Abstract
Previous studies show that aberrant tryptophan catabolism reduces maternal immune tolerance and adversely impacts pregnancy outcomes. Tryptophan depletion in pregnancy is facilitated by increased activity of tryptophan-depleting enzymes [i.e. the indolamine-2,3 dioxygenase (IDO)1 and IDO2) in the placenta. In mice, inhibition of IDO1 activity during pregnancy results in fetal loss; however, despite its important role, regulation of Ido1 gene transcription is unknown. The current study shows that the Ido1 and Ido2 genes are imprinted and maternally expressed in mouse placentas. DNA methylation analysis demonstrates that nine CpG sites at the Ido1 promoter constitute a differentially methylated region that is highly methylated in sperm but unmethylated in oocytes. Bisulfite cloning sequencing analysis shows that the paternal allele is hypermethylated while the maternal allele shows low levels of methylation in E9.5 placenta. Further study in E9.5 placentas from the CBA/J X DBA/2 spontaneous abortion mouse model reveals that aberrant methylation of Ido1 is linked to pregnancy loss. DNA methylation analysis in humans shows that IDO1 is hypermethylated in human sperm but partially methylated in placentas, suggesting similar methylation patterns to mouse. Importantly, analysis in euploid placentas from first trimester pregnancy loss reveals that IDO1 methylation significantly differs between the two placenta cohorts, with most CpG sites showing increased percent of methylation in miscarriage placentas. Our study suggests that DNA methylation is linked to regulation of Ido1/IDO1 expression and altered Ido1/IDO1 DNA methylation can adversely influence pregnancy outcomes.
Introduction
Genomic imprinting is a naturally occurring biological phenomenon resulting in parent-of-origin specific expression of genes. Many imprinted genes are clustered in a locus consisting of both maternally and paternally expressed genes (7). The organization of imprinted genes reflects that there is a common cis-acting mechanism regulating imprinting of the entire locus. The best characterized imprinting mark is DNA methylation; many imprinting loci contain a differentially methylated region (DMR) that serves as the imprinting control region (7). Imprinted genes play crucial roles during fetal, placental and postnatal development in mammals, and aberrant imprinting regulation has been linked to human disorders related to growth, metabolism and neurobehavioral development (27). Despite their important developmental roles, imprinted genes represent <1% of mammalian genes, and to date, ∼150 genes have been identified as imprinted in mice and humans. The initial discovery of imprinted genes was performed using classical genetic and molecular approaches (8,10,18,42). In recent years, however, multiple approaches including comparative methodologies for genomic features (33), gene expression (4,34) and DNA methylation (16,24) of various tissues have contributed to identification of novel imprinted genes.
In an ongoing study characterizing the impact of tryptophan catabolism on pregnancy health, our laboratory recently discovered that the genes encoding tryptophan-catabolizing enzymes, the indolamine-2,3 dioxygenase (IDO)1 and IDO2, are imprinted in mouse extra-embryonic tissue. An essential amino acid, tryptophan is not synthesized de novo in mammals and must be acquired from dietary sources. The majority of tryptophan is catabolized to produce nicotinamide adenine dinucleotide or NAD (15). The rate limiting step of the pathway is the conversion of tryptophan into kynurenine, mediated by the IDO1 and IDO2 enzymes; IDO1 is the major enzyme due to its higher affinity for tryptophan (15). Sharing ∼83% homology to human, the mouse Ido1 gene is located on chromosome 8 approximately 20.6 kb upstream of Ido2. The Ido1 gene is expressed in the uterus, placenta, decidua, lymph node and spleen (56). IDO1 protein depletes tryptophan from the local microenvironment to produce bioactive kynurenine catabolites (15,56).
In human pregnancy, maternal tryptophan plasma levels naturally decline by 30–40% from first to third trimester (1,32,45). Increased plasma or tissue tryptophan levels are more frequently observed in women with miscarriages or pathological pregnancies (6,47), demonstrating that its proper catabolism could influence pregnancy outcomes. High levels of kynurenine contribute to immune response suppression (15). In the placenta, tryptophan catabolism protects the allogeneic fetus by suppressing maternal immune responses (2,37). Previous studies show that pharmacological inhibition of mouse IDO1 results in fetal resorption in allogeneic pregnancies (37), demonstrating that tryptophan catabolism plays a role in pregnancy maintenance.
Despite its link to pregnancy maintenance, it is unclear how Ido1 is transcriptionally regulated in the placenta during early development. Through analysis of allele-specific transcriptional and epigenetic patterns of the Ido1 locus in the placenta and germline, we report that during fetal development, the Ido1 and Ido2 genes exhibit expression patterns and DNA methylation marks consistent with the locus being imprinted in the mouse placenta. We provide evidence that suggests the DNA methylation patterns are conserved in the human IDO1 gene. Furthermore, our studies suggest that altered DNA methylation of the mouse Ido1 and human IDO1 gene is linked to pregnancy loss.
Results
The Ido1 and Ido2 genes are expressed in the extra-embryonic tissue during early mouse development
Previous studies in Institute of Cancer Research (ICR) and CBA mice showed that the Ido1 transcript was first detected in the whole conceptus starting at embryonic day (E) 7.5 (37,51), and its expression diminished at E12.5 (51). To determine the spatial and temporal expression patterns of the Ido1 and Ido2 genes in C57BL/6 (B6) mice, we isolated E6.5 whole conceptus and E7.5–15.5 embryonic and extra-embryonic tissues and measured total Ido1 gene expression using real-time quantitative polymerase chain reaction (RT-qPCR). Our results demonstrate that Ido1 mRNA is detected in the extra-embryonic tissues starting at E7.5 and its expression peaks at E9.5 and reduces afterwards, although is still detectable at E15.5 (Fig. 1A). Analysis of E9.5 embryos and placentas demonstrated that the Ido1 mRNA was exclusively detected in the placenta and absent in the embryo (Fig. 1B). Western blot analysis of protein extracts from E9.5 embryos and placentas revealed a similar pattern of protein expression (Fig. 1C). The Ido2 gene and protein are expressed in similar temporal and spatial patterns (Supplementary Material, Fig. S1A– C). Our results show that the Ido1 and Ido2 genes are highly expressed in E9.5 placentas in B6 mice.
Figure 1.
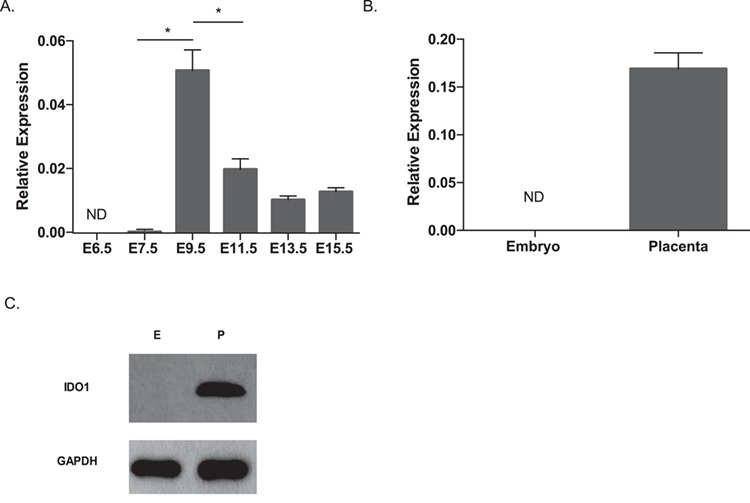
Measurement of mRNA and protein expression of the Ido1 gene in mouse placenta. RT-qPCR analysis was performed to measure Ido1 mRNA expression (A) between E6.5 to E15.5, and (B) in the E9.5 embryo and placentas. Samples tested in the E6.5 and E7.5 groups were whole conceptus and ectoplacental cone, respectively. For the time course study, 4–11 conceptuses from three dams per time point were analyzed. For the tissue-specific study, 4–5 conceptuses from two dams were analyzed. Ido1 gene expression measurement was normalized to expression of the reference genes, Arpp0 and Gapdh. All values in A and B were normalized to the housekeeping genes. No significant differences were detected among E11.5–15.5 samples. (C) Western Blot analysis of E9.5 embryos and placenta using GAPDH as a loading control. Result represented analysis of three independent conceptuses. Data were analyzed using one-way ANOVA followed by Dunnett’s multiple comparison test (A) or student t-test (B) *P < 0.05 for all statistical data. Error bars represent standard error of the mean (SEM).
The Ido1 and Ido2 genes are maternally expressed in the mouse placenta
A previous study of Ido1 KO mice demonstrates that the IDO1 protein is detected in trophoblast giant cells only when the gene is inherited maternally (5). These observations suggest that the gene is potentially imprinted; however, its allele-specific transcription pattern has not been tested. To test whether the Ido1 gene is expressed in a parent-of-origin manner, we isolated E9.5 placentas from F1 hybrid mice generated from reciprocal mating of B6 and PWD/PhJ (PWD) mice. The presence of single nucleotide polymorphisms (SNPs) between the B6 and PWD mice resulted in restriction fragment length polymorphism that allowed us to measure allele-specific transcription of the Ido1 gene (Materials and Methods). Our results indicate that the Ido1 gene is almost exclusively maternally expressed, with the maternal allele contributing >92.3% of its total expression (Fig. 2). For the Ido2 gene, we observed that its expression was also derived from the maternal allele (Supplementary Material, Fig. S1D).
