SUMMARY
The proteins that form the permeation pathway of mechanosensory transduction channels in inner-ear hair cells have not been definitively identified. Genetic, anatomical, and physiological evidence support a role for transmembrane channel-like protein (TMC) 1 in hair cell sensory transduction, yet the molecular function of TMC proteins remains unclear. Here, we provide biochemical evidence suggesting TMC1 assembles as a dimer, along with structural and sequence analyses suggesting similarity to dimeric TMEM16 channels. To identify the pore region of TMC1, we used cysteine mutagenesis and expressed mutant TMC1 in hair cells of Tmc1/2-null mice. Cysteine-modification reagents rapidly and irreversibly altered permeation properties of mechanosensory transduction. We propose that TMC1 is structurally similar to TMEM16 channels and includes ten transmembrane domains with four domains, S4–S7, that line the channel pore. The data provide compelling evidence that TMC1 is a pore-forming component of sensory transduction channels in auditory and vestibular hair cells.
In Brief
Pan et al. used biochemistry, structural modeling, and hair-cell physiology to demonstrate that TMC1 residues line the pore of hair cell sensory transduction channels.
INTRODUCTION
Exquisite sensitivity to sound has provided a powerful evolutionary advantage for countless life forms on earth. Despite the prevalence and critical nature of sound sensitivity, the genes and proteins that mediate sensory transduction in the auditory system have not been definitively identified. Among vertebrates, there is an evolutionarily conserved gene (Keresztes et al., 2003; Davies et al., 2012) known as transmembrane channel-like 1 (Tmc1) that has recently gained attention for its contributions to sensory function in the inner ears of humans, mice, and zebrafish (Kurima et al., 2002; Vreugde et al., 2002; Kawashima et al., 2011; Pan et al., 2013; Maeda et al., 2014; Erickson et al., 2017). Although multiple lines of evidence support a role for TMC1 as a part of the channel complex mediating sensory transduction (Corey and Holt, 2016), alternative interpretations exist (Wu and Müller, 2016) and definitive evidence is lacking. In addition, whereas the multiple-transmembrane topology of TMC1 resembles an ion channel, the region of the protein that forms the ion channel pore has not been identified; hence, the protein retains the name “channel-like.”
Sensory transduction in inner-ear hair cells converts mechanical information, in the form of sound or head movements, into electrical signals, which are relayed to the brain via the eighth cranial nerve. Hair cells utilize a mechanosensitive apparatus that rests atop bundles of 60–100 modified microvilli known as stereocilia. According to the prevailing model, there are an estimated 60–200 mechanosensory transduction channels per hair bundle, 1 or 2 per stereocilium. These mechanosensitive transduction channels perform the fundamental transformation, converting mechanical motion into electrical current in response to stereocilia bundle deflections. Although a biophysical model for hair cell mechanosensory transduction was proposed 35 years ago (Corey and Hudspeth, 1983), the search for the molecular identity of the hair cell transduction channel has yielded numerous dead ends and false leads (Corey and Holt, 2016). We hypothesized that TMC1, and closely related TMC2, are pore-forming components of the elusive sensory transduction channel required for auditory and vestibular transduction in inner-ear hair cells (Pan et al., 2013). Here, we use a variety of approaches to test this hypothesis and present a structural model for the channel pore.
RESULTS
TMC Proteins Assemble as Dimers
To investigate the stoichiometry of the TMC family of proteins, we expressed GFP- and mCherry-tagged variants of mouse TMC (mmTMC) in CHO cells and measured fluorescence resonance energy transfer between them. Among a subset of mmTMC homologs, we observed a robust fluorescence resonance energy transfer (FRET) signal (~0.15) between GFP and mCherry (Figures S1A and S1B). A Förster distance R0 ~ 51–54 Å (Stryer and Haugland, 1967) indicates a proximity of ~70 Å between GFP and mCherry tags on co-expressed TMCs. There was no observed FRET signal with a negative control, the α4 acetylcholine receptor subunit, for either mmTMC1 or mmTMC2. These data suggest that mmTMCs assemble as multimeric complexes—both homomeric and heteromeric—in CHO cells.
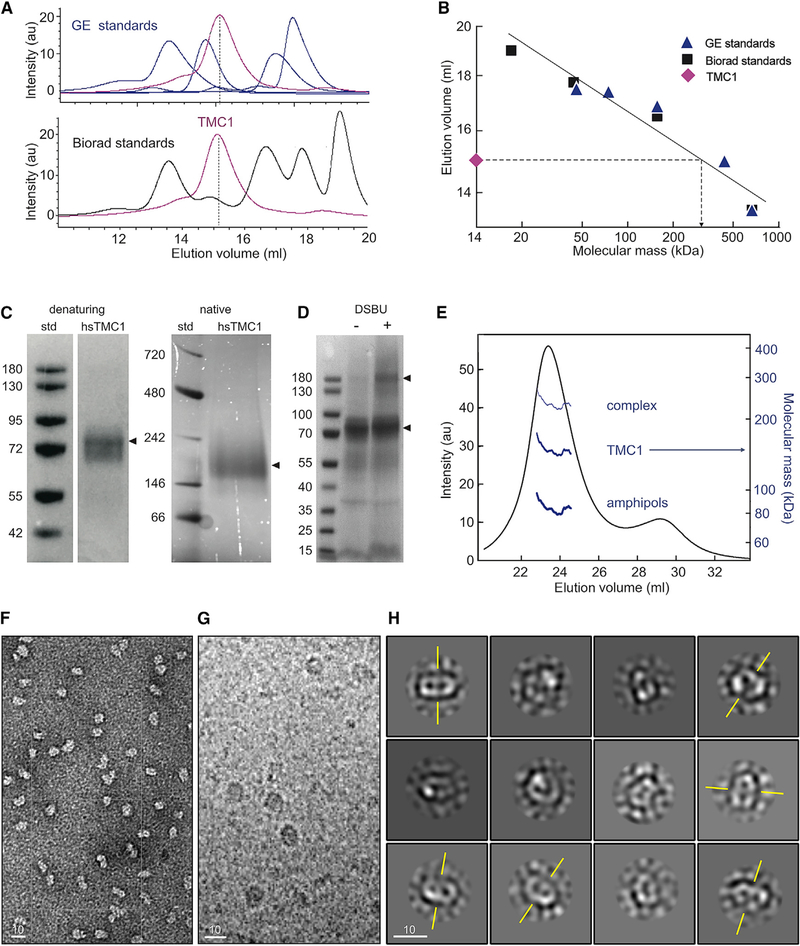
Next, we performed fluorescence size-exclusion chromatography (FSEC) on detergent-solubilized HEK cell lysates containing human TMC1 (hsTMC1) fused to GFP (Figure 1A). To obtain an initial estimate for the molecular weight of the protein, we created a calibration curve for our chromatography column (see STAR Methods; Wu, 2004). Specifically, we plotted the elution volume of established globular protein standards against their known molecular masses (Fasman, 1989); from that, we estimated that the detergent-solubilized GFP-tagged hsTMC1 elution peak volume corresponds to a mass of ~310 kDa (Figure 1B). Assuming an apparent mass for the surrounding detergent micelle in the range of ~100 kDa (Slotboom et al., 2008), the estimated protein mass is ~210 kDa, which agrees best with a dimer of 115.4 kDa (87.7 + 28.7 kDa) GFP fusion proteins. We also performed FSEC on GFP-mmTMEM16a, which is a known dimer of 139 kDa (110.9 + 28.7 kDa) subunits. The elution volume of GFP-mmTMEM16a was consistently close to that of GFP-TMC1 (Figure S1C). As a control, we performed FSEC on HEK293FT cells transfected with a plasma-membrane-targeting GFP vector, which did not show a protein peak in this position (Figure S1C).
Figure 1. Dimerization of TMC1.
(A) FSEC elution profile of TMC1-GFP (magenta) relative to the SEC profile of General Electric (GE, top) and Biorad (bottom) molecular weight standards.
(B) SEC elution volumes of protein standards plotted against their molecular weights. Protein standards (symbols) were fitted with a linear equation (line). The TMC1 elution volume predicts a molecular mass of ~310 kDa for protein and surfactant.
(C) Denaturing gel and native-PAGE with a Bis-Tris gel show the molecular weight of the purified hsTMC1 complex in denatured (~90kDa) and native (~200 kDa) states.
(D) Denaturing Coomassie-stained gel analysis of purified mmTMC1 proteins before (left lane) and after (right lane) cross-linking with 0.2 mM DSBU.</p/> (E) Astra SEC-MALS analysis of purified proteins. The UV-absorbance elution profile of the SEC-MALS sample is overlaid with the molecular mass estimates for the proteins (~150 kDa), amphipols (80 kDa), and the combined complex (230 kDa). The molecular masses are plotted on a log scale (right y axis) for the selected peak and shown as a function of column elution volume.
(F) A representative negative stain image shows a homogeneous population of purified hsTMC1 proteins, free of aggregates.
(G) Cryo-EM micrograph of purified hsTMC1 protein reconstituted into amphipols.
(H) Class averages of TMC particles. The axis of symmetry for selected class averages with two-fold symmetry is indicated in yellow.
In (F)–(H), scale bars represent 10 nm.
To perform more rigorous biochemical tests of stoichiometry, we purified TMC proteins. We expressed mouse and human TMC1 constructs with a C-terminal hemagglutinin (HA) tag preceded by an HRV3C cleavage site in HEK293FT cells. TMC1-HA was purified by binding to an HA-agarose resin and eluted via cleavage of the HA tag from the protein (see STAR Methods). To improve the stability of the purified proteins, TMCs were reconstituted into amphipols (Popot et al., 2011). The purity of the TMC1 proteins was analyzed by SDS-PAGE (Figure S1D). The circular dichroism spectrum of purified TMC1 in amphipols showed dipping peaks near 210 and 222 nm, indicating a folded protein of high α-helical content (Figure S1E). Finally, the identity of the purified proteins was confirmed by mass spectrometry sequencing of the Coomassie-stained gel band of GFP-TMC1 followed by in-gel trypsin digestion into peptide fragments (Figure S2). We found that 99.85% of the peptide signal intensity in our sample band was assigned to GFP-TMC1, with the next highest scoring protein at 0.006%. Because oligomerization state could be altered as a membrane protein is purified and transferred to detergents or amphipathic polymers that are more amenable for high-resolution structure determination, we monitored stoichiometry of TMC1 reconstituted in amphipols using FSEC. Both mouse and human TMC1 constructs eluted on SEC as single peaks (Figure S1F). Importantly, after purification, the FSEC profiles of purified GFP-TMC1-HA proteins were similar to those before purification (Figure S1G), suggesting that the purification procedure does not alter subunit stoichiometry. Consistent with what one would expect for dimers, native-PAGE experiments showed protein bands corresponding to molecular weights of ~200 kDa for untagged hsTMC1-HA (Figure 1C) and ~240 kDa for GFP-hsTMC1-HA (not shown). We then cross-linked purified proteins with disuccinimidyl dibutyric urea (DSBU) and assessed mass with denaturing PAGE (see STAR Methods). The cross-linked subunits are expected to migrate according to their combined molecular mass on denaturing gels. Bands were apparent at the predicted molecular masses of both monomers and dimers of hsTMC1 (Figure 1D).
Finally, we used SEC-multi-angle static light scattering (MALS) analysis (see STAR Methods) to estimate the mass of the amphipol-protein complex. The combined mass of protein and amphipols was ~230 kDa (±3%; Figure 1E). To determine the contribution of the amphipol to the complex, we performed a protein-conjugate analysis. We calculated a contribution of ~150 kDa from the protein (±3%) and ~80 kDa from amphipols (±8%; Figure 1E). The protein mass is roughly twice that of the untagged mmTMC1 (~88 kDa), indicating that mmTMC1 subunits assemble as a dimer.
We then purified hsTMC1 proteins and viewed them with negative stain as well as cryo-electron microscopy (cryo-EM). Images confirm that the preparations contained a fairly homogeneous population of TMC1 proteins, free of aggregation (Figures 1F and 1G). The class averages did not reveal structural details, suggesting conformational mobility and disorder. Assuming a protein density of ~1.35 g/cm3, however, their ~7- to 10-nm diameter indicates a protein of 200–220 kDa, consistent with a dimer of 88-kDa subunits. In addition, the images show a bilobed structure, but not trimeric symmetry, which is also consistent with a dimeric complex (Figure 1H). We conclude that TMC1 assembles as a dimer.
TMC Proteins Share Structural Similarity to TMEM16 Ion Channels
Most ion channel families assemble as trimers (P2Xs, ASICs, and piezos), tetramers (voltage-gated potassium channels and TRPs), pentamers (acetylcholine receptors [AChRs], GABAA receptors, glycine receptors, and 5-HT3 receptors), or hexamers (connexins). Mechanosensitive channels in bacteria assemble as tetramers, pentamers, or heptamers (Haswell et al., 2011). In each case, the multimer creates a single, central pore through which ions can pass. There are a few families that assemble as dimers; notable examples are the CLC chloride channels and transporters and the TMEM16 family of anion channels and lipid scramblases. Both of these families have two separate ion conduction pathways, one in each subunit of the dimer (Middleton et al., 1996; Brunner et al., 2014; Whitlock and Hartzell, 2016). We wondered whether TMC proteins shared structural similarity with either of these dimeric channels.
To understand the transmembrane architecture of mmTMC1, we plotted its hydrophobicity (Figure S3A). Six or seven domains (indicated with boxes 1–5 and 9 and 10) are strongly hydrophobic, but additional domains (indicated as boxes 6–8) are ambiguous (Keresztes et al., 2003; Labay et al., 2010). We used additional transmembrane prediction algorithms on the CCTOP prediction server (Dobson et al., 2015), which aggregates predictions from multiple algorithms, including TMHMM (Krogh et al., 2001), Octopus (Viklund and Elofsson, 2008), Philius (Reynolds et al., 2008), and SCAMPI (Peters et al., 2016). For mmTMC1, different algorithms did not always predict the same transmembrane domains but tended to predict 8–10 domains, with intracellular N and C termini (Figure S3B). We also used CCTOP to analyze the sequences of mmTMEM16A, an anion channel, and btCLC-K, a chloride channel. Structures for both have been solved (Park et al., 2017; Paulino et al., 2017a). CCTOP predicted 8 transmembrane domains for mmTMEM16A, which is known to have 10 transmembrane domains (Paulino et al., 2017a). As with mmTMC1, the predictions of different algorithms did not always agree. The solved transmembrane domains S1, S2, S3, S4, S9, and S10 were strongly predicted, whereas S5, S6, S7, and S8—also known to be transmembrane—showed mixed prediction. Notably, however, the regions of strong prediction and mixed prediction for mmTMC1 matched those for mmTMEM16. It is clear why predictions for S5–S8 of mmTMEM16A were mixed: these contain some charged and polar residues not normally found in lipid-facing transmembrane domains. In TMEM16A, these transmembrane regions line the channel pore (Paulino et al., 2017b; Dang et al., 2017); they may also in TMC1. S7 and S8 are short with few hydrophilic residues separating them and are always predicted as a single-transmembrane segment. Instead, a conserved PxxP sequence between S7 and S8 domains in TMEM16 provides a kink that is necessary for the hairpin structure in this region. The presence of this sequence in most TMCs supports the prediction that there are two distinct transmembrane domains in this segment: S7 between amino acids 570 and 591 in the mouse sequence and S8 between amino acids 596 and 616. In contrast, the transmembrane prediction for btCLC-K showed 14 transmembrane domains, consistent with the known structure (Park et al., 2017) but very different from the topology of TMCs (Figure S3B). Overall, the predicted transmembrane topology of TMCs matched that of TMEM16s.
