A heterogeneous distribution of oligomers complicated the structure determination of the complement regulator properdin. Using TEV protease excision from recombinant oligomers, two types of properdin monomers could be crystallized.
Keywords: complement, properdin, modular proteins, crystallization
Abstract
The 54 kDa protein properdin, also known as factor P (FP), plays a major role in the complement system through the stabilization of the alternative pathway convertases. FP circulates in the blood as cyclic dimers, trimers and tetramers, and this heterogeneity challenges detailed structural insight into the mechanism of convertase stabilization by FP. Here, the generation of an intact FP monomer and a variant monomer with the third thrombospondin repeat liberated is described. Both FP monomers were excised from recombinant full-length FP containing internal cleavage sites for TEV protease. These FP monomers could be crystallized, and complete data sets extending to 2.8 Å resolution for the intact FP monomer and to 3.5 Å resolution for the truncated variant were collected. The principle of specific monomer excision and domain removal by the insertion of a protease cleavage site may be broadly applicable to structural studies of oligomeric, flexible and modular proteins.
1. Introduction
The complement system plays an important role in innate immunity and homeostasis, where it is involved in the clearance of invading pathogens, immune complexes and dying host cells (Ricklin et al., 2016 ▸; Bajic et al., 2015 ▸). The system is activated by molecular patterns presented by pathogens, dying host cells and large macromolecular complexes through three distinct pathways. Prominent examples are antibody–antigen complexes for the classical pathway, carbohydrate or acetyl groups for the lectin pathway, and specific microbial surfaces and spontaneous activation for the alternative pathway (AP). Each pathway leads to the assembly of proteolytically active C3 convertases that cleave C3 into the anaphylatoxin C3a and the opsonin C3b, the major effector molecule of complement. Upon binding to the zymogen factor B, C3b may form the proconvertase C3bB, which is activated to the AP C3 convertase C3bBb by factor D cleavage. FP is a positive regulator of the alternative pathway, as it increases the C3bBb half-life by fivefold to tenfold (Fearon & Austen, 1975 ▸). In serum FP circulates as dimers, trimers and tetramers, but higher oligomers may be formed during purification and storage (Pangburn, 1989 ▸). FP oligomerization has excluded crystallization and made electron microscopy (EM) and small-angle X-ray scattering (SAXS) studies of FP and FP–ligand complexes challenging (Alcorlo et al., 2013 ▸; Sun et al., 2004 ▸). Recently, we described a strategy to prepare a functional monomeric FP excised from recombinant oligomeric FP. This allowed us to determine the molecular envelope of FP bound to a stabilized form of the AP C3 convertase at 6 Å resolution using crystallography (Pedersen et al., 2017 ▸). However, in the absence of an atomic model of FP, we were only able to describe the overall binding site of FP on the convertase. Here, we report that the previously described FPc (Pedersen et al., 2017 ▸) and a new FPcΔ3 variant can be crystallized and that complete diffraction data can be obtained for both proteins.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Design of the FPc and FPcΔ3 constructs
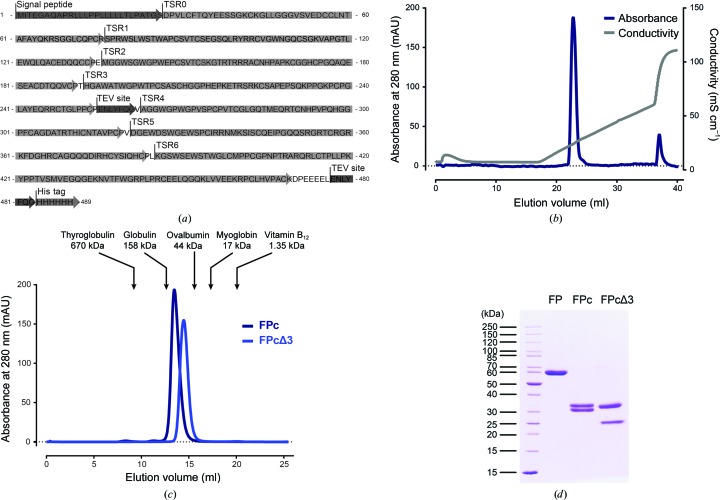
Human FP-encoding DNA was synthesized (GenScript) with the endogenous signaling peptide and a C-terminal 6×His tag (Table 1 ▸). The final constructs were generated by the insertion of TEV protease sites using site-directed mutagenesis. For FPc, TEV protease sites were inserted between TSR3 and TSR4 and C-terminally upstream from the 6×His tag (Fig. 1 ▸ a). For FPcΔ3, an additional TEV protease site was inserted between TSR2 and TSR3.
