Abstract
Commensal gut microbiota–host immune responses are experimentally delineated via gnotobiotic animal models or alternatively by antibiotic perturbation of gut microbiota. Osteoimmunology investigations in germ-free mice, revealing that gut microbiota immunomodulatory actions critically regulate physiologic skeletal development, highlight that antibiotic perturbation of gut microbiota may dysregulate normal osteoimmunological processes. We investigated the impact of antibiotic disruption of gut microbiota on osteoimmune response effects in postpubertal skeletal development. Sex-matched C57BL/6T mice were administered broad-spectrum antibiotics or vehicle-control from the age of 6 to 12 weeks. Antibiotic alterations in gut bacterial composition and skeletal morphology were sex dependent. Antibiotics did not influence osteoblastogenesis or endochondral bone formation, but notably enhanced osteoclastogenesis. Unchanged Tnf or Ccl3 expression in marrow and elevated tumor necrosis factor-α and chemokine (C-C motif) ligand 3 in serum indicated that the pro-osteoclastic effects of the antibiotics are driven by increased systemic inflammation. Antibiotic-induced broad changes in adaptive and innate immune cells in mesenteric lymph nodes and spleen demonstrated that the perturbation of gut microbiota drives a state of dysbiotic hyperimmune response at secondary lymphoid tissues draining local gut and systemic circulation. Antibiotics up-regulated the myeloid-derived suppressor cells, immature myeloid progenitor cells known for immunosuppressive properties in pathophysiologic inflammatory conditions. Myeloid-derived suppressor cell–mediated immunosuppression can be antigen specific. Therefore, antibiotic-induced broad suppression of major histocompatibility complex class II antigen presentation genes in bone marrow discerns that antibiotic perturbation of gut microbiota dysregulates critical osteoimmune cross talk.
The gut is colonized by diverse microorganisms (ie, bacteria, fungi, and viruses) that collectively form a microbial community known as the gut microbiota. Early life host-microbe interactions direct the development of immunity and the establishment of a stable complex microbial community, commonly referred to as the commensal microbiota.1, 2, 3, 4, 5 Extensive research has focused on commensal gut microbiota–host immune response effects in the context of protection against pathogenic gut microbes6, 7, 8 and the pathophysiology of chronic inflammatory/autoimmune gastrointestinal disease states.9, 10, 11 Timely investigations have delineated that indigenous gut microbiota immunomodulatory effects not only influence pathologic conditions centered in the gut, but also at distant anatomical sites (ie, liver, brain, heart, and skeleton).12, 13, 14, 15, 16, 17, 18, 19 Not well understood and central to this report, the immunomodulation of commensal gut microbiota critically influences the normal growth and development of extragastrointestinal tissues.4, 5, 20, 21
After birth, microbial colonization of the infant gut is primarily determined by the method of delivery and the mother's indigenous microbiota.22, 23, 24 Throughout life, exogenous factors (ie, lifestyle, hygiene, diet, and medications) induce shifts/changes in the gut microbiota composition, which have indirect effects on host immunity and physiology.5, 25, 26, 27 In line with this premise, antibiotic perturbation of the indigenous gut microbiota induces lasting alterations in the host immune response, which has implications for health and disease.28, 29, 30, 31 Antibiotics can induce hyperimmune proinflammatory response states, which have been purported to be driven by dysbiotic shifts in the gut microbiota composition, increased intestinal permeability, translocation of gut microbes/microbial ligands, and/or overgrowth of drug-resistant opportunistic pathogens.21, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43
The field of osteoimmunology has shown that the immune cell interactions with bone cells regulate skeletal development and homeostasis, under physiological and pathophysiologic conditions.44, 45, 46, 47, 48 Proinflammatory immune response states can suppress osteoblast-mediated bone formation and/or enhance osteoclast-mediated bone resorption, having detrimental effects on the accrual of bone mass in the growing skeleton and the maintenance of bone mass in the mature adult skeleton.49, 50, 51 Recent osteoimmunology reports in the germ-free mouse model, disclosing that gut microbiota immunomodulatory actions potently regulate and promote the development and homeostasis of skeletal tissues,52, 53, 54, 55 highlight that antibiotic perturbation of commensal gut microbiota may dysregulate normal osteoimmunological processes. Considering that antibiotic disruption of the indigenous gut microbiota has been reported to induce proinflammatory hyperimmune response states,21, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43 antibiotic administration could notably have unintended pathophysiologic effects impairing the attainment and maintenance of peak skeletal bone mass.
Low-dose (subtherapeutic) antibiotics have been administered to agricultural animals since the 1940s for growth-promoting effects, which have been largely attributed to gut microbiota alterations in nutrient metabolism.56, 57, 58, 59 Despite decades of agricultural research evaluating antibiotic perturbation of the gut microbiota in relation to animal growth effects, the initial skeletal outcomes were reported by a recent early-life antibiotic treatment study in female C57BL/6J mice.60 This timely investigation evaluated antibiotic alterations in gut microbiota composition, relative to host metabolism and growth outcomes.60 Continuous low-dose antibiotic administration, beginning at age 28 days of life, transiently increased whole animal bone mineral density scores in 7-week–old mice.60 Eleven-week–old antibiotic- versus vehicle-treated mice had similar whole-animal bone mineral density scores.60
Broad-spectrum antibiotic cocktails (combination of two or more different antibiotic drugs) are commonly administered by researchers evaluating gut microbiota: host response effects in experimental animal models. Broad-spectrum antibiotic cocktail formulation protocols have been developed, which either deplete or disrupt bacterial communities in the murine gut.61, 62, 63, 64, 65 Experimental depletion protocols are commonly used as an alternative to the germ-free mouse model, whereas experimental disruption protocols are used to delineate how perturbations in the indigenous gut microbiota impact host physiology. Highly relevant to the current study, Guss et al66 published the first known report evaluating broad-spectrum antibiotic disruption of gut microbiota effects on skeletal outcomes. Continuous broad-spectrum antibiotic administration in male C57BL/6J mice (from 4 to 16 weeks of age) decreased whole bone mechanical properties, which were unrelated to inferior cortical bone morphologic parameters.66
Antibiotic perturbation of the normal gut microbiota during postnatal skeletal development has been shown to influence bone mass accrual60, 66 and bone mechanical properties,66 yet the osteoimmune mechanisms linking antibiotic effects on skeletogenesis are unknown. This broad-spectrum antibiotic investigation in sex-matched postpubertal C57BL/6T mice delineates that antibiotic disruption of the indigenous gut microbiota impairs trabecular bone mass/microarchitecture properties. Antibiotic perturbation of gut microbiota induced a proinflammatory hyperimmune response at secondary lymphoid tissues draining local gut and systemic circulation, which potently up-regulated osteoclastogenesis at distant skeletal sites. Intriguingly, antibiotic treatment up-regulated myeloid-derived suppressor cells (MDSCs) and suppressed major histocompatibility complex (MHC) class II antigen processing/presentation in the bone marrow. These findings provide novel mechanistic insight revealing that antibiotic disruption of gut microbiota dysregulates osteoimmune cross talk in postpubertal skeletal development.
Materials and Methods
Mice
C57BL/6T sex-matched mice were bred and housed under specific pathogen-free conditions. Mice were administered antibiotics [vancomycin (500 mg/L), imipenem/cilastatin (500 mg/L), and neomycin (1 g/L)]67 or vehicle-control in drinking water from the age of 6 to 12 weeks. Animal research was approved by the Medical University of South Carolina Animal Protocols Review Board and was performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals.68
μCT
Tibiae were fixed in 10% phosphate-buffered formalin for 24 hours at room temperature and stored in 70% ethanol. Specimens were scanned with Scanco Medical Microcomputed Tomography (μCT) 40 Scanner (Scanco Medical, Brüttisellen, Switzerland), using the following acquisition parameters: X-ray tube potential = 55 kVp; X-ray intensity = 145 μA; integration time = 200 milliseconds; and isotropic voxel size = 8 μm3. Calibrated three-dimensional images were reconstructed. Tibiae trabecular and cortical bone morphology was analyzed using Analyze 12.0 Bone Microarchitecture Analysis software (Analyze Direct, Seattle, WA). For trabecular analysis, axial CT slices were analyzed beginning 250 μm distal to the proximal growth plate and extending 1200 μm distally; a fixed threshold of 1750 Hounsfield units was used to discriminate mineralized tissue. For cortical analysis, transverse CT slices were analyzed in a 1000-μm segment of the middiaphysis; a fixed threshold of 2500 Hounsfield units was used to discriminate mineralized tissue. Data are reported in accordance with standardized nomenclature,69 as previously described.55, 70
Histomorphometry
Tibiae were fixed in 10% phosphate-buffered formalin for 24 hours at room temperature, decalcified in 10% EDTA for 21 days at room temperature, and submitted for histologic processing. Proximal tibiae were embedded in paraffin, serial frontal sections (5 μm thick) were cut, and staining was performed with hematoxylin and eosin and tartrate-resistant acid phosphatase (TRAP) stains. Growth plate chondrocyte zone analysis was performed in hematoxylin and eosin–stained proximal tibia sections. Five measurements of the proliferative zone and the hypertrophic zone were performed in the central two-thirds of the growth plate to assess endochondral bone formation.70 Osteoclast cellular end point analyses were performed in TRAP-stained (with fast green counterstain) proximal tibia sections. TRAP+ multinucleated (three or more nuclei) cells lining bone were considered osteoclasts. Static and dynamic histomorphometric analyses were performed in tibiae fixed in 10% phosphate-buffered formalin, dehydrated in graded ethanol and xylene, and embedded undecalcified in modified methylmethacrylate.55, 70 Sections (4 μm thick) were stained with toluidine blue for osteoblast cellular analysis. Unstained sections (8 μm thick) were used for dynamic indexes of bone formation. Calcein (20 mg/kg) was administered via i.p. injection 5 and 2 days before sacrifice.55, 70 Analyses were limited to the secondary spongiosa, beginning 250 μm distal to the growth plate and extended 1000 μm distally (50 μm from endocortical surfaces).55, 70 Images were acquired via an Olympus BX61 microscope (Olympus America, Inc., Center Valley, PA) and Visiopharm software version 3.2.9.0 (Visiopharm, Hoersholm, Denmark). Blinded histomorphometric analysis (J.D.H.-S.) was performed semiautomatically using Visiopharm software. Data are reported in accordance with standardized nomenclature,71 as previously described.55, 70
Histopathology
Left kidneys and medial liver lobes were fixed in 10% phosphate-buffered formalin for 24 hours at room temperature and submitted for histologic processing. Specimens were embedded in paraffin, serial frontal sections (7 μm thick) were cut, and hematoxylin and eosin stains were performed. Images were acquired via an Olympus BX61 microscope and Visiopharm software. Blinded histopathological scoring was performed by a medical pathologist in the Medical University of South Carolina Laboratory Center for Oral Health Research.
In Vitro Osteoclast-Precursor Assays
First-passage nonadherent hematopoietic cells isolated from bone marrow were washed and incubated with CD11b microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). An AutoMACS Sorter (Miltenyi Biotec) was used to separate CD11bneg hematopoietic cells, as previously described.55 Cells were washed, counted, and plated for assays in α-minimal essential medium, 10% fetal bovine serum (Hyclone, South Logan, UT), and 1% PSG (penicillin, streptomycin, glutamine). Cells were plated at 1.5 × 105 cells/cm2 in 96-well plates to test TRAP+ osteoclastic cell outcomes and in 96-well Osteo Assay plates (Corning, Lowell, MA) to assess osteoclast resorption/pit formation. At initial plating, cells were primed for 36 hours with 10 ng/mL CSF1 colony stimulating factor 1 (CSF1; R&D Systems, Minneapolis, MN) to drive cells toward the preosteoclastic lineage. Cells were then treated with 25 ng/mL CSF1 alone (control cultures) or CSF1 with 50 ng/mL receptor activator of NF-κB ligand (RANKL; R&D Systems) (treatment cultures) medium for 6 days; medium was changed every other day. Day 6 control and treatment cultures were stained via the TRAP method, as previously reported.55 Day 6 Osteo Assay cultures were processed, per manufacturer's protocol. The TRAP stain assay and Osteo Assay were performed in triplicate cultures; two fields of view were analyzed per technical replicate culture.
Serum Biochemical Assays
Whole blood was collected via cardiac puncture at euthanasia. Serum was isolated and stored at −80°C for use. Osteocalcin (OCN; Alfa Aesar, Haverhill, MA), tumor necrosis factor (TNF)-α, chemokine (C-C motif) ligand 3 (CCL3), and CCL4 (Quantikine; R&D Systems) were evaluated by enzyme-linked immunosorbent assay, per manufacturer’s instructions.
