SUMMARY
Transcription factors have recently been shown to colocalize in hotspot regions of the genome, which are further clustered into super-enhancers. However, the detailed molecular organization of transcription factors at hotspot regions is poorly defined. Here, we have used digital genomic footprinting to precisely define factor localization at a genome-wide level during the early phase of 3T3-L1 adipocyte differentiation, which allows us to obtain detailed molecular insight into how transcription factors target hotspots. We demonstrate the formation of ATF-C/EBP heterodimers at a composite motif on chromatin, and we suggest that this may be a general mechanism for integrating external signals on chromatin. Furthermore, we find evidence of extensive recruitment of transcription factors to hotspots through alternative mechanisms not involving their known motifs and demonstrate that these alternative binding events are functionally important for hotspot formation and activity. Taken together, these findings provide a framework for understanding transcription factor cooperativity in hotspots.
INTRODUCTION
The recent increase in the number of genome-wide maps of transcription factor binding has made it increasingly clear that transcription factors do not work alone but rather collaborate with other transcription factors at genomic target regions. Colocalization of two cooperating factors has been observed in several different systems, e.g., activating protein 1 (AP1) has been shown to regulate the chromatin accessibility at glucocorticoid receptor (GR)-binding sites in mammary cells (Biddie et al., 2011), FoxA1 regulates the estrogen receptor (ER)-activated gene program by directly targeting ER-binding sites (Carroll et al., 2005; Hurtado et al., 2011), PU.1 cooperates with several different factors at specific target sites including CCAAT/enhancer-binding protein β (C/EBPβ) (Heinz et al., 2010), and peroxisome proliferator-activated receptor γ and C/EBPα-binding profiles overlap extensively in mature adipocytes (Lefterova et al., 2008; Nielsen et al., 2008) and cooperate in gaining access to chromatin (Madsen et al., 2014). In addition to cooperation between transcription factor pairs, we (Boergesen et al., 2012; Siersbæk et al., 2011; 2014, this issue of Cell Reports) and others (Chen et al., 2008; Gerstein et al., 2012; He et al., 2011) have demonstrated that multiple transcription factors can target the same genomic regions that we refer to as transcription factor hotspots (Siersbæk et al., 2011). Cooperative binding of transcription factors is also the basis for modeling of cis-regulatory modules determining cell specificity (Frith et al., 2003; Krivan and Wasserman, 2001). However, chromatin immunoprecipitation sequencing (ChIP-seq), which is the most widely used method to map transcription factor binding, has a relatively low resolution and does not discriminate between direct protein binding to DNA and indirect recruitment to chromatin through protein-protein interactions. The molecular architecture (i.e., the organization) of transcription factors at hotspots is therefore poorly understood.
Here, we have used digital genomic footprinting to precisely define protein localization for several adipogenic transcription factors at a genome-wide level. In combination with ChIP-seq data, these analyses reveal molecular insight into the organization of transcription factors at hotspot regions, which provides a framework for understanding transcription factor cooperativity on chromatin.
RESULTS
Digital Genomic Footprinting Reveals Precise Protein Footprints at a Genome-wide Level
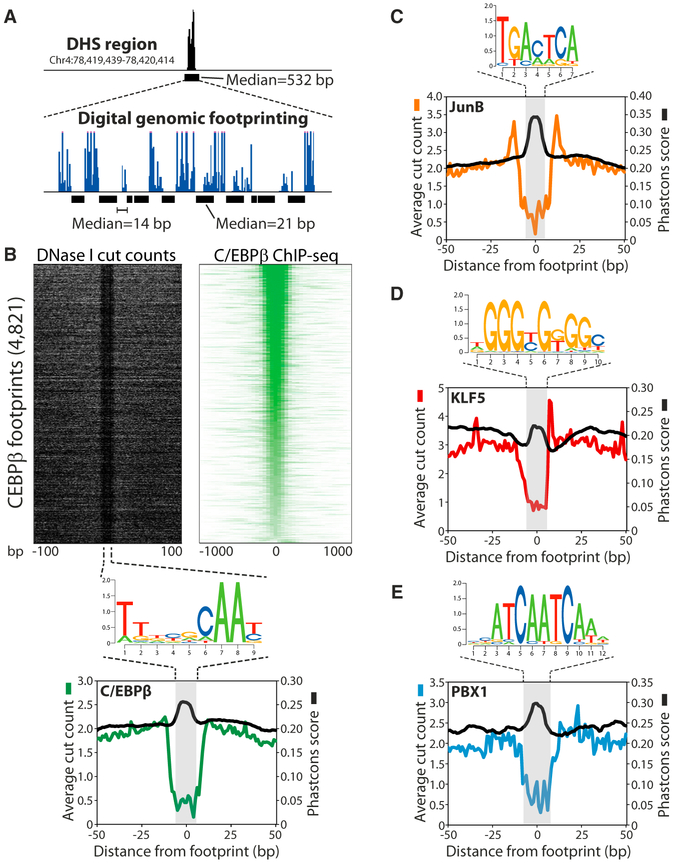
