5 × 10−8 for traditional GWAS is not applicable. Based on previous permutation analyses using HCHS/SOL data, the significance level was determined to be 5.7 × 10−5, corresponding to a family-wise error rate at 0.05 level (27). We defined a suggestive region as admixture mapping signals of P < 10−3.
Single SNP association analysis
We further evaluated three regions with significant admixture signals to identify SNPs in those regions that were significantly associated with OSA traits, using genotyped and 1000G imputed SNPs with minor AF >0.01 and imputation information scores >0.4 (67). The boundary of those significant regions was defined with admixture mapping P < 10−3. Linear mixed models were used with the same set of covariates as in the primary admixture mapping analysis. We estimated the number of independent tests in admixture mapping and the single SNP association analyses in each region (accounting for one trait at 2q37 and 16q12-21, and two traits at 18q21) using Li and Ji’s method (13,28), resulting in a Bonferroni significance level of P = 8 × 10−6. For 18q21 region significantly associated with AHI and percentage time SaO2 < 90%, we performed multiple trait analysis by combining the summary statistics for each trait and incorporating the trait correlation using CPASSOC software (58), which provides a single test P-value of association for multiple traits.
Joint SNP and local ancestry analysis
To examine whether an SNP within a candidate region could explain the admixture mapping signal, we performed a joint SNP and local ancestry analysis, simultaneously modeling local ancestry and SNP association. We compared the association P-value between the local ancestry and the OSA trait with and without adjusting for that SNP. This analysis was able to indicate whether a SNP contributes to the admixture signal.
Replication analysis
Single SNP replication analysis was conducted using eight independent cohorts comprising individuals of EA, AA and HA (Supplementary Methods). Inverse-variance fixed-effect meta-analyses were performed to combine the results in each ancestry group.
Epigenetic annotation and eQTL analysis
We used Haploreg (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) to examine the epigenetic annotations of SNPs (68). eQTL evidence and gene expression were queried using the GTEx Portal (https://www.gtexportal.org/home/) (45). We further tested the association for OSA traits on the gene expression level of the genes associated with leading SNPs (NARS and FECH) using Multi-ethnic Study of Atherosclerosis (MESA) data (online supplement), adjusting for age, sex and BMI in each population group.
Supplementary Material
Acknowledgements
The Sleep Reading Center of Brigham and Women’s Hospital has been supported by National Institutes of Health grants (5-R01-HL046380-15 and 5-KL2-RR024990-05).
The baseline examination of the Hispanic Community Health Study/Study of Latinos was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236) and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Neurological Disorders and Stroke (NINDS) and NIH Institution Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Provision of genotyping services was supported in part by the National Center for Advancing Translational Sciences (NCATS) CTSI grant UL1TR000124 and NIDDK DRC grant DK063491.
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C).
The Framingham Heart Study is conducted and supported by NHLBI in collaboration with Boston University (Contract No. N01-HC-25195). Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL- 64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Sleep Heart Health Study was provided by NIH/NHLBI grant U01 HL 53941.
The Jackson Heart Study (JHS) is supported by contracts HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C from the NHLBI and the National Institute on Minority Health and Health Disparities. Dr Wilson is supported by U54GM115428 from the National Institute of General Medical Sciences. We thank the JHS participants and staff for their contributions to this work.
The Multi Ethnic Study of Atherosclerosis (MESA) and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA). Funding support for the Sleep Polysomnography dataset was provided by grant HL56984. Provision of genotyping services supported in part by NCATS CTSI grant UL1-TR-001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
The Osteoporotic Fractures in Men (MrOS) Study is supported by NIH funding. The following institutes provide support: the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), NCATS and NIH Roadmap for Medical Research under the following grant numbers U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160 and UL1 TR000128. The NHLBI provides funding for the MrOS Sleep ancillary study `Outcomes of Sleep Disorders in Older Men’ under the following grant numbers: R01 HL071194, R01 HL070848, R01 HL070847, R01 HL070842, R01 HL070841, R01 HL070837, R01 HL070838 and R01 HL070839. The NIAMS provides funding for the MrOS ancillary study `Replication of candidate gene associations and bone strength phenotype in MrOS’ under the grant number R01 AR051124. The NIAMS provides funding for the MrOS ancillary study `GWAS in MrOS and SOF’ under the grant number RC2 AR058973.
