Abstract
Bacteria belonging to the Burkholderia cepacia complex (Bcc) are among the most important pathogens isolated from cystic fibrosis (CF) patients and in hospital acquired infections (HAI). Accurate identification of Bcc is questionable by conventional biochemical methods. Clonal typing of Burkholderia is also limited due to the problem with identification. Phenotypic identification methods such as VITEK2, protein signature identification methods like VITEK MS, Bruker Biotyper, and molecular targets such as 16S rRNA, recA, hisA and rpsU were reported with varying level of discrimination to identify Bcc. rpsU and/or 16S rRNA sequencing, VITEK2, VITEK MS and Bruker Biotyper could discriminate between Burkholderia spp. and non-Burkholderia spp. Whereas, Bcc complex level identification can be given by VITEK MS, Bruker Biotyper, and 16S rRNA/rpsU/recA/hisA sequencing. For species level identification within Bcc hisA or recA sequencing are reliable. Identification of Bcc is indispensable in CF patients and HAI to ensure appropriate antimicrobial therapy.
Keywords: Burkholderia cepacia complex, Cystic fibrosis, Hospital acquired infections, recA, hisA, rpsU
Background
Burkholderia cepacia is generally an environmental plant pathogen, second common to ESKAPE pathogens in humans. They were uncovered due to natural calamities and construction activities due to lack of infrastructure in developing nations. Differentiation of species within the B. cepacia complex (Bcc) can be particularly problematic, even with an extended panel of biochemical tests [1], as they are phenotypically very similar and most commercial bacterial identification systems cannot reliably distinguish between them. Further, reliable differentiation of these species from other related taxa, such as Ralstonia, Cupriavidus, Pandoraea, Achromobacter, Brevundimonas, Comamonas and Delftia species is challenging.
In most cases from developing nations, BCC has been misidentified as non-fermentative Gram-negative bacilli (NFGNB) especially Pseudomonas spp. [2, 3]. Due to which, reports on Bcc infections are rare in India [4].
Selection of literature for review
The articles were searched using PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) and Google Scholar. Multiple keywords were used for the literature search in combination or in alone. Some of the important keywords used for literature search were Burkholderia cepacia complex (Bcc), hospital acquired infections, phenotypic identification of Bcc, molecular identification of Bcc. B. cepacia, B. cenocepacia, phoenix, VITEK2, VITEK MS, Bruker biotyper, recA, hisA, rspU, 16S rRNA and WGS of B. cepacia/Bcc.
Main text
Classification of Burkholderia cepacia complex
Basic taxonomy
Walter H. Burkholder described a phytopathogenic bacterium causing onion rot in New York State in the mid-1940s and named the species ‘cepacia’ [5]. This was initially known as Pseudomonas cepacia, later in 1992 included in the Betaproteobacteria class, with Burkholderiales order and Burkholderiaceae family as Burkholderia cepacia [6]. Burkholderia includes former rRNA group II pseudomonads (Pseudomonas gladioli, Pseudomonas mallei, Pseudomonas pseudomallei, and Pseudomonas caryophylli), except Pseudomonas pickettii and Pseudomonas solanacearum, which were later grouped under the genus Ralstonia [7]. Burkholderia species were known as plant pathogens and soil bacteria, except B. mallei and B. pseudomallei, which are humans and animal pathogens [8].
The genus now includes 22 validly described species: B. cepacia (the type species), Burkholderia caryophylli, Burkholderia mallei, Burkholderia pseudomallei, Burkholderia gladioli, Burkholderia plantarii, Burkholderia glumae, Burkholderia vietnamiensis, Burkholderia andropogonis, Burkholderia multivorans, Burkholderia glathei, Burkholderia pyrrocinia, Burkholderia thailandensis, Burkholderia graminis, Burkholderia phenazinium, Burkholderia caribensis, Burkholderia kururiensis, Burkholderia ubonensis, Burkholderia caledonica, Burkholderia fungorum, Burkholderia stabilis, and Burkholderia ambifaria [9].
Since the mid-1990s, heterogeneity was noted among the B. cepacia strains isolated from different ecological niches. This caused problems in accurate identification of B. cepacia isolates, and evaluation of the techniques used showed that they were either not very sensitive, not very specific, or neither sensitive nor specific [10–13].
