Abstract
Purpose: We aimed to investigate the expression of FOXM1 and to determine the relationships between FOXM1 expression and clinicopathologic characteristics in patients with PCa. Furthermore, we reconfirmed the prognostic impact of FOXM1 in different cohorts using already published data.
Patients and Methods: Formalin-fixed, paraffin-embedded tissues were collected from patients with low- (n=17), intermediate- (n=36), and high-risk (n=29) disease, from patients with CRPC (n=2) and from patients with BPH (n=28). To analyze FOXM1 expression, we performed IHC analyses. Also, we analyzed gene expression data from cBioPortal to evaluate the associations between FOXM1 alteration and prognosis of PCa.
Results: FOXM1 expression measured using Allred score differed between patients with BPH, and low-, intermediate-, and high-risk PCa (0.3, 1.5, 4.8, and 6.2, respectively; p<0.001). Patients with high FOXM1 expression had higher preoperative PSA levels (p=0.023), more advanced tumor stages (p=0.047), and higher pathologic Gleason score (p<0.001) than those with low FOXM1 expression. ROC curve analysis indicated that FOXM1 expression was a useful marker for discriminating PCa from BPH (AUC 0.851, 95% CI 0.783-0.920) and for discriminating high-risk PCa from low- and intermediate-risk PCa (AUC 0.807, 95% CI 0.719-0.894). In multivariate analyses, high FOXM1 expression was an independent predictor of BCR. Finally, in the TCGA dataset, FOXM1 alteration was associated with poor overall (p=4.521e-4) and disease-free survival (p=0.0108).
Conclusions: In patients with PCa, high FOXM1 expression was associated with advanced tumor stages, high Gleason score, and poor prognosis. These data suggest a role of FOXM1 in biologically and clinically aggressive PCa.
Keywords: Prostatic Neoplasms, Forkhead Box protein M1, Biomarkers
Introduction
Prostate cancer (PCa) ranks first in the most common newly diagnosed cancers, and ranks third in the leading cause of cancer-related death in men in the United States 1. Although most patients with PCa who undergo curative treatment have a relatively good prognosis, those with aggressive PCa with recurrence after treatment or distant metastasis have a poor prognosis 2. Identification of markers associated with clinical outcomes of PCa would aid in proper clinical decision-making. However, despite the large number of studies, very few biomarkers for predicting PCa recurrence have been identified and validated in different cohorts using different techniques.
Forkhead Box M1 (FOXM1) is a member of the Forkhead box family that shares a 100 amino acid long winged-helix DNA-binding domain 3. It is a crucial transcription factor that promotes tumorigenesis and enhances metastasis and invasion capacity in several solid cancers 4, 5. FOXM1 is mainly responsible for growth and maturation during embryogenesis as well as homeostasis and repair of adult tissues in normal cells 6, however, in cancer cells, it contributes to all the hallmarks of cancer such as sustaining proliferation signaling, activating invasion and metastasis, and enabling replicative immortality 3, 4. Several studies showed that high FOXM1 expression was associated with poor prognosis in several solid tumors including breast cancer 7, colorectal cancer 8, and renal cell cancer 9. Recent meta-analyses to investigate the prognostic role of FOXM1 expression consistently reported that patients with high FOXM1 expression had advanced tumor stage and dismal prognosis in malignant solid tumors 10. However, the role of FOXM1 in PCa has not been clearly elucidated.
In this study, we aimed to investigate the expression of FOXM1 in prostate tissues and to determine the relationships between FOXM1 expression and clinicopathologic characteristics in Korean PCa patients. Furthermore, we reconfirmed the prognostic impact of FOXM1 in different cohorts using already published data from cBioPortal (http://www.cbioportal.org) 11, 12.
Materials and Methods
Patients
After institutional review board approval (IRB No. MC17SESI0079), we obtained formalin-fixed, paraffin-embedded (FFPE) tissues from patients with low- (n=17), intermediate- (n=36), and high-risk (n=29) PCa according to the National Comprehensive Cancer Network risk group,13 from patients with castration-resistant prostate cancer (CRPC, n=2) and from patients with benign prostatic hyperplasia (BPH, n=28) from the Korea Prostate Bank. To ensure a homogeneous cohort to evaluate immunohistochemistry (IHC) results, patients were excluded if they had received neoadjuvant treatment. Baseline demographics, pathologic and follow-up data were collected retrospectively from medical records.