In addition, our mass spectrometry of purified hsTMC1 is consistent with 10 transmembrane domains. Mass spectrometry of trypsin-digested membrane proteins often does not detect hydrophobic peptide segments. Mass spectrometry analysis of purified, trypsin-digested hsTMC1 resulted in ~45% peptide coverage, and the missing sequences were consistently within the predicted transmembrane domains (Figure S2A). Mass spectrometry after chymotrypsin digestion, in contrast, showed more extensive coverage. Both indicated that the sample was full-length hsTMC1 (Figure S2B).
Secondary Structure Prediction
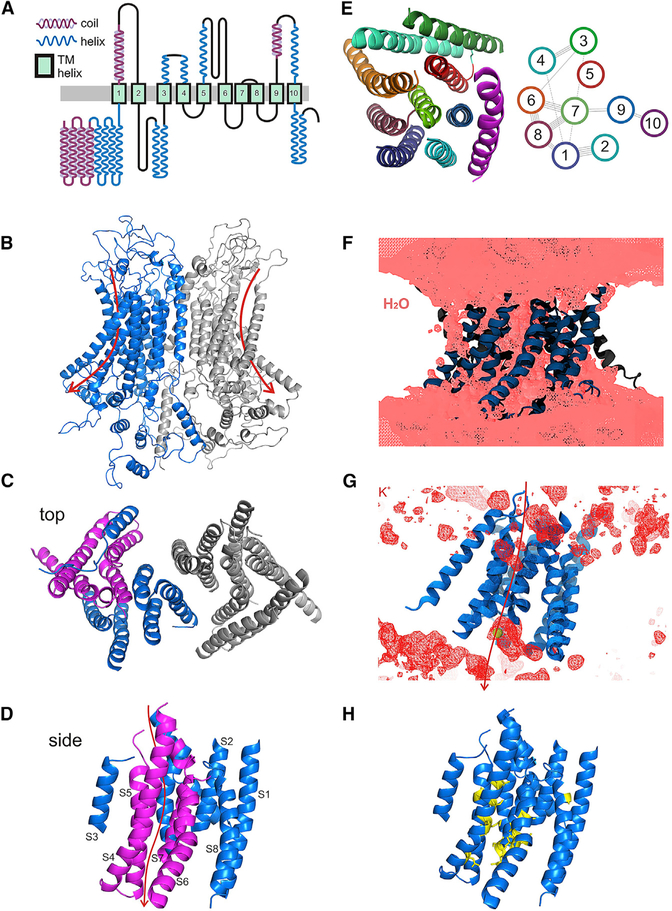
We also compared predicted secondary structure of mmTMC1 with that of mmTMEM16 using PSSpred (Yan et al., 2013). Predictions for both indicate coiled-coil and helical structure in the N terminus before S1, a helix in the C terminus after S10, and very little beta strand structure (Figure S3C), consistent with the circular dichroism spectrum of purified TMC1 (Figure S1E). They show ten helical segments that match transmembrane segments predicted from hydrophobicity. The mmTMC1 protein has an additional helix between S2 and S3, not seen in mmTMEM16, but otherwise the length and placement of helical segments is similar. The similarity in predicted secondary structure further suggests that TMCs may have a structure similar to TMEM16s. A schematic illustration of predicted transmembrane topology and secondary structure for mmTMC1, drawn from the combined transmembrane and secondary structure predictions, is shown in Figure 2A.
Figure 2. Predicted Architecture of TMC1 and a Putative Pore Region.
(A) Predicted transmembrane topology for mmTMC1, based on an alignment of many TMCs with TMEM16s, on the known transmembrane structure of mmTMEM16A, and on secondary structure predicted by PSSpred.
(B–D) Possible structure for dimeric TMC1 after 100 ns of MD simulations; model generated by I-TASSER based on the structure of mmTMEM16A (PDB: 5OYB). (B) View from within the plane of the membrane, (C) top view from outside the cell, and (D) side view of one subunit from within the membrane.
(E) Proposed arrangement of the transmembrane helices of one subunit, top view (left). EV-coupling analysis (right) reveals predicted contacts between residues, which are represented as lines connecting transmembrane helices.
(F) Isosurface of average water density computed over the last 50.5 ns of MD simulation of an I-TASSER-, mmTMEM16A-based model. Protein is shown in ribbon representation. Water permeation through the open groove of F1 can be observed.
(G) Isosurface of average potassium density computed and shown as in (F) for F1 subunit. Arrow indicates the putative permeation path.
(H) Seventeen cysteine substitutions (yellow) mapped onto the TMC1 structural model with S1–S10 shown in blue.
Post-translational Modification
Membrane proteins can be phosphorylated and glycosylated. Our mass spectrometry experiments detected six sites that were phosphorylated and located primarily in the intracellular N-terminal section of the protein (Figure S2A). In addition, the S5/S6 loop, predicted to be extracellular, bears an asparagine (N472 in mmTMC1) with the canonical NxT/S recognition sequence for N-linked glycosylation (N472 in mmTMC1), suggesting that this loop is likely to be extracellular.
Sequence Alignment of TMCs with TMEM16s
TMCs have previously been reported to have sequence similarity with the TMEM16 family (Hahn et al., 2009; Medrano-Soto et al., 2018) and are identified as part of the same genetic clan according to the PFAM database (Finn et al., 2008). We used ClustalX2 to align amino acid sequences of TMCs and TMEM16s from many species. Although similarity was low overall between TMCs and TMEM16s, there were regions of high similarity, especially for transmembrane domains S1 and a short segment preceding it, S2 and a segment following, S4, S5, S7, S8, S9, S10, and a short segment after S10 (Figure S4; Data S1). Based on this alignment, we used ClustalX2 to understand the phylogenetic relationship between these two families. Each family falls into a distinct group in an unrooted tree (Figure S5) with the fungal Nectria hematococca TMEM16 (nhTMEM16) toward the middle, but the phylogenetic distance between families is not much more than distances between some members within families.
Structure Prediction by Homology to TMEM16s
Phyre2 (Kelley and Sternberg, 2009) and I-TASSER (Yang and Zhang, 2015) servers use algorithms that identify relationships among protein families by comparing amino acid sequences of near homologs. These algorithms identify the most conserved regions and then weight those regions more heavily in a search for more distant relatives. To find proteins related to TMCs—specifically those for which structures have been determined—we submitted many TMC sequences to the Phyre2 and I-TASSER servers (see STAR Methods). In almost all cases, they identified nhTMEM16 or mmTMEM16A as the most closely related homolog with a known structure. Both Phyre2 and I-TASSER then used the nhTMEM16 or mmTMEM16A structures solved with X-ray crystallography or cryo-EM (Brunner et al., 2014; Paulino et al., 2017a, 2017b; Dang et al., 2017) and regions of reasonable alignment to create predicted structures of TMCs. The alignments were monomer-to-monomer, even though TMEM16s are dimers, so we created dimeric models of mmTMC1 and hsTMC1 by aligning the TMC monomer models to the mmTMEM16A dimer structure (Figure 2B). Extracellular and intracellular loops were not consistently modeled, and sequence similarity between TMEM16s and TMCs is weak in the loops, so we focused only on the arrangement of transmembrane helices, which are shown in several views (Figures 2C and 2D).
Evolutionary Coupling Analysis
We then sought an independent test of this arrangement. An emerging approach to predict protein structure is to use information contained within sequence variation among the members of large protein families (Marks et al., 2011). Residues that are closely apposed tend to co-vary. Using EVcoupling software (Marks et al., 2011), we identified and aligned about 3,500 sequences for TMCs in different species. The predicted contact map for hsTMC1, for 108 couplings with >80% confidence, is shown in Figure S6. We focused especially on couplings between residues in predicted transmembrane helices. This analysis predicted a number of adjacencies that are characteristic of parallel and anti-parallel transmembrane helix arrangements and—with some exceptions—are consistent with the predicted structure. Specifically, residues assigned to domains S6, S7, and S8 show evolutionary coupling (EC) signatures expected from their positions in the predicted structure, as do residues in S1 and S2, S3 and S4, and S1 and S8. These are schematized in a view of one subunit from the extracellular side (Figures 2E and S6).
Molecular Dynamics Equilibration
Structures derived by threading the sequence of one homolog onto the structure of a second are approximations at best, especially when sequence identity is low. Following a strategy used for prestin (Gorbunov et al., 2014; Geertsma et al., 2015), we reasoned that we could derive more accurate structural predictions by equilibrating a mmTMEM16A-based homology model of hsTMC1 using molecular dynamics simulations (Karplus and Petsko, 1990; Khalili-Araghi et al., 2009; see STAR Methods). The dimeric mmTMEM16A structure (PDB: 5OYB; Paulino et al., 2017b), corresponding to a putative open state, was used as a template to generate the hsTMC1 dimer, which was embedded in a hydrated bilayer for simulation (~357K atoms for 100 ns). In one subunit of the hsTMC1 dimer, the transmembrane helices were constrained (C1), and all atoms of the other hsTMC1 subunit were free to move (F1). The constraints on C1 forced transmembrane helices to stay in the conformation seen in the mmTMEM16 structure with a semi-open groove facing the lipids (Paulino et al., 2017b). Analysis of the simulation trajectories revealed that subunits remained together in a dimeric conformation. The equilibrated dimer has dimensions that are consistent with the bi-lobed structure of hsTMC1 observed using cryo-EM imaging. Although the initial conditions of the simulation were based on the putative open state of TMEM16A, the duration of the simulation was insufficient for channel closure.
Further support for an ion conduction pathway through the lipid-facing groove formed by S4–S7 of the hsTMC1 model comes from an analysis of water and potassium ion density throughout the simulation. Water molecules were excluded from the lipid, as expected, but were observed forming an aqueous pore. Most notably, water molecules were observed all along the S4–S7 groove (Figure 2F), similar to the solvent-accessible S4–S7 pathway in mmTMEM16A that is thought to constitute the pore (Dang et al., 2017). We also plotted the position of potassium ions during the simulation. Potassium was observed at the inner and outer mouth of the S4–S7 groove (Figure 2G). Both water and potassium movement during simulations is consistent with the idea that the S4–S7 groove can be an ion conduction pathway.
Cysteine Substitution Yields Viable Sensory Transduction
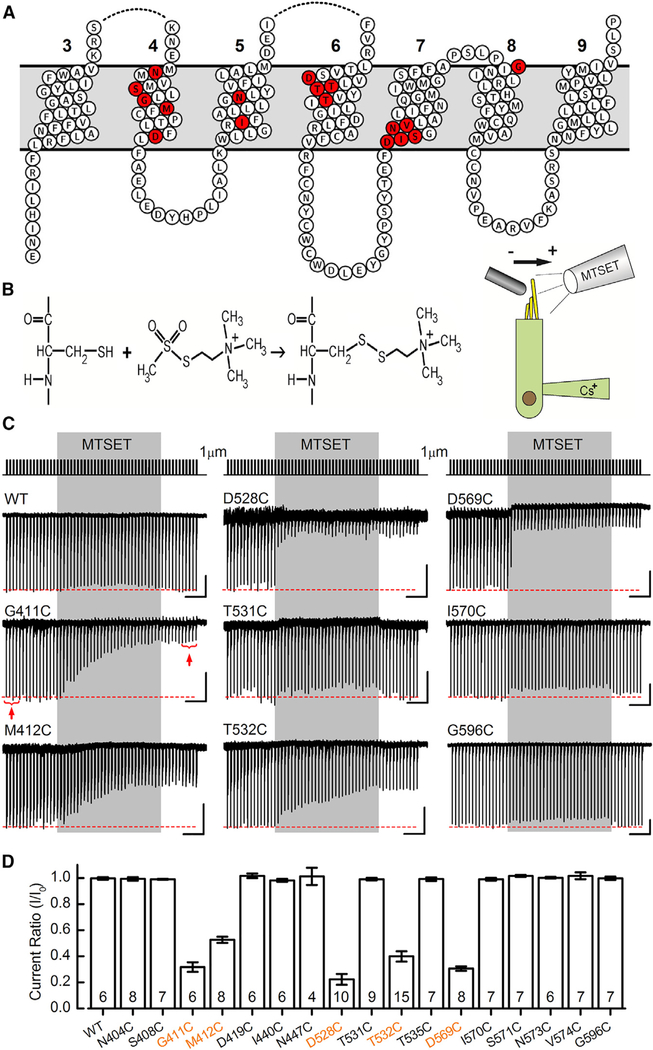
To investigate the transmembrane topology of TMC1 and identify the region and amino acid residues that govern permeation properties of hair cell sensory transduction, we generated 18 AAV2/1 vectors; each carried one of 18 mouse Tmc1ex1 sequences coding for wild-type TMC1 or TMC1 with a cysteine substitution at one of 17 different amino acid positions. Three of the sites, G411, M412, and D569, were chosen because the orthologous positions in the human TMC1 sequence are known to cause dominant, progressive hearing loss (Figure S7; DFNA36; Kawashima et al., 2015). Additional sites were selected in the four transmembrane domains that line the putative pore of TMC1 as proposed above. The sites are between N404 and G596, with several substitutions in the highly conserved TMC domains. All 17 cysteine substitutions were mapped onto the wild-type (WT) TMC1 ten-transmembrane topology (Figures 2H and 3A), 16 are in S4–S7, and final site was selected as a control in S8.
Figure 3. Cysteine Substitution in TMC1 Yields Viable Sensory Transduction Current in Cochlear Inner Hair Cells.
(A) Transmembrane topology of mouse TMC1,featuring transmembrane domains 3–9, as predicted by the homology with TMEM16. Seventeen sites were targeted for cysteine substitution using site-directed mutagenesis. The sites of cysteine substitution are shown in red.