To begin to understand how transcription factors communicate in hotspots, it is essential to know how the factors are organized at these regions, including which factors are engaged in direct DNA interactions. We therefore employed high-resolution digital genomic footprinting (Figure 1A), a recently developed method to identify protein footprints (i.e., areas of restricted nuclease access) within DNase I hypersensitive (DHS) regions based on ultradeep sequencing (>100 M sequence tags) of DHS-seq libraries (Boyle et al., 2011; Hesselberth et al., 2009; Neph et al., 2012). We used an algorithm that we have recently developed to efficiently identify protein footprints at a genome-wide level with high sensitivity and specificity (M.-H.S., M. Guertin, S.B., and G.L.H., unpublished data). Based on analysis of 129,028 DHS regions detected 4 hr after induction of 3T3-L1 adipogenesis, we identify ~330,000 protein footprints (false discovery rate = 1%) using this algorithm. These footprints have a median size of 21 bp but are mostly relatively short (8–12 bp) and long (25–30 bp) (Figure S1B), and they are located with a median distance of 14 bp from each other within DHS regions (Figure S1C). The small sizes of the identified footprints clearly demonstrate the increased precision with which footprinting analysis can map protein binding to DNA compared to regular DHS-seq analyses, which identify accessible regions with a median size of 532 bp (Figure S1A).
Figure 1. Digital Genomic Footprinting Reveals Transcription Factor Footprints at a Genome-wide Level.
(A) Schematic overview of digital genomic footprinting. Extensive sequencing reveals small protected areas of 8–30 bp within overall DNase I hypersensitive regions corresponding to protein footprints. The median size of DHS regions and footprints as well as the median distance from one footprint to the nearest neighbor obtained from Figure S1 are indicated.
(B) DNase I cut counts (left) and C/EBPβ ChIP-seq signal (right) in the vicinity of C/EBPβ footprints (top). These regions were defined as footprints containing a C/EBP predicted site that overlap a ChIP-seq peak for C/EBPβ. Note the different scales used for visualization of DNase I cut counts and ChIP-seq data. Average DNase I cut counts and phastCons score in the vicinity of C/EBPβ footprints are shown at the bottom. (C–E) Average DNase I cut counts as in (B)for JunB
(C), KLF5 (D), and PBX1 (E).
See also Figure S1.
To assign the identified footprints to specific transcription factors, we combined motif analyses with ChIP-seq data for several transcription factors involved in the early phase of adipocyte differentiation (Siersbæk et al., 2011, 2014). Footprints were assigned to specific factors based on the presence of a ChIP-seq peak for that factor as well as a predicted binding site using the known position weight matrix for the same factor (unless otherwise noted, predicted sites discussed below will always refer to predictions made by the corresponding canonical motifs in this way—see Supplemental Experimental Procedures for more details). As illustrated in Figure 1, this approach revealed strong footprints for several factors. The localized decrease in DNase I digestion at footprints is highly correlated with a colocalized peak in sequence conservation (Figures 1B–1E), indicating that generally these footprints occur at functionally important sites.
C/EBP-ATF Footprints at a Composite Motif Demonstrate Heterodimer Formation on Chromatin
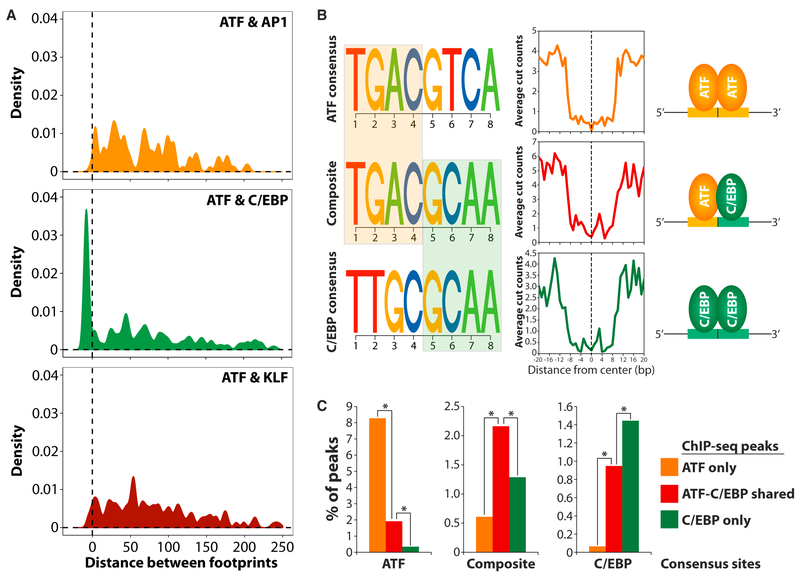