Gene Expression Analysis
Quantitative Real-Time PCR
Femur bone marrow was flushed with TRIzol (Invitrogen, Carlsbad, CA), and calvariae were flash frozen, pulverized, and homogenized in TRIzol. RNA was isolated by TRIzol method following manufacturer's protocol. Total RNA was quantified via NanoDrop 1000 (Thermo Scientific, Waltham, MA). cDNA was synthesized using TaqMan Random Hexamers and Reverse Transcription Reagents (Applied Biosystems, Foster City, CA), according to the manufacturer's protocol. cDNA was amplified using TaqMan gene expression primers/probes and Universal PCR Master Mix, via the StepOnePlus System (Applied Biosystems). Gapdh was used as an endogenous control. Relative quantification of data was performed via the ΔΔCT method,72 as previously described.55
qPCR
Fecal pellets were collected at euthanasia, and bacterial DNA was isolated using the Qiagen QIAamp DNA Mini Kit, as directed by the manufacturer (Qiagen, Hilden, Germany). Total bacterial load was tested using quantitative PCR (qPCR), as described previously.73 Bacterial load was determined as amplification of 16S rDNA target gene using the universal forward primer (5′-AAACTCAAAKGAATTGACGG-3′) and reverse primer (5′-CTCACRRCACGAGCTGAC-3′).73 Universal primers and twofold serial dilutions ranging from 200 to 1 μg/mL of bacterial DNA standard (ZymoBIOMICS, Irvine, CA) were used in a standard curve, and relative quantification of data was performed via the ΔΔCT method.72 The qPCR protocol was completed on ABI StepOne (Applied Biosystems), as described.73 Briefly, qPCRs contained 2× Fast SYBR Green Master Mix (Applied Biosystems), 0.3 μmol/L primers, and 150 μg/mL DNA template. Primers tested were α-proteobacteria, 5′-CIAGTGTAGAGGTGAAATT-3′ (forward) and 5′-CCCCGTCAATTCCTTTGAGTT-3′ (reverse); γ-proteobacteria, 5′-TCGTCAGCTCGTGTYGTGA-3′ (forward) and 5′-CGTAAGGGCCATGATG-3′ (reverse); Bacteroidetes, 5′-CRAACAGGATTAGATACCCT-3′ (forward) and 5′-GGTAAGGTTCCTCGCGTAT-3′ (reverse); Firmicutes, 5′-TGAAACTYAAAGGAATTGACG-3′ (forward) and 5′-ACCATGCACCTGTC-3′ (reverse); and actinobacteria, 5′-TACGGCCGCAAGGCTA-3′ (forward) and 5′-TCRTCCCCACCTTCCG-3′ (reverse) (Integrated DNA Technologies, Coralville, IA). Samples underwent an initial denaturing step at 95°C for 5 minutes, followed by 30 cycles of 95°C for 15 seconds, 61.5°C for 15 seconds, and 72°C for 30 seconds, and ending with a final elongation step of 72°C for 5 minutes. Samples were normalized to the Universal 16S gene and presented as percentage composition/abundance of the specific phylum.73
NanoString
nCounter Mouse Immunology Panel for mouse gene expression (NanoString Technologies, Seattle, WA) was applied to assess cytokine and antigen presentation gene expression in femoral bone marrow. Hybridization was performed, and products were run on the nCounter preparation station, according to the manufacturer's protocol. Data were normalized to the geometric means of spiked-in positive controls, negative controls, and internal control genes. Absolute quantification of RNA was reported as normalized RNA counts, as previously described.55
Flow Cytometry
Femur bone marrow, spleen, and mesenteric lymph node (mLN) cells were isolated, washed, and counted. Live cells were treated with Fc block and stained for T-cell hematopoiesis: anti–CD3–antigen-presenting cell (APC; Miltenyi Biotec; clone 145-2C11), anti–CD8-phosphatidylethanolamine (PE) (Miltenyi Biotec; clone 53-6.7), anti–CD4–fluorescein isothiocyanate (FITC; Miltenyi Biotec; clone GK1.5); natural killer (NK) cells: anti–CD3-FITC (eBioscience, San Diego, CA; clone PK136), anti–NK1.1-PE (eBioscience; clone 145-2C11), and anti–CD11b-APC (Miltenyi Biotec; clone M1/70.15.11.5); activated/memory B cells: anti–B220-FITC (Miltenyi Biotec; clone RA3-6B2), anti–IgD-eF450 (eBioscience; clone 11-26c), anti–IgM-PE (eBioscience; clone eB121-15F9), and anti–CD40-APC (eBioscience; clone 1C10); inflammatory monocytes/MDSCs: anti–CD11b-APC (Miltenyi Biotec; clone M1/70.15.11.5), anti–Ly6G-PacB (Biolegend, San Diego, CA; clone 145-2C11), anti–F4/80-PE (eBioscience; clone BM8), and anti–Ly6C-FITC (Novus Biologicals, Littleton, CO; clone HK1.4); plasmacytoid dendritic cells (DCs): anti–CD11c-APC (Miltenyi Biotec; clone REA754), anti–MHC II–PE (eBioscience; clone M5/114.15.2), and anti–CD317-FITC (eBioscience; clone 129c); and M1/M2 macrophages: anti–CD11b-APC (Miltenyi Biotec; REA592), anti–CD11c-PE-Vio770 (Miltenyi Biotec; clone REA754), anti–MHC class II–FITC (Miltenyi Biotec; clone REA528), anti–CD64-APC-Vio770 (Miltenyi Biotec; clone REA286), anti–B220-Vioblue (Miltenyi Biotec; clone REA755), and anti–CD206-PE (eBioscience; clone MR6F3). Fixed cells were treated with Fc block stained for regulatory T (TREG) cells: anti–CD3-PE-Vio770 (Miltenyi Biotec; clone REA641), anti–CD4-FITC (Miltenyi Biotec; clone GK1.5), anti–CD25-PE (eBioscience; clone PC61.5), and anti–FoxP3-APC (eBioscience; clone FJK-16s); type 1 helper (Th1) cells: anti–CD3-PE-Vio770 (Miltenyi Biotec; clone REA641), anti–CD4-FITC (Miltenyi Biotec; clone REA604), anti–CD183-PE (Miltenyi Biotec; clone CXCR3-173), anti–T-bet–APC (Miltenyi Biotec; clone REA102); and Th17 cells: anti–CD3-APC-Vio770 (Miltenyi Biotec; clone REA641), anti–CD4-FITC (Miltenyi Biotec; clone REA604), anti–RORγT-APC (Miltenyi Biotec; clone REA278), anti–AHR-PE-Vio770 (eBioscience; clone 4MEJJ). Dead cells were excluded from analysis via propidium iodide viability dye (Miltenyi Biotec) or e450 viability dye (Invitrogen). A minimum of 10,000 gated cells were analyzed per sample. Data were acquired by the MACSQuant System (Miltenyi Biotec) and analyzed by FlowJo 11.0 software (TreeStar, Ashland, OR).
Immunofluorescence
MDSC immunofluorescence analysis was performed in serial frontal sections (5 μm thick) of paraffin-embedded proximal tibiae and stained for CD11b with Ly6G/C. Samples were deparaffinized with mixed xylenes, rehydrated in graded ethanols, and then briefly washed. Antigen retrieval was performed in 0.2 mol/L boric acid (pH 7.0) at 60°C overnight. The following day, samples were cooled to room temperature, washed with 10 mmol/L phosphate-buffered saline (pH 7.4), and blocked with 10% normal goat serum (Kirkegaard & Perry Lab Inc., Gaithersburg, MD) for 30 minutes. Sections were then incubated with 1:400 dilution of CD11b polyclonal antibody (Novus Biologicals; clone X-6) and 1:50 dilution of Ly6G/C monoclonal rat antibody (Novus Biologicals; clone NIMP-R14) in antibody diluent (Thermo Scientific) overnight at 4°C. Sections were washed with phosphate-buffered saline and then incubated with a 1:400 dilution of Alexa Fluor 594–conjugated goat anti-rat IgG (Abcam, Cambridge, UK) and a 1:2000 dilution of FITC-goat anti-rabbit (Abcam) for 1 hour at room temperature (protected from light). Sections were washed with phosphate-buffered saline, mounted via ProLong Diamond Antifade Mountant with DAPI (Life Technologies, Carlsbad, CA), and coverslipped. Negative control was performed with a section of each specimen via treating with normal 10% goat serum in antibody diluent. Images were collected with an Olympus BX-61 fluorescence microscope and Visiopharm software. Acquired images were overlaid via Adobe Photoshop CS5 Extended version 12.0 (Adobe Systems, San Jose, CA), and MDSC immunofluorescence analysis performed in the proximal tibia was limited to the secondary spongiosa, beginning 250 μm distal to the growth plate and extended 1000 μm distally (50 μm from endocortical surfaces). Relative fluorescence was calculated in arbitrary units via ImageJ software version 1.51j8 (NIH, Bethesda, MD; https://imagej.nih.gov/ij).
Statistical Analysis
Unpaired t-tests were performed using GraphPad 7.0 (GraphPad Software, La Jolla, CA). Data are presented as means ± SEM; P < 0.05 was considered significant.
Results
Antibiotic Perturbation of Indigenous Gut Microbiota Is Sex Dependent
Antibiotics or vehicle-control treatment was administered via drinking water to postpubertal sex-matched C57BL/6T mice, from the age of 6 to 12 weeks (Figure 1A). To broadly disrupt the indigenous gut microbiota, the antibiotic cocktail of vancomycin, imipenem/cilastatin, and neomycin targeted Gram (+), Gram (−), and anaerobic bacteria.67 Treatment was initiated at the age of 6 weeks, the developmental age when the murine immune system is considered mature,74, 75 to avoid stunting immune development. Mice were euthanized at the age of 12 weeks because this is the developmental age when bone modeling (growth) is considered principally complete in the C57BL/6 strain.76, 77 Recognizing that the late pubertal/adolescent phase is associated with robust bone modeling/remodeling, which accounts for approximately 40% of peak bone mass accrual,78, 79, 80, 81 the experimental design provides the opportunity to evaluate antibiotic disruption of gut microbiota–mediated osteoimmunomodulatory effects during a critical window of skeletal development.
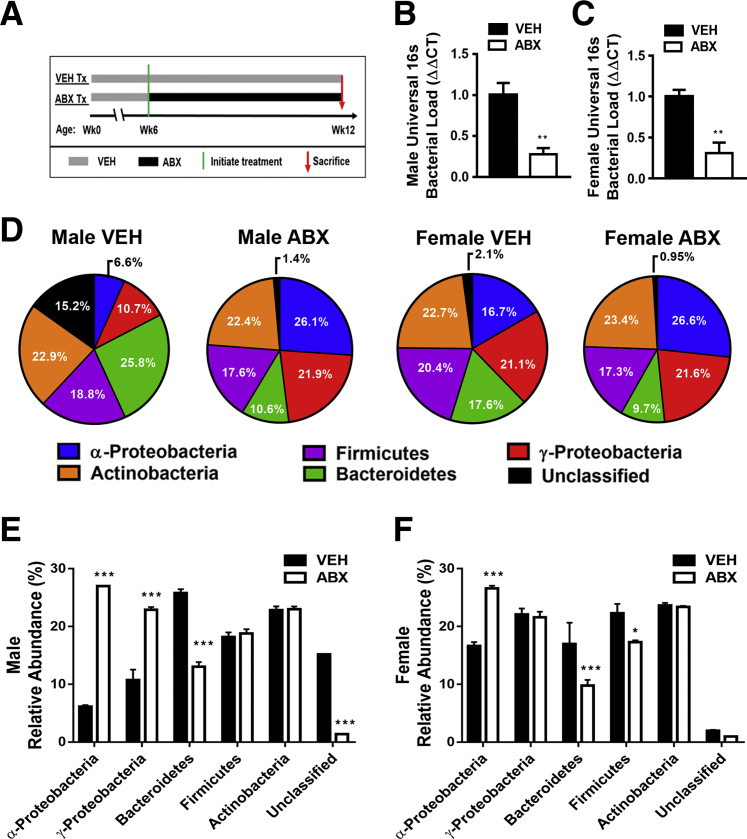
Figure 1.
Antibiotic (ABX) perturbation of indigenous gut microbiota. A: Experimental timeline of the broad-spectrum antibiotic cocktail of vancomycin (500 mg/L) targeting Gram (+) bacteria, imipenem/cilastatin (500 mg/L) targeting Gram (+), Gram (−), and anaerobes, and neomycin (1000 mg/L) targeting Gram (+) and Gram (−) bacteria. B–F: 16S rDNA analysis of fecal specimens from 12-week–old mice. B and C: Universal 16S gene analysis for bacterial load in males (B) and females (C). D–F: Phylum-level composition in males and females, displayed in pie chart (D) and bar graph (E and F) format. Phylum-level real-time quantitative PCR analysis was normalized to the universal 16S gene expression. Unpaired t-test was used. Data are expressed as means ± SEM. n = 5 per group (B and C). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus vehicle. Tx, treatment; VEH, vehicle; wk, week.
Antibiotic perturbation of gut microbiota was validated through 16S rDNA qPCR analysis (Figure 1, B–F). Antibiotic treatment reduced the bacterial load in both male antibiotic versus vehicle-treated mice (Figure 1B) and female antibiotic versus vehicle-treated mice (Figure 1C). Phylum level alterations in bacterial composition were sex dependent (Figure 1, D–F). Male antibiotic versus vehicle-treated mice demonstrated significant increases in α-proteobacteria and γ-proteobacteria and a decrease in Bacteroidetes (Figure 1E). Female antibiotic versus vehicle-treated mice showed a significant increase in α-proteobacteria and decreases in Bacteroidetes and Firmicutes (Figure 1F).