Figure 2.
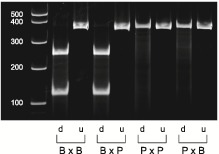
Restriction fragment length polymorphism analysis to show the allele-specific expression of Ido1 in E9.5 placentas. The Ido1 gene was amplified using PCR followed by restriction digest analysis. For each cross, we included both the digested (`d’) and undigested (`u’) PCR products. The B6-specific restriction fragments are 111 and 254 base pair (bp), while the PWD-specific fragment is 365 bp. Lane 1 is a 1 kb + ladder. Lanes 2–3 are B6 female mated to B6 male (B × B), lanes 4–5; B6 female mated to PWD male (B × P), lanes 6–7; P × P and lanes 8–9; P × B. Quantification of the digested samples revealed that the maternal allele contributes to 99.3 and 93.9% of total Ido1 expression in the BXP and PXB crosses, respectively. The figure is a representative of five independent experiments involving analysis of six mice per cross in each experiment.
Both the Ido1 and Ido2 genes have two different transcription start sites. For the Ido1 gene (Supplementary Material, Fig. S2A), the first transcript variant or transcript 1 (shown above to be imprinted) encodes a full-length IDO1 protein (407 amino acids). The second transcript variant, transcript 2, encodes a shorter (i.e. 316 amino acids) protein product. As imprinted genes have alternative transcripts of distinct imprinting status (e.g. (13,36)), we next determined the allele-specific expression of Ido1 transcript 2. Our allele-specific expression analysis shows that Ido1 transcript 2 is also imprinted and maternally expressed (Supplementary Material, Fig. S2B). The Ido2 transcript also has two variants which differ in exons 1 and 2 sequences (Supplementary Material, Fig. S2A) with the full-length transcript 1 shown above to be imprinted. Unfortunately, we were unable to test for the imprinting status of Ido2 trancript 2 as there is no available SNPs at exons 1 and 2 in the B6 and PWD mice. Overall, our data demonstrate for the first time that the Ido1 locus is imprinted and the Ido1 and Ido2 genes are transcribed from the maternal allele.
The Ido1 locus is hypermethylated on the paternal allele and has low methylation on the maternal allele
DMRs at imprinted loci show ∼50% total methylation in somatic tissues reflecting that DNA strands originating from one parental allele are fully methylated and the other unmethylated. Our whole-genome bisulfite sequencing analysis in E18.5 B6 fetal liver revealed a total methylation of 53.7% at six CpG sites within a 247 base pair (bp) region of the Ido1 gene (chr8: 24,594,682-24,594,928; mm10; unpublished data), suggesting that this region may harbor DMRs. To search for candidate DMRs at the Ido1 locus, we conducted bisulfite pyrosequencing analysis to measure total DNA methylation of 37 CpG sites spanning the promoter and exons 1–2 of the Ido1 gene in E9.5 placenta (Fig. 3A). Among these sites, we observed that nine CpG sites (sites 2 to 10; Fig. 3A) in the promoter (chr8: 24,597,072-24,597,474; mm10) and 10 CpG sites (sites 28 to 37; Fig. 3A) within the gene body (chr8: 24,593,164-24,594,955; mm10) display an average methylation of around 50% (30–77% and 43–68%, respectively; Fig. 3B) in E9.5 placenta, suggesting that these regions are potential candidates for DMRs. In contrast, the methylation levels in E9.5 embryos are at levels of ∼90% (data not shown).
Figure 3.
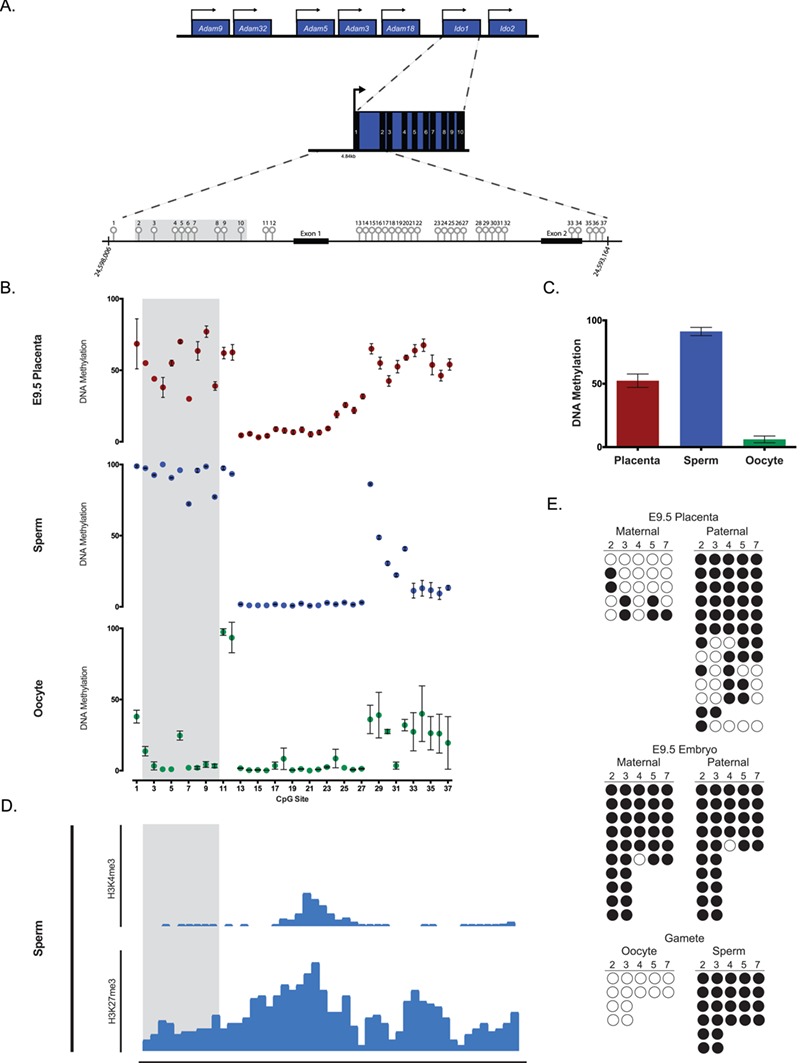
DNA methylation analysis of the mouse Ido1 locus using bisulfite pyrosequencing and cloning sequencing. (A) Schematic of the mouse Ido1 locus on chromosome 8 showing the full-length 12.8 kb transcript containing 10 exons. Unfilled light grey lollipops represent the 37 CpG sites assayed within the mouse Ido1 locus. Arrows designate direction of gene expression. Shaded region in grey highlights the DMR. (B) Methylation levels of the 37 CpG sites in E9.5 placentas (red), sperm (blue) and oocytes (green). Each data point represents average DNA methylation from five mice. (C) Average DNA methylation levels for CpG sites 2 to 10 in E9.5 placentas (n = 5), sperm (n = 5) and oocytes (n = 5). (D) Published genome-wide ChIP-seq data set at Ido1 locus using sperm replicate 2 (19). In sperm, Ido1 is depleted of H3K4me3 but relatively enriched in H3K27me3. Shaded region represents the Ido1 DMR. (E) Ido1 DMR methylation profile at CpG sites 2–7 of E9.5 placenta and embryo, and gametes by bisulfite cloning sequencing. The gametes demonstrate differential methylation with oocytes hypomethylated (0%) and sperm hypermethylated (100%). The placenta shows similar differential between maternal (28.0%) versus paternal (72.5%) alleles, while the embryo is hypermethylated on both alleles (97.3 and 97.1%, respectively). Placenta and embryos were generated from B6xPWD and PWDxB6 hybrid crosses, while gametes are of B6 origin. Each circle represents an individual CpG site, while each row represents an independent original DNA strand. The solid and open circles represent methylated and unmethylated CpG sites, respectively. Maternal and paternal alleles were distinguished by SNPs between B6 and PWD mice. CpG site 6 was excluded from the analysis due to an existing SNP between B6 (C) and PWD (T) making it indistinguishable to determine if the CpG was unmethylated (`TG’) or a strand of PWD origin (`TG’). All error bars represent SEM. Data were analyzed using ANOVA followed by Dunnett’s multiple comparison test. P < 0.05 for all statistical analysis.