Table 1. DNA constructs used for the expression of recombinant oligomeric FP.
| FPc | FPcΔ3 | |
|---|---|---|
| Source organism | Homo sapiens | Homo sapiens |
| DNA source | Synthesized (GenScript) | Synthesized (GenScript) |
| Forward primer | None | None |
| Reverse primer | None | None |
| Cloning vector | None | None |
| Expression vector | pCEP4 | pCEP4 |
| Expression host | HEK293F | HEK293F |
| Complete amino-acid sequence of the construct produced† | MITEGAQAPRLLLPPLLLLLTLPATGSDPVLCFTQYEESSGKCKGLLGGGVSVEDCCLNTAFAYQKRSGGLCQPCRSPRWSLWSTWAPCSVTCSEGSQLRYRRCVGWNGQCSGKVAPGTLEWQLQACEDQQCCPEMGGWSGWGPWEPCSVTCSKGTRTRRRACNHPAPKCGGHCPGQAQESEACDTQQVCPTHGAWATWGPWTPCSASCHGGPHEPKETRSRKCSAPEPSQKPPGKPCPGLAYEQRRCTGLPPCPENLYFQGVAGGWGPWGPVSPCPVTCGLGQTMEQRTCNHPVPQHGGPFCAGDATRTHICNTAVPCPVDGEWDSWGEWSPCIRRNMKSISCQEIPGQQSRGRTCRGRKFDGHRCAGQQQDIRHCYSIQHCPLKGSWSEWSTWGLCMPPCGPNPTRARQRLCTPLLPKYPPTVSMVEGQGEKNVTFWGRPLPRCEELQGQKLVVEEKRPCLHVPACKDPEEEELENLYFQGHHHHHH | MITEGAQAPRLLLPPLLLLLTLPATGSDPVLCFTQYEESSGKCKGLLGGGVSVEDCCLNTAFAYQKRSGGLCQPCRSPRWSLWSTWAPCSVTCSEGSQLRYRRCVGWNGQCSGKVAPGTLEWQLQACEDQQCCPEMGGWSGWGPWEPCSVTCSKGTRTRRRACNHPAPKCGGHCPGQAQESEACDTQQVCPENLYFQGTHGAWATWGPWTPCSASCHGGPHEPKETRSRKCSAPEPSQKPPGKPCPGLAYEQRRCTGLPPCPENLYFQGVAGGWGPWGPVSPCPVTCGLGQTMEQRTCNHPVPQHGGPFCAGDATRTHICNTAVPCPVDGEWDSWGEWSPCIRRNMKSISCQEIPGQQSRGRTCRGRKFDGHRCAGQQQDIRHCYSIQHCPLKGSWSEWSTWGLCMPPCGPNPTRARQRLCTPLLPKYPPTVSMVEGQGEKNVTFWGRPLPRCEELQGQKLVVEEKRPCLHVPACKDPEEEELENLYFQGHHHHHH |
The underlined residues are released upon TEV cleavage. The signal peptide is shown in italics.
Figure 1.
Expression and purification of FPc and FPcΔ3. (a) Amino-acid sequence (single-letter code) showing the location of the signal peptide and the TSR domains of the FPc construct and the TEV protease cleavage sites. In the vector used for the expression of FPcΔ3 a third TEV protease site was inserted between TSR2 and TSR3. (b) Purification of monomeric FPc on a 1 ml Mono S column. (c) SEC analysis of FPc and FPcΔ3 on a 24 ml Superdex 200 Increase column. Elution volumes and molecular weights of standard proteins are marked by arrows. (d) SDS–PAGE analysis (reduced) of purified uncleaved FP, FPc and FPcΔ3. FP and its fragments appear to be significantly larger than predicted by the protein sequence (Bern et al., 2018 ▸) owing to extensive post-translational glycosylation, including 14 C-linked (Trp) mannosylations, four O-linked (Ser/Thr) glycans and one N-linked (Asn) glycan.