The high resolution of genomic footprinting allows us to obtain information about protein localization within ChIP-seq peak regions (see Figure 1B for an example of the high resolution of footprinting analyses compared to ChIP-seq. Note the different scales used for ChIP-seq and DNase I cut count visualization). Detailed investigation of the distances between footprints for different factors revealed that ATF footprints overlap specifically with C/EBP footprints, whereas minimal overlap was observed between ATF footprints and footprints for KLFs and AP1 (Figure 2A). Interestingly, the exact same composite motif containing a half site from the ATF motif in the 5′ end and a half site from the C/EBP motif in the 3′ end was found at almost all these regions, where ATF and C/EBP footprints overlap (Figure 2B, left). C/EBP-ATF heterodimers have previously been shown to bind to this DNA sequence in vitro (Mann et al., 2013; Shuman et al., 1997; Vinson et al., 1993), and our data provide direct evidence that such heterodimers also bind to this element in a chromatin context in cells. Importantly, the composite DNA element is significantly enriched at shared ATF-C/EBP regions identified by ChIP-seq (Figure 2C), indicating that it specifically directs ATF-C/EBP heterodimer formation. Consistent with heterodimer formation on this composite DNA element, ATF7 and C/EBPβ simultaneously occupy all three investigated predicted sites in re-ChIP experiments (Figure S2). Taken together, this suggests that alternative heterodimer formation on composite binding sites may be an important mechanism through which C/EBPs and ATFs are targeted to specific genomic regions and thereby integrate different signaling pathways on chromatin.
Figure 2. Footprint at a Composite Motif Demonstrates ATF-C/EBP Heterodimer Formation on Chromatin.
(A)Distance between footprints for different transcription factor pairs (i.e., ATFs and AP1, ATFs and C/EBPs, and ATFs and KLFs) found within the same DHS sites. Negative distances mean that the footprints for the two types of factors overlap. Here, footprints refer to predicted binding sites found in footprint regions overlapping a ChIP-seq peak.
(B) Consensus binding sites for ATFs and C/EBPs (i.e., the core predicted binding sites that best fit the position weight matrices for these factors and which have previously been shown to be strong binding sequences for ATF and C/EBP homodimers; Mann et al., 2013) as well as a composite DNA element found at the overlapping footprints identified in (A) containing an ATF and a C/EBP half site (left). Average DNase I cut counts in the vicinity of the predicted binding sites for ATF and C/EBP as well as the composite ATF-C/EBP DNA element found in footprint regions (middle). Schematic view of transcription factor binding to the different types of predicted sites (right).
(C) Percentage of ATF only, C/EBP only, and shared ATF-C/EBP ChIP-seq peaks that contain the different types of predicted consensus sites shown in (B). *p < 0.01 as determined by Fisher’s exact test.
See also Figure S2.
Extensive Transcription Factor Recruitment to Hotspots through Alternative Mechanisms
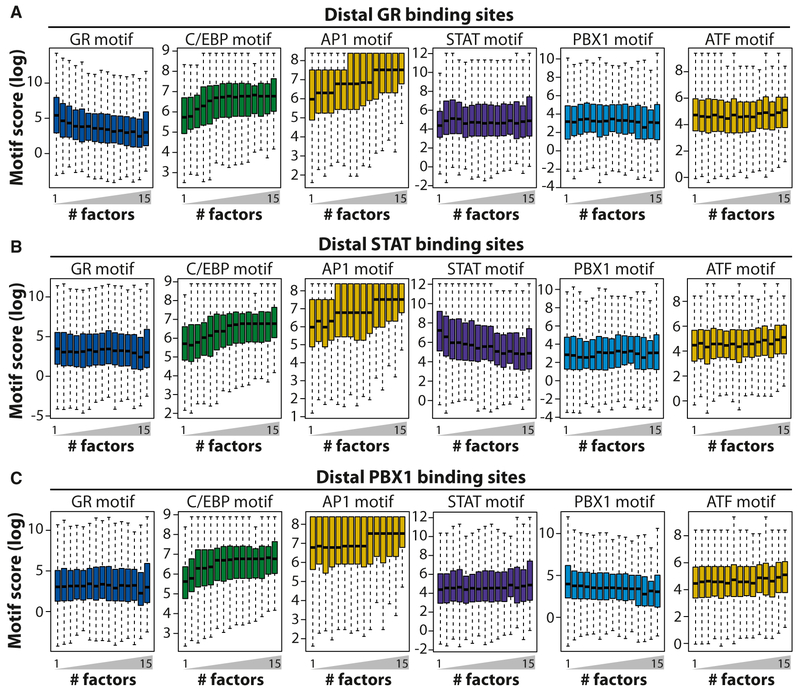
Intriguingly, investigation of the GR motif occurrence at distal GR-binding sites (>2 kb away from the transcription start site [TSS]) revealed that the binding site prediction score (i.e., the degree of resemblance of a predicted site to the GR motif) decreased with the number of co-occupying factors (Figure 3A). This suggests that the sequence-specific constraints for GR binding are relaxed when many other factors bind to the site. This is not due to a general relaxation of binding constraints to open chromatin in hotspots, because binding site prediction scores for STAT, PBX1, and ATF are unaffected and prediction scores for C/EBP and AP1 increase with the number of factors at these distal GR-binding sites. The fact that prediction scores for C/EBP and AP1 increase at these sites furthermore suggests that C/EBP and AP1 factors may play a direct role in facilitating GR binding. A similar pattern of prediction scores was observed for distal STAT- and PBX1-binding sites (Figures 3B and 3C), indicating that, also for these factors, binding is less constrained than their motifs indicate and more dependent on AP1 and C/EBP sites when binding in hotspots.