Primary DMRs at imprinted loci are derived from distinct DNA methylation events in sperm versus oocytes (29). Thus, total DNA methylation levels at the candidate Ido1 DMRs were examined in sperm and oocytes. As a positive control for differential methylation and to validate the purity of our collected cells, we first examined DNA methylation levels at the paternal H19 DMR and maternal Snrpn DMR in sperm and oocytes. Consistent with well-established evidence that the H19 and Snprn genes are paternally and maternally methylated genes, respectively, our analysis showed that the H19 DMR in sperm and Snrpn DMR in oocytes were hypermethylated (Supplementary Material, Fig. S3A and B). At the Ido1 gene, analysis of the region containing CpG sites 2–10 (Fig. 3A) revealed high methylation in sperm (i.e. >90% methylation levels in all sites except for sites 7 and 10, which were 72.3% and 77.3% methylated, respectively) and low methylation in oocytes (i.e. <2% methylation in oocytes except for CpG sites 2 and 6 that were 13.7% and 24.7% methylated, respectively; Fig. 3B). When average methylation of CpG sites 2 to 10 was calculated (Fig. 3C), we found that E9.5 placentas displayed partial methylation (52.4% ± 5.28) while sperm and oocytes were hypermethylated and hypomethylated, respectively (91.2% ± 3.27 and 6.1% ± 2.65, respectively). These data suggest that CpG sites 2–10 constitute a candidate primary DMR. In contrast, we did not detect significant differences in DNA methylation levels at CpG sites 23–37 of the Ido1 gene in sperm versus oocytes (Fig. 3A and B) suggesting this region is not likely to be a primary DMR.
Pre-implantation embryos undergo genome-wide epigenetic reprogramming at the time when majority of DNA methylation is erased and subsequently reestablished in the developing embryo (29). Additionally, the vast majority of DNA methylation in sperm is erased during pre-implantation development (39). Imprinted loci are protected from this genome-wide reprogramming event and their parent-of-origin DNA methylation is thus maintained in the developing embryo. To determine if sperm- and oocyte-specific methylation patterns are maintained at the Ido1 locus in E9.5 placenta, we performed bisulfite cloning sequencing for CpG sites 2–7 in F1 hybrid mice generated from reciprocal mating of B6 and PWD mice. This method allowed us to determine the parent-of-origin DNA methylation levels at these CpG sites. Our results show that the paternal allele has significantly higher percent levels of methylation relative to maternal allele (Fig. 3E), suggesting that the parent-of-origin methylation pattern at CpG sites 2–7 in E9.5 placenta is mostly maintained. In contrast to the placenta, E9.5 embryos has levels of methylation close to 100% in both paternal and maternal alleles (Fig. 3E). Consistent with the pyrosequencing results, CpG sites 2–7 are fully methylated in sperm and unmethylated in oocytes (Fig. 3E). Thus, our collective data suggest that DNA methylation patterns of CpG sites 2–10 of the Ido1 gene are consistent with those of a paternally methylated primary DMR.
Sperm is enriched in H3K27me3 and depleted of H3K4me3 at the Ido1 locus
Although DNA methylation is the most well characterized imprinting mark, increasing evidence have demonstrated that posttranslational histone modifications are present in an allele-specific manner at imprinted loci (14,20,48,55). Previous studies at the maternally expressed H19 gene, for example, reveal that repressive histone marks including histone (H)3, lysine (K)9, trimethylation (me3) and H3K27me3 are enriched on the methylated paternal allele (19,55). More recently, genome-wide ChIP-seq analysis in mouse sperm showed that the H19 gene is depleted in the activating histone mark H3K4me3 but moderately enriched in H3K27me3 [(19); Supplementary Material, Fig. S3C]. Using this published ChIP-seq data set, we asked whether the Ido1 gene in mouse sperm showed enrichment or depletion of these histone marks. Interestingly, we found that similar to H19, the Ido1 gene is enriched in H3K27me3 but depleted in H3K4me3 (Fig. 3D). These data show that differential enrichment of repressive and activating histone marks is linked to the imprinted expression of Ido1.
Ido1 methylation is significantly altered in conceptuses from the spontaneous abortion CBA/J X DBA/2 mouse model.
Previous studies have shown that the mouse Ido1 gene contributes to maternal fetal immune tolerance during pregnancy and that inhibition of IDO1 activity causes fetal loss (37). Consistent with this, pregnancy loss in the CBA/J X DBA/2 spontaneous abortion mouse model is partly rescued by increasing IDO1 expression (31). Due to the newly discovered role of DNA methylation in regulating Ido1 gene expression, we asked if altered DNA methylation of the Ido1 gene is linked to fetal loss in this spontaneous abortion model. CBA/J females mated to DBA/2 males (i.e. the CBA X DBA2) have increased fetal loss relative to C57BL/6 mated CBA/J females (i.e. CBA X B6), partly due to reduced maternal fetal immune tolerance (12). We compared DNA methylation patterns of E9.5 placentas from the CBA X DBA2 versus CBA X B6 pregnancies. Because only a subset of fetuses in the CBA X DBA2 pregnancies result in resorption, we tested whether CBA X DBA2 placentas showed differences in variance of DNA methylation using a homogeneity of variance test (Materials and Methods). For this analysis, we measured methylation levels at CpG sites 4 to 7 at the center of the DMR (Fig. 3A). The variance of DNA methylation levels at CpG sites 4 and 7 were significantly different between the CBAX B6 and CBA X DBA2 placentas (Fig. 4; P-values = 0.048 and 0.008, respectively). No significant differences in DNA methylation levels were detected at CpG sites 5 and 6 (data not shown). These data suggest that altered DNA methylation of the Ido1 gene is observed in a spontaneous pregnancy loss mouse model.
Figure 4.
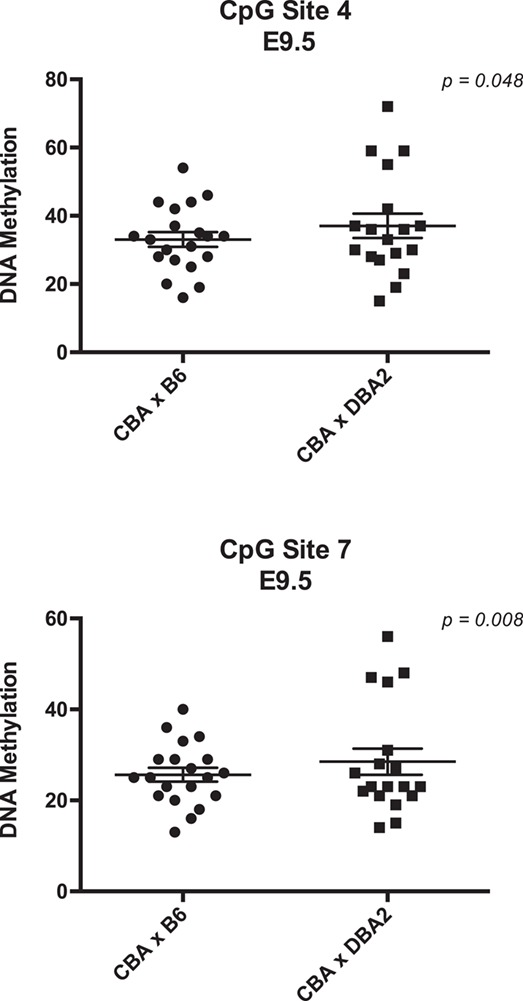
Analysis of DNA methylation of Ido1 in a spontaneous abortion mouse model. Distribution of methylation levels for CBA X DBA2 (n = 18) and CBA X B6 (n = 20) at CpG sites 4 and 7. Each data point represents one placenta. All error bars represent SEM. Data were analyzed using F variance ratio test. P < 0.05 for all statistical analysis.
Human sperm and first trimester placentas show patterns of DNA methylation consistent with the IDO1 locus being imprinted
Although species-specific differences exist, imprinting is a conserved developmental feature among mammalian species including mouse and human (7). The human IDO1 gene has been widely implicated in cancer, immunity and inflammation (56); however, its role during early development is less well understood. Located on chromosome 8, the human IDO1 gene contains 12 exons (Fig. 5A). To determine whether the human IDO1 gene shows evidence of imprinting, we studied DNA methylation patterns in sperm. We focused on regions that are homologous to the mouse, and analyzed 19 CpG sites in the IDO1 promoter and exons 1–2 regions (Fig. 5A). Analysis of average DNA methylation across all 19 CpG sites reveals that the IDO1 locus is hypermethylated in human sperm and that sites 4 to 19 is methylated at levels of 84.5–99.3% (Fig. 5B). These data suggest that similar to mouse, the IDO1 locus is highly methylated in human sperm. Methylation analysis of the paternally methylated H19 gene (10) in our sperm samples showed the expected high methylation levels at >95% (Supplementary Material, Fig. S3A).
Figure 5.
