Abstract
CYP46A1 is the cytochrome P450 enzyme that converts cholesterol to 24-hydroxycholesterol, a cholesterol elimination product and a potent liver X receptor (LXR) ligand. We conducted retinal characterizations of Cyp46a1−/− mice that had normal fasting blood glucose levels but up to a 1.8-fold increase in retinal cholesterol. The retina of Cyp46a1−/− mice exhibited venous beading and tortuosity, microglia/macrophage activation, and increased vascular permeability, features commonly associated with diabetic retinopathy. The expression of Lxrα and Lxrβ was increased in both the whole Cyp46a1−/− retina and retinal macroglia/macrophages. The LXR-target genes were affected as well, primarily in activated microglial cells and macrophages. In the latter, the LXR-transactivated genes (Abca1, Abcg1, Apod, Apoe, Mylip, and Arg2) were up-regulated; similarly, there was an up-regulation of the LXR-transrepressed genes (Ccl2, Ptgs2, Cxcl1, Il1b, Il6, Nos2, and Tnfa). For comparison, gene expression was investigated in bone marrow–derived macrophages from Cyp46a1−/− mice as well as retinal and bone marrow–derived macrophages from Cyp27a1−/− and Cyp27a1−/−Cyp46a1−/− mice. CYP46A1 expression was detected in retinal endothelial cells, and this expression was increased in the proinflammatory environment. Retinal Cyp46a1−/− phosphoproteome revealed altered phosphorylation of 30 different proteins, including tight junction protein zonula occludens 1 and aquaporin 4. Collectively, the data obtained establish metabolic and regulatory significance of CYP46A1 for the retina and suggest pharmacologic activation of CYP46A1 as a potential therapeutic approach to dyslipidemia-induced retinal damage.
Changes in retinal microcirculation are the early manifestations of diabetic retinopathy, the most common microvascular complications in type 1 diabetes and type 2 diabetes.1, 2, 3 These changes include retinal microaneurysms, capillary nonperfusion and degeneration, venous beading and looping, intraretinal microvascular abnormalities (large-caliber shunt vessels within nonperfused regions of the capillary bed), excessive vasopermeability, retinal edema, and impairment of neural function.1, 3 Remarkably, increased vascular permeability, dilation, nonperfusion, capillary degeneration, and arteriovenous shunts were observed in the retina of Cyp27a1−/−Cyp46a1−/− mice4 but not Cyp27a1−/− mice, the two genotypes that had normal blood glucose levels but increased total retinal cholesterol.4
Cytochrome P450 27A1 (CYP27A1) is a sterol 27-hydroxylase,5 whereas cytochrome P450 46A1 (CYP46A1) catalyzes cholesterol 24-hydroxylation.6 Both P450s are expressed in the retina7, 8, 9 and are important for retinal cholesterol elimination.4, 10 CYP27A1 is ubiquitous and is highly abundant in the photoreceptor inner segments, Muller cells, and retinal pigment epithelium (RPE).11 CYP46A1 is less abundant in the retina7 and is mainly found in the neurons of the ganglion cell layer with a lower expression in the RPE.12 CYP46A1 and CYP27A1 produce 24-hydroxycholesterol (24HC) and 27-hydroxycholesterol (27HC), respectively, oxysterols, which are the transport forms of cholesterol9, 13 from the retina to the systemic circulation. In addition, 24HC and 27HC are biologically active molecules that can interact with different regulatory proteins, including the liver X receptors (LXRs), a family of transcription factors.14 24HC is a more potent LXR agonist than 27HC14; hence, we hypothesized that some of the abnormalities in Cyp27a1−/−Cyp46a1−/− mice were due to deficiency in CYP46A1 and a lack of 24HC activation of LXR. Indeed, Lxrα/β−/− mice showed vascular changes similar to those observed in the diabetic retina with increases in acellular capillaries despite the lack of metabolic dysfunction.15 Also, the expression of Lxr was shown to be significantly decreased in diabetic human retinas and in a type 2 diabetes animal model.16 Conversely, LXR activation by synthetic ligands was found to prevent retinal inflammation and diabetic retinopathy in diabetic animal models15, 16 and to reduce proinflammatory macrophage activity.16
There are two LXR isoforms, LXRα and LXRβ, that share high sequence identity (approximately 80%) and are activated by the same ligands, typically oxygenated metabolites of cholesterol (eg, 24HC and 27HC) as well as cholesterol precursor desmosterol.14, 17, 18 LXRβ is ubiquitous, whereas LXRα is tissue specific and is highly expressed in the liver, intestine, kidney, adipose tissue, and macrophages.19, 20, 21 Activation of LXRs leads either to gene transactivation or to gene transrepression. In transactivation, the basal condition is gene silencing by a complex of LXR with retinoid X receptor and corepressors bound to the promoter of the target gene.22 Ligand binding to LXR leads to release of corepressors in exchange for coactivators, thus initiating gene transcription.23 Many of the cholesterol-related genes [eg, ATP-binding cassette subfamily members A1 (Abca1) and G1 (Abcg1), apolipoprotein E (Apoe), inducible degrader of low-density lipoprotein receptor (Idol; official name, Mylip), and sterol regulatory element-binding protein 1c (Srebp1c)] along with the genes involved in fatty acid synthesis [eg, elongation of long chain fatty acids family member 5 (Elovl5), fatty acid synthase (Fasn), fatty acid desaturase 2 (Fads2), and stearoyl–coenzyme A desaturases 1 and 2 (Scd1 and Scd2, respectively)] as well as other processes [eg, apoptosis inhibitor of macrophages (Aim), apolipoprotein D (Apod), arginase 2 (Arg2), glucose transporter type 4 (Glut4), and Mer receptor tyrosine kinase (Mertk)] are regulated by this mechanism.24, 25 In transrepression, ligand binding leads to LXR sumoylation, which prevents the corepressor release and gene expression.26 This mechanism, shown to be operative in the liver and macrophages, blocks the transcription of NF-κB and other transcription factors and thereby the expression of the proinflammatory genes [eg, C-C motif chemokine 2 (Ccl2), prostaglandin G/H synthase 2 (Cox-2; official name, Ptgs2), C-X-C motif chemokine ligand 1 (Cxcl1), IL-1β (Il1b), IL-6 (Il6), inducible nitric oxide synthase (iNos), and tumor necrosis factor-α (Tnfa)], which are controlled by these factors.25, 26, 27, 28
LXRα and LXRβ appear to be largely interchangeable in transactivation, with their contribution determined by relative expression level.29 However, in macrophages, LXRα may be more robust at transactivation and LXRβ more potent at basal target repression.29 LXRα and LXRβ are both present in the neural retina and RPE and were immunolocalized to retinal layers: LXRβ appears to be ubiquitous, and LXRα seems to be expressed in the cells of the ganglion cell layer and RPE.8, 30 However, knowledge of cell-specific retinal LXR localizations is still lacking, including retinal endothelial cells and microglia, which may be missed on retinal cross sections because of the small numbers of these cells. Herein, we characterized the ocular phenotype of Cyp46a1−/− mice and obtained evidence that both metabolic and regulatory CYP46A1 activities are of significance for the retina and retinal blood vessels. We also performed multicolor immunohistochemistry labeling of vascular endothelial cells and CYP46A1. Our findings suggest that CYP46A1 may represent a new pharmacologic target for early-stage diabetic retinopathy treatment.
Materials and Methods
Animals
Animals were 6- to 9-month–old female or male mice. In both sexes, retinal vascular abnormalities on fluorescein angiography (FA) were detected starting from the age of 6 months. Furthermore, electroretinography (ERG) responses, conducted for both sexes at 6 months of age, were similar, as were the levels of mouse retinal and serum sterols. Hence, all subsequent experiments used male mice. Cyp46a1+/− mice (on the mixed C57BL/6J; 129S6/SvEv background) were provided by Dr. David Russell (UT Southwestern, Dallas, TX).31 Cyp27a1+/− mice (on the C57BL/6J background) were provided by Dr. Sandra Erickson (University of California, San Francisco, San Francisco, CA).32 The heterozygous animals obtained were crossed to generate the Cyp46a1−/−, Cyp46a1+/+, Cyp27a1−/−, and Cyp27a1+/+ breeding pairs, which established the Cyp46a1−/−, Cyp46a1+/+, Cyp27a1−/−, and Cyp27a1+/+ colonies. The Cyp27a1−/−Cyp46a1−/− strain and Cyp27a+/+Cyp46a1+/+ controls were generated by crossing Cyp27a1+/− and Cyp46a1+/− mice.4 All mice were free of the Crblrd8 mutation, which was bred out of our colonies. Cyp46a1−/− mice had normal fertility, body weight, food consumption, and appearance/weight of difference organs (data are not shown), consistent with previous genotype characterizations.31, 33 All animals were maintained on a standard 12-hour light (approximately 10 lux)–dark cycle and were fed standard rodent chow and water ad libitum. All animal procedures were approved by the Case Western Reserve University (Cleveland, OH) Institutional Animal Care and Use Committee and conformed to recommendations of the American Veterinary Association Panel on Euthanasia and the Association for Research in Vision and Ophthalmology.
ERG
ERG was performed as previously described.4 The scotopic a- and b-waves or the photopic b-wave was recorded in response to strobe flash stimuli presented after overnight dark adaptation (scotopic) or adaptation to a steady adapting field (photopic).
FA
FA was as described10 after a bolus (0.1-mL) i.p. injection of 1.6% sodium fluorescein in phosphate-buffered saline (PBS).
Serum Chemistry and Glucose Tolerance Test
Mice were fasted overnight and anesthetized via i.p. injection of 80 mg/kg ketamine (Fort Dodge Animal Health, Overland Park, KS) and 15 mg/kg xylazine (Akorn Inc., Lake Forest, IL) in PBS, pH 7.4. Blood was withdrawn via cardiac puncture and used for serum isolation, as described.34 Serum was analyzed for total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, triglycerides, and free fatty acids by Marshfield Labs (Marshfield, WI). For the glucose tolerance test, mice were fasted overnight and injected with a solution of 50% d-glucose (2 g/kg body weight; Hospira, Lake Forest, IL) into the peritoneum. The blood was withdrawn from the tail vein and assayed for glucose by an Elite XL Glucometer (Bayer Contour, Parsippany, NJ) before and after the injection (30, 60, 120, and 150 minutes).
Retinal Isolation and Sterol Quantifications
Retinal isolation and processing were as described,4 as are subsequent sterol quantifications by isotope dilution gas chromatography–mass spectroscopy.9
Isolation and Treatment of RMMs and BMDMs
Primary retinal microglia/macrophages (RMMs) and bone marrow–derived macrophages (BMDMs) were prepared from 1.5- to 2.5-month–old 8 to 12 male and female mice of the same genotype, as described35 with the following modifications. After isolation, mouse eyes were first placed in PBS containing 1% of penicillin/streptomycin (ThermoFisher, Waltham, MA) and then transferred to the ice-cold Dulbecco's modified Eagle's medium (DMEM; ThermoFisher) supplemented with 1% of penicillin/streptomycin. Next, eyes were cleaned from the connectives tissues and incubated for 30 minutes at 37°C and 60 rpm shaking in DMEM containing 2% dispase (Invitrogen, Waltham, MA) and 100 mg/mL collagenase IV (Invitrogen). The cornea, lens, and vitreous were removed, and the retinas were isolated from the eye cup under a surgical microscope (High Illuminator NI-5; Nikon Instruments Inc., Melville, NY) using tweezers. The retinas were then placed back in DMEM containing 2% dispase and 100 mg/mL collagenase IV and incubated for additional 15 minutes at 37°C and 60 rpm shaking. The tissue was transferred to a culture dish to peel off the RPE. The remaining neural retinas were homogenized by pipetting and cultured in T25 or T75 flasks (Falcon, Waltham, MA) for 7 days at 37°C in DMEM containing 1% penicillin/streptomycin and 20% fetal bovine serum (FBS; ThermoFisher). After 1 week, the culture medium was changed and the remaining adherent cells were left to replicate for another 7 to 10 days. The medium was changed every other day. After the last medium change, cells adherent to the plastic surface were treated with 0.05% trypsin (Invitrogen) for 2 to 3 minutes, and the less adhesive cells were collected as RMM. A small portion of RMM (approximately 25,000 cells) was plated on a μ-slide (Ibidi, Fitchburg, WI) and kept overnight in DMEM containing 1% penicillin/streptomycin and 20% FBS. These cells were used for the characterization of preparation homogeneity. The remaining RMMs were plated (no less than 250,000 cells per well) in 6-well tissue culture plates (Falcon) and allowed to adhere for 5 to 6 hours. Cells were then starved overnight in DMEM containing 1% penicillin/streptomycin without FBS; the next morning, they were stimulated for 6 hours with or without 100 ng/mL lipopolysaccharide (LPS; Escherichia coli O111:B4; InvivoGen, San Diego, CA). At the end of stimulation, cells were washed with ice-cold PBS and lysed with TRIzol reagent (ThermoFisher) for RNA isolation. The isolation of BMDM was as described.35 Briefly, bone marrows from femurs and tibias were incubated for 7 days in RPMI medium 1640 (ThermoFisher) containing 20% FBS and 30% L929-conditioned medium (obtained as described36) to differentiate them into macrophages. Subsequent cell processing, characterization, and treatment were the same as for RMM. The RMM and BMDM homogeneity was >90%, as assessed by morphology and staining for ionized calcium binding adaptor molecule 1 (Iba1).
Isolation and Treatment of HRECs, BRECs, and RPE
Human retinal endothelial cell (HREC), bovine retinal endothelial cell (BREC), and RPE isolation and characterization were as described16, 37 using donor retinas38 provided by the National Disease Research Interchange and eyes donated by the Michigan State University Meat Laboratory, respectively. Before treatment, BREC and RPE were serum starved in 1% FBS complete medium. Cells were then treated with TNF-α (R&D Systems, Minneapolis, MN), a diabetic relevant stimulus, for 24 hours (10 ng/mL) as described,39 followed by RNA isolation.
Quantitative Real-Time PCR
Total RNA (1 μg) from pooled samples of mouse retinas, RMM, and BMDM was isolated as described4 using the TRIzol Reagent (ThermoFisher). This RNA was then converted to cDNA by SuperScript III reverse transcriptase (Invitrogen), according to manufacturer's instructions. PCRs were performed in triplicate and were normalized to β-actin. The isolation of total RNA (1 μg) from HREC, BREC, and RPE was as described16 using the RNeasy minikit (Qiagen, Valencia, CA). First-strand cDNA was synthesized using SuperScript II reverse transcription. All PCRs were performed in triplicate and normalized to cyclophilin A (CypA). The primer sequences are shown in Table 1. Changes in relative mRNA level were calculated by the 2−ΔΔCt method.40
Table 1.