Immunohistochemical staining
The expression of FOXM1 was analyzed by IHC staining. Tissue sections (4-µm-thick) were dewaxed in xylene and rehydrated gradually with graded ethanol. After incubation in a microwave oven in Tris-EDTA buffer solution (pH 9.0) for 20 min, endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide in methanol, and nonspecific immunoglobulin binding was blocked by incubation with 10% normal horse serum for 15 min. After that, tissue slides were incubated with anti-rabbit FOXM1 (dilution 1:100, GeneTex, Irvine, CA) at 4°C overnight and then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG for 30 min, followed by reaction with 3,3'-diaminobenzidine tetrahydrochloride for 3 min and counterstaining with hematoxylin. The images were obtained using a microscope (Olympus Optical Co. Ltd., Tokyo, Japan).
The slides were analyzed in parallel by two investigators unaware of the patient's clinical data. The Allred scoring system was used to evaluate staining intensity and proportion. The intensity of the most predominant area was recorded as 0, 1, 2, or 3 for none, light, medium, or dark staining, respectively. The proportion of stained cells was divided into 6 categories (0: no staining; 1: <1% positive; 2: 1-10% positive; 3: 11-33% positive; 4: 34-66% positive; and 5: 67-100% positive). Addition of the two values provides the total Allred score, yielding scores of 0-8 14.
FOXM1 in published datasets
Gene expression data were downloaded and analyzed from cBioPortal (http://www.cbioportal.org) to evaluate the association between FOXM1 alterations and clinical variables of PCa using independent sample tests 11, 12. We selected three studies that included gene expression data from RNAseq or gene expression microarray: 1) Prostate Adenocarcinoma (The Cancer Genome Atlas [TCGA], Provisional) 15, 2) Metastatic Prostate Cancer, SU2C/PCF Dream Team (Robinson et al., Cell 2015) 16, and 3) Neuroendocrine Prostate Cancer (Trento/Cornell/Broad 2016) 17.
Statistical analyses