The Starr County Health Studies is supported in part by grants R01 DK073541,
U01 DK085501, R01 AI085014 and R01 HL102830 from the National Institutes of Health, and funds from The University of Texas Health Science Center at Houston. Graeme I. Bell was supported in part by grant P30 DK020595 and a gift from the Kovler Family Foundation.
The Cardiovascular Health Study (CHS) was supported by contract numbers N01-HC- 85079, N01-HC-85080, N01-HC-85081, N01-HC-85082, N01-HC-85083, N01-HC- 85084, N01-HC-85085, N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC- 55222, N01-HC-75150, N01-HC-45133, N01-HC-85239 and HHSN268201200036C; grant numbers U01 HL080295 from NHLBI and R01 AG-023629 from NIA, with additional contribution from NINDS. Support for the Whole Genome Study was provided by NHLBI grant HL087652. Additional support for infrastructure was provided by HL105756 and additional genotyping among the African-American cohort was supported in part by HL085251. DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by National Center for Research Resources grant UL1RR033176, now at NCATS CTSI grant UL1TR000124; in addition to NIDDK grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Conflict of Interest statement. None declared.
Funding
National Institutes of Health, the National Heart, Lung, Blood Institute (R01HL113338 to S.R., R35HL135818 to S.R., R01HL098433 to S.R., R01HL46380 to S.R. and K01HL135405 to B.E.C); National Human Genome Research Institute (HG003054 to X.Z. B.E.C. K01 award is from NHLBI); Sleep Research Society Foundation Career Development Award 018-JP-18 (to H.W.); American Thoracic Society Foundation Unrestricted Grant for Sleep (to B.E.C.).
References
- 1. Somers V.K., White D.P., Amin R., Abraham W.T., Costa F., Culebras A., Daniels S., Floras J.S., Hunt C.E., Olson L.J. et al. (2008) Sleep apnea and cardiovascular disease: an American Heart Association/American College of Cardiology Foundation Scientific Statement from the American Heart Association Council for High Blood Pressure Research Professional Education Committee, Council on Clinical Cardiology, Stroke Council, and Council on Cardiovascular Nursing. J. Am. Coll. Cardiol., 52, 686–717. [DOI] [PubMed] [Google Scholar]
- 2. Peppard P.E., Young T., Barnet J.H., Palta M., Hagen E.W. and Hla K.M. (2013) Increased prevalence of sleep-disordered breathing in adults. Am. J. Epidemiol., 177, 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gottlieb D.J., Yenokyan G., Newman A.B., O'Connor G.T., Punjabi N.M., Quan S.F., Redline S., Resnick H.E., Tong E.K., Diener-West M. et al. (2010) Prospective study of obstructive sleep apnea and incident coronary heart disease and heart failure: the sleep heart health study. Circulation, 122, 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Redline S., Yenokyan G., Gottlieb D.J., Shahar E., O'Connor G.T., Resnick H.E., Diener-West M., Sanders M.H., Wolf P.A., Geraghty E.M. et al. (2010) Obstructive sleep apnea-hypopnea and incident stroke: the sleep heart health study. Am. J. Respir. Crit. Care. Med., 182, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Punjabi N.M., Caffo B.S., Goodwin J.L., Gottlieb D.J., Newman A.B., O'Connor G.T., Rapoport D.M., Redline S., Resnick H.E., Robbins J.A. et al. (2009) Sleep-disordered breathing and mortality: a prospective cohort study. PLoS Med., 6, e1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Larkin E.K., Patel S.R., Goodloe R.J., Li Y., Zhu X., Gray-McGuire C., Adams M.D. and Redline S. (2010) A candidate gene study of obstructive sleep apnea in European Americans and African Americans. Am. J. Respir. Crit. Care. Med., 182, 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Patel S.R., Goodloe R., De G., Kowgier M., Weng J., Buxbaum S.G., Cade B., Fulop T., Gharib S.A., Gottlieb D.J. et