Abstract
Mammalian hibernators store fat extensively in white adipose tissues (WATs) during pre-hibernation period (Pre-HIB) to prepare for hibernation. However, the molecular mechanisms underlying the pre-hibernation remodeling of WAT have not been fully elucidated. Syrian hamsters, a food-storing hibernator, can hibernate when exposed to a winter-like short day photoperiod and cold ambient temperature (SD-Cold). Animals subjected to prolonged SD-Cold had smaller white adipocytes and beige-like cells within subcutaneous inguinal WAT (iWAT). Time-course analysis of gene expression with RNA-sequencing and quantitative PCR demonstrated that the mRNA expression of not only genes involved in lipid catabolism (lipolysis and beta-oxidation) but also lipid anabolism (lipogenesis and lipid desaturation) was simultaneously up-regulated prior to hibernation onset in the animals. The enhanced capacity of both lipid catabolism and lipid anabolism during hibernation period (HIB) is striking contrast to previous observations in fat-storing hibernators that only enhance catabolism during HIB. The mRNA expression of mTORC1 and PPAR signaling molecules increased, and pharmacological activation of PPARs indeed up-regulated lipid metabolism genes in iWAT explants from Syrian hamsters. These results suggest that the Syrian hamster rewires lipid metabolisms while preparing for hibernation to effectively utilize body fat and synthesize it from food intake during HIB.
Keywords: hibernation, Syrian hamsters, white adipose tissue (WAT), beige adipocytes, lipid metabolism
Introduction
Hibernation is an adaptive strategy that allows animals to persist in environments with seasonal or unpredictable decreases in food availability (Heldmaier et al., 2004; Geiser, 2013; Nowack et al., 2017). In small-bodied mammals (e.g., hamster, ground squirrel, and chipmunk), HIB involves multiday hypothermic deep torpor bout (HIB-DT) and normothermic periodic arousal (HIB-PA) (Figure 1A). HIB-DT is characterized by the profound suppression of metabolism, body temperature, heart rate, food intake, and locomotive activity (Lyman, 1958; Melvin and Andrews, 2009; Ruf and Geiser, 2015). The drastic alterations in physiology associated with deep torpor bout - PA cycles would lead to multiple organ dysfunction and death in non-hibernators such as mice and humans, whereas hibernators can tolerate these physiological extremes. Interestingly, however, this amazing tolerance is only operational during the hibernation season in several strictly seasonal hibernators (Kondo and Shibata, 1984; Carey et al., 2003; Kurtz et al., 2006; Andres-Mateos et al., 2013). These lines of evidence suggest that hibernators prepare for hibernation by undergoing “fall transition” or in other words, “pre-hibernation remodeling,” through which their bodies are converted from the summer phenotype to the winter phenotype during fall, the pre-HIB (Kondo, 1987; Grabek et al., 2011; Jinka et al., 2011; Hindle and Martin, 2014). However, the molecular bases underlying the induction and regulation of pre-hibernation remodeling, as well as hibernation itself, remain poorly understood (Jastroch et al., 2016).
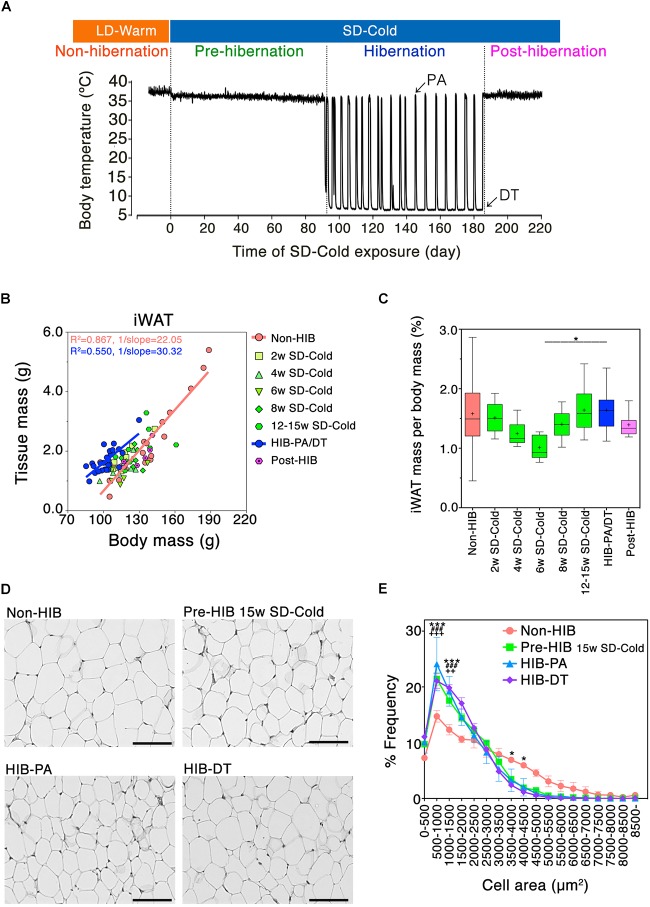
FIGURE 1.
Mass and morphological changes of iWAT during the transitional period from non-hibernation state to hibernation state. (A) A representative change in core body temperature of hamsters measured by intraperitoneally implanted core body temperature loggers. At Post-hibernation, animals spontaneously terminated hibernation while still being kept in SD-Cold conditions. (B) Positive correlation of adipose tissue mass with body mass of hamsters: orange line; Non-HIB group, R2 = 0.867, 1/slope = 22.05, P < 0.0001: blue line; HIB-PA/DT group; R2 = 0.550, 1/slope = 30.32, P < 0.0001. Non-HIB, non-hibernation group under LD-Warm conditions (n = 20); 2/4/6/8/12–15 weeks SD-Cold, pre-hibernation group at different time points after the transfer to an SD-Cold environment (n = 8, 8, 5, 8, and 8, respectively); HIB-PA/DT, hibernation group combing both PA and DT (n = 21); Post-HIB, post-hibernation group (n = 7). (C) Changes of iWAT-to-body mass ratio over time. Horizontal lines and crosses indicate medians and means of iWAT-to-body mass ratio, respectively. Boxes enclose the interquartile ranges and whiskers show the minimum and maximum iWAT-to-body mass ratio of each experimental group. ∗p < 0.05 between the “6 weeks SD-Cold” and “HIB-PA/DT,” assessed by one-way ANOVA followed by analysis using Tukey’s multiple comparison tests. (D,E) H&E staining (D) and the frequency distribution of adipocyte cell size (E) in iWAT from the four different states (n = 3 for each condition). The results are expressed as mean ± SEM. P-value was derived using two-way ANOVA and Bonferroni post-tests. Significant differences are marked by ∗ between “HIB-DT” and “Non-HIB,” # between “HIB-PA” and “Non-HIB,” and by + between “Pre-HIB” and “Non-HIB”. ∗, #, +p < 0.05, ∗∗, ##, ++p < 0.01, ∗∗∗, ###, +++p < 0.005.
Nutritional and environmental changes induce “WAT remodeling,” which is defined as alterations in the number, shape, type, and function of the adipose resident cells (Choe et al., 2016). For instance, chronic cold stimuli can recruit beige adipocytes to the WAT of non-hibernators such as mice, rats, and humans (Wu et al., 2012; Jimenez-Aranda et al., 2013). Beige adipocytes resemble brown adipocytes in the heat-producing organ, brown adipose tissue (BAT); they contain enriched mitochondria and multilocular lipid droplets, and express mitochondrial uncoupling protein 1 (Ucp1), a key contributor to heat generation in beige and brown adipocytes in those species. These properties enable the beige and brown adipocytes to produce heat via a non-shivering thermogenesis, and the recruitment of beige adipocyte leads to the acquisition of thermogenic properties in WAT, just as BAT (Enerback et al., 1997; Harms and Seale, 2013; Okamatsu-Ogura et al., 2013; Bartesaghi et al., 2015; Kajimura et al., 2015). Although it is not yet known to our knowledge whether beige adipocytes are recruited in hibernators, WAT is expected to play a key role during multi-day fasting and the re-warming process in hibernation (Moreau-Hamsany et al., 1988; Carneheim et al., 1989; Florant et al., 2004).
During the pre-hibernation remodeling, strictly seasonal hibernators, such as marmots, dormice, ground squirrels, and bears, fatten rapidly in order to store fat reserves for survival during HIB (Armitage et al., 1976; Swenson et al., 2007; Sheriff et al., 2013; Giroud et al., 2014; Arnold et al., 2015). The expression and activity of lipogenic enzymes in WAT is increased during the pre-HIB, but once winter hibernation begins, lipogenesis is weakened, and lipolysis is dominantly activated in WAT in these strictly seasonal hibernators (Mostafa et al., 1993; Wang et al., 1997; Hampton et al., 2011; Arnold et al., 2015; Shimozuru et al., 2016). This is possibly because most of these animals studied are fat-storing hibernators; they become hyperphagia from late summer to fall, and then completely stop feeding and largely subsist on the accumulated fat as substrate for energy metabolism in winter, where respiratory quotient values are ∼0.7 (Wilson et al., 1992; Melvin and Andrews, 2009; Toien et al., 2011). In contrast to the aforementioned fat-storing hibernators, Syrian (golden) hamsters (Mesocricetus auratus) are food-storing hibernators that eat food stored in the nest during PA. It was reported that they maintain a high lipogenic capacity of WAT in both Pre-HIB and HIB in response to prolonged cold (Baumber and Denyes, 1963). Despite this difference between fat-storing and food-storing hibernators, the above studies suggest that dynamic reorganization of lipid metabolic pathways is a key feature of pre-hibernation remodeling in mammalian hibernators examined so far, whereas it is yet unclear how the reorganization is achieved.
To study pre-hibernation remodeling and hibernation in the laboratory, Syrian hamster is a good experimental animal model for the following reasons. Syrian hamsters can initiate hibernation throughout the year if they are maintained under appropriate winter-like conditions, and are therefore classified as opportunistic (facultative) hibernators (Jansky et al., 1984; Tamura et al., 2005; Lewis et al., 2012; Geiser, 2013; Chayama et al., 2016; Trefna et al., 2017). In the laboratory, after transfer from summer-like conditions [LD photoperiod and warm ambient temperature (LD-Warm)] to winter-like conditions [SD photoperiod and cold ambient temperature (SD-Cold)], most Syrian hamsters require a long pre-HIB (typically 2–3 months) before initiating hibernation (Figure 1A) (Jansky et al., 1984; Chayama et al., 2016). This requirement of a long induction period in SD-Cold implies that the pre-hibernation remodeling process, involving changes in the thermoregulatory system and the regulation of body mass, is present in Syrian hamsters, as in strictly seasonal hibernators (Arnold et al., 2011; Olson et al., 2013; Sheriff et al., 2013; Drew et al., 2017). In fact, during the pre-HIB, the female Syrian hamsters underwent physiological body remodeling; they gradually reduce their body mass and the set-point of core body temperature (Arai et al., 2005; Chayama et al., 2016). After the body mass dips lower than the threshold for hibernation, the hamsters begin to hibernate (Supplementary Figure S1A) (Chayama et al., 2016). However, little is known about molecular and cellular aspects of the pre-hibernation remodeling; differences between summer-like non-hibernating body conditions and winter-like hibernating body conditions, and mechanisms underlying the regulation of those pre-hibernation remodeling in Syrian hamsters.