Primers for Quantitative Real-Time PCR
| Gene | Forward primer | Reverse primer | Species |
|---|---|---|---|
| Abca1 | 5′-AGGCCGCACCATTATTTTGTC-3′ | 5′-GGCAATTCTGTCCCCAAGGAT-3′ | M |
| Abcg1 | 5′-ATTTCATCGTCCTGGGCATCT-3′ | 5′-CGGATTTTGTATCTGAGGACGAA-3′ | M |
| Actβ | 5′-TGTTACCAACTGGGACGACATG-3′ | 5′-TTGTAGAAGGTGTGGTGCCAGA-3′ | M |
| Aim | 5′-CGTTAGAAGAAGAAGGTCGTTGGA-3′ | 5′-AGATATGCAAGCGACCCATCTAC-3′ | M |
| Apod | 5′-TGAAGCCAAACAGAGCAACGT-3′ | 5′-GGCATCAACGGGAAGAACTG-3′ | M |
| Apoe | 5′-GGCCCAGGAGAATCAATGAG-3′ | 5′-CCTGGCTGGATATGGATGTTG-3′ | M |
| ArgII | 5′-GACCACAGCCTGGCAATAGGT-3′ | 5′-TCAACCCAGATGACACAGAGATCT-3′ | M |
| Ccl2 | 5′-TCACTGAAGCCAGCTCTCTCTTC-3′ | 5′-GTGAACAGCAGGCCCAGAA-3′ | M |
| Cox-2 | 5′-CACCTCAAGAACATCCAGAGCTT-3′ | 5′-CTCGCGACCATTCTTGAGTGT-3′ | M |
| Cxcl1 | 5′-CACCTCAAGAACATCCAGAGCTT-3′ | 5′-CTCGCGACCATTCTTGAGTGT-3′ | M |
| CYPA | 5′- GGTCCCAAAGACAGCAGAAA-3′ | 5′-GTCACCACCGTCACACATAAA-3′ | H |
| CypA | 5′-GAGCACTGGAGAGAAAGGATTT-3′ | 5′-GACTTGCCACCAGTACCATTAT-3′ | B |
| CYP27A1 | 5′-CACAAACTCCCGGATCATAGAA-3′ | 5′-CACATAGTGGAACACAAAC-3′ | H |
| Cyp27a1 | 5′-AGTAGACACGACATCCAACAC-3′ | 5′-CACACCCACCACTTCCTTAT-3′ | B |
| CYP46A1 | 5′-GAGTCCTGAGTCGCTTAAGAAG-3′ | 5′-CGAAGAGTCTCTCACCAAACA-3′ | H |
| Cyp46a1 | 5′-CAAGCCCAAGTTCACCTACT-3′ | 5′-TCACCTCCATCTGAGCAAAC-3′ | B |
| Glut4 | 5′-ACCGGATTCCATCCCACAA-3′ | 5′-CATGCCACCCACAGAGAAGA-3′ | M |
| Idol | 5′-GGAGCATGTCCAGCACGTCTA-3′ | 5′-GTGCAGGACGCATCAGATGA-3′ | M |
| Il-1β | 5′-GGTCAAAGGTTTGGAAGCAG-3′ | 5′-TGTGAAATGCCACCTTTTGA-3′ | M |
| Il-6 | 5′-AGTTGCCTTCTTGGGACTGA-3′ | 5′-TCCACGATTTCCCAGAGAAC-3′ | M |
| iNOS | 5′-GCCACCAACAATGGCAACA-3′ | 5′-CGTACCGGATGAGCTGTGAA-3′ | M |
| Lxrα | 5′-AGCGTCCATTCAGAGCAAGTG-3′ | 5′-CACTCGTGGACATCCCAGATCT-3′ | M |
| Lxrβ | 5′-ACTCGGAGCAGGTCTTTGCAT-3′ | 5′-CCTACTCGTGCACATCCCAGAT-3′ | M |
| Mertk | 5′-AAAGGTCCCCGTCTGTCCTAA-3′ | 5′-TGGACACCGTCAGTCCTTTGT-3′ | M |
| Srebp1a | 5′-ATGGACGAGCCACCCTTCA-3′ | 5′-AAGTCACTGTCTTGGTTGTTGATGA-3′ | M |
| Srebp1c | 5′-ACGGAGCCATGGATTGCA-3′ | 5′-AAGTCACTGTCTTGGTTGTTGATGA-3′ | M |
| Tnfα | 5′-GGTCTGGGCCATAGAACTGA-3′ | 5′-CAGCCTCTTCTCATTCCTGC-3′ | M |
B, bulls; H, humans; M, mice.
Histochemistry
Retinal stains for unesterified cholesterol (UC) and esterified cholesterol (EC) were performed as described8 using filipin (Cayman Chemical, Ann Arbor, MI).
Immunohistochemistry
The preparation of frozen retinal sections was as described.10 Before the experiment, retinal sections were warmed to room temperature for 30 minutes and washed three times for 5 minutes with PBS containing 0.05% Tween 20. Sections were then permeabilized for 20 minutes with PBS containing 0.5% Triton X-100 and blocked for 1 hour with 5% normal goat serum (PCN5000; Invitrogen) in PBS containing 0.05% Tween 20 (blocking buffer). Sections were incubated overnight at 4°C with either rabbit anti-albumin antibodies (ab207327; Abcam, Cambridge, MA; 1:500 dilutions in blocking buffer) or blocking buffer. The next morning, slides were washed three times for 5 minutes with PBS containing 0.05% Tween 20 and incubated for 1 hour in the dark with either Alexa Fluor 647–conjugated goat anti-rabbit IgG (111-605-144; Jackson ImmunoResearch, West Grove, PA; 1:200 dilutions in blocking buffer) or blocking buffer. Sections were washed three times for 5 minutes with PBS, then dipped three times in distilled water, and next covered with ProLong Gold antifade reagent with DAPI (P36935; Molecular Probes, Eugene, OR) and protected with a glass coverslip. After image acquisition, a coverslip was removed, and sections were washed three times for 5 minutes with PBS, followed by staining at room temperature for 30 minutes with isolectin B4 (121413; Invitrogen; 1:100 dilutions in PBS). Slides were washed three times with PBS, then two times with distilled water, blotted dry, and mounted with ProLong Gold antifade reagent without DAPI (P36965; Molecular Probes).
Staining of retinal cross sections for CYP46A1, CD31, and neuronal-specific nuclear protein was conducted similarly using the following sources and dilutions of primary and secondary antibodies: polyclonal rabbit anti-CYP46A1 IgG (OD280 = 0.6; 1:200 dilutions; generated in the laboratory of I.A.P.) and donkey anti-rabbit AF 488 antibody (A21206; Invitrogen; 1:500 dilution); monoclonal mouse CD31 antibody (sc-81158; Santa Cruz Biotechnology, Dallas, TX; 1:100 dilution); donkey anti-mouse AF 555 antibody (A31570; Invitrogen; 1:500 dilution); monoclonal mouse neuronal-specific nuclear protein antibody (MAB377; Millipore Sigma, St. Louis, MO; 1:100 dilution); and donkey anti-mouse AF 555 antibody (A31570; Invitrogen; 1:500 dilution).
The anti-Iba1 staining of RMM and BMDM was performed after cell medium was discarded and cells were washed with ice-cold PBS. Cells were then fixed for 15 to 20 minutes in 4% paraformaldehyde, washed three times with PBS containing 0.05% Tween 20, and treated with rabbit anti-Iba1 antibodies (019-19741; Wako, Richmond, VA; 1:1000 dilutions), followed by the incubation with Alexa Fluor 647–conjugated goat anti-rabbit IgG (4414; Cell Signaling, Danvers, MA; 1:200 dilution), as described above for retinal sections.
The preparation of retinal flat mounts was as described.41 Briefly, mice were sacrificed and their eyes were enucleated, followed by fixation in 4% paraformaldehyde for 15 minutes at room temperature (for F4/80 staining) or 4 hours at 4°C (for CYP46A1 and CD31 staining). Eyes were then washed three times with PBS and stored in PBS until processing for flat mounts, which included the cornea, lens, and vitreous removal, and four incisions from outside of the eye cup toward the center. Next, retinal flat mounts were permeabilized and blocked for 24 hours at 4°C with PBS containing 0.5% Triton X-100, 5% bovine serum albumin, and either 5% normal goat serum (for F4/80 staining) or 2% donkey serum (for CYP46A1 and CD31 staining). Subsequent incubation with primary rat antibodies against F4/80 (MCA497GA; AbD Serotec, now BioRad, Hercules, CA; 1:100 dilutions in PBS) was overnight at 4°C, followed by a 6-hour incubation at 4°C with Hoechst (33342; Invitrogen; 1:1000 dilutions in PBS). The incubation with the polyclonal rabbit anti-CYP46A1 IgG (OD280 = 0.6; 1:200 dilutions; generated in the laboratory of I.A.P.) and monoclonal mouse CD31 antibodies (sc-81158; Santa Cruz Biotechnology; 1:100 dilutions in PBS) was for 24 hours at 4°C. Retinal flat mounts were then washed for 1 hour at 4°C with four changes of PBS and incubated for 1 hour at room temperature in 2% bovine serum albumin, followed by an overnight incubation at 4°C in PBS. The next morning, either anti-rat Alexa 647 IgG (4418; Cell Signaling, Danvers, MA; 1:200 dilutions) was added (for F4/80 staining) or donkey anti-rabbit AF 488 and donkey anti-mouse AF 555 (A21206 and A31570, respectively; Invitrogen; 1:500 dilutions) were added (for CYP46A1 and CD31 staining, respectively) and incubated for 24 hours at 4°C. Retinal flat mounts were washed for 1 hour at 4°C with four changes of PBS and then mounted with either ProLong Gold without DAPI (for F4/80 staining) or VECTASHIELD mounting media with DAPI (H-1200; Vector Laboratories, Inc., Burlingame, CA) (for CYP46A1 and CD31 staining). The F4/80 staining was imaged on an inverted microscope (DMI 6000 B; Leica Microsystems, Buffalo Grove, IL) using a Retiga EXi-Fast camera (QImaging, Surrey, BC, Canada), and the CYP46A1 and CD32 staining was imaged on a Nikon Eclipse Ti with C2+ confocal system (Nikon Instruments Inc., Melville, NY).
Retinal Phosphoproteomics
Retinal processing was the same as for the brain42 and used three Cyp46a1−/− and three Cyp46a1+/+ samples, each containing two retinas from the same animal, an 8- to 10-month–old female mouse. All chemicals were from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. Briefly, each sample was lysed in 0.5 mL of 20 mmol/L HEPES buffer, pH 8.0, containing 9 mol/L urea and 1× Halt phosphatase inhibitor cocktail (ThermoFisher Scientific, Waltham, MA) and alkylated with 30 mmol/L iodoacetamide after reduction with 10 mmol/L dithiothreitol. A total of 0.25 mg of protein from each sample was digested with trypsin and subjected to phosphoserine and phosphothreonine enrichment using the ThermoFisher Scientific Pierce TiO2 Phosphopeptide Enrichment and Clean-up Kit. Enriched peptide samples were subjected to cleanup by C18 cartridges (Waters Corp., Milford, MA) and were analyzed by a Finnigan LTQ-Obitrap Elite hybrid mass spectrometer (ThermoFisher Scientific) connected to a Dionex Acclaim Pepmap C18 reversed-phase capillary chromatography column (ThermoFisher Scientific). All collision-induced dissociation spectra obtained were searched for the mouse reference sequence using Sequest that is bundled in Proteome Discoverer 1.4 (ThermoFisher Scientific); oxidized methionine, along with the serine, threonine, and tyrosine phosphorylation, was considered as a dynamic modification. Carbamidomethylation of cysteine residues was considered as a static modification. The resulting search results were filtered on the basis of Sequest XCorr scores >1.5 (+1), 2.0 (+2), 2.25 (+3), and 2.5 (+4). The quantitation of the phosphorylated peptides was by a label-free method, which involves aligning the liquid chromatography runs and comparing the extracted ion chromatograms for each identified peptide by SIEVE (ThermoFisher Scientific). Data were normalized to the total ion chromatogram. All differentially expressed phosphopeptides were manually validated using a second normalization step to the synthetic phosphopeptides added as internal standards. Differentially abundant phosphorylated proteins in the Cyp46a1−/− retina were analyzed manually using Molecular function and Biological process annotations of UniProt (http://www.uniprot.org, last accessed July 12, 2018).
Statistical Analysis
The quantitative data represent either the means ± SEM or the means ± SD; the number of animals (n) is indicated in each figure or figure legend. All images are representative of studies in three to five animals per genotype. Data were analyzed by either a two-tailed, unpaired t-test or the two-way analysis of variance with Bonferroni correction. P ≤ 0.05 was considered statistically significant.
Results
Retinal and Serum Lipids in Cyp46a1−/− Mice
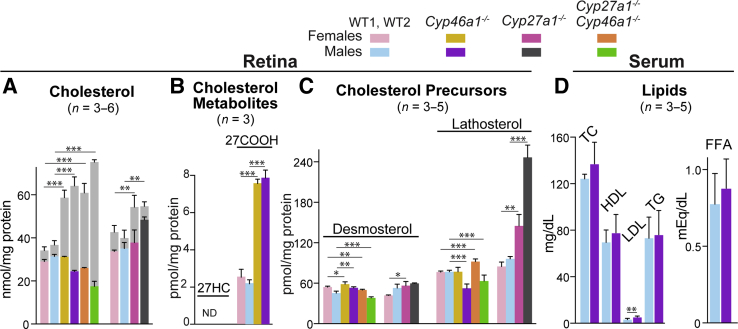
Retinal samples for sterol quantifications were composed of both the neural retina and RPE. Total retinal cholesterol (a sum of UC and EC) was increased 1.7- and 1.8-fold in male and female Cyp46a1−/− mice, respectively, whereas UC either remained unchanged (Cyp46a1−/− female mice) or was reduced 1.2-fold (Cyp46a1−/− male mice) (Figure 1A), suggesting that cholesterol excess in the Cyp46a1−/− genotype was converted to EC, as was previously observed in the Cyp27a1−/−Cyp46a1−/− genotype and Cyp27a1−/− female mice.4, 10 In addition, a lack of metabolic ability of the Cyp46a1−/− retina to convert cholesterol to 24HC led to a compensatory increase in retinal cholesterol elimination via 27-hydroxylation by CYP27A1. However, 27HC was not detected in the Cyp46a1−/− retina; instead, there was a 3.5-fold increase in 5-cholestenoic acid (27COOH), a metabolic product generated from 27HC by CYP27A1 (Figure 1B). Retinal cholesterol biosynthesis was affected only slightly in both neurons and astrocytes of Cyp46a1−/− mice, as indicated by the levels of cholesterol precursors lathosterol and desmosterol, respectively. Yet, a 1.5-fold reduction of lathosterol was observed in Cyp46a1−/− male mice (Figure 1C). Serum lipids were mostly unchanged in Cyp46a1−/− mice, except a 1.8-fold increase in low-density lipoprotein cholesterol, whose fraction in mice is small compared with high-density lipoprotein cholesterol (Figure 1D). Collectively, these data support that changes in total cholesterol in the Cyp46a1−/− retina reflect disturbances in local retinal cholesterol homeostasis rather than in systemic lipids.
Figure 1.