Antibiotic Disruption of Gut Microbiota Alters Bone Mineral Density and Trabecular Bone Morphology
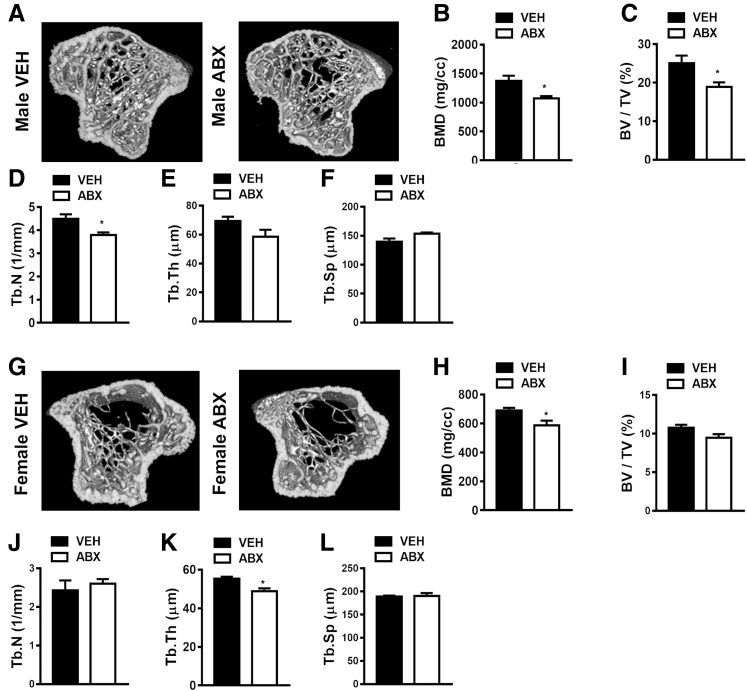
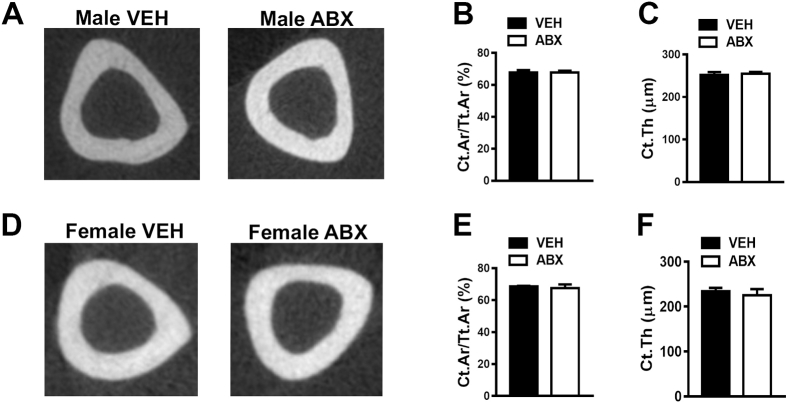
μCT analysis was performed to investigate antibiotic-induced tissue level alterations in cortical (Supplemental Figure S1) and trabecular (Figure 2) bone properties. Cortical bone area fraction and cortical thickness were similar in male antibiotic versus vehicle-treated mice (Supplemental Figure S1, A–C) and female antibiotic versus vehicle-treated mice (Supplemental Figure S1, D–F). To the contrary, antibiotics suppressed the accrual of bone mass in the trabecular bone compartment of both male (Figure 2, A and B) and female (Figure 2, G and H) mice. μCT analysis of trabecular parameters demonstrated that antibiotic treatment induced a more profound inferior trabecular bone phenotype in the proximal tibia of male mice. Male antibiotic- versus vehicle-treated mice had decreased trabecular bone volume fraction (Figure 2C), which was attributed to reduced trabecular number (Figure 2D) and trends toward decreased trabecular thickness (P < 0.09) (Figure 2E) and increased trabecular separation (P < 0.07) (Figure 2F). Female antibiotic- versus vehicle-treated mice had decreased trabecular thickness (Figure 2K), but there were no differences in other trabecular microarchitecture parameters (Figure 2, J and L) or overall trabecular bone volume fraction (Figure 2I). Although antibiotic treatment induced an osteopenic trabecular bone phenotype in both male (Figure 2B) and female (Figure 2H) mice, based on the microarchitecture differences being more profound in male (Figure 2, C–F) versus female (Figure 2, I–L) mice, the subsequent analyses were limited to male antibiotic- versus vehicle-treated mice.
Figure 2.
Antibiotic (ABX) disruption of gut microbiota effects on bone mineral density (BMD) and trabecular bone morphology. Twelve-week–old male vehicle (VEH)– and ABX-treated mice and female VEH- and ABX-treated mice were euthanized; specimens were harvested for analysis. A–F: Micro–computed tomographic (μCT) analyses of the proximal tibiae trabecular bone in male VEH- and ABX-treated mice. A: Representative reconstructed cross-sectional images, extending 360 μm distally from where analysis was initiated, in male VEH- and ABX-treated mice. B: Male trabecular BMD. C: Male trabecular bone volume fraction (BV/TV). D: Male trabecular number (Tb.N). E: Male trabecular thickness (Tb.Th). F: Male trabecular separation (Tb.Sp). G–L: μCT analyses of the proximal tibiae trabecular bone in female VEH- and ABX-treated mice. G: Representative reconstructed cross-sectional images, extending 360 μm distally from where analysis was initiated, in female VEH- and ABX-treated mice. H: Female BMD. I: Female BV/TV. J: Female Tb.N. K: Female Tb.Th. L: Female Tb.Sp. Unpaired t-test was used. Data are expressed as means ± SEM. n = 4 per group (A–L). ∗P < 0.05 versus vehicle. cc, cubic centimeters.
Antibiotic Treatment Does Not Induce Toxicity Effects, Disrupt Normal Somatic Growth Processes, or Affect Endochondral Bone Formation
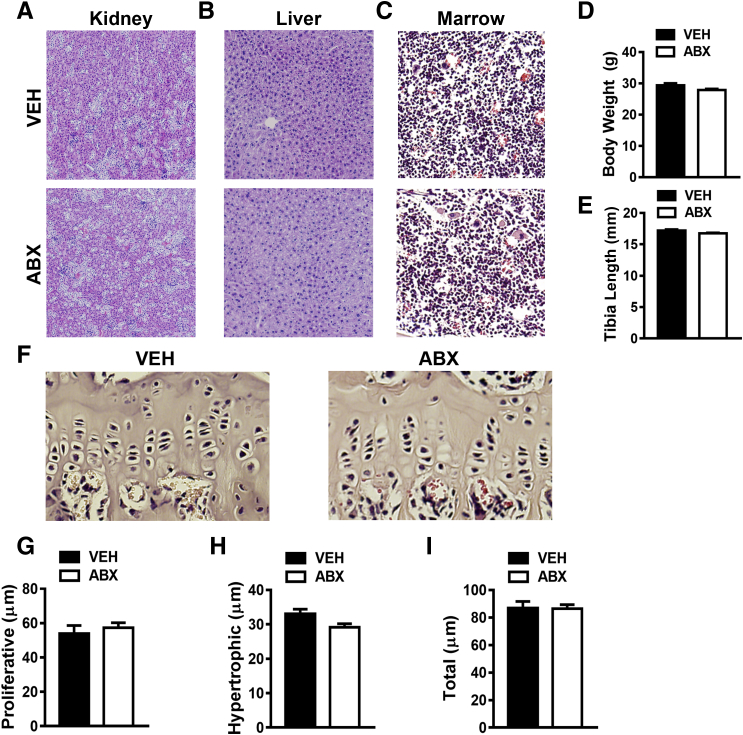
Histopathology investigations performed in kidney (Figure 3A), liver (Figure 3B), and bone marrow (Figure 3C) tissue sections ruled out antibiotic-mediated toxicity effects on host mammalian cells. Lack of differences in animal body weight (Figure 3D) and tibia length (Figure 3E) in antibiotic- versus vehicle-treated mice further support the notion that antibiotic treatment did not induce harmful effects on normal somatic growth processes.
Figure 3.
Antibiotic (ABX) treatment impact on tissue histology and endochondral bone formation. Twelve-week–old male vehicle (VEH)– and ABX-treated mice were euthanized; specimens harvested for analysis. A–C: Histologic assessment of kidney (A), liver (B), and tibia (C) bone marrow. D: Animal body weights. E: Tibia length (mm). F–I: Tibial growth plate morphology and height were evaluated in hematoxylin and eosin–stained proximal tibia sections. F: Representative images of tibial growth plate. G: Proliferative zone height. H: Hypertrophic zone height. I: Total growth plate height. Unpaired t-test was used. Data are expressed as means ± SEM. n = 4 to 5 per group (D, E, and G–I). Original magnification, ×200 (A–C and F).
To validate no changes in tibia length, growth-plate chondrocyte zone morphology and height were assessed in proximal tibiae to evaluate alterations in endochondral bone formation (Figure 3F). Antibiotic- versus vehicle-treated mice had similar growth plate chondrocyte zone morphology, which lacked differences in proliferative zone height (Figure 3G), hypertrophic zone height (Figure 3H), and total growth plate height (Figure 3I). The findings that antibiotic- versus vehicle-treated mice had similar growth plate morphology (Figure 3, F–I) and tibia length (Figure 3E) suggest that antibiotic perturbation of gut microbiota does not disrupt endochondral bone formation.
Antibiotic Perturbation of Gut Microbiota Does Not Alter Osteoblastogenesis
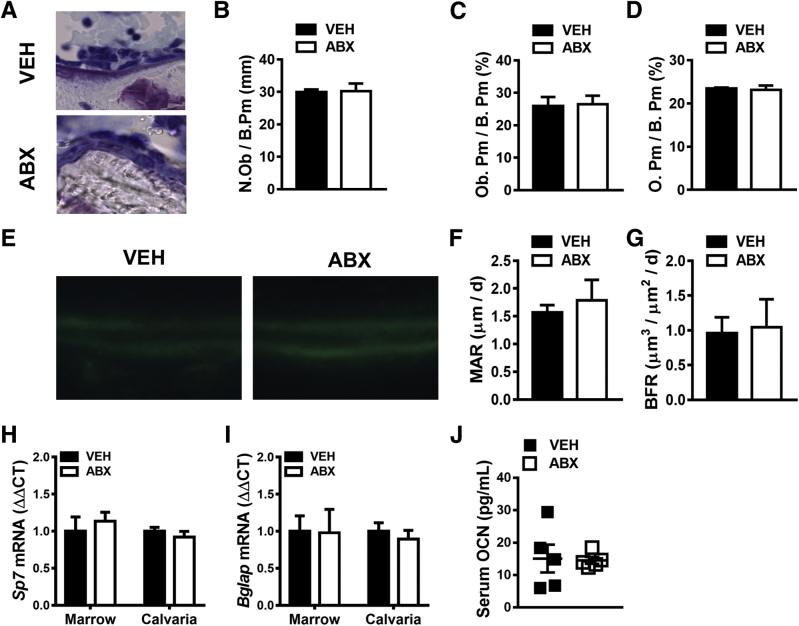
Static histomorphometric analysis was performed on toluidine blue–stained undecalcified tibia sections to investigate antibiotic effects on osteoblast cellular end points (Figure 4, A–D). Antibiotic- and vehicle-treated mice had similar osteoblast numbers (Figure 4B), no differences in osteoblast perimeter per bone perimeter (Figure 4C), and no changes in osteoid per bone perimeter (Figure 4D).
Figure 4.
Impact of antibiotic (ABX) treatment on osteoblastogenesis. Twelve-week–old male vehicle (VEH)– and ABX-treated mice were euthanized; specimens were harvested for analysis. A–D: Histomorphometric analyses of osteoblast cellular end points were performed in the trabecular bone secondary spongiosa of toluidine blue–stained proximal tibia sections. A: Representative images of toluidine blue–stained secondary spongiosa. B: Osteoblast number per bone perimeter (N.Ob/B.Pm). C: Osteoblast perimeter per bone perimeter (Ob.Pm/B.Pm). D: Osteoid per bone perimeter (O.Pm/B.Pm). E–G: Dynamic histomorphometric analysis of bone formation indexes in proximal tibia trabecular bone; calcein was administered 5 and 2 days before sacrifice. E: Representative images of calcein-labeled secondary spongiosa. F: Mineral apposition rate (MAR). G: Bone formation rate (BFR). H and I: Real-time quantitative RT-PCR (RT-qPCR) analysis of mRNA in femur bone marrow and calvaria. Relative quantification of mRNA was performed via the comparative CT method (ΔΔCT); Gapdh was used as an internal control. Data are expressed as fold difference relative to VEH. H: RT-qPCR analysis of Sp7 (osterix) mRNA in femur bone marrow and calvaria. I: RT-qPCR analysis of Bglap [osteocalcin (OCN)] mRNA in femur bone marrow and calvaria. J: Serum was isolated from whole blood; enzyme-linked immunosorbent assay analysis of intact OCN levels. Unpaired t-test was used. Data are expressed as means ± SEM. n = 4 per group (A–G); n = 4 to 5 per group (H and I); n = 5 per group (J). Original magnifications: ×200 (A); ×400 (E).
Considering investigations in the C57BL/6 germ-free mouse model have shown that the normal gut microbiota can blunt bone modeling/remodeling,52, 55 dynamic bone formation indexes were analyzed (Figure 4, E–G) to elucidate whether antibiotic disruption of gut microbiota alters mineral apposition and formation. Mineral apposition rate (Figure 4F) and bone formation rate (Figure 4G) were not different in the proximal tibia of vehicle- and antibiotic-treated mice, which demonstrates that antibiotic perturbation of gut microbiota does not alter osteoblast function.
To further corroborate antibiotic-induced lack of changes in osteoblastogenesis, alterations in Sp7 (osterix) (Figure 4H), a transcription factor essential for osteoblastic cell differentiation and maturation, and Bglap (OCN) (Figure 4, I and J), a noncollagenous matrix factor produced by mature osteoblasts, were evaluated in marrow and calvaria. The rationale for evaluating two distinct skeletal sites was to discern systemic osteoblast effects. In addition, calvaria has a more homogeneous stromal-osteoblastic composition better reflecting osteoblast-specific gene expression. Vehicle- versus antibiotic-treated mice had similar Sp7 (osterix) (Figure 4H) and Bglap (OCN) gene expression in femoral bone marrow and calvaria (Figure 4I). To further rule out systemic alterations in osteoblastogenesis, intact OCN protein levels were evaluated in serum (Figure 4J). Importantly, the serum enzyme-linked immunosorbent assay excluded detection of OCN fragments derived from osteoclast-mediated bone resorption. Vehicle- and antibiotic-treated mice had similar intact OCN serum levels (Figure 4J), validating that antibiotic disruption of gut microbiota does not influence osteoblast-mediated bone formation.
Antibiotic Alteration of Gut Microbiota Enhances Osteoclastogenesis and Local/Systemic Proinflammatory Cytokines
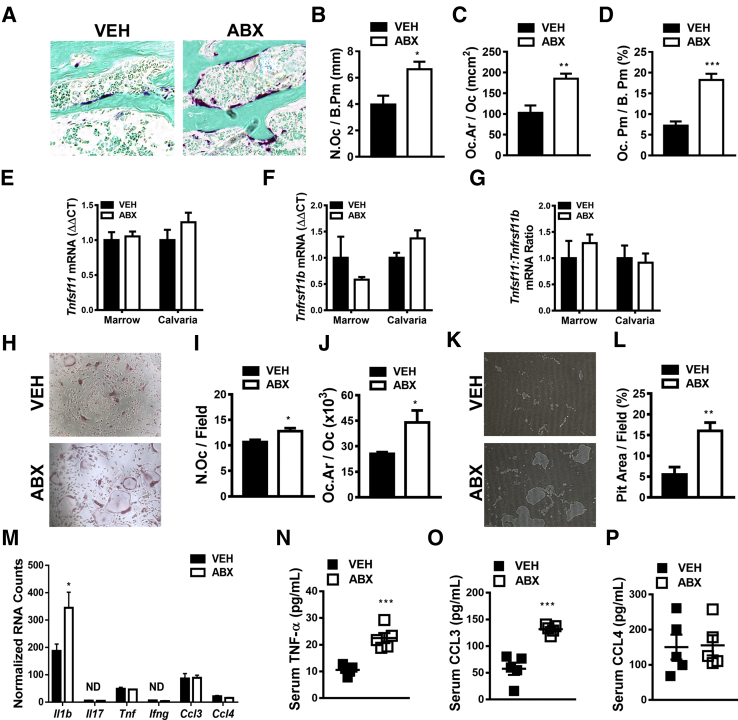
Recognizing that bone modeling/remodeling occurs through dual osteoclast-osteoblast actions, studies were performed to elucidate the impact of antibiotic perturbation of gut microbiota on osteoclastogenesis. Histomorphometric analysis of TRAP-stained proximal tibia sections was performed to investigate antibiotic effects on osteoclastogenesis (Figure 5, A–D). Antibiotic- versus vehicle-treated mice had 2.5× greater osteoclast perimeter per bone perimeter (Figure 5D), which was attributed to a 1.75× increase in osteoclast size (Figure 5C) and a 1.67× increase osteoclast numbers (Figure 5B).