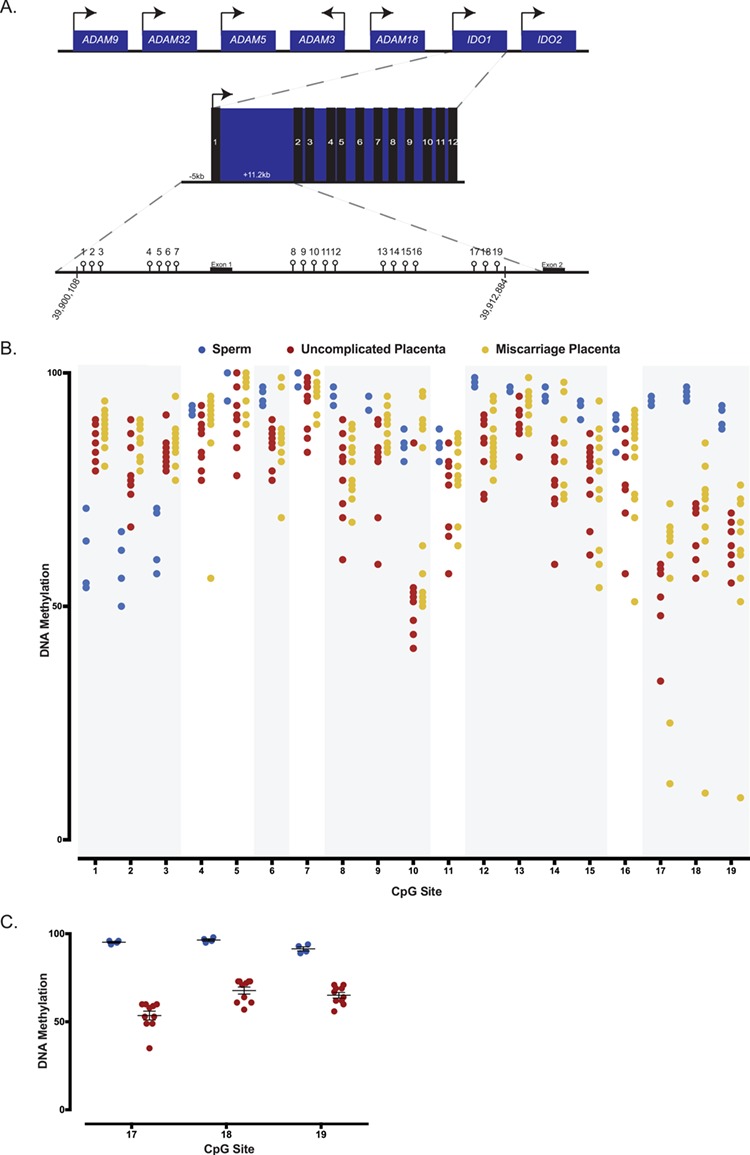
DNA methylation analysis of human sperm and first trimester placentas from uncomplicated and pregnancy loss using bisulfite pyrosequencing. (A) Schematic representation of the human IDO1 locus on chromosome 8 containing 12 exons, and of the 19 CpG sites assayed within the human IDO1 locus. Arrows designate direction of gene expression. (B) Average DNA methylation levels for human sperm, uncomplicated first trimester placentas and euploid first trimester miscarriage placentas for all 19 CpG sites. A total of four sperm samples, 10 uncomplicated placentas and 13 miscarriage placentas are analyzed and presented. Global analysis of average CpG sites reveals significant differences in methylation between sperm and uncomplicated placentas. Individual CpG sites analysis shows that CpG sites 1–3, 6, 8–10, 12–15 and 17–19 significantly differed between sperm and uncomplicated placentas (regions shaded in grey). Comparing uncomplicated to miscarriage placentas, average methylation significantly differ when the CpG sites are analyzed globally. (C) Average DNA methylation of CpG sites 17, 18 and 19 in human sperm (blue) and placentas from uncomplicated pregnancies (red). Each data point represents an individual. CpG sites represent a candidate imprinted DMR as they are highly methylated in sperm but partially methylated in the placentas. Differences in percent or variance in methylation for individual CpGs were assessed using a t-test or homogeneity test of variance, and the corresponding P-values were adjusted for multiple comparisons using the Benjamini–Hochberg method. P-values and FDR of <0.05 were considered to be statistically significant.
Due to restricted accessibility to human oocytes, we were unable to determine DNA methylation levels in the oocytes. Genome-wide methylome data in human oocytes, however, have been published (22,38,57). Data analysis of published reduced representation bisulfite sequencing study in human oocytes (38) revealed DNA methylation levels of 12.07% at the human IDO1 gene. Combined with our analyses, these data suggest that similar to mouse, the human IDO1 gene is hypermethylated in sperm and hypomethylated in oocytes.
To determine if human first trimester placentas show DNA methylation patterns consistent with the IDO1 gene being imprinted, we performed methylation analysis on placentas obtained from elective termination of uncomplicated pregnancies. As a positive control, we tested methylation levels at the H19 locus and found that all samples display the expected ∼50% levels of DNA methylation (Supplementary Material, Fig. S4B). When the IDO1 locus was analyzed, we found that it was generally hypermethylated in first trimester placentas (Figs 5B) consistent with published human placental methylome data (46). Global analysis of DNA methylation across all CpG sites revealed highly significant differences in sperm versus uncomplicated placentas (P < 2.2e-16). Analysis of individual CpG sites demonstrated that these differences are due to altered methylation at 14 sites including CpG sites 1–3, 6, 8–10, 12–15 and 17–19 (False discovery rate or FDR <0.05; shaded grey regions in Fig. 5B; Table 1). Among these sites, all CpG sites showed higher percent DNA methylation in sperm relative to placentas, except CpG sites 1–3 had lower methylation (Fig. 5B; Table 1).
Table 1.
Significance of differences in percent methylation at individual CpG sites between sperm and uncomplicated placentas
| Estimate | P-value | Adj P-value | |
|---|---|---|---|
| CpG 1 | −1.189 | 0.000 | 0.000 |
| CpG 2 | −0.888 | 0.000 | 0.000 |
| CpG 3 | −1.024 | 0.000 | 0.000 |
| CpG 4 | 0.773 | 0.131 | 0.131 |
| CpG 5 | 1.742 | 0.108 | 0.114 |
| CpG 6 | 1.294 | 0.010 | 0.014 |
| CpG 7 | 2.169 | 0.108 | 0.114 |
| CpG 8 | 1.643 | 0.001 | 0.003 |
| CpG 9 | 1.398 | 0.001 | 0.003 |
| CpG 10 | 1.576 | 0.006 | 0.010 |
| CpG 11 | 0.597 | 0.047 | 0.059 |
| CpG 12 | 2.226 | 0.001 | 0.003 |
| CpG 13 | 1.070 | 0.006 | 0.010 |
| CpG 14 | 1.779 | 0.001 | 0.003 |
| CpG 15 | 1.276 | 0.013 | 0.018 |
| CpG 16 | 0.790 | 0.080 | 0.095 |
| CpG 17 | 2.693 | 0.000 | 0.001 |
| CpG 18 | 2.356 | 0.004 | 0.008 |
| CpG 19 | 1.674 | 0.002 | 0.005 |
Because our mouse data demonstrate that the Ido1 DMR is hypermethylated in sperm and partially methylated in the placenta (Fig. 3B), we searched whether a similar candidate imprinted DMR is present among CpG sites 6, 8–10, 12–15 and 17–19. Based on results from the mouse Ido1 gene (Fig. 3B), we defined arbitrary cutoff methylation levels of >90% as `highly methylated’ and 30–70% as `partially methylated’. We found that CpG sites 17, 18 and 19 (Supplementary Material, Table S2) fulfill these criterias; the sites were hypermethylated in sperm (i.e. 94.3% ± 0.48, 95.5% ± 0.65 and 90.5% ± 1.19, respectively) and partially methylated in the placenta (i.e. 50.5% ± 2.98, 67.5% ± 2.09 and 63.8% ± 1.84, respectively; Fig. 5C). Our DNA methylation analysis, combined with published data, suggest that the human IDO1 locus shows methylation patterns consistent with it being imprinted as it is hypermethylated in sperm, hypomethylated in oocytes and partially methylated in first trimester placentas and that CpG sites 17–19 constitute a candidate imprinted DMR.
Euploid human placentas from first trimester miscarriages show significant differences in IDO1 methylation patterns relative to placentas from uncomplicated pregnancies
Although studies have supported that Ido1 plays a role in pregnancy maintenance in mice, and our data show that its methylation pattern is altered among conceptuses in a spontaneous abortion model, it is unclear whether the human IDO1 gene is involved in pregnancy success. To gain insights into the potential epigenetic contribution of the human IDO1 gene on pregnancy success, we performed DNA methylation analysis in placentas obtained from first trimester miscarriages. Because aneuploidy is a major cause for human spontaneous abortion (25), we studied only chromosomally normal or euploid placentas. Analysis of the H19 gene shows that these tissues exhibit the expected ∼50% DNA methylation level (Supplementary Material, S3C). To explore differences between placentas from uncomplicated pregnancies and miscarriages, we compared average DNA methylation across CpG sites 1–19 and found that placentas from the two cohorts were significantly different (Fig. 5B; P = 7.6e-4). We subsequently focused on CpG sites 1–3, 6, 8–10, 12–15 and 17–19 which differed between sperm and uncomplicated placentas (shaded grey regions in Fig. 5B). Average percent methylation of these CpG sites differed significantly between the two cohorts of placentas (P < 8.5e-4; Fig. 5B) due to increased DNA methylation levels in the miscarriage placentas. When analyzed individually, however, only two CpG sites (i.e. 9 and 10) showed higher methylation in the miscarriage placentas relative to uncomplicated placentas (Table 2; FDR < 0.05), suggesting that the individual CpG sites were not independent entities and that the statistical differences in average percent methylation was derived from CpG*group interaction.