Retinal and serum lipids in mice of different genotypes. A–C: Colored bars indicate unesterified sterols in female and male mice of different genotypes; gray bars indicate esterified cholesterol in animals of both sexes. D: Blue and ink bars indicate serum lipids in wild-type (WT) and Cyp46a1−/− male mice. The results are the measurements in samples from individual animals (n), whose number is indicated in parenthesis on each panel. Only 5-cholestenoic acid (27COOH) was measured in pooled samples of 8 to 10 retinas from four to five mice; the results are triplicate measurements. Cyp46a1−/− and Cyp27a1−/−Cyp46a1−/− mice were on a background (C57BL/6J;129S6/SvEv) different from that of Cyp27a1−/− mice (C57BL/6J); hence, sterols were measured in WT animals on two different backgrounds, WT1 and WT2. Data are expressed as means ± SD. ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001 versus WT mice of the same sex (two-tailed, unpaired t-test). FFA, free fatty acid; HDL, high-density lipoprotein, LDL, low-density lipoprotein; ND, not determined; 27-OH, 27-hydroxycholesterol; TC, total cholesterol; TG, triglyceride.
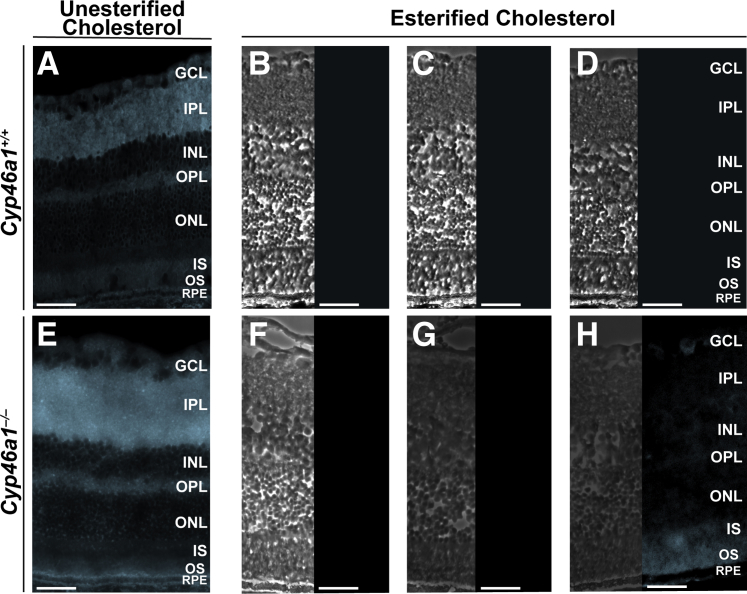
Retinal distribution of UC and EC in Cyp46a1−/− mice was investigated by staining of retinal cross sections with a fluorescent compound filipin. The staining pattern for UC was similar in wild-type and Cyp46a1−/− mice (Figure 2); the only difference was a diffuse filipin fluorescence observed within the RPE layer in Cyp46a1−/− animals. EC was not detectable in the retina of wild-type mice (Figure 2), whereas in Cyp46a1−/− mice, it was mainly localized to the photoreceptors and basal membrane of the RPE (Figure 2). Compared with the Cyp27a1−/−Cyp46a1−/− retina,43 there was an additional accumulation of EC in the inner segments of Cyp46a1−/− mice. Conversely, the inner Cyp46a1−/− retina had only a faint filipin fluorescence for EC.
Figure 2.
Retinal detection of unesterified and esterified cholesterol with filipin. Representative images are shown. A and E: Stains (cyan) for unesterified cholesterol (these sections were treated with filipin). B and F: Control stains for completeness of removal of unesterified cholesterol (these sections were extracted with 70% aqueous ethanol and treated then with filipin). C and G: Control stains for background fluorescence (these sections were extracted with ethanol but not treated with filipin). D and H: Stains (cyan) for esterified cholesterol (these sections were extracted with 70% ethanol and then sequentially treated with cholesterol esterase and filipin). B–D and F–H: Panels for esterified cholesterol consist of phase contrast images (left panels) and histochemistry images (right panels). n = 3 (A–H). Scale bars = 50 μm (A–H). GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS, photoreceptor inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; OS, photoreceptor outer segment; RPE, retinal pigment epithelium.
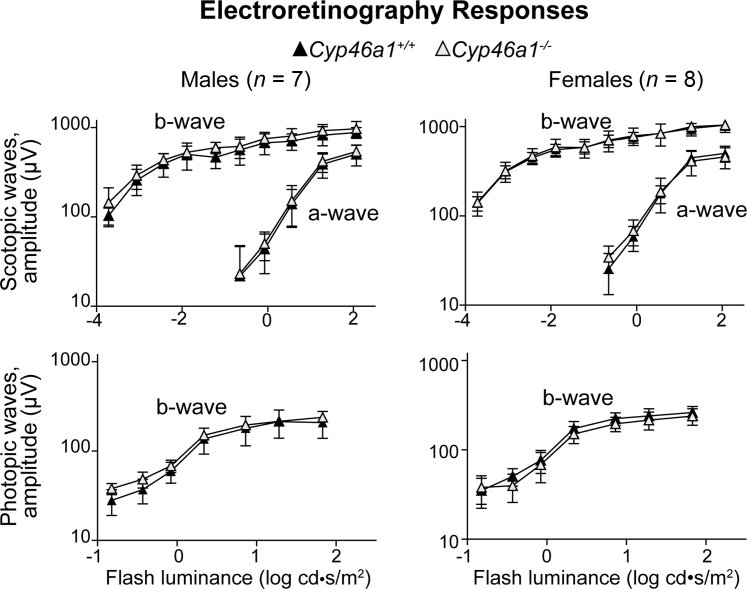
Overall Retinal Function in Cyp46a1−/− Mice
ERGs were recorded under dark- and light-adapted conditions from female and male mice (Figure 3). Neither dark-adapted (scotopic) ERGs nor light-adapted (photopic) ERGs of Cyp46a1−/− mice were significantly different from those of Cyp46a1+/+ mice. This is different from Cyp27a1−/−Cyp46a1−/− mice, which at the same age had sex-specific amplitude reductions in both scotopic and photopic ERGs.43
Figure 3.
Electroretinography responses in 6-month–old Cyp46a1−/− mice. The results are the measurements in seven male and eight female mice. Analysis of variance with repeated measures was used for statistical analysis, and no significant changes (P ≤ 0.05) were found in Cyp46a1−/− mice compared with Cyp46a1+/+ mice. Data are expressed as means ± SEM.
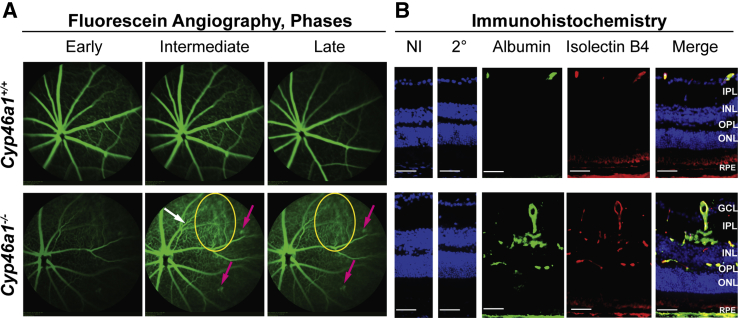
Retinal Vasculature in Cyp46a1−/− Mice
The retinal vasculature was evaluated using FA, an in vivo imaging technique. The beam of the laser scanning ophthalmoscope was focused on the superficial/intermediate plexi of the retinal vasculature to detect retinal capillaries, and fundus images were acquired during early, intermediate, and late phases of FA (Figure 4A). Initial assessments were conducted on 6-week–old mice, and no vascular abnormalities were detected (data not shown). However, when mice were reevaluated at 6 months of age or later, retinal venous beading and tortuosity, typical features of diabetic retinopathy in humans, were observed.1, 3 In addition, bright fluorescent spots were seen, which were particularly visible during intermediate and late FA stages; the appearance of these spots was similar to that of capillary microaneuryms. Furthermore, diffuse margins of retinal capillaries during intermediate and late FA stages were indicative of a marked increase in vascular permeability. All of the observed abnormalities were present in the Cyp27a1−/−Cyp46a1−/− retina,43 but in Cyp46a1−/− mice, these abnormalities were less prominent than in the double-knockout animals.
Figure 4.
Vascular abnormalities and vascular leakage in the Cyp46a1−/− retina, as assessed by fluorescein angiography and immunohistochemistry. A: Representative images of blood vessel tortuosity (white arrow), microaneurysm-like fluorescent spots (pink arrows), and area of vascular leakage (yellow ovals). B: Representative images of increased vascular permeability, as assessed by stains for albumin (green) and isolectin B4 (red). Nuclei were stained with DAPI (blue). n = 3 (A and B). Scale bars = 50 μm (B). 2°, Control stain using secondary antibody only; GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS, photoreceptor inner segment; NI, control stain using nonimmune serum; ONL, outer nuclear layer; OPL, outer plexiform layer; OS, photoreceptor outer segment; RPE, retinal pigment epithelium.
Increased vasopermeability in the Cyp46a1−/− retina was further confirmed by staining for albumin, normally remaining restricted within blood vessels.44 Yet, retinal vessels become permeable to small molecules in diabetic retinopathy, thus enabling lipids and proteins, such as albumin, to leak into surrounding tissues.44 Albumin extravasation from both large and small blood vessels was detected in the Cyp46a1−/− retina, stained also with isolectin B4 to delineate blood vessels (Figure 4B). By contrast, the Cyp46a+/+ retina had the anti-albumin immunoreactivity only within the large blood vessels of the ganglion cell layer with no detectable albumin staining in the inner nuclear layer and outer plexiform layer containing retinal capillaries.
Serum Glucose and Glucose Tolerance in Cyp46a1−/− Mice
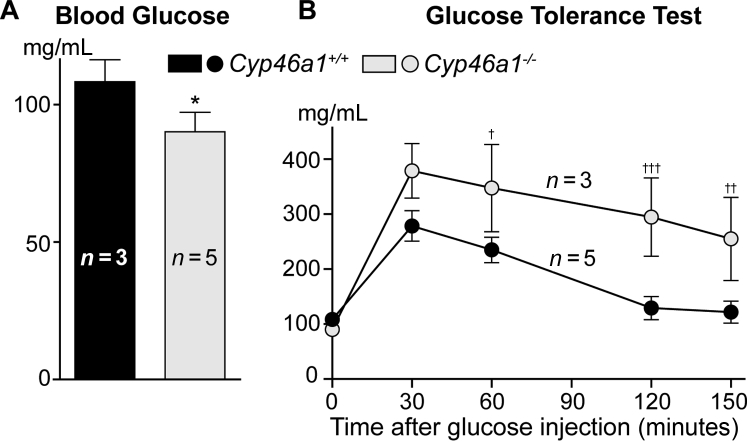
The fasting blood glucose levels were slightly decreased in Cyp46a1−/− mice compared with Cyp46a1+/+ mice (Figure 5A). However, during a glucose tolerance test, the sugar levels were always higher in Cyp46a1−/− mice (Figure 5B), even 150 minutes after injection, the time when in wild-type mice, serum glucose returned to the preinjection level (Figure 5B). Thus, the Cyp46a1−/− genotype seems to have delayed clearance of glucose.
Figure 5.
Fasting blood glucose and glucose tolerance test in Cyp46a1−/− mice. The results are the measurements in samples from individual animals (n), whose number is indicated on each panel. Data are expressed as means ± SEM (A and B). ∗P ≤ 0.05 versus Cyp46a1+/+ mice (two-tailed, unpaired t-test); †P ≤ 0.05, ††P ≤ 0.01, and †††P ≤ 0.001 versus Cyp46a1+/+ mice (two-way analysis of variance, followed by the Bonferroni correction).
Retinal Gene Expression in Cyp46a1−/− Mice
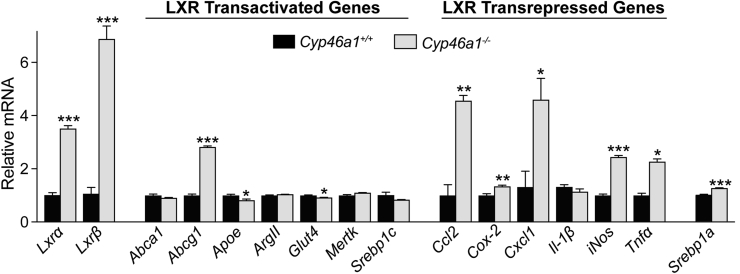
Mainly LXRs and their target genes were evaluated. The expression of Lxrα and Lxrβ was significantly up-regulated (up to sevenfold) in the Cyp46a1−/− retina compared with the wild-type retina (Figure 6). Nevertheless, there was a substantial up-regulation of only one (Abcg1) of the seven tested genes, which are normally transactivated by LXRs (Figure 6). The expression of the other transactivated targets was either unchanged (Abca1, ArgII, and Mertk) or slightly decreased (Apoe, Glut4, and Srebpc1, up to 1.2-fold) in the Cyp46a1−/− retina. Conversely, the expression of the genes that are normally suppressed by LXR activation via the inhibition of NF-κB (Ccl2, Cox2, Cxcl1, iNos, and Tnfa) was mostly increased in the Cyp46a1−/− retina compared with the wild-type retina (up to 4.6-fold) (Figure 6). Thus, a lack of 24HC and increased Lxr expression in the Cyp46a1−/− retina did not seem to affect the LXR transactivating activities but decreased the LXR transrepressing activities and thereby up-regulated the expression of key proinflammatory genes. The latter is consistent with the marked increase in vascular permeability observed in the Cyp46a1−/− retina (Figure 4B).
Figure 6.
Retinal expression of Lxrs and LXR target genes in the Cyp46a−/− retina. The results are triplicate measurements in a pooled sample of 8 to 10 retinas from four to five mice. Data are expressed as means ± SEM. ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001 versus the Cyp46a1+/+ retina (two-tailed, unpaired t-test).
NF-κB regulates the expression of not only proinflammatory genes (Figure 6) but also of many other genes, including Srebp1a, a potent activator of all SREBP-responsive genes,45 and of Nlrp1a, which encodes a core inflammasome component.46 The expression of Srebp1a was increased only 1.25-fold in the Cyp46a1−/− retina (Figure 6), suggesting that retinal inflammasome complex as well as retinal expression of the canonical SREBP target genes (the pathways of cholesterol and fatty acid synthesis) were not significantly affected by a lack of 24HC.