Figure 5.
Antibiotic (ABX) perturbation of gut microbiota effects on osteoclastogenesis and local/systemic proinflammatory cytokines. Twelve-week–old male vehicle (VEH)– and ABX-treated mice were euthanized; specimens were harvested for analysis. A–D: Histomorphometric analyses of osteoclast cellular end points were performed in the trabecular bone secondary spongiosa of tartrate-resistant acid phosphatase (TRAP)–stained proximal tibia sections; TRAP+ cell lining bone with three or more nuclei designated an osteoclast. A: Representative images of TRAP-stained (fast green counterstain) secondary spongiosa. B: Osteoclast number per bone perimeter (N.Oc/B.Pm). C: Osteoclast area/size (Oc.Ar/Oc). D: Osteoclast perimeter per bone perimeter (Oc.Pm/B.Pm). E–G: Real-time quantitative RT-PCR analysis in bone marrow and calvaria to assess alterations in the RANKL/OPG axis: Tnfsf11(Rankl) mRNA (E), Tnfrsf11b(Opg) mRNA (F), and Tnfsf11(Rankl):Tnfrsf11b(Opg) ratio (G). Relative quantification of mRNA was performed via the comparative CT method (ΔΔCT); Gapdh was used as an internal control. Data are expressed as fold difference relative to VEH. H–J: Hematopoietic cells were isolated from marrow, and CD11b sorting was performed to enrich for CD11bneg OCP cells. CD11bneg OCP cells were stimulated with 25 ng/mL colony stimulating factor 1 (CSF1) and 50 ng/mL RANKL to induce osteoclastogenesis. Cytomorphometric cellular end points were analyzed in TRAP-stained cultures. TRAP+ cells with three or more nuclei were considered osteoclasts. H: Representative images of OCP cultures stimulated with CSF1 and RANKL media. I: Number of osteoclasts per field of view (N.Oc/Field). J: Average osteoclast size (Oc.Ar/Oc). K and L: Hematopoietic cells were isolated from marrow, and CD11b sorting was performed to enrich for CD11bneg OCP cells. CD11bneg OCP cells were stimulated with 25 ng/mL CSF1 and 50 ng/mL RANKL to induce osteoclastogenesis. Osteoclast resorption activity was assessed by quantifying pit area in Osteo Assay plates. K: Representative images of pit area. L: Pit area per field area (percentage). M: NanoString analysis was performed to assess proinflammatory cytokines in femur bone marrow. Data were normalized to the geometric means of spiked-in positive controls and internal control genes. Absolute quantification of RNA expressed as normalized RNA counts. N–P: Serum was isolated from whole blood; enzyme-linked immunosorbent assay analysis of TNF-α (N), CCL3 (O), and CCL4 (P). Unpaired t-test was used. Data are expressed as means ± SEM. n = 4 per group (A–D and H–M); n = 4 to 5 per group (E–G); n = 5 per group (N–P). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus vehicle. Original magnification, ×200 (A, H, and K). ND, not detectable.
The Tnfsf11(Rankl):Tnfrsf11b(Opg) axis was evaluated (Figure 5, E–G) to discern whether alterations in critical osteoclastic signaling factors mediate the pro-osteoclastic phenotype in antibiotic-treated mice. RANKL, which signals at the RANK receptor on preosteoclast/osteoclast cells, is required for osteoclast differentiation and function. Because osteoprotegerin (OPG) functions as the RANK decoy receptor, the RANKL/OPG ratio must be assessed when evaluating potential alterations in RANKL-mediated osteoclastogenesis.45, 50, 82, 83 Tnfsf11(Rankl) (Figure 5E), Tnfrsf11b(Opg) (Figure 5F), and Tnfsf11(Rankl):Tnfrsf11b(Opg) ratio (Figure 5G) outcomes were similar in the femoral bone marrow and calvaria of antibiotic- versus vehicle-treated mice, which indicates that antibiotic perturbation of gut microbiota does not alter levels of unbound RANKL available to activate RANK signaling.
Osteoclast-precursor (OCP) assays (Figure 5, H–L) were performed in vitro to confirm antibiotic perturbation of gut microbiota effects on osteoclast differentiation/maturation and to evaluate osteoclast function. Hematopoietic cells were isolated from marrow, and CD11b magnetic cell sorting was performed to enrich for CD11bneg OCP cells, having a high osteoclastic potential. CD11bneg OCP cells were stimulated with 25 ng/mL CSF1 and 50 ng/mL RANKL to induce osteoclastogenesis. Osteoclast differentiation and maturation were defined via cytomorphometric analyses of TRAP+ cell (≥3 nuclei) numbers and size (Figure 5, H–J). Corroborating in vivo osteoclast cellular outcomes (Figure 5, B and C), OCP cultures derived from antibiotic-treated mice displayed increased osteoclast numbers (per field) (Figure 5I) and size (Figure 5J). Osteoclast resorption activity was analyzed by quantifying pit area in Osteo Assay plates (Figure 5, K and L). In line with the pro-osteoclastic cellular end points found in TRAP-stained OCP cultures from antibiotic- versus vehicle-treated mice (Figure 5, H–J), pit area per field area was more pronounced in OCP cultures derived from antibiotic-treated mice (Figure 5, K and L). The enhanced osteoclast maturation phenotype (Figure 5, H–J) and resorptive function (Figure 5, K and L) shown in OCP cultures derived from antibiotic-treated mice suggests that antibiotic disruption of gut microbiota induces immunostimulatory effects that up-regulate the osteoclastic potential of marrow OCP cells.
Proinflammatory cytokines enhance RANKL-mediated osteoclastogenesis via modulating the Tnfsf11(Rankl):Tnfrsf11b(Opg) ratio and through synergistic effects on RANKL signaling.45, 50, 82, 83 Therefore, alterations in proinflammatory cytokines known to enhance RANKL-mediated osteoclastogenesis were investigated. Gene expression analysis demonstrated that Il1b was the only characteristic proinflammatory/osteoclastic cytokine locally up-regulated in the marrow of antibiotic- versus vehicle-treated mice (Figure 5M). Il17 and Ifng were nondetectable, and there were no differences in Tnf, Ccl3, or Ccl4 expression in the marrow of antibiotic- versus vehicle-treated mice (Figure 5M).
Recognizing that endocrine signaling effects can modulate RANKL-mediated osteoclastogenesis in the local bone marrow environment, it is imperative to evaluate circulating proinflammatory cytokine levels. Cytokines expressed at detectable levels in the bone marrow (ie, Tnf, Ccl3, and Ccl4) (Figure 5M) were evaluated via serum enzyme-linked immunosorbent assay analysis (Figure 5, N–P). Circulating TNF-α (Figure 5N) and CCL3 (Figure 5O) levels were enhanced 2× fold in antibiotic- versus vehicle-treated mice, whereas CCL4 levels were similar (Figure 5P). The lack of differences in Ccl3 and Tnf expression locally in the bone marrow (Figure 5M), and the finding that CCL3 and TNF-α were substantially up-regulated in the circulation of antibiotic- versus vehicle-treated mice (Figure 5, N and O), supports the notion that antibiotic pro-osteoclastic actions are secondary to immune response effects occurring in local gut or systemic lymphoid tissues.
Antibiotic Disruption of Gut Microbiota Substantially Alters the Innate and Adaptive Immune Cell Profile in mLNs and Spleen
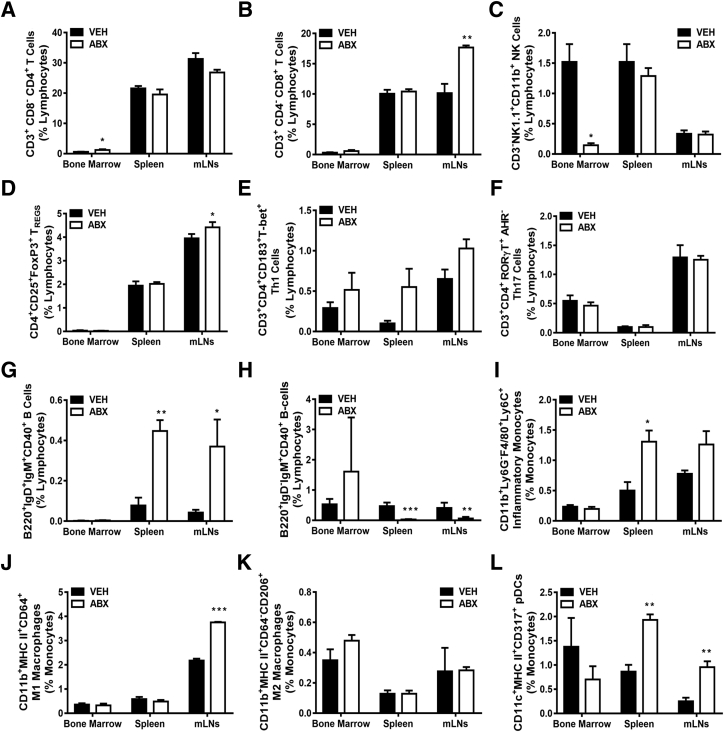
Appreciating that adaptive and innate immune cells are critical regulators of osteoclastogenesis and bone modeling/remodeling processes,44, 45, 46, 47, 48 flow cytometric analysis of immune cells was performed in femur bone marrow and secondary lymphoid tissues draining local gut and systemic circulation (Figures 6 and 7). mLN cells were evaluated to determine local gut immune response effects, and spleen cells were assessed to discern systemic immune response effects. The frequency of CD3+CD4+CD8− (helper) T cells was increased in the bone marrow of antibiotic-treated mice (Figure 6A), whereas the frequency of CD3−NK1.1+CD11b+ NK cells was decreased in antibiotic-treated bone marrow (Figure 6C). Astonishingly, other T- and B-cell lymphocyte population differences were centered in the mLNs and spleens of antibiotic- versus vehicle-treated mice. The frequencies of CD3+CD4−CD8+ (cytotoxic) T cells (Figure 6B) and CD3+CD4+CD25+FOXP3+ TREG cells (Figure 6D) were up-regulated in the mLNs, whereas CD3+CD4+CD183+T-bet+ Th1 cells (Figure 6E) were approaching significance in spleens (P < 0.06) and mLNs (P < 0.08) of antibiotic-treated mice. However, there were no differences observed in frequency of CD3+CD4+RORγT+AHR− Th17 cells (Figure 6F) in antibiotic-treated mice compared with vehicle-treated mice. Furthermore, the frequency of B220+IgD+IgM+CD40+ (activated) B cells was enhanced (Figure 6G), whereas the frequency of B220+IgD−IgM+CD40+ (memory) B cells was suppressed (Figure 6H), in the mLNs and spleens of antibiotic-treated mice.
Figure 6.
Antibiotic (ABX) alteration of gut microbiota impact of innate and adaptive immune cell profile. Twelve-week–old male vehicle (VEH)– and ABX-treated mice were euthanized; specimens were harvested for analysis. A–H: Flow cytometric analysis of lymphocyte immune cell populations in bone marrow, spleen, and mesenteric lymph nodes (mLNs). Percentages of CD3+CD8−CD4+ (helper) T cells (A); CD3+CD4−CD8+ (cytotoxic) T cells (B); CD3−NK1.1+CD11b+ NK cells (C); CD3+CD4+CD25+FoxP3+ TREG cells (D); CD3+CD4+CD183+T-bet+ Th1 cells (E); CD3+CD4+RORγT+AHR− Th17 cells (F); B220+IgD+IgM+CD40+ (activated) B cells (G); and B220+IgD−IgM+CD40+ (memory) B cells (H). Percentages are expressed relative to total live gated lymphocytes. I–L: Flow cytometric analysis of innate immune cell populations in bone marrow, spleen, and mLNs. Percentages of CD11b+Ly6G−F4/80+Ly6C+ (proinflammatory) monocytes (I); CD11b+ MHC-class II+CD64+ M1 macrophages (J); CD11b+MHC II+CD64−CD206+ M2 macrophages (K); and CD11c+MHC-class II+CD317+ (activated plasmacytoid DCs; L). Cell percentages are expressed relative to total live-gated monocyte cells. Unpaired t-test was used. Data are expressed as means ± SEM (A–L). n = 4 to 5 per group (A–L). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus vehicle.
Figure 7.