al. (2012) Association of genetic loci with sleep apnea in European Americans and African-Americans: the Candidate Gene Association Resource (CARe). PLoS One, 7, e48836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun J., Hu J., Tu C., Zhong A. and Xu H. (2015) Obstructive sleep apnea susceptibility genes in Chinese population: a field synopsis and meta-analysis of genetic association studies. PLoS One, 10, e0135942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cade B.E., Chen H., Stilp A.M., Gleason K.J., Sofer T., Ancoli-Israel S., Arens R., Bell G.I., Below J.E., Bjonnes A.C. et al. (2016) Genetic associations with obstructive sleep apnea traits in Hispanic/Latino Americans. Am. J. Respir. Crit. Care. Med., 194, 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen H., Cade B.E., Gleason K.J., Bjonnes A.C., Stilp A.M., Sofer T., Conomos M.P., Ancoli-Israel S., Arens R., Azarbarzin A. et al. (2018) Multi-ethnic meta-analysis identifies RAI1 as a possible obstructive sleep apnea related quantitative trait locus in men. Am. J. Respir. Cell. Mol. Biol., 58, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu X., Luke A., Cooper R.S., Quertermous T., Hanis C., Mosley T., Gu C.C., Tang H., Rao D.C., Risch N. et al. (2005) Admixture mapping for hypertension loci with genome-scan markers. Nat. Genet., 37, 177–181. [DOI] [PubMed] [Google Scholar]
- 12. Zhu X., Tang H. and Risch N. (2008) Admixture mapping and the role of population structure for localizing disease genes. Adv. Genet., 60, 547–569. [DOI] [PubMed] [Google Scholar]
- 13. Zhu X., Young J.H., Fox E., Keating B.J., Franceschini N., Kang S., Tayo B., Adeyemo A., Sun Y.V., Li Y. et al. (2011) Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum. Mol. Genet., 20, 2285–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reich D., Patterson N., De Jager P.L., McDonald G.J., Waliszewska A., Tandon A., Lincoln R.R., DeLoa C., Fruhan S.A., Cabre P. et al. (2005) A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat. Genet., 37, 1113–1118. [DOI] [PubMed] [Google Scholar]
- 15. Mersha T.B. (2015) Mapping asthma-associated variants in admixed populations. Front. Genet., 6, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young T., Shahar E., Nieto F.J., Redline S., Newman A.B., Gottlieb D.J., Walsleben J.A., Finn L., Enright P., Samet J.M. et al. (2002) Predictors of sleep-disordered breathing in community-dwelling adults: the Sleep Heart Health Study. Arch. Intern. Med., 162, 893–900. [DOI] [PubMed] [Google Scholar]
- 17. Redline S., Tishler P.V., Schluchter M., Aylor J., Clark K. and Graham G. (1999) Risk factors for sleep-disordered breathing in children. Associations with obesity, race, and respiratory problems. Am. J. Respir. Crit. Care. Med., 159, 1527–1532. [DOI] [PubMed] [Google Scholar]
- 18. Rosen C.L., Palermo T.M., Larkin E.K. and Redline S. (2002) Health-related quality of life and sleep-disordered breathing in children. Sleep, 25, 657–666. [PubMed] [Google Scholar]
- 19. Weinstock T.G., Rosen C.L., Marcus C.L., Garetz S., Mitchell R.B., Amin R., Paruthi S., Katz E., Arens R., Weng J. et al. (2014) Predictors of obstructive sleep apnea severity in adenotonsillectomy candidates. Sleep, 37, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehra R., Stone K.L., Blackwell T., Ancoli Israel S., Dam T.T., Stefanick M.L. Redline S. and Osteoporotic Fractures in Men Study (2007) Prevalence and correlates of sleep-disordered breathing in older men: osteoporotic fractures in men Sleep study. J. Am. Geriatr. Soc., 55, 1356–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen X., Wang R., Zee P., Lutsey P.L., Javaheri S., Alcantara C., Jackson C.L., Williams M.A. and Redline S. (2015) Racial/Ethnic differences in sleep disturbances: the Multi-Ethnic Study of Atherosclerosis (MESA). Sleep, 38, 