In this article, it is shown that the structure of the apo ligand-binding domain (LBD) of the retinoic X receptor, which was historically the first structure of a nuclear receptor LBD to be reported and which is still used as a reference today, contained minor inaccuracies that could be corrected using modern structure-refinement tools.
Keywords: nuclear receptors, retinoic acid, ligand binding domains, retinoic X receptor α
Abstract
The retinoic X receptor (RXR) plays a crucial role in the superfamily of nuclear receptors (NRs) by acting as an obligatory partner of several nuclear receptors; its role as a transcription factor is thus critical in many signalling pathways, such as metabolism, cell development, differentiation and cellular death. The first published structure of the apo ligand-binding domain (LBD) of RXRα, which is still used as a reference today, contained inaccuracies. In the present work, these inaccuracies were corrected using modern crystallographic tools. The most important correction concerns the presence of a π-bulge in helix H7, which was originally built as a regular α-helix. The presence of several CHAPS molecules, which are visible for the first time in the electron-density map and which stabilize the H1–H3 loop, which contains helix H2, are also revealed. The apo RXR structure has played an essential role in deciphering the molecular mode of action of NR ligands and is still used in numerous biophysical studies. This refined structure should be used preferentially in the future in interpreting experiments as well as for modelling and structural dynamics studies of the apo RXRα LBD.
1. Introduction
The homodimer of the ligand-binding domain (LBD) of human retinoid X receptor α (RXRα) was the first crystal structure of a nuclear receptor LBD to be published (Bourguet et al., 1995 ▸). This was soon followed by crystal structures of the active form of the retinoic acid receptor γ (RARγ) LBD (Renaud et al., 1995 ▸; Bourguet et al., 2000 ▸). These structures marked the start of a new era for the nuclear receptor community. Acting as ligand-dependent factors, nuclear receptors control a large number of physiological processes by regulating gene transcription, and they are targets of prime interest for the development of new drugs (Gronemeyer et al., 2004 ▸). The structural work provided unprecedented insight into the molecular mechanisms of ligand control of nuclear receptor signalling and paved the way for therapeutic ligand design (for reviews see, for example, Bain et al., 2007 ▸; Pawlak et al., 2012 ▸).
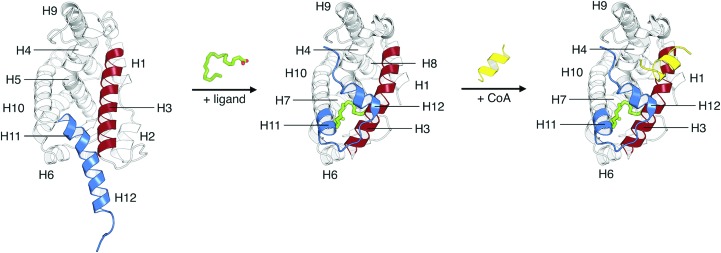
RXR plays a crucial role in the superfamily of nuclear receptors (NRs) by acting as an obligatory heterodimeric partner of an entire class of NRs, including the peroxisome proliferator-activated receptor (PPAR), the farnesoid X receptor (FXR), the liver X receptor (LXR), the vitamin D receptor (VDR), the thyroid hormone receptor (TR) and RAR, as well as forming homodimers (Brelivet et al., 2004 ▸; Billas & Moras, 2013 ▸; Renaud et al., 1995 ▸). Consequently, its role as a transcription factor is critical in many signalling pathways, such as metabolism, cell development, differentiation and cellular death (Germain et al., 2006 ▸). It thus plays a key role in various human diseases, including cancer (Altucci et al., 2007 ▸). RXRs have the same architecture as all other NRs, with a variable N-terminal domain (NTD), a highly conserved DNA-binding domain (DBD) and a ligand-binding domain (LBD) (Renaud & Moras, 2000 ▸). Among these domains, the LBD, which is the most studied domain in NRs, contains the ligand-binding pocket (LBP) that is associated with the ligand-dependent function (AF-2), the main dimerization interface for homodimerization and heterodimerization, and an interaction surface for co-regulatory proteins. Structurally, the LBD is composed of 12 α-helices organized as an antiparallel helical sandwich, with a small β-sheet between helices H5 and H6 (see Fig. 1 ▸). This architecture is conserved in all NR LBDs. The transcriptional activity is activated by the binding of an agonist ligand, such as 9-cis-retinoic acid, in an L-shaped hydrophobic pocket located in the core of the domain (Egea et al., 2000 ▸, 2002 ▸). The activation of the domain is accompanied by structural changes upon ligand binding, originally described as the ‘mouse-trap’ mechanism (Wurtz et al., 1996 ▸; Renaud et al., 1995 ▸). Compared with the ligated forms of the TRβ and RARα LBDs (Wagner et al., 1995 ▸; Renaud et al., 1995 ▸), it was suggested that upon ligand binding helix H12, which is also known as the activating domain (AD) for the AF-2 function, packs against the core of the LBD, acting as a ‘lid’ over the LBP. This movement is accompanied by the repositioning of H11 to be continuous with H10, and by the bending of the N-terminal part of H3 towards the ligand (Fig. 1 ▸). The new position of H12 forms hydrophobic contacts with H3 and H4, to which coactivator proteins (CoAs) can bind and initiate the formation of the transcription complex (Fig. 1 ▸). The influence of ligand binding on the structure and dynamics of NR LBDs has been confirmed by several experimental methods (for a review, see Nagy & Schwabe, 2004 ▸), yet X-ray crystallography remains the method of choice for characterizing the interactions of natural and synthetic ligands with LBDs. To date, over 800 experimental structures of nuclear receptor LBDs are available in the PDB.