(B) Chemical reaction between the cysteine side chain and the MTS reagent 2-(trimethylammonium)-ethyl methanethiosulfonate (MTSET) yields a covalent linkage that can modify protein function (left). Schematic diagram illustrating the experimental configuration for recording from cochlear inner hair cells (right) is shown. The stimulation pipette is oriented to deflect the bundle along the sensitive axis (arrow), and the drug application pipette is oriented perpendicular to the sensitive axis to eliminate fluid motion artifact.
(C) Hair bundles were mechanically stimulated with 1-μm step deflections at 3 Hz for 20 s (top trace). MTSET was applied for 10 s (gray shaded region) and washed out. Sensory transduction current traces recorded from cochlear inner hair cells are shown for WT TMC1 and eight representative TMC1 cysteine substitutions as indicated. The scale bars represent 2 s (horizontal) and 100 pA (vertical) for each trace, except D528C (50 pA vertical).
(D) Mean current ratios (±SEM) plotted for WT TMC1 and seventeen cysteine mutant TMC1s. Current ratio was calculated as the mean response to five bundle steps before (I0) and after (I) application of MTSET, as indicated in (C) for the G411C trace (red brackets and arrows). Number of cells, from 2–5 mice/substitution, is shown below.
Because TMC1 expressed in heterologous cell lines is not targeted to the membrane (Labay et al., 2010; Kawashima et al., 2011; Guo et al., 2016), we opted to examine the consequences of cysteine substitutions in native hair cells of organotypic explants from virally transduced mouse cochleas and utricles. Hair cells from Tmc1Δ/Δ;Tmc2Δ/Δ mice were exposed to AAV2/1-Tmc1 viral vectors. For cochlear sensory epithelia, viral vectors were injected into the inner ears at postnatal day (P)1 using the round window approach (Askew et al., 2015), excised at P5–P7, and placed in culture for an additional 3–13 days, the equivalent of P10–P20. Utricles were excised at P0, exposed to vectors, and cultured for 8–10 days (see STAR Methods).
Because hair cells of Tmc1Δ/Δ;Tmc2Δ/Δ mice lack sensory transduction, they do not take up the transduction-channel permeable dye FM1–43 (Kawashima et al., 2011; Askew et al., 2015). To identify Tmc1Δ/Δ;Tmc2Δ/Δ inner hair cells transduced with AAV2/1-Tmc1, we used brief (10 s) application of 50 nM FM1–43, followed by epifluorescence microscopy with an FM1–43 filter set. FM1–43-positive hair cells (Figure S8A), presumably expressing exogenous Tmc1 and functional sensory transduction channels, were selected for single-cell recording using the whole-cell, tight-seal technique and stiff glass probes to deflect their sensory hair bundles.
WT TMC1 and fifteen of seventeen TMC1 cysteine substitutions yielded sensory transduction currents with maximal amplitudes that ranged from 250 to 600 pA. Figure S8B shows mean maximal sensory transduction currents from 566 AAV2/1-transduced cochlear inner hair cells from Tmc1Δ/Δ;Tmc2Δ/Δ mice, binned according to the eighteen different constructs. Two of the constructs, N447C and D528C, yielded only very small sensory transduction currents (30–100 pA). This was probably not due to low expression levels, because the viral titers for AAV2/1-TMC1-N447C and AAV2/1-TMC1-D528C were similar to those of the other 15 AAVs. Rather, we suspect these two cysteine substitutions resulted in sensory transduction channels that had significantly attenuated function relative to WT and the other 15 cysteine mutants.
MTS Reagent 2-(Trimethylammonium)-Ethyl Methanethiosulfonate Inhibits Sensory Transduction in TMC1 Cysteine Mutants
To evaluate the accessibility of the TMC1-cysteine substitutions, we used local extracellular perfusion of methanethiosulfonate (MTS) reagents. Because cysteines contain sulfur-bearing side chains, cysteine residues exposed to aqueous solution may be available to react with MTS reagents, forming covalent disulfide bonds (Figure 3B, left). If the cysteines occur within the permeation pathway of an ion channel, covalent modification of those sites may cause acute, irreversible changes in the biophysical properties of the ionic currents that pass through the channels (Akabas et al., 1992).
We began with rapid, 10-s puff application of 2 mM MTS reagent 2-(trimethylammonium)-ethyl methanethiosulfonate (MTSET) (molecular weight [mol wt]: 198 g/mol) while delivering a repeating series of 1-μm hair bundle deflections (Figure 3B, right). Covalent modification of cysteine side chains with MTSET adds a positive charge, which may alter the flux of cations through an ion channel pore. Figure 3C shows WT and eight representative traces from the cysteine substitutions. Hair cells expressing WT TMC1 constructs showed no change in the amplitude of sensory transduction currents following MTSET application, suggesting that native cysteines in the WT sequence (Figure S7A) were either unable to react with MTSET or inconsequential for whole-cell currents. Twelve of seventeen mutant TMC1 constructs also showed no change in the amplitude of sensory transduction currents following MTSET application, suggesting these cysteine substitutions were also inaccessible or inconsequential for whole-cell currents. However, five sites with cysteine substitutions—G411C, M412C, D528C, T532C, and D569C—showed an irreversible decline in current amplitude in cochlear inner hair cells (Figure 3C) following application of MTSET. Mean sensory transduction current ratios, before and after MTSET application, are shown in Figure 3D for all 18 TMC1 constructs. For the five reactive cysteines, the currents were reduced 50%–80% on average (Figure 3D). Because even very large compounds, such as gentamicin-Texas Red, can pass through the channel, it is not surprising that the relatively small MTS reagents do not completely inhibit ion flux. However, we did note that two of the mutations, N447C and D528C, inhibited current almost completely and, for the latter, application of MTSET reduced current by 98% relative to WT TMC1. The current amplitude did not recover even after several minutes of washout, consistent with covalent modification of the cysteine side chains (Figures S8C, top trace, and S11F). To confirm the effect was a specific consequence of cysteine modification, we used the compound DTT, which breaks disulfide bonds. Within 2 or 3 min of 10 mM DTT application externally, we observed a full recovery of transduction current amplitude (Figures S8C, middle traces, and S11G). There was no recovery when DTT was introduced internally via the recording pipette (Figure S8C, bottom trace). The time course of MTSET inhibition varied among the cysteine mutants (Figure S9). D569C reacted fastest with a time constant of 191 ± 62 ms (n = 7), suggesting it was most accessible to aqueous solutions, and M412C reacted slowest with a time constant of 17.3 ± 16.1 s (n = 5).
MTSET did not affect the sensitivity of the mechanosensory response. Stimulus-response (I(X)) curves before and after MTSET application had similar steepness (Figure S10), suggesting that the relationship between force and open probability was not altered by the manipulation. Thus, the most salient effect of MTSET in cochlear inner hair cells was reduction of the mean maximal transduction current amplitudes by half (M412C) to three-quarters (D528C; Figure 3D). In type II vestibular hair cells of the mouse utricle, the reduction was similar (Figure S11) for M412C and D569C (other mutations were not tested in utricle cells). We found that the rate of the reaction was dose dependent, with higher MTSET concentrations causing faster reductions in current amplitude for three of the sites tested (Figures S12A–S12F).
Because these five sites (G411C, M412C, D528C, T532C, and D569C) reacted acutely and irreversibly, producing a change in ion permeation, we conclude these sites must be accessible to aqueous solutions and that they must be within the permeation path for sensory transduction channels. Since MTSET is a charged, polar compound, it is not membrane permeable; thus, the reactive cysteines within transmembrane domains must be facing the channel’s aqueous pore and not the interface with the lipid membrane or other transmembrane domains.
Cysteine Modification Alters Calcium Selectivity
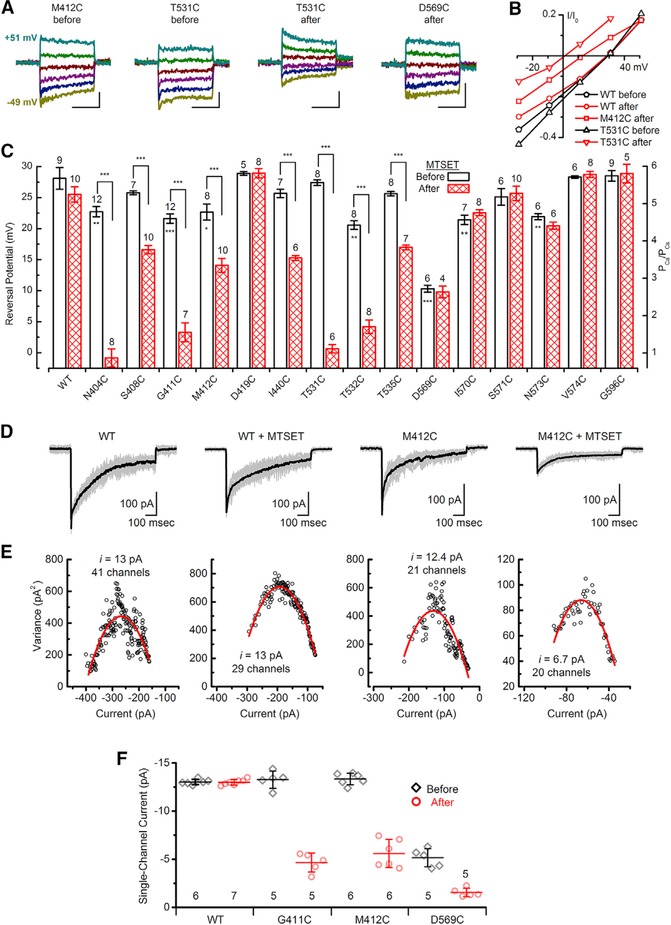
Because MTSET diminished but did not eliminate current for several of the cysteine substitutions, we wondered whether ionic selectivity was altered by the substitutions themselves or MTSET. Cells were bathed in 100 mM external Ca2+ with 140 mM Cs+ internal as the only charge carriers as described previously (Pan et al., 2013). Saturating bundle deflections were delivered at membrane potentials that ranged between −49 mV and +51 mV (Figure 4A). Peak transduction currents at each potential were plotted versus membrane voltage to generate current-voltage relations for each substitution (Figure 4B), and the x-intercept was taken as the reversal potential. Mean reversal potentials were measured from 245 inner hair cells and were plotted for WT and each of 15 TMC1 constructs before and after application of MTSET (Figure 4C). The analysis revealed seven sites for which cysteine substitution alone (N404C, G411C, M412C, T532C, D569C, I570C, and N573C) had a statistically significant shift in reversal potential, relative to WT TMC1, prior to addition of MTSET. Eight of the sites (N404C, S408C, G411C, M412C, I440C, T531C, T532C, and T535C) had statistically significant reversal potential shifts that were evoked within seconds after application of MTSET. For four cysteine sites (N404C, G411C, T531C, and T532C) reversal potential shifted more than 15 mV. Because all shifts were in the negative direction, covalent modification of the cysteine residues with positively charged MTSET led to reduced calcium selectivity, as indicated by reduction of the calcium permeability ratio relative to cesium. Together with the Beethoven mutation (M412K), the eleven cysteine substitutions presented here demonstrate that four different TMC1 transmembrane domains, between residues 404 and 573 (S4–S7), are intimately associated with ionic selectivity, a fundamental property of an ion channel pore. Furthermore, for eight sites, reversal potential was altered within seconds following MTSET application, providing very strong evidence that these sites are within the permeation pathway in hair cell sensory transduction channels.
Figure 4. Reversal Potentials and Single-Channel Currents for WT TMC1 and Cysteine Mutants.
(A) Representative families of current evoked by 20-ms, 1-μm step hair bundle deflections measured at membrane potentials between −49 mV and 51 mV for IHCs expressing M412C, T531C, or D569C before or after MTSET as indicated. Cells were recorded with 100 mM Ca2+ extracellular and 140 mM Cs+ intracellular ion concentrations as the only charge carriers. The scale bars represent 10 ms (horizontal) and 25 pA (vertical).
(B) Current-voltage relationships were generated from the peak currents and plotted as function of membrane potential for the five TMC1 conditions indicated.
(C) Summary plots of mean (± SEM) reversal potentials for WT TMC1 and 15 cysteine mutants before and after treatment with 2 mM MTSET. Number of cells, from 2–4 mice/substitution, and the cysteine substitutions are indicated above and below each bar, respectively. Stars within bars indicate statistically significant differences relative to WT TMC1. Stars above brackets indicate statistically significant differences before and after MTSET. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; two population t tests.
(D) Mean sensory transduction current (black) for 14–28 individual current traces (gray) evoked by identical 1-μm bundle deflections for cochlear IHCs expressing WT TMC1 or M412C, before and after application of 2 mM MTSET.
(E) Plots of variance as a function of current amplitude (symbols) fitted with a parabolic function (red curve; see STAR Methods). Axes were adjusted to allow better visualization of the data and the quality of the fit.
(F) Mean (±SD) single-channel current estimates for WT and three mutants before and after application of MTSET. Number of cells, from 2 or 3 mice/substitution, is listed below.
MTSET Reduces Single-Channel Current in TMC1 Mutants
Because mutations at three of the sites (G411, M412, and D569) have been linked with dominant progressive hearing loss in humans, we characterized these sites in greater detail. In this and subsequent sections, we focus on the G411C, M412C, and D569C substitutions with the goal of gaining further insight into the structure, function, and dysfunction of TMC1.
To determine whether the reduction in current amplitude (Figure 3D) was due to complete block of a fraction of the channels or partial block of some or all of the channels, we performed nonstationary noise analysis (Sigworth, 1980) on the whole-cell current records obtained from cochlear inner hair cells. We generated mean sensory transduction current traces (Figure 4D, black trace) from 14–28 individual current traces (gray traces) evoked by identical 1-μm bundle deflections for cochlear inner hair cells (IHCs) expressing WT TMC1 or M412C, before and after application of 2 mM MTSET. Variance was calculated from the square of the difference between the mean trace and each individual trace and was plotted as a function of whole-cell current amplitude (Figure 4E). The data were fit with a parabolic function (see STAR Methods), which revealed single channel currents of ~13 pA (at −84 mV) in inner hair cells expressing WT TMC1 before and after application of MTSET. However, following application of MTSET to cells expressing G411C or M412C there was a 65% and 58% decrease in the single-channel current, respectively, and little or no change in the number of functional channels (Figure 4F). The D569C substitution had reduced single-channel currents relative to WT prior to MTSET application, suggesting that substitution of a negatively charged aspartate with a neutral cysteine had a direct effect on cation permeation. Adding a positive charge with application of 2 mM MTSET further reduced single-channel current in D569C cells by ~70% without significantly affecting the number of channels. The data for all three substitutions are consistent with the interpretation that MTSET reduced whole-cell current by partial inhibition of current through single channels rather than a complete block of a fraction of the channels. Importantly, as suggested by Wu and Müller (2016), changes in single-channel conductance link a protein closely to the pore of a channel. Whereas it remains possible that the cysteine sites are outside of the pore region and that binding of MTSET induces a conformational change that reduces single-channel current, below we present data that argue against that possibility (see section titled Pore Blockers Impede Access to Cysteine Residues).