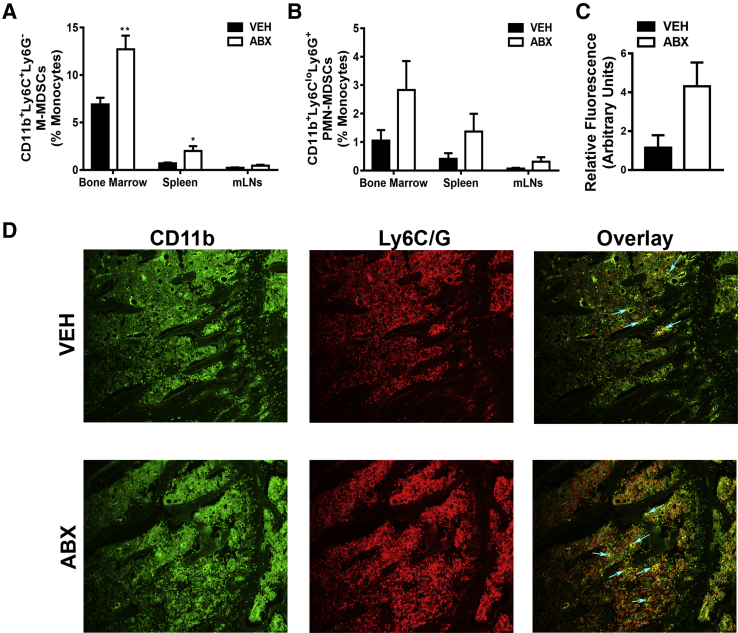
Antibiotic (ABX) perturbation of gut microbiota influence on myeloid-derived suppressor cells (MDSCs). Twelve-week–old male vehicle (VEH)– and ABX-treated mice were euthanized; specimens were harvested for analysis. A and B: Flow cytometric analysis of MDSC subset populations in bone marrow, spleen, and mesenteric lymph nodes (mLNs). Percentages of CD11b+Ly6C+Ly6G− (monocytic) MDSCs (M-MDSCs; A) and CD11b+Ly6CloLy6G+ (polymorphonuclear) MDSCs (PMN-MDSCs; B). Cell percentages are expressed relative to total gated monocyte cells. C and D: MDSC immunofluorescence analysis was performed on proximal tibia (secondary spongiosa) sections. C: Relative fluorescence of CD11b+Ly6C+Ly6G+ MDSCs. D: Representative MDSC immunofluorescence-labeled proximal tibia sections (green, CD11b-FITC; red, Ly6C/G-rhodamine); arrows indicate dual-labeled MDSCs. Unpaired t-test was used. Data are expressed as means ± SEM (A–C). n = 4 per group (A–D). ∗P < 0.05, ∗∗P < 0.01 versus vehicle. Original magnification, ×200 (D).
Dysregulation in adaptive immune cell populations indicates that improper interactions with innate immune cells may exist within the lymphoid tissue microenvironment. Innate immune cell differences were evaluated by performing flow cytometric analysis of proinflammatory monocytes (Figure 6I), M1 macrophages (Figure 6J), M2 macrophages (Figure 6K), and activated plasmacytoid dendritic cells (Figure 6L). Corroborating the altered T-cell (Figure 6, B, D, and E) and B-cell (Figure 6, G and H) populations in the mLNs and spleen, the frequencies of CD11b+Ly6G−F4/80+Ly6C+ (proinflammatory) monocytes (Figure 6I), CD11b+CD11c−CD64+B220−MHC II+ M1 macrophages (Figure 6J), and CD11c+MHCII+CD317+ (activated plasmacytoid dendritic) cells (Figure 6L) were enhanced in the mLNs and spleens of antibiotic-treated mice, whereas there were no differences in CD11b+CD11c−CD64−B220−MHC II+CD206+ M2 macrophages across tissues (Figure 6K). There were no differences in percentage of CD11b+Ly6G−F4/80+Ly6C+ (proinflammatory) monocytes (Figure 6I), percentage of CD11b+CD11c−CD64+B220−MHC II+ M1 macrophages (Figure 6J), or percentage of CD11c+MHCII+CD317+ (activated plasmacytoid dendritic) cells (Figure 6L) in the marrow of antibiotic- versus vehicle-treated mice.
Antibiotic Perturbation of Gut Microbiota Up-Regulates MDSCs
To further explore the role of innate immunity in antibiotic perturbation of gut microbiota–induced immune response effects, MDSCs were assessed in bone marrow, spleen, and mLNs (Figure 7). MDSCs are a heterogeneous population of myeloid precursor cells, which regulate the innate and adaptive immune response.84, 85 Although MDSCs were initially described in cancer, research advances have elucidated that MDSCs regulate immune responses and the pathogenesis of many pathologic inflammatory conditions.84, 85 MDSCs primarily consist of two subtypes, monocytic MDSCs and polymorphonuclear MDSCs, which is based on cell phenotypic and immunosuppressive features.85, 86
Flow cytometric analysis demonstrated that the frequency of CD11b+Ly6C+Ly6G− monocytic MDSCs was significantly enhanced in the bone marrow and spleen and marginally increased in the mLNs (P < 0.07), of antibiotic- versus vehicle-treated mice (Figure 7A). Moreover, flow cytometric analysis revealed that percentage of CD11b+Ly6CloLy6G+ polymorphonuclear MDSCs was marginally increased in the marrow of antibiotic-treated mice (P < 0.08) (Figure 7B). The MDSC differences found in the bone marrow of antibiotic-treated mice were unexpected, considering our prior flow cytometry outcomes demonstrating that antibiotic-induced immune cell alterations were centered in the mLNs and spleens. As a means to validate the up-regulated frequency of MDSCs found in bone marrow by flow cytometry (Figure 7, A and B), immunofluorescence staining for total (CD11b+Ly6C+Ly6G+) MDSCs was performed in proximal tibia bone marrow sections (Figure 7, C and D). Consistent with the flow cytometry outcomes, in situ immunofluorescence analysis demonstrated marginally increased total (CD11b+Ly6C+Ly6G+) MDSCs in the marrow of antibiotic- versus vehicle-treated mice (P < 0.06) (Figure 7, C and D).
MHC Class II Antigen Presentation Pathway Is Altered in Antibiotic Disruption of Gut Microbiota
Prior reports have demonstrated that antibiotic perturbation of the gut microbiota affects immune recognition and cross talk pathways.87, 88, 89 Given the antibiotic-induced up-regulation of MDSCs in the bone marrow (Figure 7), MDSC-related immune response mechanisms were investigated. Because MDSC-mediated immunosuppression can occur in an antigen (Ag)–specific manner,90, 91, 92 the impact of antibiotic perturbation of the gut microbiota on MHC class II Ag presentation pathways was evaluated. MHC class II Ag processing and presentation mechanisms were assessed in the marrow via nCounter gene expression analysis (Table 1). Antibiotic treatment marginally down-regulated gene variants for the class II complex (H2-Aa, H2-DMb2, H2-Eb1, and H2-Ob) as well as the MHC class II transactivator (Ciita). Cd74, which is further along the class II pathway and functions to encode for invariant chain (Ii) and class II–associated Ii peptide, was suppressed in the marrow of antibiotic-treated mice. Ctss and Ctsc, which play key roles in MHC class II Ag processing, were reduced in the marrow of antibiotic- versus vehicle-treated mice. Further validating the effect of antibiotic perturbation of the gut microbiota on the MHC class II Ag presentation pathway, Cd86, a member of the B7 costimulatory family, was also down-regulated in the marrow of antibiotic-treated mice.
Table 1.
MHC Class II Gene Expression Modified by Antibiotic Treatment
| MHC class II antigen presentation–associated genes | |||
|---|---|---|---|
| Gene | VEH | ABX | P value |
| H2-Aa | 5717.26 ± 849 | 3988.75 ± 987 | 0.037 |
| H2-DMb2 | 317.52 ± 135 | 182.45 ± 57 | 0.057 |
| H2-Eb1 | 1082.71 ± 349 | 703.77 ± 211 | 0.056 |
| H2-Ob | 304.93 ± 103 | 219.61 ± 45 | 0.090 |
| Ciita | 173.74 ± 82 | 104.58 ± 20 | 0.076 |
| Cd74 | 7260.18 ± 2589 | 4401.80 ± 1085 | 0.044 |
| Ctss | 3139.61 ± 405 | 2309.16 ± 287 | 0.015 |
| Ctsc | 1118.08 ± 130 | 873.48 ± 117 | 0.031 |
| Cd86 | 209.11 ± 54 | 121.94 ± 19 | 0.023 |
Gene expression tested by nCounter analysis, represented as total RNA counts. n = 4 per group. Unpaired t-test was used. Data are expressed as means ± SEM. Significant values (P < 0.05) are in bold.
ABX, antibiotic; VEH, vehicle.
Discussion
This osteoimmunology study in sex-matched growing C57BL/6T mice demonstrates that antibiotic perturbation of the gut microbiota has catabolic effects on the accrual of bone mass during postpubertal skeletal development. Although prior investigations have shown that antibiotic perturbation of the gut microbiota influences bone mass60, 66 and bone mechanical properties,66 the current report reveals that antibiotic disruption of the gut microbiota critically alters normal osteoimmune processes. Broad-spectrum antibiotics induce sex-dependent alterations in the gut microbiota composition, which result in a more profound suppression of trabecular bone parameters in male versus female mice. Osteoimmunological studies discerned that antibiotic perturbation of the gut microbiota has pro-osteoclastic effects, which appear to be mediated by increased circulating levels of CCL3 and TNF. Antibiotic-induced proinflammatory changes in adaptive and innate immune cells primarily occurred in the mLNs and spleen, which suggests that elevated circulating proinflammatory cytokines are due to hyperimmune response effects at secondary lymphoid tissues draining local gut and systemic circulation. Antibiotic induced up-regulation of MDSCs and suppressed MHC class II antigen processing/presentation in the bone marrow implies that the antibiotic disruption of the gut microbiota critically alters osteoimmune cross talk.
Whereas previous reports have demonstrated that antibiotic perturbation of the gut microbiota influences bone mass accrual in the postnatal developing skeleton,60, 66 the current investigation is the first known study to discern sex-dependent effects. A low-dose antibiotic administration study initiated in 4-week–old female C57BL/6J mice showed that continuous antibiotics elevated the Firmicutes/bacteria ratio in feces and transiently increased whole animal bone mineral density scores at the age of 7 weeks.60 Guss et al66 administered broad-spectrum antibiotics to male C57BL/6J mice from the age of 4 to 16 weeks. The continuous broad-spectrum antibiotic therapy induced a phylum level shift from a Bacteroidetes- to a Proteobacteria-dominated fecal microbiota and resulted in an inferior cortical bone phenotype.
This broad-spectrum antibiotic administration study initiated in 6-week–old sex-matched C57BL/6T mice reveals that continuous antibiotics induced sex-dependent phylum-level shifts in fecal microbiota composition, which resulted in a more drastic inferior trabecular bone phenotype in 12-week–old male versus female mice. Similar to the prior broad-spectrum antibiotic treatment study performed in 4- to 16-week–old male C57BL/6J mice,66 it was found that broad-spectrum antibiotic administration in 6- to 12-week–old male C57BL/6T mice resulted in a phylum-level shift from a Bacteroidetes- to a Proteobacteria-dominated fecal microbiota. Shifts in the gut microbiota favoring the Proteobacteria phylum have been associated with increased incidence of dysbiotic proinflammatory states,93, 94 metabolic syndrome,95 and gut epithelial dysfunction.96 Taking into consideration that increased prevalence of the Proteobacteria phylum is associated with enhanced inflammation,93, 94 and that gastrointestinal inflammation suppresses bone modeling/remodeling processes in the developing skeleton,97, 98, 99 the increased prevalence of the Proteobacteria phylum is a candidate mediator of the antibiotic-induced osteopenic skeletal phenotype. Although the current study shows that sex-dependent antibiotic changes in gut bacterial composition induced a more prominent osteopenic trabecular bone phenotype in male versus female mice, future investigations are necessary to delineate underlying sex-specific osteoimmunulogical mechanisms. Outside the scope of the current report, the authors acknowledge that sex-dependent antibiotic alterations in gut microbiota composition could modulate sex hormones that importantly contribute to sex-specific osteoimmune response effects.
Although Guss et al66 found that administering broad-spectrum antibiotics to male C57BL/6J mice, from 4 to 16 weeks of age, impaired cortical bone morphologic parameters, the current report shows that administering broad-spectrum antibiotics to male C57BL/6T mice, from 6 to 12 weeks of age, suppressed trabecular bone morphologic parameters. The osteopenic cortical bone phenotype reported by Guss et al66 versus the osteopenic trabecular bone phenotype found herein demonstrates the complex nature of antibiotic perturbation of gut microbiota effects on postnatal skeletal development. Although unclear, differential study outcomes in antibiotic-treated growing male mice may be related to the following differences in study design: antibiotic cocktail: 1 g/L ampicillin and 0.5 g/L neomycin66 versus 0.5 g/L vancomycin, 0.5 g/L imipenem/cilastatin, and 1 g/L neomycin; mouse substrain: C57BL/6J mice66 versus C57BL/6T mice; and developmental period: prepubertal 4-week–old mice treated to 16 weeks of age66 versus postpubertal 6-week–old mice treated to 12 weeks of age.
The current study substantially advances knowledge about the role of osteoimmunological mechanisms in antibiotic perturbation of gut microbiota effects on postnatal skeletal development. This is the first known report to elucidate antibiotic alteration of gut microbiota effects on bone cell (osteoclast and osteoblast) outcomes. The increased TRAP+ osteoclast cell numbers and size found in vivo and in vitro, as well as enhanced OCP cell resorptive activity shown in vitro, demonstrate that broad-spectrum antibiotics enhance osteoclastogenesis. The lack of static and dynamic histomorphometric differences in osteoblast cellular outcomes, no alterations in surrogate osteoblastic gene expression in marrow and calvaria, and the lack of differences in circulating intact OCN levels imply that broad-spectrum antibiotics do not alter osteoblastogenesis. These findings suggest that the osteopenic trabecular bone phenotype in antibiotic- versus vehicle-treated mice is secondary to antibiotic perturbation of gut microbiota–mediated pro-osteoclastogenic effects.
Although there was no difference in the Tnfsf11(Rankl):Tnfrsf11b(Opg) axis in antibiotic- versus vehicle-treated mice, it is important to understand that proinflammatory cytokines enhance RANKL-mediated osteoclastogenesis.45, 50, 82, 83 Antibiotic treatment up-regulated the proinflammatory cytokine Il1b in the marrow. IL-1β enhances osteoclastogenesis in the bone marrow environment by up-regulating the expression of RANKL and other proinflammatory mediators.100, 101 Considering that there was no difference in the expression of Tnfsf11(Rankl) or other proinflammatory cytokines in the marrow of antibiotic- versus vehicle-treated mice, the up-regulated Il1b expression may function as a compensatory mechanism to sustain normal bone metabolic processes in antibiotic-treated mice. Antibiotic treatment increased the circulating levels of CCL3 and TNF, which are proinflammatory cytokines that have potent synergistic effects on RANKL-signaling–induced osteoclastogenesis.50, 82, 102, 103, 104 Considering these findings that Ccl3 and Tnf expression was not altered locally in the bone marrow, but CCL3 and TNF were substantially up-regulated in the circulation of antibiotic- versus vehicle-treated mice, led us to postulate that antibiotic-induced pro-osteoclastogenic actions are secondary to immune response effects occurring in gut or systemic lymphoid tissues.