Figure 1.
Structural changes associated with the mechanism of activation of the RXRα LBD by an agonist ligand. On the left, the apo form of the RXRα LBD (re-refined apo structure using the density map from PDB entry 1lbd; Bourguet et al., 1995 ▸) is shown in cartoon representation. The secondary-structural elements that are most affected by ligand binding, namely H3 and H11/H12, are highlighted in red and blue, respectively. Upon binding of an agonist ligand (exemplified by the agonist docosahexaenoic acid, represented in green), the RXRα LBD experiences conformational changes that stabilize the ligand-dependent function (AF-2), localized in helix H12, against the helical core (the structure displayed is PDB entry 1mv9; Egea et al., 2002 ▸). The holo form of the RXRα LBD with the newly formed hydrophobic surface, composed of helices H3, H4 and H12, favours the recruitment of coactivator proteins (CoAs), represented here by a co-activator peptide harbouring the LXXLL motif (represented in yellow; Egea et al., 2002 ▸).
In this study, we perform a re-refinement of the original data from the first crystal structure of the apo form of the RXRα LBD homodimer using modern crystallographic tools. This was motivated by the fact that it remains the only crystal structure that is available for the apo RXRα LBD homodimer. In addition to correcting some construction errors, such as the π-bulge in helix H7, we also reveal the presence of several CHAPS molecules, which are visible for the first time in the electron-density map, around the H1–H3 loop that contains helix H2 and which stabilize its conformation. As apo RXRα is one of the few apo structures of an NR LBD, it is a reference structure for biophysical studies of NR activation mechanisms. The corrections to this historically important structure that are presented here will allow these mechanistic studies to start from a structure refined using current protocols and into which a number of structural corrections that are important for proper analysis have been introduced.
2. Materials and methods
As the original diffraction images were no longer available, it was not possible to reprocess them to recover missing low-resolution data or to extend the data to higher resolution. Therefore, the structure of the apo RXRα LBD dimer (PDB entry 1lbd; Bourguet et al., 1995 ▸) and the corresponding diffraction data were imported from the PDB using PHENIX (Adams et al., 2010 ▸). 5% of the reflections were assigned to the test set, as the original test set could not be recovered. The B factors were reset to the Wilson B factor (55 Å2) before refinement. The structure was refined using BUSTER (Bricogne et al., 2016 ▸) and was manually rebuilt using Coot (Emsley et al., 2010 ▸). The final model quality was verified using MolProbity (Chen et al., 2010 ▸). A comparison of the final refined model statistics with those of the original structure (PDB entry 1lbd) is shown in Table 1 ▸. The coordinates have been deposited in the Protein Data Bank (PDB entry 6hn6).
Table 1. Refinement statistics of the original and re-refined structures.
The original data-collection statistics are taken from Bourguet et al. (1995 ▸). Values in parentheses are for the outer shell.
| Original (PDB entry 1lbd) | After re-refinement (PDB entry 6hn6) | |
|---|---|---|
| Space group | P6322 | |
| a, b, c (Å) | 110.80, 110.80, 109.90 | |
| α, β, γ (°) | 90, 90, 120 | |
| No. of unique reflections | 9167 | |
| Completeness (%) | 85 | |
| R merge(I) (%) | 6.9 | |
| Resolution used for refinement (Å) | 5.999–2.71 (2.80–2.71) | 7.99–2.71 (3.03–2.71) |
| No. of reflections used in refinement | 8067 (587) | 9164 (2597) |
| No. of reflections used for R free | 791 (60) | 459 (130) |
| R cryst (%) | 23.0 | 17.3 (24.9) |
| R free (%) | 34.2 | 23.1 (32.1) |
| No. of non-H atoms | ||
| Total | 1870 | 2090 |
| Protein | 1870 | 1882 |
| CHAPS | 168 | |
| Water | 40 | |
| R.m.s.d. from ideal values | ||
| Bond lengths (Å) | 0.02 | 0.010 |
| Bond angles (°) | 2.48 | 1.23 |
| Average B factors (Å2) | ||
| Protein | 34.3 | 62.4 |
| CHAPS | 94.6 | |
| Water | 61.4 | |
| Ramachandran statistics | ||
| Favoured (%) | 88.98 | 96.58 |
| Allowed (%) | 8.05 | 2.99 |
| Outliers (%) | 2.97 | 0.43 |
X-ray structures that are described as containing at least one apo RXRα monomer (for a list, see the supporting information) were systematically compared using PSS (Gaillard et al., 2013 ▸) and clustering was achieved using the dihedral backbone angles from the common residues only. The structures were visualized and the r.m.s.d. values were calculated using PyMOL (v.1.8; Schrödinger).