Voltage Dependence of Cysteine Modification
To explore localization of three sites (G411C, M412C, and D596C), we investigated the voltage dependence of the MTSET reaction by depolarizing cells to +80 mV and then applying 2 mM MTSET (Figure S12G). The current ratio measured before and after the MTSET application revealed that modification of the M412C site was highly voltage dependent, reacting at −80 mV (Figures 3C and 3D), but not at +80 mV during the 10-s MTSET exposure (Figures S12G and S12H). D569C was the least voltage dependent (Figure S12H), reacting at both potentials but with slower kinetics at +80 mV (Figure S9E). G411C did not react with 2 mM MTSET at +80 mV but did react with 10 mM MTSET, indicating intermediate voltage dependence. The data suggest that all three sites are within the electric field of the ion channel pore. Although the reaction was slowed by depolarization, D569C seems to be the most accessible of the three sites, perhaps facing directly into the mouth of the aqueous pore.
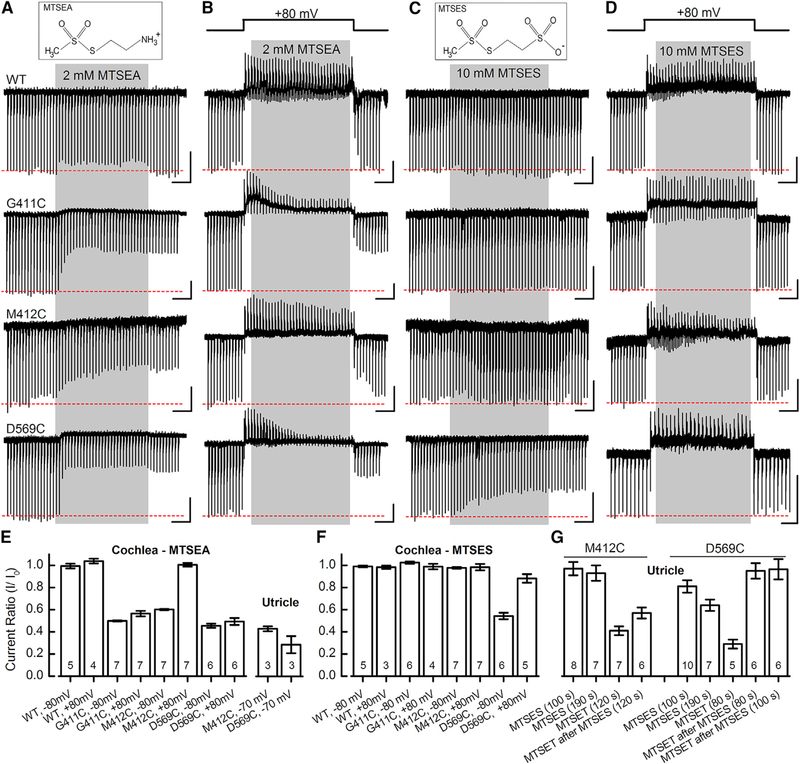
MTSEA and MTSES Modify Current in Cysteine Mutants
Next, we wondered whether different MTS reagents might differ in their effects on the three reactive cysteine residues (G411C, M412C, and D596C). Figures 5A and 5B show the consequences of 2-Aminoethyl MethaneThioSulfonate (MTSEA), a smaller, positively charged, membrane-permeable MTS reagent (mol wt: 155 g/mol), at −80 and +80 mV, respectively. We found no effect on WT TMC1, but we found that, at −80 mV, MTSEA evoked faster (Figure S9E) inhibition of current amplitude than MTSET for all three sites. At +80 mV, M412C did not react significantly with 2 mM MTSEA (Figures 5B and 5E). Both M412C and D569C also reacted with MTSEA when expressed in utricle hair cells (Figure 5E). In general, the MTSEA data are consistent with the MTSET data and support the conclusion that modification of D569C is the least voltage sensitive and that M412C is the most voltage sensitive of the three sites.
Figure 5. Effects of MTSEA and MTSES on WT TMC1 and Cysteine Mutants in IHCs and Vestibular Hair Cells.
(A) Structure of MTSEA (top). Sensory transduction currents recorded from IHCs during 10 s application of 2 mM MTSEA (gray) at −80 mV holding potential. The scale bars represent 2 s (horizontal) and 100 pA (vertical) for each trace (A–D).
(B) IHC currents recorded at +80 mV during 10 s application of 2 mM MTSEA (gray).
(C) Structure of negatively charged MTSES (top). Representative IHC currents after 10 s, 10 mM MTSES application (gray) at −80 mV are shown.
(D) IHC currents recorded at +80 mV during 10 s application of 10 mM MTSES (gray).
(E and F) Mean (± SEM) current ratios before and after 2 mM MTSEA (E) or 10 mM MTSES (F) application in cochlea IHC and utricle type II cells. Substitution, membrane potential, and number of cells, from 2–10 mice/condition, are indicated at the bottom.
(G) Mean (± SEM) current ratio for utricle type II cells expressing M412C or D569C after exposure to 10 mM MTSES alone or before 2 mM MTSET application. MTSES was applied for 80–190 s as indicated below.
Sodium (2-sulfonatoethyl) MethaneThioSulfonate (MTSES), a negatively charged MTS reagent (mol wt: 219 g/mol) did not inhibit current for WT, G411C, or M412C TMC1 mutants but did slowly inhibit D569C current (Figures 5C, 5F, 5G, and S9E). Although we predicted that depolarization might drive a negatively charged molecule into the cell and promote reaction with the cysteine mutants, we found that MTSES applied at +80 mV had no effect on WT, G411C, or M412C (Figure 5D) and only 10% inhibition on D569C in cochlear cells (Figure 5F) at +80 mV. Interestingly, in utricle hair cells expressing D569C, we found that application of MTSET after application of MTSES had little effect on current amplitude. Whereas MTSET alone reduced current by 70%, application after MTSES reduced current by only a few percent (Figure 5G). There was no similar protective effect for the M412C site. We interpret this result to mean that MTSES can access and react with the D569C site, blocking subsequent reaction with MTSET. It must have reacted with a significant fraction of the channels because MTSET was no longer able to induce the characteristic robust inhibition. Although the D569C substitution significantly reduced the single-channel current (Figure 4F), restoring the negative charge of the aspartate side chain by covalent attachment of negatively charged MTSES did not increase but rather further decreased current amplitude (Figure 5F), suggesting the effect was steric.
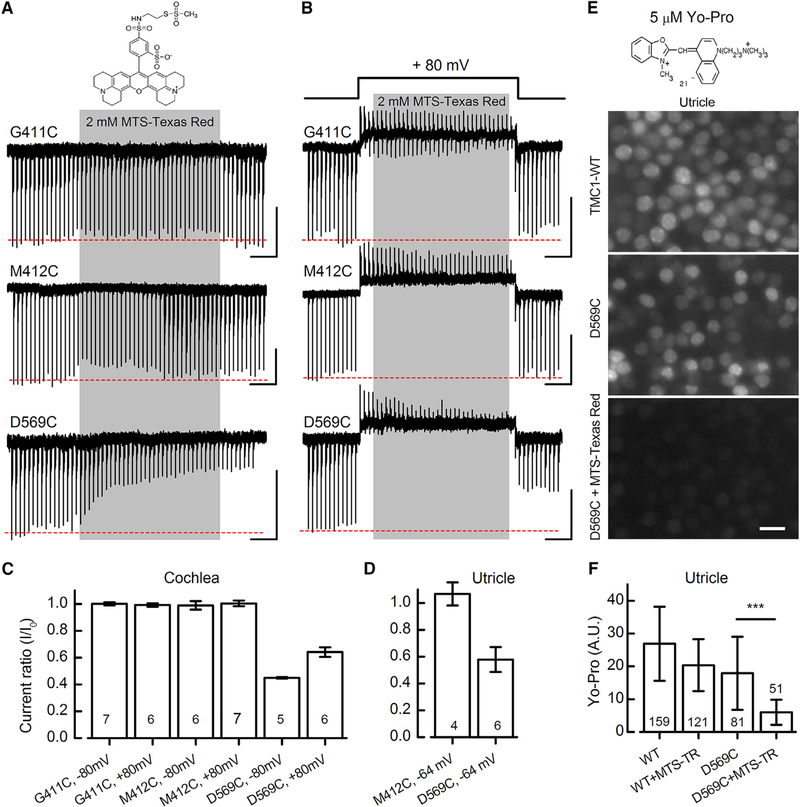
Consequences of MTS-Texas Red on Sensory Transduction
To investigate whether large compounds can access the three cysteine substitutions within the pore region of TMC1, we used MTS conjugated to a bulky side group, Texas Red (mol wt: 744 g/mol). 2 mM MTS-Texas Red did not inhibit currents at −80 mV or +80 mV when applied to inner hair cells expressing G411C or M412C (Figures 6A and 6B), suggesting these sites are inaccessible to the bulky reagent. However, when applied to cells expressing D569C, transduction currents were slowly inhibited by ~50% and ~40% at −80 mV or +80 mV, respectively (Figures 6A–6C and S9E). Utricle hair cells behaved in a similar fashion (Figure 6D), again suggesting greater steric accessibility for D569C than M412C.
Figure 6. MTS-Texas Red Has Limited Access to G411C and M412C, but Not D569C.
(A and B) Structure of MTS-Texas Red (top). Transduction currents from IHCs after 10 s MTS-Texas Red application at −80 mV (A) and +80 mV (B) are shown. The scale bars represent 2 s (horizontal) and 100 pA (vertical).
(C and D) Summary plot of mean (± SEM) current ratios (I/I0) following exposure to MTS-Texas Red for cochlear IHCs (C) and utricle type II hair cells (D). Substitution, membrane potential, and number of cells, from 2–6 mice/condition, are shown below.
(E) Structure of YO-PRO-1 iodide (MW 629, top). Fluorescence images of YO-PRO-1 uptake in Tmc1/Tmc2 mutant utricle hair cells expressing WT TMC1 (top) or D569C (middle and bottom) are shown. The tissue was exposed to 5 μM YO-PRO-1 for 60 s followed by four bath exchanges during a five-minute wash. For the bottom panel, the tissue was exposed to 2 mM MTS-Texas Red for 60 s prior to application of YO-PRO-1. All images were acquired with identical gain and contrast settings. The scale bar represents 10 μm.
(F) Mean (±SD) YO-PRO-1 fluorescence quantified in A.U. (arbitrary units) for each condition shown at the bottom. Number of cells, from 2–3 mice/condition, is indicated in each bar. The experiment was replicated twice for WT tissue and three times for D569C. ***p = 1.3 × 10−11 two population t test.
Although gentamicin-Texas Red has previously been shown to pass through the hair cell transduction channel (Wang and Steyger, 2009), we did not observe accumulation of MTS-Texas Red in hair cells. However, we did find that brief application of the large cationic fluorescent molecule, YO-PRO-1 (Santos et al., 2006), like FM1–43, rapidly entered hair cells expressing WT TMC1 or the D569C substitution (Figure 6E) but did not enter Tmc1Δ/Δ;Tmc2Δ/Δ double mutant cells. Interestingly, application of MTS-Texas Red significantly inhibited YOPRO-1 uptake in D569C-expressing cells, but not in WT cells (Figures 6E and 6F), suggesting the bulky Texas Red side group impeded permeation of YO-PRO. This provides another line of evidence that modification of D569 alters the permeation pathway.
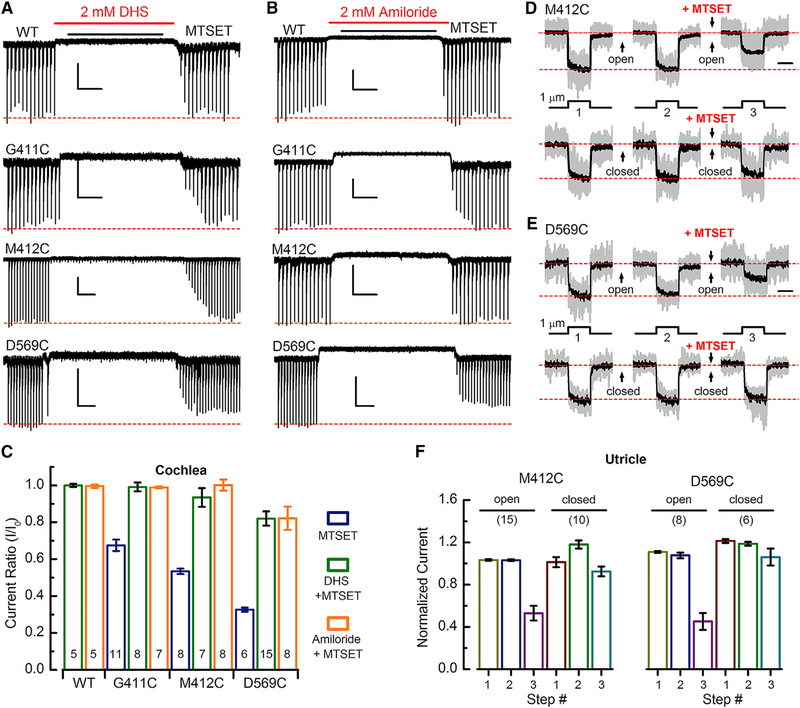
Pore Blockers Impede Access to Cysteine Residues
Dihydro-streptomycin (DHS) is an established open-channel blocker of hair cell sensory transduction channels (Marcotti et al., 2005) and binds within the channel’s electric field (Farris et al., 2004). Recently, Corns et al. (2016) demonstrated that the Bth mutation (M412K) reduces the binding affinity for DHS by 8-fold, suggesting the site is within the permeation path. In utricle type II hair cells, we found that the D569C substitution reduced the affinity for extracellular DHS (Figure S13) by >4-fold, providing another line of evidence that the substitution itself alters the permeation pathway.
If the G411, M412, and D569 sites line the channel pore, we reasoned that open-channel blockers may prevent access of MTS reagents. To test this hypothesis, we applied 2 mM DHS followed by 2 mM MTSET to cells expressing either WT TMC1, G411C, M412C, or D569C substitutions. Unlike application of MTSET alone, when the DHS and MTSET cocktail was washed out, there was a full recovery of sensory transduction currents in cells expressing G411C or M412C (Figures 7A and 7C), indicating that DHS blocked access of MTSET and “protected” these sites from reacting. However, in cells expressing D569C, there was only partial protection. Following washout of DHS and MTSET, the currents in D569C-expressing cells did not recover the full amplitude, indicating that D569C may be more accessible to externally applied MTS reagents. Reapplication of MTSET alone confirmed that the cysteine substitutions were still available to react after washout of DHS (Figure S14).