Flow cytometric analysis of immune cells was performed in bone marrow, mLNs, and spleen, to discern whether the immunomodulatory effects of antibiotic-altered gut microbiota occurred in the marrow or at secondary lymphoid tissues draining local gut and systemic circulation. Whereas Guss et al66 assessed broad-spectrum antibiotic–induced alterations in total CD3+ T cells and total CD20+ B cells in the spleen, we evaluated alterations in B-cell subsets, T-cell subsets, and innate immune cell subsets across bone marrow, mLNs, and spleen. Spleen and mLN flow cytometry outcomes demonstrating that broad-spectrum antibiotics increased the frequencies of cytotoxic T cells, TREG cells, Th1 cells, activated B cells, activated plasmacytoid dendritic cells, proinflammatory monocytes, M1 macrophages, and polymorphonuclear MDSCs show that antibiotic perturbation of gut microbiota induced a proinflammatory hyperimmune response at systemic and local gut draining lymphoid tissues. Although broad-spectrum antibiotics altered the gut microbiota composition at the phylum level, antibiotic-induced proinflammatory hyperimmune response effects may be attributed to dysbiotic shifts in gut microbiota composition, increased intestinal permeability, translocation of gut microbes/microbial ligands, and/or overgrowth of drug-resistant opportunistic pathogens.21, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43
Josefsdottir et al105 recently demonstrated that antibiotic depletion of the gut microbiota impairs T-lymphocyte progenitor maintenance and granulocyte maturation in mice. This timely investigation used an antibiotic-depletion cocktail (0.5 g/L vancomycin, 1 g/L neomycin, 1 g/L ampicillin, and 1 g/L metronidazole),105 which is inherently different from the antibiotic-perturbation protocol used in this study. Josefsdottir et al105 importantly performed serum enzyme-linked immunosorbent assay studies demonstrating that antibiotic depletion of the gut microbiota did not impact systemic proinflammatory cytokine levels (ie, granulocyte colony-stimulating factor, interferon-α, interferon-γ, IL-6, and TNF). Although this seminal work delineates that antibiotic depletion of the gut microbiota suppresses bone marrow hematopoiesis,105 the current osteoimmunology study importantly begins to describe antibiotic perturbation of the gut microbiota effects on bone marrow hematopoiesis.
MDSCs, an immature myeloid cell population derived in bone marrow, comprise a significant portion of normal bone marrow cells.84, 106 More important, MDSCs are substantially up-regulated in the marrow during disease states, such as neoplasia, systemic lupus erythematosus, and inflammatory arthritis.107, 108, 109 MDSCs have two subtypes; monocytic MDSCs have more robust immunosuppressive properties, whereas polymorphonuclear MDSCs are the prevalent circulating population during tumorigenesis.86, 92, 110 Although these cells have been well characterized in several cancers and autoimmune diseases, their role in osteoimmune processes is unclear.
Under inflammatory/oncogenic conditions, MDSCs travel to the spleen, an important lymphoid organ for hematopoiesis that supports the development of myeloid cells from bone marrow–derived precursors.111 MDSCs are then recruited from the spleen to accumulate at sites of inflammation, where they contribute to a hyperinflammatory microenvironment and tissue damage via several mechanisms. Primarily, MDSCs function in blocking T-cell proliferation,107 but they also modulate the TREG cell population through MHC class II108 and suppress NK cell functions.112, 113 This study shows that antibiotic perturbation of gut microbiota induced changes in MDSC populations in the marrow and spleen, which we speculate drove a significant reduction in NK cell frequency in marrow and up-regulated TREG cell frequency in mLNs.
In addition to their specific role in immunosuppression, MDSCs are the progenitor population of several myeloid-derived immune cells, such as macrophages, plasmacytoid DCs, inflammatory monocytes, and granulocytes. Inflammation-derived proinflammatory soluble factors (ie, IL-1β, S100A8, and S100A9) initiate the immunosuppressive pathways that commit immature myeloid cells to become MDSCs and then further promote differentiation toward immunosuppressive macrophages and DCs.114, 115 Furthermore, cytokines, such as TNF, CCL3, and CCL4, aid MDSC-immunosuppressive function and chemotaxis to sites of inflammation (ie, spleen and mLNs) for maturation and tissue destruction.111, 116 The findings demonstrating that antibiotics elevated Il1b in marrow and TNF and CCL3 in serum suggest that antibiotic disruption of gut microbiota may drive the development and recruitment of MDSCs to exacerbate inflammatory tissue damage.
Plasmacytoid DCs and M1 macrophages, much like their MDSC precursors, can contribute proinflammatory cytokines to dysregulate MHC class II Ag presentation117 and promote RANKL-induced osteoclastogenesis.118 Recognizing that MDSCs can function as osteoclast progenitors in proinflammatory pathologic states,109, 119, 120, 121 the up-regulated MDSCs in the marrow of antibiotic-treated mice may contribute to the increased number of osteoclastic cells found in this study. Considering that pro-MDSC cytokines, such as IL-1β, TNF, and CCL3, have pro-osteoclastic actions,102, 103, 104 antibiotic up-regulation of these factors may further support the pro-osteoclastic phenotype found in antibiotic- versus vehicle-treated mice.
MDSCs have been reported to have important roles in Ag-specific versus nonspecific immunosuppression.108, 110, 122 Moreover, MDSCs may induce the conversion of naïve CD4 T cells to TREG cells.123 Because of this cell population's capacity to suppress Ag-specific CD4+ and CD8+ T-cell–mediated adaptive immune responses,107, 108 we postulate that MDSCs modulate the MHC class II Ag processing and presentation pathway toward a proinflammatory/pro-osteoclastic phenotype in antibiotic disruption of gut microbiota.
Ag presentation is a critical component of innate/adaptive immune cross talk as APCs present self-peptides/non–self-peptides to the adaptive immune system. It is well understood that bone marrow–derived APCs are necessary for thymic T-cell education for immune tolerance and T-cell interactions in lymphoid tissues in health and immune challenges.45, 46 Because MHC class II complexes are expressed on APCs (DCs, macrophages, B cells, and MDSCs), we propose a mechanism for MDSC-mediated immune suppression when gut microbiota are disrupted by antibiotics (Figure 8).
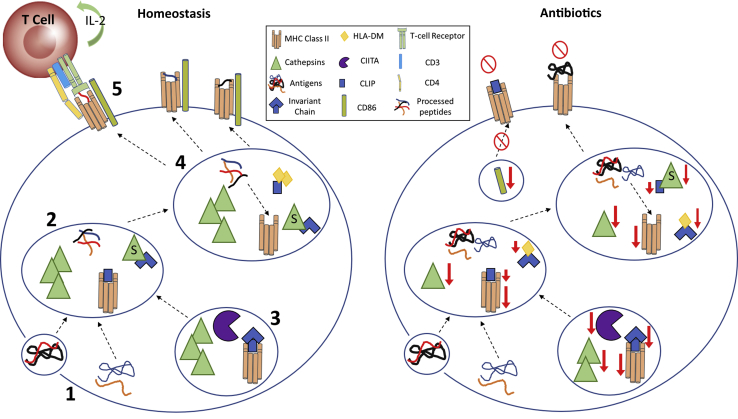
Figure 8.
Schematic of antibiotic effects on MHC class II Ag processing/presentation. 1: Under homeostatic conditions, Ags of exogenous/endogenous nature are taken up by APCs and processed by cathepsins within the endosomal/lysosomal compartments. 2: Cathepsins B, C, and D process Ags by unfolding and splicing these Ags into smaller, functionally active peptides. 3: Simultaneously, the MHC class II transactivator (CIITA) regulates MHC class II α/β heterodimer transcription, leading to class II components being synthesized within the endoplasmic reticulum to form a complex with invariant chain (Ii). This complex is further processed in the endosomal/lysosomal compartments by cathepsins S and L to degrade Ii, leaving class II–associated Ii peptide (CLIP) in the class II binding groove to block low-affinity peptides from prematurely binding to class II complexes. 4: The nonclassic class II molecule, human leukocyte antigen (HLA)-DM, chaperones the removal of CLIP from the peptide binding groove and catalyzes the loading of high-affinity peptides, leading to a stable MHC class II–peptide complex. 5: This complex is then transported to the cell surface for CD4+ T-cell recognition, with costimulation by proteins such as CD80/CD86. However, when antibiotics alter the gut microbiota, dysregulation is detected in several genes responsible for proper MHC class II–Ag processing and presentation. Without proper Ag processing and presentation, T cells may not develop tolerance to self-Ag, and APCs are less likely to clear long-term immune challenges.
Under homeostatic conditions, Ags of exogenous/endogenous nature are taken up by APCs and processed by cysteinyl and aspartyl cathepsins within the endosomal/lysosomal compartments (Figure 8). Specifically, cathepsins B, C, and D process Ags by unfolding and splicing these Ags into smaller, functionally active peptides (Figure 8).124, 125, 126 Antibiotic alteration of gut microbiota disrupts Ag processing with a marked decrease in cathepsin C gene expression in the marrow. Improper Ag processing can lead to low-affinity peptide loading onto class II complexes and suppressed helper T-cell interaction. Simultaneously, the MHC class II transactivator regulates MHC class II α/β heterodimer transcription, leading to class II components being synthesized within the endoplasmic reticulum to form a complex with Ii (Figure 8).127, 128, 129 This complex is further processed in the endosomal/lysosomal compartments by acidic proteases, such as cathepsins S and L, to degrade Ii, leaving class II–associated Ii peptide in the class II binding groove to block low-affinity peptides from prematurely binding to class II complexes (Figure 8).125, 130, 131 However, when antibiotic treatment perturbs the gut microbiota, alterations in class II variants (H2-Aa, H2-DMb2, H2-Eb1, and H2-Ob), cathepsin S (Ctss), and Ii/class II–associated Ii peptide (Cd74) are observed, which can result in incomplete Ag processing and improper loading. The nonclassic class II molecule, human leukocyte antigen-DM, chaperones the removal of class II–associated Ii peptide from the peptide binding groove and catalyzes the loading of high-affinity peptides, leading to a stable MHC class II–peptide complex (Figure 8).132, 133, 134 This complex is then transported to the cell surface for CD4+ T-cell recognition, with costimulation by proteins such as CD80/CD86 (Figure 8). However, when antibiotics alter the gut microbiota, dysregulation is detected in several genes responsible for proper MHC class II–Ag processing and presentation. Without proper Ag processing and presentation, T cells may not develop tolerance to self-Ag, and APCs are less likely to clear long-term immune challenges.
Breakdown of osteoimmune cross talk may contribute to the proinflammatory/pro-osteoclastic phenotype presented in antibiotic-treated mice. Prior reports have shown altered skeletal phenotypes when different components of the MHC class II pathway (ie, MHC II, cathepsin S, and Ii) are knocked out.135, 136, 137 MHC II gene deletion significantly delays bone development,135 whereas cathepsin S and Ii knockouts display reduced cortical thickness and increased osteoclast size and numbers.136, 137 Considering that the MHC class II Ag pathway supports normal skeletal development,135, 136, 137 our findings support the notion that antibiotic perturbation of gut microbiota disrupts osteoimmune cross talk in the postpubertal developing skeleton.
This research demonstrates that antibiotic disruption of gut microbiota composition alters host immune response effects, which critically modulates normal osteoimmune processes in the postpubertal developing skeleton. More important, this report discerns that exogenous perturbation of gut microbiota immunomodulation impacts normal growth and development at sites distant to the gut. Previously unrecognized, antibiotic-induced up-regulation of MDSCs in marrow and secondary lymphoid tissues highlights that MDSCs are a critical modulator of gut microbiota host immune response effects. Unanticipated findings showing that antibiotics dysregulate MDSCs and the MHC class II Ag pathway in bone marrow intriguingly reveal that antibiotic perturbation of gut microbiota disrupts osteoimmune cross talk.
Acknowledgments
We thank the Medical University of South Carolina Laboratory Center for Oral Health Research for histopathological scoring services, and Drs. Keith L. Kirkwood and Lixia Zhang for intellectual discussions and financial support.
C.M.N. conceived the study; J.D.H.-S., H.M.S., and C.M.N. designed the research; J.D.H.-S., H.M.S., M.B.C., N.A.P., J.E.K., M.E.C., E.H., J.I.A., and C.M.N. collected the data; J.D.H.-S., N.A.P., J.E.K., M.E.C., and C.M.N. analyzed the data; J.D.H.-S., A.V.A., J.I.A., and C.M.N. interpreted the data; J.D.H.-S., A.V.A., and C.M.N. wrote the manuscript; all authors approved the manuscript.
Footnotes
Supported by the American Society for Bone and Mineral Research Rising Star Award (C.M.N.); the NIH/National Institute of Dental and Craniofacial Research grants K08DE025337 (C.M.N.), R01DE021423 (K.L.K.), T32DE017551 (Medical University of South Carolina), and R25DE022677 (Medical University of South Carolina); and NIH/National Institute of General Medical Sciences grant P30GM103331 (Medical University of South Carolina).
Disclosures: None declared.
See related Commentary on page 229
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.10.017.
Supplemental Data
Supplemental Figure S1.
Antibiotic (ABX) disruption of gut microbiota does not affect cortical bone. Twelve-week–old male vehicle (VEH)– and ABX-treated mice and female VEH- and ABX-treated mice were euthanized; specimens were harvested for analysis. A–C: Micro–computed tomographic (μCT) analyses of the tibia middiaphysis cortical bone in male VEH- and ABX-treated mice. A: Representative reconstructed cross-sectional images in male VEH- and ABX-treated mice. B: Male cortical area fraction (Ct.Ar/Tt.Ar). C: Male cortical thickness (Ct.Th). D–F: μCT analyses of the tibia middiaphysis cortical bone in female VEH- and ABX-treated mice. D: Representative reconstructed cross-sectional images in female VEH- and ABX-treated mice. E: Female Ct.Ar/Tt.Ar. F: Female Ct.Th. Unpaired t-test was used. Data are expressed as means ± SEM (B, C, E, and F). n = 4 per group (A–F).