In the present study, we hypothesized that pre-hibernation remodeling would be induced in WAT, the central organ in regulation of the whole body-energy homeostasis and body mass, under prolonged SD-Cold condition before initiating hibernation. To examine this at the cellular and molecular levels, we conducted morphological analyses of adipocytes and comprehensive RNA-seq profiling on the iWAT of female Syrian hamsters. In particular, we focused on (i) alterations in adipocyte size, and emergence of distinct types of adipocytes in iWAT, and (ii) lipid metabolism remodeling in iWAT from pre-HIB to HIB.
Materials and Methods
Ethics Statement
All animal work has been conducted according to the ethics guidelines of the University of Tokyo and Hokkaido University, and was approved by the Ethics Committee of the University of Tokyo (Ethical Approval no. P24–34) and of the Hokkaido University (Ethical Approval no. 18-0140).
Animals and Housing
Female Syrian hamsters (Mesocricetus auratus) from a closed breeding colony were purchased from SLC, Inc., Japan. Animals were housed in groups of 3 or 4 per cage with ad libitum access to diets (MR standard diet, Nihon Nosan, Japan) and water under LD-Warm conditions (light condition = 16L:8D cycle, lights on 05:00–21:00, ambient temperature = 24–25°C). For hibernation induction, animals were reared under LD-Warm conditions until most animals weighed over 100–120 g. The body mass of animals and amounts of food consumption were measured weekly when cages were changed.
One to 2 weeks before the transfer to SD-Cold conditions, core body temperature (Tb) loggers (iButton®, Maxim Integrated, United States, #DS1992 L-F5 model; operating temperature range from -40 to +70°C) coated with rubber (Plasti Dip, Performix®; total mass ∼3.5 g, 1.8–3.5% of animal mass) were implanted intraperitoneally under inhalation anesthesia with 4% isoflurane (DS Pharma Animal Health, Japan) and intraperitoneal injection of pentobarbital sodium (65 mg/kg, diluted with phosphate-buffered saline; Kyoritsu Seiyaku, Japan). We did not calibrate iButtons as we assumed linearity and no drift of the temperature signal based on our previous experience. After 1–2 weeks of recovery, animals were transferred to SD-Cold conditions (8L:16D cycle, lights on 10:00–18:00, ambient temperature = 5°C). Animals were individually housed in polypropylene cages, and the Tb of animals were measured every 90 min with an accuracy of 0.5°C. The onset of hibernation was detected comprehensively by the characteristic postures of animals (rolled into ball, Supplementary Figure S1A), the reduced activity and consumption of food when cages were changed. The ‘saw-dust method’ was also used for confirming that animals successfully hibernated; wood chips placed on the back of hibernating individuals remained in place until the animals experienced HIB-PA (Jansky et al., 1984). The cage replacement and body mass measurement of animals in HIB-DT were skipped to avoid disturbing HIB-DT. HIB-DT is defined as prolonged hypothermia, with Tb lowered to 15°C or less (Ruf and Geiser, 2015).
Tb loggers were recovered from animals sacrificed by decapitation after they were subjected to 10–15 min anesthesia with intraperitoneal injection of pentobarbital sodium (65 mg/kg) and inhalation of 4% isoflurane.
iWAT Collection
To examine the time-course of remodeling in iWAT during the pre-HIB, iWAT of hamsters was collected at different time-points: Non-HIB (euthermic and active animals at the LD-Warm condition), Pre-HIB (2, 4, 6, 8, and 12–15 weeks of the SD-Cold), HIB-PA [animals that spontaneously aroused from HIB-DT to euthermic state (Tb > 35°C) for 10–15 h under SD-Cold], HIB-DT (Tb < 15°C over 6 h), and post-hibernation group (Post-HIB: euthermic and active after spontaneous termination of hibernation under the SD-Cold). All animals were sacrificed between 13:00 and 15:00, by decapitation 10–15 min under anesthesia with intraperitoneal injection of pentobarbital sodium (65 mg/kg) and 4% isoflurane. Whole iWAT pads were dissected from the surrounding tissues, and cleaned of any contaminating tissue (Supplementary Figure S1B). The wet tissue mass of both sides of iWAT was measured for each hamster, and the average of these measurements was calculated for analysis. The correlation of iWAT mass with body mass was analyzed using Pearson’s correlation coefficient. The iWAT-to-body mass ratio for each hamster was calculated and expressed as a percentage of body mass. P-value was derived using one-way ANOVA and Tukey’s multiple comparison tests using the Graph Pad Prism 5.0 software (GraphPad, San Diego, CA, United States).
Measurement of Adipocyte Size
iWAT were obtained from animals sacrificed at four collection points: Non-HIB (18 weeks old), Pre-HIB at 15 weeks of the SD-Cold condition (24 weeks old), HIB-PA (25–26 weeks old), and HIB-DT (24–26 weeks old). Three animals for each group were analyzed. One side of the iWAT was dissected into four parts (Supplementary Figure S1B), and fixed in 4% of phosphate buffered paraformaldehyde (PFA) for more than 2 days at 4°C, dehydrated, and embedded in paraffin as previously described (Berry et al., 2014). Regions 2 and 3 of iWAT were then cut in 6-μm sections and stained using hematoxylin (Sigma-Aldrich) and eosin (Wako).
Tile-scanned images of each section were obtained at 20× magnification using an AF6000 microscope (Leica Microsystems). Adipocyte cell size was semi-automatically determined using the FIJI plug-in Adiposoft (Galarraga et al., 2012). Measurements were obtained from three animals per group; two sections for each hamster were analyzed, and the mean of these measurements was used for the calculation. The frequency distribution of adipocyte sizes in iWAT was calculated and expressed as a percentage of total adipocytes counted. The results are expressed as mean ± SEM. P-value was derived from two-way ANOVA and Bonferroni’s multiple comparison tests using the Graph Pad Prism 5.0 software.
Immunohistochemical Analysis
Six-micrometer sections from paraffin-embedded iWAT were deparaffinized and rehydrated, and submitted to heat antigen retrieval using a microwave oven and citrate buffer, pH 6.0 for 15 min. The endogenous peroxidase activity was quenched by exposing sections to 3% hydrogen peroxide in methanol overnight at 4°C. After rinsing extensively using Tris-buffered saline with 0.1% Tween20 (WAKO) (TBS-T), the sections were blocked using 20% ImmunoBlock (DS Pharma, Osaka, Japan) in TBS-T for 30 min at room temperature, and incubated with rabbit anti-human UCP1 polyclonal antibody (Abcam, ab23841, lot#GR116982-1) at 1:100 dilution in Immunoreaction Enhancer Solution (Can Get Signal A solution, TOYOBO) with 10% ImmunoBlock at 4°C overnight. Mouse anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Abcam, ab99702) was applied at 1:200 dilution in the Immunoreaction Enhancer Solution with 10% ImmunoBlock for 60 min at room temperature. After rinsing with Tris-buffered saline (TBS), slides were incubated with diaminobenzidine (Sigma Aldrich) and hydrogen peroxide in TBS (pH 7.2) for 15 min, and then rinsed in tap water. After counterstaining with hematoxylin (Sigma-Aldrich), the sections were dehydrated, coverslipped, and imaged at 20×/40× magnification with an AF6000 microscope (Leica Microsystems). The numbers of animals for immunohistochemical analysis were as follows; Non-HIB (n = 6), Pre-HIB (n = 6), HIB-PA (n = 3), and HIB-DT (n = 3).
RNA Isolation and Real-Time Quantitative PCR
A whole iWAT pad dissected as described above was minced and incubated in RNA stabilization solution (RNA later, Ambion), and stored at -80°C until RNA purification. Total RNA was purified from a whole iWAT using Qiazol Lysis Reagent and RNeasy Mini Kit (Qiagen).
Total RNA samples from individual animals were quantified with a Nanodrop (Thermo Scientific). Total RNA (100–250 ng) was reverse-transcribed using a Prime Script RT reagent with a gDNA eraser (TAKARA). Primers were designed using an online tool, Primer-BLAST1 based on known Syrian hamster sequences (Mesocricetus auratus, taxid:10036). Primer sequences are depicted in Supplementary Table S6.
Quantitative real-time PCR (qPCR) was performed using a SYBR Premix Ex Taq II (Tli RnaseH Plus) (TAKARA) and a Light Cycler 480 real-time PCR system (Roche), according to the manufacturer’s instructions. The specificity of PCR product was checked using melting curve analysis. The relative mRNA expression of each gene was quantified using a standard curve. The most stable reference gene was determined from potential reference genes by geNorm analysis (Otis et al., 2010), resulting in the selection of Peptidylprolyl isomerase A (Ppia) as an internal control gene for normalization. Results of relative quantification are expressed as mean ± standard errors (SEM), or box-and-whisker plot. P-value was derived from one-way ANOVA and Tukey’s multiple comparison tests using Graph Pad Prism 5.0 software.
RNA Isolation for Illumina Sequencing Analysis
For RNA-sequencing analysis, three animals were sacrificed per collection point (Non-HIB, Pre-HIB, HIB-PA, and HIB-DT, 12 animals) (Figure 3A) and processed for tissue collection. Total RNA was purified from whole iWAT as described above. Total RNA samples from individual animals were quantified on a Nanodrop (Thermo Scientific) and Qubit RNA BR assay kit (Invitrogen). Genomic DNA remaining in purified RNA samples was measured using a Qubit dsDNA HS assay kit (Invitrogen), and confirmed to be lower than 10% of total RNA. Quality of total RNA was evaluated using capillary electrophoresis (Agilent Bioanalyzer 2100). All the 12 samples were verified as high quality (RNA Integrity Number > 7.9) and thus were converted to Illumina sequencing libraries.
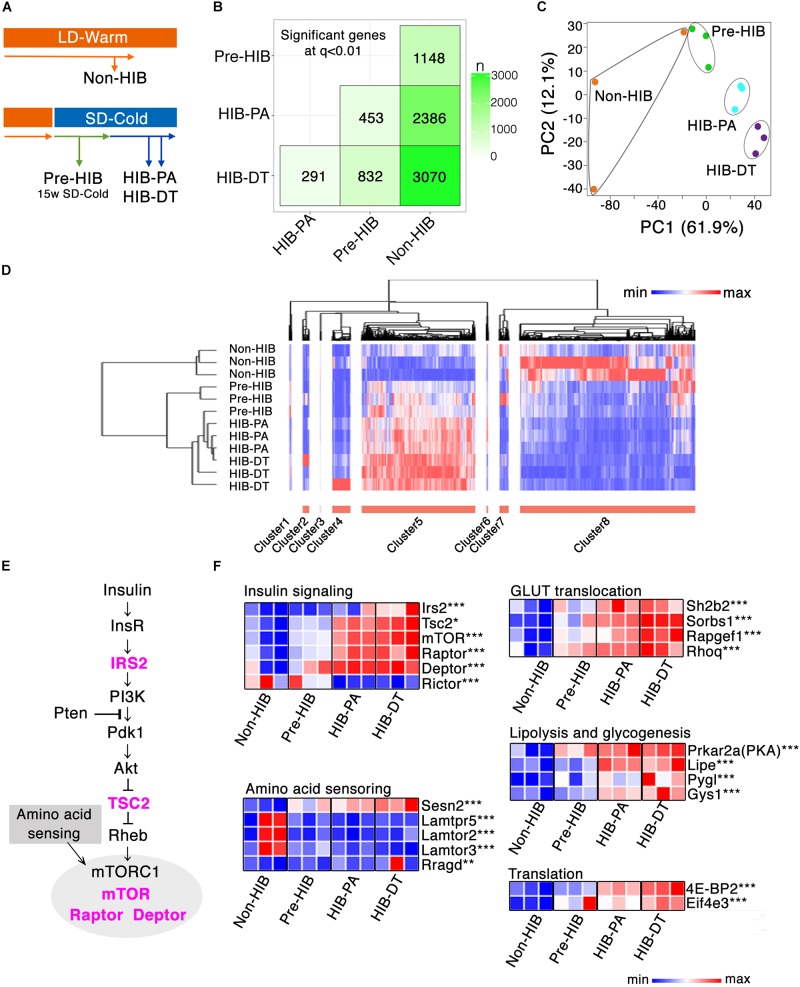
FIGURE 3.