Table 2.
Significance of differences in percent methylation between uncomplicated and miscarriage placentas at individual CpG sites that differed between sperm and uncomplicated placentas (as shown in Table 1)
| Estimate | P-value | Adj P-value | |
|---|---|---|---|
| CpG 1 | 0.364 | 0.022 | 0.078 |
| CpG 2 | 0.439 | 0.008 | 0.039 |
| CpG 3 | 0.107 | 0.422 | 0.613 |
| CpG 6 | 0.278 | 0.228 | 0.532 |
| CpG 8 | 0.154 | 0.438 | 0.613 |
| CpG 9 | 0.692 | 0.002 | 0.030 |
| CpG 10 | 0.910 | 0.007 | 0.039 |
| CpG 12 | 0.156 | 0.412 | 0.613 |
| CpG 13 | 0.411 | 0.041 | 0.114 |
| CpG 14 | 0.225 | 0.278 | 0.557 |
| CpG 15 | −0.0541 | 0.818 | 0.880 |
| CpG 17 | 0.137 | 0.556 | 0.708 |
| CpG 18 | 0.026 | 0.921 | 0.921 |
| CpG 19 | −0.067 | 0.777 | 0.880 |
At the candidate imprinted DMR (i.e. CpG sites 17–19), we did not observe significant differences in average methylation when individual CpG sites were tested between the placenta cohorts (data not shown). Closer examinations of methylation levels at these CpG sites revealed that while a subset of miscarriage placentas showed hypermethylation (Fig. 5B, Supplementary Material, Table S2), another subset were hypomethylated (i.e. placenta 101 at CpG sites 17–19 and placenta 108 at CpG site 17), resulting in significant differences in variances between the miscarriages and uncomplicated placentas (FDR < 0.05). Collectively, our data demonstrate that the overall DNA methylation patterns at the IDO1 gene significantly differ in placentas from first trimester miscarriages relative to uncomplicated placentas, and that methylation in the candidate imprinted DMR shows significant inter-individual variations in the miscarriage cohort.
Discussion
Imprinted genes play well-established roles in regulating the growth and development of the mammalian fetus and placenta. In humans, expression levels of several imprinted genes including PHLDA2, IGF2 and GRB10 have been associated with differences in birth weight, crown-rump length and head circumference (35). In the case of PHLDA2, for example, elevated expression of this maternally expressed imprinted gene is associated with fetal growth restriction and/or low birth weight (28). Importantly, overexpression of the mouse Phlda2 also results in fetal growth restrictions (53,54), suggesting that the associations in human pregnancies are causal. These studies suggest that aberrant imprinting regulation can adversely impact fetal growth potential. Thus, elucidating mechanisms underlying imprinting regulation and identifying factors that perturb imprinting may lead to the development of intervention strategies that improve fetal health and survival.
In the current study, we have found that the mouse Ido1 and Ido2 genes are imprinted in extra-embryonic tissues and exhibit mRNA expression patterns consistent with maternal-specific activation. The Ido1 promoter contains a DMR which acquires methylation imprint during spermatogenesis as it is devoid of methylation in oocytes but is highly methylated in sperm. Importantly, Ido1 paternal-specific DNA methylation is mostly maintained in E9.5 placenta suggesting that it is a primary DMR. We noted, however, that DNA methylation level of the paternal allele in the placenta is lower than that of sperm, suggesting that the region is not fully protected from the genome wide demethylation event that occurs in pre-implantation embryos. Of the primary DMRs characterized so far in imprinted loci, only three are paternal DMRs (i.e. acquire methylation during spermatogenesis) including the H19/Igf2, DLk1-Gtl2 and Rasgrf1 (29). A fourth locus has been described, the Gpr1/Zdbf2, but its regulatory role on imprinted expression is unknown (26). Our study shows that germline and allele-specific DNA methylation patterns at the Ido1 DMR exhibits features of a paternal DMR. Similar to other paternal DMRs (46), the Ido1 DMR has low CpG content. It is, however, distinct relative to other paternal DMRs which are all intergenic (46) because of its position at the promoter. The functional role of the Ido1 DMR as well as its precise size remain to be determined. Identification of known consensus binding sites reveals that the Ido1 DMR is a binding site for XPF-1 and CP2, transcription factors known to be associated with imprinted genes in human and mouse placenta (49); whether these are elements regulating imprinting of the locus should be determined in the future. Furthermore, as most imprinted loci contain multiple genes which are maternally and paternally expressed (7), it is possible that imprinting at the Ido1 locus extends beyond the maternally expressed Ido1 and Ido2 genes. More detailed studies are thus needed to further characterize the imprinting regulation of this locus.
Enzymatic activity of the Ido1 gene product plays an important role during pregnancy. IDO1 activation is the rate-limiting step in catabolism of tryptophan that produces kynurenine which supports maternal immune tolerance (52). One proposed mechanism is that increased kynurenine at the maternal-fetal interface promotes aryl hydrocarbon receptor-dependent T cell differentiation into regulatory T cells or `Tregs’, a subset of T cells that maintain peripheral self-tolerance (56). Both IDO1 inhibition and Tregs depletion have been linked to fetal loss (2,37). It was previously unknown, however, whether changes in transcriptional regulation of the Ido1 gene is linked to pregnancy success in mice. To gain insights into the role of DNA methylation of Ido1 in pregnancy maintenance, we compared the CBA X DBA2 and CBA X B6 conceptuses. Early studies revealed a high rate of fetal loss in DBA/2-mated CBA/J females, but not B6-mated CBA/J females (17). Starting at E7.5, loss of cellular contact between decidual and ectoplacental cone cells was observed in a significant proportion of CBA X DBA2 conceptuses, and complete resorption occurred between E10.5 and E12.5 (21). It has been postulated that the CBA X DBA2 pregnancies are deficient in maternal immune-suppressive factors that are essential for fetal protection (17). Our studies reveal that ∼20% of CBA X DBA2 placentas have significantly higher DNA methylation levels at CpG sites 4 and 7 of the Ido1 gene relative to average methylation in the CBA X B6 placentas (Fig. 4). Although we have not determined whether increased DNA methylation is causatively linked to pregnancy loss, the proportion is consistent with previous reports of the 20–30% fetal loss rate in the CBA X DBA2 pregnancy (12). Future studies to causatively link expression and methylation of the Ido1 gene would benefit from the functional characterization of the DMR.
Published studies in human oocytes (38) and our current study of human sperm and first trimester placentas from uncomplicated pregnancies suggest that the human IDO1 is also imprinted. Interestingly, our human studies show that placentas from first trimester pregnancy loss have significantly different IDO1 DNA methylation levels relative to placentas from uncomplicated pregnancies mostly due to increased methylation in the miscarriage group. Future studies should determine whether altered DNA methylation of the IDO1 gene is directly linked to changes in IDO1 gene and protein expression, and whether altered expression is causatively linked to first trimester human pregnancy loss. At least seventeen genetic variants of the human IDO1 gene have been identified (3) and two variants (i.e. an Arg77His SNP and a 9 bp deletion in exon 7) result in significantly reduced IDO1 protein expression and loss of enzymatic activity. Thus, molecular characterization of the human IDO1 gene will further elucidate genetic factors that are linked to increased susceptibilities to pregnancy loss.
Despite these interesting observations, we noted at least two limitations in our study. First, mice with engineered deletion of the Ido1 gene are viable and fertile (45). These observations suggest that in the absence of Ido1, other mechanisms that are involved in maternal-fetal immune tolerance compensate. Furthermore, although our study is the first suggestion of an imprinted locus on mouse chromosome 8, we are unaware of studies reporting lethality in mice carrying uniparental disomy for chromosome 8. Second, although our DNA methylation analysis in human sperm and first trimester placentas support that imprinting of Ido1 is conserved in humans, we have not directly tested IDO1 parental-specific expression to validate this observation.
In summary, our studies have revealed that Ido1 is a novel and potentially conserved imprinted locus. We demonstrate that altered DNA methylation of the Ido1/IDO1 gene is linked to pregnancy loss in both mice and humans. Although maternal immunological incompatibilities underlie a significant proportion of human infertility and miscarriages (23,30,40,52), elucidation of the underlying mechanisms remains one of the most challenging questions in reproductive immunology. Our studies suggest that epigenetic alteration of the IDO1 gene may contribute to some cases of human pregnancy loss, although the precise mechanisms remain to be determined. Future studies should also determine whether the Ido1 DMR represents a cis-acting regulatory element at the locus and elucidate whether genetic and environmental factors perturb its regulation, leading to reduced pregnancy success.