References
- 1.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanov, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brestoff J.R., Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sommer F., Backhed F. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 6.Buffie C.G., Pamer E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamada N., Chen G.Y., Inohara N., Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumler A.J., Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamada N., Seo S.U., Chen G.Y., Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 10.Dalal S.R., Chang E.B. The microbial basis of inflammatory bowel diseases. J Clin Invest. 2014;124:4190–4196. doi: 10.1172/JCI72330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickard J.M., Zeng M.Y., Caruso R., Nunez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung C., Rivera L., Furness J.B., Angus P.W. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol. 2016;13:412–425. doi: 10.1038/nrgastro.2016.85. [DOI] [PubMed] [Google Scholar]
- 13.Yu L.X., Schwabe R.F. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14:527–539. doi: 10.1038/nrgastro.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berer K., Mues M., Koutrolos M., Rasbi Z.A., Boziki M., Johner C., Wekerle H., Krishnamoorthy G. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 15.Sampson T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E., Challis C., Schretter C.E., Rocha S., Gradinaru V., Chesselet M.F., Keshavarzian A., Shannon K.M., Krajmalnik-Brown R., Wittung-Stafshede P., Knight R., Mazmanian S.K. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 2016;167:1469–1480.e1412. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Troseid M., Ueland T., Hov J.R., Svardal A., Gregersen I., Dahl C.P., Aakhus S., Gude E., Bjorndal B., Halvorsen B., Karlsen T.H., Aukrust P., Gullestad L., Berge R.K., Yndestad A. Microbiota-dependent metabolite trimethylamine-N-oxide is associated with disease severity and survival of patients with chronic heart failure. J Intern Med. 2015;277:717–726. doi: 10.1111/joim.12328. [DOI] [PubMed] [Google Scholar]
- 17.Jie Z., Xia H., Zhong S.L., Feng Q., Li S., Liang S. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H.J., Ivanov, Darce J., Hattori K., Shima T., Umesaki Y., Littman D.R., Benoist C., Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J.Y., Chassaing B., Tyagi A.M., Vaccaro C., Luo T., Adams J., Darby T.M., Weitzmann M.N., Mulle J.G., Gewirtz A.T., Jones R.M., Pacifici R. Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J Clin Invest. 2016;126:2049–2063. doi: 10.1172/JCI86062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroeder B.O., Backhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–1089. doi: 10.1038/nm.4185. [DOI] [PubMed] [Google Scholar]
- 21.Blander J.M., Longman R.S., Iliev I.D., Sonnenberg G.F., Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol. 2017;18:851–860. doi: 10.1038/ni.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Backhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H., Khan M.T., Zhang J., Li J., Xiao L., Al-Aama J., Zhang D., Lee Y.S., Kotowska D., Colding C., Tremaroli V., Yin Y., Bergman S., Xu X., Madsen L., Kristiansen K., Dahlgren J., Wang J. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Chu D.M., Ma J., Prince A.L., Antony K.M., Seferovic M.D., Aagaard K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho I., Blaser M.J. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ananthakrishnan A.N. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 27.Sommer F., Anderson J.M., Bharti R., Raes J., Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 28.Keeney K.M., Yurist-Doutsch S., Arrieta M.C., Finlay B.B. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol. 2014;68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 29.Blaser M.J. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ianiro G., Tilg H., Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 31.Becattini S., Taur Y., Pamer E.G. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med. 2016;22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knoop K.A., McDonald K.G., Kulkarni D.H., Newberry R.D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016;65:1100–1109. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D., Bittinger K., Bailey A., Friedman E.S., Hoffmann C., Albenberg L., Sinha R., Compher C., Gilroy E., Nessel L., Grant A., Chehoud C., Li H., Wu G.D., Bushman F.D. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lichtman J.S., Ferreyra J.A., Ng K.M., Smits S.A., Sonnenburg J.L., Elias J.E. Host-microbiota interactions in the pathogenesis of antibiotic-associated diseases. Cell Rep. 2016;14:1049–1061. doi: 10.1016/j.celrep.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grasa L., Abecia L., Forcen R., Castro M., de Jalon J.A., Latorre E., Alcalde A.I., Murillo M.D. Antibiotic-induced depletion of murine microbiota induces mild inflammation and changes in toll-like receptor patterns and intestinal motility. Microb Ecol. 2015;70:835–848. doi: 10.1007/s00248-015-0613-8. [DOI] [PubMed] [Google Scholar]
- 36.Belkaid Y., Harrison O.J. Homeostatic immunity and the microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Underhill D.M., Iliev I.D. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abt M.C., McKenney P.T., Pamer E.G. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol. 2016;14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrello C., Garavaglia F., Cribiu F.M., Ercoli G., Bosari S., Caprioli F., Facciotti F. Short-term oral antibiotics treatment promotes inflammatory activation of colonic invariant natural killer T and conventional CD4(+) T cells. Front Med. 2018;5:21. doi: 10.3389/fmed.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng M.Y., Inohara N., Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edelblum K.L., Sharon G., Singh G., Odenwald M.A., Sailer A., Cao S., Ravens S., Thomsen I., El Bissati K., McLeod R., Dong C., Gurbuxani S., Prinz I., Mazmanian S.K., Turner J.R. The microbiome activates CD4 T-cell-mediated immunity to compensate for increased intestinal permeability. Cell Mol Gastroenterol Hepatol. 2017;4:285–297. doi: 10.1016/j.jcmgh.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tulstrup M.V., Christensen E.G., Carvalho V., Linninge C., Ahrne S., Hojberg O., Licht T.R., Bahl M.I. Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar rats dependent on antibiotic class. PLoS One. 2015;10:e0144854. doi: 10.1371/journal.pone.0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romick-Rosendale L.E., Legomarcino A., Patel N.B., Morrow A.L., Kennedy M.A. Prolonged antibiotic use induces intestinal injury in mice that is repaired after removing antibiotic pressure: implications for empiric antibiotic therapy. Metabolomics. 2014;10:8–20. doi: 10.1007/s11306-013-0546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Toraldo G., Li A., Yang X., Zhang H., Qian W.P., Weitzmann M.N. B cells and T cells are critical for the preservation of bone homeostasis and attainment of peak bone mass in vivo. Blood. 2007;109:3839–3848. doi: 10.1182/blood-2006-07-037994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenzo J., Horowitz M., Choi Y. Osteoimmunology: interactions of the bone and immune system. Endocr Rev. 2008;29:403–440. doi: 10.1210/er.2007-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takayanagi H. Osteoimmunology and the effects of the immune system on bone. Nat Rev Rheumatol. 2009;5:667–676. doi: 10.1038/nrrheum.2009.217. [DOI] [PubMed] [Google Scholar]
- 47.Pacifici R. T cells: critical bone regulators in health and disease. Bone. 2010;47:461–471. doi: 10.1016/j.bone.2010.04.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh M.C., Takegahara N., Kim H., Choi Y. Updating osteoimmunology: regulation of bone cells by innate and adaptive immunity. Nat Rev Rheumatol. 2018;14:146–156. doi: 10.1038/nrrheum.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 50.Redlich K., Smolen J.S. Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 2012;11:234–250. doi: 10.1038/nrd3669. [DOI] [PubMed] [Google Scholar]
- 51.Weitzmann M.N., Ofotokun I. Physiological and pathophysiological bone turnover: role of the immune system. Nat Rev Endocrinol. 2016;12:518–532. doi: 10.1038/nrendo.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sjogren K., Engdahl C., Henning P., Lerner U.H., Tremaroli V., Lagerquist M.K., Backhed F., Ohlsson C. The gut microbiota regulates bone mass in mice. J Bone Miner Res. 2012;27:1357–1367. doi: 10.1002/jbmr.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarzer M., Makki K., Storelli G., Machuca-Gayet I., Srutkova D., Hermanova P., Martino M.E., Balmand S., Hudcovic T., Heddi A., Rieusset J., Kozakova H., Vidal H., Leulier F. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science. 2016;351:854–857. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 54.Yan J., Herzog J.W., Tsang K., Brennan C.A., Bower M.A., Garrett W.S., Sartor B.R., Aliprantis A.O., Charles J.F. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci U S A. 2016;113:E7554–E7563. doi: 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novince C.M., Whittow C.R., Aartun J.D., Hathaway J.D., Poulides N., Chavez M.B., Steinkamp H.M., Kirkwood K.A., Huang E., Westwater C., Kirkwood K.L. Commensal gut microbiota immunomodulatory actions in bone marrow and liver have catabolic effects on skeletal homeostasis in health. Sci Rep. 2017;7:5747. doi: 10.1038/s41598-017-06126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaskins H.R., Collier C.T., Anderson D.B. Antibiotics as growth promotants: mode of action. Anim Biotechnol. 2002;13:29–42. doi: 10.1081/ABIO-120005768. [DOI] [PubMed] [Google Scholar]
- 57.Cox L.M., Blaser M.J. Antibiotics in early life and obesity. Nat Rev Endocrinol. 2015;11:182–190. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stokstad E.L. Antibiotics in animal nutrition. Physiol Rev. 1954;34:25–51. doi: 10.1152/physrev.1954.34.1.25. [DOI] [PubMed] [Google Scholar]
- 59.Taylor J.H. Antibiotics and other growth-promoting substances. Proc Nutr Soc. 1962;21:73–80. doi: 10.1079/pns19620013. [DOI] [PubMed] [Google Scholar]
- 60.Cho I., Yamanishi S., Cox L., Methe B.A., Zavadil J., Li K., Gao Z., Mahana D., Raju K., Teitler I., Li H., Alekseyenko A.V., Blaser M.J. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reikvam D.H., Erofeev A., Sandvik A., Grcic V., Jahnsen F.L., Gaustad P., McCoy K.D., Macpherson A.J., Meza-Zepeda L.A., Johansen F.E. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kostic A.D., Howitt M.R., Garrett W.S. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013;27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ericsson A.C., Franklin C.L. Manipulating the gut microbiota: methods and challenges. ILAR J. 2015;56:205–217. doi: 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansen A.K., Krych L., Nielsen D.S., Hansen C.H. A review of applied aspects of dealing with gut microbiota impact on rodent models. ILAR J. 2015;56:250–264. doi: 10.1093/ilar/ilv010. [DOI] [PubMed] [Google Scholar]
- 65.Franklin C.L., Ericsson A.C. Microbiota and reproducibility of rodent models. Lab Anim (NY) 2017;46:114–122. doi: 10.1038/laban.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guss J.D., Horsfield M.W., Fontenele F.F., Sandoval T.N., Luna M., Apoorva F., Lima S.F., Bicalho R.C., Singh A., Ley R.E., van der Meulen M.C., Goldring S.R., Hernandez C.J. Alterations to the gut microbiome impair bone strength and tissue material properties. J Bone Miner Res. 2017;32:1343–1353. doi: 10.1002/jbmr.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S., Dai R.M., Kiu H., Cardone M., Naik S., Patri A.K., Wang E., Marincola F.M., Frank K.M., Belkaid Y., Trinchieri G., Goldszmid R.S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Committee for the Update of the Guide for the Care and Use of Laboratory Animals. National Research Council . Eighth Edition. National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 69.Bouxsein M.L., Boyd S.K., Christiansen B.A., Guldberg R.E., Jepsen K.J., Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 70.Novince C.M., Michalski M.N., Koh A.J., Sinder B.P., Entezami P., Eber M.R., Pettway G.J., Rosol T.J., Wronski T.J., Kozloff K.M., McCauley L.K. Proteoglycan 4: a dynamic regulator of skeletogenesis and parathyroid hormone skeletal anabolism. J Bone Miner Res. 2012;27:11–25. doi: 10.1002/jbmr.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dempster D.W., Compston J.E., Drezner M.K., Glorieux F.H., Kanis J.A., Malluche H., Meunier P.J., Ott S.M., Recker R.R., Parfitt A.M. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28:2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 73.Bacchetti De Gregoris T., Aldred N., Clare A.S., Burgess J.G. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods. 2011;86:351–356. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 74.Landreth K.S. Critical windows in development of the rodent immune system. Hum Exp Toxicol. 2002;21:493–498. doi: 10.1191/0960327102ht287oa. [DOI] [PubMed] [Google Scholar]
- 75.Dietert R.R., Etzel R.A., Chen D., Halonen M., Holladay S.D., Jarabek A.M., Landreth K., Peden D.B., Pinkerton K., Smialowicz R.J., Zoetis T. Workshop to identify critical windows of exposure for children's health: immune and respiratory systems work group summary. Environ Health Perspect. 2000;108 Suppl 3:483–490. doi: 10.1289/ehp.00108s3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferguson V.L., Ayers R.A., Bateman T.A., Simske S.J. Bone development and age-related bone loss in male C57BL/6J mice. Bone. 2003;33:387–398. doi: 10.1016/s8756-3282(03)00199-6. [DOI] [PubMed] [Google Scholar]
- 77.Glatt V., Canalis E., Stadmeyer L., Bouxsein M.L. Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res. 2007;22:1197–1207. doi: 10.1359/jbmr.070507. [DOI] [PubMed] [Google Scholar]
- 78.Bonjour J.P., Theintz G., Buchs B., Slosman D., Rizzoli R. Critical years and stages of puberty for spinal and femoral bone mass accumulation during adolescence. J Clin Endocrinol Metab. 1991;73:555–563. doi: 10.1210/jcem-73-3-555. [DOI] [PubMed] [Google Scholar]