Global gene expression profiling of iWAT. (A) Schematic overview of the four experimental groups that were used for RNA-seq analysis of iWAT (n = 3 per group). (B) SigMatrix plot indicating the number of DEGs among each combination of groups at 0.01 q-value threshold. (C) Principle component analysis on 3804 genes. PC1, principle component 1; PC2, principle component 2. (D) Hierarchal clustering analysis on the screened 2831 DEGs, which exhibited more than twofold change between any groups with q-value ≤ 0.01. Relative mRNA expression level among four groups are denoted in red (high expression) to blue (low expression) gradient. (E,F) mRNA expression changes of genes involved in insulin signaling. (E) Insulin-Akt-mTORC1 signaling cascade. The genes colored in magenta denote the significant up-regulation either in HIB-PA or HIB-DT compared to the Non-HIB group. (F) Heat map representation of relative mRNA expression change in the Insulin-Akt-mTORC1 signaling pathway and its downstream signaling pathways. ∗q < 0.05, ∗∗q < 0.01, ∗∗∗q < 0.005.
Library Creation
Conversion of RNA samples to sequencing libraries was performed using TruSeq Sample Preparation Kit v2 (Illumina; RS-122-2001) according to manufacturer’s protocol, with minor modifications: all reactions were carried out at half scale, fragmentation of mRNA was performed for 4 min, and PCR reduced to 8 cycles to minimize PCR biases. One microgram of total RNA was used for each library. The size distribution of the libraries was validated using capillary electrophoresis (Agilent Bioanalyzer 2100) and quantified using KAPA library quantification kits (KAPA Biosystems) using a Light Cycler 480 real-time PCR system (Roche).
Sequencing
Twelve multiplexed libraries were sequenced in one lane using HiSeq1500 (Illumina) with 101-bp paired-end readings. Raw data processing, base calling, and quality control were performed according to manufacturer’s standard protocol using RTA, OLB, and CASAVA software (Illumina). All raw Illumina sequences are available for download at the DDBJ Sequence Read Archive (DRA) under the accession number PRJDB6278.
Bioinformatic Analysis
Sequencing using Illumina HiSeq1500 platform yielded 141.3 million 101-bp paired-end sequence reads (Supplementary Table S1). Quality of the raw sequence data was validated by FastQC. Sequence reads were mapped to a Syrian hamster genomic scaffold and annotation2 using Tophat v2.0.10 (Supplementary Table S2). To quantify gene expression and determine genes that are differentially expressed across the four groups, the q-values (adjusted P-value) of test statistics for each gene were computed using Cufflinks v2.2.1. The Cufflinks outputs were further analyzed and visualized using cummeRbund and Morpheus3 (Broad Institute). Hierarchical clustering was performed by one minus pearson correlation.
Adipose Tissue Explants Culture
iWATs were dissected out from hamsters of Non-HIB group (euthermic and active at the LD-Warm condition, 28–31 weeks old, n = 5) and HIB-DT group (heterothermic and inactive at the SD-Cold condition, 28–31 weeks old, n = 5). Adipose explant cultures were established by modification of a reported procedure (Toyoda et al., 2008). The tissues were minced into small fragments in cold PBS, then were transferred to a tube containing 30 mL of Dulbecco’s Modified Eagle’s Medium (DMEM; D5921, Sigma-Aldrich) with 8 mM glutamine (Sigma-Aldrich), 1% (v/v) penicillin/streptomycin (pe/st) solution (WAKO), and 1% (w/v) bovine serum albumin (BSA) (A7030; Sigma-Aldrich), and were gently pre-cultured with rotation for 6 h in a CO2 incubator (37°C, 5% CO2). The tissue fragments were placed into 24-well plates containing 800 μL of DMEM (8 mM glutamine, 1% pe/st, 1% BSA), and treated with or without 0.1% dimethyl sulfoxide (DMSO) (WAKO), 100 μM fenofibrate (Wako), 100 μM L-165041 (Cayman), and 100 μM rosiglitazone (Tokyo Kasei), for 48 h in the CO2 incubator. The culture medium was daily replaced with fresh agonist-containing medium. Four technical replicates were used for each treatment condition. After the treatment with PPAR agonists, tissue fragments were collected in RNA stabilizing solution, and used for RNA analysis as described above.
Mean of fold change of the four technical replicates was calculated using Microsoft Excel. Standardization among five biological replicates was performed as previously described (Willems et al., 2008). Boxes enclose the interquartile ranges and whiskers show the minimum and maximum values of log2-fold change of each experimental group against non-treated group. P-value was derived from one-way ANOVA and Tukey’s multiple comparison tests using Graph Pad Prism 5.0 software (GraphPad).
Results
Utilization and Preservation of White Adipose Tissues During the Pre-hibernation and Hibernation Periods
To examine how WAT is remodeled during the pre-HIB in Syrian hamsters, we focused on iWAT (Supplementary Figure S1B). Previously, we demonstrated that prolonged SD-Cold condition reduces the body mass of female Syrian hamsters raised in LD-Warm condition below the threshold for initiating hibernation under ad lib feeding (Chayama et al., 2016). We found that iWAT mass strongly correlated with the body mass of hamsters under LD-Warm conditions (Figure 1B, Non-HIB group). Under SD-Cold conditions, iWAT-to-body mass ratio tended to decrease, and minimized 6 weeks after the transition (Figure 1C). Interestingly, however, this ratio did not continue to fall during the SD-Cold period; rather, an increase was observed in animals exposed to SD-Cold for 8–15 weeks and which had entered hibernation (Figure 1C). This trend was also observed in intraperitoneal WATs (Supplementary Figure S1C). These data indicate that iWAT mass was lost extensively at first under SD-Cold conditions, but was then retained relative to overall body mass loss.
We then examined the histological changes of iWAT during hibernation. The proportion of small adipocytes (area: 500–1500 μm2) was significantly greater in hibernating animals (HIB-PA and HIB-DT groups) than in the Non-HIB group (Figures 1D,E). This increased number of small white adipocytes was already evident in the Pre-HIB group (Figures 1D,E). Because adipocyte size can reflect changes in the mobilization or accumulation of stored lipids, these data suggest that Syrian hamsters alter lipid metabolism in iWAT under prolonged SD-Cold conditions prior to hibernation.
UCP1-Positive, Beige-Like Multilocular Adipocytes Emerge in iWAT During the Pre-hibernation Period
Prolonged cold exposure induces beige adipocyte formation in WAT of non-hibernators including mice, rats, and humans. However, few studies have addressed whether hibernators recruit beige adipocytes to their WAT. Our immunohistochemical analysis on WAT for UCP1 identified a small proportion of multilocular beige-like cells positive for UCP1 in iWAT of pre-hibernation and hibernating hamsters under prolonged SD-Cold conditions (Figure 2A). Time-course analysis using qPCR revealed that expression of Ucp1 mRNA increased drastically 2 weeks after initiation of SD-Cold conditions (Figures 2B,C). It was then gradually reduced by the onset of hibernation (Figure 2C), although it still remained higher than levels found under LD-Warm conditions (Non-HIB).
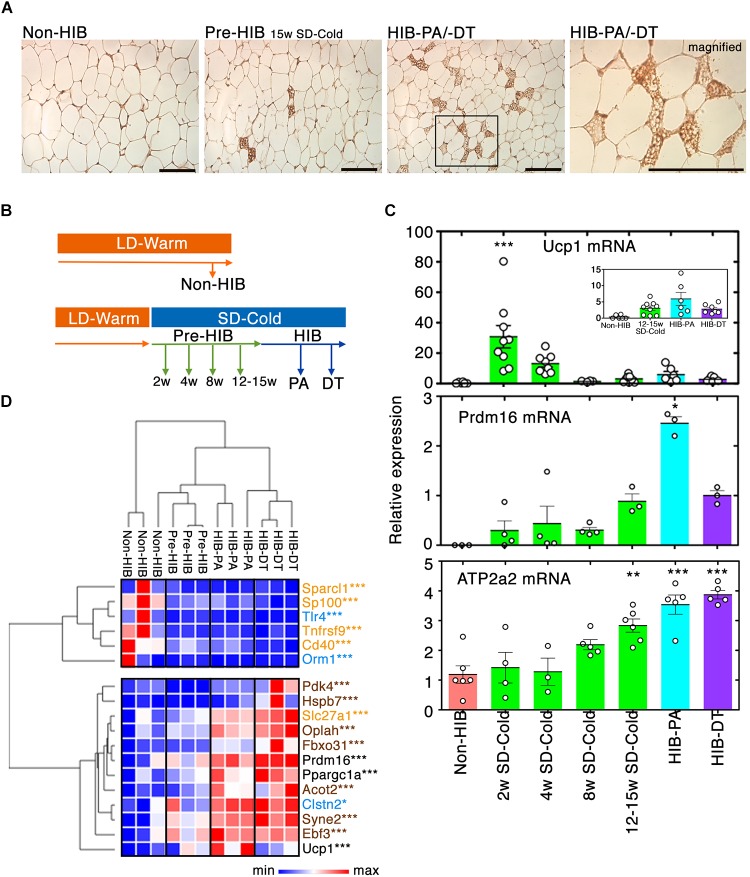
FIGURE 2.
Recruitment of beige-like cells to iWAT by prolonged SD-Cold exposure. (A) Representative images of UCP1 staining of iWAT from each condition. The numbers of iWAT with beige-like cells in Non-HIB, Pre-HIB, HIB-PA, and HIB-DT were 0/6, 5/6, 2/3, and 2/3, respectively. Scale bars, 100 μm. (B,C) Time-course analysis of Ucp1 and Prdm16 mRNA expression in iWAT by qPCR. (B) Schematic overview of time-course analysis. (C) Relative expression of Ucp1, Prdm16, and Atp2a2 mRNA expression. The numbers of samples analyzed for Ucp1 mRNA were Non-HIB: n = 6, 2/4/8/12–15 weeks SD-Cold: n = 9, 8, 4, and 8, respectively, HIB-PA: n = 6, and HIB-DT: n = 7. Those for Prdm16 mRNA were Non-HIB: n = 3, 2/4/8/12–15 weeks SD-Cold: n = 4, 4, 4, and 3, respectively, HIB-PA: n = 3, and HIB-DT: n = 3. Those for Atp2a2 mRNA were Non-HIB: n = 6, 2/4/8/12–15 weeks SD-Cold: n = 4, 3, 5, and 6, respectively, HIB-PA: n = 5, and HIB-DT: n = 5. The results are expressed as mean ± SEM. White circles show each animal. P-value was derived using one-way ANOVA and Tukey’s multiple comparison tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005 against the “Non-HIB group.” (D) Heat map representation of relative expression changes of genes in the beige/brown adipocyte signature as differentially expressed with q-value ≤ 0.01 and more than twofold change between any groups. Blue, orange, and brown characters indicate marker genes for WAs, beige adipocytes, and brown adipocytes, respectively. Pre-HIB here indicates pre-hibernation group following 15 weeks of exposure to SD-Cold conditions. ∗q < 0.05, ∗∗q < 0.01, ∗∗∗q < 0.005.