3. Results
3.1. Correction of the initial crystal structure
An examination of the re-refined electron-density map allowed us to correct construction errors, either minor corrections involving the side-chain positions of some residues or larger changes concerning the backbone of helix H7 within the dimerization interface.
Among the side-chain corrections, Arg316, which is localized in the C-terminal part of H5, is noteworthy. Belonging to the LBP, which is mainly composed of hydrophobic residues located in helices H5, H7, H11 and H12, as well as the β-sheet (Bourguet et al., 2000 ▸), Arg316 is one of the highly conserved charged residues involved in ligand recognition. Indeed, it participates in the anchoring of acidic ligands into the LBP through a network of ionic and hydrogen bonds between the carboxyl moiety of the ligand and the backbone of Ala327 in the β-sheet region of the LBD (Egea et al., 2002 ▸).
A comparison of PDB entry 1lbd with the other apo forms of the LBD shows that in these latter structures the Arg316 side chains are oriented towards the centre of the LBP, making hydrogen bonds to the backbone of Leu325 in the β-sheet. The re-refined density map of the apo RXRα LBD shows that Arg316 also has a similar position in this structure (Fig. 2 ▸ a). Instead of being exposed to the solvent, Arg316 now points towards the ligand-binding pocket; however, the quality of the electron-density map for this side chain was not sufficient to identify the intramolecular interactions between the side chain of Arg316 and the rest of the LBD with precision. Nevertheless, the proximity to the backbone of Leu325, the side chain of Ser312 and a water molecule suggests the existence of hydrogen bonds between them. Another conserved residue with an alternative conformation after refinement is Trp305, which switches from a p90 rotamer to a p-90 rotamer.
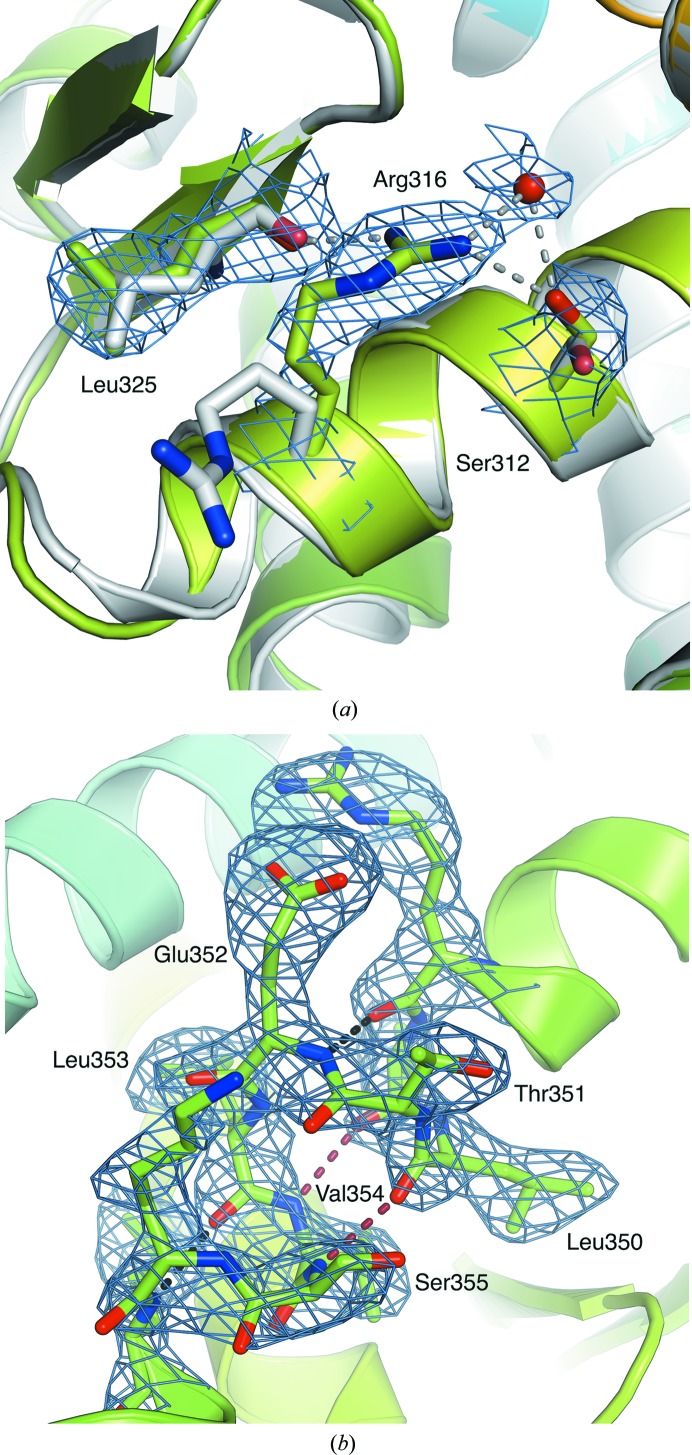
Figure 2.