Figure 7. Pore Blockers and Closed Channels Impede Access to Cysteine Mutations.
(A) Representative IHC sensory transduction currents for WT, G411C, M412C, and D569C in response to dual application of 2 mM dihydrostreptomycin (DHS) and MTSET as indicated above. MTSET was applied at 2 mM for G411C, 10 mM for M412C, and 0.2 mM for D569C, selected as the minimal concentration required to achieve the maximal response during the 8-s application. The scale bars represent 2 s (horizontal) and 100 pA (vertical).
(B) IHC responses for dual application of 2 mM amiloride and MTSET. Scale bars indicate 2 s (horizontal) and 100 pA (vertical).
(C) Mean (±SEM) current ratios for WT, G411C, M412C, and D569C in response to MTSET alone, DHS + MTSET, or amiloride + MTSET. Number of cells, from 2–5 mice/condition, is shown at the bottom of each bar.
(D) Mean sensory transduction current traces (black) superimposed on 10–15 normalized individual traces (gray) recorded from utricle type II hair cells expressing M412C. Deflection protocol is shown below. Between each step, the bundle was deflected 1.7 μm (top row) or −0.5 μm (bottom row) for 6 s to open or close channels, respectively. 2 mM MTSET was applied between the 2nd and 3rd steps as indicated. The scale bar represents 25 ms.
(E) Mean sensory transduction traces (black) superimposed on 6–8 normalized individual traces (gray) recorded from utricle type II hair cells expressing D569C.The same stimulation protocol was used to evoke the responses shown in (D) and (E) except the bundle was deflected for 2 s and the MTSET concentration was 0.5 mM.
(F) Mean (±SEM) normalized M412C and D569C currents for each step (1–3) shown in (D) and (E) for bundles held in the open and closed positions. Number of cells, from 2–5 mice/condition, is shown above the bars.
To further investigate this phenomenon, we used a second, smaller, open-channel blocker, amiloride (Rüsch et al., 1994). In a manner similar to DHS, 2 mM amiloride protected the G411C and M412C sites from reacting with MTSET but did not fully protect the D569C site (Figures 7B and 7C). Thus, both open-channel blockers prevented access of MTSET to the G411C and M412C sites, suggesting they are deep within the permeation pathway, whereas D569C, which was only partially protected by both blockers, may be more exposed, even in the presence of blockers.
Closed Channels Prevent MTS Reactions
Because open-channel blockers prevent MTSET from reacting with the mutant cysteine residues, we wondered whether channel closure during normal gating might block MTSET access to the cysteine sites. Because negative deflections that promote prolonged channel closure are experimentally feasible for vestibular hair bundles, we examined utricle hair cells expressing M412C or D569C. Currents were measured during three identical test steps. In the interval between each step, prolonged positive (>1.5 μm) or negative (−0.5 μm) bundle offsets were imposed, which held channels in either the open or closed states. We found no difference between current amplitudes measured during the first and second test steps, indicating that prolonged bundle offsets did not lead to current rundown. In the interval between the second and third test steps, MTSET was applied during the prolonged bundle deflections. We found for both M412C and D569C (Figures 7D and 7E) that MTSET applied while channels were held open led to inhibition of current amplitude, consistent with earlier data (Figures 3C and 3D). However, when the bundles were deflected in the negative direction, inducing prolonged channel closure, MTSET application did not yield current inhibition. Mean current amplitudes, summarized in Figure 7F, revealed a significant difference between MTSET applied to closed versus open channels for both cysteine substitutions. The data suggest that M412C and D569C sites are inaccessible when transduction channels are closed, perhaps protected by a gating mechanism that prevents entry of MTS reagents.
DISCUSSION
A comprehensive understanding of sensory transduction by hair cells requires knowledge of all the protein components in the transduction complex and how they assemble. Central to the complex are TMC1 and TMC2, which are required for auditory and vestibular function (Kawashima et al., 2011). Whether TMC1 and TMC2 are pore-forming subunits of the hair cell transduction channel has been a subject of debate (Corey and Holt, 2016; Wu and Müller, 2016). Here, we used biochemical and computational methods to suggest a dimeric stoichiometry and an arrangement of transmembrane helices for TMC1 based on known structures of the related TMEM16 channels. The permeation pathway of the mouse TMEM16A anion channel is formed by transmembrane domains S4–S7 (Yu et al., 2012; Peters et al., 2016; Paulino et al., 2017a; Dang et al., 2017), suggesting that—with substitutions that would confer cation selectivity—S4–S7 may form the pore of TMC1. Based on this arrangement and on the location of deafness-causing mutations, we individually replaced 17 residues in mmTMC1 with cysteines. We used AAV2/1 vectors to deliver the mutant coding sequence to the cochleas of neonatal mice lacking endogenous TMC1 and TMC2 and, after 8–13 days, recorded the sensory transduction currents evoked by hair bundle deflections before and after application of cysteine-modifying reagents.
Sensory transduction channels in hair cells are highly selective for calcium (Corey and Hudspeth, 1979; Lumpkin et al., 1997; Beurg et al., 2006), and their positive reversal potential in high extracellular calcium is a measure of calcium selectivity. For eight of the cysteine substitutions in TMC1, addition of MTSET evoked a significant shift in reversal potential within seconds. Of these, four sites (N404C, G411C, M412C, and T532C) had both a shift in reversal potential due to the cysteine substitution alone and a further shift upon application of MTSET. In total, 11 of 15 sites tested showed a significant change in reversal potential caused by these manipulations. All reversal potential shifts were in the negative direction, indicating a decrease in calcium selectivity, by 5- to 6-fold in some cases. Thus, acutely modifying TMC1 residues alters a fundamental property of the hair cell mechanosensory ion channel pore.
In addition, five of the TMC1 cysteine mutants showed a rapid, irreversible decline in current amplitude following application of MTSET. The addition of a bulky, positively charged MTSET group to a residue lining the pore would be expected to inhibit cation flux due to steric hindrance and charge repulsion. Consistent with reduction in whole-cell currents, noise analysis for three of the mutants revealed that single-channel currents were reduced by 60%–70% following application of MTSET. MTSET acting on TMC1 cysteine mutants evidently inhibits receptor current by partially impeding cation flux through single channels rather than by blocking all current through a subset of channels.
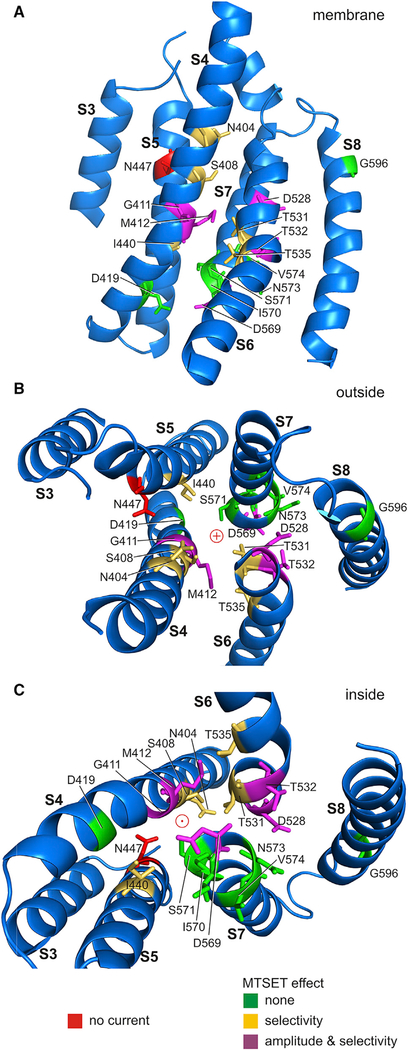
Unlike ion channel structures that have a linear permeation path on an axis of radial symmetry, TMEM16A has a curved and poorly defined pore within each monomer of the dimeric channel. By analogy, TMC1 may have a similarly tortuous permeation path (red arrow, Figure 2D). To understand which residues participate in the permeation pathway, we mapped the sites affected by MTSET onto the predicted helices of TMC1. Figure 8 and Video S1 highlight helices S3–S8 for a single TMC1 monomer; Figures 8A–8C show S3–S8 viewed from within and outside and inside the membrane, respectively. Side chains are shown for the cysteine-substituted amino acids and are colored according to MTSET effect: sites that did not react with MTSET are shown in green, sites where MTSET changed reversal potential are yellow, and sites with a change in both reversal potential and current amplitude are magenta. One mutation, N447C, eliminated current almost entirely before MTSET and is shown in red. All residues that altered permeation properties face the predicted pore of the channel where they would be available to alter ion selectivity and flux. Importantly, non-reactive residues are largely facing away or are at some distance from the pore. Because all eleven of the cysteine mutants that affected the transduction current face the permeation pathway predicted from the mmTMEM16A structure, the physiology data provide strong qualitative support for the idea that S4–S7 line the pore of TMC1 and support the proposed arrangement of transmembrane helices.
Figure 8. TMC1 Homology Model with Proposed Permeation Pathway.
Homology model for TMC1 transmembrane domains S3–S8 with cysteine substitutions mapped onto the structure. The model shows the human sequence with F1 helix positions after 100 ns molecular dynamics equilibration of side chains. This may represent an open conformation, as channel closure is unlikely during the simulation time.
(A) A single TMC1 monomer shown from within the cell membrane. The various cysteine substitutions are color coded as follows: green substitutions had no effect; gold residues altered calcium selectivity after MTSET application; and magenta residues altered both calcium selectivity and current amplitudes after MTSET application. Cysteine substitution of the one red residue (N447) eliminated current entirely, even without MTSET application.
(B) View of domains 3–8 from outside the cell.
(C) View from inside the cell.
Labay et al. (2010) explored the topology of TMC1 by testing the accessibility of epitope tags, an approach made challenging by the retention of TMC1 in the endoplasmic reticulum when expressed in heterologous cells and by the need to distinguish cytoplasmic from endoplasmic reticulum (ER) lumen locations with detergents of different strength. The six-transmembrane model they proposed differs from the 10-transmembrane model suggested here; specifically, their model lacks S4 and S6, the domains that have ambiguous prediction in CCTOP and that are thought to contribute to a polar pore, and it lacks S7 and S8, short helices in a hairpin that barely reach the extracellular side (Figure S15). MTS reagent effects explored in detail for three sites (G411, M412, and D569) help resolve this difference. In particular, the Labay et al. (2010) topology places G411 and M412 within an extracellular loop, whereas five observations from our experiments indicate that G411 and M412 are within the membrane: (1) both G411C and M412C reacted with MTSET more slowly than would be expected for residues that are freely accessible to the extracellular solution. Based on reaction rates of 105 M−1 s−1 for MTSET with cysteines in exposed positions (Liu et al., 1997), we predict the reaction should reach saturation in less than 100 ms. However, the reactions for G411C and M412C were much slower and required 5–10 s to reach saturation (Figures 3C and S9), suggesting the sites are less accessible. (2) The MTSET reactions with G411C and M412C were highly voltage dependent, reacting at −80 mV (Figures 3C and 3D), but not +80 mV (Figures S12G and S12H), suggesting these sites are deep within the electric field of the channel pore. (3) The negatively charged MTSES did not react with G411C or M412C (Figures 5C and 5F), consistent with the idea that negative reagents may not easily enter the pore of a cation channel. (4) G411C and M412C did not react with the bulky MTS-Texas Red (Figures 6A–6C), supporting the notion that the sites are inaccessible to large compounds. (5) Both pore blockers, DHS and amiloride, protected G411C and M412C from reacting with MTSET (Figures 7A–7C), suggesting that the blockers prevented MTSET from accessing residues deep within the pore region. Based on these observations, we propose G411C and M412C are within the fourth transmembrane domain deep within the ion channel pore.
The Labay et al. (2010) topology also suggested that D569 is part of a large intracellular loop, where it would be inaccessible to the membrane-impermeant MTSET. The following observations suggest that D569C is within the aqueous pore, in a location that is rapidly accessible to MTS reagents applied extracellularly: (1) MTSET and MTSEA altered current amplitude with D569C within less than a second (Figures 3D and 5A). (2) MTS-Texas Red was able to react with D569C (Figure 6), suggesting that even bulky reagents can access the site. (3) The D569C substitution reduced affinity for the pore blocker, DHS (Figure S13). (4) DHS and amiloride only partially protected D569C from reacting with MTSET (Figures 7A–7C), perhaps because the reaction rates for DHS and MTSET (Figures S9B and S9E) were within a similar range, again suggesting the D569 site is rapidly accessible to aqueous solutions. (5) Because the reaction rates were voltage-dependent (Figure S9E), the D569 site likely resides within the channel’s electric field. Furthermore, because both D569C and M412C did not react with MTSET when hair bundles were deflected in the negative direction (Figures 7D–7F), we conclude that these residues must be within the permeation path in a location that is inaccessible when channels are in the closed conformation but exposed when channels are open.
Biochemical assays and cryo-EM strongly suggest that TMC1 occurs as a dimer, which has interesting implications for the stoichiometry of a transduction complex. Channels are at the lower ends of tip links (Beurg et al., 2009), which are thought to include a homodimer of PCDH15 (Ahmed et al., 2006; Gillespie et al., 2005; Kazmierczak et al., 2007). PCDH15 binds to both TMC1 and TMC2 (Maeda et al., 2014). Although graphic representations of the transduction apparatus often depict the individual strands of PCDH15 as binding to distinct channel complexes (Fettiplace, 2016; Kazmierczak and Müller, 2012), this has never been conceptually satisfying, as it is unclear how a single PCDH15 would associate with a multimeric channel. An attractive possibility arising from this model of TMC1 structure is that a dimer of PCDH15 associates with a dimer of TMC1 or TMC2, with each monomer of PCDH15 regulating the opening of one pore in each TMC monomer. The dimeric assembly and identification of the pore region of TMC1 have additional implications for hair cell sensory transduction (Corey et al., 2018).