Basal Gene Expression in Macrophages
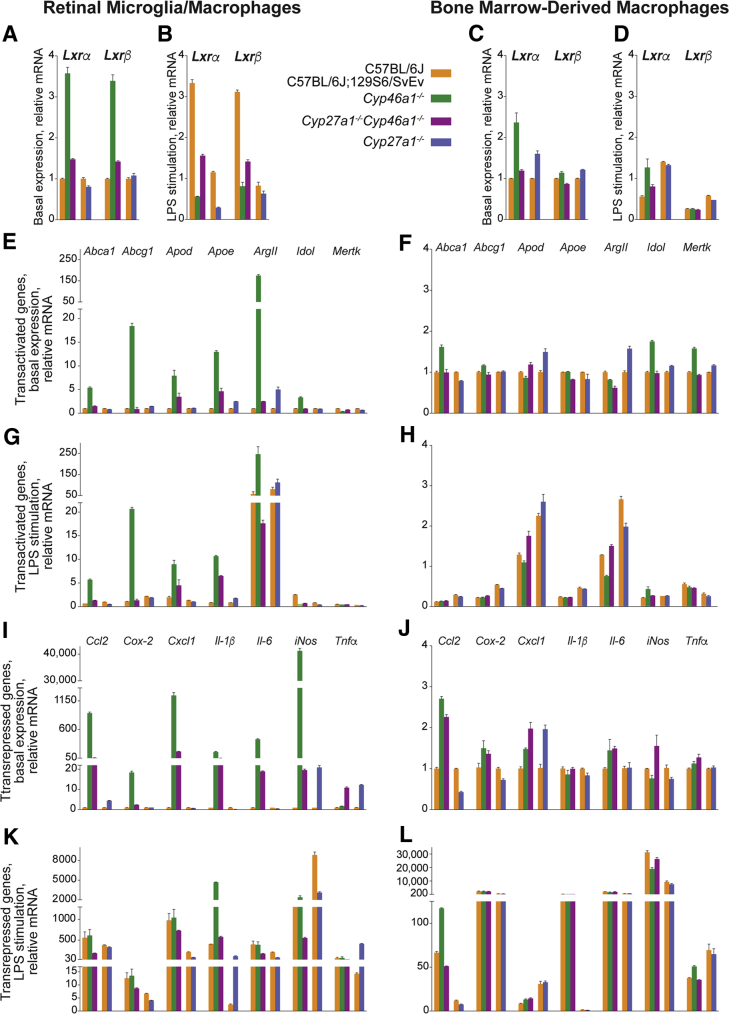
The up-regulated expression in the Cyp46a1−/− retina of some of the inflammatory genes prompted studies of the gene expression in RMM, innate immune cells either known (microglia) or suggested (macrophages from infiltrating monocytes) to be activated in diabetes in both humans and animal models.47, 48, 49, 50 Macrophages are the most studied cells with respect to the LXR actions on immunity and were the first cell types in which the anti-inflammatory effects of LXRs were recognized.28, 51 To ascertain whether retinal environment and knockout genotypes have specific effects on the gene expression in microglia/macrophages, both RMM and BMDM from five genetic lines were evaluated: two wild-type strains (C57BL/6J and C57BL/6J;129S6/SvEv) and three knockout genotypes (Cyp46a1−/−, Cyp27a1−/−, and Cyp27a1−/−Cyp46a1−/−) (Figure 7).
Figure 7.
Expression of Lxrs and LXR target genes in mice of different genotypes. A, B, E, G, I, and K: Retinal microglia/macrophages. C, D, F, H, J, and L: Bone marrow–derived macrophages. The results are duplicate measurements in two to three different cell preparations. A, C, E, F, I, and J: Nonstimulated cells. B, D, G, H, K, and L: LPS-stimulated cells. A two-tailed, unpaired t-test was used to assess statistical significance in knockout genotypes relative to the corresponding wild-type cells; LPS-stimulated cells were also compared for statistically significant changes to nonstimulated cells (basal expression). All statistical significance data are summarized in Supplemental Tables S1 and S2. Data are expressed as means ± SEM.
Lxrs
In the Cyp46a1−/− genotype, RMM exhibited increased Lxrα and Lxrβ expression (3.6- and 3.4-fold, respectively). A lower (1.3-fold, Lxrα) or unchanged gene expression (Lxrβ) was found in the RMM from the Cyp27a1−/− genotype (Figure 7A). Accordingly, in the Cyp27a1−/−Cyp46a1−/− RMM, the Lxr expression was increased 1.5-fold for Lxrα and 1.4-fold for Lxrβ, representing a smaller increase than in the Cyp46a1−/− RMM. In BMDM, the Lxr expression was increased in both Cyp46a1−/− and Cyp27a1−/− genotypes, with increases being higher for the α isoform (up to 2.4-fold) than the β isoform (up to 1.2-fold) and for the Cyp46a1−/− genotype than the Cyp27a1−/− genotype (Figure 7C). In the Cyp27a1−/−Cyp46a1−/− BMDM, the Lxr expression was isoform specific, with a slight (1.2-fold) increase in the Lxrα expression and essentially unchanged Lxrβ expression. Thus, the Cyp46a1−/− and Cyp27a1−/− genotypes demonstrated different effects on the Lxr expression in RMM but not BMDM, and changes in the expression of Lxrα were more pronounced in RMM than BMDM.
Genes that Are Transactivated by LXRs
In RMM, all of the tested genes, except Mertk, were up-regulated in the Cyp46a1−/− genotype (up to 175-fold, ArgII) (Figure 7E). In contrast, in the Cyp27a1−/− RMM, changes in the gene expression were highly variable (ie, gene specific), with the gene expression being unchanged (Apod and Idol), decreased (Abca1 and Mertk, up to 1.4-fold), or increased (Abcg1, Apoe, and ArgII, up to fivefold). Consequently, in the Cyp27a1−/−Cyp46a1−/− RMM, the gene expression was mostly increased (up to 5.3-fold, Apoe) with the exception of Idol, which had an unchanged expression, and Merk, whose expression was decreased 1.3-fold. Similar to the Cyp46a1−/− RMM, the Cyp46a1−/− BMDM had an up-regulated expression of more than half of the tested genes (four of seven), yet the extent of up-regulation was much smaller, only up to 1.8-fold (Figure 7F). Also, some of the genes that showed increased expression in the Cyp46a1−/− RMM (Apod, Apoe, and ArgII) were not up-regulated in the Cyp46a1−/− BMDM and vice versa (Mertk). In the Cyp27a1−/− BMDM, the gene expression was unchanged (Abcg1 and Apoe), decreased (Abca1, up to 1.3-fold), or increased (Apod, ArgII, Idol, and Mertk, up to 1.6-fold), like in the Cyp27a1−/− RMM, except the genes that showed the same direction of a change did not completely overlap in the Cyp27a1−/− RMM and Cyp27a1−/− BMDM. The latter likely led to a different pattern of gene changes in the Cyp27a1−/−Cyp46a1−/− BMDM relative to the Cyp27a1−/−Cyp46a1−/− RMM: in the former, only one gene was slightly (1.2-fold) up-regulated (Apod), and the expression of the remaining genes was either unchanged (Abc1, Abcg1, and Idol) or decreased (Apoe, ArgII, and Mertk). Thus, changes in the gene expression were not identical in RMM and BMDM; however, in both, the Cyp46a1−/− genotype up-regulated the expression of the LXR transactivated gene, although to a different extent. A higher gene up-regulation in the Cyp46a1−/− RMM than Cyp46a1−/− BMDM could be due to a higher up-regulation of the Lxr expression in the Cyp46a1−/− RMM than Cyp46a1−/− BMDM. Also possible, in both Cyp46a1−/− RMM and Cyp46a1−/− BMDM, there were oxysterol ligands (likely 27HC and/or 27COOH) that compensated for a lack of 24HC. In contrast, in both Cyp27a1−/− RMM and Cyp27a1−/− BMDM, the effect of the knockout genotype was gene specific.
Genes that Are Transrepressed by LXRs
In the Cyp46a1−/− RMM, all of the tested genes were up-regulated (up to 41,199-fold, iNos), whereas in the Cyp27a1−/− RMM, only three genes (Ccl2, iNos, and Tnfa) had an increased expression (up to 21-fold) (Figure 7I). The levels of the remaining four genes were either unchanged (Cox-2) or decreased (Cxcl1, Il-1β, and Il-6, up to fivefold). Nevertheless, in the Cyp27a1−/−Cyp46a1−/− RMM, all of the tested genes were up-regulated as well (up to 176-fold, Cxcl1), as observed in the Cyp46a1−/− genotype. In BMDM, the pattern of changes in the gene expression was in general similar to that in RMM, with an increase in the gene expression in the Cyp46a1−/− genotype and gene-specific responses in the Cyp27a1−/− genotype (Figure 7J). The differences from RMM were both the smaller number of up-regulated genes and low magnitude of gene up-regulation. Indeed, only four genes in the Cyp46a1−/− BMDM (Ccl2, Cox-2, Cxcl1, and Il-6), one gene in the Cyp27a1−/− BMDM (Cxcl1), and five genes in the Cyp27a1−/−Cyp46a1−/− BMDM (Ccl2, Cox-2, Cxcl1, Il-6, and iNos) had an increased expression ranging from 1.3-fold (Tnfa in the Cyp27a1−/−Cyp46a1−/− BMDM) to 2.7-fold (Ccl2 in the Cyp46a1−/− BMDM). Thus, within each knockout genotype, the pattern of changes in the expression of the LXR transrepressed genes was similar in RMM and BMDM and was similar to that of the LXR transactivated genes. The latter was unexpected because normally, LXR activation has opposite effects on the expression of the transactivated and transrepressed genes.52 Yet, this was not the case for the Cyp46a1−/− RMM and BMDM, suggesting that in the Cyp46a1−/− genotype, LXR-mediated gene transrepression is dissociated from LXR-mediated gene transactivation.
Gene Expression in the LPS-Activated Macrophages
Studies have shown that in macrophages with unchanged LXR expression, the induction of the LXR transactivated genes is profoundly inhibited by bacterial and viral pathogens as well as their components, which act through toll-like receptors 3 and 4 and an interferon regulatory factor 3–dependent pathway.52, 53 Conversely, signaling through toll-like receptor 4 or receptors for Il-1β and Tnfa is attenuated by cell or animal treatments with synthetic LXR ligands or genetic manipulations to achieve a constitutively active LXR.28, 54, 55 Hence, it was tested whether a lack of CYP46A1 or/and CYP27A1 affected LXR activity in cultured macrophages activated by LPS, a toll-like receptor 4 ligand.56 Data analysis included a comparison of the gene expression in stimulated cells of the wild-type genotypes versus that in the corresponding nonstimulated cells (Supplemental Tables S1 and S2). Also, stimulated cells of the knockout genotypes were compared with the stimulated wild-type cells and then with the corresponding nonstimulated cells.
Lxrs
The levels of Lxrα and Lxrβ were increased greater than threefold in the stimulated wild-type C57BL/6J;129S6/SvEv RMM and essentially unchanged (<1.2-fold difference in the gene expression) in the stimulated wild-type C57BL/6J RMM (Figure 7B). Nevertheless, the stimulated RMM of the Cyp46a1−/− and Cyp27a1−/− genotypes showed a decrease in the Lxr expression relative to that in the stimulated wild-type cells (up to 5.5-fold) as well as the corresponding nonstimulated cells (up to sixfold). The effect of LPS on the Lxr expression in BMDM was isoform specific (Figure 7D). In both stimulated wild-type and knockout cells, Lxrβ but not Lxrα showed a uniform down-regulation of the expression relative to that in the corresponding nonstimulated cell. Thus, compared with the Lxr expression in the stimulated wild-type cells, only in the stimulated RMM but not BMDM, the Cyp46a1−/− and Cyp27a1−/− genotypes had a clear down-regulating effect on the gene expression.
Genes that Are Transactivated by LXRs
The effect of LPS treatment of RMM from wild-type C57BL/6J;129S6/SvEv mice was gene specific (Figure 7G). The gene expression was unchanged (Abcg1 and Apoe), decreased (up to twofold, Abca1 and Merk), or increased [moderately (up to 2.5-fold, Apod and Idol) or substantially (58-fold, ArgII)]. Gene-specific responses were also observed in the stimulated RMM from wild-type C57BL/6J mice. Yet, in the stimulated Cyp46a1−/− and Cyp46a1−/−Cyp27a1−/− RMM, the LPS effect was more uniform [ie, mostly led to an increase in the gene expression (up to 18.8-fold, Abcg1) relative to that in the stimulated wild-type cells and comparable gene expression (within a 1.5-fold change) relative to that in the corresponding nonstimulated cells]. The two exceptions were Idol and Mertk. In contrast, the gene expression in the stimulated Cyp27a1−/− RMM was moderately decreased (up to twofold, Abca1 and Idol) relative to that in the stimulated wild-type cells; however, it was comparable to that in the corresponding nonstimulated cells. The two exceptions were Apoe and ArgII. In BMDM, all of the genotypes responded to the LPS treatment similarly [namely, had a canonical down-regulation of the gene expression (up to 10-fold, Abca1) relative to that in the corresponding nonstimulated cells] (Figure 7H). Notably, this down-regulated gene expression was comparable between wild-type and knockout gene expression. The two exceptions were Apod and ArgII. Thus, in the stimulated RMM, the Cyp46a1−/− and Cyp27a1−/− genotypes mostly had opposite, up- and down-regulating effects, respectively, on the expression of the transactivated genes relative to that in the stimulated wild-type cells. Apparently, the Cyp46a1−/− genotype ablated the suppressing LPS effect on the LXR transactivating activities. In contrast, the Cyp27a1−/− genotype retained this effect; despite stimulated RMM, it had the Lxr expression significantly down-regulated in both knockout genotypes compared with that in the stimulated wild-type cells. In the stimulated BMDM, the knockout genotype effect cannot be assessed because it could be masked by a significant reduction in the expression of most of the tested genes.
Genes that Are Transrepressed by LXRs
In RMM from both wild-type genotypes, the LPS stimulation led to an expected substantial up-regulation of all of the studied genes: up to 1860-fold (iNos) in the C57BL/6J;129S6/SvEv cells and up to 378-fold (Ccl2) in the C57BL/6J cells (Figure 7K). Similarly, an up-regulated gene expression (comparable to that in the stimulated wild-type RMM and the corresponding nonstimulated cells) was also observed in the stimulated Cyp46a1−/− RMM. The three exceptions were Il-1β, iNos, and Tnfa. In the stimulated Cyp27a1−/− and Cyp46a1−/−Cyp27a1−/− RMM, the gene expression was either unchanged or lower (up to threefold, Ccl2) than that in the stimulated wild-type RMM and much higher (up to 120-fold, Cxcl1) than the gene expression in the corresponding nonstimulated cells. The exceptions in these genotypes were Il-1β and Tnfa. Like the stimulated wild-type RMM, the stimulated wild-type BDDM also had uniform increases in the gene expression: up to 31,303-fold (iNos) in the C57BL/6J;129S6/SvEv cells and up to 9335-fold (iNos) in the C57BL/6J cells (Figure 7L). Yet, there was no knockout genotype effect on the gene expression in the stimulated BMDM; the levels of most of the genes were comparable to that in the stimulated wild-type cells and much higher than the gene expression in the corresponding nonstimulated cells. The two exceptions were Ccl2 and iNos. Thus, in RMM and BMDM, the transrepressed genes in the knockout genotypes responded differently to the LPS stimulation. In the stimulated RMM, only the Cyp27a1−/− genotype had a down-regulating effect on the expression of the transrepressed genes relative to that in the stimulated wild-type cells. This suggests that the LXR activity that antagonizes gene transrepression was higher in the Cyp27a1−/− RMM than the Cyp46a1−/− RMM. In contrast, in the stimulated BMDM, there was no effect of the knockout genotype on the gene expression, which was always much higher than that in the corresponding nonstimulated cells; the knockout genotype effect in these cells could be masked by already significantly up-regulated gene expression in the stimulated wild-type cells.