Continuous variables are presented as mean (± standard deviation [SD]), and dichotomous variables are presented as number of patients (proportions). Comparison of demographic, clinical, and pathologic data was performed with independent t-test or one-way analysis of variance (one-way ANOVA) test for continuous variables and with chi-square test for dichotomous variables. Receiver operator characteristic (ROC) curves were plotted to evaluate the prognostic significance of FOXM1 and area under the ROC curve by analyzing the area under the curve (AUC) value. Kaplan-Meier analysis with log-rank test was performed to estimate biochemical recurrence (BCR)-free survival. The association between the FOXM1 expression and BCR was assessed using multivariate Cox proportional hazard regression analysis in a forward stepwise regression after adjusting for known important clinicopathologic features. All statistical analyses were performed with SPSS version 24.0 (SPSS Inc., Chicago, IL, USA) with a two-tailed p-value <0.05 indicating statistical significance.
Results
FOXM1 expression in BPH and PCa
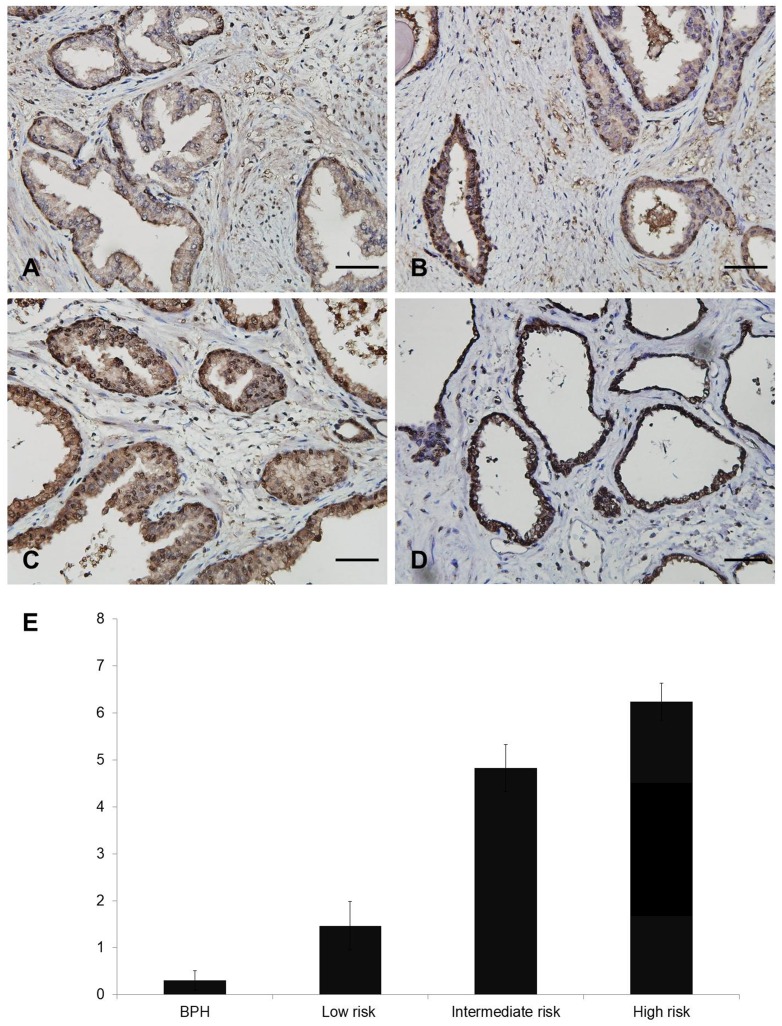
Representative figures showing FOXM1 expression are displayed in Figure 1A-D. Nuclear FOXM1 expression was observed only in 7.1 % (2 of 28) of BPH tissues, whereas nuclear FOXM1 expression was more frequently observed in low- (35.2%, 6 of 17), intermediate- (75%, 27 of 36), and high-risk (93.1%, 27 of 29) PCa tissues. The strongest diffuse nuclear FOXM1 positivity was detected in CRPC (100%, 2 of 2) tissues. The Allred score of FOXM1 was significantly different among patients with BPH, and low-risk, intermediate-risk, and high-risk PCa (0.3 ± 0.2, 1.5 ± 0.5, 4.8 ± 0.5, 6.2 ± 0.4, respectively, p<0.001; Fig. 1E). These results indicate that nuclear FOXM1 expression is associated with PCa aggressiveness.
Figure 1.
Different nuclear staining of FOXM1 in human prostate tissue. A-D. Representative images of FOXM1 (Scale bar, 50 μm): A, Allred score 0 in BPH tissue. B, Allred score 5 in PCa tissue. C, Allred score 8 in PCa tissue. D, Allred score 8 in CRPC tissue. E. Quantitative analysis of nuclear expression of FOXM1 in BPH and PCa patients.
Baseline characteristics according to FOXM1 expression
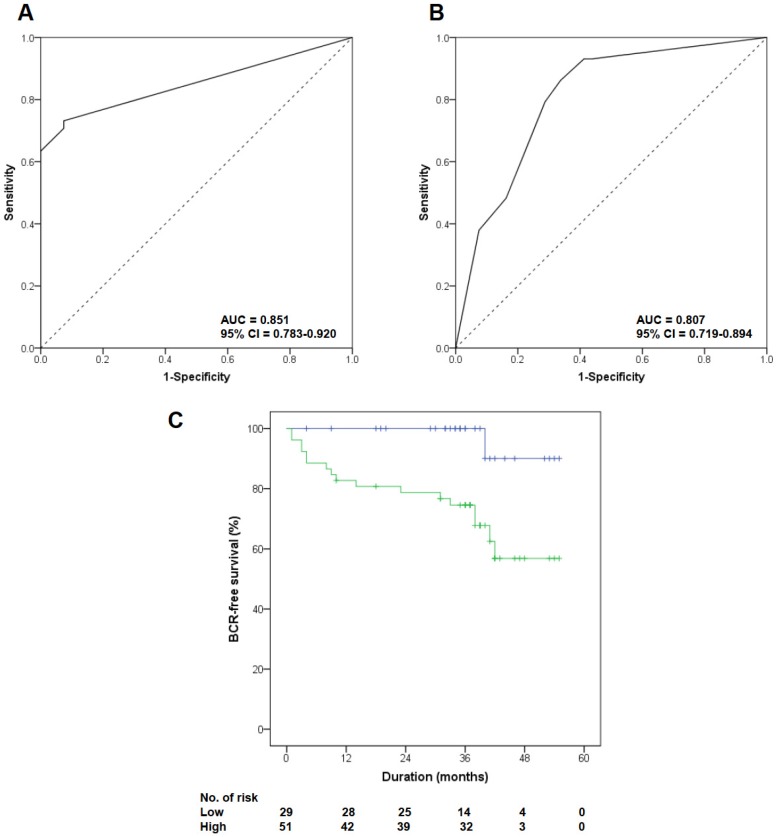