Summary and Conclusions
Taken together, our data demonstrate that TMC1 assembles as a dimer and contains ten transmembrane domains. Four transmembrane domains (S4–S7) include at least 12 amino acid residues that line the pore of hair cell sensory transduction channels. Eleven cysteine substitutions altered calcium selectivity either on their own or within seconds following application of MTSET. Substitutions at five sites caused an immediate reduction of whole-cell current in response to MTS reagents; three of the substitutions, examined in greater detail, caused a reduction in single-channel current. One site, N447C, caused a near complete loss of current and another, D528C, caused an 80% reduction in current when exposed to MTSET, corresponding to a 98% reduction in current amplitude relative to WT TMC1. Placing these 12 amino acid residues within the homology model of TMC1 revealed localizations consistent with them lining the permeation pathway. In particular, N404, S408, G411C, and M412C are in the fourth transmembrane domain; I440 and N447 are in the fifth domain; D528, T531, T532, and T535 are in the sixth; and D569, I570, and N573C are in the seventh domain. Cysteine substitution at additional sites may identify other residues that line the pore and provide further tests of residue localization and the TMEM16–TMC1 homology model. Whereas the model presents a general TMC1 transmembrane architecture, further data from cryo-EM or X-ray crystallography may help clarify additional structural details of TMC1, including the pore region, the location of the gate, and perhaps the key force-dependent transformation that occurs as TMC1 channels transition from closed to open states.
Lastly, because multiple TMC1 amino acid sites affect core permeation properties (whole-cell and single-channel current amplitude and reversal potential), and because these properties are altered within seconds upon exposure to MTS reagents, the data provide clear evidence that TMC1 residues are part of the permeation pathway for hair cell sensory transduction channels. Thus, we conclude that TMC1 is a major pore-forming component of the hair cell sensory transduction channel in mammalian auditory and vestibular hair cells. Although TMC1 includes four transmembrane domains that line the permeation path, it is not yet clear whether TMC1 forms the entire pore. Our data do not rule out contributions from lipids or other proteins. For example, there may be a role for membrane lipids, as with some TMEM16 channels (Whitlock and Hartzell, 2016) or other transmembrane proteins, such as TMHS/LHFPL5, TMIE, or PCDH15 (Wu and Müller, 2016).Nonetheless, together with prior evidence (Corey and Holt, 2016; Corey et al., 2018), it is now clear that TMC1 is an ion channel protein mediating sound sensitivity in animals ranging from fish to mice to humans and perhaps broadly throughout the animal kingdom.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Jeffrey R. Holt (jeffrey. holt@childrens.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cultured Cells
TMC1 orthologs (Genscript) were subcloned into a pcDNA3.1(+1) vector containing a mammalian cytomegalovirus promoter (CMV) and a C-terminal HRV3C cleavage site followed by the human influenza hemagglutinin (HA) tag, and expressed in HEK293FT cells. For protein expression, the cells were grown at 37°C in a tissue culture incubator and transfected when cell density reached ~90% confluency. To boost protein expression, sodium butyrate was added to the culture 24 h after transfection to a final concentration of 10 mM. Cells were collected 48 h after transfection. The cell pellets were either kept at −80°C or used immediately.
Mice
Double mutant mice with targeted deletions of critical exons in Tmc1 and Tmc2 (Kawashima et al., 2011) were used for all experiments. All experiments were performed in accord with protocols approved by the Boston Children’s Hospital Animal Care and Use Committee (protocols 2878, 3396). Both male and female mice were used in approximately equal proportions. Tmc1Δ/Δ/Tmc2Δ/Δ genotypes were confirmed as described in Pan et al. (2013). Genomic DNA was prepared with Proteinase K (final concentration 1 mg/ml) and a Tail Lysis reagent (Viagen). One hundred fifty microliters of the mixture of Proteinase K and Lysis reagent was added per sample, and tubes were incubated overnight at 55°C. Once digestion was complete, the temperature was increased to 85°C for 50 min. For each sample, two separate PCR reactions were set up; one for Tmc1, and one for Tmc2. All reactions were prepared using GoTaq Green Master Mix 2X (Promega) with 2 mL of genomic DNA and four primers per gene (final concentration 0.2 nM each). Both PCR reactions were performed at 95°C for 2 min, (95°C for 30 s, 56C for 30 s, 72°C for 45 s) 3 35 cycles, 72°C for 5 min, hold at 4°C. Primers Tmc1Exon9L2 (50-GATGAACATT TTGGTACCCTTCTACTA-30) and Tmc1Exon9R2 (50-CACACTTTGACACG TACAGTCTTTT AT-30) specifically amplified a 557-base pair fragment of the wild-type Tmc1 allele. Primers Tmc1KO50ConfF2 (50-TCTGAGCTTCTTAATCTCTGGTAGAAC-30) and Tmc1KO50ConfR2 (50-ATA
CAGTCCTCTTCACATCCATGCT-30) amplified a 408 base pair fragment of the targeted deletion allele of Tmc1. Primers Tmc2–7L08 (50-CGGTTCTTCTGTGGCATCTTACTT-30) and Tmc2-7R08 (50-ACCAGGCAATT GACATGAATA-30) amplified a 401 base pair fragment of wild-type Tmc2.
Tmc2KO50L08 (50-CTGCCTTCTGGTTAGATCACTTCA-30) and Tmc2KO50R08 (50-GTGTTTTAAGTG TACCCACGGTCA-30) amplified a 621 base pair fragment of the targeted deletion allele of Tmc2. To genotype Tmc1Bth mice PCR reactions were set up as described above. BthMutF2 (50-CTAATCATACCAAGGAAACATATGGAC-30) and BthMutR2 (50-TAGACTCACCTTGTTGT TAATCTCA TC −30) were used to amplify a 376 base pair product which was purified and sequenced.
METHOD DETAILS
Confocal FRET imaging
FRET was detected by donor recovery after acceptor photobleaching (Drenan et al., 2008; Nashmi et al., 2003), in which dequenching of GFP (donor) during incremental photo-destruction of mCherry (acceptor) was recorded on a confocal microscope. CHO cells were plated on 35-mm glass-bottomed dishes, transfected with Tmc cDNAs, and they were imaged live 15–24 hours after transfection at ambient temperature. Before an imaging session, cell culture medium was replaced with phenol red-free Leibovitz L-15 medium, and the cells/media were equilibrated to room temperature for 15–20 minutes.
Fluorescence was measured on an Olympus FV1000 confocal microscope with a 60X, 1.42-NA oil objective. Sequential images of GFP and mCherry fluorescence signals were acquired by excitation with 488 and 559 nm excitation, respectively. The fluorescence signal was detected by splitting the green and red emission with a dichroic filter and then directing the signal from each emissive species through the appropriate optical bandpass filters. All images were 512 × 512 pixels at 16 bits per pixel.
For detection of FRET by donor recovery after acceptor photobleaching, a rectangular area of interest enclosing a single cell was selected for bleaching at 559 nm with 75% output level. Images were acquired prior to (t = 0) and at 1, 2, 5, 10, and 20 s of mCherry bleaching. GFP and mCherry mean fluorescence intensities were calculated only for the areas in which GFP and mCherry signals colocalized, by selecting the perimeter as the boundary of region of interests using ImageJ software (National Institutes of Health). Fluorescence intensities of GFP and mCherry at various time points after photobleaching were normalized to t = 0 (100%).
FRET efficiency (E) was calculated as E = 1/(IDA/ID), where IDA represents the normalized fluorescence intensity of GFP (100%) in the presence of both donor (GFP) and acceptor (mCherry), and ID represents the normalized fluorescence intensity of GFP in the presence of donor only (complete photo-destruction of mCherry). The ID value was extrapolated from a scatterplot of the fractional increase of GFP versus the fractional decrease of mCherry for each cell. Data are reported as mean ± SEM. The reported E values were averages of 5–9 cells from 2–3 batches of cell preparation. Note that we found no correlation between fluorescence intensity of the GFP or mCherry signal and FRET efficiency.
FSEC/SEC estimation of molecular weight
HEK293FT cells (Invitrogen) were pelleted after 24–48 hours of TMC1 expression and suspended in buffer containing 400 mM NaCl, 20 mM HEPES (pH = 7), and 2× HALT protease inhibitor cocktail (Thermo Fisher), solubilized with FOS13 or CHS-DDM detergents (Anatrace) and run on a Superose6 column (GE) in a Shimadzu HPLC system with an in-line fluorescence detector. We also ran purified proteins through the same column.
In order to have a rough estimate for the molecular mass of GFP-TMC1, we have exploited the near linear relationship between the logarithm of the molecular weight of the proteins and their elution time (Whitaker, 1963). Linear fitting was performed in GraphPad Prism software. The GE standards (plotted in blue) included thyroglobin (670 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44kDa); Biorad standards (black) including thyroglobin (670 kDa), gamma-globin (158 kDa), ovalbumin (44 kDa) and myoglobin (17 kDa). We also included BSA (66 kDa) in the analysis.
Protein purification
HEK293FT cells (Invitrogen) were pelleted after 24–48 hours of TMC1 expression and suspended in buffer containing 400 mM NaCl, 20 mM HEPES (pH = 7) and 2× HALT protease inhibitor. The cell pellets were broken by passing through a dounce homogenizer in resuspension buffer containing 400 mM NaCl, 20 mM HEPES pH = 7, and 2× HALT. Membranes were collected by ultracentrifugation (50000g × 1 h) and solubilized in the same buffer supplemented with 1% FOS13 (Anatrace). Detergent-insoluble material was removed by ultracentrifugation (40,000g × 45 min) and the supernatant was mixed with HA-tag antibody agarose conjugate resin (Thermo Fisher) in buffer A (200 mM NaCl, 20 mM HEPES at pH = 7.4, 0.5 mM TCEP, 2% glycerol) supplemented with 1 mM DDM and equilibrated for 2 h at 4°C. The resin (50 μL per 10 cm plate) was washed with 20× resin volume of buffer A with DDM at 4°C to free the protein from the resin-bound HA tag. The bound protein was equilibrated overnight with 1 units of HRV3C protease solution (Thermo Fisher) per cell plate. The concentration of the eluted protein was measured by absorbance at 280 nm using a NanoDrop instrument (Thermo Scientific) and its purity confirmed by Coomassie-stained denaturing gel. Subsequently, the eluted protein was mixed with A8–35 (Affymetrix) at approximately 1:1 (w/w) ratio with gentle agitation for 6 hours at 4°C. Detergent was removed with 15 mg/ml of Bio-Beads SM-2 (Bio-Rad) overnight at 4°C, and the beads were subsequently removed over a disposable polyprep column. The eluent was cleared by centrifugation before further purification on a Superose 6 increase column (GE Health-care) in buffer containing 150 mM NaCl and 20 mM HEPES at pH = 7.4. The peak corresponding to TMC1 was collected for analysis.
Crosslinking experiments
Purified proteins at 1 mg/ml were reacted with the crosslinking reagent disuccinimidyl dibutyric urea (DSBU; Thermo Fisher) at the indicated concentrations and incubated for 2 hours at 4°C. The cross-linked and solubilized proteins were examined with denaturing 4%–12% NUPAGE bis-tris gels (Thermo Fisher) in NUPAGE MOPS SDS running buffer (Thermo Fisher). Prestained protein ladders (Thermo Fisher) were used to estimate molecular weights.
Circular Dichroism
Protein samples were analyzed with a Jasco J-815 Circular Dichroism Spectropolarimeter at 4°C in a 1-mm path-length quartz cuvette. Five sequential scans were taken at 0.5-nm increments from 200–260 nm. The data were plotted with the online analysis tool CAPITO (Wiedemann et al., 2013).
SEC-MALS
Protein samples were analyzed with SEC-MALS (size-exclusion chromatography coupled to multi-angle static light scattering) using an Agilent AdvanceBio300 column attached to an Agilent 1260 Chromatography System in-line with the following detectors: Dawn Heleos II for MALS, Optilab rEX for refractive-index detection and QELS for dynamic light scattering (Wyatt Technology; (Wyatt, 1993). Molar masses were calculated with the ASTRA software package (Wyatt Technology) using the ‘protein conjugate’ module. The dn/dc value was set to 0.187 ml/g for protein and 0.1424 ml/g for A8–35 amphipols (Gohon et al., 2008). The extinction coefficient (1.695 ml/mg/cm) for monomeric TMC1 was calculated using the ProtParam tool on the ExPaSy server. Bovine serum albumin (Thermo Fisher) was used to correct for band-broadening effects.
Native PAGE Gel Analysis
We mixed purified samples with NativePAGE sample buffer, performed electrophoresis using NativePAGE 4%–16% Bis-Tris gels (Thermo Fisher), and performed Coomassie staining.
Negative stain and cryo-EM imaging
For negative stain EM experiments, we applied 3 μL of purified TMC protein at ~0.02 mg/ml to glow-discharged carbon grids (FCF400-CU, EMS) stained with uranyl-formate (0.75%) solution. Imaging was performed on a Phillips CM10 machine (HMS-EM facility) at 75K magnification. Relion software was used for selecting and averaging single particle images, in order to recognize symmetry and subunit organization of the TMC complex in 2D images.
For cryo-EM experiments, 2.5 μL of purified sample at 0.75 mg/ml concentration was applied to Quantifoil R1.2/1.3, 400 Mesh copper grids (Electron Microscopy Sciences), blotted for 2.5 s and plunged into liquid ethane with a CP3 set-up, and immediately stored in liquid nitrogen. Cryo-EM imaging was performed with a Talos-Arctica microscope (200 kV) at the Cryo-EM facility at the University of Massachusetts Medical School. About 200,000 particles were picked from 800 images and class averages were obtained with Relion.
Sequence alignment
Sequences were aligned, and a phylogeny plot generated using ClustalX2. Sequences included hsTMC1, mmTMC1, ggTMC1, ×lTMC1, hsTMC2, mmTMC2, ggTMC2, ×lTMC2, hsTMC3, mmTMC3, ggTMC3, ×lTMC3, hsTMC4, hsTMC5, hsTMC6, hsTMC7, hsTMC8, ceTMC1, ceTMC2, nhTMEM16, hsTMEM16a, mmTMEM16a, hsTMEM16b, hsTMEM16c, hsTMEM16d, hsTMEM16e, hsTMEM16f, hsTMEM16 g, hsTMEM16h, hsTMEM16j and hsTMEM16k, where hs is human, mm is mouse, gg is chicken, ×l is Xenopus and nh is Nectria. Sequence similarity was calculated by the Sequence Identity and Similarity server (SIAS; http://imed.med.ucm.es/Tools/sias.html).
Structural modeling
We used I-TASSER (Iterative Threading ASSEmbly Refinement) (Yang and Zhang, 2015) as well as Phyre2 (Protein Homology/analogY Recognition Engine V 2.0) (Kelley et al., 2015) to perform a protein structure prediction for TMC1. We submitted the sequences of hsTMC1, mmTMC1, ggTMC1, ×lTMC1, hsTMC2, mmTMC2, ggTMC2, ×lTMC2, hsTMC3, mmTMC3, ggTMC3, ×lTMC3, hsTMC4, hsTMC5, hsTMC6, hsTMC7, hsTMC8, ceTMC1, ceTMC2 for structure prediction.