RMM Activation in Vivo
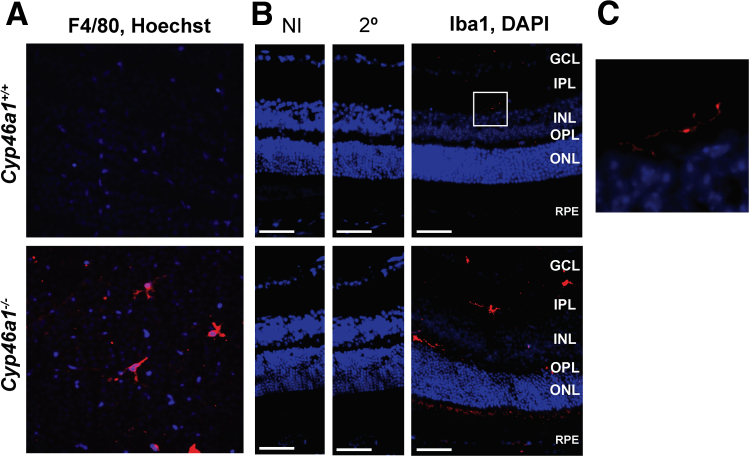
Retinal flat mounts and cross sections were stained for F4/80 and Iba1, respectively, markers for microglia/macrophage activation.57 The F4/80-positive cells were detected in the inner retina of Cyp46a1−/− mice but not anywhere in the wild-type retina (Figure 8A). The detected F4/80-positive cells had stout bodies and short processes, indicative of the activated state. Similarly, the Iba1-positive cells were clearly detected in retinal cross sections of Cyp46a1−/− mice (Figure 8B). Yet in wild-type mice (Figure 8C), there were only few Iba1-positive cells, thus suggesting that RMMs are activated in vivo in the Cyp46a1−/− genotype.
Figure 8.
Macrophage/microglia activation in the Cyp46a1−/− retina. A and B: Representative stains of retinal flat mounts for F4/80 (red; A) and retinal cross sections for Iba1 (red; B); nuclei were stained with Hoechst (blue) and DAPI (blue), respectively. C: Enlargement of the boxed area in B showing wild-type retina with Iba1-positive cells. n = 3 (A and B). Scale bars = 50 μm (B). Original magnification, ×200 (A). 2°, Control stain using secondary antibody only; GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; NI, control stain using nonimmune serum; ONL, outer nuclear layer; OPL, outer plexiform layer; RPE, retinal pigment epithelium.
Gene Expression in Retinal Endothelial Cells and RPE
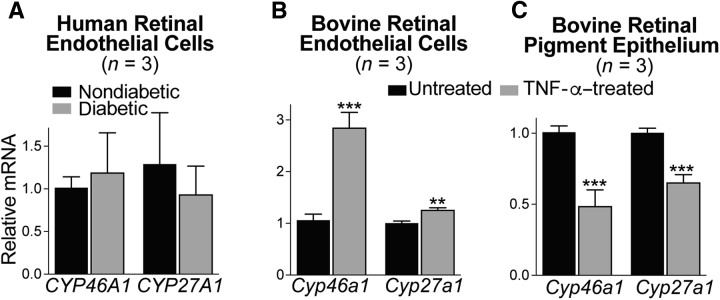
Both chronic inflammation and dyslipidemia contribute to vascular damage in diabetes,1, 58 consistent with the data on RMM activation (Figure 7), disturbance of retinal cholesterol homeostasis (Figure 1), and retinal vascular abnormalities in Cyp46a1−/− mice (Figure 4). In addition, retinal endothelial cells are involved, which are affected by the proinflammatory environment in the diabetic retina, and whose injury in diabetes plays a key role in retinal vascular degeneration.59 Hence, CYP46A1 gene expression was first assessed in primary HRECs from nondiabetic and diabetic donors and we also evaluated the expression of CYP27A1 (Figure 9), which is known to be abundant in human aortic endothelium.60 In both nondiabetic and diabetic donors, there was a significant interdonor variability in primary HRECs with respect to the Ct numbers for CYP46A1 and CYP27A1: the mean Ct for CYP46A1 was approximately 27.0, and for CYP27A1, approximately 30.0. These numbers suggest that the transcript abundance is intermediate for CYP46A1 and lower for CYP27A1. Yet, a high interdonor variability in Ct numbers for both CYPs precluded a conclusion about changes (if any) in HREC expression of CYP46A1 and CYP27A1 in diabetes. Hence, the Cyp expression was next evaluated in BRECs, which were stimulated by Tnfa to mimic the proinflammatory environment in the retina. In BRECs, the treatment with Tnfa up-regulated Cyp46a1 twofold and did not affect the expression of Cyp27a1.
Figure 9.
Cyp46a1 and Cyp27a1 expression in different cell types. The results are triplicate measurements of cells from three different donors (A) or three different cell preparations (B and C). Asterisks are significant changes relative to untreated cells (B and C), as assessed by a two-tailed, unpaired t-test. Data are expressed as means ± SEM. ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001 versus untreated cells.
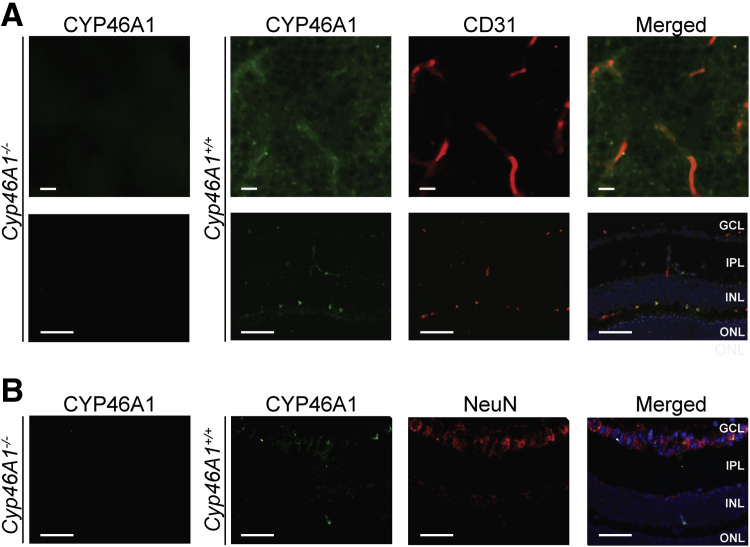
RPE, which expresses both CYP46A1 and CYP27A1,8, 11, 12 was of interest to us as well to gain insight into whether CYP46A1 and CYP27A1 expression is affected similarly in different cell types by the proinflammatory stimulus. In Tnfa-treated RPE, the Cyp46a1 expression was reduced greater than threefold, and the expression of Cyp27a1 was reduced 1.3-fold. Apparently, Tnfa-induced changes in the cytochrome P450 expression are P450 and cell specific. Finally, CYP46A1 immunolocalization was conducted in retinal flat mounts and cross sections, which were also stained for CD31, a marker of vascular endothelial cells, or neuronal-specific nuclear protein, a DNA-binding protein that identifies many populations of mature neurons.61, 62 In both flat mounts and cross sections, the immunoreactivity for CYP46A1 overlapped in part with that for CD31 (Figure 10A). There was also CYP46A1 immunoreactivity outside retinal blood vessels, which could reflect CYP46A1 expression in some of the retinal neurons. The latter was confirmed by double stains for CYP46A1 and neuronal-specific nuclear protein, which identified partial overlaps in immunoreactivity (Figure 10B). Thus, CYP46A1 seems to be expressed in some, but not all, retinal vascular endothelial cells as well as neurons. Neuronal CYP46A1 expression in the retina is consistent with that in the brain, where CYP46A1 was found to be distributed throughout the cell bodies and dendrites of multiple neurons.12
Figure 10.
Immunohistochemical localization of CYP46A1 in the retina. A: Representative stains of retinal flat mounts (top row) and cross sections (bottom row) for CYP46A1 (green) and CD31 (red). B: Representative stains of retinal cross sections for CYP46A1 (green) and neuronal-specific nuclear protein (NeuN; red). The Cyp46a1−/− retina was used as a negative control. Nuclei were stained with DAPI (blue). n = 3 (A and B). Scale bars: 10 μm (A, flat mounts); 20 μm (A and B, cross sections). GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer.
Retinal Phosphoproteomics in Cyp46a1−/− Mice
LXRα can be modified by phosphorylation, which affects its transcriptional activities.63, 64, 65, 66 Also, in Cyp46a1−/− mice, the phosphorylation of 146 proteins was altered in the brain.42 Hence, retinal phosphoproteome was studied. A total of 31 phosphopeptides from 30 distinct proteins showed a statistically significant difference in abundance between the Cyp46a1−/− and wild type retinas (Table 2). Of these peptides, 28 phosphopeptides from 27 proteins were more abundant in the knockout retina than the wild-type retina, including 12 phosphopeptides that were found only in the Cyp46a1−/− retina. Fewer phosphopeptides (only three from different proteins: Ras GTPase-activating protein-binding protein 1, putative RNA-binding protein Luc7-like 2, and dihydropyrimidinase-related protein 3) were more abundant in the Cyp46a1+/+ than Cyp46a1−/− retina. Two of the differentially phosphorylated retinal proteins were protein kinases: calcium/calmodulin-dependent protein kinase kinase 1 and brain-specific serine/threonine-protein kinase 2. The remaining proteins were pertinent to at least five general biological processes: i) information transfer (gene transcription and translation, mRNA binding, stability, and processing: 11 proteins); ii) neuritogenesis (six proteins); iii) cytoskeleton maintenance (five proteins); iv) membrane vesicle trafficking (three proteins); and v) signal transduction (three proteins).
Table 2.
Phosphoproteins with Different Phosphopeptide Abundance in the Cyp46a1−/− Retina Compared with the Wild-Type Retina
| Protein | Peptide sequence | Phosphorylation site | Peptide ratio, KO/WT |
|---|---|---|---|
| Information transfer (gene transcription and translation, mRNA binding, stability, and processing) | |||
| NOLC1 | ESEEEEEEEETEEK | S2 | 8.4 |
| HTATSF1 | EFEEDSDEKEEEGDDDEEEVVYER | S6 | 7.0 |
| PURA | GPGLGSTQGQTIALPAQGLIEFR | T7 | 6.6 |
| PABPC1L2B | DFDDDSDDEATFR | S6 | 5.1 |
| NUCKS1 | EMoLLEDVGSEEEPEEDDEAPFQEK | S9 | 4.3 |
| SRSF10 | SFDYNYR | S1 | 2.5 |
| SRRM2 | SPVPSAFSDQSR | S1 | 2.4 |
| YTHDC1 | GISPIVFDR | S3 | 2.3 |
| GTF2F1 | GTSRPGTPSAEAASTSSTLR | T2, T7 | 2.2 |
| G3BP1 | STSPAPADVAPAQEDLR | S3 | 0.5 |
| LUC7L2 | SEDRRSSEEREAGEI | S6, S7 | 0.3 |
| Neuritogenesis | |||
| GPRIN1 | VRPGSVLAAALAPQEATEPVR | S5 | KO only |
| PCLO | RFSLNLGGIADAPK | S3 | KO only |
| RTN4 | RGSGSVDETLFALPAASEPVIPSSAEK | S3, S5 | KO only |
| RP1L1 | RPASVECLPSVSVPYQVAQK | S4 | KO only |
| BRSK2 | SISGASSGLSTSPLSSPR | S1 | 6.8 |
| AQP4 | SQVETEDLILKPGVVHVIDIDRGEEK | S1 | 3.2 |
| Cytoskeleton maintenance | |||
| SLC9A1 | IGSDPLAYEPK | S3 | KO only |
| TJP1 (ZO-1) | DDISEIQSLASDHSGR | S11, S14 | KO only |
| MARCKS | ESGEGAEAEGATAEGAK | S2 | 11.1 |
| MAP1B | SPCDSGYSYETIEK | S1 | 6.1 |
| DPYSL3 | GGTPAGSTRGSPTRPNPPVR | T3, S7 S11 |
0.4 |
| Membrane vesicle trafficking | |||
| ARFGAP3 | LTNTSFTEIEK | S5 | KO only |
| SYNRG | QLSLEGAGLAMEEFK | S3 | 13.8 |
| TOM1L2 | GIEFPMoADLDALSPIHTPQR | S13, T17 | 4.5 |
| Signal transduction | |||
| CAMKK1 | SFGNPFEPQAR | S1 | KO only |
| PCP2 | VTVNSLPGFQPIGPK | S5 | KO only |
| PCP2 | AGSPDQEGFFNLLTHVQGDR | S3 | 3.0 |
| RAP1GAP | SSAIGIENIQEVQEK | S2 | KO only |
Involvement in only one biological process is indicated for each protein, although many proteins participate in more than one biological process.
AQP4, aquaporin-4 isoform M23A; ARFGAP3, ADP-ribosylation factor GTPase-activating protein 3; BRSK2, brain-specific serine/threonine-protein kinase 2; CAMKK1, calcium/calmodulin-dependent protein kinase kinase 1; DPYSL3, dihydropyrimidinase-related protein 3; G3BP1, Ras GTPase-activating protein-binding protein 1; GPRIN1, G-protein–regulated inducer of neurite outgrowth 1 isoform X1; GTF2F1, general transcription factor IIF subunit 1 isoform X1; HTATSF1, HIV Tat-specific factor 1 homolog; KO, Cyp46a1−/− retina; LUC7L2, putative RNA-binding protein Luc7-like 2; MAP1B, microtubule-associated protein 1B isoform X1; MARCKS, myristoylated alanine-rich C-kinase substrate; Mo, oxidized methionine; NOLC1, nucleolar and coiled-body phosphoprotein 1 isoform B; NUCKS1, nuclear ubiquitous casein and cyclin-dependent kinase substrate 1 isoform 1; PABPC1L2B, polyadenylate-binding protein 1–like 2; PCLO, protein piccolo isoform X4; PCP2, Purkinje cell protein 2 isoform 1; PURA, transcriptional activator protein Pur-α isoform X1; RAP1GAP, rap1 GTPase-activating protein 1 isoform 3; RP1L1, retinitis pigmentosa 1–like 1 protein; RTN4, reticulon-4 isoform A; SLC9A1, sodium/hydrogen exchanger 1 isoform X2; SRRM2, serine/arginine repetitive matrix protein 2; SRSF10, serine/arginine-rich splicing factor 10 isoform 2; SYNRG, synergin γ; TJP1, tight junction protein ZO-1 isoform 2; TOM1L2, target of Myb protein 1 isoform X1; WT, wild-type retina; YTHDC1, YTH domain-containing protein 1 isoform X6; ZO-1, zonula occludens 1.