Figure 3. The Motif Score for GR, STATs, and PBX1 Decreases as a Function of the Number of Co-occupying Factors.
The motif score for the best-fit predicted site in all distal (>2 kb away from the TSS) GR- (A), STAT- (B), and PBX1- (C) binding sites occupied by 1 (only the factor itself) through 15 factors. The motif score is a measure of how well a given predicted site resembles the motif found in the JASPAR database (Mathelier et al., 2014; Sandelin et al., 2004).
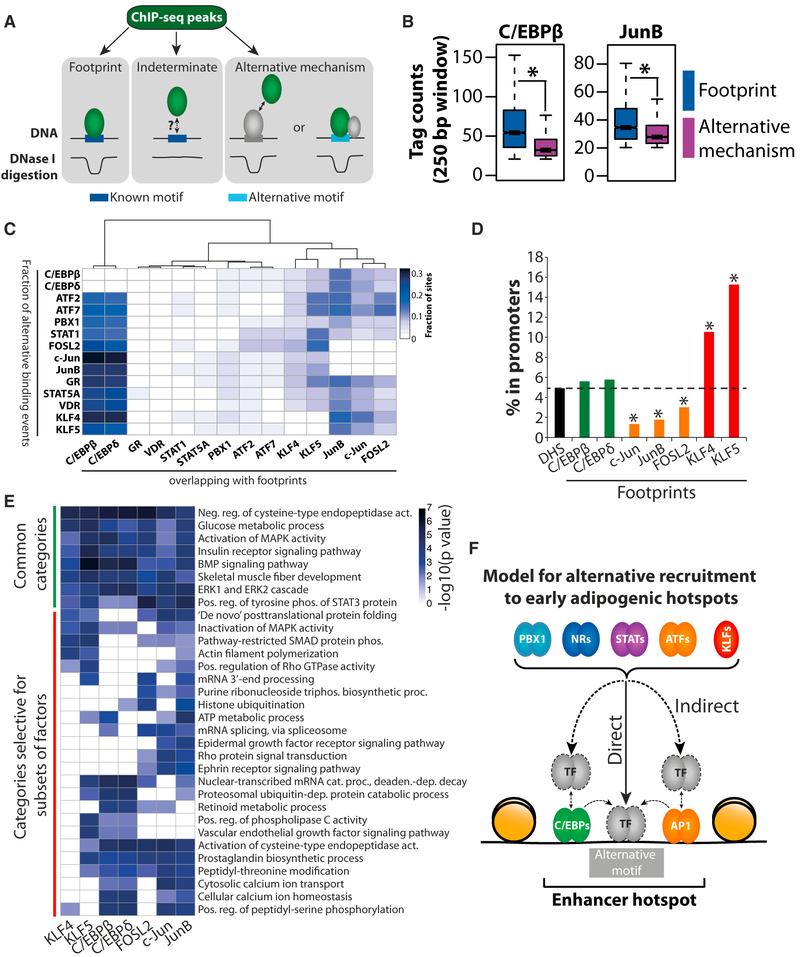
To further investigate the mechanisms of binding of transcription factors to hotspots, we took advantage of the information obtained from our footprinting analyses that allows us to identify direct binding events of transcription factors to DNA. Using this approach, we subdivided ChIP-seq peaks for each factor into three different categories (Figure 4A). First, peaks characterized by the presence of a footprint with the corresponding predicted transcription factor site could be classified as transcription factor footprints (i.e., regions where the factor binds directly to its predicted site). Second, peaks characterized by the presence of a predicted binding site for that factor but the absence of an associated footprint in the DNase I digestion pattern were classified as indeterminate binding events. Although the presence of a predicted binding site would indicate that the factor binds directly to DNA, the reason for the absence of a footprint could be many, including short residence time of the protein on DNA (Voss and Hager, 2014). Third, peaks characterized by the absence of a predicted binding site for the corresponding factor were classified as alternative binding events. Alternative binding events may stem from indirect binding to DNA via tethering to other factors or from direct binding of factors to alternative sites in the DNA possibly assisted by other DNA bound factors (Figure 4A). As expected, and consistent with previous findings (Neph et al., 2012), transcription factor footprints have a significantly higher ChIP-seq signal compared to alternative binding events (Figures 4B and S3A). Based on the classification of binding events for each factor described above, we can correlate these different mechanisms of transcription factor binding for all transcription factor pairs. Indeterminate binding events are omitted from these analyses as it is not clear whether or not the corresponding predicted binding site is involved in recruitment of the factor to chromatin. Interestingly, footprints for C/EBPs, KLFs, and AP1 factors (i.e., c-Jun, JunB, and FOSL2) are highly associated with alternative binding events for all other factors (Figure 4C). Taken together with the motif analyses described above (Figure 3), this indicates that these factors may function through direct interactions with their known binding sites to facilitate recruitment of other factors either through indirect binding (tethering) or through direct DNA binding to sites not corresponding to their canonical motifs by dynamic assisted loading (Voss et al., 2011).