2.1.2. Expression of FPcΔ3 and FPc
The transient transfection of HEK293F cells (ThermoFisher) with the FPc-encoding plasmid and expression were performed as described by Pedersen et al. (2017 ▸). Stably transfected HEK293F cell lines expressing FPcΔ3 were generated to facilitate its production. Prior to transfection, HEK 293F cells were split to 0.5 × 106 cells per millilitre in serum-free FreeStyle 293 medium and incubated for 24 h at 37°C at 8% CO2 with shaking. The cells were collected by centrifugation and were resuspended in fresh medium to give 2.77 × 106 cells per millilitre. The cells were incubated for 1 h with shaking and transfected using 1 µg DNA per 1 ml final culture volume and a 2:1(w:w) ratio of polyethylenimine (25 kDa; Polysciences) to DNA. After 4 h, the cells were diluted to 106 cells per millilitre in FreeStyle 293 medium and the cells were incubated overnight at 37°C at 8% CO2 with shaking. The following day, 106 cells were transferred to a Petri dish containing 10 ml FreeStyle 293 medium supplemented with 10%(v/v) fetal bovine serum (FBS) and incubated statically overnight at 37°C at 8% CO2 to allow the cells to adhere. Selection pressure was applied by the addition of fresh medium containing 10%(v/v) FBS and 150–250 µg ml−1 hygromycin B. The medium was changed every second day to remove dying cells. Selection pressure was maintained for ten days or until single colonies started to appear. Single colonies were transferred to a 24-well plate and grown statically for six weeks in FreeStyle 293 medium supplemented with FBS and hygromycin B. Over this period, the cell cultures were expanded and transferred to six-well plates, in which the FBS concentration was gradually reduced from 10 to 0%(v/v). At 0%(v/v) FBS the cells were resuspended in 5 ml FreeStyle 293 medium supplemented with hygromycin B and transferred into 125 ml flasks. The cells were cultured as recommended by the manufacturer until a cell viability of >90% was achieved.
2.1.3. Purification of FPc and FPcΔ3
The cells were harvested by centrifugation and the conditioned medium was filtered (0.2 µm) and adjusted to pH 8.0. Oligomeric FP in the medium was purified on a 5 ml HisTrap Excel column (GE Healthcare) equilibrated in 0.1 M Tris, 500 mM NaCl, 10 mM imidazole pH 8.0. The column was washed with equilibration buffer until the baseline was stable, and bound protein was eluted with equilibration buffer containing 500 mM imidazole. FP was concentrated to approximately 0.5–1 mg ml−1 and TEV protease was added to a TEV protease:FP ratio of 1:4(w:w). TEV protease cleavage was carried out without reducing agent for 3–4 days at 4°C. Cleavage of oligomeric FP to monomeric FPc or FPΔ3 was followed by SDS–PAGE analysis, where the fragments of FPc (TSR0–TSR3, 25.5 kDa; TSR4–TSR6, 24.6 kDa) and FPcΔ3 (TSR0–TSR2, 18.6 kDa; TSR4–TSR6, 24.6 kDa) could be separated under reducing conditions. To remove TEV protease and other impurities, FPc and FPcΔ3 were subsequently dialyzed against 50 mM sodium phosphate buffer pH 6.0, 2.5 mM EDTA. FPc and FPcΔ3 were loaded onto a 1 ml Mono S column (GE Healthcare) and eluted with a 20 ml linear gradient from 0 to 500 mM NaCl in 50 mM sodium phosphate pH 6. Finally, FPc and FPcΔ3 were purified by size-exclusion chromatography (SEC) on a 24 ml Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated in 10 mM HEPES pH 7.2, 100 mM NaCl.
2.2. Crystallization
Crystallization experiments were set up with a Mosquito (TTP Labtech) or an Oryx4 (Douglas Instruments) robot. Initial crystallization conditions for FPc were optimized with respect to pH, ammonium sulfate concentration and temperature using Mimer (Brodersen et al., 2013 ▸). Additional optimization was performed using additive and detergent screens (Hampton Research) in combination with microseeding. Initial FPcΔ3 crystals were obtained at 19°C in sitting drops with 1.5 M lithium sulfate, 0.1 M sodium acetate pH 4.6 using the SaltRx screen (Hampton Research). Crystals were optimized with respect to pH and lithium sulfate concentration using Mimer and were further optimized using an additive screen (Hampton Research) and microseeding (Table 2 ▸).