Further, Vandamme et al. [14] evaluated a polyphasic taxonomic approach to demonstrate that presumed “B. cepacia” from CF patients and other sources were different and belonged to five distinct genomovars (phenotypically similar genomic species). This includes, B. cepacia genomovar I, B. multivorans genomovar II, genomovar III, B. stabilis genomovar IV and B. vietnamiensis genomovar V. Initially these five genomic species were collectively referred to as the B. cepacia complex (Bcc). Subsequent polyphasic taxonomic studies identified genomovar VI and B. ambifaria genomovars VII which added to Bcc [15, 16]. In addition, B. pyrrocinia was added to Bcc [17].
Ralstonia, Cupriavidus, Pandoraea, Achromobacter, Brevundimonas, Comamonas and Delftia are the most common genus those are closely related to the Burkholderia and cause problems in accurate identification of Bcc. These are hitherto referred as non-Burkholderia spp. in this manuscript. Similarly, Burkholderia spp. (B. humptydooensis and B. pseudomallei complex) which interferes in correct identification of Bcc are referred as non-Bcc.
Molecular phylogeny
Previously, different species within the B. cepacia complex had shown to have DNA–DNA hybridisation values between 30 and 60%, while strains of same species showed values > 70%. Whereas, values obtained with non-Bcc Burkholderia were below 30% [14–16, 18–20]. The DNA relatedness is rated as high (> 70%) in strains of same species, low (30–60%) but significant below the species level, and non-significant (< 30%).
Coenye et al. [15], has compared the 16S rDNA sequences of B. cepacia complex and related species, where, the similarities of strains within B. cepacia complex were higher (> 97.7%) compared to other Burkholderia species (< 97.0%).
Biochemical reactions
Different media composition were in use for years to selectively isolate B. cepacia complex from samples of CF patients. This includes, P. cepacia medium (PC agar) (300 U of polymyxin B/ml and 100 µg of ticarcilline/ml) [21]; Oxidation-fermentation agar with lactose and polymyxin B (OFPBL agar) (300 U of polymyxin B/ml and 0.2 U of bacitracin/ml) [22], and B. cepacia selective agar (BCSA) (1% lactose and 1% sucrose in an enriched base of casein and yeast extract with 600 U of polymyxin B/ml, 10 µg of gentamicin/ml, and 2.5 µg of vancomycin/ml) [23]. BCSA was proven effective than the other two in recovering B. cepacia complex from CF respiratory specimens by inhibiting growth of other organisms [24]. Though, B. gladioli and Ralstonia spp. are exceptions which could grow on BCSA. On isolation, few biochemical reactions used to differentiate B. cepacia complex, B. gladioli, Pandoraea spp., R. pickettii, A. xylosoxidans, and S. maltophilia are enlisted in Table 1.
Table 1.
Biochemical characteristics to differentiate B. cepacia complex, B. gladioli, Pandoraea spp., R. pickettii, A. xylosoxidans, and S. maltophilia
| B. cepacia | B. gladioli | Pandoraea species | R. pickettii | A. xylosoxidans | S. maltophilia | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomovar I | Genomovar II | Genomovar III | Genomovar IV | Genomovar V | Genomovar VI | Genomovar VII | ||||||
| Oxidase | + | + | + | + | + | + | + | ‒ | v | + | + | ‒ |
| Oxidation of: | ||||||||||||
| Sucrose | v | ‒ | v | ‒ | + | ‒ | + | ‒ | ‒ | ‒ | ‒ | v |
| Adonitol | v | + | v | v | – | + | + | + | – | – | – | – |
| Lactose | v | + | v | + | + | + | + | ‒ | ‒ | ‒ | ‒ | + |
| Lysine decarboxylase | + | v | + | + | + | ‒ | + | ‒ | ‒ | ‒ | ‒ | + |
| Ornithinie decarboxylase | v | ‒ | v | + | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ | ‒ |
| Gelatine liquefaction | v | ‒ | v | + | ‒ | ‒ | + | v | ‒ | ‒ | ‒ | + |
| Aesculine hydrolysis | v | ‒ | v | ‒ | ‒ | ‒ | v | v | ‒ | v | ‒ | + |
| β-Galactosidase activity | + | + | + | ‒ | + | + | + | + | ‒ | ‒ | ‒ | + |
| Growth at 42 °C | v | + | v | ‒ | + | + | v | ‒ | v | v | NK | v |
| β-Hemolysis | ‒ | ‒ | ‒ | ‒ | v | ‒ | v | ‒ | ‒ | ‒ | NK | NK |
Recent developments had led to invention of automated/commercial test systems for pathogen identification. However, there are several reports pertaining to inability of these commercial systems to identify or differentiate B. cepacia complex isolates from other Burkholderia spp. [25].
