Abstract
Cervical cancer is the most common and lethal gynaecological tumor. Long noncoding RNAs (lncRNAs) have critical roles in various cancers, including cervical cancer. However, few studies investigated the diagnostic value of lncRNAs for cervical cancer. In this study, we investigated the expression pattern of a recently identified lncRNA GIHCG in cervical cancer tissues, cell lines, and serums by qRT-PCR. Furthermore, we explored the roles of GIHCG in cervical cancer using gain-of-function and loss-of-function assays. Our results revealed that GIHCG is up-regulated in cervical cancer tissues and cell lines compared with adjacent normal tissues and normal cervical epithelial cell line, respectively. Furthermore, serum GIHCG is significantly up-regulated in cervical cancer patients compared with healthy controls. ROC curve analysis revealed that serum GIHCG could accurately discriminate cervical cancer patients from healthy controls. Functionally, we found that overexpression of GIHCG promotes cell proliferation, inhibits cell apoptosis, and promotes cell migration of cervical cancer cells. Conversely, depletion of GIHCG inhibits cell proliferation, induces cell apoptosis, and inhibits cell migration of cervical cancer cells. Mechanistically, we found that GIHCG represses the expression of miR-200b. The expression of miR-200b is inversely correlated with the expression of GIHCG in cervical cancer tissues. Moreover, overexpression of miR-200b attenuates the roles of GIHCG in promoting cervical cancer tumor growth in vivo. In summary, this study demonstrated that GIHCG functions as an oncogene in cervical cancer via repressing miR-200b. This study also suggested that GIHCG may be a non-invasive diagnostic biomarker and a potential therapeutic target for cervical cancer.
Keywords: long noncoding RNA, cervical cancer, oncogene, diagnosis, microRNA
Introduction
Cervical cancer is the most common gynaecological tumor, with estimated 527,600 new cases and 265,700 deaths in 2012 worldwide 1. Most early cervical cancers could be cured by surgery 2. However, for those cervical cancer patients in later stages, there are no effective therapeutic strategies and the survival rate of these cervical cancer patients is only about 15% 3. Therefore, novel early diagnostic and therapeutic approaches are urgently needed.
Serum non-invasive diagnostic biomarkers have been commonly used for early diagnosis of various cancers, such as α-fetoprotein (AFP) for liver cancer, prostate-specific antigen (PSA) for prostate cancer 4, 5. Although several serum biomarkers are also used for diagnosis of cervical cancer, such as carbohydrate antigen 125 (CA125) and squamous cell carcinoma antigen (SCC Ag), the diagnostic sensitivity and specificity are not satisfactory and limit their utility 6, 7. Thus, identifying novel sensitive and specific serum biomarkers are critical for the early diagnosis of cervical cancer.
Except for these well-known proteins AFP, PSA, CA125 and so on, many long noncoding RNAs (lncRNAs) are also detected in the serum and could be used as diagnostic biomarkers for cancers 8-12. LncRNA is a class of RNA molecules with no protein coding potential and longer than 200 nucleotides in length 13-15. Next-generation sequencings of human genome and transcriptome have demonstrated that the number of lncRNAs is much larger than that of mRNAs, with about 58,000 different lncRNAs and 21,000 different mRNAs 16, 17. Accumulating evidences displayed that many lncRNAs are involved in various pathophysiological processes, including cancers 18-21. Many lncRNAs are frequently dysregulated in various cancer tissues 22-26. Furthermore, several lncRNAs have been found in the serum and could be used as diagnostic biomarkers for cancers, such as PVT1 for cervical cancer 9; UCA1 and WRAP53 for liver cancer 10; H19 and PTPEN1 for gastric cancer 8, 11; XIST and HIF1A-AS1 for non-small cell lung cancer 12. In addition, a variety of lncRNAs function as oncogenes or tumor-suppressors in cancers 27-29. They promote or inhibit cell proliferation, cell apoptosis, cell cycle, cell migration, cell invasion, drug resistance and so on 30-32. Targeting these lncRNAs represent novel therapeutic strategies for cancers.
LncRNA GIHCG is first identified in liver cancer, which is up-regulated in liver cancer and promotes liver cancer cell proliferation and migration, and liver cancer growth and metastasis in vivo 33. Our previous study found that GIHCG is also up-regulated in renal cell carcinoma (RCC) tissues and depletion of GIHCG represses RCC cell proliferation and migration 34. Furthermore, we identified the existence of GIHCG in human serum and found that GIHCG is up-regulated in the serum of RCC patients and could serve as a diagnostic biomarker for RCC 34. However, the expression, roles, and clinical significances of GIHCG in cervical cancer are still unknown.
In this study, we investigated the expression of GIHCG in cervical cancer tissues and cell line, and also in the serum of cervical cancer patients and healthy controls. Using gain-of-function and loss-of-function assays, we investigated the roles of GIHCG in cervical cancer cell proliferation, apoptosis, and migration in vitro and tumor growth in vivo. Moreover, the underlying mechanisms mediating the roles of GIHCG in cervical cancer are also explored.