All PCa patients underwent radical prostatectomy, and their median follow-up duration was 38 months (range 6 to 55). Table 1 lists the baseline clinicopathologic characteristics of the patients, stratified by FOXM1 expression level. We set the median Allred score of FOXM1 as the cutoff value to divide patients into two groups (median Allred score 4.0). Patients with high FOXM1 expression had higher preoperative PSA levels (p=0.023), more advanced pathological tumor stages (p=0.047), and higher pathologic Gleason scores (p<0.001) than those with low FOXM1 expression. ROC curve analysis indicated that FOXM1 expression was a useful marker for discriminating PCa from BPH (AUC 0.851, 95% CI 0.783-0.920; Fig. 2A) and for discriminating high-risk PCa from low- and intermediate-risk PCa (AUC 0.807, 95% CI 0.719-0.894; Fig. 2B).
Table 1.
Clinicopathologic characteristics according to FOXM1 expression
| Variables | Overall | FOXM1 expression | ||
|---|---|---|---|---|
| Low | High | p-value | ||
| Age (years)* | 68.8, 70.0 (± 7.2) | 69.7 (± 7.3) | 68.1 (± 7.0) | 0.242 |
| BMI (kg/m2)* | 23.3, 22.9 (± 2.6) | 23.3 (± 2.6) | 23.3 (± 2.7) | 0.922 |
| Preoperative PSA (ng/mL)* | 11.0, 6.8 (± 16.7) | 7.5 (± 8.2) | 14.8 (± 22.2) | 0.023 |
| Prostate volume (mL)*† | 43.7, 35.0 (± 24.1) | 53.3 (± 26.9) | 31.9 (± 12.9) | <0.001 |
| Pathologic T stage (%) | 0.047 | |||
| 2 | 58 (70.7) | 26 (86.7) | 32 (61.5) | |
| 3a | 10 (12.2) | 1 (3.3) | 9 (17.3) | |
| 3b | 14 (17.1) | 3 (10.0) | 11 (21.2) | |
| Pathologic N stage (%) | 0.004 | |||
| X | 55 (67.1) | 26 (86.7) | 29 (55.8) | |
| 0 | 27 (32.9) | 4 (13.3) | 23 (44.2) | |
| 1 | 0 (0) | 0 (0) | 0 (0) | |
| Pathologic Gleason score (%) | <0.001 | |||
| ≤ 6 | 17 (20.7) | 15 (50.0) | 2 (3.8) | |
| 7 (3+4) | 30 (36.6) | 7 (23.3) | 23 (44.2) | |
| 7 (4+3) | 22 (26.8) | 6 (20.0) | 16 (30.8) | |
| ≥ 8 | 13 (15.9) | 2 (6.7) | 11 (21.2) | |
| Positive surgical margin (%) | 31 (37.8) | 9 (30.0) | 22 (42.3) | 0.268 |
| Lymphovascular invasion (%) | 3 (3.7) | 1 (3.3) | 2 (3.8) | 0.905 |
| Perineural invasion (%) | 55 (67.1) | 17 (56.7) | 38 (73.1) | 0.128 |
* Values are expressed as mean, median (±SD).
† Prostate volume was measured by transrectal ultrasonography.
BMI, body mass index; PSA, prostate-specific antigen; FOXM1, Forkhead box M1
Figure 2.
Clinical significance of FOXM1 alteration in patients with PCa in the Korean Prostate Bank cohort. (A, B) Receiver operator characteristic (ROC) curves were plotted to evaluate the predictive accuracy of FOXM1 expression for overall PCa and high-risk PCa. (C) Biochemical recurrence-free survival according to FOXM1 expression level was quantified by Kaplan-Meier analysis.