(a) Comparison of the new and initial positions (in green and white, respectively) of Arg316 located in the C-terminal part of the H5 helix of the RXRα LBD. Possible hydrogen bonds between the new coordinates of Arg316 and the side chain of Ser312, the backbone of Leu325 and a water molecule are represented in grey. The 2F o − F c electron-density map, contoured at 1.5σ, is coloured blue. (b) Helix H7 exhibits a π-helix conformation between residues Val349 and Ser355. The α-type and π-type hydrogen bonds are coloured black and red, respectively. The 2F o − F c electron-density map, contoured at 1.5σ, is coloured blue.
A more important correction to the original structure concerns helix H7. The model constructed in PDB entry 1lbd shows H7 as a straight α-helix, with the sequence out of register by one position compared with all other structures of the LBD, which show the presence of a π-helix conformation from residues Val349 to Ser355 of H7. In PDB entry 1lbd, this has the consequence of fully burying some hydrophilic residues in the LBD, such as Glu352 and Lys356, and conversely exposing hydrophobic residues, such as Leu352 and Val354, to the solvent. The reconstruction of helix H7 from the newly refined density maps shows that the structure of this helix indeed harbours a π-helix turn in its centre, in agreement with the subsequent crystal structures of RXR LBDs (Bourguet et al., 2000 ▸; Gampe, Montana, Lambert, Wisely et al., 2000 ▸; Fig. 2 ▸ b). The presence of π-helices, also known as π-bulges (Cartailler & Luecke, 2004 ▸) or α-bulges (Hardy et al., 2000 ▸), in proteins is often associated with functional advantages (Fodje & Al-Karadaghi, 2002 ▸; Cooley et al., 2010 ▸). Structurally, π-helices are defined by the occurrence of at least two sequential π-type hydrogen bonds between residues that are five positions apart in sequence, owing to the insertion of a single residue into an α-helix (Fodje & Al-Karadaghi, 2002 ▸; Cooley et al., 2010 ▸). In the context of RXR, the presence of a π-helix (π-bulge or p-helix conformation) within H7 could be related to its ability to dimerize. In order to interact with other nuclear receptors, RXRα possesses two dimerization interfaces, one located in the DBD and the other in the LBD. The dimerization interface located in the LBD mainly involves residues located in helix H10 (Bourguet et al., 1995 ▸), and to a lesser extent in H7, H9 and loop H8–H9, and, in the heterodimeric RAR–RXR complex, loop H9–H10 (Bourguet et al., 2000 ▸). Structures of the RXRα LBD dimer within the tetramer (Gampe, Montana, Lambert, Wisely et al., 2000 ▸) indicate that the insertion of Glu352 could facilitate its dimerization by increasing the surface area of the dimerization interface and stabilize the dimer through a series of hydrogen bonds compared with the original structure with PDB code 1lbd. A π-helix containing a glutamate residue is also found in the same position in HNF4α, an NR that belongs to the RXR subfamily, and mutation or deletion of this residue strongly impairs the dimerization of HNF4α in solution (Eeckhoute et al., 2003 ▸). Therefore, these results emphasize the importance of the presence of this π-helix in H7 and its role in the dimerization function of RXRα. At the same time, a new model was constructed of loop H9–H10 (residues Cys404–Arg414) that was very similar to that in other apo crystal structures.
3.2. Stabilization of the H1–H3 loop by CHAPS molecules
Of all the crystal structures available in the PDB (Berman et al., 2000 ▸) that contain an LBD of RXRα (67 in total in June 2018), only seven structures contain a complete H1–H3 connection with the presence of helix H2. Nevertheless, examination of the re-refined electron-density maps shows that only the data deposited for PDB entry 1lbd contains sufficient information to reconstruct the loop completely. No clear density is seen in this region for PDB entries 1fm6, 1fm9 (Gampe, Montana, Lambert, Miller et al., 2000 ▸), 1g1u (Gampe, Montana, Lambert, Wisely et al., 2000 ▸) and 1xls (Suino et al., 2004 ▸), while the structure factors have not been deposited for PDB entries 1g5y (Gampe, Montana, Lambert, Wisely et al., 2000 ▸) and 1k74 (Xu et al., 2001 ▸).