Bcc in cystic fibrosis
Most often, cases with fulminating pneumonic infection along with fever and respiratory failure, occasional association with septicaemia, is known as “cepacia syndrome” [26]. The overwhelming B. cepacia complex infections in cystic fibrosis patients have prompted an unusual number of studies and variety of data. B. cepacia was also frequently encountered in nosocomial outbreaks due to contaminated disinfectants, nebulizer solutions, mouth wash, medical devices and intravenous solutions due to contamination of lipid emulsion stoppers [27]. Though, B. multivorans and B. cenocepacia were reported predominant amongst CF patients than non-CF patients as reported from United States, Canada, Italy and Australia [16, 28, 29].
Problems in accurate identification of Burkholderia spp.
Phenotypic tests either manual or automated commercial systems were in use to identify Bcc in routine clinical laboratories. Though, species level identification is not achieved due to high similarity of biochemical results between species. Automated identification systems including Phoenix, VITEK 2, VITEK MS and Bruker identifies Bcc, non-Bcc and non-Burkholderia spp. at different specificities (Table 2) [30–33].
Table 2.
Biochemical and molecular identification of Burkholderia cepacia complex in hospital acquired infections
| Methods | Target | Identification | Remarks | |
|---|---|---|---|---|
| Phenotypic methods | Conventional biochemical method | Catalase, Gluconate, Malate, Phenylacetate, leucine arylamidase activity | Overlapping biochemical profiles for Bcc, Ralstonia spp. and Pandoraea spp. | Bcc and non-Bcc cannot be distinguished |
| Phoenix | Biochemicals—automated | Cannot identify Ralstonia pickettii | Misidentification rate for Bcc is 23% | |
| VITEK 2 | Biochemicals—automated | Can identify Ralstonia pickettii (83%) | Misidentification rate for Bcc is 12% | |
| Protein signature | VITEK MS | Mass spectrogram of the protein | Genus level identification of B. cepacia—55–63% R. pickettii identification—85–100% Pandoraea spp.—87% |
Species within Bcc cannot be distinguished |
| Bruker Biotyper | Mass spectrogram of the protein | Agreement between Bruker Biotyper and recA sequencing Genus level—100% Species level—76.9% B. cenocepacia—95.8–100% B. multivorans—78.5% B. contaminans—0% B. vietnamiensis—100% B. cepacia—30–33.3% |
Can identify and discriminate Bcc from non-Burkholderia spp. Few species within Bcc cannot be distinguished |
|
| Molecular targets | recA | DNA recombinase enzyme for DNA repair | Promising for differentiation of Burkholderia species including Bcc | Non-Bcc cannot be distinguished |
| hisA | Encodes for an enzyme involved in histidine biosynthesis | Could discriminate among the Bcc species | Non-Bcc cannot be distinguished | |
| rspU | Coding for a ribosomal protein S21 | Burkholderia spp. and non-Burkholderia spp. can be distinguished | Species within Bcc cannot be distinguished | |
| 16S rRNA | Component of the 30S small subunit of a prokaryotic ribosome | Burkholderia spp. and non-Burkholderia spp. can be distinguished | Unacceptable for discrimination of Bcc intra-species |
There is considerable interest in recent days on the reliability of MALDI-TOF MS for accurate bacterial identification. It is based on the spectral analysis of bacterial proteins, mainly ribosomal proteins, ionized by laser irradiation of the bacterial cell. Fehlberg et al. [25] has evaluated the performance of MALDI-TOF MS for species identification of Bcc clinical isolates in comparison to recA sequencing. MALDI-TOF MS results were 100% in concordant with recA sequencing for genus level identification (n = 91), while 76.9% (n = 70) concordance was seen for species level identification. Another study by Gautam et al. [34], has compared MALDI-TOF MS with an expanded MLST and recA sequencing for Bcc identification. MALDI-TOF MS exhibited 100% concordance for genus identification and 82% for species level identification.