Materials and methods
Clinical samples
Fifty-eight pairs of cervical cancer tissues and adjacent normal tissues were obtained from cervical cancer patients who received surgery at Fuzhou General Hospital, Fuzhou, Fujian, China from December 2014 to December 2017. All the tissues were frozen in liquid nitrogen immediately after surgery and stored at -80°C until use. All samples were confirmed by pathological examination. Venus blood was collected in serum separator tubes from 80 cervical cancer patients before surgery and 80 age-matched healthy controls at The People's Hospital of Yichun City, Yichun, Jiangxi, China from December 2014 to December 2017. Then, the tubes were centrifugated at 3000 rpm for 15 min at 4°C to collect serum. The serum samples were divided into multiple aliquots of 500 µl each and stored at -80°C until use. All the clinical samples were acquired with informed consent from the participants. The Ethics Review Committee of Fujian Medical University reviewed and approved the use of clinical samples.
Cell culture
The human normal cervical epithelial cell line HCerEpiC was acquired from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in Cervical Epithelial Cell Growth Supplement (ScienCell). The cervical cancer cell lines HeLa, SiHa, C-33A, and CaSki were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). HeLa, SiHa, and C-33A cells were cultured in Eagle's Minimum Essential Medium (Gibco, Carlsbad, CA, USA). CaSki cells were cultured in RPMI-1640 Medium (Gibco). All the cells were grown in the above described medium supplemented with 10% fetal bovine serum (Gibco) at an incubator with a humidified atmosphere of 5% CO2 at 37°C.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from indicated tissues and cells with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's protocol. First-strand cDNA was generated using the extracted RNA and the M-MLV Reverse Transcriptase (Invitrogen) in accordance with the manufacturer's protocol. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the SYBR Premix Ex Taq™ II (Takara, Dalian, Liaoning, China) on StepOnePlus™ Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA) with the standard SYBR-Green PCR protocols. β-actin was employed as an endogenous control for quantification of GIHCG. The primer sequences are as follows: for GIHCG, 5'-CTTTCAAGAAGTTTGGCTGTC-3' (sense) and 5'-GCTCATTCAACGGATAAGTC-3' (anti-sense); for β-actin, 5'-TCCTCTCCCAAGTCCACACA-3' (sense) and 5'-GCACGAAGGCTCATCATTCA-3' (anti-sense). For the quantification of miR-200b, qRT-PCR was performed using TaqMan microRNA assays (Applied Biosystems) in accordance with the manufacturer's protocol. U6 was employed as an endogenous control for quantification of miR-200b. The expression of GIHCG and miR-200b was calculated with the comparative cycle threshold (CT; 2-ΔΔCT) method.
Vectors construction, small interfering RNA (siRNA) synthesis and transfection
GIHCG overexpression vector pcDNA3.1-GIHCG was constructed as previously described 33. Briefly, the cDNA encoding GIHCG was PCR-amplified with the Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher, Waltham, MA, USA) and subcloned into the Hind III and BamH I sites of pcDNA3.1 empty vector (Invitrogen). The primers sequences are as follows: 5'-CCCAAGCTTGGAGGAGTTACGCAGGCAGGGTT-3' (sense) and 5'-CGGGATCCGATTTAATGATGTATACTTCTATATTAG-3' (anti-sense).
The siRNAs specifically targeting GIHCG and control siRNA were acquired from GenePharma (Shanghai, China) and have been used in our previous report 34. The transfection of vectors and siRNAs was performed with Lipofectamine 3000 (Invitrogen) in accordance with the manufacturer's protocol.
Stable cell lines construction
To construct GIHCG stably overexpressed HeLa cells, pcDNA3.1-GIHCG or pcDNA3.1 was transfected into HeLa cells. Seventy-two hours after transfection, the cells were selected with neomycin for four weeks. To construct miR-200b and GIHCG concurrently stably overexpressed HeLa cells, GIHCG stably overexpressed HeLa cells were infected with miR-200b overexpression lentivirus (GeneChem, Shanghai, China) and selected with puromycin for four weeks.
Cell proliferation assays
Cell proliferation was evaluated by Glo cell viability assay and Ethynyl deoxyuridine (EdU) incorporation assay. For Glo cell viability assay, 3000 indicated cervical cancer cells were seeded per well in 96-well plates. At the indicated time cell viability was detected with the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) in accordance with the manufacturer's protocol. The luminescence value at each time point was acquired and used to plot cell proliferation curves. EdU incorporation assay was carried out using the EdU kit (Roche, Mannheim, Germany) in accordance with the manufacturer's protocol. The results were acquired and quantified using Zeiss photomicroscope (Carl Zeiss, Oberkochen, Germany).
Cell apoptosis assay
Cell apoptosis was evaluated by terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) assay. After being treated with 25 ng/ml doxorubicin (Selleck, Houston, TX, USA) for 24 hours, apoptosis of indicated cervical cancer cells was detected using TUNEL Cell Apoptosis Detection Kit (Beyotime, Jiangsu, China) in accordance with the manufacturer's protocol. The results were acquired and quantified using Zeiss photomicroscope (Carl Zeiss).
Cell migration assay
Cell migration was evaluated by transwell assay. Indicated cervical cancer cells suspended in serum-free medium with 1 mg/ml Mitomycin C to inhibit cell proliferation were seeded in the upper chamber of a transwell (24-well insert, 8 µm pore size, Millipore, Bedford, MA, USA). Medium containing 20% fetal bovine serum was added to the lower chamber. After 48 hours incubation, the cells that migrated into the lower surface of the transwell insert were fixed, stained, photographed, and quantified with at least ten random fields.