Besides, the mRNA expression of PR-domain containing 16 (Prdm16), a master regulator of beige adipocyte differentiation in WAT (Cohen et al., 2014), was induced 15 weeks after SD-Cold exposure, and significantly up-regulated in hibernating animals (Figure 2C). Moreover, Atp2a2 gene (encoding SERCA2) was significantly up-regulated in iWAT after SD-Cold exposure (Figure 2C). Recently, SERCA2 was shown to be up-regulated in iWAT from transgenic mice overexpressing Prdm16 in an adipose tissue-selective manner (Ikeda et al., 2017). Importantly, it was further shown to be crucial for a non-canonical UCP1-independent thermogenic mechanism through Ca2+ cycling in beige adipocyte of mice, pigs, and humans (Ikeda et al., 2017). Thus, the beige-like cells might generate heat during hibernation by activating the enhanced SERCA2 pathway rather than UCP1 pathway in iWAT.
Further evidence that beige-like cells emerged in iWAT after SD-Cold exposure, was provided by a comprehensive gene expression analysis following RNA-seq (see below); a ‘beige/brown adipocyte’ signature (Wu et al., 2012; Harms and Seale, 2013; Long et al., 2014; de Jong et al., 2015; Shinoda et al., 2015) was indeed induced in hibernating animals (Figure 2D). Altogether, these data indicate that beige-like cells are recruited to the iWAT of Syrian hamsters after prolonged exposure to SD-Cold, and during the HIB.
Global Gene Expression Profiling of White Adipose Tissue Discriminates Hibernation State From Non-hibernation State
To further identify differences in iWAT between the non-hibernation and hibernation states, we conducted RNA-seq analysis (Figure 3A). More than 8 million high-quality reads of transcripts were obtained in all the groups, and ≥70% of them were successfully mapped on hamster’s genome and quantified (Supplementary Tables S1, S2). Pairwise comparison of transcripts profile showed the positive correlations of most gene expressions between each of the groups and subsets of the genes differentially expressed (Supplementary Figures S2, S3). Total 3804 genes were identified as differentially expressed genes (DEGs) between each of the groups, with a q-value of 0.01. The number of DEGs between two groups is shown in Figure 3B and Supplementary Figure S3.
To visualize the variation and patterns of gene expression, principal component analysis (PCA) and hierarchal clustering analysis of the DEGs were performed. Principal component 1 (PC1) clearly described the transition from non-hibernation to hibernation states (Figure 3C). Among the genes that contributed the most to PC1, those involved in insulin signaling (e.g., Rapgef1, Calm2, Exoc7, mTOR, Prkar2a, Rhoq, Rps6; Gene name for each gene symbol described hereafter is provided in Supplementary Table S7) were identified as significantly enriched upon Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Supplementary Table S3). Hierarchical clustering analysis of 2831 DEGs, which exhibited more than twofold changes of expression between at least any two groups, also identified distinct transcript profiles between groups (Figure 3D). The Pre-HIB group was closer to the cluster of hibernation groups (HIB-PA and HIB-DT groups) than was the Non-HIB group (Figure 3D). These data demonstrate that the iWAT gene expression profile in Syrian hamsters is altered during pre-hibernation remodeling. To gain further insight into pre-hibernation remodeling, we focused on clusters 5 and 8, which discriminate the non-hibernation state from the others under prolonged SD-Cold conditions (Pre-HIB, HIB-PA, and HIB-DT; Figure 3D).
Cluster 8, which was down-regulated in hibernation groups, consisted of 1528 genes including a number of ribosomal protein genes and genes related to immune cell signaling (e.g., primary immunodeficiency, chemokine signaling, cytokine–cytokine receptor interaction; Supplementary Table S4). The down-regulation of immune-cell signature genes might be indicative of changes in the abundance of resident cells (e.g., pre-adipocytes, macrophages, T-cells, and B-cells) in iWAT during the transition period (Osborn and Olefsky, 2012; Choe et al., 2016).
The 990 genes of cluster 5, which was up-regulated in hibernation groups, contained many genes involved in cell–cell interaction (e.g., focal adhesion, ECM-receptor interaction, adherens junctions, and vascular smooth muscle contraction), which may reflect progression of angiogenesis in iWAT during the pre-HIB (Supplementary Table S5). Other enriched pathways in cluster 8 include insulin signaling, peroxisome proliferator-activated receptor (PPAR) signaling, and adipocytokine signaling pathways. These pathways are known to regulate the transcription of genes related to energy metabolism, adipogenesis, mitochondrial biogenesis, and ribosome biogenesis (Cai et al., 2016). Together, these results show that the expression profiles of genes involved in several pathways are concomitantly regulated with the progression of pre-hibernation remodeling, and these profiles can be used as a predictive signature to discriminate between non-hibernation and hibernation states.
Among the genes involved in the insulin signaling that were enriched in cluster 8, mechanistic target of rapamycin complex 1 (mTORC1) (mTOR, Rptor, and Dptor), which responds to nutrient signals through amino acid sensing pathway and insulin-PI3K-Akt signaling pathway, were significantly up-regulated in hibernation groups (HIB-PA and HIB-DT) than in the non-hibernation group (non-HIB) (Figures 3E,F). Among the key molecular players in the amino acid sensing pathway (Ricoult and Manning, 2013; Lee et al., 2017), Sestrin2 (Sesn2) was significantly up-regulated in the hibernation groups (Figure 3F). Furthermore, the downstream targets of PI3K-Akt and mTORC1 signaling networks were also up-regulated by prolonged exposure to SD-Cold conditions. Genes involved in GLUT translocation (Sh2b2, Sorbs1, Rapgef1, and Rhoq), lipolysis and glycogenesis (Prkar2a, Lipe, Pygl, and Gys1), and translation (Eif4e3 and 4E-BP2) were mostly up-regulated in hibernation groups (Figure 3F). In sum, these changes in gene expression appear to enhance the functional capacity of the mTORC1 pathway and its downstream signaling networks.
Lipid Catabolism and Anabolism Are Both Increased in iWAT Prior to Hibernation
Activation of mTORC1 mutually affects the expression and activity of PPAR molecules in WAT (Laplante and Sabatini, 2009). PPARs are important nutrient sensors that regulate transcriptions of genes involved in lipid metabolism (Nelson et al., 2009; Ahmadian et al., 2013; Kersten, 2014). Genes involved in transcriptional co-regulation of PPARs (Ppara, Ppargc1a/b, and Rxra/g) and adipocytokine signaling (Adipoq, Adipor2, and Camkk2) were significantly higher in hibernation groups than in the non-hibernation group (Figure 4A).
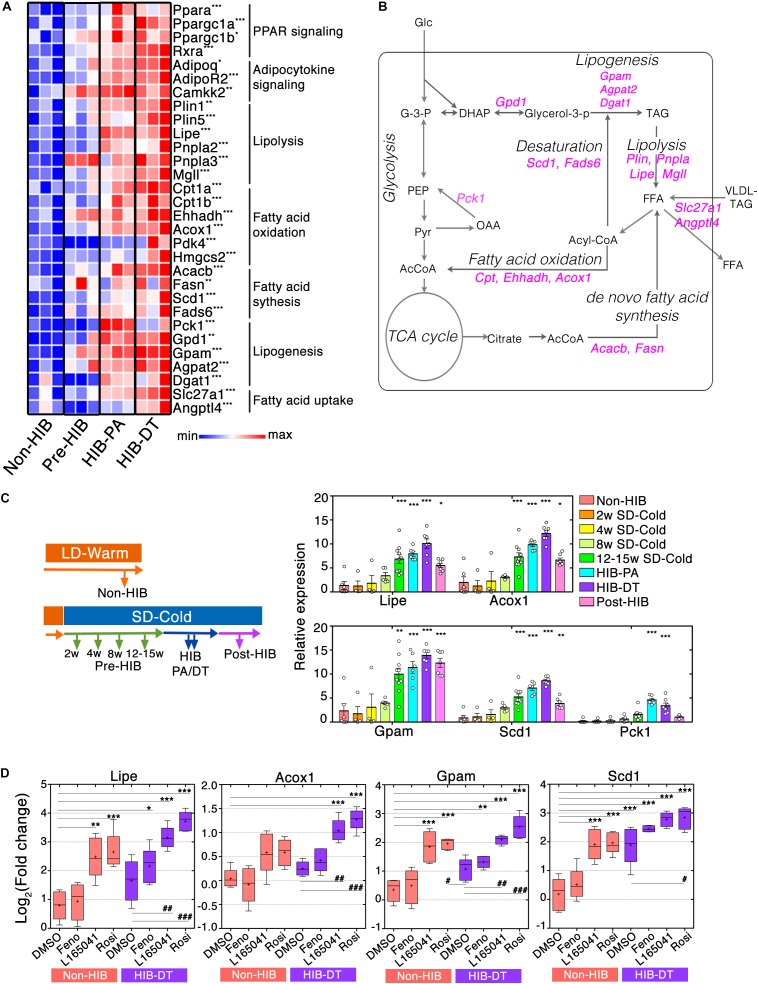
FIGURE 4.
PPAR signaling augments mRNA expression of genes required for lipid catabolism and anabolism in iWAT during hibernation. (A) Heat maps of fold changes in mRNA expression of genes related to PPAR signaling pathways. ∗q < 0.05, ∗∗q < 0.01, ∗∗∗q < 0.005. (B) A schematic representation of the cellular biochemical networks of lipid and glucose metabolism in WAT. Genes that encode enzymes for chemical reaction and that are up-regulated during HIB are shown in magenta. AcCoA (acetyl-CoA), DHAP (dihydroxyacetone phosphate), FFA (free fatty acid), G-3-P (glyceraldehyde 3-phosphate), Glc (glucose), Glycerol-3-P (glycerol 3-phosphate), OAA (oxaloacetate), PEP (phosphoenolpyruvate), Pyr (pyruvate), TAG (triacylglyceride), VLDL-TAG (very-low-density lipoprotein triacylglyceride). (C) Time-course analysis of the lipid metabolism genes that were up-regulated during HIB. qPCR analysis of relative mRNA expression levels of lipid metabolism genes at the different time points. The results are expressed as mean ± SEM. White circles show each animal. P-value was derived from one-way ANOVA and Tukey’s multiple comparison tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005 against the “Non-HIB group.” (D) Ex vivo experiments of adipose explant cultures. iWAT explants from Syrian hamsters were treated with 0.1% dimethyl sulfoxide (DMSO), 100 μM fenofibrate (Feno), 100 μM L165041 (L165041), and 100 μM rosiglitazone (Rosi) for 48 h. Relative mRNA expression level of the lipid metabolism genes in the different conditions was determined using qPCR. Horizontal lines and crosses indicate medians and means of relative mRNA expression, respectively. Boxes enclose the interquartile ranges and whiskers show the minimum and maximum relative mRNA expression of each experimental condition. P-value was derived using one-way ANOVA and Tukey’s multiple comparison tests. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005 against the DMSO-treated condition of the Non-HIB group. #p < 0.05, ##p < 0.01, ###p < 0.005 against the DMSO-treated condition of HIB-DT group.