The accuracy in the identification and differentiation of Burkholderia spp. in clinical specimens with the close neighbours Pandoraea, Cupriavidus and Ralstonia is essential for the treatment of patients. These three are the most prevalent genera identified outside Burkholderia genus. Most of the time these are phenotypically misidentified as Bcc.
Pandoraea species have been reported from both cystic fibrosis (CF) and non-CF patients. The invasive potential of this genus can be understood through various reported cases of Pandoraea bacteraemia caused by P. pnomenusa, P. apista, P. pulmonicola and P. sputorum [35–40], where identification was a major setback when conventional biochemical methods were used.
The Ralstonia genus includes R. pickettii and R. solanacearum (formerly Burkholderia pickettii and B. solanacearum), R. insidiosa, and R. mannitolilytica, where R. pickettii is still regarded as the main pathogenic species [41]. Though R. pickettii, is considered with minor clinical significance, many instances of infections are reported in the literature. Due to high similarity between R. pickettii and Bcc, many of the Bcc cases might have been misidentified which are actually R. pickettii [42]. Contaminated solutions including water for injection, saline solutions made with purified water, and sterile drug solutions were regarded as the cause of R. pickettii infections in many of the cases. Major conditions associated with R. pickettii infection are bacteraemia/septicaemia and respiratory infections/pneumonia [41, 43, 44].
Very often, Ralstonia and Pandaroeae are misidentified as Bcc. These genus are very closely related to Burkholderia spp., such that they cannot be distinguished by standard biochemical method [35]. The species include Bcc (B. cepacia, B. multivorans, B. cenocepacia, B. vietnamiensis, B. stabilis, B. ambifaria, B. dolosa, B. anthina, B. pyrrocinia and B. ubonensis), B. humptydooensis, Cupriavidus spp., Pandoraea spp., and B. pseudomallei.
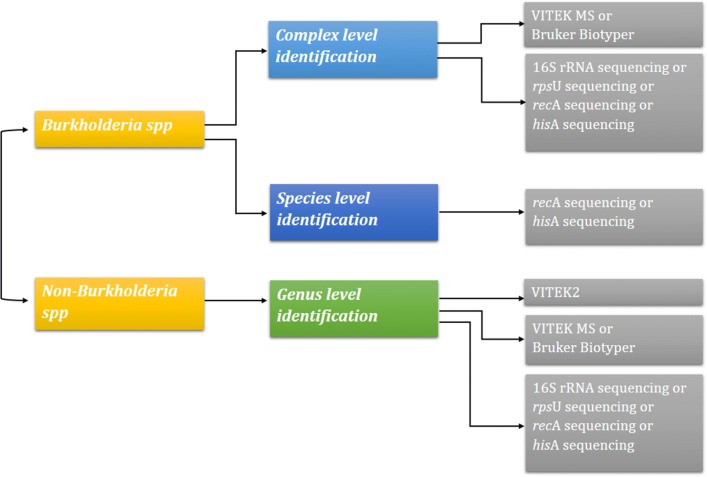
Bcc and non-Burkholderia spp. could not be distinguished by conventional biochemical methods. Due to problem with identification, clonal typing of Burkholderia is questionable. Molecular targets such as 16S rRNA, recA, hisA and rpsU were reported to increase the discrimination of Bcc. Representation of various techniques and its ability to accurately identify Bcc is given in Fig. 1.
Fig. 1.
Algorithm depicting the methods for accurate identification of Burkholderia at genus level (near neighbouring genus), cepacia complex level (Bcc) and species level (within Bcc)
Need for molecular identification of Bcc
Isolates from CF patients with persistent pathogenic colonization often lose their characteristic phenotypes or growth conditions which leads to difficulty in accurate identification of Bcc. To overcome this, molecular identification is required to distinguish species within Bcc and from the related genus/species. Though molecular targets for identification are not reliable when used individually, a multi-target approach is essential to improve the identification of Bcc and non-Bcc organisms. Some of the reported molecular targets are hisA, rpsU, recA and 16S rRNA. Discriminating ability on genus/complex/species level using these targets were listed in Table 2.
hisA and rpsU gene sequencing
Sequencing of hisA gene, encodes for an enzyme involved in histidine biosynthesis was reported to distinguish species within Bcc [45]. Neighbour-joining method analysis of 134 Bcc organisms revealed high degree of sequence similarity between strains of same species. Meanwhile, each species was clearly separated from each other. The hisA based analysis separated 17 Bcc species in different clusters (including 4 lineage divisions of B. cenocepacia) with high bootstrap values (> 75%) [45]. Burkholderia strains used for hisA based analysis were previously identified using a polyphasic taxonomy or recA sequencing [46, 47].