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liang J., Cade B.E., Wang H., Chen H., Gleason K.J., Larkin E.K., Saxena R., Lin X., Redline S. and Zhu X. (2016) Comparison of heritability estimation and linkage analysis for multiple traits using principal component analyses. Genet. Epidemiol., 40, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sorlie P.D., Aviles-Santa L.M., Wassertheil-Smoller S., Kaplan R.C., Daviglus M.L., Giachello A.L., Schneiderman N., Raij L., Talavera G., Allison M. et al. (2010) Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol., 20, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lavange L.M., Kalsbeek W.D., Sorlie P.D., Aviles-Santa L.M., Kaplan R.C., Barnhart J., Liu K., Giachello A., Lee D.J., Ryan J. et al. (2010) Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol., 20, 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernandez-Rhodes L., Justice A.E., Graff M. et al. (2016) Genetic diversity and association studies in US Hispanic/Latino Populations: applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet., 98, 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhatia G., Tandon A., Patterson N., Aldrich M.C., Ambrosone C.B., Amos C., Bandera E.V., Berndt S.I., Bernstein L., Blot W.J. et al. (2014) Genome-wide scan of 29,141 African Americans finds no evidence of directional selection since admixture. Am. J. Hum. Genet., 95, 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schick U.M., Jain D., Hodonsky C.J., Morrison J.V., Davis J.P., Brown L., Sofer T., Conomos M.P., Schurmann C., McHugh C.P. et al. (2016) Genome-wide association study of platelet count identifies ancestry-specific loci in Hispanic/Latino Americans. Am. J. Hum. Genet., 98, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J. and Ji L. (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb), 95, 221–227. [DOI] [PubMed] [Google Scholar]
- 29. Lappalainen T., Sammeth M., Friedlander M.R., Hoen P.A., Monlong J., Rivas M.A., Gonzalez-Porta M., Kurbatova N., Griebel T., Ferreira P.G. et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature, 501, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guindalini C., Colugnati F.A., Pellegrino R., Santos-Silva R., Bittencourt L.R. and Tufik S. (2010) Influence of genetic ancestry on the risk of obstructive sleep apnoea syndrome. Eur. Respir. J., 36, 834–841. [DOI] [PubMed] [Google Scholar]
- 31. Halder I., Matthews K.A., Buysse D.J., Strollo P.J., Causer V., Reis S.E. and Hall M.H. (2015) African genetic ancestry is associated with sleep depth in older African Americans. Sleep, 38, 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaditis A.G., Gozal D., Khalyfa A., Kheirandish-Gozal L., Capdevila O.S., Gourgoulianis K., Alexopoulos E.I., Chaidas K., Bhattacharjee R., Kim J. et al. (2014) Variants in C-reactive protein and IL-6 genes and susceptibility to obstructive sleep apnea in children: a candidate-gene association study in European American and Southeast European populations. Sleep Med., 15, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kripke D.F., Kline L.E., Nievergelt C.M., Murray S.S., Shadan F.F., Dawson A., Poceta J.S., Cronin J., Jamil S.M., Tranah G.J. et al. (2015) Genetic variants associated with sleep disorders. Sleep Med., 16, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baik I., Seo H.S., Yoon D., Kim S.H. and Shin C. (2015) Associations of sleep apnea, NRG1 polymorphisms, alcohol consumption, and cerebral white matter hyperintensities: analysis with genome-wide association data. Sleep, 38, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cronin R.M., Field J.R., Bradford Y., Shaffer C.M., Carroll R.J., Mosley J.D., Bastarache L., Edwards T.L., Hebbring S.J., Lin S. et al. (2014) Phenome-wide association studies demonstrating pleiotropy of genetic