To further address how lipid metabolism pathways are altered during pre-hibernation remodeling, we analyzed the mRNA expression of catabolic genes involved in lipolysis (Plin1/5, Lipe, Pnpla2/3, and Mgll) and fatty acid (FA) oxidation (FAO) (Cpt1a/b, Acadvl, Ehhadh, and Acox1) by RNA-seq data and qPCR. This analysis showed that they were significantly elevated in hibernation groups (Figures 4A,B and Supplementary Figure S4). Interestingly, anabolic genes involved in lipogenesis (Gpd1, Gpam, Agpat2, and Dgat1), FA synthesis (Acacb and Fasn), and desaturation (Scd1 and Fads6) were also highly expressed in those groups (Figure 3A and Supplementary Figure S4). In addition, the mRNA expression of genes involved in FA uptake (Slc27a1 and Angptl4) was also higher in hibernating animals (Figure 4A). These results suggest that the capacities of both lipid catabolism and anabolism are simultaneously enhanced during hibernation, which contributes to an efficient regulation of the utilization and storage of lipid fuels in iWAT (Figure 4B).
To further identify the time at which remodeling of lipid metabolic pathways initiates during pre-hibernation remodeling, we performed a time course qPCR analysis of lipid metabolism genes (Figure 4C). Four genes that are crucial for each lipid metabolic process (Lipe for lipolysis, Acox1 for FAO, Gpam for lipogenesis, and Scd1 for FA desaturation) were selected as readout of those pathways. This analysis revealed that no changes in the mRNA expression of all four genes were observed after 8 weeks’ exposure to SD-CA conditions (Pre-HIB of 2, 4, and 8 weeks after initiation of SD-Cold conditions), but they were significantly up-regulated after 12–15-week exposure to SD-CA conditions (Pre-HIB 12–15 weeks SD-Cold). The mRNA expression of these genes peaked during hibernation (HIB-PA and HIB-DT) (Figure 4C). Thus, up-regulation of lipid metabolism genes preceded the onset of hibernation. Interestingly, after an average of 4–6 months of hibernation, the mRNA expression of Lipe, Acox1, and Scd1 fell to the same level as that observed in the Pre-HIB group by 12–15 weeks after initiation of SD-Cold conditions, whereas Gpam mRNA expression remained at higher level (Figure 4C, Post-HIB). At this timepoint, all the animals examined spontaneously terminated hibernation and started to gain body mass, even though they were still maintained under SD-Cold conditions (Figure 1A, “Post-hibernation”). These results suggest that the elevated mRNA expression of genes involved in lipolysis, FAO, and FA desaturation is a phenomenon closely associated with the winter-like body state that allows hibernation, and is not just a consequence of simple cold acclimation.
The gene encoding phosphoenolpyruvate carboxykinase 1 (Pck1), a key enzyme that promotes glyceroneogenesis in WAT (Beale et al., 2004), was present in cluster 5 (Figure 3D), and exhibited a “hibernation-specific” expression pattern, in that it significantly increased only in hibernation groups, but not in pre-hibernation groups (Figure 4C). After the HIB, the Pck1 mRNA returned to the same level as that observed in the non-hibernation and pre-hibernation groups. Such hibernation-specific induction of Pck1 may enhance the rate of glyceroneogenesis and FA recycling in order to maintain triglyceride homeostasis during hibernation (Figure 4C).
Involvement of PPAR Signaling in Up-Regulation of Both Lipid Catabolism and Anabolisms Genes
We then explored potential mechanisms that elicit an enhanced mRNA expression of lipid metabolism genes. Isotypes of the PPAR family (PPAR-α, -β/δ, and -γ) have different expression patterns, ligand specificities, and co-regulator interactions (Gross et al., 2017). Because PPAR signaling molecules were up-regulated during the pre-HIB, we addressed whether they might be responsible for induction of the lipid metabolism genes that were highly expressed in iWAT during hibernation. PPAR agonists were applied to cultured iWAT explants obtained from non-hibernating hamsters (Non-HIB at LD-Warm) and hibernating hamsters (HIB-DT at SD-Cold). The explants from Non-HIB animals exhibited clear responses to PPAR-β/δ or -γ agonists (L-165041 or rosiglitazone, respectively), resulting in significant up-regulation of Lipe, Gpam, and Scd1 mRNA and, to a lesser extent, Acox1, when compared to control (DMSO) (Figure 4D). In contrast, the PPAR-α agonist, fenofibrate did not affect the expression of any of these genes (Figure 4D). Likewise, the iWAT explants from hibernating animals (HIB-DT) exhibited significant increases of Lipe, Acox1, and Gpam mRNAs in response to PPAR-β/δ or -γ agonists, but the PPAR-α agonist. The basal mRNA expression of these genes was similar in DMSO-treated HID-DT and Non-HIB explants, suggesting that ex vivo manipulation of iWAT may mask elevated PPAR-dependent signaling in tissues from hibernating animals as shown in Figure 4. However, the basal expression of Scd1 mRNA was significantly higher in HIB-DT explants when compared to their Non-HIB counterparts. We infer that the mechanisms that underlie up-regulation of Scd1 mRNA during hibernation are resistant to experimental procedures, and/or that Scd1 mRNA expression may be regulated differently than the mRNA expression of Lipe, Acox1, and Gpam. Consistent with this, Scd1 mRNA in HIB-DT was further increased only by the PPAR-γ agonist rosiglitazone, but not by other agonists. These data demonstrate that the dual up-regulation of genes involved in both lipid catabolism and anabolism we observed is dependent on several PPAR isotypes in Syrian hamsters.
Discussion
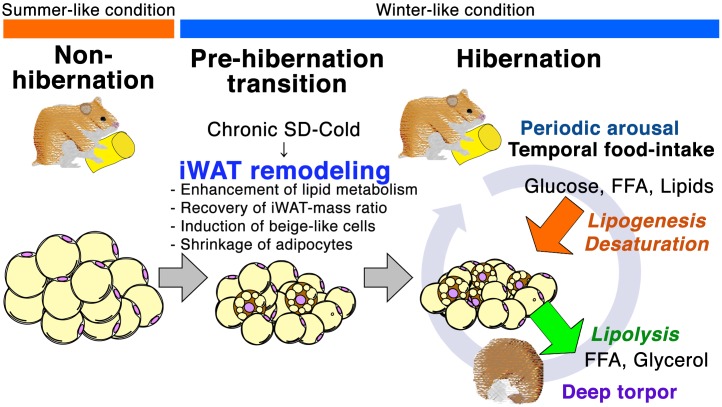
In the present study, we identified that transferal of female Syrian hamsters to winter-like conditions triggers pre-hibernation remodeling of iWAT at the cellular and molecular level (Figure 5). This initially manifests as a reduction in iWAT mass, although the iWAT-to-body mass ratio stabilizes and then increases over the following 2 months. This biphasic response of iWAT is likely explained by our finding that mRNA expression of lipogenic genes is induced with similar timing. The up-regulation of Gpam, a rate-limiting enzyme of triglyceride synthesis, may prevent over-shrinkage of white adipocytes and limit lipodystrophy by promoting lipogenesis before the onset of hibernation and during HIB. The simultaneous induction of lipid catabolism and anabolism genes in iWAT may be a phenotype that is unique to hamster hibernation (Baumber and Denyes, 1963); Syrian hamsters do not fatten during the pre-HIB under SD-Cold, and reduce their body mass below approximately 140 g (Chayama et al., 2016). They hoard food before the onset of hibernation (Larkin et al., 2002), and during PA states they do ingest small amounts of food. This food intake enables them to replenish glucose and FAs, which are then used to synthesize triglycerides. Thus, Syrian hamsters would require enhancement of lipogenesis to quickly synthesize and maintain sufficient lipids in WAT as a fuel during HIB. This is in striking contrast to the fat-storing species (ground squirrels, bears, marmots, dormice, etc.), which fatten by hyperphagia from late summer to fall and become anorexic in winter to survive without hoarding and eating food (Boswell et al., 1994; Dark, 2005; Melvin and Andrews, 2009).
FIGURE 5.
Schematic representation of iWAT remodeling for hibernation in Syrian hamster. Prolonged SD-Cold stimuli induce iWAT remodeling prior to initiating hibernation in Syrian hamsters. The morphology and cell population of adipocytes in iWAT changes during the transition from the non-hibernation to the hibernation state. Small adipocyte population increases, and beige-like cells emerge during Pre-HIB and HIB. Prolonged SD-Cold stimuli also alter the mRNA expression of genes in the insulin-PPAR signaling pathway. Up-regulation of lipid metabolism genes before the onset of hibernation could enhance the capacity for both lipid catabolism and anabolism in iWAT to prepare for metabolic challenges during the HIB. Thus, seasonal cues mediated by the ambient temperature and photoperiod alter the mTORC1-PPAR signaling network and induce iWAT remodeling prior to hibernation.
Several possible mechanisms of simultaneous enhancement of lipid catabolism and anabolism pathway in iWAT can be considered. From Pre-HIB to HIB, a part of cells may activate lipid catabolism to supply fatty acids and generate heat, and the others may be anabolic to replenish lipid storage in iWAT. Previous reports showed that sympathetic nervous system and humor factors can temporarily activate the lipolysis and FAO in WAT (Braun et al., 2018; Caron et al., 2018). Such regulatory mechanism might switch lipid anabolic state to catabolic state in a spatiotemporally manner within iWAT, synchronizing to the torpor-arousal cycles during hibernation. Alternatively, lipid catabolism and anabolism pathways might be enhanced in the same cells. One possible explanation for simultaneous enhancement of lipogenesis and lipolysis comes from a recent study in a mouse model demonstrating that adipocytes are protected from lipid-induced endoplasmic reticulum (ER) stress by a re-esterification of fatty acids to triglyceride, which is mediated by diacylglycerol O-acyltransferase 1 (DGAT1) during lipolysis (Chitraju et al., 2017). In fact, we found that expression of Dgat1 mRNA was significantly up-regulated during HIB (Figure 4A). Enhancement of re-esterification of fatty acids might also contribute to lowering oxidative stress levels, which is speculated to increase during hibernation, notably during PA because of the preferentially and highly retained polyunsaturated fatty acids (PUFA) during HIB (Arnold et al., 2011, 2015). Thus, hibernation accompanying active lipolysis may require protective mechanisms from lipotoxicity.