Surprisingly, a re-examination of the density map around the H1–H3 loop in the re-refined apo RXRα LBD structure reveals the presence of four CHAPS molecules (Fig. 3 ▸ a). These CHAPS molecules interact with residues located in H2, loop H2–H3, H3, the β-sheet and H12. The most buried CHAPS molecule (A501) is surrounded by all of the listed structural elements. The nature of these interactions is mainly hydrophobic. Nevertheless, several hydrogen bonds formed by the hydroxyl and carbonyl groups of the CHAPS steroid core complete the set of interactions: bonds are formed between the hydroxyl O4 atom and the side-chain and backbone O atoms of Ser260, located in the H2–H3 loop, between the hydroxyl O3 atom and the backbone O atom of Ala252 in H2, and between the carbonyl O1 atom and the side chain of Glu453 in H12 (Fig. 3 ▸ b). Two other CHAPS molecules (A502 and A503) are located between H2 and H12 and are oriented perpendicularly to the buried CHAPS molecule. They interact with each other through hydrophobic contacts, and with the domain and the buried CHAPS molecule. For those two CHAPS molecules, the hydrophobic interactions are completed by hydrogen bonds: one between the O4 atom of the first CHAPS molecule (A502) and the side chain of Glu456 in H12, and one between the O4 atom of the second CHAPS molecule (A503) and the backbone O atom of Asn253 located in H2. The last CHAPS molecule (A504), which is the most exposed, is located along helix H2 and makes hydrophobic contacts with Thr246, Glu247, Val250, Met254, Leu256 and Asn257 (Fig. 3 ▸ c). CHAPS is known to be a protein stabilizer (Gall et al., 2003 ▸), and PDB entry 1lbd is the only RXRα structure in which it was present in the crystallization conditions. The presence of CHAPS could therefore explain why the H1–H3 region forms a short α-helix in this structure only and not in other crystal structures. The H1–H3 connection is indeed not part of the α-helical sandwich fold of the LBD and does not form extensive interactions with the core of the domain. Overall, this region is rather flexible, but the presence of CHAPS molecules seems to be sufficient to stabilize the loop. Interestingly, all four CHAPS molecules are involved in crystal contacts around the twofold crystallographic symmetry axis, mostly through hydrophobic interactions, although one hydrogen bond is formed between O3 of CHAPS A502 and O2 of the symmetry-related CHAPS A503.
Figure 3.
(a) Wall-eyed stereoview of the CHAPS molecules A501, A502, A503 and A504 (represented as white sticks) that interact with the RXRα LBD (represented in cartoon mode). (b) 2F o − F c electron-density map, contoured at 1.0σ, of the loop between the H2–H3 helices and Glu453 located on helix H12, with which a CHAPS molecule interacts. (c) 2F o − F c electron-density map, contoured at 1.0σ, of the CHAPS molecule along the H2 helix. The electron-density maps for the CHAPS molecules and the RXRα LBD are coloured cyan and blue, respectively. Hydrogen bonds are shown in black.
3.3. Alternative side-chain positioning in different apo structures of the RXRα LBD
A comparison of apo RXRα structures (see also the supporting information) reveals that PDB entry 1lbd is the only crystal structure in which both LBDs of RXRα interact with neither a ligand nor a co-activator or co-repressor peptide. Indeed, in several RXR LBD dimer structures one monomer contains a ligand while the other is in the apo form. Moreover, for some of the monomers indicated as being in the apo form there is weak density in the ligand-binding pocket. For example, despite the absence of a ligand in PDB entry 1g1u, the structure is superimposable with PDB entry 1g5y (with an overall r.m.s.d. of 0.186 Å) that contains a non-activating retinoic acid (Supplementary Figs. S1 and S2; Gampe, Montana, Lambert, Wisely et al., 2000 ▸). A careful examination of the LBPs of the four monomers in PDB entry 1g1u reveals weak density to differing extents, indicating that a ligand may possibly be partially bound. In PDB entry 3nsp neither monomer shows a ligand, but the crystallization conditions included the antagonist danthron, and chain B is in a characteristic antagonist conformation and is superimposable with the antagonist-bound PDB entry 3nsq (chain B; r.m.s.d. of 0.306 Å; Supplementary Figs. S1 and S3; Zhang, Zhou et al., 2011 ▸). However, an examination of the density of the chain B LBP of both PDB entries 3nsp and 3nsq reveals that although some density is present, in neither case does it seem to be consistent with the molecular structure of the antagonist danthron. Chains A of PDB entries 3nsp and 3nsq, on the other hand, do not show density in the LBP and can be considered to be in the apo form, as are chains B and C of PDB entries 4n5g and 4n8r (Chen et al., 2014 ▸).