Figure 4. Extensive Recruitment of Transcription Factors to Hotspots through Alternative Mechanisms Not Involving Their Predicted Sites or Canonical Binding Preferences.
(A) Illustration of the classification of ChIP-seq regions into three different types: (1) regions containing a footprint for the factor, (2) indeterminate binding events characterized by the presence of a predicted binding site that is not found in a footprint region, and (3) alternative mechanisms of recruitment, where no predicted binding site for the factor is found at all.(B) Boxplots show the binding intensity (i.e., the number of ChIP-seq tags) for C/EBPβ and JunB at footprints and binding sites where these factors are recruited through alternative mechanisms. See also Figure S3A.
(C) Colocalization of footprints and alternative binding events for each transcription factor pair. The fraction of alternative binding events for the factors on the y axis that colocalizes with footprints for the factors on the x axis is shown.
(D) Percentage of DHS sites (black) and footprints for C/EBPs (green), AP1 factors (yellow), and KLFs (red) located in promoters of annotated genes. The proximal promoter is defined as the 1 kb region immediately upstream of the TSS. *p < 0.01 as determined by Fisher’s exact test.
(E) Genes that were induced during the first 4 hr of differentiation (Siersbæk et al., 2014) were assigned to a given factor if a hotspot where that factor formed a footprint was located within 50 kb of their TSS. The genes assigned to C/EBP, KLF, and AP1 factors were then subjected to GO enrichment analysis, and the p values for the significant GO terms are illustrated in the heatmap. GO term-enrichment analysis was performed using GeneAnswers (Feng et al., 2010).
(F) Schematic model of the alternative mechanisms through which transcription factors can be recruited to hotspots. In all cases, alternative mechanisms of recruitment do not involve the factor’s canonical motif. Instead, transcription factors can be recruited indirectly to DNA through mechanisms involving proteinprotein interactions that are facilitated by DNA-bound factors (e.g., C/EBPs and AP1 factors) or they can be directly recruited to DNA through alternative motifs, which may also be facilitated by other factors.
See also Figure S3.
Importantly, the footprints for C/EBPs, KLFs, and AP1 factors are significantly enriched in the vicinity of genes that are induced during the first 4 hr of differentiation (Figure S3B), strongly suggesting that these regions are in fact involved in regulating gene transcription. Interestingly, KLF footprints are significantly enriched in the proximal promoter region of annotated genes relative to all DHS sites, whereas footprints for the AP1 factors JunB, c-Jun, and FOSL2 are depleted in promoter regions relative to DHS sites (Figure 4D). This indicates that KLF factors function as facilitating factors that preferentially bind directly at promoters in addition to distal elements, whereas AP1 factors almost exclusively bind directly to their predicted binding sites in distal enhancers. Interestingly, assigning genes that are induced during the first 4 hr of differentiation to hotspots where different factors form a footprint reveals that these hotspots are associated with distinct sets of Gene Ontology (GO) categories (Figure 4E). Thus, different facilitating factors appear to drive the formation of hotspots that control different gene programs.
Taken together, these detailed footprinting analyses suggest that alternative mechanisms of transcription factor recruitment not involving the known factor motifs are central for hotspot formation and that especially C/EBPs, AP1, and KLF proteins may be involved in facilitating alternative mechanisms of recruitment of other factors (Figure 4F).