Several lines of evidence indicate that seasonal alteration in composition of lipids and FAs in the cellular membrane and stored fat is an important feature of mammalian hibernation. Increased dietary intake of unsaturated FAs can affect the length of torpor, metabolic rates, and minimum Tb tolerated by hibernators (reviewed in Arnold et al., 2015). It is proposed that high proportions of linoleic acid (C18:2 n-6) in phospholipids of sarcoplasmic reticulum (SR) in the heart improve cardiac function at low body temperature by enhancing the SR Ca2+ ATPase 2a (SERCA2a) activity during hibernation in Syrian hamsters and garden dormice (Arnold et al., 2011; Giroud et al., 2013, 2018). These studies also demonstrated diet-independent remodeling of tissue lipid composition, which can be in line with our finding that Syrian hamsters enhance de novo synthesis of unsaturated FAs and lipids in WAT by up-regulation of FA desaturases, Scd1 and Fads6 during Pre-HIB and HIB (Figure 3A). Such enhancement of de novo synthesis of unsaturated FAs from saturated FAs in iWAT might contribute to systemic alteration of FA composition in cellular membrane, and increased tolerance of cells and tissues under low body temperature during hibernation (Arnold et al., 2011). It could also help thermogenesis in BAT during hibernation, as BAT preferentially utilizes specific unsaturated FAs stored in WAT as an energy source for thermogenesis during rewarming from torpor (Carneheim et al., 1989; Jefimow and Wojciechowski, 2014). Furthermore, specific unsaturated FAs and lipids could work as lipokines, endogenous ligands for PPARγ in WAT (Mottillo et al., 2014). Further analysis will be needed for better understandings of the significance of enhanced de novo synthesis of unsaturated FAs in iWAT during Pre-HIB and HIB in Syrian hamsters.
The mTORC1-PPAR signaling network seems to be an important regulatory mechanism to adapt energy metabolism for hibernation (Buck et al., 2002; Eddy and Storey, 2003; El Kebbaj et al., 2009). Our data indicated that PPAR-γ agonist treatment simultaneously induced mRNAs of lipid catabolism and anabolism genes in iWAT explants of Syrian hamsters. The up-regulation of mTORC1 complexes during the period from pre-hibernation to hibernation that we observed may increase PPAR-γ levels, and consequently levels of its downstream target genes, and prevent excessive loss of fat mass (Polak et al., 2008; Laplante and Sabatini, 2009; Blanchard et al., 2012; Cai et al., 2016). However, it was reported that plasma insulin levels decrease under short photoperiodic conditions and during hibernation in Syrian hamsters (Weitten et al., 2013; Chakir et al., 2015). These lines of evidence imply that mTORC1 may be activated independently of insulin-PI3K-Akt signaling during Pre-HIB and HIB in iWAT of Syrian hamsters. Although future studies are required in order to elucidate the mechanisms responsible for the enhancement of mTORC1 activity, there are several possible candidates reported in the literature. Amino acids such as leucine can activate mTORC1 in an insulin-independent manner (Roh et al., 2003); leucine directly binds sestrin2 and suppresses its inhibitory effect on mTORC1 (Wolfson et al., 2016). Alternatively, serotonin may activate mTORC1 and PPARs in peripheral tissues of Syrian hamster during hibernation (Talaei et al., 2011; Talaei, 2014), because serotonin is reported to be an endogenous ligand for PPAR-γ (Waku et al., 2010) and the activation of serotonin receptor promotes lipogenesis in WAT of mice (Oh et al., 2015). Moreover, previous studies suggest that melatonin transduces photoperiodic stimulation to a neuroendocrine signal and could therefore control sexual activity, body mass, and hibernation in Syrian hamsters (Bartness and Wade, 1984; Vanecek et al., 1984; Pitrosky et al., 2003), which might be an upstream machinery of iWAT remodeling during the pre-HIB.
Another important finding in the present study is the emergence of beige-like cells in the iWAT of Syrian hamsters after prolonged exposure to SD-Cold stimuli (Figure 2). The beige-like cells in WAT may contribute to local thermogenesis (Okamatsu-Ogura et al., 2013; Bartesaghi et al., 2015). The enhancement of mTORC1 and PPAR signaling pathways may also recruit and activate beige-like cells in iWAT (Harms and Seale, 2013; Liu et al., 2016). Because lipolysis and FAO are required for cold-induced thermogenesis of brown and beige adipocytes in mice and humans (Lee et al., 2015; Blondin et al., 2017), the up-regulation of lipid catabolic genes in iWAT may reflect the emergence and activation of beige-like cells in the tissue during hibernation.
A recent study reported that beige cells can generate heat through UCP1-independent mechanism by activating SERCA2 pathway (Ikeda et al., 2017); SERCA2 pathway highly depends on glucose oxidation and activates Ca2+ cycling in endoplasmic reticulum for thermogenesis, although the UCP1 pathway mainly utilizes FAs and dissipate chemical energy in mitochondria as heat. Our data demonstrated that the Atp2a2 mRNA (SERCA2) significantly increased after prolonged SD-Cold and during hibernation, while the Ucp1 mRNA gradually decreased toward the onset of hibernation after prolonged SD-Cold in iWAT (Figure 2). The enhancement of SERCA2 pathway could be an effective strategy for simultaneously activating thermogenesis and FA synthesis in iWAT during PA; the FAs and triglycerides synthesized during PA may be saved in iWAT for the next torpor, by rewiring energy metabolism from FAO toward glucose oxidation for thermogenesis through SERCA2 pathway in beige-like cells during PA.
In Syrian hamsters, only a small proportion of cells were recruited to beige-like cells within iWAT, as is evident from our histological analysis compared to that of mice under similar environmental conditions (de Jong et al., 2015). One possible explanation is that Syrian hamsters preferentially retain white adipocytes to store a fuel for hibernation, rather than converting them into beige-like cells which may consume energy for heat-production. Indeed, hamsters establish large amount of BAT for hibernation (Kitao and Hashimoto, 2012), and may not require extra thermogenic cells in iWAT.
Our data also demonstrates Syrian hamsters provide a suitable model for monitoring the molecular physiology associated with pre-hibernation remodeling during the transition from a summer-like, non-hibernation state to a winter-like, hibernation state. Tracing the timing of the onset of body remodeling in these animals can be achieved simply by controlling the environmental condition (photoperiod and ambient temperature) (Jansky et al., 1984; Chayama et al., 2016), independent of circannual rhythms that define the timing of body remodeling and hibernation in some strictly seasonal hibernators (Kondo, 1987; Grabek et al., 2011; Olson et al., 2013; Hindle and Martin, 2014). We anticipate that this model will help elucidate the molecular pathways involved in pre-hibernation remodeling of lipid metabolism and whole body homeostasis. In addition to providing new insight into the amazing powers of hibernators, such information will also further our understanding of the metabolism and physiology of non-hibernators, including humans.
Author Contributions
YC and YY conceived the experiments. YC, LA, DA, YS, TF, HT, and YT performed the experiments. YC, LA, YS, and YY analyzed data and wrote the manuscript. SS supervised RNA-seq analysis. MM and YY supervised the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to T. Takahashi, S. Suzuki, and H. Makino for experimental assistance. We thank the Functional Genomics Facility, and NIBB Core Research Facilities for the technical support. We also thank Y. Gotoh, and all the members of the Miura Laboratory and JST PRESTO “Elucidation and regulation in the dynamic maintenance and transfiguration of homeostasis in living body” for the helpful discussions and comments.
Abbreviations
- BAT
brown adipose tissue
- HIB
hibernation period
- HIB-DT
deep torpor during HIB
- HIB-PA
periodic arousal during HIB
- iWAT
subcutaneous inguinal WAT
- LD
Long day
- Non-HIB
non-hibernation period
- PA
periodic arousal
- Post-HIB
post-hibernation period
- Pre-HIB
pre-hibernation period
- SD
short day
- WAT
white adipose tissue
Funding. This work was supported by PRESTO from the Japan Science and Technology Agency (JPMJPR12M9 to YY), the Japanese Society for the Promotion of Science, and the Ministry of Education, Culture, Sports, Science, and Technology in Japan (JP26110005, JP18K19321, JP16K15114, and JP16H05127 to YY, and JP16H06385 to MM), and AMED-CREST from Japan Agency for Medical Research and Development, AMED (JP17gm0610004 and JP18gm5010001 to MM). This work was also supported by grants (to YY) from the Cell Science Foundation, Sekisui Chemical Innovations Inspired by Nature Research Support Program, the Takeda Science Foundation, the Kanae Foundation for the promotion of medical science, and NIBB Collaborative Research Program (16-408). YC was supported by the Graduate Program for Leaders in Life Innovation, The University of Tokyo Life Innovation Leading Graduate School from MEXT, Japan. DA is a research fellow of the Japan Society for the Promotion of Science.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01973/full#supplementary-material
References
- Ahmadian M., Suh J. M., Hah N., Liddle C., Atkins A. R., Downes M., et al. (2013). PPARgamma signaling and metabolism: the good, the bad and the future. Nat. Med. 19 557–566. 10.1038/nm.3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Mateos E., Brinkmeier H., Burks T. N., Mejias R., Files D. C., Steinberger M., et al. (2013). Activation of serum/glucocorticoid-induced kinase 1 (SGK1) is important to maintain skeletal muscle homeostasis and prevent atrophy. EMBO Mol. Med. 5 80–91. 10.1002/emmm.201201443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S., Hanaya T., Sakurai T., Ikeda M., Kurimoto M. (2005). A novel phenomenon predicting the entry into a state of hibernation in Syrian hamsters (Mesocricetus auratus). J. Vet. Med. Sci. 67 215–217. 10.1292/jvms.67.215 [DOI] [PubMed] [Google Scholar]
- Armitage K. B., Downhower J. F., Svendsen G. E. (1976). Seasonal-changes in weights of marmots. Am. Midl. Nat. 96 36–51. 10.2307/2424566 [DOI] [Google Scholar]
- Arnold W., Giroud S., Valencak T. G., Ruf T. (2015). Ecophysiology of omega Fatty acids: a lid for every jar. Physiology 30 232–240. 10.1152/physiol.00047.2014 [DOI] [PubMed] [Google Scholar]
- Arnold W., Ruf T., Frey-Roos F., Bruns U. (2011). Diet-independent remodeling of cellular membranes precedes seasonally changing body temperature in a hibernator. PLoS One 6:e18641. 10.1371/journal.pone.0018641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi S., Hallen S., Huang L., Svensson P. A., Momo R. A., Wallin S., et al. (2015). Thermogenic activity of UCP1 in human white fat-derived beige adipocytes. Mol. Endocrinol. 29 130–139. 10.1210/me.2014-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartness T. J., Wade G. N. (1984). Photoperiodic control of body weight and energy metabolism in Syrian hamsters (Mesocricetus auratus): role of pineal gland, melatonin, gonads, and diet. Endocrinology 114 492–498. 10.1210/endo-114-2-492 [DOI] [PubMed] [Google Scholar]