Table 2. Crystallization of FPc and FPcΔ3.
| FPc | FPcΔ3 | |
|---|---|---|
| Method | Sitting drop | Sitting drop |
| Plate type | 2-drop Swissci | 2-drop Swissci |
| Temperature (K) | 292 | 292 |
| Protein concentration (mg ml−1) | 5.5 | 5.8 |
| Buffer composition of protein solution | 10 mM HEPES pH 7.2, 100 mM NaCl | 10 mM HEPES pH 7.2, 100 mM NaCl |
| Composition of reservoir solution | 1.1 M ammonium sulfate, 0.1 M sodium acetate pH 6.0, 1.2%(w/v) myo-inositol | 1.0 M lithium sulfate, 0.1 M sodium acetate pH 4.0, 0.1 M barium chloride |
| Volume and ratio of drop | 200:190:10 nl protein:reservoir:seed solution | 200:190:10 nl protein:reservoir:seed solution |
| Volume of reservoir (µl) | 50 | 50 |
2.3. Data collection and processing
Crystals were cryoprotected by soaking in reservoir solution supplemented with 25%(v/v) glycerol (FPc) or 17.5%(v/v) ethylene glycol (FPcΔ3) prior to flash-cooling in liquid nitrogen. Two wedges of diffraction data were collected from a single FPc crystal using the continuous helical data-collection mode on I02 at Diamond Light Source (DLS). For FPcΔ3, diffraction data were collected from a single crystal at PETRA III using the standard oscillation mode. Data processing and scaling was performed with XDS (Kabsch, 2010 ▸).
3. Results and discussion
After initial capture by nickel-affinity chromatography, the two different recombinant oligomeric FPs could be completely cleaved by TEV protease to generate monomeric FPc and FPcΔ3. Both types of monomer were further purified by cation-exchange and size-exclusion chromatography (Figs. 1 ▸ b, 1 ▸ c and 1 ▸ d). Purified FPc was concentrated to 5–6 mg ml−1 and was used for crystallization. Optimization of the crystallization conditions (Table 2 ▸) resulted in single plate-shaped crystals with dimensions of approximately 200 × 50 × 10 µm (Fig. 2 ▸ a). The presence of both polypeptide chains, TSR0–TSR3 and TSR4–TSR6, was verified by SDS–PAGE analysis of washed crystals (Fig. 2 ▸ b). Approximately 200 crystals were tested with synchrotron X-ray radiation; the majority showed diffraction to 3.5–5 Å resolution, but one crystal diffracted to a maximum resolution of 2.8 Å (Fig. 2 ▸ e). Two data sets were collected from this crystal; they had a correlation of 0.996 according to XSCALE and were therefore merged (Table 3 ▸). Assuming the presence of one FPc molecule (50 kDa for the protein sequence; 54 kDa including all post-translational modifications) per asymmetric unit, the Matthews coefficient (V M; Matthews, 1968 ▸) was calculated to be 4.18 Å3 Da−1, corresponding to a solvent content of 71%. With two FPc molecules in the asymmetric unit, the solvent content would be 41% and a higher maximum resolution of the diffraction pattern would have been expected.
Figure 2.
Crystals of recombinant FPc and FPcΔ3. (a) Optimized FPc crystals. (b) Washed FPc crystals analyzed by reducing SDS–PAGE, demonstrating the presence of both TSR0–TSR3 (25.5 kDa) and TSR4–TSR6 (24.6 kDa). (c) Optimized FPcΔ3 crystals. (d) Reducing SDS–PAGE analysis of washed FPcΔ3 crystals, demonstrating the presence of both TSR0–TSR2 (18.6 kDa) and TSR4–TSR6 (24.6 kDa). (e) Diffraction image from an FPc crystal. The image was created by merging 20 frames of 0.1° oscillation with the MERGE2CBF program.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| FPc | FPcΔ3 | |
|---|---|---|
| Diffraction source | I02, DLS | P13, PETRA III |
| Wavelength (Å) | 0.9795 | 0.9794 |
| Temperature (K) | 100 | 100 |
| Detector | PILATUS 6MF | PILATUS 6M |
| Crystal-to-detector distance (mm) | 624.56 | 620.935 |
| Rotation range per image (°) | 0.1 | 0.1 |
| Total rotation range (°) | 320 | 180 |
| Exposure time per image (s) | 0.04 | 0.04 |
| Space group | C2 | I4 |
| a, b, c (Å) | 184.0, 47.1, 122.9 | 223.4, 223.4, 47.3 |
| α, β, γ (°) | 90.0, 119.4, 90.0 | 90, 90, 90 |
| Mosaicity (°) | 0.135 | 0.108 |