In vivo xenograft experiment
Indicated cervical cancer cells were subcutaneously injected into the flanks of female athymic BALB/c nude mice (Laboratory Animal Resources, Chinese Academy of Sciences, Shanghai, China). Subcutaneous tumor growth was measured every seven days using a caliper. Tumor volume was calculated according to the equation V = a*b*b/2 (a, long axes; b, short axes). The Ethics Review Committee of Fujian Medical University reviewed and approved the use of animals.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (version 5.0). For comparisons, Student's t-test, Wilcoxon signed-rank test, Mann-Whitney U test, receiver operating characteristic (ROC) curve analysis, and Pearson correlation analysis were carried out as indicated. P < 0.05 was considered as statistically significant.
Results
GIHCG is up-regulated in cervical cancer tissues and cell lines
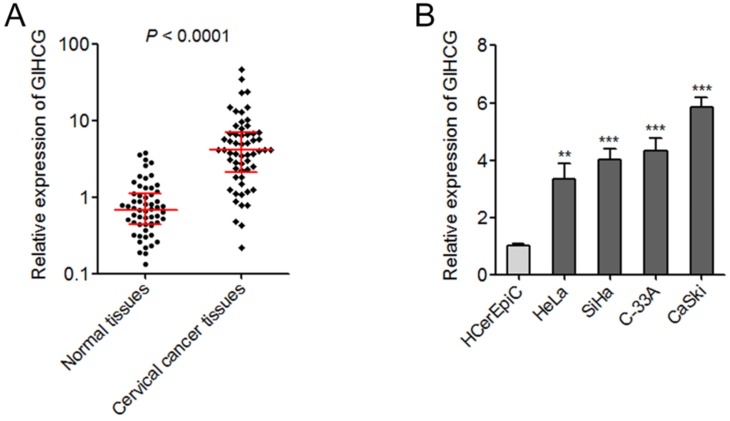
The expression of GIHCG in 58 pairs of cervical cancer tissues and adjacent normal tissues was measured by qRT-PCR. As shown in Figure 1A, the expression of GIHCG was significantly up-regulated in cervical cancer tissues compared with adjacent normal tissues (P < 0.0001). In addition, the expression of GIHCG in human normal cervical epithelial cell line (HCerEpiC) and cervical cancer cell lines (HeLa, SiHa, C-33A, and CaSki) was also measured by qRT-PCR. In accordance, the expression of GIHCG was significantly up-regulated in cervical cancer cell lines compared with normal cervical epithelial cell line (Figure 1B). These results revealed the up-regulation of GIHCG in cervical cancer.
Figure 1.
GIHCG is up-regulated in cervical cancer tissues and cell lines. (A) The expression of GIHCG in 58 pairs of cervical cancer tissues and adjacent normal tissues was measured by qRT-PCR. P < 0.0001 by Wilcoxon signed-rank test. (B) the expression of GIHCG in human normal cervical epithelial cell line (HCerEpiC) and cervical cancer cell lines (HeLa, SiHa, C-33A, and CaSki) was measured by qRT-PCR. Results are displayed as mean ± SD based on three independent biological replicates. **P < 0.01, ***P < 0.001 by Student's t-test.
GIHCG is up-regulated in the serum of cervical cancer patients and may serve as a novel diagnostic biomarker for cervical cancer
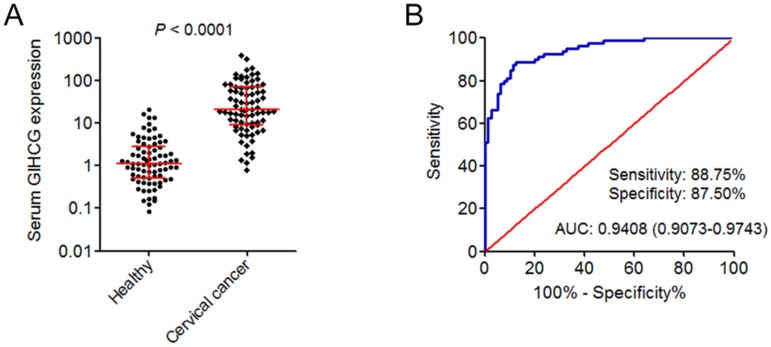
Next, the expression of GIHCG in the serum of 80 cervical cancer patients and 80 age-matched healthy controls was measured by qRT-PCR. As shown in Figure 2A, the expression of serum GIHCG was also significantly up-regulated in cervical cancer patients compared with healthy controls (P < 0.0001). To investigate whether serum GIHCG could serve as a non-invasive diagnostic biomarker for cervical cancer, receiver operating characteristic (ROC) curve analyses were performed. ROC curve showed accurate discrimination between cervical cancer patients and healthy controls, with an area under the ROC curve (AUC) of 0.9408 (95% CI: 0.9073-0.9743), a sensitivity of 88.75%, and a specificity of 87.50% (Figure 2B). These results revealed the up-regulation of serum GIHCG in cervical cancer patients and suggested that serum GIHCG may serve as a novel non-invasive diagnostic biomarker for cervical cancer.
Figure 2.
GIHCG is up-regulated in the serum of cervical cancer patients and may serve as a novel diagnostic biomarker for cervical cancer. (A) The expression of GIHCG in the serum of 80 cervical cancer patients and 80 age-matched healthy controls was measured by qRT-PCR. P < 0.0001 by Mann-Whitney U test. (B) ROC curve analysis of serum GIHCG for discrimination between cervical cancer patients and healthy controls (AUC: 0.9408, sensitivity: 88.75%, specificity: 87.50%). P < 0.0001.