Discussion
The present study led to several major findings. First, some of the features of early-stage diabetic retinopathy were observed in Cyp46a1−/− mice (Figure 4), which exhibited a slight decrease in fasting blood glucose levels but had delayed glucose clearance in a glucose tolerance test (Figure 5). In addition, Cyp46a1−/− mice showed up to a 1.8-fold increase in total retinal cholesterol (Figure 1A). Second, the Cyp46a1−/− retina had an up-regulated expression of both Lxrα/Lxrβ and several of the proinflammatory genes (Ccl2, Cox-2, Cxcl1, iNos, and Tnfa) that are normally suppressed by LXRs (Figure 6). Similarly, Lxrs and many of the proinflammatory genes (Ccl2, Cox-2, Cxcl1, Il-1β, Il-6, iNos, and Tnfa) were up-regulated in the Cyp46a1−/− RMM, suggesting a decrease in LXR-transrepressing activities (Figure 7I). Yet, the LXR-transactivating activities were increased in the Cyp46a1−/− RMM (Figure 7E) and affected both cholesterol-related genes (Abca1, Abcg1, Apod, Apoe, and Idol) and a gene of macrophage function (ArgII). Third, changes in the gene expression in the Cyp46a1−/− genotype were much more pronounced in the RMM than BMDM. Finally, CYP46A1 appears to be expressed in retinal vascular endothelial cells, and CYP46A1 gene expression is increased in proinflammatory conditions.
Recent evidence suggests that changes to retinal microglia are an early feature of diabetic retinopathy.48 Microglia are also altered in experimental models of diabetic retinopathy.67 Yet, the causes and consequences of microglial changes in diabetic retinopathy are not fully understood. The causes might be retinal formation of molecules produced as a result of hyperglycemia48, 68 and/or a systemic low-grade inflammation present chronically in diabetes.1, 69 The consequences could be neuronal death48, 70 and retinal vascular dysfunction.59, 67 The latter was inferred from the findings that microglia play an important role in formation and maintenance of the retinal vasculature71 and are present in diabetic retinas near dilated veins, microaneurysms, vitreal neovascularizations, hemorrhages, and cotton-wool spots.47 Activated microglia produce cytokines and thus can contribute to vascular and neural damage in diabetic retina.59 Cyp46a1−/− mice have changes to both, RMM and retinal vasculature, and hence support the notion that microglia (and infiltrating macrophages) contribute to retinal vascular dysfunction. In addition, Cyp46a1−/− mice represent an interesting model of RMM activation (Figures 7 and 8) when there is no serum hyperglycemia (Figure 5). Physically, Cyp46a1−/− mice are outwardly normal,31, 33 likely because tissue expression of CYP46A1 is largely confined to the brain and retina.12, 33, 72
There are probably several causes of RMM activation in Cyp46a1−/− mice: i) a significantly increased retinal cholesterol content and/or a lack of retinal 24HC; ii) an increased Lxr expression in both, the whole retina and RMM, assuming this increase enhances LXR activation by oxysterols; and iii) distinct roles of 24- and 27-oxysterols in LXR transactivation and transrepression in RMM. Indeed, normally, microglia are actively monitoring the retinal microenvironment and may be activated in Cyp46a1−/− mice by the increased cholesterol content of the retina (Figure 1A). In addition, the Cyp46a1−/− RMM might be activated by a lack of 24HC. This would be similar to retinal glial cells, shown to be activated by a decrease in retinal 24HC as a result of pharmacologic CYP46A1 inhibition with voriconazole.73 Atypical (nonneuronal) CYP46A1 expression in brain astrocytes and microglia seems to be a common feature of various pathologic conditions in the brain.12, 74, 75, 76, 77 It is possible that the retina could have a similar feature, and RMM activation in response to different stimuli (eg, disturbed sterol content) triggers CYP46A1 expression in these cells. In the Cyp46a1−/− RMM, LXR-transactivating activity (and the corresponding gene expression) could be increased because of increased Lxr expression and/or the presence of the activating ligands other than 24HC (eg, desmosterol, 27OH, 27COOH, and/or other oxysterols). LXR-transrepressing activity might be decreased because 24HC, the major activating ligand, is absent, and/or LXR activation by 27HC, 27COOH, and/or other sterols does not suppress the inflammatory gene expression.
Different effects of oxysterol/sterol ligands on LXR transactivation and transrepression in the RMM explain the dissociation of the two LXR activities in these cell types and are supported by the gene expression pattern in the knockout genotypes. A much lower expression of the LXR-transactivated genes in the Cyp27a1−/−Cyp46a1−/− RMM than Cyp46a1−/− RMM suggests an important contribution of 27HC or 27COOH to the RMM LXR transactivation; unchanged or increased gene expression in the Cyp27a1−/− RMM highlights the involvement in this process of 24HC and perhaps other ligands (Figure 7E). Conversely, 27HC or 27COOH in the Cyp46a1−/− RMM was generally not sufficient for LXR transrepression, emphasizing the importance of 24HC (and possibly other ligands) for the suppression of inflammatory genes in the activated RMM. The 24HC importance is also supported by unchanged (or even decreased) expression of Cox-2, Cxcl1, Il-1β, and Il-6 but not Ccl2, iNos, and Tnfa in the Cyp27a1−/− RMM (Figure 7I). Thus, 27HC, 27COOH, 24HC, and possibly other sterols/oxysterols appear to be important for LXR transactivation, whereas 24HC (but not 27HC and 27COOH) seems to be important for LXR transrepression. This explanation is consistent with previous studies showing that 25HC and 27HC act in thioglycolate-elicited macrophages as transactivators only, whereas 22R-hydroxycholesterol, 24,25-epicholesterol, and 24HC induce both LXR transactivation and transrepression.26 Moderate changes in the expression of LXR-dependent genes in BMDM (Figure 7, F and J) suggest that either CYP46A1 and CYP27A1 are not expressed in these cells or oxysterols produced by these P450s are not involved in the LXR activation in BMDM. Apparently, the retina is much more sensitive to a lack of CYP46A1 than the bone marrow, thus establishing the importance of CYP46A1 for the retina, in addition to enzyme roles in brain cholesterol maintenance and higher-order brain functions.78
A significant up-regulation of Lxrα and Lxrβ in the whole Cyp46a1−/− retina (Figure 6) and RMM (Figure 7A) is an unexpected finding. Lxrα was also found to be up-regulated in the brain of Cyp46a1−/−.42 Transcriptional regulation of LXRs includes Lxrα autoregulation79 and regulation by fatty acids,80 thyroid hormone,81 certain cytokines (TNF-α and Il-1β),82, 83 and interferon-γ.83 In addition, Lxrα can be alternatively spliced, leading to the production of three LXRα isoforms (LXRα1 to 3), with LXRα2 having a reduced transcriptional activity and LXRα3 being transcriptionally inactive.84 LXRβ could also be alternatively spliced and produce the LXRBSV variant, which selectively enhances LXRβ-mediated transactivation.85 The expression of Tnfa and Il-1β was increased in the Cyp46a1−/− RMM (Figure 7I), but it is not clear whether this increase is the cause or consequence of Lxrα up-regulation. Also, this increase does not explain the up-regulation of Lxrβ. Remarkably, studies of the retinal Cyp46a1−/− phosphoproteomics identified altered phosphorylation of 11 different proteins (the largest number of proteins per functional group) involved in transmission of genetic information (Table 2). It is conceivable that some of these proteins are involved in the control of the Lxr levels, and their altered phosphorylation increases the levels of Lxrα and Lxrβ in the Cyp46a1−/− retina. Of course, this is only a speculation, and future studies are required to identify the unknown factors that up-regulate the expression of Lxrα and Lxrβ in the Cyp46a1−/− retina and brain. Regardless of the cause of Lxr up-regulation, the present work and previous studies of Cyp27a1−/−, Cyp27a1−/−Cyp46a1−/−,4, 10 and Lxrα−/−Lxrβ−/− mice15 suggest that aberrant LXR signaling plays an important role in the development of vascular abnormalities typical for early-stage diabetic retinopathy. Moreover, the development of these changes does not require such features of diabetes as hyperglycemia and chronic systemic inflammation.
Studies of retinal phosphoproteomics indicate that microglia activation might not be the only factor contributing to retinal vascular abnormalities in Cyp46a1−/− mice. The modulation of protein phosphorylation could contribute as well because oxysterols and cholesterol were shown to modulate membrane function and receptor molecules associated with plasma and subcellular membranes.86, 87, 88 The phosphorylation of tight junction protein zonula occludens 1, or ZO-1 (TJP1), and aquaporin 4 (AQP4) (Table 2) may be of particular relevance to retinal vascular abnormalities in Cyp46a1−/− mice. ZO-1 is a key scaffolding protein necessary for the formation of tight junctions in various tissues.89, 90 In the retina, ZO-1 is a part of tight junctions of the outer retinal blood barrier (formed by the RPE) and the inner retinal blood barrier (formed by the blood vessels of the neural retina).91 Deletion of ZO-1 is embryonically lethal because of impaired formation of vascular trees and defective chorioallantoic fusion in yolk sac.92 Studies show that reduced expression of tight junction proteins, including ZO-1, leads to increased blood-retinal barrier permeability, which is closely linked to angiogenesis.93, 94 Vascular changes in diabetic retinopathy are due, at least in part, to elevated vascular endothelial growth factor expression,94 and ZO-1 was found to be phosphorylated after vascular endothelial growth factor stimulation of bovine aortic endothelial cells.95 This phosphorylation was suggested to inactivate ZO-1, whereas ZO-1 down-regulation was established to enhance endothelial cell proliferation.95 In the present work, ZO-1 seems to be only phosphorylated in the Cyp46a1−/− retina, thus raising a possibility that this phosphorylation contributes, at least in part, to increased vascular permeability and other retinal vascular abnormalities in Cyp46a1−/− mice.
AQP4 is another protein in Cyp46a1−/− mice, whose increased retinal phosphorylation could be of pertinence to the Cyp46a1−/− retinal phenotype (Table 2). AQP4 is a member of a large family of membrane protein channels that facilitate transport of water and small solutes.96 AQP4 is the predominant aquaporin in mammalian retina with a prominent localization around blood vessels (it is mainly present in the perivascular and end feet of Muller cells and astrocytes).96, 97 Both expression and localization of AQP4 are altered in experimental diabetes,98, 99, 100, 101 and AQP4 knockdown was found to exacerbate streptozotocin-induced diabetic retinopathy (retinal vascular permeability, retinal thickness, and vascular endothelial growth factor expression) through increased inflammatory response.101 Studies of Aqp4−/− mice report the following: mild ERG changes with no changes in retinal ultrastructure102; increased retinal glutamate levels103; low-level retinal inflammation along with increased retinal susceptibility to osmotic stress104; and the dysfunction of the blood-retinal barrier as well as reactive gliosis.105 Phosphorylated AQP4 was found in several tissues but not the retina; hence, physiological significance of retinal AQP4 phosphorylation is not yet clear. Nevertheless, a role of AQP4 in water transport,96 expression around retinal blood vessels,97 and changes in expression and localization in experimental diabetes98, 99, 100, 101 raise a possibility that increased phosphorylation of AQP4 in the Cyp46a1−/− retina is of relevance to the changes of retinal vasculature. Future research is required to ascertain retinal significance of ZO-1 and AQP4 phosphorylation.
Overall, the Cyp46a1−/− retina had much less differentially phosphorylated peptides than the Cyp46a1−/− brain (31 retinal phosphopeptides versus 185 brain phosphopeptides), with 28 retinal peptides having increased phosphorylation (Table 2). In contrast, in the brain, 151 phosphopeptides were less abundant in Cyp46a1−/− mice.42 Also, only two proteins (microtubule-associated protein 1B isoform X1 and myristoylated alanine-rich C-kinase substrate) and three protein isoforms (dihydropyrimidinase-related protein 3, rap1 GTPase-activating protein 1 isoform 3, and reticulon) were common in retinal and brain phosphoproteomics, suggesting that a lack of CYP46A1 elicits organ-specific changes in protein phosphorylation.
Beside effects on protein phosphorylation and microglia activation, CYP46A1 could be directly involved in maintenance of retinal vasculature, as indicated by the CYP46A1 mRNA and protein expression in retinal vascular endothelial cells (Figures 9 and 10). Indeed, CYP46A1 is a cholesterol-eliminating enzyme, and clinical studies support the role of lipids in vascular damage in diabetic retinopathy; moreover, they suggest that retinal lipid levels may be more critical than the circulating lipid levels.58 Increased Cyp46a1 levels in BREC in response to TNF-α stimulation could be protective because this Cyp46a1 up-regulation may increase the cellular 24HC concentration and LXR activation and thereby down-regulate the expression of the proinflammatory genes. Thus, in addition to metabolism of cholesterol and LXR regulation in RMM, CYP46A1 could potentially mediate (via LXRs) the anti-inflammatory effects in retinal vascular endothelial cells and be even involved in cellular antiadhesive properties, as demonstrated for artery endothelial cells.106
The present work and previous studies of LXRs15 suggest that increasing LXR activities would ameliorate retinal inflammation. A tiny dose of the anti-HIV drug efavirenz activates CYP46A1 and increases tissue 24HC concentrations in mouse brain107, 108 and retina (data not shown). Efavirenz treatment also ameliorates the manifestations of Alzheimer disease in a mouse model of this disease (5XFAD mice).108 Perhaps, efavirenz should be tested on a mouse model of diabetic retinopathy for the effect on retinal vascular abnormalities.
In summary, ablation of Cyp46a1 in mice led to some of the manifestations of early-stage diabetic retinopathy, retinal increase in total cholesterol, and a significant up-regulation of key proinflammatory genes in both the whole retina and RMM but not BMDM. These data establish CYP46A1 roles in retinal cholesterol homeostasis and regulation of retinal immune response. In addition, CYP46A1 may have other roles, as suggested by studies of retinal endothelial cells and retinal phosphoproteome. The present work supports the importance of LXRs in the retina and brings attention to CYP46A1 as a potential new player in diabetic retinopathy and a target that can be activated pharmacologically.
Acknowledgments
We thank Joseph Lin for contributions to retinal phosphoproteomics; the Visual Sciences Research Center Core Facilities for assistance with mouse breeding (Heather Butler and Kathryn Franke), animal genotyping (John Denker), tissue sectioning (Catherine Doller), and microscopy (Anthony Gardella); Yan Levitsky (Michigan State University) for image acquisition; Dr. Belinda Willard (Proteomics Laboratory, Cleveland Clinic Foundation) for conducting retinal phosphoproteomics; Dr. David Russell (UT Southwestern) for providing Cyp46a1+/− mice; and Dr. Sandra Erickson (University of California, San Francisco) for providing Cyp27a1+/− mice.
Footnotes
Supported in part by NIH grants EY025383 (M.B.G. and J.V.B.), EY018383 (I.A.P.), and EY11373 (I.A.P.). The Visual Sciences Research Center Core Facilities was supported by NIH grant P30 EY11373.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2018.10.013.