variants within FTO with and without adjustment for body mass index. Front. Genet., 5, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grilo A., Ruiz-Granados E.S., Moreno-Rey C., Rivera J.M., Ruiz A., Real L.M. and Saez M.E. (2013) Genetic analysis of candidate SNPs for metabolic syndrome in obstructive sleep apnea (OSA). Gene, 521, 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu H., Guan J., Yi H. and Yin S. (2014) A systematic review and meta-analysis of the association between serotonergic gene polymorphisms and obstructive sleep apnea syndrome. PLoS One, 9, e86460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qin B., Sun Z., Liang Y., Yang Z. and Zhong R. (2014) The association of 5-HT2A, 5-HTT, and LEPR polymorphisms with obstructive sleep apnea syndrome: a systematic review and meta-analysis. PLoS One, 9, e95856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Varvarigou V., Dahabreh I.J., Malhotra A. and Kales S.N. (2011) A review of genetic association studies of obstructive sleep apnea: field synopsis and meta-analysis. Sleep, 34, 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Falk R.E. and Casas K.A. (2007) Chromosome 2q37 deletion: clinical and molecular aspects. Am. J. Med. Genet. C Semin. Med. Genet., 145C, 357–371. [DOI] [PubMed] [Google Scholar]
- 41. Williams S.R., Aldred M.A., Der Kaloustian V.M., Halal F., Gowans G., McLeod D.R., Zondag S., Toriello H.V., Magenis R.E. and Elsea S.H. (2010) Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am. J. Hum. Genet., 87, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kendzerska T., Gershon A.S., Hawker G., Leung R.S. and Tomlinson G. (2014) Obstructive sleep apnea and risk of cardiovascular events and all-cause mortality: a decade-long historical cohort study. PLoS Med., 11, e1001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oldenburg O., Wellmann B., Buchholz A., Bitter T., Fox H., Thiem U., Horstkotte D. and Wegscheider K. (2016) Nocturnal hypoxaemia is associated with increased mortality in stable heart failure patients. Eur. Heart. J., 37, 1695–1703. [DOI] [PubMed] [Google Scholar]
- 44. Gellen B., Canoui-Poitrine F., Boyer L., Drouot X., Le Thuaut A., Bodez D., Covali-Noroc A., D'Ortho M.P., Guendouz S., Rappeneau S. et al. (2016) Apnea-hypopnea and desaturations in heart failure with reduced ejection fraction: are we aiming at the right target? Int. J. Cardiol., 203, 1022–1028. [DOI] [PubMed] [Google Scholar]
- 45. Consortium G.T. (2013) The Genotype-Tissue Expression (GTEx) project. Nat. Genet., 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiabrando D., Vinchi F., Fiorito V., Mercurio S. and Tolosano E. (2014) Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol., 5, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gozzelino R. (2016) The pathophysiology of heme in the brain. Curr. Alzheimer Res., 13, 174–184. [DOI] [PubMed] [Google Scholar]
- 48. Wegiel B., Nemeth Z., Correa-Costa M., Bulmer A.C. and Otterbein L.E. (2014) Heme oxygenase-1: a metabolic nike. Antioxid. Redox Signal., 20, 1709–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prabhakar N.R. (2013) Sensing hypoxia: physiology, genetics and epigenetics. J. Physiol., 591, 2245–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peng Y.J., Zhang X., Gridina A., Chupikova I., McCormick D.L., Thomas R.J., Scammell T.E., Kim G., Vasavda C., Nanduri J. et al. (2017) Complementary roles of gasotransmitters CO and H2S in sleep apnea. Proc. Natl. Acad. Sci. U. S. A., 114, 1413–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang X., Fang X. and Wang F. (2015) Pleiotropic actions of iron balance in diabetes mellitus. Rev. Endocr. Metab. Disord., 16, 15–23. [DOI] [PubMed] [Google Scholar]