GIHCG promotes cervical cancer cell proliferation, inhibits cell apoptosis, and promotes cell migration
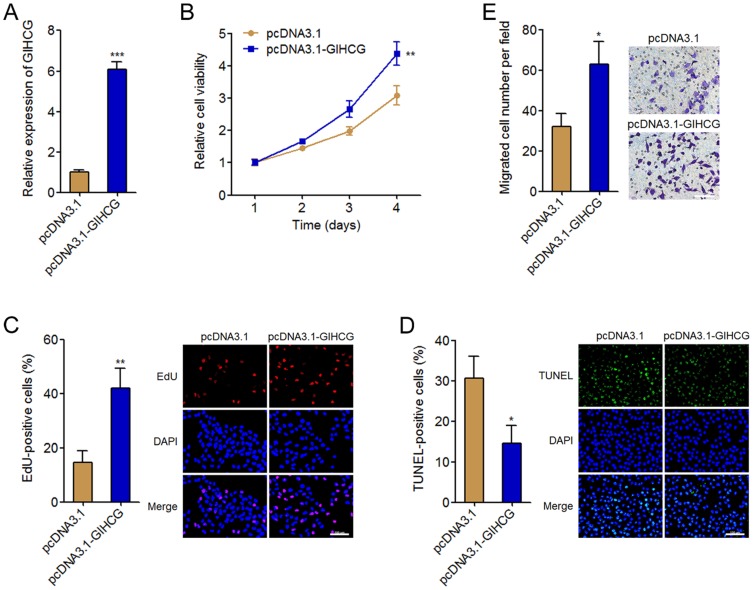
To investigate the biological roles of GIHCG in cervical cancer, we stably overexpressed GIHCG in HeLa cells by transfecting GIHCG overexpression vectors. The successful overexpression of GIHCG in HeLa cells was confirmed by qRT-PCR (Figure 3A). Cell proliferation was assessed by Glo cell viability assays and EdU incorporation assays. Glo cell viability assays displayed that enhanced expression of GIHCG increased cell viability of HeLa cells (Figure 3B). In accordance, EdU incorporation assays further displayed that enhanced expression of GIHCG promoted cell proliferation of HeLa cells (Figure 3C). Next, cell apoptosis was assessed by TUNEL assays. The results displayed that enhanced expression of GIHCG inhibited cell apoptosis of HeLa cells (Figure 3D). Moreover, cell migration was assessed by transwell assays. The results displayed that enhanced expression of GIHCG promoted cell migration of HeLa cells (Figure 3E). These results revealed that GIHCG functions as an oncogene in cervical cancer via promoting cell proliferation and migration, and inhibiting cell apoptosis.
Figure 3.
GIHCG promotes cervical cancer cell proliferation, inhibits cell apoptosis, and promotes cell migration. (A) The expression of GIHCG in GIHCG stably overexpressed and control HeLa cells was measured by qRT-PCR. (B) Cell viability of GIHCG stably overexpressed and control HeLa cells at indicated time was measured by Glo cell viability assays. (C) Cell proliferation of GIHCG stably overexpressed and control HeLa cells was measured by EdU incorporation assays. Red color indicates EdU-positive proliferative cells. Scale bars, 100 μm. (D) Cell apoptosis of GIHCG stably overexpressed and control HeLa cells was measured by TUNEL assays. Green color indicates TUNEL-positive apoptotic cells. Scale bars, 100 μm. (E) Cell migration of GIHCG stably overexpressed and control HeLa cells was measured by transwell migration assays. Scale bars, 100 μm. Results are displayed as mean ± SD based on three independent biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001 by Student's t-test.
Silencing GIHCG inhibits cervical cancer cell proliferation, induces cell apoptosis, and represses cell migration
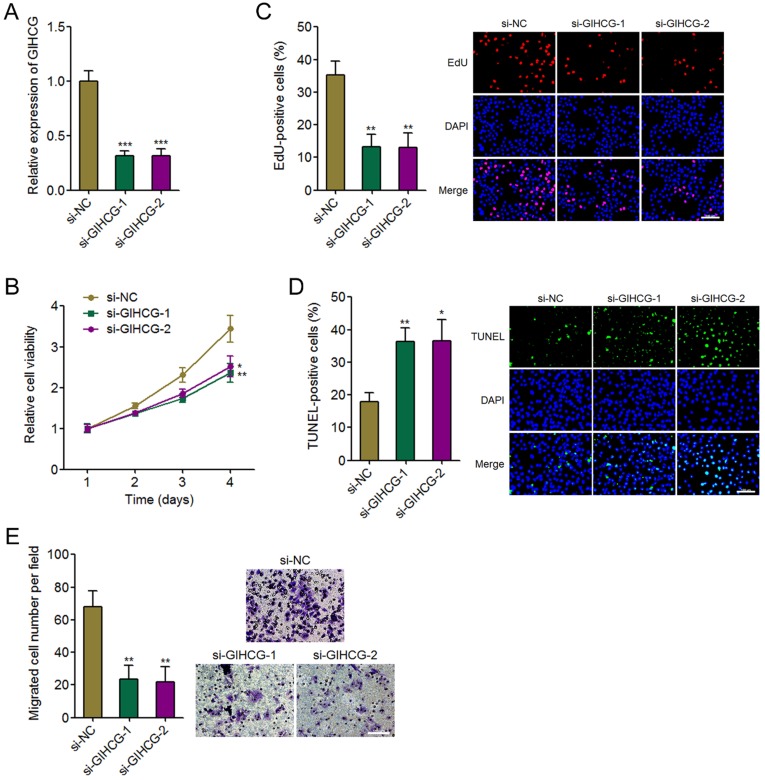
To further confirm the oncogenic roles of GIHCG in cervical cancer, we silenced GIHCG expression in CaSki cells by transfecting two independent GIHCG specific siRNAs. The successful knockdown of GIHCG in CaSki cells was confirmed by qRT-PCR (Figure 4A). Glo cell viability assays displayed that silencing GIHCG by two independent siRNAs both reduced cell viability of CaSki cells (Figure 4B). In accordance, EdU incorporation assays further displayed that silencing GIHCG by two independent siRNAs both inhibited cell proliferation of CaSki cells (Figure 4C). TUNEL assays displayed that silencing GIHCG by two independent siRNAs both induced cell apoptosis of CaSki cells (Figure 4D). Transwell migration assays displayed that silencing GIHCG by two independent siRNAs both inhibited cell migration of CaSki cells (Figure 4E). These results revealed that silencing GIHCG has tumor suppressive roles in cervical cancer via inhibiting cell proliferation and migration and inducing cell apoptosis.