Impact of FOXM1 expression on BCR in Korean Prostate Bank specimens
During the follow-up period, 22.0% (18 of 82) of patients experienced BCR after radical prostatectomy, including 3.3% (1 of 30) of patients with low FOXM1 expression levels and 32.7% (17 of 52) of patients with high FOXM1 expression levels (p=0.002). Kaplan-Meier analysis showed a significant association between high FOXM1 expression and shorter BCR-free survival (Fig. 2C, p=0.004). High FOXM1 expression was significantly associated with an increased risk of BCR (HR 10.524, 95% CI 1.400-79.087, p=0.022), and this association remained significant after adjusting for various known prognostic factors in the multivariate Cox regression analysis (HR 8.819, 95% CI 1.166-66.711, p=0.035; Table 2).
Table 2.
Univariate and multivariate Cox proportional hazard regression analysis of clinicopathologic features for biochemical recurrence
| Factors | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | ||
| Age (years) | 1.083 | 1.001-1.171 | 0.048 | 1.084 | 0.968-1.172 | 0.064 | |
| BMI (kg/m2) | 1.008 | 0.841-1.208 | 0.933 | - | - | - | |
| Preoperative PSA (ng/mL) | 1.027 | 1.013-1.042 | <0.001 | 1.022 | 1.006-1.038 | 0.006 | |
| Prostate volume (mL) | 0.998 | 0.963-1.035 | 0.927 | - | - | - | |
| Pathologic T stage | |||||||
| 2 | Reference | - | - | - | |||
| 3a | 3.991 | 1.205-13.217 | 0.023 | - | - | - | |
| 3b | 7.258 | 2.417-21.797 | <0.001 | - | - | - | |
| Pathologic Gleason score | |||||||
| ≤ 6 | Reference | - | - | - | |||
| 7 (3+4) | 1.246 | 0.129-12.072 | 0.850 | - | - | - | |
| 7 (4+3) | 4.577 | 0.565-37.051 | 0.154 | - | - | - | |
| ≥ 8 | 8.162 | 0.979-68.048 | 0.052 | - | - | - | |
| Positive surgical margin | 4.053 | 1.516-10.836 | 0.005 | 3.039 | 1.099-8.401 | 0.032 | |
| Lymphovascular invasion | 1.372 | 0.182-10.328 | 0.759 | - | - | - | |
| Perineural invasion | 4.086 | 0.939-17.781 | 0.061 | - | - | - | |
| High FOXM1 expression | 10.524 | 1.400-79.087 | 0.022 | 8.819 | 1.166-66.711 | 0.035 | |
HR, hazard ratio; CI, confidence interval; BMI, body mass index; PSA, prostate-specific antigen
Impact of FOXM1 mutation on prognosis in published datasets
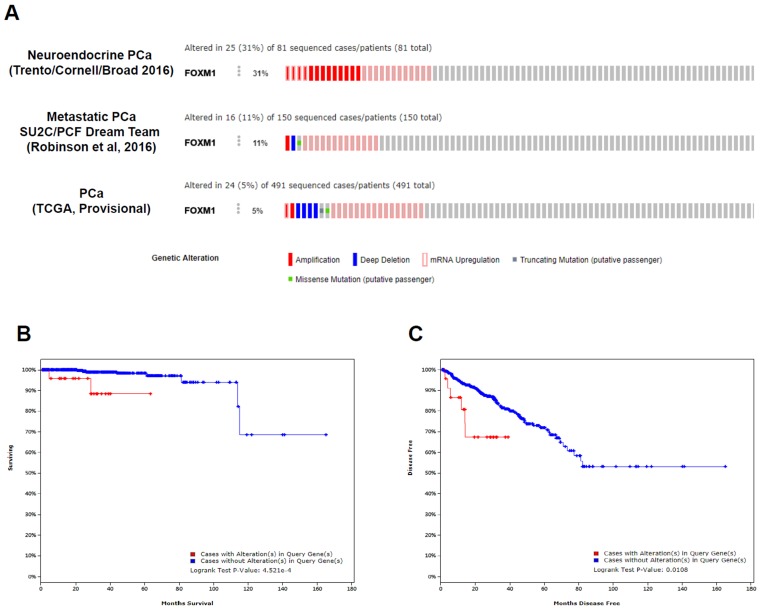
There are currently 13 datasets of prostate cancer genomics in the cBioPortal 11, 12, and these datasets cover primary, metastatic, and neuroendocrine PCa. Among 491 prostate adenocarcinomas 15, 150 metastatic PCas 16, and 81 neuroendocrine PCas 17, the FOXM1 gene is altered in 5%, 11%, and 31% of patients, respectively (p<0.001; Fig. 3A), demonstrating increased alterations of the FOXM1 gene from primary PCa to metastatic or neuroendocrine PCa. FOXM1 mRNA upregulation and amplification is the most frequently identified alterations in these datasets.