- 79.Cheung W.W., Zhan J.Y., Paik K.H., Mak R.H. The impact of inflammation on bone mass in children. Pediatr Nephrol. 2011;26:1937–1946. doi: 10.1007/s00467-010-1733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCormack S.E., Cousminer D.L., Chesi A., Mitchell J.A., Roy S.M., Kalkwarf H.J., Lappe J.M., Gilsanz V., Oberfield S.E., Shepherd J.A., Winer K.K., Kelly A., Grant S.F.A., Zemel B.S. Association between linear growth and bone accrual in a diverse cohort of children and adolescents. JAMA Pediatr. 2017;171:e171769. doi: 10.1001/jamapediatrics.2017.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weaver C.M., Gordon C.M., Janz K.F., Kalkwarf H.J., Lappe J.M., Lewis R., O'Karma M., Wallace T.C., Zemel B.S. The National Osteoporosis Foundation's position statement on peak bone mass development and lifestyle factors: a systematic review and implementation recommendations. Osteoporos Int. 2016;27:1281–1386. doi: 10.1007/s00198-015-3440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boyce B.F., Xing L. Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem Biophys. 2008;473:139–146. doi: 10.1016/j.abb.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Walsh M.C., Choi Y. Biology of the RANKL-RANK-OPG system in immunity, bone, and beyond. Front Immunol. 2014;5:511. doi: 10.3389/fimmu.2014.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Veglia F., Perego M., Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., Rodriguez P.C., Sica A., Umansky V., Vonderheide R.H., Gabrilovich D.I. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hua Z., Hou B. TLR signaling in B-cell development and activation. Cell Mol Immunol. 2013;10:103–106. doi: 10.1038/cmi.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balmer M.L., Schurch C.M., Saito Y., Geuking M.B., Li H., Cuenca M., Kovtonyuk L.V., McCoy K.D., Hapfelmeier S., Ochsenbein A.F., Manz M.G., Slack E., Macpherson A.J. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J Immunol. 2014;193:5273–5283. doi: 10.4049/jimmunol.1400762. [DOI] [PubMed] [Google Scholar]
- 89.Gonzalez-Perez G., Lamouse-Smith E.S. Gastrointestinal microbiome dysbiosis in infant mice alters peripheral CD8+ T cell receptor signaling. Front Immunol. 2017;8:265. doi: 10.3389/fimmu.2017.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stoll S., Delon J., Brotz T.M., Germain R.N. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science. 2002;296:1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 91.Nagaraj S., Gabrilovich D.I. Regulation of suppressive function of myeloid-derived suppressor cells by CD4+ T cells. Semin Cancer Biol. 2012;22:282–288. doi: 10.1016/j.semcancer.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Movahedi K., Guilliams M., Van den Bossche J., Van den Bergh R., Gysemans C., Beschin A., De Baetselier P., Van Ginderachter J.A. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 93.Mirpuri J., Raetz M., Sturge C.R., Wilhelm C.L., Benson A., Savani R.C., Hooper L.V., Yarovinsky F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rizzatti G., Lopetuso L.R., Gibiino G., Binda C., Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed Res Int. 2017;2017:9351507. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shin N.R., Whon T.W., Bae J.W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 96.Litvak Y., Byndloss M.X., Tsolis R.M., Baumler A.J. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol. 2017;39:1–6. doi: 10.1016/j.mib.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 97.Walters T.D., Griffiths A.M. Mechanisms of growth impairment in pediatric Crohn's disease. Nat Rev Gastroenterol Hepatol. 2009;6:513–523. doi: 10.1038/nrgastro.2009.124. [DOI] [PubMed] [Google Scholar]
- 98.Sanderson I.R. Growth problems in children with IBD. Nat Rev Gastroenterol Hepatol. 2014;11:601–610. doi: 10.1038/nrgastro.2014.102. [DOI] [PubMed] [Google Scholar]
- 99.Rosen M.J., Dhawan A., Saeed S.A. Inflammatory bowel disease in children and adolescents. JAMA Pediatr. 2015;169:1053–1060. doi: 10.1001/jamapediatrics.2015.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rossa C., Ehmann K., Liu M., Patil C., Kirkwood K.L. MKK3/6-p38 MAPK signaling is required for IL-1beta and TNF-alpha-induced RANKL expression in bone marrow stromal cells. J Interferon Cytokine Res. 2006;26:719–729. doi: 10.1089/jir.2006.26.719. [DOI] [PubMed] [Google Scholar]
- 101.Ruscitti P., Cipriani P., Carubbi F., Liakouli V., Zazzeroni F., Di Benedetto P., Berardicurti O., Alesse E., Giacomelli R. The role of IL-1beta in the bone loss during rheumatic diseases. Mediators Inflamm. 2015;2015:782382. doi: 10.1155/2015/782382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han J.H., Choi S.J., Kurihara N., Koide M., Oba Y., Roodman G.D. Macrophage inflammatory protein-1alpha is an osteoclastogenic factor in myeloma that is independent of receptor activator of nuclear factor kappaB ligand. Blood. 2001;97:3349–3353. doi: 10.1182/blood.v97.11.3349. [DOI] [PubMed] [Google Scholar]
- 103.Watanabe T., Kukita T., Kukita A., Wada N., Toh K., Nagata K., Nomiyama H., Iijima T. Direct stimulation of osteoclastogenesis by MIP-1alpha: evidence obtained from studies using RAW264 cell clone highly responsive to RANKL. J Endocrinol. 2004;180:193–201. doi: 10.1677/joe.0.1800193. [DOI] [PubMed] [Google Scholar]
- 104.Yu X., Huang Y., Collin-Osdoby P., Osdoby P. CCR1 chemokines promote the chemotactic recruitment, RANKL development, and motility of osteoclasts and are induced by inflammatory cytokines in osteoblasts. J Bone Miner Res. 2004;19:2065–2077. doi: 10.1359/JBMR.040910. [DOI] [PubMed] [Google Scholar]
- 105.Josefsdottir K.S., Baldridge M.T., Kadmon C.S., King K.Y. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129:729–739. doi: 10.1182/blood-2016-03-708594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao E., Xu H., Wang L., Kryczek I., Wu K., Hu Y., Wang G., Zou W. Bone marrow and the control of immunity. Cell Mol Immunol. 2012;9:11–19. doi: 10.1038/cmi.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nagaraj S., Gabrilovich D.I. Myeloid-derived suppressor cells. Adv Exp Med Biol. 2007;601:213–223. doi: 10.1007/978-0-387-72005-0_22. [DOI] [PubMed] [Google Scholar]
- 108.Watanabe S., Deguchi K., Zheng R., Tamai H., Wang L.X., Cohen P.A., Shu S. Tumor-induced CD11b+Gr-1+ myeloid cells suppress T cell sensitization in tumor-draining lymph nodes. J Immunol. 2008;181:3291–3300. doi: 10.4049/jimmunol.181.5.3291. [DOI] [PubMed] [Google Scholar]
- 109.Charles J.F., Hsu L.Y., Niemi E.C., Weiss A., Aliprantis A.O., Nakamura M.C. Inflammatory arthritis increases mouse osteoclast precursors with myeloid suppressor function. J Clin Invest. 2012;122:4592–4605. doi: 10.1172/JCI60920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dolcetti L., Peranzoni E., Ugel S., Marigo I., Fernandez Gomez A., Mesa C., Geilich M., Winkels G., Traggiai E., Casati A., Grassi F., Bronte V. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. Eur J Immunol. 2010;40:22–35. doi: 10.1002/eji.200939903. [DOI] [PubMed] [Google Scholar]
- 111.Gabrilovich D.I., Ostrand-Rosenberg S., Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li H., Han Y., Guo Q., Zhang M., Cao X. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J Immunol. 2009;182:240–249. doi: 10.4049/jimmunol.182.1.240. [DOI] [PubMed] [Google Scholar]
- 113.Stiff A., Trikha P., Mundy-Bosse B., McMichael E., Mace T.A., Benner B., Kendra K., Campbell A., Gautam S., Abood D., Landi I., Hsu V., Duggan M., Wesolowski R., Old M., Howard J.H., Yu L., Stasik N., Olencki T., Muthusamy N., Tridandapani S., Byrd J.C., Caligiuri M., Carson W.E. Nitric oxide production by myeloid-derived suppressor cells plays a role in impairing Fc receptor-mediated natural killer cell function. Clin Cancer Res. 2018;24:1891–1904. doi: 10.1158/1078-0432.CCR-17-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Song X., Krelin Y., Dvorkin T., Bjorkdahl O., Segal S., Dinarello C.A., Voronov E., Apte R.N. CD11b+/Gr-1+ immature myeloid cells mediate suppression of T cells in mice bearing tumors of IL-1beta-secreting cells. J Immunol. 2005;175:8200–8208. doi: 10.4049/jimmunol.175.12.8200. [DOI] [PubMed] [Google Scholar]
- 115.Kennedy D.E., Knight K.L. Inhibition of B lymphopoiesis by adipocytes and IL-1-producing myeloid-derived suppressor cells. J Immunol. 2015;195:2666–2674. doi: 10.4049/jimmunol.1500957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Z., Liu Y., Zhang Y., Shang Y., Gao Q. MDSC-decreasing chemotherapy increases the efficacy of cytokine-induced killer cell immunotherapy in metastatic renal cell carcinoma and pancreatic cancer. Oncotarget. 2016;7:4760–4769. doi: 10.18632/oncotarget.6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Young L.J., Wilson N.S., Schnorrer P., Proietto A., ten Broeke T., Matsuki Y., Mount A.M., Belz G.T., O'Keeffe M., Ohmura-Hoshino M., Ishido S., Stoorvogel W., Heath W.R., Shortman K., Villadangos J.A. Differential MHC class II synthesis and ubiquitination confers distinct antigen-presenting properties on conventional and plasmacytoid dendritic cells. Nat Immunol. 2008;9:1244–1252. doi: 10.1038/ni.1665. [DOI] [PubMed] [Google Scholar]
- 118.Alnaeeli M., Penninger J.M., Teng Y.T. Immune interactions with CD4+ T cells promote the development of functional osteoclasts from murine CD11c+ dendritic cells. J Immunol. 2006;177:3314–3326. doi: 10.4049/jimmunol.177.5.3314. [DOI] [PubMed] [Google Scholar]
- 119.Zhuang J., Zhang J., Lwin S.T., Edwards J.R., Edwards C.M., Mundy G.R., Yang X. Osteoclasts in multiple myeloma are derived from Gr-1+CD11b+myeloid-derived suppressor cells. PLoS One. 2012;7:e48871. doi: 10.1371/journal.pone.0048871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sawant A., Deshane J., Jules J., Lee C.M., Harris B.A., Feng X., Ponnazhagan S. Myeloid-derived suppressor cells function as novel osteoclast progenitors enhancing bone loss in breast cancer. Cancer Res. 2013;73:672–682. doi: 10.1158/0008-5472.CAN-12-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang H., Wang S., Huang Y., Wang H., Zhao J., Gaskin F., Yang N., Fu S.M. Myeloid-derived suppressor cells are proinflammatory and regulate collagen-induced arthritis through manipulating Th17 cell differentiation. Clin Immunol. 2015;157:175–186. doi: 10.1016/j.clim.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nagaraj S., Nelson A., Youn J.I., Cheng P., Quiceno D., Gabrilovich D.I. Antigen-specific CD4(+) T cells regulate function of myeloid-derived suppressor cells in cancer via retrograde MHC class II signaling. Cancer Res. 2012;72:928–938. doi: 10.1158/0008-5472.CAN-11-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Serafini P., Mgebroff S., Noonan K., Borrello I. Myeloid-derived suppressor cells promote cross-tolerance in B-cell lymphoma by expanding regulatory T cells. Cancer Res. 2008;68:5439–5449. doi: 10.1158/0008-5472.CAN-07-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Moss C.X., Villadangos J.A., Watts C. Destructive potential of the aspartyl protease cathepsin D in MHC class II-restricted antigen processing. Eur J Immunol. 2005;35:3442–3451. doi: 10.1002/eji.200535320. [DOI] [PubMed] [Google Scholar]
- 125.Rudensky A., Beers C. Lysosomal cysteine proteases and antigen presentation. Ernst Schering Res Found Workshop. 2006;56:81–95. doi: 10.1007/3-540-37673-9_5. [DOI] [PubMed] [Google Scholar]
- 126.Goldstein O.G., Hajiaghamohseni L.M., Amria S., Sundaram K., Reddy S.V., Haque A. Gamma-IFN-inducible-lysosomal thiol reductase modulates acidic proteases and HLA class II antigen processing in melanoma. Cancer Immunol Immunother. 2008;57:1461–1470. doi: 10.1007/s00262-008-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brown J.H., Jardetzky T.S., Gorga J.C., Stern L.J., Urban R.G., Strominger J.L., Wiley D.C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 128.Bikoff E.K., Huang L.Y., Episkopou V., van Meerwijk J., Germain R.N., Robertson E.J. Defective major histocompatibility complex class II assembly, transport, peptide acquisition, and CD4+ T cell selection in mice lacking invariant chain expression. J Exp Med. 1993;177:1699–1712. doi: 10.1084/jem.177.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roche P.A., Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990;345:615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- 130.Shi G.P., Villadangos J.A., Dranoff G., Small C., Gu L., Haley K.J., Riese R., Ploegh H.L., Chapman H.A. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 1999;10:197–206. doi: 10.1016/s1074-7613(00)80020-5. [DOI] [PubMed] [Google Scholar]
- 131.Driessen C., Bryant R.A., Lennon-Dumenil A.M., Villadangos J.A., Bryant P.W., Shi G.P., Chapman H.A., Ploegh H.L. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J Cell Biol. 1999;147:775–790. doi: 10.1083/jcb.147.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mosyak L., Zaller D.M., Wiley D.C. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity. 1998;9:377–383. doi: 10.1016/s1074-7613(00)80620-2. [DOI] [PubMed] [Google Scholar]
- 133.Denzin L.K., Robbins N.F., Carboy-Newcomb C., Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 134.Rocha N., Neefjes J. MHC class II molecules on the move for successful antigen presentation. EMBO J. 2008;27:1–5. doi: 10.1038/sj.emboj.7601945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Simske S.J., Bateman T.A., Smith E.E., Ferguson V.L., Chapes S.K. Effects of major histocompatibility complex class II knockout on mouse bone mechanical properties during development. Biomed Sci Instrum. 2002;38:47–52. [PubMed] [Google Scholar]
- 136.Rauner M., Foger-Samwald U., Kurz M.F., Brunner-Kubath C., Schamall D., Kapfenberger A., Varga P., Kudlacek S., Wutzl A., Hoger H., Zysset P.K., Shi G.P., Hofbauer L.C., Sipos W., Pietschmann P. Cathepsin S controls adipocytic and osteoblastic differentiation, bone turnover, and bone microarchitecture. Bone. 2014;64:281–287. doi: 10.1016/j.bone.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 137.Mun S.H., Won H.Y., Hernandez P., Aguila H.L., Lee S.K. Deletion of CD74, a putative MIF receptor, in mice enhances osteoclastogenesis and decreases bone mass. J Bone Miner Res. 2013;28:948–959. doi: 10.1002/jbmr.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.