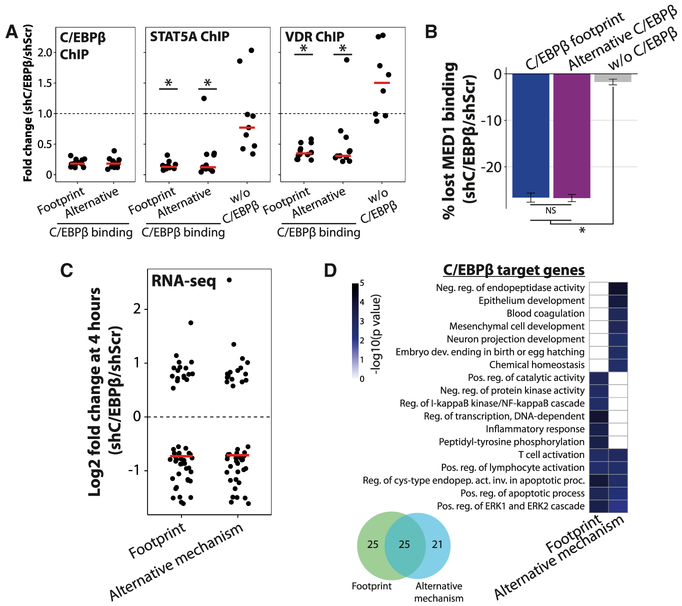
Alternative Mechanisms of C/EBPβ Recruitment Are Functionally as Important as Direct Binding to Predicted C/EBP Sites
To investigate the relative functional importance of footprints and alternative binding events for hotspot formation, we first analyzed the effect of C/EBPβ depletion on recruitment of transcription factors to hotspots where C/EBPβ binds directly to its predicted binding sites in DNA relative to hotspots where C/EBPβ binds through alternative mechanisms. Surprisingly, alternative STAT5A and VDR recruitment to these two types of hotspots is impaired to the same extent by C/EBPβ depletion (Figure 5A). Importantly, STAT5A and VDR recruitment to sites without C/EBPβ binding is largely unaffected by C/EBPβ knockdown. This indicates that alternative mechanisms of C/EBPβ binding to hotspots are functionally as important as direct binding to its predicted sites for the recruitment of additional transcription factors. Consistent with a functionally important role of alternative binding mechanisms, MED1 recruitment is similarly affected at C/EBPβ footprints and alternative C/EBPβ-binding regions by knockdown of C/EBPβ, whereas MED1 recruitment to binding regions without C/EBPβ is unaffected (MED1 ChIP-seq data were obtained from Siersbæk et al., 2014) (Figure 5B). Furthermore, RNA-seq in 3T3-L1 cells depleted of C/EBPβ revealed that early induced genes in the vicinity of hotspots where C/EBPβ binds through alternative mechanisms are affected to the same extent by the C/EBPβ knockdown as induced genes in the vicinity of hotspots, where C/EBPβ binds directly to its predicted sites (Figure 5C). Interestingly, however, hotspots where C/EBPβ is recruited through alternative mechanisms are associated with a different gene program than the hotspots with a C/EBPβ footprint (Figure 5D), which is consistent with the observation that hotspots occupied by different facilitating factors are associated with distinct gene programs (Figure 4E). Taken together, this demonstrates that, despite the lower ChIP-seq signal, C/EBPβ recruitment to chromatin by alternative mechanisms is as important as direct C/EBPβ binding to its predicted sites for additional transcription factor recruitment, enhancer formation, and gene activation.
Figure 5. Alternative Mechanisms of C/EBPβ Recruitment Are Functionally as Important as Direct Binding to Predicted C/EBP Sites.
(A)Fold change in C/EBPβ, STAT5A, and VDR ChIP as determined by qPCR upon depletion of C/EBPβ at hotspots, where C/EBPβ binds directly to its predicted sites in DNA (n = 12) or through alternative mechanisms (n = 11). STAT5A and VDR are recruited through alternative mechanisms not involving their predicted binding sites to all investigated hotspots. Binding sites where STAT5A or VDR bind without C/EBPβ cobinding are included as control sites (n = 9 for STAT5A and n = 8 for VDR). The red lines represent the medians of the data. *Student’s t test p < 0.05 for difference between fold change at control sites and the investigated hotspot group. Results are representative of two independent experiments.
Loss of MED1 binding as determined by ChIP-seq upon depletion of C/EBPβ at binding sites where C/EBPβ binds directly to its predicted sites or through alternative mechanisms. The loss of MED1 at binding sites without C/EBPβ (Siersbæk et al., 2014) is shown as a control. Error bars show the 95% confidence interval around the mean. *p < 2.2 × 10−16, Student’s t test. MED1 ChIP-seq data were obtained from Siersbæk et al. (2014).
(B)The induced genes that have a hotspot where C/EBPβ binds through alternative mechanisms or directly to its predicted sites within 50 kb of their TSS, which are significantly regulated by C/EBPβ TSS, which are significantly regulated by C/EBPβ knockdown as determined by RNA-seq, were identified. Induced genes were defined as genes that are significantly induced during the first 4 hr of 3T3-L1 adipogenesis in our 4sU-RNA-seq experiment published in this issue of Cell Reports (Siersbæk et al., 2014) as well as in the RNA-seq data presented here. The change in expression of each gene upon C/EBPβ knockdown at 4 hr of differentiation is illustrated in a dot plot. The red lines represent the medians of the data.