Materials and Methods
Mouse information
Studies to measure expression of both parental alleles (i.e. designated as total expression) were conducted using the C57BL/6 J strain, DNA methylation studies using the B6, CBA/J (CBA), and/or DBA/2 J (DBA2), and allele-specific expression with the B6 and PWD/PhJ (PWD). Mice were purchased from JAX (Bar Harbor, ME). Virgin, 6–10 week females were used for all matings. Females were time-mated and euthanized between embryonic day (E) 6.5 and 15.5. At these stages, we carefully removed maternal decidua, and isolated embryonic and extraembryonic tissues for analysis. For allele-specific expression and bisulfite cloning sequencing analysis, E9.5 F1 hybrid progeny were obtained through reciprocal matings between B6 and PWD. For studies on the spontaneous abortion mouse models, we generated E9.5 F1 hybrid progeny from the matings of CBA females with DBA2 or B6 males. This stage was selected as Ido1 expression in extraembryonic tissue peaks at E9.5 [Fig. 1A; (51)]. Furthermore, the embryos and placentas are sufficiently separated at this stage, so that contamination of the placenta with maternally derived decidua is minimized. In the allele-specific expression and bisulfite cloning sequencing studies, SNPs between the two strains were used to distinguish the parental origin of expression. For sperm analysis, we collected mature sperm from the cauda epidydimus of 10–12 week old male mice. For oocyte analysis, germinal vesicle-stage oocytes were retrieved from 4-week-old female mice. Once isolated, tissues or cells were immediately frozen and stored at −80 °C until analysis. All mouse work was carried out with the approval of the University Committee on Animal Resources of the University of Rochester School of Medicine and Dentistry.
Human tissues collections
For the uncomplicated pregnancy cohort: Tissues were collected at the University of Rochester Department of Obstetrics and Gynecology. The villi of first trimester (6–12 weeks) placentas (n = 10) were collected anonymously from elective pregnancy terminations. Placentas from pregnant women with known complications or abnormalities, including sexually transmitted infections, diabetes, hypertension, kidney disease, genetic abnormalities and cigarette or illicit drug use were excluded from this study. All tissues collected for this study were from informed and consented women. Collection of tissues was approved by the Institutional Human Subjects Review Board at the University of Rochester. For the miscarriage cohort: The villi of placentas from first trimester miscarriages (n = 13) were collected by the Yale Cytogenetics Department. Microarray or Comparative Genomic Hybridization was performed at the Yale Cytogenetics Department on DNA extracted from these tissues for clinical purposes prior to performing the current study. For sperm: sperm (n = 4) DNA was provided by Yale Fertility Center Andrology Laboratory. As first trimester miscarriages and sperm were obtained from discarded tissues, the study was exempt from the Institutional Review Board human subject regulations at Yale School of Medicine.
Gene expression studies
RNA was extracted using the RNeasy mini kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol and quantified using the Nano Drop spectrophotometer. cDNA was prepared by RT-PCR using Superscript IV reverse transcriptase and random hexamers (Invitrogen, Waltham, MA). To test that RNA was free of DNA contamination, a minus reverse transcriptase control was included.
Total gene expression
Total RNA was extracted from E6.5 whole conceptus, E7.5 ectoplacental cones and E9.5, E11.5, E13.5 and E15.5 placentas as described above. To measure total gene expression, RT-qPCR analysis was conducted using an Applied Biosystems QuantStudio 5 Real-Time PCR system with the following protocol: 5 μL of Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 0.08 μL of reverse and forward primers, and 3.84 μL of water and 1 μL of cDNA (5 ng) with annealing temperature of 64°C. The following primers were used: 10 μM 5′AGTCGGAAGAGCCCTCAAAT3′ (forward) and 5′GGTGTTTTCTGTGCCCTGAT3′ (reverse) for Ido1 with amplicon size of 152 bp; (25 μM) 5′TTAATTGTGGCTCCCACCTC3′ (forward) and 5′GGGTATCTCCGACTTGTCCA3′ (reverse) for Ido2 with amplicon size of 145 bp. Samples were set up in duplicate and analyzed using QuantStudio design and analysis software (Applied Biosystems). Relative expression of the Ido1 and Ido2 genes was calculated using the comparative Ct method and measurements were normalized to expression of two reference genes (i.e. Arpp0 and Gapdh).
Allele-specific expression
Allele-specific Ido1 and Ido2 expression assays were conducted on E9.5 placental cDNA. Using GoTaq DNA Polymerase (Promega Corporation, Madison, WI), PCR was performed on the cDNA, using 25 μM of each primer, followed by digestion using restriction enzymes. The PCR condition is as follows: 95°C for 3 min, followed by 35 cycles at 95°C for 30 s, 60°C for 1 min, 72°C for 1 min and a final extension of 72°C for 5 min). The primers for Ido1 transcript 1, forward (5′-GTAGACAGCAATGGCACTCA3′), and reverse (5′-AGAGCTCGCAGTAGGGAACA-3′), amplified a 365 bp fragment containing a polymorphism between B6(A) and PWD(G) (chr 8: 24,593,416; mm10). Restriction digestion with Hpy99I (New England Biolabs, Ipswich, MA) resulted in 111 and 254 bp fragments in B6, whereas the PWD amplicon was undigested. Ido1 transcript 2 primers, forward (5′-AGGGTTTAGCGCTGCAGTT-3′) and reverse (5′-AGAGCTCGCAGTAGGGAACA-3′), amplified a 396 bp amplicon containing a polymorphism between B6 (A) and PWD (G). Restriction digestion with Hpy99I (New England Biolabs) resulted in 156 and 240 bp fragments in B6 while the PWD amplicon was undigested. The Ido2 transcript 1 primers, forward (5′-CAGAGACCTCACGCGAAAAT-3′) and reverse (5′-TTGTCAGCACCAGGTCAGAG-3′) amplified a 430 bp fragment containing a polymorphism between B6(A) and PWD(G) (ch8: 24,576,177; mm10). Restriction digestion with BseRI resulted in 92 and 338 bp fragments in PWD while B6 was undigested. The digested PCR products were resolved by electrophoresis on 12% polyacrylamide gels. Parental allele contribution was determined by quantifying relative band intensities of each parental expression [i.e. B6 versus PWD-specific fragment(s)] to total expression (i.e. B6 plus PWD fragments) using a Gel Doc XR+ Molecular Imager (Bio-Rad, Hercules, CA) and ImageJ software. Because a limitation of this quantification method includes detection of background signal from the silent parental allele, we define samples that show ≥90% of total expression derived from one allele to be considered as `imprinted’ or `almost exclusively expressed’ from the allele.
Protein expression
Tissues were homogenized in ice-cold RIPA lysis and extraction buffer (Thermo Fisher Scientific, Rockford, IL; Cat #: 89900) containing Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Cat #: 78440) using the Omni Bead Rupter (Omni-International, Kennesaw, GA). Total protein concentrations were determined with the Bradford protein detection assay (Bio-Rad, Hercules, CA). Proteins (20 μg) were fractionated on 12% polyacrylamide gels and transferred onto 0.45 μm Immobilon–polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA). Membranes were blocked with 5% blotting-grade blocker (Bio-Rad, Hercules, CA) in 1x Tris-buffered saline containing 0.1% Tween-20 and probed for IDO1 (1:1000; rat monoclonal anti-IDO (mIDO-48), Cat #: sc-53978, Santa Cruz Biotechnology, Santa Cruz, CA), IDO2 [1:1000; mouse monoclonal anti-IDOL1 (C-9), Cat #: sc-374159; Santa Cruz Biotechnology, Santa Cruz, CA], and GAPDH (1:1000; mouse monoclonal anti–GAPDH, Cat #: CB1001, EMD Millipore) overnight at 4°C. Membranes were then probed with the appropriate horseradish peroxidase-conjugated secondary antibody (1:10,000; Jackson Immunoresearch; West Grove, PA) for 1 h at room temperature. Proteins were visualized using Immobilon Western chemiluminescent horseradish peroxidase substrate (EMD Millipore). Chemiluminescent signals were captured using a ChemiDoc MP imaging system (Bio-Rad, Hercules, CA).