Figure 3.
Clinical significance of FOXM1 alterations in patients with PCa in published datasets. (A) The OncoPrint of genetic alterations of FOXM1, including amplifications, deep deletions, mRNA upregulation, truncation mutations, and missense mutations, are shown in PCa using cBioPortal. Only representative cases are shown. (B, C) Kaplan-Meier analysis of overall survival and disease-free survival is shown from TCGA data from cBioPortal.
Using the TCGA dataset that contained follow-up data for PCa recurrence in 91 patients and PCa-related mortality in 10 cases, we conducted statistical analyses to determine whether FOXM1 alterations were associated with adverse pathologic features or poor prognosis. The results of the analysis of the TCGA dataset revealed that patients with FOXM1 alteration had more advanced tumor stage (p < 0.001) and higher Gleason score (p < 0.001). Figure 3B and 3C show a statistically significant prognostic value of FOXM1 alteration for the overall and disease-free survival in the TCGA dataset. Patients with FOXM1 alteration were significantly associated with poor overall survival (p=4.521e-4) and poor disease-free survival (p=0.0108).
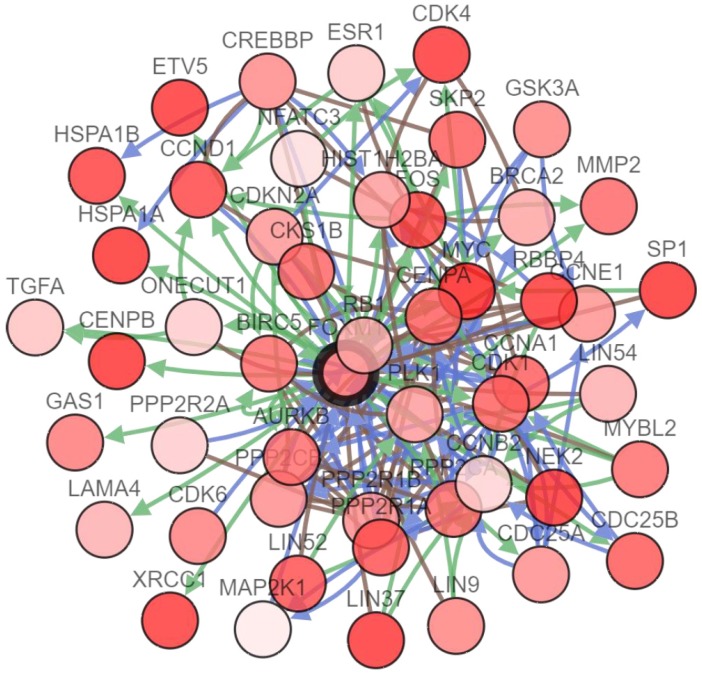
To further study the interactions between FOXM1 and several frequently altered neighboring genes, a FOXM1 gene network was drawn consisting of the seed node FOXM1 and 49 linker nodes, which were generated using the cBioPortal system (Fig. 4). The network reflected clear interactions between FOXM1 and several major pathways: notably, a group of cell cycle genes (CCNA1, CCNB2, CCND1, and so on) and DNA damage repair genes (BRCA2 for homologous recombination and XRCC1 for base excision repair). Moreover, several proto-oncogenes including FOS, MYC, and SKP2 were demonstrated to have connections with FOXM1.
Figure 4.
Network analysis of FOXM1 in patients with PCa. The network analysis of genetic alterations in FOXM1 is shown in PCa from TCGA data using cBioPortal.
Discussion
In this study, we explored the association between nuclear FOXM1 expression level and prognosis in patients with PCa. Our findings demonstrated that higher nuclear FOXM1 expression is associated with PCa aggressiveness and shorter BCR-free survival. Moreover, the public dataset showed that genetic alteration of FOXM1 is associated with poor overall and disease-free survival. Our results suggest that nuclear FOXM1 expression level is a potential prognostic marker for PCa.