Figure 4.
Silencing GIHCG inhibits cervical cancer cell proliferation, induces cell apoptosis, and represses cell migration. (A) The expression of GIHCG in CaSki cells 48h after GIHCG siRNAs transfection was measured by qRT-PCR. (B) Cell viability of CaSki cells at indicated time after GIHCG siRNAs transfection was measured by Glo cell viability assays. (C) Cell proliferation of CaSki cells 48h after GIHCG siRNAs transfection was measured by EdU incorporation assays. Red color indicates EdU-positive proliferative cells. Scale bars, 100 μm. (D) Cell apoptosis of CaSki cells 48h after GIHCG siRNAs transfection was measured by TUNEL assays. Green color indicates TUNEL-positive apoptotic cells. Scale bars, 100 μm. (E) Cell migration of CaSki cells 48h after GIHCG siRNAs transfection was measured by transwell migration assays. Scale bars, 100 μm. Results are displayed as mean ± SD based on three independent biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001 by Student's t-test.
GIHCG represses the expression of miR-200b in cervical cancer cells
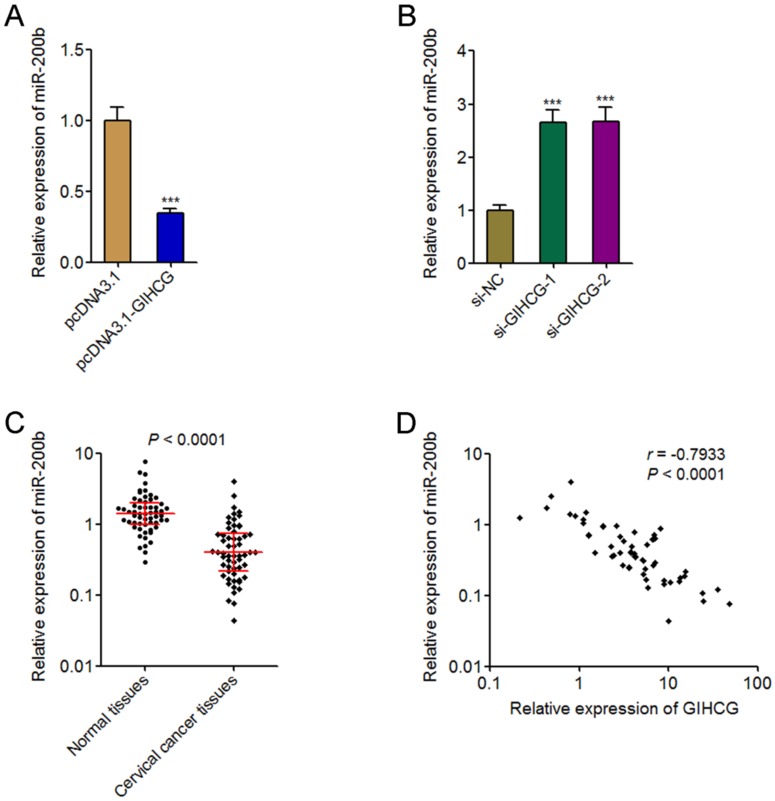
GIHCG is reported to repress the expression of miR-200b in hepatocellular carcinoma cells 33. miR-200b is a well-known tumor suppressor in many cancers, including cervical cancer 35. Therefore, we further investigated whether GIHCG also repressed miR-200b in cervical cancer cells and whether miR-200b is the mediator of the oncogenic roles of GIHCG in cervical cancer. The expression of miR-200b in GIHCG stably overexpressed and controlled HeLa cells was measured by qRT-PCR. As shown in Figure 5A, the expression of miR-200b was significantly reduced in GIHCG overexpressed cells compared with that in control cells. Reciprocally, silencing GIHCG using two dependent siRNAs both up-regulated the expression of miR-200b in CaSki cells (Figure 5B). To investigate whether the regulation of miR-200b by GIHCG also exists in vivo, the expression of miR-200b in the same 58 pairs of cervical cancer tissues and adjacent normal tissues used in Figure 1A was measured by qRT-PCR. The results displayed that the expression of miR-200b was significantly down-regulated in cervical cancer tissues compared with adjacent normal tissues (P < 0.0001, Figure 5C). The correlation between miR-200b and GIHCG expression levels in these 58 cervical cancer tissues was examined by Pearson correlation analysis. The results displayed a statistically significant inverse correlation between miR-200b and GIHCG expression levels (r = -0.7933, P < 0.0001, Figure 5D). Thus, these results revealed the repression of miR-200b by GIHCG in cervical cancer.