DNA methylation analysis in mouse
For placental tissues, DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen). Sperm DNA extraction was performed using the phenol chloroform method as previously described (9). DNA was quantified using the Nano Drop spectrophotometer. We performed bisulfite treatment of DNA using the EpiTect Bisulfite kit (Qiagen) in all tissues, except for E6.5–7.5 tissues and oocytes in which we used Epitech Fast LyseAll Bisulfite Kit (Qiagen). For pyrosequencing: genes of interest were amplified by PCR using the PyroMark PCR kit (Qiagen). PCR conditions were: 95°C for 15 min followed by 45 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 30 s, and a final extension of 72°C for 10 min. PCR products (10 μL) were sequenced using the PyroMark Advanced Reagents kit (Qiagen). Pyrosequencing was run using the PyroMark Advanced Q24 (Qiagen) and analyzed using Qiagen’s PyroMark Q24 Advanced software. For the list of mouse pyrosequencing primers, see Supplementary Table S1. Methylation analysis of the mouse H19 and Snrpn genes has been described previously (50). For bisulfite cloning sequencing: bisulfite cloning sequencing was performed to analyze the allele-specific methylation of the Ido1 DMR. Fifty ng of bisulfite-treated DNA was subjected to two rounds of PCR amplification. Using GoTaq DNA Polymerase (Promega Corporation), we used 25 μm primers described below. The first-round primers are forward, 5′- AAAGTGGATATAAATTTAGGAG-3′ and reverse, 5′-ACAACCAAACCACAAAAAAT-3′. The first-round PCR conditions were: 94°C for 2 min followed by 40 cycles of 94°C for 30 s, 55°C (or 57°C for paternal allele) with ramping 1°C/s, and 72°C for 1 min. Two μl of first-round PCR was used for second-round PCR amplification. The second-round primer sequences were as follows: forward 5′- AAAGTGGATATAAATTTAGGAG-3′ and reverse, 5′-CCCATCAATAATTTAAATAACCAC-3′. The second-round PCR conditions were 94°C for 2 min followed by 40 cycles of 94°C for 30 s, 55°C (or 57°C for paternal allele) with ramping 1°C/s, and 72°C for 1 min, and a final step of 72°C for 15 min. Second-round PCR products were gel extracted and purified (Qiagen) and ∼5.63 ng of purified product was used for TA Cloning Kit with TOP10 competent cells (Thermo Fisher, Rockford, IL). Colonies were harvested, miniprepped (Qiagen), and sequenced (Genewiz, South Plainfield, NJ). Parent-of-origin methylation was determined by confirmation of the three SNPs present between C57BL/6J and PWD/PhJ mice as follows: G/T (Ch8:24,597,831mm10), G/T (Ch8:24,597,746 mm10) and G/A (Ch8:24,597,672 mm10). For this study, we used E9.5F1 hybrid placentas and embryos from the reciprocal crosses of B6 and PWD mice, and B6 sperm and oocytes.
DNA methylation analysis in human
DNA from placentas in the uncomplicated pregnancy cohort was extracted using the Qiagen DNeasy Blood and Tissue kit. DNA from sperm and miscarriage cohort was provided by Yale Cytogenetics Department and Yale Fertility Center Andrology Laboratory, respectively. Prior to analysis, we quantified DNA using a NanoDrop spectrophotometer. We conducted pyrosequencing analysis to measure DNA methylation of 19 CpG sites spanning the promoter and intron 1 regions of the human IDO1 gene (chr8: 39,913,809 – 39,928,790). PCR and sequencing were performed as described above for methylation studies in mice. Human pyrosequencing primers are listed in Supplementary Table S1. Human H19 pyrosequencing assay has been previously published (41,58).
Statistical Analysis
For mouse studies: to evaluate differences in Ido1 and Ido2 mRNA expression, we performed unpaired t-test analysis (i.e. embryo versus placenta) or one-way analysis of variance (ANOVA) and Dunnett’s posttest (i.e. developmental expression). To compare average DNA methylation across placentas, sperm and oocytes, we used ANOVA followed by Dunnett’s multiple comparisons test. To compare differences in DNA methylation variances in CBAXB6 versus CBAXDBA/2 pregnancies, we used the F variance ratio test. For human studies: a quasibinomial generalized linear model with a logit link was used to model differences in percent methylation of CpG sites between sperm, uncomplicated placentas and miscarriage placentas. Nested models were compared using a deviance F-test. Differences in percent or variance in methylation for individual CpGs were assessed using a t-test or homogeneity test of variance, and the corresponding P-values were adjusted for multiple comparisons using the Benjamini–Hochberg method (11). P-values and FDR of <0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgements
We thank Marisa Bartolomei for the discussion and helpful comments on the manuscript. We also thank Erica Sloma for her technical assistance in elective termination tissue collection for this study.
Conflict of Interest statement. None declared.
Funding
National Institute of Environment Health Sciences (R00ES022244 and P30 ES001247 to M.S., T32 ES007026 to J.M.R.); National Institute of General Medical Sciences (K12 GM106997 to S.E.L.).
Author contributions
P.S. contributed to the writing of the manuscript, running the majority of the mouse studies and analyzing the data. S.E.L. contributed to the writing of the manuscript, running the human studies and analyzing the data. J.M.R. contributed to the writing of the manuscript and some aspects of the mouse studies (i.e. gene expression analysis and histone modifications marks). A.F. conducted the bisulfite cloning sequencing analysis. B.B. performed the initial mouse imprinting studies and data analysis. X.L. and M.N.M. performed biostatistical analysis of the human data and contributed to the writing of this section of the manuscript. S.P.M. contributed to the writing of the manuscript and experimental design for the human studies and provided samples for the uncomplicated pregnancy cohort. W.M. contributed to the writing of the manuscript, experimental design for the human studies and provided samples for the sperm and pregnancy loss cohort studies. M.S. conceived the ideas, was the prime initiator of the concept and studies, supervised across the different aspects of the studies, planned the experiments, interpreted data and wrote the manuscript.
References
- 1. Alegre E., López A.S., Díaz-Lagares A. and González A. (2008) Study of the plasmatic levels of tryptophan and kynurenine throughout pregnancy. Clin. Chim. Acta, 393, 132–133. [DOI] [PubMed] [Google Scholar]
- 2. Aluvihare V.R., Kallikourdis M. and Betz A.G. (2004) Regulatory T cells mediate maternal tolerance to the fetus. Nat. Immunol., 5, 266–271. [DOI] [PubMed] [Google Scholar]
- 3. Arefayene M., Philips S., Cao D., Mamidipalli S., Desta Z., Flockhart D.A., Wilkes D.S. and Skaar T.C. (2009) Identification of genetic variants in the human indoamine 2,3-dioxygenase (IDO1) gene, which have altered enzyme activity. Pharmacogenet. Genomics, 19 (6), 464–476. [DOI] [PubMed] [Google Scholar]
- 4. Babak T., DeVeale B., Tsang E.K., Zhou Y., Li X., Smith K.S., Kukurba K.R., Zhang R., Li J.B., Kooy D. et al. (2015) Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat. Genet., 47, 544–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baban B., Chandler P., McCool D., Marshall B., Munn D.H. and Mellor A.L. (2004) Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. J. Reprod. Immunol., 61, 67–77. [DOI] [PubMed] [Google Scholar]
- 6. Badawy A.A.-B. (2014) The tryptophan utilization concept in pregnancy. Obstet. Gynecol. Sci. 57, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barlow D.P. and Bartolomei M.S. (2014) Genomic imprinting in mammals. Cold. Spring. Harb. Perspect. Biol., 6, a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barlow D.P., Stöger R., Herrmann B.G., Saito K. and Schweifer N. (1991) The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature, 349, 84–87. [DOI] [PubMed] [Google Scholar]
- 9. Bartolomei M.S., Webber A.L., Brunkow M.E. and Tilghman S.M. (1993) Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev., 7, 1663–1673. [DOI] [PubMed] [Google Scholar]
- 10. Bartolomei M.S., Zemel S. and Tilghman S.M. (1991) Parental imprinting of the mouse H19 gene. Nature, 351, 153–155. [DOI] [PubMed] [Google Scholar]
- 11. Benjamini Y. and Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B, 57, 289–300. [Google Scholar]
- 12. Bonney E.A. and Brown S.A. (2014) To drive or be driven: the path of a mouse model of recurrent pregnancy loss. Reproduction, 147, R153–R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buiting K., Kanber D., Horsthemke B. and Lohmann D. (2010) Imprinting of RB1 (the new kid on the block). Brief. Funct. Genomics, 9, 347–353. [DOI] [PubMed] [Google Scholar]
- 14. Carr M.S., Yevtodiyenko A., Schmidt C.L. and Schmidt J.V. (2007) Allele-specific histone modifications regulate expression of the Dlk1–Gtl2 imprinted domain. Genomics, 89, 280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cervenka I., Agudelo L.Z. and Ruas J.L. (2017) Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science, 357, eaaf9794. [DOI] [PubMed] [Google Scholar]
- 16. Choufani S., Shapiro J.S., Susiarjo M., Butcher D.T., Grafodatskaya D., Lou Y., Ferreira J.C., Pinto D., Scherer S.W., Shaffer L.G. et al. (2011) A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res., 21, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark D.A., McDermott M.R. and Szewczuk M.R. (1980) Impairment of host-versus-graft reaction in pregnant mice. II. Selective suppression of cytotoxic T-cell generation correlates with soluble suppressor activity and with successful allogeneic pregnancy. Cell. Immunol., 52, 106–118. [PubMed] [Google Scholar]