Molecular dynamics simulations
For molecular dynamics equilibration, two copies of the hsTMC1 homology model generated by I-TASSER (Zhang, 2008) were aligned independently to each of the protomers in the dimeric mmTMEM16A structure (PDB: 5oyb; Paulino et al., 2017b) using VMD (Humphrey et al., 1996). The final model was embedded in a hydrated POPC membrane using the VMD membrane plug-in, and the resulting system was solvated with explicit water molecules and ions (150 mM KCl with overall system neutral) to create a ~357,000-atom system.
All simulations were performed using NAMD 2.12 with the CHARMM36 force field for proteins and lipids, the CMAP correction, and the TIP3P water model (12 Å cutoff with switching function starting at 10 Å) (Best et al., 2012; Klauda et al., 2010; Phillips et al., 2005). A time step of 2 fs was assumed together with SHAKE. Long-range electrostatic interactions were computed every other time step using the Particle Mesh Ewald method with a grid density > 1 Å−3 and periodic boundary conditions. Langevin dynamics was used to keep temperature constant (310 K during production runs; damping coefficient set to 0.1 ps−1), while constant pressure was maintained using the hybrid Nosé-Hoover Langevin piston method (period 200 fs; damping timescale 50 fs). An initial simulation melting the lipid tails for 0.5 ns (all but lipid tails fixed) was followed by dynamics with protein atoms constrained and water molecules forced out of the protein-lipid interface for 0.5 ns. The resulting system was used for a simulation in which the transmembrane domain of one subunit was constrained with a harmonic potential (k = 1 kcal/mol/Å2) to its initial conformation (C1 constrained, F1 free; 0.5 ns with water molecules forced out, 98.5 ns without any forces applied to water molecules). Coordinates of all atoms were saved every 2 ps in stored trajectories. RMSD was computed using VMD and Cα atoms of the entire protein subunit or of its transmembrane helices as indicated. Volumetric density maps were generated using VMD’s VolMap tool using default settings and trajectory frames at 20 ps intervals over the last 50.5 ns.
EVcoupling analysis
Using the EVcoupling software (evfold.org), multiple sequence alignments to mouse TMC1 residues 140–735 were generated using the Markov model-based sequence-search tool ‘JackHMMER’ (Johnson et al., 2010) with five iterations. Alignments were built with Uniref datasets (Suzek et al., 2015). After removing sequences with more than 80% identity and no more than 30% gaps, the alignment included ~3500 TMC sequences representing different TMC orthologs from different species. Residue pairs with high probability of co-evolution were referenced to the mmTMC1 or hsTMC1 sequence.
AAV vector production
Custom oligonucleotide primers were designed to generate cysteine codons (TGC) at desired locations in mouse Tmc1ex1 cDNA using PCR based site-directed mutagenesis. To be consistent with prior literature, we adopted the numbering scheme for Tmc1ex2 (Vreugde et al., 2002; Pan et al., 2013; Askew et al., 2015; Corns et al., 2016). We generated mutations in Tmc1ex1 that coded for following amino acid substitutions: G411C, M412C, D569C, I570C, S571C N573C, V574C, G596C. AAV2 plasmids were generated that encoded either WT Tmc1ex1 (AAV2-CMV-Tmc1ex1) or mutants with one of eight cysteine substitutions. Briefly, mouse Tmc1 cDNA was transferred using SalI and XbaI into an AAV2 shuttle plasmid, which was modified from pAAV2-MCS by deleting beta globin intron and alternating the multi cloning sites. For N404C, S408C, D419C, I440C, N447C, D528C, T531C, T532C, T535C, gene fragments containing TGC changes were synthesized (GenScript, Piscataway NJ, USA) and cloned into a pUC57 shuttle vector. For cysteine changes between amino acid positions 393 and 535, flanking restriction sites for AvrII and BstAPI (all enzymes were purchased from New England Biolabs Inc., Ipswich, MA) were added to the ends to permit subcloning into pAAV2-CBA-Tmc1ex1 vector. For amino acid positions between 567 and 596, BastAPI and ApaI enzymes were used for subcloning. Constructs were validated by Sanger sequencing using the following primers: GGAGAACATCCACCTCATCAG and TGTTCCTCACACTTGGTGCA for mutants encoding for a cysteine between position 393 and 535 and GTTCTGCAATTACTGCTGGTG for mutants encoding for cysteine between position of 567 and 596. Constructs were screened with SmaI digest to check for ITR (inverted terminal repeat) integrity before packaging into AAV vectors. Viral vectors were packaged with AAV2 ITRs into the AAV1 capsid where the transgene cassette was driven by a CMV promoter. The following AAV2/1 vectors were generated by the Gene Transfer Vector Core at Boston Children’s Hospital: AAV2/1-CMV-Tmc1ex1-WT (6 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-N404C (2.9 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-S408C (6.5 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-G411C (2.5 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-M412C (7.9 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-D419C (1.1 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-I440C (9.4 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-N447C (5.7 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-D528C (1.3 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-T531C (5.8 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-T532C (8.3 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-T535C (8.7 × 1012gc/ml); AAV2/1-CMV-Tmc1ex1-D569C (1.2 × 1014gc/ml); AAV2/1-CMV-Tmc1ex1-I570C (7.3 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-S571C (1 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-N573C (3.5 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-V574C (4.6 × 1013gc/ml); AAV2/1-CMV-Tmc1ex1-G596C (2.2 × 1014gc/ml). Vectors were aliquotted and stored at →80°C.
Tissue preparation
Tmc1Δ/Δ/Tmc2Δ/Δ mouse pups postnatal (P) day 1 to P2 were injected with ~1.0 μL of AAV vectors manually via the round window membrane (RWM) using glass micropipettes, as previously described (Askew et al., 2015). Prior to injection mice were anesthetized using hypothermia at 2°C for ~2 min. A post-auricular incision was made to expose the otic bulla and visualize the cochlea. Following injection pups were returned to their cages and allowed to recover. At P5 to P7 organs of Corti were excised from injected ears for electrophysiological studies and the tectorial membrane was removed. Utricles were excised at the same time and the otolithic membrane was removed following 15-minute treatment in 0.1 mg/mL protease XXIV at room temperature. The sensory epithelia were mounted on round glass coverslips in a flat position. Tissues were kept in culture in the presence of 1% FBS and maintained for 3 to 13 days, or the equivalent of P10 to P20. The cultures were bathed in media containing DMEM supplemented with 10 mM HEPES, 0.05 mg/mL ampicillin, and 10 mg/L Ciprofloxacin (NaOH to pH 7.4) at 37°C in 5% CO2 and media was changed every other day.
Drug preparation and perfusion
The MTS reagents, 2-(trimethylammonium)ethyl MTS (MTSET; mol wt: 198 g/mol), 2-Aminoethyl MethaneThioSulfonate (MTSEA 155 g/mol), Sodium (2-sulfonatoethyl) MethaneThioSulfonate (MTSES; 219 g/mol) and Sulforhodamine methanethiosulfonate (MTS-Texas Red; 744 g/mol), were obtained from Toronto Research Chemicals (Ontario, Canada). Dihydro-streptomycin (DHS), amiloride and Dithiothreitol (DTT) were obtained from Sigma-Aldrich. For utricle experiments, MTS reagents were dissolved in external solutions immediately before application via bath exchange. For MTS-Texas Red and for channel open/closed experiments, a working solution was kept on ice and filled into a glass pipette coupled to a Picospritzer III (Parker Hannifin, NJ). Bath was exchanged and control solution was flushed through perfusion lines before subsequent cells (although all cells in bath were assumed to have been exposed to the reagents). Wash in of drug was monitored with a series of near-maximal deflections of the bundle at 1–10 s intervals, as indicated. Wash in data were normalized by dividing by the amplitude of the sweep immediately preceding drug perfusion (t = 0). DTT effect was normalized to amplitude preceding application of DTT (but after MTSET application). For cochlea, MTS solutions were applied at the apical surface of the hair cell using a glass perfusion pipettes with ~5–10 μm tips, oriented perpendicular to the sensitive axis of the hair bundle. Pipette pressure was controlled with a PV830 Pneumatic PicoPump (WPI). In some experiments two apical puff pipettes were used for MTSET application with or without 2 mM DHS or Amiloride as described in Figures 7 and S14.
Electrophysiology
The tissue was visualized on a Zeiss Axioskop FS at 63× equipped with DIC and epifluorescence with filtersets to detect FM1–43 and YO-PRO-1 fluorescence (Chroma Technologies). To identify Tmc1Δ/Δ/Tmc2Δ/Δ hair cells with functional transduction after exposure to AAV-Tmcs we used 300 nM FM1–43 application for 50 s for utricles and 50 nM FM1–43 for 10 s for cochlea. Since FM1–43 permeates functional sensory transduction channels, FM1–43 fluorescent cells were targeted for recording using the whole-cell, tight-seal recording technique. Sensory epithelia were bathed in external solution containing (in mM): 137 NaCl, 5.8 KCl, 0.7 NaH2PO4, 10 HEPES, 1.3 CaCl2, 0.9 MgCl2, 5.6 Glucose, and vitamins and essential amino acids (ThermoFisher Scientific, Waltham, MA), adjusted to pH 7.4 with NaOH, ~310 mmol/kg. To determine relative permeability of Ca2+, we used a high Ca2+ solution containing: 100 CaCl2, 30 NMDG, 6 Tris, 10 Glucose, adjusted to pH 7.4 with HCl, ~305 mOsm). Recording electrodes were pulled from R6 capillary glass (King Precision Glass). The intracellular solution containing (in mM): 137 CsCl, 5 EGTA, 5 HEPES, 2.5 Na2-ATP, 0.1 CaCl2, 3.5 MgCl2, adjusted to pH 7.4 with CsOH, ~290 mmol/kg. Sensory transduction currents were recorded under whole-cell voltageclamp configuration using an Axopatch 200B (Molecular Devices) amplifier. Cells were held at –80 mV (cochlea) or –60 mV (utricle) unless noted otherwise. Data were low-pass filtered at 1–5 kHz (Bessel filter), then sampled at 20 kHz with a 16-bit acquisition board (Digidata 1322A). Data were corrected for a −4 mV liquid junction potential in standard extracellular solutions and −9 mV in the high calcium solution. Cochlea inner hair cell bundles were deflected using stiff glass probes mounted on a PICMA chip piezo actuator (Physik Instruments) driven by an LPZT amplifier (Physik Instruments) and filtered with an 8-pole Bessel filter at 40 kHz to eliminate residual pipette resonance. Fire-polished stimulus pipettes with 4–5 μm tip diameter were designed to fit into the concave aspect of inner hair cell bundle as previously described (Stauffer and Holt, 2007). Utricle hair bundles were stimulated by gently drawing the kinocilium into a pipette coupled to a piezoelectric bimorph controlled by a piezo driver (ThorLabs, model MDT694) and filtered at 1 kHz (utricle) as previously described (Stauffer et al., 2005). Hair bundle deflections were monitored using a C2400 CCD camera (Hamamatsu, Japan).
Noise analysis
We adapted the non-stationary noise analysis described by Höger and French (1999) and Holton and Hudspeth (1986) as follows. The mean response was subtracted from each individual response and the difference was squared to generate variance, which was plotted as function of whole-cell current amplitude for each of the four conditions. To ensure that all portions of the variance plot were equally represented we averaged the current and variance within 1-pA bins, as described by Traynelis and Jaramillo (1998) and Heinemann and Conti (1992). The data were fit with the following equation: σ2 = σ02 + iI - (I2/N), where σ2 is the variance, i is the single-channel current, I is the whole-cell current and N is the number of channels. Cells were included in the analysis if they met the following inclusion criteria: 1) Less than a 5% shift in baseline leak current between the first and last stimulus presentation; 2) Hair bundles were well-coupled to the probe throughout the duration of the recording; 3) The plot of σ2 versus I yielded a parabolic dataset that was well-fit by the equation above. Of 32 cells analyzed, 23 (72%) met these criteria.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed with Clampfit and ORIGIN 2015 (OriginLab) and are presented as means ± SEM as noted in the text and figure legends. Number (n) of hair cells recorded is indicated for each mean value given in the text and figures legends. Number of mice used is indicated in the figures legends. One-way analysis of variance (ANOVA) or Student’s t test was used to compare biophysical properties of sensory transduction currents using the statistical function in Origin 2015 (OriginLab). Statistical significance is indicated as p values.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCES | SOURCE | IDENTIFIER |
| Antibodies | ||
| HA Tag Antibody, mAb, Mouse | Genscript | Cat. No. A01244; RRID:AB_1289306 |
| Goat Anti-Mouse IgG (H+L), highly cross-adsorbed CF770 | Biotium | Cat. No. 20077; RRID:AB_10559194 |
| Bacterial and Virus Strains | ||
| One Shot TOP10 Competent E.coli | Thermo Fisher | Cat. No. C404006 |
| AAV2/1 | BCH Viral Core | http://www.childrenshospital.org/research/centers-departmental-programs/fm-kirbyneurobiology-center/cores/viral-core |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Salts, acids, bases | Sigma-Aldrich | stock |
| DDM | Anatrace | Cat. No. D310 |
| FOS13 | Anatrace | Cat. No. F310 |
| DMEM Media | GIBCO | Cat. No. 11995040 |
| Sodium Butyrate | Acros Organics | Cat. No. 156-54-7 |
| Recombinant DNA | ||
| hsTMC1 | Genscript | Accession number NM_138691.2 |
| Software and Algorithms | ||
| GraphPad PRISM 6 | GraphPad Software | N/A |
| Relion | Scheres, 2012 | https://www2.mrc-lmb.cam.ac.uk/relion/index.php?title=main_page |
| The PyMOL Molecular Graphics System | Schrodinger | https://pymol.org/2/ |
| Clustal Omega sequence alignment software | European Bioinformatics Institute | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| I-TASSER | Zhang, 2008 | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ |
| NAMD | Phillips et al., 2005 | http://www.ks.uiuc.edu/Research/namd/ |
| VMD | Humphrey et al., 1996 | http://www.ks.uiuc.edu/Research/vmd/ |
| SIAS | N/A | http://imed.med.ucm.es/Tools/sias.html |
Highlights.