(C)Different mechanisms of C/EBPβ binding are associated with distinct gene programs. The overlap between the gene groups identified in (C) is shown in a Venn diagram (bottom left). GO categories associated with these gene groups are shown in a heatmap (right). Enrichment analysis of GO terms was done using GeneAnswers (Feng et al., 2010).
DISCUSSION
The functional importance of hotspots (Siersbæk et al., 2014) along with the intriguing finding that multiple transcription factors associate with a relatively small stretch of DNA of a few hundred base pairs at hotspots prompted us to investigate how transcription factors are organized at these regions.
We demonstrate that digital genomic footprinting combined with ChIP-seq and motif analyses can be used to identify binding of specific heterodimers to accessible chromatin. The findings that a specific composite ATF-C/EBP motif is found in footprints and enriched within shared C/EBP and ATF ChIP-seq peaks strongly suggest that these sites direct ATF-C/EBP heterodimer formation on chromatin. It has previously been demonstrated that ATF2 and ATF4 can form heterodimers with C/EBPβ that bind to the composite site also identified here (i.e., TGACGCAA) in vitro (Mann et al., 2013; Shuman et al., 1997; Vinson et al., 1993). Our data suggest that this is a general feature of C/EBPs and ATFs, and we provide direct evidence of ATF-C/EBP heterodimer formation in a chromatin context within cells. In addition to the composite ATF-C/EBP motif described here, it is likely that there is an additional repertoire of motifs that can direct ATF-C/EBP heterodimer formation on chromatin, as has been described previously in vitro (Mann et al., 2013; Vallejo et al., 1993). Taken together, these findings indicate that the assembly of alternative dimers may be a general way to integrate external signals on the chromatin template.
Interestingly, our genomics analyses also revealed that alternative binding of transcription factors not involving their regular binding sites is a key mechanism through which transcription factors are recruited to hotspots. This is consistent with a recent study demonstrating that a large fraction of ChIP-seq peaks for several different factors is associated with a binding site for another factor, but not a site for the factor itself (Wang et al., 2012). Whether these alternative binding events occur via indirect binding or assisted loading to nonconsensus motifs remains unclear.
Our footprinting analyses demonstrate that C/EBPs, KLFs, and AP1 factors make many strong footprints that are highly associated with alternative mechanisms of binding of other factors, indicating that these factors may be involved in facilitating recruitment of additional factors to hotspots through mechanisms not involving their known motifs. Interestingly, however, we have observed that different factors have different abilities to make footprints, with nuclear receptors being the least efficient, consistent with a recent study (He et al., 2014). It is therefore likely that the other factors we have investigated, including nuclear receptors, can also bind directly to some hotspots and be involved in facilitating recruitment of additional factors through alternative mechanisms as described here for C/EBPs, AP1, and KLFs, even though we do not identify a footprint for these factors in this study. The basis for the varying efficiency of footprint formation by different factors is unclear but may relate to the molecular details of how factors bind to DNA or the dynamics of the protein-DNA interactions. In favor of the latter is the finding that nuclear receptors have a very short residence time on DNA (McNally et al., 2000; Voss et al., 2011) and a poor ability to make footprints (M.-H.S., M. Guertin, S.B., and G.L.H., unpublished data; this study; He et al., 2014).
Surprisingly, we find that alternative binding events for C/EBPβ appear to be functionally as important as direct binding to its predicted sites for the formation of hotspots, recruitment of coactivators, and gene activation, even though alternative binding events are associated with significantly lower ChIP-seq signal than footprints at predicted sites. Intriguingly, the finding that C/EBPβ footprints and alternative binding of C/EBPβ are associated with distinct gene programs suggests that it may be possible to design drugs to target specific gene programs by specifically targeting one, but not the other, of these binding mechanisms of a particular transcription factor.
In conclusion, we present molecular insight into how transcription factors are organized and function at shared target regions, thereby providing a framework for understanding transcription factor cooperativity on chromatin.
EXPERIMENTAL PROCEDURES
Cell Culture and Knockdown
3T3-L1 cells were grown and induced to differentiate as described in Siersbæk et al. (2014). Knockdown of C/EBPβ was done using the pSicoR PGK puro (Addgene; 12084) system as described in Siersbæk et al. (2014).
ChIP
ChIP was performed as described (Siersbæk et al., 2011) using antibodies against C/EBPβ (C-19, sc-150; Santa Cruz Biotechnology), VDR (C-20, sc-1008; Santa Cruz), and STAT5A (L-20, sc-1081; Santa Cruz). Purified DNA was analyzed by quantitative PCR (qPCR).