Figure 5.
GIHCG represses the expression of miR-200b in cervical cancer. (A) The expression of miR-200b in GIHCG stably overexpressed and control HeLa cells was measured by qRT-PCR. (B) The expression of miR-200b in CaSki cells 48h after GIHCG siRNAs transfection was measured by qRT-PCR. For A-B, results are displayed as mean ± SD based on three independent biological replicates. ***P < 0.001 by Student's t-test. (C) The expression of miR-200b in 58 pairs of cervical cancer tissues and adjacent normal tissues was measured by qRT-PCR. P < 0.0001 by Wilcoxon signed-rank test. (D) The correlation between miR-200b and GIHCG expression level in 58 cervical cancer tissues was analyzed. r = -0.7933, P < 0.0001 by Pearson correlation analysis.
miR-200b reverses the oncogenic roles of GIHCG in cervical cancer in vivo
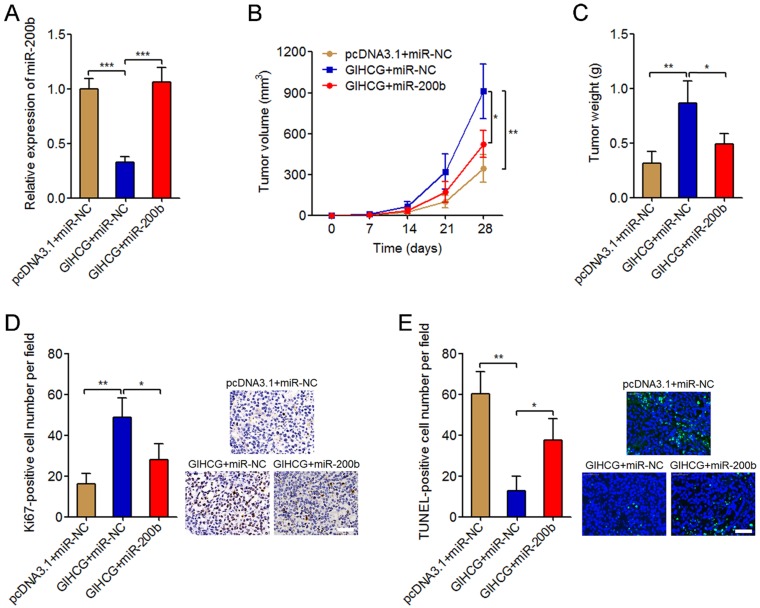
To investigate whether miR-200b is the mediator of the oncogenic roles of GIHCG in cervical cancer, we stably overexpressed miR-200b in GIHCG stably overexpressed HeLa cells via infecting miR-200b overexpression lentivirus. The successful overexpression of miR-200b in GIHCG stably overexpressed HeLa cells was confirmed by qRT-PCR (Figure 6A). These stably overexpressed and control HeLa cells were subcutaneously injected into immune-compromised nude mice. Tumor growth was monitored every seven days, and the results displayed that enhanced expression of GIHCG significantly promoted subcutaneous tumor growth, which was attenuated by miR-200b overexpression (Figure 6B). At 4-weeks after injection, the mice were sacrificed and the subcutaneous tumors were resected and weighed. The results displayed that enhanced expression of GIHCG significantly increased tumor weights, which was also attenuated by miR-200b overexpression (Figure 6C). Immunohistochemical staining of proliferation marker Ki67 in the subcutaneous tumors displayed that enhanced expression of GIHCG increased the number of Ki67-positive cells, which was attenuated by miR-200b overexpression (Figure 6D). TUNEL staining of the subcutaneous tumors displayed that enhanced expression of GIHCG reduced the number of TUNEL-positive apoptotic cells, which was also attenuated by miR-200b overexpression (Figure 6E). Thus, these results revealed the oncogenic roles of GIHCG in cervical cancer in vivo via promoting tumor growth, which could be reversed by miR-200b.
Figure 6.
Overexpression of miR-200b attenuates the oncogenic roles of GIHCG in cervical cancer in vivo. (A) The expression of miR-200b in GIHCG and miR-200b concurrently overexpressed and control HeLa cells was measured by qRT-PCR. Results are displayed as mean ± SD based on three independent biological replicates. ***P < 0.001 by Student's t-test. (B) GIHCG and miR-200b stably overexpressed and control HeLa cells were subcutaneously injected into nude mice. Tumor volumes were monitored every seven days. (C) The subcutaneous tumor weights were measured at 4-weeks after injection. (D) Immunohistochemical staining of Ki67 in subcutaneous tumors formed by GIHCG and miR-200b stably overexpressed and control HeLa cells. Scale bars, 50 μm. (E) TUNEL staining of subcutaneous tumors formed by GIHCG and miR-200b stably overexpressed and control HeLa cells. Scale bars, 50 μm. For B-E, results are displayed as mean ± SD based on n = 5 mice in each group. *P < 0.05, **P < 0.01 by Mann-Whitney U test.