The crystal structures reported in PDB entries 3r29 and 3r2a are truly apo, but are co-crystallized with co-repressor peptides and their presence has a significant impact on their structures (Zhang, Chen et al., 2011 ▸).
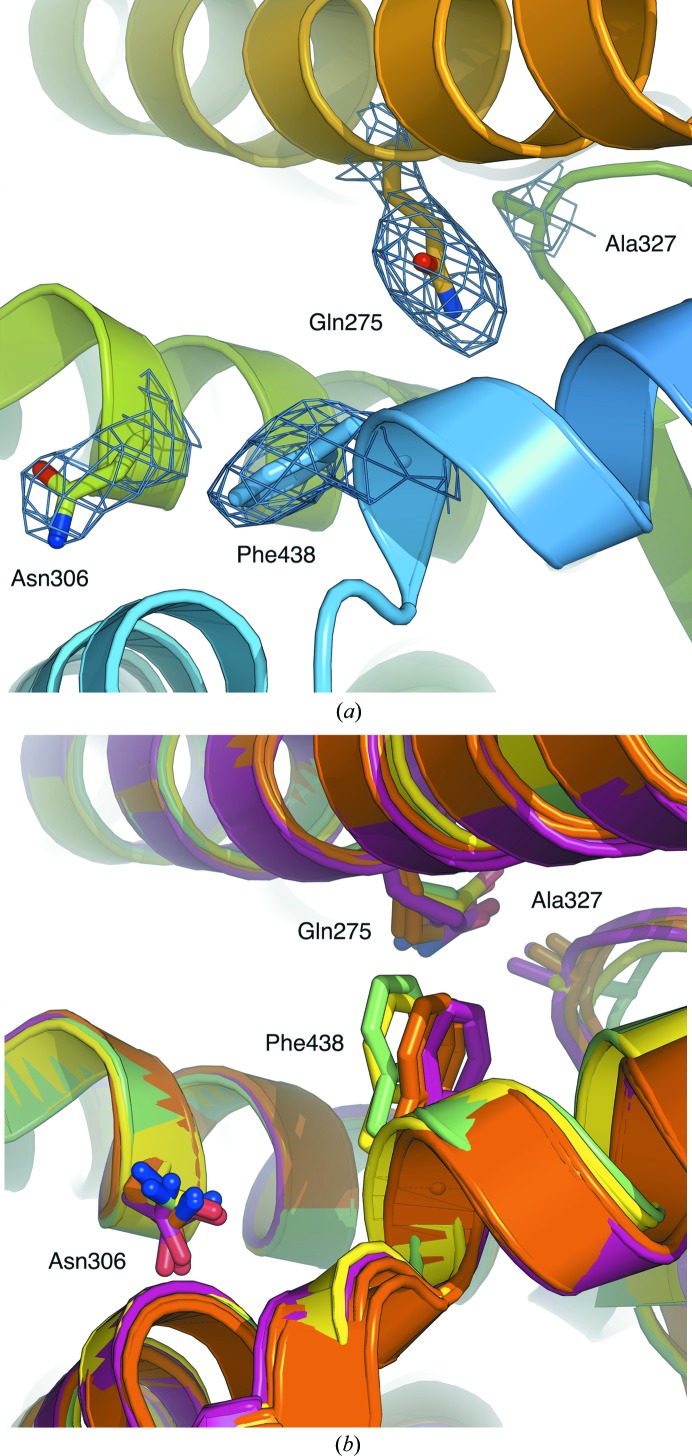
The re-refined apo RXRα LBD structure was compared with the apo partners of the RXRα LBD structures in PDB entries 3nsp (chain A), 3nsq (chain A), 4n5g (chains B and C) and 4n8r (chains B and C). The analysis reveals alternative side-chain positions for residues Gln275 (H3), Asn306 (H5) and Phe438 (H11) at the interface of helices H3, H5 and H11 in the LBP (Figs. 4 ▸ a and 4 ▸ b). The presence of Gln275 at the interface in the re-refined apo RXRα LBD structure forces Phe438 towards H5 and in turn pushes Asn306 towards H10 (Fig. 4 ▸ a). The rotation of Gln275 prevents unfavourable polar–hydrophobic contacts with Ala327 located in the β-sheet. Alternatively, in PDB entries 3nsp, 3nsq, 4n5g and 4n8r Gln275 is displaced towards the β-sheet, leaving room for Phe438 and Asn306 at the interface, which is also linked to a movement of H10 and H11 towards the centre of the LBP (Fig. 4 ▸ b).
Figure 4.