RNA-Seq
Cells were harvested in Isol-RNA lysis reagent (5 PRIME) and purified according to the manufacturer. Purified RNA was then prepared for sequencing (Illumina) according to the instructions of the manufacturer. Sequence reads were mapped to the mouse genome (mm9) as described in Siersbæk et al. (2014). Mapped reads in exons were counted using HOMER (Heinz et al., 2010), and differentially expressed genes (adjusted p < 0.05) were identified using DESeq (Anders and Huber, 2010).
Digital Genomic Footprinting
Using ultradeep sequencing of our previously published DHS-seq library (Siersbæk et al., 2011), we obtained 137 million mapped sequence tags. These were analyzed using a footprinting algorithm we developed, DNase2TF, to obtain footprints of 8–30 bp (M.-H.S., M. Guertin, S.B., and G.L.H., unpublished data). All footprints were scanned with JASPAR motifs (Mathelier et al., 2014; Sandelin et al., 2004) to predict binding sites for the investigated factors. See Supplemental Experimental Methods for more details.
Genomics Analyses
Mapped ChIP-seq (Siersbæk et al., 2011, 2014) and RNA-seq (this study; Siersbæk et al., 2014) were analyzed by HOMER (Heinz et al., 2010) and BEDTools (Quinlan and Hall, 2010).
ACCESSION NUMBERS
Sequencing data are available from the Gene Expression Omnibus (GEO) under accession number GSE57415. Additional information is available in the Supplemental Experimental Procedures online.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Mandrup and Hager groups for valuable discussions. In particular, we thank Bjørk D. Larsen for experimental assistance. The work was in part carried out at the Villum Center for Bioanalytical Sciences, Department of Biochemistry and Molecular Biology, SDU, supported by the Villum Foundation. Work in the Mandrup laboratory was supported by grants from the Danish Independent Research Council|Natural Sciences and the Novo Nordisk Foundation; work in the Jensen laboratory was financed by the Danish National Research Foundation grant number DNRF82 to the Center for Epigenetics; work in the Hager laboratory was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research; and work in the Sandelin group was supported by the Lundbeck foundation and Novo Nordisk Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and three figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.04.043.
REFERENCES
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung M-H, Trump S, Lightman SL, et al. (2011). Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boergesen M, Pedersen TÅ, Gross B, van Heeringen SJ, Hagenbeek D, Bindesbøll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, et al. (2012). Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol 32, 852–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Song L, Lee B-K, London D, Keefe D, Birney E, Iyer VR, Crawford GE, and Furey TS (2011). High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 21, 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. (2005). Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. (2008). Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117. [DOI] [PubMed] [Google Scholar]
- Feng G, Du P, Krett NL, Tessel M, Rosen S, Kibbe WA, and Lin SM (2010). A collection of bioconductor methods to visualize gene-list annotations. BMC Res. Notes 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Li MC, and Weng Z (2003). Cluster-Buster: Finding dense clusters of motifs in DNA sequences. Nucleic Acids Res 31, 3666–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M,Landt SG,Yan K-K,Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. (2012). Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, and Pu WT (2011). Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. USA 108, 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Hu SS, Chen M-W, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, et al. (2014). Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods 11, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC,Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. (2009). Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, and Carroll JS (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet 43, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan W, and Wasserman WW (2001). A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 11, 1559–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ Jr., Liu XS, and Lazar MA (2008). PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 22, 2941–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen MS, Siersbæk R, Boergesen M, Nielsen R, and Mandrup S (2014). Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol. Cell. Biol 34, 939–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann IK, Chatterjee R, Zhao J, He X, Weirauch MT, Hughes TR, and Vinson C (2013). CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 23, 988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. (2014). JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42 (Database issue), D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally JG, Müller WG, Walker D, Wolford R, and Hager GL (2000). The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265. [DOI] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. (2012). An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Børgesen M, Francoijs K-J, Mandrup S, and Stunnenberg HG (2008). Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22, 2953–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engström P, Wasserman WW, and Lenhard B (2004). JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32 (Database issue), D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman JD, Cheong J, and Coligan JE (1997). ATF-2 and C/EBPalpha can form a heterodimeric DNA binding complex in vitro. Functional implications for transcriptional regulation. J. Biol. Chem 272, 12793–12800. [DOI] [PubMed] [Google Scholar]
- Siersbæk R, Nielsen R, John S, Sung M-H, Baek S, Loft A, Hager GL, and Mandrup S (2011). Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 30, 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, Poulsen LLC, Rogowska-Wrzesinska A, Jensen ON, and Mandrup S (2014). Transcription factor cooperativity in early adipogenic hotspots and superenhancers. Cell Rep. 7, this issue, 1443–1455. [DOI] [PubMed] [Google Scholar]
- Vallejo M, Ron D, Miller CP, and Habener JF (1993). C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc. Natl. Acad. Sci. USA 90, 4679–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson CR, Hai T, and Boyd SM (1993). Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 7, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Voss TC, and Hager GL (2014). Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet 15, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung M-H, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, and Hager GL (2011). Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146, 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. (2012). Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.