Discussion
The alterations of protein-coding genes and signaling pathways in the initiation and progression of cervical cancer have been well recognized 36, 37. The implications of lncRNAs in cervical cancer are relative much less recognized. Although several lncRNAs have been reported to be associated with cervical cancer, such as NCK1-AS1, GAS5, UFC1, SNHG16, and CCHE1, the expression, roles, and clinical significances of lncRNAs in cervical cancer remain largely unclear 38-42.
Recently, lncRNA GIHCG is reported to have oncogenic roles in liver cancer, tongue squamous cell carcinoma, and renal cell carcinoma (RCC) 33, 34, 43. Sui et al reported that GIHCG is upregulated in liver cancer tissues and cell lines compared with adjacent normal liver tissues and normal liver cell lines. GIHCG promotes liver cancer proliferation and migration via inhibiting miR-200b/a/429 33. Ma et al reported that GIHCG is upregulated in tongue squamous cell carcinoma tissues and cell lines. GIHCG promotes tongue squamous cell carcinoma cell cycle, proliferation, and migration via inhibiting miR-429 43. Our previous study reported the up-regulation of GIHCG in RCC tissues compared with adjacent normal renal tissues. Furthermore, we identified the existence of GIHCG in the serum, and found that GIHCG is increased in the serum of RCC patients and could serve as a non-invasive diagnostic biomarker for RCC 34. However, the clinical significances of GIHCG in cervical cancer are still unknown.
In this study, we first investigated the expression pattern of GIHCG in cervical cancer. We discovered that the expression of GIHCG is notably up-regulated in cervical cancer tissues compared with corresponding normal tissues, and also significantly up-regulated in cervical cancer cell lines compared with normal cervical epithelial cell line. Furthermore, we collected 80 cervical cancer patients' serums before surgery and 80 serums from age-matched healthy controls. We discovered that serum GIHCG is remarkably up-regulated in cervical cancer patients compared with healthy controls. ROC curve analyses displayed that serum GIHCG could efficiently discriminate cervical cancer patients from healthy controls and suggested that serum GIHCG could serve as a non-invasive diagnostic biomarker for cervical cancer. Thus, this study provided a novel biomarker for the diagnosis of cervical cancer. Combining our previous study about the diagnostic significance of GIHCG for RCC, we proposed that the diagnostic values of GIHCG for cancers may be cancer-popular, but not cancer type-specific. More serum from various cancers patients are needed to elucidate this proposal.
The functions of GIHCG in cervical cancer are explored by in vitro and in vivo gain-of-function and loss-of-function assays. Our data revealed that overexpression of GIHCG promotes cervical cancer cell proliferation and migration and inhibits cervical cancer cell apoptosis. Depletion of GIHCG inhibits cervical cancer cell proliferation and migration and promotes cervical cancer cell apoptosis. Overexpression of GIHCG also promotes cervical cancer tumor growth in vivo. These data supported the oncogenic roles of GIHCG in cervical cancer and implied that targeting GIHCG may be a potential therapeutic strategy for cervical cancer.
Previous report showed that GIHCG epigenetically silenced miR-200b/a/429 expression in liver cancer 33. miR-200b is a well-known tumor suppressor in many cancers via directly repressing several critical oncogenes 35. In this study, we also found that GIHCG repressed the expression of miR-200b in cervical cancer. The expression of miR-200b is inversely correlated with the expression of GIHCG in cervical cancer tissues. Moreover, overexpression of miR-200b attenuated the oncogenic roles of GIHCG in promoting cervical cancer tumor growth in vivo. Thus, our data suggested that miR-200b at least partially mediated the roles of GIHCG in cervical cancer.
In conclusion, this study displayed that lncRNA GIHCG is up-regulated in cervical cancer tissues, cell lines, and serums. Serum GIHCG may serve as a novel non-invasive diagnostic biomarker for cervical cancer. Functionally, GIHCG promotes cervical cancer cell proliferation and migration, inhibits cervical cancer cell apoptosis, and promotes cervical cancer tumor growth in vivo via repressing miR-200b. Therefore, GIHCG functions as an oncogene and may serve as a potential therapeutic target in cervical cancer.