(a) Positions of the Gln275 (H3), Asn306 (H5), Ala327 (β-sheet) and Phe438 (H12) side chains, represented as sticks, in the ligand-binding pocket of the re-refined apo RXRα LBD structure. The 2F o − F c electron-density map, contoured at 1.5σ, is coloured blue. (b) Positions of the Gln275 (H3), Asn306 (H5), Ala327 (β-sheet) and Phe438 (H12) side chains, represented as sticks, in the ligand-binding pocket of the apo RXRα LBD in PDB entries 3nsq (chain A; green), 3nsp (chain A; yellow), 4n8r (chains B and C; pink) and 4n5g (chains B and C; orange).
Amino acids located in the dimerization interface, cofactor-binding surface and LBP have been proposed to belong to a putative allosteric communication network identified through statistical coupling analysis (SCA; Shulman et al., 2004 ▸). It remains to be established whether the alternative positions of these residues observed in the re-refined apo RXRα LBD structure with respect to PDB entries 3nsp, 3nsq, 4n5g and 4n8r could also be linked to this allosteric mechanism owing to the presence of a ligand inside the LBP of the neighbouring monomer in the latter structures.
4. Conclusion
Since the first publications of crystallographic structures of nuclear receptor ligand-binding domains in the 1990s (Renaud et al., 1995 ▸; Bourguet et al., 1995 ▸, 2000 ▸), over 800 structures of LBDs have been deposited in the Protein Data Bank, and this remarkable interest has been fuelled by the therapeutic value of modulating NR activity using natural and synthetic ligands. While the structure of apo LBDs is important for a mechanistic understanding of activation mechanisms, and ultimately for the informed design of active ligands, there are comparatively few of these with respect to ligand-bound structures. In this article, we show that the structure of the apo RXRα LBD, which was the first to be published and which is still used as a reference today, contained minor inaccuracies that could be corrected using modern structure-refinement tools. The corrected structure displays features that are coherent with newer structures, but still retains some genuine differences in side-chain orientations, a feature that could be related to the absence of ligand in both monomers of the dimeric architecture. The new refined structure should be used in the future for modelling and structural dynamics studies of the apo RXRα LBD.
Supplementary Material
PDB reference: human RXRα, ligand-binding domain, 6hn6
Supplementary Figures.. DOI: 10.1107/S2053230X18018022/ow5011sup1.pdf
Acknowledgments
We thank Dr Roland H. Stote for helpful discussions. The authors acknowledge the support and the use of the resources of the French Infrastructure for Integrated Structural Biology FRISBI ANR-10-INBS-05 and of Instruct-ERIC.
Funding Statement
This work was funded by Centre National de la Recherche Scientifique grant . Institut National de la Santé et de la Recherche Médicale grant . Université de Strasbourg grant . Agence Nationale de la Recherche grants ANR-10-LABX-0030-INRT, ANR-10-IDEX-0002-02 , and ANR-IAB-2011-BIP:BIP (ANR-10-BINF-0003). French Infrastructure for Integrated Structural Biology grant ANR-10-INBS-05 . INSTRUCT ERIC grant .
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Altucci, L., Leibowitz, M. D., Ogilvie, K. M., de Lera, A. R. & Gronemeyer, H. (2007). Nature. Rev. Drug Discov. 6, 793–810. [DOI] [PubMed]
- Bain, D. L., Heneghan, A. F., Connaghan-Jones, K. D. & Miura, M. T. (2007). Annu. Rev. Physiol. 69, 201–220. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Billas, I. & Moras, D. (2013). J. Mol. Biol. 425, 2317–2329. [DOI] [PubMed]
- Bourguet, W., Ruff, M., Chambon, P., Gronemeyer, H. & Moras, D. (1995). Nature (London), 375, 377–382. [DOI] [PubMed]
- Bourguet, W., Vivat, V., Wurtz, J.-M., Chambon, P., Gronemeyer, H. & Moras, D. (2000). Mol. Cell, 5, 289–298. [DOI] [PubMed]
- Brelivet, Y., Kammerer, S., Rochel, N., Poch, O. & Moras, D. (2004). EMBO Rep. 5, 423–429. [DOI] [PMC free article] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2016). BUSTER v.2.10.1. Global Phasing Ltd, Cambridge, England.