FOXM1 is a regulator of overall biological processes including angiogenesis, apoptosis, invasion, metastasis, metabolism, and inflammation, as well as cell proliferation and cell cycle progression.3, 4 Moreover, it is upregulated in various solid tumors, including colorectal cancer, gastric cancer, hepatic cancer, lung cancer, and ovarian cancer, and high expression indicates poor prognosis 10, 18. A few studies have evaluated the prognostic potential of FOXM1 in PCa. Chandran et al. reported that FOXM1 mRNA was highly expressed in metastatic PCa samples obtained from 4 patients 19. Other researchers reported that increased FOXM1 expression was associated with accelerated development of PCa in transgenic mice 20, 21 and induced epithelial-mesenchymal transition in PCa cell lines 22. These studies are limited due to a small number of patients and lack of clinical data. In our study, FOXM1 protein expression was also significantly higher in PCa tissues, which is consistent with previous studies. Our study clearly showed that nuclear FOXM1 expression is significantly correlated with advanced tumor stage and poor prognosis using a large number of PCa patients with meaningful clinical data.
FOXM1 increases mRNA and protein expression of the androgen receptor regardless of the presence of androgens. FOXM1 directly binds to the forkhead motif of the PSA promoter/enhancer regions to regulate PSA gene transcription. Interestingly, the binding of FOXM1 markedly increased in androgen-independent cells compared to androgen-dependent cells. These results indicate that FOXM1 promotes PCa progression in both androgen-dependent and androgen-independent ways as an oncogenic transcription factor 23. Our gene network analysis suggests the potential of FOXM1 as an oncogenic transcription factor regardless of androgen-dependence.
A gene network analysis of FOXM1 in PCa showed that FOXM1 interacts with many genes related to the cell cycle and DNA damage repair (Fig 4). FOXM1 is well-known as a key regulator of cell cycle control. It targets the transcription of genes that regulate the G1-S transition including cyclin D1 (CCND1) and cell division cycle 25A (Cdc25A), and regulates genes involved in the G2-S transition such as CCNB1, CCNB2, cyclin-dependent kinase 1 (CDK1), polo-like kinase 1 (PLK1), and Cdc25B. Moreover, genes required for M phase progression, such as aurora B kinase (AURKB), centromere protein A (CENPA), CENPB, and CENPF, are under the transcriptional control of FOXM1 24. Therefore, FOXM1 stimulates proliferation by promoting G1-S and G2-M transition and M phase progression.
In addition, FOXM1 induces DNA repair, particularly homologous recombination, and this process involves the transcriptional control of DNA repair genes. For example, FOXM1 directly binds to the promoters of DNA repair genes such as base excision repair factor X-ray cross-complementing group 1 (XRCC1), breast cancer associated gene 2 (BRCA2), Rad51, and exonuclease 1 (EXO1) and enhances their expression 25-27.
Furthermore, FOXM1 is implicated in all stages of tumorigenesis from initiation to metastasis 28. The c-Myc oncogene is a multifunctional transcription factor that plays an important role in various cellular processes including proliferation, differentiation, and apoptosis 29. Interestingly, FOXM1 binds to the c-Myc promoter, and c-Myc also binds to the FOXM1 promoter. As a result, a positive feedback loop between FOXM1 and c-Myc promotes cell proliferation 30.
Our study has some limitations. First of all, this is a retrospective study that could be biased due to confounding factors. However, as factors that could influence the results were controlled using multivariate analysis, it is unlikely that the apparent survival benefit associated with low FOXM1 expression was an artifact. Moreover, this is a single national study composed of one ethnicity, and it may be difficult to generalize the results to other ethnic groups. Although we electronically validated the prognostic value of FOXM1 in another independent cohort using different biological methods from cBioPortal, further studies are warranted to validate the results of our study. Finally, since FOXM1 might be connected to several important pathways such as the cell cycle and DNA damage repair, the exact regulation mechanisms of FOXM1 and associations between these pathways require further investigation.