Acknowledgments
This work was supported by grants from Science and Technology Department of Jiangxi Province, China (No. 2015ZBBG70020) and Science and Technology Bureau of Yichun City, Jiangxi, China (No. JXYC2016KSA012).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Testing for cervical cancer. new recommendations from the American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology. CA Cancer J Clin. 2012;62:211–2. doi: 10.3322/caac.21135. [DOI] [PubMed] [Google Scholar]
- 3.Burki TK. Cervical cancer: screening and risk with age. Lancet Oncol. 2014;15:e107. doi: 10.1016/s1470-2045(14)70032-4. [DOI] [PubMed] [Google Scholar]
- 4.Heyward WL, Lanier AP, Bender TR, McMahon BJ, Kilkenny S, Paprocki TR. et al. Early detection of primary hepatocellular carcinoma by screening for alpha-fetoprotein in high-risk families. A case-report. Lancet. 1983;2:1161–2. doi: 10.1016/s0140-6736(83)91214-x. [DOI] [PubMed] [Google Scholar]
- 5.Cuzick J, Thorat MA, Andriole G, Brawley OW, Brown PH, Culig Z. et al. Prevention and early detection of prostate cancer. Lancet Oncol. 2014;15:e484–92. doi: 10.1016/S1470-2045(14)70211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamani N, Modares Gilani M, Zamani F, Zamani MH. Utility of Pelvic MRI and Tumor Markers HE4 and CA125 to Predict Depth of Myometrial Invasion and Cervical Involvement in Endometrial Cancer. Journal of family & reproductive health. 2015;9:177–83. [PMC free article] [PubMed] [Google Scholar]
- 7.Salvatici M, Achilarre MT, Sandri MT, Boveri S, Vanna Z, Landoni F. Squamous cell carcinoma antigen (SCC-Ag) during follow-up of cervical cancer patients: Role in the early diagnosis of recurrence. Gynecol Oncol. 2016;142:115–9. doi: 10.1016/j.ygyno.2016.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JP, Yang XJ, Xiao L, Wang Y. Long noncoding RNA PVT1 as a novel serum biomarker for detection of cervical cancer. Eur Rev Med Pharmacol Sci. 2016;20:3980–6. [PubMed] [Google Scholar]
- 10.Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AHF. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res. 2016;168:134–45. doi: 10.1016/j.trsl.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu QH. et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137:1128–35. doi: 10.1002/ijc.29484. [DOI] [PubMed] [Google Scholar]
- 12.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–95. [PMC free article] [PubMed] [Google Scholar]
- 13.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF. et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L. et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–24. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD. et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–40. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP. et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–32. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W. et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 2018;33:690–705. doi: 10.1016/j.ccell.2018.03.014. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F. et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol. 2017;19:1400–11. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B. et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16:111. doi: 10.1186/s12943-017-0681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739–54. doi: 10.1111/febs.13839. [DOI] [PubMed] [Google Scholar]
- 24.Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C. et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238–51. doi: 10.1038/ncb3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK. et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–15. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizrahi I, Mazeh H, Grinbaum R, Beglaibter N, Wilschanski M, Pavlov V. et al. Colon Cancer Associated Transcript-1 (CCAT1) Expression in Adenocarcinoma of the Stomach. J Cancer. 2015;6:105–10. doi: 10.7150/jca.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y. et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell. 2018;33:706–20. doi: 10.1016/j.ccell.2018.03.006. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L. et al. Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell. 2018;33:417–34. doi: 10.1016/j.ccell.2018.01.020. e7. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS. et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–86. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 30.Li C, Wang S, Xing Z, Lin A, Liang K, Song J. et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19:106–19. doi: 10.1038/ncb3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD. et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492–502. doi: 10.1038/s41556-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 32.Parasramka M, Yan IK, Wang X, Nguyen P, Matsuda A, Maji S. et al. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Cancer. 2017;16:22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sui CJ, Zhou YM, Shen WF, Dai BH, Lu JJ, Zhang MF. et al. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med (Berl) 2016;94:1281–96. doi: 10.1007/s00109-016-1442-z. [DOI] [PubMed] [Google Scholar]
- 34.He ZH, Qin XH, Zhang XL, Yi JW, Han JY. Long noncoding RNA GIHCG is a potential diagnostic and prognostic biomarker and therapeutic target for renal cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:46–54. doi: 10.26355/eurrev_201801_14099. [DOI] [PubMed] [Google Scholar]
- 35.Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124:649–58. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 36.Prasad SB, Yadav SS, Das M, Govardhan HB, Pandey LK, Singh S. et al. Down Regulation of FOXO1 Promotes Cell Proliferation in Cervical Cancer. J Cancer. 2014;5:655–62. doi: 10.7150/jca.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padilla-Quirarte HO, Trejo-Moreno C, Fierros-Zarate G, Castaneda JC, Palma-Irizarry M, Hernandez-Marquez E. et al. Interferon-Tau has Antiproliferative effects, Represses the Expression of E6 and E7 Oncogenes, Induces Apoptosis in Cell Lines Transformed with HPV16 and Inhibits Tumor Growth In Vivo. J Cancer. 2016;7:2231–40. doi: 10.7150/jca.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Jia Y, Cheng J, Liu G, Song F. LncRNA NCK1-AS1 promotes proliferation and induces cell cycle progression by crosstalk NCK1-AS1/miR-6857/CDK1 pathway. Cell Death Dis. 2018;9:198. doi: 10.1038/s41419-017-0249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W, Hong L, Xu X, Wang Q, Huang J, Jiang L. LncRNA GAS5 suppresses the tumorigenesis of cervical cancer by downregulating miR-196a and miR-205. Tumour Biol. 2017;39:1010428317711315. doi: 10.1177/1010428317711315. [DOI] [PubMed] [Google Scholar]
- 40.Xi J, Feng J, Zeng S, Huang P. Long noncoding RNA UFC1 is activated by E2F1 and exerts oncogenic properties by functioning as a ceRNA of FOXP3. Cancer medicine; 2018. doi: 10.1002/cam4.1556. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Zhu H, Zeng Y, Zhou CC, Ye W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys. 2018;637:1–8. doi: 10.1016/j.abb.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015;36:7615–22. doi: 10.1007/s13277-015-3465-4. [DOI] [PubMed] [Google Scholar]
- 43.Ma L, Wang Q, Gong Z, Xue L, Zuo Z. Long noncoding RNA GIHCG enhanced tongue squamous cell carcinoma progression through regulating miR-429. J Cell Biochem; 2018. doi: 10.1002/jcb.27164. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]