- Cartailler, J.-P. & Luecke, H. (2004). Structure, 12, 133–144. [DOI] [PubMed]
- Chen, L., Wang, Z.-G., Aleshin, A. E., Chen, F., Chen, J., Jiang, F., Alitongbieke, G., Zeng, Z., Ma, Y., Huang, M., Zhou, H., Cadwell, G., Zheng, J.-F., Huang, P.-Q., Liddington, R. C., Zhang, X.-K. & Su, Y. (2014). Chem. Biol. 21, 596–607. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cooley, R. B., Arp, D. J. & Karplus, P. A. (2010). J. Mol. Biol. 404, 232–246. [DOI] [PMC free article] [PubMed]
- Eeckhoute, J., Moerman, E., Bouckenooghe, T., Lukoviak, B., Pattou, F., Formstecher, P., Kerr-Conte, J., Vandewalle, B. & Laine, B. (2003). Endocrinology, 144, 1686–1694. [DOI] [PubMed]
- Egea, P. F., Mitschler, A. & Moras, D. (2002). Mol. Endocrinol. 16, 987–997. [DOI] [PubMed]
- Egea, P. F., Mitschler, A., Rochel, N., Ruff, M., Chambon, P. & Moras, D. (2000). EMBO J. 19, 2592–2601. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fodje, M. N. & Al-Karadaghi, S. (2002). Protein Eng. 15, 353–358. [DOI] [PubMed]
- Gaillard, T., Schwarz, B. B., Chebaro, Y., Stote, R. H. & Dejaegere, A. (2013). J. Chem. Inf. Model. 53, 2471–2482. [DOI] [PubMed]
- Gall, A.-L., Ruff, M. & Moras, D. (2003). Acta Cryst. D59, 603–606. [DOI] [PubMed]
- Gampe, R. T., Montana, V. G., Lambert, M. H., Miller, A. B., Bledsoe, R. K., Milburn, M. V., Kliewer, S. A., Willson, T. M. & Xu, H. E. (2000). Mol. Cell, 5, 545–555. [DOI] [PubMed]
- Gampe, R. T., Montana, V. G., Lambert, M. H., Wisely, G. B., Milburn, M. V. & Xu, H. E. (2000). Genes Dev. 14, 2229–2241. [DOI] [PMC free article] [PubMed]
- Germain, P., Chambon, P., Eichele, G., Evans, R. M., Lazar, M. A., Leid, M., De Lera, A. R., Lotan, R., Mangelsdorf, D. J. & Gronemeyer, H. (2006). Pharmacol. Rev. 58, 760–772. [DOI] [PubMed]
- Gronemeyer, H., Gustafsson, J.-A. & Laudet, V. (2004). Nature Rev. Drug Discov. 3, 950–964. [DOI] [PubMed]
- Hardy, J. A., Walsh, S. T. & Nelson, H. C. (2000). J. Mol. Biol. 295, 393–409. [DOI] [PubMed]
- Nagy, L. & Schwabe, J. W. R. (2004). Trends Biochem. Sci. 29, 317–324. [DOI] [PubMed]
- Pawlak, M., Lefebvre, P. & Staels, B. (2012). Curr. Top. Med. Chem. 12, 486–504. [DOI] [PMC free article] [PubMed]
- Renaud, J.-P. & Moras, D. (2000). Cell. Mol. Life Sci. 57, 1748–1769. [DOI] [PMC free article] [PubMed]
- Renaud, J.-P., Rochel, N., Ruff, M., Vivat, V., Chambon, P., Gronemeyer, H. & Moras, D. (1995). Nature (London), 378, 681–689. [DOI] [PubMed]
- Shulman, A. I., Larson, C., Mangelsdorf, D. J. & Ranganathan, R. (2004). Cell, 116, 417–429. [DOI] [PubMed]
- Suino, K., Peng, L., Reynolds, R., Li, Y., Cha, J.-Y., Repa, J. J., Kliewer, S. A. & Xu, H. E. (2004). Mol. Cell, 16, 893–905. [DOI] [PubMed]
- Wagner, R. L., Apriletti, J. W., McGrath, M. E., West, B. L., Baxter, J. D. & Fletterick, R. J. (1995). Nature (London), 378, 690–697. [DOI] [PubMed]
- Wurtz, J.-M., Bourguet, W., Renaud, J.-P., Vivat, V., Chambon, P., Moras, D. & Gronemeyer, H. (1996). Nature Struct. Biol. 3, 87–94. [DOI] [PubMed]
- Xu, H. E., Lambert, M. H., Montana, V. G., Plunket, K. D., Moore, L. B., Collins, J. L., Oplinger, J. A., Kliewer, S. A., Gampe, R. T. Jr, McKee, D. D., Moore, J. T. & Willson, T. M. (2001). Proc. Natl Acad. Sci. USA, 98, 13919–13924. [DOI] [PMC free article] [PubMed]
- Zhang, H., Chen, L., Chen, J., Jiang, H. & Shen, X. (2011). J. Biol. Chem. 286, 24593–24598. [DOI] [PMC free article] [PubMed]
- Zhang, H., Zhou, R., Li, L., Chen, J., Chen, L., Li, C., Ding, H., Yu, L., Hu, L., Jiang, H. & Shen, X. (2011). J. Biol. Chem. 286, 1868–1875. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human RXRα, ligand-binding domain, 6hn6
Supplementary Figures.. DOI: 10.1107/S2053230X18018022/ow5011sup1.pdf