| Resolution range (Å) | 50–2.80 (2.87–2.80) | 50–3.50 (3.59–3.50) |
| Total No. of reflections | 133020 | 100344 |
| No. of unique reflections | 22273 (1171) | 15199 (1113) |
| Completeness (%) | 96.1 (69.8) | 99.8 (99.9) |
| Multiplicity | 5.97 (5.44) | 6.6 (7.01) |
| 〈I/σ(I)〉 | 11.3 (1.0) | 9.2 (0.7) |
| R meas (%) | 12.8 (178.5) | 12.6 (315.4) |
| Overall B factor from Wilson plot (Å2) | 71.4 | 143.5 |
SEC analysis showed that FPcΔ3 eluted significantly later compared with FPc (14.4 and 13.3 ml, respectively), which is in agreement with the removal of TSR3 in FPcΔ3 and a more compact molecule (Fig. 1 ▸ c). The best FPcΔ3 crystals were approximately 50 × 30 × 30 µm in size (Table 2 ▸). SDS–PAGE analysis demonstrated that both the TSR0–TSR2 and TSR4–TSR6 fragments were present in washed crystals (Figs. 2 ▸ c and 2 ▸ d). Diffraction data from crystals of FPcΔ3 extended to 3.5 Å resolution (Table 3 ▸). Assuming the presence of either one or two FPcΔ3 molecules (43 kDa for the protein sequence; 46 kDa including all post-translational modification) per asymmetric unit, the solvent content would be 80% or 60%, respectively. With three FPc molecules in the asymmetric unit, the solvent content would be 41% and a diffraction pattern extending to a higher maximum resolution would have been anticipated. Attempts to determine the structures of FPc and FPcΔ3 by a combination of molecular replacement, anomalous scattering and isomorphous replacement are in progress, but are complicated owing to a lack of search models with high sequence homology and the presence of at least six homologous TSR domains in FPc.
In conclusion, our results with respect to crystallization and diffraction appear to enable the determination of an atomic structure of an FP monomer. Furthermore, both FPc and FPcΔ3 offer opportunities for atomic structures of complexes of FP with ligands, including C3bP, the proconvertase C3bBP and the AP C3 convertase C3bBbP, to be obtained at medium or high resolution by crystallography and cryo-EM. Such structures will allow detailed structural insight into the function of FP as a positive regulator of the complement system.
Acknowledgments
We would like to thank the beamline staff at the Diamond Light Source, the European Synchrotron Radiation Facility and the EMBL Outstation at PETRA III for support during data collection, and Karen Magrethe Nielsen for invaluable technical assistance.
Funding Statement
This work was funded by Lundbeckfonden grant BRAINSTRUC to Gregers Rom Andersen. Natur og Univers, Det Frie Forskningsråd grant DFF-4181-00137 to Gregers Rom Andersen. Novo Nordisk Fonden grant 29880 to Gregers Rom Andersen.
References
- Alcorlo, M., Tortajada, A., Rodríguez de Córdoba, S. & Llorca, O. (2013). Proc. Natl Acad. Sci. USA, 110, 13504–13509. [DOI] [PMC free article] [PubMed]
- Bajic, G., Degn, S. E., Thiel, S. & Andersen, G. R. (2015). EMBO J. 34, 2735–2757. [DOI] [PMC free article] [PubMed]
- Bern, M., Caval, T., Kil, Y. J., Tang, W., Becker, C., Carlson, E., Kletter, D., Sen, K. I., Galy, N., Hagemans, D., Franc, V. & Heck, A. J. R. (2018). J. Proteome Res. 17, 1216–1226. [DOI] [PMC free article] [PubMed]
- Brodersen, D. E., Andersen, G. R. & Andersen, C. B. F. (2013). Acta Cryst. F69, 815–820. [DOI] [PMC free article] [PubMed]
- Fearon, D. T. & Austen, K. F. (1975). J. Exp. Med. 142, 856–863. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Pangburn, M. K. (1989). J. Immunol. 142, 202–207. [PubMed]
- Pedersen, D. V., Roumenina, L., Jensen, R. K., Gadeberg, T. A., Marinozzi, C., Picard, C., Rybkine, T., Thiel, S., Sørensen, U. B., Stover, C., Fremeaux-Bacchi, V. & Andersen, G. R. (2017). EMBO J. 36, 1084–1099. [DOI] [PMC free article] [PubMed]
- Ricklin, D., Reis, E. S. & Lambris, J. D. (2016). Nature Rev. Nephrol. 12, 383–401. [DOI] [PMC free article] [PubMed]
- Sun, Z., Reid, K. B. & Perkins, S. J. (2004). J. Mol. Biol. 343, 1327–1343. [DOI] [PubMed]