Supplemental Data
References
- 1.Tang J., Kern T.S. Inflammation in diabetic retinopathy. Prog Retin Eye Res. 2011;30:343–358. doi: 10.1016/j.preteyeres.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durham J.T., Herman I.M. Microvascular modifications in diabetic retinopathy. Curr Diab Rep. 2011;11:253–264. doi: 10.1007/s11892-011-0204-0. [DOI] [PubMed] [Google Scholar]
- 3.Stitt A.W., Lois N., Medina R.J., Adamson P., Curtis T.M. Advances in our understanding of diabetic retinopathy. Clin Sci (Lond) 2013;125:1–17. doi: 10.1042/CS20120588. [DOI] [PubMed] [Google Scholar]
- 4.Saadane A., Mast N., Charvet C., Omarova S., Zheng W., Huang S.S., Kern T.S., Peachey N.S., Pikuleva I.A. Retinal and non-ocular abnormalities in Cyp27a1-/- Cyp64a1-/- mice with dysfunctional metabolism of cholesterol. Am J Pathol. 2014;184:2403–2419. doi: 10.1016/j.ajpath.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wikvall K. Hydroxylations in biosynthesis of bile acids: isolation of a cytochrome P-450 from rabbit liver mitochondria catalyzing 26-hydroxylation of C27-steroids. J Biol Chem. 1984;259:3800–3804. [PubMed] [Google Scholar]
- 6.Lund E.G., Guileyardo J.M., Russell D.W. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci U S A. 1999;96:7238–7243. doi: 10.1073/pnas.96.13.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao W.L., Heo G.Y., Dodder N.G., Reem R.E., Mast N., Huang S., Dipatre P.L., Turko I.V., Pikuleva I.A. Quantification of cholesterol-metabolizing P450s CYP27A1 and CYP46A1 in neural tissues reveals a lack of enzyme-product correlations in human retina but not human brain. J Proteome Res. 2011;10:241–248. doi: 10.1021/pr1008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng W., Reem R.E., Omarova S., Huang S., DiPatre P.L., Charvet C.D., Curcio C.A., Pikuleva I.A. Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS One. 2012;7:e37926. doi: 10.1371/journal.pone.0037926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mast N., Reem R., Bederman I., Huang S., DiPatre P.L., Bjorkhem I., Pikuleva I.A. Cholestenoic acid is an important elimination product of cholesterol in the retina: comparison of retinal cholesterol metabolism with that in the brain. Invest Ophthalmol Vis Sci. 2011;52:594–603. doi: 10.1167/iovs.10-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omarova S., Charvet C.D., Reem R.E., Mast N., Zheng W., Huang S., Peachey N.S., Pikuleva I.A. Abnormal vascularization in mouse retina with dysregulated retinal cholesterol homeostasis. J Clin Invest. 2012;122:3012–3023. doi: 10.1172/JCI63816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.W., Fuda H., Javitt N.B., Strott C.A., Rodriguez I.R. Expression and localization of sterol 27-hydroxylase (CYP27A1) in monkey retina. Exp Eye Res. 2006;83:465–469. doi: 10.1016/j.exer.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez D.M., Andersson S., Russell D.W. Neuronal expression and subcellular localization of cholesterol 24-hydroxylase in the mouse brain. J Comp Neurol. 2008;507:1676–1693. doi: 10.1002/cne.21605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meaney S., Bodin K., Diczfalusy U., Bjorkhem I. On the rate of translocation in vitro and kinetics in vivo of the major oxysterols in human circulation: critical importance of the position of the oxygen function. J Lipid Res. 2002;43:2130–2135. doi: 10.1194/jlr.m200293-jlr200. [DOI] [PubMed] [Google Scholar]
- 14.Janowski B.A., Willy P.J., Devi T.R., Falck J.R., Mangelsdorf D.J. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 15.Hazra S., Rasheed A., Bhatwadekar A., Wang X., Shaw L.C., Patel M., Caballero S., Magomedova L., Solis N., Yan Y., Wang W., Thinschmidt J.S., Verma A., Li Q., Levi M., Cummins C.L., Grant M.B. Liver X receptor modulates diabetic retinopathy outcome in a mouse model of streptozotocin-induced diabetes. Diabetes. 2012;61:3270–3279. doi: 10.2337/db11-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammer S.S., Beli E., Kady N., Wang Q., Wood K., Lydic T.A., Malek G., Saban D.R., Wang X.X., Hazra S., Levi M., Busik J.V., Grant M.B. The mechanism of diabetic retinopathy pathogenesis unifying key lipid regulators, sirtuin 1 and liver X receptor. EBioMedicine. 2017;22:181–190. doi: 10.1016/j.ebiom.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W., Chen G., Head D.L., Mangelsdorf D.J., Russell D.W. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab. 2007;5:73–79. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spann N.J., Garmire L.X., McDonald J.G., Myers D.S., Milne S.B., Shibata N., Reichart D., Fox J.N., Shaked I., Heudobler D., Raetz C.R., Wang E.W., Kelly S.L., Sullards M.C., Murphy R.C., Merrill A.H., Jr., Brown H.A., Dennis E.A., Li A.C., Ley K., Tsimikas S., Fahy E., Subramaniam S., Quehenberger O., Russell D.W., Glass C.K. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willy P.J., Umesono K., Ong E.S., Evans R.M., Heyman R.A., Mangelsdorf D.J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 20.Auboeuf D., Rieusset J., Fajas L., Vallier P., Frering V., Riou J.P., Staels B., Auwerx J., Laville M., Vidal H. Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator-activated receptors and liver X receptor-alpha in humans: no alteration in adipose tissue of obese and NIDDM patients. Diabetes. 1997;46:1319–1327. doi: 10.2337/diab.46.8.1319. [DOI] [PubMed] [Google Scholar]
- 21.Apfel R., Benbrook D., Lernhardt E., Ortiz M.A., Salbert G., Pfahl M. A novel orphan receptor specific for a subset of thyroid hormone-responsive elements and its interaction with the retinoid/thyroid hormone receptor subfamily. Mol Cell Biol. 1994;14:7025–7035. doi: 10.1128/mcb.14.10.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner B.L., Valledor A.F., Shao G., Daige C.L., Bischoff E.D., Petrowski M., Jepsen K., Baek S.H., Heyman R.A., Rosenfeld M.G., Schulman I.G., Glass C.K. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol. 2003;23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glass C.K., Rosenfeld M.G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 24.Spann N.J., Glass C.K. Sterols and oxysterols in immune cell function. Nat Immunol. 2013;14:893–900. doi: 10.1038/ni.2681. [DOI] [PubMed] [Google Scholar]
- 25.Calkin A.C., Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghisletti S., Huang W., Ogawa S., Pascual G., Lin M.E., Willson T.M., Rosenfeld M.G., Glass C.K. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakobsson T., Treuter E., Gustafsson J.A., Steffensen K.R. Liver X receptor biology and pharmacology: new pathways, challenges and opportunities. Trends Pharmacol Sci. 2012;33:394–404. doi: 10.1016/j.tips.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 28.Joseph S.B., Castrillo A., Laffitte B.A., Mangelsdorf D.J., Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- 29.Bischoff E.D., Daige C.L., Petrowski M., Dedman H., Pattison J., Juliano J., Li A.C., Schulman I.G. Non-redundant roles for LXRalpha and LXRbeta in atherosclerosis susceptibility in low density lipoprotein receptor knockout mice. J Lipid Res. 2010;51:900–906. doi: 10.1194/jlr.M900096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwyer M.A., Kazmin D., Hu P., McDonnell D.P., Malek G. Research resource: nuclear receptor atlas of human retinal pigment epithelial cells: potential relevance to age-related macular degeneration. Mol Endocrinol. 2011;25:360–372. doi: 10.1210/me.2010-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund E.G., Xie C., Kotti T., Turley S.D., Dietschy J.M., Russell D.W. Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J Biol Chem. 2003;278:22980–22988. doi: 10.1074/jbc.M303415200. [DOI] [PubMed] [Google Scholar]
- 32.Dubrac S., Lear S.R., Ananthanarayanan M., Balasubramaniyan N., Bollineni J., Shefer S., Hyogo H., Cohen D.E., Blanche P.J., Krauss R.M., Batta A.K., Salen G., Suchy F.J., Maeda N., Erickson S.K. Role of CYP27A in cholesterol and bile acid metabolism. J Lipid Res. 2005;46:76–85. doi: 10.1194/jlr.M400219-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Russell D.W., Halford R.W., Ramirez D.M., Shah R., Kotti T. Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu Rev Biochem. 2009;78:1017–1040. doi: 10.1146/annurev.biochem.78.072407.103859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mast N., Shafaati M., Zaman W., Zheng W., Prusak D., Wood T., Ansari G.A., Lovgren-Sandblom A., Olin M., Bjorkhem I., Pikuleva I. Marked variability in hepatic expression of cytochromes CYP7A1 and CYP27A1 as compared to cerebral CYP46A1: lessons from a dietary study with omega 3 fatty acids in hamsters. Biochim Biophys Acta. 2010;1801:674–681. doi: 10.1016/j.bbalip.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohno H., Chen Y., Kevany B.M., Pearlman E., Miyagi M., Maeda T., Palczewski K., Maeda A. Photoreceptor proteins initiate microglial activation via Toll-like receptor 4 in retinal degeneration mediated by all-trans-retinal. J Biol Chem. 2013;288:15326–15341. doi: 10.1074/jbc.M112.448712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Earle W.R., Schilling E.L., Stark T.H., Straus N.P., Brown M.F., Shelton E. Production of malignancy in vitro, IV: the mouse fibroblast cultures and changes seen in the living cells. J Natl Cancer Inst. 1943;4:165–212. [Google Scholar]
- 37.Jaffe G.J., Earnest K., Fulcher S., Lui G.M., Houston L.L. Antitransferrin receptor immunotoxin inhibits proliferating human retinal pigment epithelial cells. Arch Ophthalmol. 1990;108:1163–1168. doi: 10.1001/archopht.1990.01070100119046. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q., Bozack S.N., Yan Y., Boulton M.E., Grant M.B., Busik J.V. Regulation of retinal inflammation by rhythmic expression of MiR-146a in diabetic retina. Invest Ophthalmol Vis Sci. 2014;55:3986–3994. doi: 10.1167/iovs.13-13076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Han X., Wittchen E.S., Hartnett M.E. TNF-alpha mediates choroidal neovascularization by upregulating VEGF expression in RPE through ROS-dependent beta-catenin activation. Mol Vis. 2016;22:116–128. [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tual-Chalot S., Allinson K.R., Fruttiger M., Arthur H.M. Whole mount immunofluorescent staining of the neonatal mouse retina to investigate angiogenesis in vivo. J Vis Exp. 2013;77:e50546. doi: 10.3791/50546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mast N., Lin J.B., Anderson K.W., Bjorkhem I., Pikuleva I.A. Transcriptional and post-translational changes in the brain of mice deficient in cholesterol removal mediated by cytochrome P450 46A1 (CYP46A1) PLoS One. 2017;12:e0187168. doi: 10.1371/journal.pone.0187168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saadane A., Mast N., Dao T., Ahmad B., Pikuleva I.A. Retinal hypercholesterolemia triggers cholesterol accumulation and esterification in photoreceptor cells. J Biol Chem. 2016;291:20427–20439. doi: 10.1074/jbc.M116.744656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radu M., Chernoff J. An in vivo assay to test blood vessel permeability. J Vis Exp. 2013;73:e50062. doi: 10.3791/50062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Im S.S., Yousef L., Blaschitz C., Liu J.Z., Edwards R.A., Young S.G., Raffatellu M., Osborne T.F. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011;13:540–549. doi: 10.1016/j.cmet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng H.Y., Green W.R., Tso M.O. Microglial activation in human diabetic retinopathy. Arch Ophthalmol. 2008;126:227–232. doi: 10.1001/archophthalmol.2007.65. [DOI] [PubMed] [Google Scholar]
- 48.Grigsby J.G., Cardona S.M., Pouw C.E., Muniz A., Mendiola A.S., Tsin A.T., Allen D.M., Cardona A.E. The role of microglia in diabetic retinopathy. J Ophthalmol. 2014;2014:705783. doi: 10.1155/2014/705783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tesch G.H. Role of macrophages in complications of type 2 diabetes. Clin Exp Pharmacol Physiol. 2007;34:1016–1019. doi: 10.1111/j.1440-1681.2007.04729.x. [DOI] [PubMed] [Google Scholar]
- 50.Duh E.J., Sun J.K., Stitt A.W. Diabetic retinopathy: current understanding, mechanisms, and treatment strategies. JCI Insight. 2017;2:e93751. doi: 10.1172/jci.insight.93751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waddington K.E., Jury E.C., Pineda-Torra I. Liver X receptors in immune cell function in humans. Biochem Soc Trans. 2015;43:752–757. doi: 10.1042/BST20150112. [DOI] [PubMed] [Google Scholar]
- 52.Zelcer N., Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Castrillo A., Joseph S.B., Vaidya S.A., Haberland M., Fogelman A.M., Cheng G., Tontonoz P. Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Mol Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- 54.Ogawa D., Stone J.F., Takata Y., Blaschke F., Chu V.H., Towler D.A., Law R.E., Hsueh W.A., Bruemmer D. Liver x receptor agonists inhibit cytokine-induced osteopontin expression in macrophages through interference with activator protein-1 signaling pathways. Circ Res. 2005;96:e59–e67. doi: 10.1161/01.RES.0000163630.86796.17. [DOI] [PubMed] [Google Scholar]
- 55.Hong C., Walczak R., Dhamko H., Bradley M.N., Marathe C., Boyadjian R., Salazar J.V., Tontonoz P. Constitutive activation of LXR in macrophages regulates metabolic and inflammatory gene expression: identification of ARL7 as a direct target. J Lipid Res. 2011;52:531–539. doi: 10.1194/jlr.M010686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beutler B., Hoebe K., Du X., Ulevitch R.J. How we detect microbes and respond to them: the Toll-like receptors and their transducers. J Leukoc Biol. 2003;74:479–485. doi: 10.1189/jlb.0203082. [DOI] [PubMed] [Google Scholar]
- 57.Santos A.M., Calvente R., Tassi M., Carrasco M.C., Martin-Oliva D., Marin-Teva J.L., Navascues J., Cuadros M.A. Embryonic and postnatal development of microglial cells in the mouse retina. J Comp Neurol. 2008;506:224–239. doi: 10.1002/cne.21538. [DOI] [PubMed] [Google Scholar]