- 52. Woo K.J., Lee T.J., Park J.W. and Kwon T.K. (2006) Desferrioxamine, an iron chelator, enhances HIF-1alpha accumulation via cyclooxygenase-2 signaling pathway. Biochem. Biophys. Res. Commun., 343, 8–14. [DOI] [PubMed] [Google Scholar]
- 53. Lahiri S., Roy A., Li J., Baby S.M., Mokashi A. and Di Giulio C. (2004) Role of Fe2+ in oxygen sensing in the carotid body. Adv. Exp. Med. Biol., 551, 59–64. [DOI] [PubMed] [Google Scholar]
- 54. Smith C.A., Nakayama H. and Dempsey J.A. (2003) The essential role of carotid body chemoreceptors in sleep apnea. Can. J. Physiol. Pharmacol., 81, 774–779. [DOI] [PubMed] [Google Scholar]
- 55. Wains S.A., El-Chami M., Lin H.S. and Mateika J.H. (2017) Impact of arousal threshold and respiratory effort on the duration of breathing events across sleep stage and time of night. Respir. Physiol. Neurobiol., 237, 35–41. [DOI] [PubMed] [Google Scholar]
- 56. Sforza E., Krieger J. and Petiau C. (1998) Nocturnal evolution of respiratory effort in obstructive sleep apnoea syndrome: influence on arousal threshold. Eur. Respir. J., 12, 1257–1263. [DOI] [PubMed] [Google Scholar]
- 57. Edwards B.A., Wellman A., Sands S.A., Owens R.L., Eckert D.J., White D.P. and Malhotra A. (2014) Obstructive sleep apnea in older adults is a distinctly different physiological phenotype. Sleep, 37, 1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhu X., Feng T., Tayo B.O., Liang J., Young J.H., Franceschini N., Smith J.A., Yanek L.R., Sun Y.V., Edwards T.L. et al. (2015) Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. Am. J. Hum. Genet., 96, 21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Westbrook P.R., Levendowski D.J., Cvetinovic M., Zavora T., Velimirovic V., Henninger D. and Nicholson D. (2005) Description and validation of the apnea risk evaluation system: a novel method to diagnose sleep apnea-hypopnea in the home. Chest, 128, 2166–2175. [DOI] [PubMed] [Google Scholar]
- 60. Redline S., Sotres-Alvarez D., Loredo J., Hall M., Patel S.R., Ramos A., Shah N., Ries A., Arens R., Barnhart J. et al. (2014) Sleep-disordered breathing in Hispanic/Latino individuals of diverse backgrounds. The Hispanic Community Health Study/Study of Latinos. Am. J. Respir. Crit. Care. Med., 189, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Louis J., Auckley D., Miladinovic B., Shepherd A., Mencin P., Kumar D., Mercer B. and Redline S. (2012) Perinatal outcomes associated with obstructive sleep apnea in obese pregnant women. Obstet. Gynecol., 120, 1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Browning S.R., Grinde K., Plantinga A., Gogarten S.M., Stilp A.M., Kaplan R.C., Aviles-Santa M.L., Browning B.L. and Laurie C.C. (2016) Local ancestry inference in a large US-based Hispanic/Latino study: Hispanic Community Health Study/Study of Latinos (HCHS/SOL). G3 (Bethesda), 6, 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Maples B.K., Gravel S., Kenny E.E. and Bustamante C.D. (2013) RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet., 93, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sofer T. (2017) Confidence intervals for heritability via Haseman–Elston regression. Stat. Appl. Genet. Mol. Biol., 16, 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Conomos M.P., Miller M.B. and Thornton T.A. (2015) Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol., 39, 276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Conomos M.P., Reiner A.P., Weir B.S. and Thornton T.A. (2016) Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet., 98, 127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Howie B., Fuchsberger C., Stephens M., Marchini J. and Abecasis G.R. (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet., 44, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ward L.D. and Kellis M. (2016) HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic. Acids. Res., 44, D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