- Baumber J., Denyes A. (1963). Acetate-1-C14 metabolism of white fat from hamsters in cold exposure and hibernation. Am. J. Physiol. 205 905–908. 10.1152/ajplegacy.1963.205.5.905 [DOI] [PubMed] [Google Scholar]
- Beale E. G., Hammer R. E., Antoine B., Forest C. (2004). Disregulated glyceroneogenesis: PCK1 as a candidate diabetes and obesity gene. Trends Endocrinol. Metab. 15 129–135. 10.1016/j.tem.2004.02.006 [DOI] [PubMed] [Google Scholar]
- Berry R., Church C. D., Gericke M. T., Jeffery E., Colman L., Rodeheffer M. S. (2014). Imaging of adipose tissue. Methods Enzymol. 537 47–73. 10.1016/B978-0-12-411619-1.00004-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard P. G., Festuccia W. T., Houde V. P., St-Pierre P., Brule S., Turcotte V., et al. (2012). Major involvement of mTOR in the PPARgamma-induced stimulation of adipose tissue lipid uptake and fat accretion. J. Lipid Res. 53 1117–1125. 10.1194/jlr.M021485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondin D. P., Frisch F., Phoenix S., Guerin B., Turcotte E. E., Haman F., et al. (2017). Inhibition of intracellular triglyceride lipolysis suppresses cold-induced brown adipose tissue metabolism and increases shivering in humans. Cell Metab. 25 438–447. 10.1016/j.cmet.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Boswell T., Woods S. C., Kenagy G. J. (1994). Seasonal changes in body mass, insulin, and glucocorticoids of free-living golden-mantled ground squirrels. Gen. Comp. Endocrinol. 96 339–346. 10.1006/gcen.1994.1189 [DOI] [PubMed] [Google Scholar]
- Braun K., Oeckl J., Westermeier J., Li Y., Klingenspor M. (2018). Non-adrenergic control of lipolysis and thermogenesis in adipose tissues. J. Exp. Biol. 221(Pt Suppl. 1):jeb165381. 10.1242/jeb.165381 [DOI] [PubMed] [Google Scholar]
- Buck M. J., Squire T. L., Andrews M. T. (2002). Coordinate expression of the PDK4 gene: a means of regulating fuel selection in a hibernating mammal. Physiol. Genomics 8 5–13. 10.1152/physiolgenomics.00076.2001 [DOI] [PubMed] [Google Scholar]
- Cai H., Dong L. Q., Liu F. (2016). Recent advances in adipose mTOR signaling and function: therapeutic prospects. Trends Pharmacol. Sci. 37 303–317. 10.1016/j.tips.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey H. V., Andrews M. T., Martin S. L. (2003). Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol. Rev. 83 1153–1181. 10.1152/physrev.00008.2003 [DOI] [PubMed] [Google Scholar]
- Carneheim C., Cannon B., Nedergaard J. (1989). Rare fatty acids in brown fat are substrates for thermogenesis during arousal from hibernation. Am. J. Physiol. 256(1 Pt 2), R146–R154. 10.1152/ajpregu.1989.256.1.R146 [DOI] [PubMed] [Google Scholar]
- Caron A., Lee S., Elmquist J. K., Gautron L. (2018). Leptin and brain-adipose crosstalks. Nat. Rev. Neurosci. 19 153–165. 10.1038/nrn.2018.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakir I., Dumont S., Pevet P., Ouarour A., Challet E., Vuillez P. (2015). Pineal melatonin is a circadian time-giver for leptin rhythm in Syrian hamsters. Front. Neurosci. 9:190. 10.3389/fnins.2015.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayama Y., Ando L., Tamura Y., Miura M., Yamaguchi Y. (2016). Decreases in body temperature and body mass constitute pre-hibernation remodelling in the Syrian golden hamster, a facultative mammalian hibernator. R. Soc. Open Sci. 3:160002. 10.1098/rsos.160002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitraju C., Mejhert N., Haas J. T., Diaz-Ramirez L. G., Grueter C. A., Imbriglio J. E., et al. (2017). Triglyceride synthesis by DGAT1 protects adipocytes from lipid-induced ER Stress during lipolysis. Cell Metab. 26 407.e3–418.e3. 10.1016/j.cmet.2017.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S. S., Huh J. Y., Hwang I. J., Kim J. I., Kim J. B. (2016). Adipose tissue remodeling: its role in energy metabolism and metabolic disorders. Front. Endocrinol. 7:30 10.3389/fendo.2016.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P., Levy J. D., Zhang Y., Frontini A., Kolodin D. P., Svensson K. J., et al. (2014). Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156 304–316. 10.1016/j.cell.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dark J. (2005). Annual lipid cycles in hibernators: integration of physiology and behavior. Annu. Rev. Nutr. 25 469–497. 10.1146/annurev.nutr.25.050304.092514 [DOI] [PubMed] [Google Scholar]
- de Jong J. M., Larsson O., Cannon B., Nedergaard J. (2015). A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Endocrinol. Metab. 308 E1085–E1105. 10.1152/ajpendo.00023.2015 [DOI] [PubMed] [Google Scholar]
- Drew K. L., Frare C., Rice S. A. (2017). Neural Signaling metabolites may modulate energy use in hibernation. Neurochem. Res. 42 141–150. 10.1007/s11064-016-2109-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. F., Storey K. B. (2003). Differential expression of Akt, PPARgamma, and PGC-1 during hibernation in bats. Biochem. Cell Biol. 81 269–274. 10.1139/o03-056 [DOI] [PubMed] [Google Scholar]
- El Kebbaj Z., Andreoletti P., Mountassif D., Kabine M., Schohn H., Dauca M., et al. (2009). Differential regulation of peroxisome proliferator-activated receptor (PPAR)-alpha1 and truncated PPARalpha2 as an adaptive response to fasting in the control of hepatic peroxisomal fatty acid beta-oxidation in the hibernating mammal. Endocrinology 150 1192–1201. 10.1210/en.2008-1394 [DOI] [PubMed] [Google Scholar]
- Enerback S., Jacobsson A., Simpson E. M., Guerra C., Yamashita H., Harper M. E., et al. (1997). Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387 90–94. 10.1038/387090a0 [DOI] [PubMed] [Google Scholar]
- Florant G. L., Porst H., Peiffer A., Hudachek S. F., Pittman C., Summers S. A., et al. (2004). Fat-cell mass, serum leptin and adiponectin changes during weight gain and loss in yellow-bellied marmots (Marmota flaviventris). J. Comp. Physiol. B 174 633–639. 10.1007/s00360-004-0454-0 [DOI] [PubMed] [Google Scholar]
- Galarraga M., Campion J., Munoz-Barrutia A., Boque N., Moreno H., Martinez J. A., et al. (2012). Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. J. Lipid Res. 53 2791–2796. 10.1194/jlr.D023788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser F. (2013). Hibernation. Curr. Biol. 23 R188–R193. 10.1016/j.cub.2013.01.062 [DOI] [PubMed] [Google Scholar]
- Giroud S., Frare C., Strijkstra A., Boerema A., Arnold W., Ruf T. (2013). Membrane phospholipid fatty acid composition regulates cardiac SERCA activity in a hibernator, the Syrian hamster (Mesocricetus auratus). PLoS One 8:e63111. 10.1371/journal.pone.0063111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroud S., Stalder G., Gerritsmann H., Kubber-Heiss A., Kwak J., Arnold W., et al. (2018). Dietary lipids affect the onset of hibernation in the garden dormouse (Eliomys quercinus): implications for cardiac function. Front. Physiol. 9:1235. 10.3389/fphys.2018.01235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroud S., Zahn S., Criscuolo F. O., Chery I., Blanc S., Turbill C., et al. (2014). Late-born intermittently fasted juvenile garden dormice use torpor to grow and fatten prior to hibernation: consequences for ageing processes. Proc. R. Soc. B Biol. Sci. 281:20141131. 10.1098/rspb.2014.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabek K. R., Karimpour-Fard A., Epperson L. E., Hindle A., Hunter L. E., Martin S. L. (2011). Multistate proteomics analysis reveals novel strategies used by a hibernator to precondition the heart and conserve ATP for winter heterothermy. Physiol. Genomics 43 1263–1275. 10.1152/physiolgenomics.00125.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B., Pawlak M., Lefebvre P., Staels B. (2017). PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 13 36–49. 10.1038/nrendo.2016.135 [DOI] [PubMed] [Google Scholar]
- Hampton M., Melvin R. G., Kendall A. H., Kirkpatrick B. R., Peterson N., Andrews M. T. (2011). Deep sequencing the transcriptome reveals seasonal adaptive mechanisms in a hibernating mammal. PLoS One 6:e27021. 10.1371/journal.pone.0027021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms M., Seale P. (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19 1252–1263. 10.1038/nm.3361 [DOI] [PubMed] [Google Scholar]
- Heldmaier G., Ortmann S., Elvert R. (2004). Natural hypometabolism during hibernation and daily torpor in mammals. Respir. Physiol. Neurobiol. 141 317–329. 10.1016/j.resp.2004.03.014 [DOI] [PubMed] [Google Scholar]
- Hindle A. G., Martin S. L. (2014). Intrinsic circannual regulation of brown adipose tissue form and function in tune with hibernation. Am. J. Physiol. Endocrinol. Metab. 306 E284–E299. 10.1152/ajpendo.00431.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Kang Q., Yoneshiro T., Camporez J. P., Maki H., Homma M., et al. (2017). UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med. 23 1454–1465. 10.1038/nm.4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansky L., Haddad G., Kahlerova Z., Nedoma J. (1984). Effect of external factors on hibernation of golden-hamsters. J. Comp. Physiol. 154 427–433. 10.1007/Bf00684450 3760293 [DOI] [Google Scholar]
- Jastroch M., Giroud S., Barrett P., Geiser F., Heldmaier G., Herwig A. (2016). Seasonal control of mammalian energy balance: recent advances in the understanding of daily torpor and hibernation. J. Neuroendocrinol. 28. 10.1111/jne.12437 [DOI] [PubMed] [Google Scholar]
- Jefimow M., Wojciechowski M. S. (2014). Effect of dietary fatty acids on metabolic rate and nonshivering thermogenesis in golden hamsters. J. Exp. Zool. A Ecol. Genet. Physiol. 321 98–107. 10.1002/jez.1840 [DOI] [PubMed] [Google Scholar]
- Jimenez-Aranda A., Fernandez-Vazquez G., Campos D., Tassi M., Velasco-Perez L., Tan D. X., et al. (2013). Melatonin induces browning of inguinal white adipose tissue in Zucker diabetic fatty rats. J. Pineal Res. 55 416–423. 10.1111/jpi.12089 [DOI] [PubMed] [Google Scholar]
- Jinka T. R., Toien O., Drew K. L. (2011). Season primes the brain in an arctic hibernator to facilitate entrance into torpor mediated by adenosine A(1) receptors. J. Neurosci. 31 10752–10758. 10.1523/JNEUROSCI.1240-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S., Spiegelman B. M., Seale P. (2015). Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 22 546–559. 10.1016/j.cmet.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S. (2014). Integrated physiology and systems biology of PPARalpha. Mol. Metab. 3 354–371. 10.1016/j.molmet.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao N., Hashimoto M. (2012). Increased thermogenic capacity of brown adipose tissue under low temperature and its contribution to arousal from hibernation in Syrian hamsters. Am. J. Physiol. Regul. Integr. Comp. Physiol. 302 R118–R125. 10.1152/ajpregu.00053.2011 [DOI] [PubMed] [Google Scholar]