TMC1 assembles as a dimer and resembles TMEM16 ion channels
Cysteine modification with MTS reagents alters hair cell sensory transduction
The data support a revised topology of TMC1 with 10 transmembrane domains
TMC1 residues line the pore of hair cell sensory transduction channels
ACKNOWLEDGMENTS
We thank Drs. Andrew Griffith, Criss Hartzell, Jonathan Cohen, Gary Yellen, and Hiroshi Suzuki and members of Holt/Géléoc and Corey laboratories for many helpful discussions and critical review of a prior version of the manuscript. We thank the Boston Children’s Hospital (BCH) Viral Core (BCH Intellectual and Developmental Disabilities Research Center [IDDRC], grant no. 1U54HD090255), Dr. Chen Xu for extensive training on electron microscopy, Dr. Kelly Arnett for SEC-MALS training and discussions, Dr. Tomaino Ross for mass spectrometry analysis, and Dr. Debora Marks for EV-coupling analysis. Molecular dynamics simulations were carried out using the Texas Advanced Computing Center (TACC)-Stampede2 (XSEDE XRAC MCB140226) and Ohio Supercomputing Center (OSC) supercomputers (PAS1037). This work was supported by NIH/NIDCD (National Institute for Deafness and Communication Disorders) grants R01-DC013521 (J.R.H.), R01-DC000304 (D.P.C.), R01 DC015271 (M.S.), and F32 DC016210 (N.A.). B.G. was an Edward R. and Anne G. Lefler Center Postdoctoral Fellow. K.K. was supported by an NIDCD intramural research fund Z01-DC000060-10 (A.J. Griffith).
Footnotes
DECLARATION OF INTERESTS
J.R.H. holds patents on Tmc1 gene therapy and is a scientific advisor to several biotech firms focused on inner-ear therapies. The authors declare no other competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 15 figures, one data file, and one video and can be found with this article online at https://doi.org/10.1016/j.neuron.2018.07.033.
REFERENCES
- Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess SM, Lilley KS, Wilcox ER, Riazuddin S, et al. (2006). The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci. 26, 7022–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akabas MH, Stauffer DA, Xu M, and Karlin A (1992). Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science 258, 307–310. [DOI] [PubMed] [Google Scholar]
- Askew C, Rochat C, Pan B, Asai Y, Ahmed H, Child E, Schneider BL, Aebischer P, and Holt JR (2015). Tmc gene therapy restores auditory function in deaf mice. Sci. Transl. Med 7, 295ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, and Mackerell AD Jr. (2012). Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Evans MG, Hackney CM, and Fettiplace R (2006). A largeconductance calcium-selective mechanotransducer channel in mammalian cochlear hair cells. J. Neurosci 26, 10992–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Fettiplace R, Nam JH, and Ricci AJ (2009). Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat. Neurosci 12, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner JD, Lim NK, Schenck S, Duerst A, and Dutzler R (2014). X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212. [DOI] [PubMed] [Google Scholar]
- Corey DP, Akyuz N, and Holt JR (2018). Function and dysfunction of TMC channels in inner hair cells. Cold Spring Harb. Perspect. Med 10.1101/cshperspect.a033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DP, and Holt JR (2016). Are TMCs the mechanotransduction channels of vertebrate hair cells? J. Neurosci 36, 10921–10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DP, and Hudspeth AJ (1979). Ionic basis of the receptor potential in a vertebrate hair cell. Nature 281, 675–677. [DOI] [PubMed] [Google Scholar]
- Corey DP, and Hudspeth AJ (1983). Kinetics of the receptor current in bullfrog saccular hair cells. J. Neurosci 3, 962–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corns LF, Johnson SL, Kros CJ, and Marcotti W (2016). Tmc1 point mutation affects Ca2+ sensitivity and block by dihydrostreptomycin of the mechanoelectrical transducer current of mouse outer hair cells. J. Neurosci 36, 336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S, Feng S, Tien J, Peters CJ, Bulkley D, Lolicato M, Zhao J, Zuberbühler K, Ye W, Qi L, et al. (2017). Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KT, Cotton JA, Kirwan JD, Teeling EC, and Rossiter SJ (2012). Parallel signatures of sequence evolution among hearing genes in echolocating mammals: an emerging model of genetic convergence. Heredity (Edinb.) 108, 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson L, Reményi I, and Tusnády GE (2015). CCTOP: a consensus constrained TOPology prediction web server. Nucleic Acids Res. 43 (W1), W408–W412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenan RM, Nashmi R, Imoukhuede P, Just H, McKinney S, and Lester HA (2008). Subcellular trafficking, pentameric assembly, and subunit stoichiometry of neuronal nicotinic acetylcholine receptors containing fluorescently labeled alpha6 and beta3 subunits. Mol. Pharmacol 73, 27–41. [DOI] [PubMed] [Google Scholar]
- Erickson T, Morgan CP, Olt J, Hardy K, Busch-Nentwich E, Maeda R, Clemens R, Krey JF, Nechiporuk A, Barr-Gillespie PG, et al. (2017). Integration of Tmc1/2 into the mechanotransduction complex in zebrafish hair cells is regulated by transmembrane O-methyltransferase (Tomt). eLife 6, e28474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris HE, LeBlanc CL, Goswami J, and Ricci AJ (2004). Probing the pore of the auditory hair cell mechanotransducer channel in turtle. J. Physiol 558, 769–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasman GD (1989). Practical Handbook of Biochemistry and Molecular Biology (Boca Raton, Fla.: CRC Press; ). [Google Scholar]
- Fettiplace R (2016). Is TMC1 the hair cell mechanotransducer channel? Biophys. J 111, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, and Bateman A (2008). The Pfam protein families database. Nucleic Acids Res. 36, D281–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertsma ER, Chang YN, Shaik FR, Neldner Y, Pardon E, Steyaert J, and Dutzler R (2015). Structure of a prokaryotic fumarate transporter reveals the architecture of the SLC26 family. Nat. Struct. Mol. Biol 22, 803–808. [DOI] [PubMed] [Google Scholar]
- Gillespie PG, Dumont RA, and Kachar B (2005). Have we found the tip link, transduction channel, and gating spring of the hair cell? Curr. Opin. Neurobiol 15, 389–396. [DOI] [PubMed] [Google Scholar]
- Gohon Y, Dahmane T, Ruigrok RW, Schuck P, Charvolin D, Rappaport F, Timmins P, Engelman DM, Tribet C, Popot JL, and Ebel C (2008). Bacteriorhodopsin/amphipol complexes: structural and functional properties. Biophys. J 94, 3523–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunov D, Sturlese M, Nies F, Kluge M, Bellanda M, Battistutta R, and Oliver D (2014). Molecular architecture and the structural basis for anion interaction in prestin and SLC26 transporters. Nat. Commun 5, 3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wang Y, Zhang W, Meltzer S, Zanini D, Yu Y, Li J, Cheng T, Guo Z, Wang Q, et al. (2016). Transmembrane channel-like (tmc) gene regulates Drosophila larval locomotion. Proc. Natl. Acad. Sci. USA 113, 7243–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y, Kim DS, Pastan IH, and Lee B (2009). Anoctamin and transmembrane channel-like proteins are evolutionarily related. Int. J. Mol. Med 24, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haswell ES, Phillips R, and Rees DC (2011). Mechanosensitive channels: what can they do and how do they do it? Structure 19, 1356–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann SH, and Conti F (1992). Nonstationary noise analysis and application to patch clamp recordings. Methods Enzymol. 207, 131–148. [DOI] [PubMed] [Google Scholar]
- Höger U, and French AS (1999). Estimated single-channel conductance of mechanically-activated channels in a spider mechanoreceptor. Brain Res. 826, 230–235. [DOI] [PubMed] [Google Scholar]
- Holton T, and Hudspeth AJ (1986). The transduction channel of hair cells from the bull-frog characterized by noise analysis. J. Physiol 375, 195–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, and Schulten K (1996). VMD: visual molecular dynamics. J. Mol. Graph 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Johnson LS, Eddy SR, and Portugaly E (2010). Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M, and Petsko GA (1990). Molecular dynamics simulations in biology. Nature 347, 631–639. [DOI] [PubMed] [Google Scholar]
- Kawashima Y, Géléoc GS, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, and Griffith AJ (2011). Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest 121, 4796–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, Kurima K, Pan B, Griffith AJ, and Holt JR (2015). Transmembrane channel-like (TMC) genes are required for auditory and vestibular mechanosensation. Pflugers Arch. 467, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak P, and Müller U (2012). Sensing sound: molecules that orchestrate mechanotransduction by hair cells. Trends Neurosci. 35, 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Müller U, and Kachar B (2007). Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91. [DOI] [PubMed] [Google Scholar]
- Kelley LA, and Sternberg MJ (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, and Heller S (2003). TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics 4, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili-Araghi F, Gumbart J, Wen PC, Sotomayor M, Tajkhorshid E, and Schulten K (2009). Molecular dynamics simulations of membrane channels and transporters. Curr. Opin. Struct. Biol 19, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD Jr., and Pastor RW (2010). Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, and Sonnhammer EL (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, et al. (2002). Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet 30, 277–284. [DOI] [PubMed] [Google Scholar]
- Labay V, Weichert RM, Makishima T, and Griffith AJ (2010). Topology of transmembrane channel-like gene 1 protein. Biochemistry 49, 8592–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Holmgren M, Jurman ME, and Yellen G (1997). Gated access to the pore of a voltage-dependent K+ channel. Neuron 19, 175–184. [DOI] [PubMed] [Google Scholar]
- Lumpkin EA, Marquis RE, and Hudspeth AJ (1997). The selectivity of the hair cell’s mechanoelectrical-transduction channel promotes Ca2+ flux at low Ca2+ concentrations. Proc. Natl. Acad. Sci. USA 94, 10997–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R, Kindt KS, Mo W, Morgan CP, Erickson T, Zhao H, Clemens-Grisham R, Barr-Gillespie PG, and Nicolson T (2014). Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proc. Natl. Acad. Sci. USA 111, 12907–12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotti W, van Netten SM, and Kros CJ (2005). The aminoglycoside antibiotic dihydrostreptomycin rapidly enters mouse outer hair cells through the mechano-electrical transducer channels. J. Physiol 567, 505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, and Sander C (2011). Protein 3D structure computed from evolutionary sequence variation. PLoS ONE 6, e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano-Soto A, Moreno-Hagelsieb G, McLaughlin D, Ye ZS, Hendargo KJ, and Saier MH Jr. (2018). Bioinformatic characterization of the Anoctamin superfamily of Ca2+-activated ion channels and lipid scramblases. PLoS ONE 13, e0192851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton RE, Pheasant DJ, and Miller C (1996). Homodimeric architecture of a ClC-type chloride ion channel. Nature 383, 337–340. [DOI] [PubMed] [Google Scholar]
- Nashmi R, Dickinson ME, McKinney S, Jareb M, Labarca C, Fraser SE, and Lester HA (2003). Assembly of alpha4beta2 nicotinic acetylcholine receptors assessed with functional fluorescently labeled subunits: effects of localization, trafficking, and nicotine-induced upregulation in clonal mammalian cells and in cultured midbrain neurons. J. Neurosci 23, 11554–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Géléoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, and Holt JR (2013). TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Campbell EB, and MacKinnon R (2017). Structure of a CLC chloride ion channel by cryo-electron microscopy. Nature 541, 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino C, Neldner Y, Lam AK, Kalienkova V, Brunner JD, Schenck S, and Dutzler R (2017a). Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. eLife 6, e26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino C, Kalienkova V, Lam AKM, Neldner Y, and Dutzler R (2017b). Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 552, 421–425. [DOI] [PubMed] [Google Scholar]
- Peters C, Tsirigos KD, Shu N, and Elofsson A (2016). Improved topology prediction using the terminal hydrophobic helices rule. Bioinformatics 32, 1158–1162. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, and Schulten K (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popot JL, Althoff T, Bagnard D, Bane` res JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, et al. (2011). Amphipols from A to Z. Annu. Rev. Biophys 40, 379–408. [DOI] [PubMed] [Google Scholar]
- Reynolds SM, Käll L, Riffle ME, Bilmes JA, and Noble WS (2008). Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput. Biol 4, e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüsch A, Kros CJ, and Richardson GP (1994). Block by amiloride and its derivatives of mechano-electrical transduction in outer hair cells of mouse cochlear cultures. J. Physiol 474, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, MacDonald G, Rubel EW, and Raible DW (2006). Lateral line hair cell maturation is a determinant of aminoglycoside susceptibility in zebrafish (Danio rerio). Hear. Res. 213, 25–33. [DOI] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth FJ (1980). The variance of sodium current fluctuations at the node of Ranvier. J. Physiol 307, 97–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotboom DJ, Duurkens RH, Olieman K, and Erkens GB (2008). Static light scattering to characterize membrane proteins in detergent solution. Methods 46, 73–82. [DOI] [PubMed] [Google Scholar]
- Stauffer EA, and Holt JR (2007). Sensory transduction and adaptation in inner and outer hair cells of the mouse auditory system. J. Neurophysiol 98, 3360–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer EA, Scarborough JD, Hirono M, Miller ED, Shah K, Mercer JA, Holt JR, and Gillespie PG (2005). Fast adaptation in vestibular hair cells requires myosin-1c activity. Neuron 47, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L, and Haugland RP (1967). Energy transfer: a spectroscopic ruler. Proc. Natl. Acad. Sci. USA 58, 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Wang Y, Huang H, McGarvey PB, and Wu CH; UniProt Consortium (2015). UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, and Jaramillo F (1998). Getting the most out of noise in the central nervous system. Trends Neurosci. 21, 137–145. [DOI] [PubMed] [Google Scholar]
- Viklund H, and Elofsson A (2008). OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24, 1662–1668. [DOI] [PubMed] [Google Scholar]
- Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Balling R, et al. (2002). Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat. Genet 30, 257–258. [DOI] [PubMed] [Google Scholar]
- Wang Q, and Steyger PS (2009). Trafficking of systemic fluorescent gentamicin into the cochlea and hair cells. J. Assoc. Res. Otolaryngol 10, 205–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker JR (1963). Determination of molecular weights of proteins by gel filtration on Sephadex. Anal. Chem 35, 1950–1953. [Google Scholar]
- Whitlock JM, and Hartzell HC (2016). A pore idea: the ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid. Pflugers Arch. 468, 455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann C, Bellstedt P, and Görlach M (2013). CAPITO—a web server-based analysis and plotting tool for circular dichroism data. Bioinformatics 29, 1750–1757. [DOI] [PubMed] [Google Scholar]
- Wu C-S (2004). Handbook of Size Exclusion Chromatography and Related Techniques, Second Edition (Marcel Dekker; ), 1 online resource (xiv, 694 p.). [Google Scholar]
- Wu Z, and Müller U (2016). Molecular identity of the mechanotransduction channel in hair cells: not quiet there yet. J. Neurosci 36, 10927–10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt PJ (1993). Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta 272, 1–40. [Google Scholar]
- Yan R, Xu D, Yang J, Walker S, and Zhang Y (2013). A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Sci. Rep 3, 2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, and Zhang Y (2015). Protein structure and function prediction using I-TASSER. Curr. Protoc. Bioinformatics 52, 5.8.1–5.8.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Duran C, Qu Z, Cui YY, and Hartzell HC (2012). Explaining calcium-dependent gating of anoctamin-1 chloride channels requires a revised topology. Circ. Res 110, 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.