- 58.Hammer S.S., Busik J.V. The role of dyslipidemia in diabetic retinopathy. Vision Res. 2017;139:228–236. doi: 10.1016/j.visres.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakravarthy H., Beli E., Navitskaya S., O'Reilly S., Wang Q., Kady N., Huang C., Grant M.B., Busik J.V. Imbalances in mobilization and activation of pro-inflammatory and vascular reparative bone marrow-derived cells in diabetic retinopathy. PLoS One. 2016;11:e0146829. doi: 10.1371/journal.pone.0146829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reiss A.B., Martin K.O., Rojer D.E., Iyer S., Grossi E.A., Galloway A.C., Javitt N.B. Sterol 27-hydroxylase: expression in human arterial endothelium. J Lipid Res. 1997;38:1254–1260. [PubMed] [Google Scholar]
- 61.Rakocevic J., Orlic D., Mitrovic-Ajtic O., Tomasevic M., Dobric M., Zlatic N., Milasinovic D., Stankovic G., Ostojic M., Labudovic-Borovic M. Endothelial cell markers from clinician's perspective. Exp Mol Pathol. 2017;102:303–313. doi: 10.1016/j.yexmp.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 62.Buckingham B.P., Inman D.M., Lambert W., Oglesby E., Calkins D.J., Steele M.R., Vetter M.L., Marsh-Armstrong N., Horner P.J. Progressive ganglion cell degeneration precedes neuronal loss in a mouse model of glaucoma. J Neurosci. 2008;28:2735–2744. doi: 10.1523/JNEUROSCI.4443-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen M., Bradley M.N., Beaven S.W., Tontonoz P. Phosphorylation of the liver X receptors. FEBS Lett. 2006;580:4835–4841. doi: 10.1016/j.febslet.2006.07.074. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto T., Shimano H., Inoue N., Nakagawa Y., Matsuzaka T., Takahashi A., Yahagi N., Sone H., Suzuki H., Toyoshima H., Yamada N. Protein kinase A suppresses sterol regulatory element-binding protein-1C expression via phosphorylation of liver X receptor in the liver. J Biol Chem. 2007;282:11687–11695. doi: 10.1074/jbc.M611911200. [DOI] [PubMed] [Google Scholar]
- 65.Wu C., Hussein M.A., Shrestha E., Leone S., Aiyegbo M.S., Lambert W.M., Pourcet B., Cardozo T., Gustafson J.A., Fisher E.A., Pineda-Torra I., Garabedian M.J. Modulation of macrophage gene expression via liver X receptor alpha serine 198 phosphorylation. Mol Cell Biol. 2015;35:2024–2034. doi: 10.1128/MCB.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang W., Ghisletti S., Saijo K., Gandhi M., Aouadi M., Tesz G.J., Zhang D.X., Yao J., Czech M.P., Goode B.L., Rosenfeld M.G., Glass C.K. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abcouwer S.F. Neural inflammation and the microglial response in diabetic retinopathy. J Ocul Biol Dis Infor. 2011;4:25–33. doi: 10.1007/s12177-012-9086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zong H., Ward M., Stitt A.W. AGEs, RAGE, and diabetic retinopathy. Curr Diab Rep. 2011;11:244–252. doi: 10.1007/s11892-011-0198-7. [DOI] [PubMed] [Google Scholar]
- 69.Zhang W., Liu H., Rojas M., Caldwell R.W., Caldwell R.B. Anti-inflammatory therapy for diabetic retinopathy. Immunotherapy. 2011;3:609–628. doi: 10.2217/imt.11.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krady J.K., Basu A., Allen C.M., Xu Y., LaNoue K.F., Gardner T.W., Levison S.W. Minocycline reduces proinflammatory cytokine expression, microglial activation, and caspase-3 activation in a rodent model of diabetic retinopathy. Diabetes. 2005;54:1559–1565. doi: 10.2337/diabetes.54.5.1559. [DOI] [PubMed] [Google Scholar]
- 71.Checchin D., Sennlaub F., Levavasseur E., Leduc M., Chemtob S. Potential role of microglia in retinal blood vessel formation. Invest Ophthalmol Vis Sci. 2006;47:3595–3602. doi: 10.1167/iovs.05-1522. [DOI] [PubMed] [Google Scholar]
- 72.Bretillon L., Diczfalusy U., Bjorkhem I., Maire M.A., Martine L., Joffre C., Acar N., Bron A., Creuzot-Garcher C. Cholesterol-24S-hydroxylase (CYP46A1) is specifically expressed in neurons of the neural retina. Curr Eye Res. 2007;32:361–366. doi: 10.1080/02713680701231857. [DOI] [PubMed] [Google Scholar]
- 73.Fourgeux C., Martine L., Acar N., Bron A.M., Creuzot-Garcher C.P., Bretillon L. In vivo consequences of cholesterol-24S-hydroxylase (CYP46A1) inhibition by voriconazole on cholesterol homeostasis and function in the rat retina. Biochem Biophys Res Commun. 2014;446:775–781. doi: 10.1016/j.bbrc.2014.01.118. [DOI] [PubMed] [Google Scholar]
- 74.Bogdanovic N., Bretillon L., Lund E.G., Diczfalusy U., Lannfelt L., Winblad B., Russell D.W., Bjorkhem I. On the turnover of brain cholesterol in patients with Alzheimer's disease: abnormal induction of the cholesterol-catabolic enzyme CYP46 in glial cells. Neurosci Lett. 2001;314:45–48. doi: 10.1016/s0304-3940(01)02277-7. [DOI] [PubMed] [Google Scholar]
- 75.Brown J., 3rd, Theisler C., Silberman S., Magnuson D., Gottardi-Littell N., Lee J.M., Yager D., Crowley J., Sambamurti K., Rahman M.M., Reiss A.B., Eckman C.B., Wolozin B. Differential expression of cholesterol hydroxylases in Alzheimer's disease. J Biol Chem. 2004;279:34674–34681. doi: 10.1074/jbc.M402324200. [DOI] [PubMed] [Google Scholar]
- 76.Lavrnja I., Smiljanic K., Savic D., Mladenovic-Djordjevic A., Tesovic K., Kanazir S., Pekovic S. Expression profiles of cholesterol metabolism-related genes are altered during development of experimental autoimmune encephalomyelitis in the rat spinal cord. Sci Rep. 2017;7:2702. doi: 10.1038/s41598-017-02638-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cartagena C.M., Ahmed F., Burns M.P., Pajoohesh-Ganji A., Pak D.T., Faden A.I., Rebeck G.W. Cortical injury increases cholesterol 24S hydroxylase (Cyp46) levels in the rat brain. J Neurotrauma. 2008;25:1087–1098. doi: 10.1089/neu.2007.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kotti T.J., Ramirez D.M., Pfeiffer B.E., Huber K.M., Russell D.W. Brain cholesterol turnover required for geranylgeraniol production and learning in mice. Proc Natl Acad Sci U S A. 2006;103:3869–3874. doi: 10.1073/pnas.0600316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitney K.D., Watson M.A., Goodwin B., Galardi C.M., Maglich J.M., Wilson J.G., Willson T.M., Collins J.L., Kliewer S.A. Liver X receptor (LXR) regulation of the LXRalpha gene in human macrophages. J Biol Chem. 2001;276:43509–43515. doi: 10.1074/jbc.M106155200. [DOI] [PubMed] [Google Scholar]
- 80.Tobin K.A., Steineger H.H., Alberti S., Spydevold O., Auwerx J., Gustafsson J.A., Nebb H.I. Cross-talk between fatty acid and cholesterol metabolism mediated by liver X receptor-alpha. Mol Endocrinol. 2000;14:741–752. doi: 10.1210/mend.14.5.0459. [DOI] [PubMed] [Google Scholar]
- 81.Hashimoto K., Matsumoto S., Yamada M., Satoh T., Mori M. Liver X receptor-alpha gene expression is positively regulated by thyroid hormone. Endocrinology. 2007;148:4667–4675. doi: 10.1210/en.2007-0150. [DOI] [PubMed] [Google Scholar]
- 82.Kim M.S., Sweeney T.R., Shigenaga J.K., Chui L.G., Moser A., Grunfeld C., Feingold K.R. Tumor necrosis factor and interleukin 1 decrease RXRalpha, PPARalpha, PPARgamma, LXRalpha, and the coactivators SRC-1, PGC-1alpha, and PGC-1beta in liver cells. Metabolism. 2007;56:267–279. doi: 10.1016/j.metabol.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hao X.R., Cao D.L., Hu Y.W., Li X.X., Liu X.H., Xiao J., Liao D.F., Xiang J., Tang C.K. IFN-gamma down-regulates ABCA1 expression by inhibiting LXRalpha in a JAK/STAT signaling pathway-dependent manner. Atherosclerosis. 2009;203:417–428. doi: 10.1016/j.atherosclerosis.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 84.Chen M., Beaven S., Tontonoz P. Identification and characterization of two alternatively spliced transcript variants of human liver X receptor alpha. J Lipid Res. 2005;46:2570–2579. doi: 10.1194/jlr.M500157-JLR200. [DOI] [PubMed] [Google Scholar]
- 85.Hashimoto K., Ishida E., Matsumoto S., Shibusawa N., Okada S., Monden T., Satoh T., Yamada M., Mori M. A liver X receptor (LXR)-beta alternative splicing variant (LXRBSV) acts as an RNA co-activator of LXR-beta. Biochem Biophys Res Commun. 2009;390:1260–1265. doi: 10.1016/j.bbrc.2009.10.132. [DOI] [PubMed] [Google Scholar]
- 86.Bielska A.A., Schlesinger P., Covey D.F., Ory D.S. Oxysterols as non-genomic regulators of cholesterol homeostasis. Trends Endocrinol Metab. 2012;23:99–106. doi: 10.1016/j.tem.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Martin M.G., Perga S., Trovo L., Rasola A., Holm P., Rantamaki T., Harkany T., Castren E., Chiara F., Dotti C.G. Cholesterol loss enhances TrkB signaling in hippocampal neurons aging in vitro. Mol Biol Cell. 2008;19:2101–2112. doi: 10.1091/mbc.E07-09-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sodero A.O., Trovo L., Iannilli F., Van Veldhoven P., Dotti C.G., Martin M.G. Regulation of tyrosine kinase B activity by the Cyp46/cholesterol loss pathway in mature hippocampal neurons: relevance for neuronal survival under stress and in aging. J Neurochem. 2011;116:747–755. doi: 10.1111/j.1471-4159.2010.07079.x. [DOI] [PubMed] [Google Scholar]
- 89.Paris L., Tonutti L., Vannini C., Bazzoni G. Structural organization of the tight junctions. Biochim Biophys Acta. 2008;1778:646–659. doi: 10.1016/j.bbamem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 90.Fanning A.S., Anderson J.M. Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann N Y Acad Sci. 2009;1165:113–120. doi: 10.1111/j.1749-6632.2009.04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ciolofan C., Li X.B., Olson C., Kamasawa N., Gebhardt B.R., Yasumura T., Morita M., Rash J.E., Nagy J.I. Association of connexin36 and zonula occludens-1 with zonula occludens-2 and the transcription factor zonula occludens-1-associated nucleic acid-binding protein at neuronal gap junctions in rodent retina. Neuroscience. 2006;140:433–451. doi: 10.1016/j.neuroscience.2006.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Katsuno T., Umeda K., Matsui T., Hata M., Tamura A., Itoh M., Takeuchi K., Fujimori T., Nabeshima Y., Noda T., Tsukita S., Tsukita S. Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol Biol Cell. 2008;19:2465–2475. doi: 10.1091/mbc.E07-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gardner T.W. Histamine, ZO-1 and increased blood-retinal barrier permeability in diabetic retinopathy. Trans Am Ophthalmol Soc. 1995;93:583–621. [PMC free article] [PubMed] [Google Scholar]
- 94.Erickson K.K., Sundstrom J.M., Antonetti D.A. Vascular permeability in ocular disease and the role of tight junctions. Angiogenesis. 2007;10:103–117. doi: 10.1007/s10456-007-9067-z. [DOI] [PubMed] [Google Scholar]
- 95.Chidiac R., Zhang Y., Tessier S., Faubert D., Delisle C., Gratton J.P. Comparative phosphoproteomics analysis of VEGF and angiopoietin-1 signaling reveals ZO-1 as a critical regulator of endothelial cell proliferation. Mol Cell Proteomics. 2016;15:1511–1525. doi: 10.1074/mcp.M115.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schey K.L., Wang Z., L Wenke J., Qi Y. Aquaporins in the eye: expression, function, and roles in ocular disease. Biochim Biophys Acta. 2014;1840:1513–1523. doi: 10.1016/j.bbagen.2013.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nagelhus E.A., Veruki M.L., Torp R., Haug F.M., Laake J.H., Nielsen S., Agre P., Ottersen O.P. Aquaporin-4 water channel protein in the rat retina and optic nerve: polarized expression in Muller cells and fibrous astrocytes. J Neurosci. 1998;18:2506–2519. doi: 10.1523/JNEUROSCI.18-07-02506.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Iandiev I., Pannicke T., Reichenbach A., Wiedemann P., Bringmann A. Diabetes alters the localization of glial aquaporins in rat retina. Neurosci Lett. 2007;421:132–136. doi: 10.1016/j.neulet.2007.04.076. [DOI] [PubMed] [Google Scholar]
- 99.Fukuda M., Nakanishi Y., Fuse M., Yokoi N., Hamada Y., Fukagawa M., Negi A., Nakamura M. Altered expression of aquaporins 1 and 4 coincides with neurodegenerative events in retinas of spontaneously diabetic Torii rats. Exp Eye Res. 2010;90:17–25. doi: 10.1016/j.exer.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 100.Kumar B., Gupta S.K., Nag T.C., Srivastava S., Saxena R., Jha K.A., Srinivasan B.P. Retinal neuroprotective effects of quercetin in streptozotocin-induced diabetic rats. Exp Eye Res. 2014;125:193–202. doi: 10.1016/j.exer.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 101.Cui B., Sun J.H., Xiang F.F., Liu L., Li W.J. Aquaporin 4 knockdown exacerbates streptozotocin-induced diabetic retinopathy through aggravating inflammatory response. Exp Eye Res. 2012;98:37–43. doi: 10.1016/j.exer.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 102.Li J., Patil R.V., Verkman A.S. Mildly abnormal retinal function in transgenic mice without Muller cell aquaporin-4 water channels. Invest Ophthalmol Vis Sci. 2002;43:573–579. [PubMed] [Google Scholar]
- 103.Li X.M., Wendu R.L., Yao J., Ren Y., Zhao Y.X., Cao G.F., Qin J., Yan B. Abnormal glutamate metabolism in the retina of aquaporin 4 (AQP4) knockout mice upon light damage. Neurol Sci. 2014;35:847–853. doi: 10.1007/s10072-013-1610-7. [DOI] [PubMed] [Google Scholar]
- 104.Pannicke T., Wurm A., Iandiev I., Hollborn M., Linnertz R., Binder D.K., Kohen L., Wiedemann P., Steinhauser C., Reichenbach A., Bringmann A. Deletion of aquaporin-4 renders retinal glial cells more susceptible to osmotic stress. J Neurosci Res. 2010;88:2877–2888. doi: 10.1002/jnr.22437. [DOI] [PubMed] [Google Scholar]
- 105.Nicchia G.P., Pisani F., Simone L., Cibelli A., Mola M.G., Dal Monte M., Frigeri A., Bagnoli P., Svelto M. Glio-vascular modifications caused by Aquaporin-4 deletion in the mouse retina. Exp Eye Res. 2016;146:259–268. doi: 10.1016/j.exer.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 106.Calkin A.C., Tontonoz P. Liver x receptor signaling pathways and atherosclerosis. Arterioscler Thromb Vasc Biol. 2010;30:1513–1518. doi: 10.1161/ATVBAHA.109.191197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mast N., Li Y., Linger M., Clark M., Wiseman J., Pikuleva I.A. Pharmacologic stimulation of cytochrome P450 46A1 and cerebral cholesterol turnover in mice. J Biol Chem. 2014;289:3529–3538. doi: 10.1074/jbc.M113.532846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mast N., Saadane A., Valencia-Olvera A., Constans J., Maxfield E., Arakawa H., Li Y., Landreth G., Pikuleva I.A. Cholesterol-metabolizing enzyme cytochrome P450 46A1 as a pharmacologic target for Alzheimer's disease. Neuropharmacology. 2017;123:465–476. doi: 10.1016/j.neuropharm.2017.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.