- Kondo N. (1987). Identification of a pre-hibernating state in myocardium from nonhibernating chipmunks. Experientia 43 873–875. 10.1007/BF01951645 [DOI] [PubMed] [Google Scholar]
- Kondo N., Shibata S. (1984). Calcium source for excitation-contraction coupling in myocardium of nonhibernating and hibernating chipmunks. Science 225 641–643. 10.1126/science.6740332 [DOI] [PubMed] [Google Scholar]
- Kurtz C. C., Lindell S. L., Mangino M. J., Carey H. V. (2006). Hibernation confers resistance to intestinal ischemia-reperfusion injury. Am. J. Physiol. Gastrointest. Liver Physiol. 291 G895–G901. 10.1152/ajpgi.00155.2006 [DOI] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M. (2009). An emerging role of mTOR in lipid biosynthesis. Curr. Biol. 19 R1046–R1052. 10.1016/j.cub.2009.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J. E., Jones J., Zucker I. (2002). Temperature dependence of gonadal regression in Syrian hamsters exposed to short day lengths. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282 R744–R752. 10.1152/ajpregu.00299.2001 [DOI] [PubMed] [Google Scholar]
- Lee J., Ellis J. M., Wolfgang M. J. (2015). Adipose fatty acid oxidation is required for thermogenesis and potentiates oxidative stress-induced inflammation. Cell Rep. 10 266–279. 10.1016/j.celrep.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. L., Jung S. M., Guertin D. A. (2017). The complex roles of mechanistic target of rapamycin in adipocytes and beyond. Trends Endocrinol. Metab. 28 319–339. 10.1016/j.tem.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C. J., Becker J. J., Manis A. D., Hamilton J. S., Horowitz J. M., Horwitz B. A. (2012). Neuroprotection supports signal processing in the hippocampus of Syrian hamsters, a facultative hibernator. Neurosci. Lett. 520 20–25. 10.1016/j.neulet.2012.05.010 [DOI] [PubMed] [Google Scholar]
- Liu D., Bordicchia M., Zhang C., Fang H., Wei W., Li J. L., et al. (2016). Activation of mTORC1 is essential for beta-adrenergic stimulation of adipose browning. J. Clin. Invest. 126 1704–1716. 10.1172/JCI83532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. Z., Svensson K. J., Tsai L., Zeng X., Roh H. C., Kong X., et al. (2014). A smooth muscle-like origin for beige adipocytes. Cell Metab. 19 810–820. 10.1016/j.cmet.2014.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman C. P. (1958). Oxygen consumption, body temperature and heart rate of woodchucks entering hibernation. Am. J. Physiol. 194 83–91. 10.1152/ajplegacy.1958.194.1.83 [DOI] [PubMed] [Google Scholar]
- Melvin R. G., Andrews M. T. (2009). Torpor induction in mammals: recent discoveries fueling new ideas. Trends Endocrinol. Metab. 20 490–498. 10.1016/j.tem.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Hamsany C., Castex C., Hoo-Paris R., Kacemi N., Sutter B. (1988). Hormonal control of lipolysis from the white adipose tissue of hibernating jerboa (Jaculus orientalis). Comp. Biochem. Physiol. A Comp. Physiol. 91 665–669. 10.1016/0300-9629(88)90945-0 [DOI] [PubMed] [Google Scholar]
- Mostafa N., Everett D. C., Chou S. C., Kong P. A., Florant G. L., Coleman R. A. (1993). Seasonal changes in critical enzymes of lipogenesis and triacylglycerol synthesis in the marmot (Marmota flaviventris). J. Comp. Physiol. B 163 463–469. [DOI] [PubMed] [Google Scholar]
- Mottillo E. P., Balasubramanian P., Lee Y. H., Weng C., Kershaw E. E., Granneman J. G. (2014). Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic beta3-adrenergic receptor activation. J. Lipid Res. 55 2276–2286. 10.1194/jlr.M050005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. J., Otis J. P., Carey H. V. (2009). A role for nuclear receptors in mammalian hibernation. J. Physiol. 587(Pt 9), 1863–1870. 10.1113/jphysiol.2008.167692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack J., Stawski C., Geiser F. (2017). More functions of torpor and their roles in a changing world. J. Comp. Physiol. B 187 889–897. 10.1007/s00360-017-1100-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C. M., Namkung J., Go Y., Shong K. E., Kim K., Kim H., et al. (2015). Regulation of systemic energy homeostasis by serotonin in adipose tissues. Nat. Commun. 6:6794. 10.1038/ncomms7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamatsu-Ogura Y., Fukano K., Tsubota A., Uozumi A., Terao A., Kimura K., et al. (2013). Thermogenic ability of uncoupling protein 1 in beige adipocytes in mice. PLoS One 8:e84229. 10.1371/journal.pone.0084229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J. M., Jinka T. R., Larson L. K., Danielson J. J., Moore J. T., Carpluck J., et al. (2013). Circannual rhythm in body temperature, torpor, and sensitivity to A(1) adenosine receptor agonist in arctic ground squirrels. J. Biol. Rhythms 28 201–207. 10.1177/0748730413490667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn O., Olefsky J. M. (2012). The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 18 363–374. 10.1038/nm.2627 [DOI] [PubMed] [Google Scholar]
- Otis J. P., Ackermann L. W., Denning G. M., Carey H. V. (2010). Identification of qRT-PCR reference genes for analysis of opioid gene expression in a hibernator. J. Comp. Physiol. B 180 619–629. 10.1007/s00360-009-0430-9 [DOI] [PubMed] [Google Scholar]
- Pitrosky B., Delagrange P., Rettori M. C., Pevet P. (2003). S22153, a melatonin antagonist, dissociates different aspects of photoperiodic responses in Syrian hamsters. Behav. Brain Res. 138 145–152. 10.1016/S0166-4328(02)00235-8 [DOI] [PubMed] [Google Scholar]
- Polak P., Cybulski N., Feige J. N., Auwerx J., Ruegg M. A., Hall M. N. (2008). Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 8 399–410. 10.1016/j.cmet.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Ricoult S. J., Manning B. D. (2013). The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO Rep. 14 242–251. 10.1038/embor.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C., Han J., Tzatsos A., Kandror K. V. (2003). Nutrient-sensing mTOR-mediated pathway regulates leptin production in isolated rat adipocytes. Am. J. Physiol. Endocrinol. Metab. 284 E322–E330. 10.1152/ajpendo.00230.2002 [DOI] [PubMed] [Google Scholar]
- Ruf T., Geiser F. (2015). Daily torpor and hibernation in birds and mammals. Biol Rev Camb Philos Soc 90 891–926. 10.1111/brv.12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheriff M. J., Fridinger R. W., Toien O., Barnes B. M., Buck C. L. (2013). Metabolic rate and prehibernation fattening in free-living arctic ground squirrels. Physiol. Biochem. Zool. 86 515–527. 10.1086/673092 [DOI] [PubMed] [Google Scholar]
- Shimozuru M., Nagashima A., Tanaka J., Tsubota T. (2016). Seasonal changes in the expression of energy metabolism-related genes in white adipose tissue and skeletal muscle in female Japanese black bears. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 19 38–47. 10.1016/j.cbpb.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Shinoda K., Luijten I. H., Hasegawa Y., Hong H., Sonne S. B., Kim M., et al. (2015). Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat. Med. 21 389–394. 10.1038/nm.3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson J. E., Adamic M., Huber D., Stokke S. (2007). Brown bear body mass and growth in northern and southern Europe. Oecologia 153 37–47. 10.1007/s00442-007-0715-1 [DOI] [PubMed] [Google Scholar]
- Talaei F. (2014). Modulation of mTOR and autophagy in hibernating hamster lung and the application of the potential mechanism to improve the recellularization process of decellularized lung scaffolds. J. Regen. Med. Tissue Eng. 3:1 10.7243/2050-1218-3-1 [DOI] [Google Scholar]
- Talaei F., Bouma H. R., Van der Graaf A. C., Strijkstra A. M., Schmidt M., Henning R. H. (2011). Serotonin and dopamine protect from hypothermia/rewarming damage through the CBS/H2S pathway. PLoS One 6:e22568. 10.1371/journal.pone.0022568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Shintani M., Nakamura A., Monden M., Shiomi H. (2005). Phase-specific central regulatory systems of hibernation in Syrian hamsters. Brain Res. 1045 88–96. 10.1016/j.brainres.2005.03.029 [DOI] [PubMed] [Google Scholar]
- Toien O., Blake J., Edgar D. M., Grahn D. A., Heller H. C., Barnes B. M. (2011). Hibernation in black bears: independence of metabolic suppression from body temperature. Science 331 906–909. 10.1126/science.1199435 [DOI] [PubMed] [Google Scholar]
- Toyoda T., Kamei Y., Kato H., Sugita S., Takeya M., Suganami T., et al. (2008). Effect of peroxisome proliferator-activated receptor-alpha ligands in the interaction between adipocytes and macrophages in obese adipose tissue. Obesity 16 1199–1207. 10.1038/oby.2008.62 [DOI] [PubMed] [Google Scholar]
- Trefna M., Goris M., Thissen C. M., Reitsema V. A., Bruintjes J. J., de Vrij E. L., et al. (2017). The influence of sex and diet on the characteristics of hibernation in Syrian hamsters. J. Comp. Physiol. B 187 725–734. 10.1007/s00360-017-1072-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanecek J., Jansky L., Illnerova H., Hoffmann K. (1984). Pineal melatonin in hibernating and aroused golden hamsters (Mesocricetus auratus). Comp. Biochem. Physiol. A Comp. Physiol. 77 759–762. 10.1016/0300-9629(84)90197-X [DOI] [PubMed] [Google Scholar]
- Waku T., Shiraki T., Oyama T., Maebara K., Nakamori R., Morikawa K. (2010). The nuclear receptor PPARgamma individually responds to serotonin- and fatty acid-metabolites. EMBO J. 29 3395–3407. 10.1038/emboj.2010.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Walter R. D., Bhat B. G., Florant G. L., Coleman R. A. (1997). Seasonal changes in enzymes of lipogenesis and triacylglycerol synthesis in the golden-mantled ground squirrel (Spermophilus lateralis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 118 261–267. 10.1016/S0305-0491(97)00102-8 [DOI] [PubMed] [Google Scholar]
- Weitten M., Robin J. P., Oudart H., Pevet P., Habold C. (2013). Hormonal changes and energy substrate availability during the hibernation cycle of Syrian hamsters. Horm. Behav. 64 611–617. 10.1016/j.yhbeh.2013.08.015 [DOI] [PubMed] [Google Scholar]
- Willems E., Leyns L., Vandesompele J. (2008). Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379 127–129. 10.1016/j.ab.2008.04.036 [DOI] [PubMed] [Google Scholar]
- Wilson B. E., Deeb S., Florant G. L. (1992). Seasonal changes in hormone-sensitive and lipoprotein lipase mRNA concentrations in marmot white adipose tissue. Am. J. Physiol. 262(2 Pt 2), R177–R181. 10.1152/ajpregu.1992.262.2.R177 [DOI] [PubMed] [Google Scholar]
- Wolfson R. L., Chantranupong L., Saxton R. A., Shen K., Scaria S. M., Cantor J. R., et al. (2016). Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351 43–48. 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Bostrom P., Sparks L. M., Ye L., Choi J. H., Giang A. H., et al. (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150 366–376. 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.