- 18. DeChiara T.M., Efstratiadis A. and Robertson E.J. (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature, 345, 78–80. [DOI] [PubMed] [Google Scholar]
- 19. Erkek S., Hisano M., Liang C.-Y., Gill M., Murr R., Dieker J., Schübeler D., Vlag J., Stadler M.B. and Peters A.H.F.M. (2013) Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol., 20, 868–875. [DOI] [PubMed] [Google Scholar]
- 20. Fournier C., Goto Y., Ballestar E., Delaval K., Hever A.M., Esteller M. and Feil R. (2002) Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J., 21, 6560–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gendron R.L. and Baines M.G. (1989) Morphometric analysis of the histology of spontaneous fetal resorption in a murine pregnancy. Placenta, 10, 309–318. [DOI] [PubMed] [Google Scholar]
- 22. Guo F., Yan L., Guo H., Li L., Hu B., Zhao Y., Yong J., Hu Y., Wang X., Wei Y. et al. (2015) The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell, 161, 1437–1452. [DOI] [PubMed] [Google Scholar]
- 23. Haller-Kikkatalo K., Salumets A. and Uibo R. (2012) Review on autoimmune reactions in female infertility: antibodies to follicle stimulating hormone. Clin. Dev. Immunol., 2012, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanna C.W., Peñaherrera M.S., Saadeh H., Andrews S., McFadden D.E., Kelsey G. and Robinson W.P. (2016) Pervasive polymorphic imprinted methylation in the human placenta. Genome Res., 26, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hassold T., Hall H. and Hunt P. (2007) The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet., 16 (Spec No. 2), R203–R208. [DOI] [PubMed] [Google Scholar]
- 26. Hiura H., Sugawara A., Ogawa H., John R.M., Miyauchi N., Miyanari Y., Horiike T., Li Y., Yaegashi N., Sasaki H. et al. (2010) A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res., 38, 4929–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ideraabdullah F.Y., Vigneau S. and Bartolomei M.S. (2008) Genomic imprinting mechanisms in mammals. Mutat. Res. Fund. Mol. Mech.Mut., 647, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jensen A.B., Tunster S.J. and John R.M. (2014) The significance of elevated placental PHLDA2 in human growth restricted pregnancies. Placenta, 35, 528–532. [DOI] [PubMed] [Google Scholar]
- 29. Kelsey G. and Feil R. (2013) New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond., B, Biol. Sci., 368, 20110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kudo Y., Boyd C.A.R., Sargent I.L. and Redman C.W.G. (2003) Decreased tryptophan catabolism by placental indoleamine 2,3-dioxygenase in preeclampsia. Am. J. Obstet. Gynecol., 188, 719–726. [DOI] [PubMed] [Google Scholar]
- 31. Li W., Li B., Fan W., Geng L., Li X., Li L., Huang Z. and Li S. (2009) CTL4Ig gene transfer alleviates abortion in mice by expanding CD4+CD25+ regulatory T cells and inducing indoleamine 2,3-dioxygenase. J. Reprod. Immunol., 80,–11. [DOI] [PubMed] [Google Scholar]
- 32. Luan H., Meng N., Liu P., Feng Q., Lin S., Fu J., Davidson R., Chen X., Rao W., Chen F. et al. (2014) Pregnancy-induced metabolic phenotype variations in maternal plasma. J. Proteome Res., 13, 1527–1536. [DOI] [PubMed] [Google Scholar]
- 33. Luedi P.P., Dietrich F.S., Weidman J.R., Bosko J.M., Jirtle R.L. and Hartemink A.J. (2007) Computational and experimental identification of novel human imprinted genes. Genome Res. 17, 1723–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Metsalu T., Viltrop T., Tiirats A., Rajashekar B., Reimann E., Kõks S., Rull K., Milani L., Acharya G., Basnet P. et al. (2014) Using RNA sequencing for identifying gene imprinting and random monoallelic expression in human placenta. Epigenetics, 9, 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore G.E., Ishida M., Demetriou C., Al-Olabi L., Leon L.J., Thomas A.C., Abu-Amero S., Frost J.M., Stafford J.L., Chaoqun Y. et al. (2015) The role and interaction of imprinted genes in human fetal growth. Philos. Trans. R. Soc. Lond., B, Biol. Sci., 370, 20140074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moore T., Constancia M., Zubair M., Baileul B., Feil R., Sasaki H. and Reik W. (1997) Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2 Proc. Natl. Acad. Sci. U. S. A., 94, 12509–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Munn D.H., Zhou M., Attwood J.T., Bondarev I., Conway S.J., Marshall B., Brown C. and Mellor A.L. (1998) Prevention of allogeneic fetal rejection by tryptophan catabolism. Science, 281, 1191–1193. [DOI] [PubMed] [Google Scholar]
- 38. Okae H., Chiba H., Hiura H., Hamada H., Sato A., Utsunomiya T., Kikuchi H., Yoshida H., Tanaka A., Suyama M. et al. (2014) Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet., 10 e1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W. and Walter J. (2000) Active demethylation of the paternal genome in the mouse zygote. Curr. Biol., 10, 475–478. [DOI] [PubMed] [Google Scholar]
- 40. Pearson H. (2002) Reproductive immunology: immunity's pregnant pause. Nature, 420, 265–266. [DOI] [PubMed] [Google Scholar]
- 41. Pliushch G., Schneider E., Weise D., El Hajj N., Tresch A., Seidmann L., Coerdt W., Müller A.M., Zechner U. and Haaf T. (2010) Extreme methylation values of imprinted genes in human abortions and stillbirths. Am. J. Pathol., 176, 1084–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rappolee D.A., Sturm K.S., Behrendtsen O., Schultz G.A., Pedersen R.A. and Werb Z. (1992) Insulin-like growth factor II acts through an endogenous growth pathway regulated by imprinting in early mouse embryos. Genes Dev., 6, 939–952. [DOI] [PubMed] [Google Scholar]
- 43. Santillan M.K., Pelham C.J., Ketsawatsomkron P., Santillan D.A., Davis D.R., Devor E.J., Gibson-Corley K.N., Scroggins S.M., Grobe J.L., Yang B. et al. (2015) Pregnant mice lacking indoleamine 2,3-dioxygenase exhibit preeclampsia phenotypes. Physiol. Rep., 3, e12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schroeder D.I., Blair J.D., Lott P., Yu H.O.K., Hong D., Crary F., Ashwood P., Walker C., Korf I., Robinson W.P. et al. (2013) The human placenta methylome. Proc. Natl. Acad. Sci. U. S. A., 110, 6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schröcksnadel H., Baier-Bitterlich G., Dapunt O., Wachter H. and Fuchs D. (1996) Decreased plasma tryptophan in pregnancy. Obstet. Gynecol., 88, 47–50. [DOI] [PubMed] [Google Scholar]
- 46. Schulz R., Proudhon C., Bestor T.H., Woodfine K., Lin C., Lin S., Prissette M., Oakey R.J. and Bourc'his D. (2010) The parental non-equivalence of imprinting control regions during mammalian development. PLoS Genetics, 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sedlmayr P., Blaschitz A. and Stocker R. (2014) The role of placental tryptophan catabolism. Front. Immunol., 5, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh P., Cho J., Tsai S.Y., Rivas G.E., Larson G.P. and Szabó P.E. (2010) Coordinated allele-specific histone acetylation at the differentially methylated regions of imprinted genes. Nucleic Acids Res., 38, 7974–7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steinhoff C., Paulsen M., Keilbasa S., Walter J. and Vingron M. (2009) Expression profile and transcription factor binding site exploration of imprinted genes in human and mouse. BMC Genomics, 10, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Susiarjo M., Sasson I., Mesaros C. and Bartolomei M.S. (2013) Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet., 9, e1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suzuki S., Toné S., Takikawa O., Kubo T., Kohno I. and Minatogawa Y. (2001) Expression of indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase in early concepti. Biochem. J., 355, 425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trowsdale J. and Betz A.G. (2006) Mother’s little helpers: mechanisms of maternal-fetal tolerance. Nat. Immunol., 7, 241–246. [DOI] [PubMed] [Google Scholar]
- 53. Tunster S.J., Tycko B. and John R.M. (2010) The imprinted Phlda2 gene regulates extraembryonic energy stores. Mol. Cell. Biol., 30, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tunster S.J., Van de Pette M. and John R.M. (2014) Isolating the role of elevated Phlda2 in asymmetric late fetal growth restriction in mice. Dis. Model. Mech., 7, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Verona R.I., Thorvaldsen J.L., Reese K.J. and Bartolomei M.S. (2008) The transcriptional status but not the imprinting control region determines allele-specific histone modifications at the imprinted H19 locus. Mol. Cell. Biol., 28, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yeung A.W.S., Terentis A.C., King N.J.C. and Thomas S.R. (2015) Role of indoleamine 2,3-dioxygenase in health and disease. Clin. Sci., 129, 601–672. [DOI] [PubMed] [Google Scholar]
- 57. Yu B., Dong X., Gravina S., Kartal Ö., Schimmel T., Cohen J., Tortoriello D., Zody R., Hawkins R.D. and Vijg J. (2017) Genome-wide, single-cell DNA methylomics reveals increased non-CpG methylation during human oocyte maturation. Stem Cell Reports, 9, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zechner U., Pliushch G., Schneider E., El Hajj N., Tresch A., Shufaro Y., Seidmann L., Coerdt W., Muller A.M. and Haaf T. (2010) Quantitative methylation analysis of developmentally important genes in human pregnancy losses after ART and spontaneous conception. Mol. Hum. Reprod., 16, 704–713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