In conclusions, high FOXM1 expression was associated with advanced tumor stages, high Gleason score, and poor prognosis in patients with PCa. These data suggest a role for FOXM1 in biologically and clinical aggressive PCa. The inclusion of FOXM1 as a prognostic factor may improve prognostic abilities and help physicians manage this population through the proper stratification of patients.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No. 2017R1D1A1B03030051).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Brawley OW. Trends in prostate cancer in the United States. J Natl Cancer Inst Monogr. 2012;2012:152–6. doi: 10.1093/jncimonographs/lgs035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–33. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gemenetzidis E, Bose A, Riaz AM, Chaplin T, Young BD, Ali M. et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS One. 2009;4:e4849. doi: 10.1371/journal.pone.0004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bella L, Zona S, Nestal de Moraes G, Lam EW. FOXM1: A key oncofoetal transcription factor in health and disease. Semin Cancer Biol. 2014;29:32–9. doi: 10.1016/j.semcancer.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Bektas N, Haaf A, Veeck J, Wild PJ, Luscher-Firzlaff J, Hartmann A. et al. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer. 2008;8:42. doi: 10.1186/1471-2407-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Wei P, Peng Z, Huang C, Tang H, Jia Z. et al. The critical role of dysregulated FOXM1-PLAUR signaling in human colon cancer progression and metastasis. Clin Cancer Res. 2013;19:62–72. doi: 10.1158/1078-0432.CCR-12-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu XR, Chen YH, Liu DM, Sha JJ, Xuan HQ, Bo JJ. et al. Increased expression of forkhead box M1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Med Oncol. 2013;30:346. doi: 10.1007/s12032-012-0346-1. [DOI] [PubMed] [Google Scholar]
- 10.Dai J, Yang L, Wang J, Xiao Y, Ruan Q. Prognostic Value of FOXM1 in Patients with Malignant Solid Tumor: A Meta-Analysis and System Review. Dis Markers. 2015;2015:352478. doi: 10.1155/2015/352478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohler J, Armstrong A, Bahnson R. National Comprehensive Cancer Network clinical practice guidelines in oncology: Prostate cancer. 2016; 2016. [Google Scholar]
- 14.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11:155–68. [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM. et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Wu D, Yu Q, Li L, Wu P. Prognostic value of FOXM1 in solid tumors: a systematic review and meta-analysis. Oncotarget. 2017;8:32298–308. doi: 10.18632/oncotarget.15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W. et al. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV. et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer research. 2006;66:1712–20. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Y, Balli D, Ustiyan V, Fulford L, Hiller A, Misetic V. et al. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. The Journal of biological chemistry. 2013;288:22527–41. doi: 10.1074/jbc.M113.455089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Yao B, Wang Y, Zhang M, Fu S, Gao H. et al. Increased FoxM1 expression is a target for metformin in the suppression of EMT in prostate cancer. International journal of molecular medicine. 2014;33:1514–22. doi: 10.3892/ijmm.2014.1707. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Liu Y, Yuan B, Yin L, Peng Y, Yu X. et al. FOXM1 promotes the progression of prostate cancer by regulating PSA gene transcription. Oncotarget. 2017;8:17027–37. doi: 10.18632/oncotarget.15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal N, Chakraborty S, Nag A. BIOLOGY OF FOXM1 AND ITS EMERGING ROLE IN CANCER THERAPY. 2014; 2014. [Google Scholar]
- 25.Zhou J, Wang Y, Wang Y, Yin X, He Y, Chen L. et al. FOXM1 modulates cisplatin sensitivity by regulating EXO1 in ovarian cancer. PloS one. 2014;9:e96989. doi: 10.1371/journal.pone.0096989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang N, Wu X, Yang L, Xiao F, Zhang H, Zhou A. et al. FoxM1 inhibition sensitizes resistant glioblastoma cells to temozolomide by downregulating the expression of DNA-repair gene Rad51. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:5961–71. doi: 10.1158/1078-0432.CCR-12-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–16. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teh M-T. FOXM1 coming of age: time for translation into clinical benefits? Frontiers in Oncology; 2012. p. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nature Reviews Cancer. 2002;2:764. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 30.Wierstra I, Alves J. FOXM1c transactivates the human c-myc promoter directly via the two TATA boxes P1 and P2. FEBS J. 2006;273:4645–67. doi: 10.1111/j.1742-4658.2006.05468.x. [DOI] [PubMed] [Google Scholar]