Similarly, rpsU was recognized to identify different species among the Burkholderia genus [48]. Frickmann et al. [48] has employed rpsU sequencing method to compare Burkholderia strains of known identity from ATCC (American Type Culture Collection, Manassas, Virginia, USA), DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany), JCM (Japan Collection of Microorganisms, Tsukuba, Ibaraki Prefecture, Japan), BCCM/LMG (Bacteria Collection, Ghent, Belgium), and NCTC (National Collection of Type Cultures, Porton Down, UK). Also few clinical strains were included in the analysis for comparison, after ensuring their identity using recA sequencing. rpsU sequences formed four clusters including B. plantarii, B. glumae, B. cocovenenans and B. gladioli in cluster I, the Burkholderia pseudomallei complex (B. mallei, B. pseudomallei, and B. thailandensis) in cluster II, B. caryophylli, B. multivorans, P. norimbergensis, B. ubonensis, B. stabilis, B. cenocepacia, B. cepacia, B. pyrrocinia, B. ambifaria, B. anthina, B. vietnamiensis and B. dolosa in cluster III, and B. sacchari, B. graminis, B. fungorum, B. phytofirmans, B. xenovorans, B. phenoliruptrix, B. phenazinium, B. caribensis, B. hospita and B. phymatum in cluster IV. B. glathei, B. caledonica and B. kururensis were observed as outliers.
Moreover, the rpsU sequence homology for Burkholderia and Pandorea was > 86%. Most of the clinical pathogens of Bcc belongs to cluster III of rpsU sequencing, where B. caryophylli, B. multivorans, and P. norimbergensis had identical sequences and B. cenocepacia clustered with B. cepacia. Limitation of rpsU sequencing is it could not reliably discriminate Burkholderia spp. at the species level as single target.
recA gene sequencing
recA is another well-known target promising for differentiation of Burkholderia species [46]. recA can differentiate the following 19 species of Burkholderia namely, B. pseudomallei, B. mallei, B. thailandensis, B. humptydooensis, B. oklahomensis, B. oklahomensis-like, B. ubonensis, B. ambifaria, B. multivorans, B. vietnamiensis, B. fungorum, B. glumae, B. cepacia, B. xenovorans, B. dolosa, B. gladioli and Bcc [49]. However, non-Burkholderia spp. cannot be distinguished by recA sequencing. Burkholderia strains used for evaluation of recA sequencing were characterised using whole-cell protein profile analysis and a polyphasic approach [47, 50].
16S rRNA sequencing
16S rRNA sequencing is one important option for differentiating non-Burkholderia spp. from Burkholderia spp. [49]. The 16S rRNA similarity between Burkholderia spp. and non-Burkholderia spp. is given in Table 3. In a study from environmental sample, the different species of Burkholderia including non-Burkholderia spp. were found in the same consortium suggesting that same environmental niche hosts the sharing of genes through lateral gene transfer [51]. Due to these challenges in identification, the clonality of these species is also difficult to investigate, as the MLST housekeeping genes are species specific. As of now, MLST database is available only for Bcc and B. pseudomallei. HAI outbreaks caused due to non-Burkholderia spp. could not be typed.
Table 3.
Similarity of 16S rRNA sequences between Burkholderia and non-Burkholderia spp.
| % Similarity of 16S rRNA | Pandoraea | B. stabilis | B. cenocepacia | B. cepacia | B. multivorans | B. pyrrocinia | B. vietnamiensis | B. ambifaria | B. anthina | B. humptydooensis | B. pseudomallei | B. mallei | B. cupriavidus | Ralstonia |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1: Pandoraea | 100 | |||||||||||||
| 2: B. stabilis | 95.16 | 100 | ||||||||||||
| 3: B. cenocepacia | 94.02 | 98.32 | 100 | |||||||||||
| 4: B. cepacia | 95.1 | 98.38 | 98.32 | 100 | ||||||||||
| 5: B. multivorans | 95.28 | 98.46 | 97.43 | 99.79 | 100 | |||||||||
| 6: B. pyrrocinia | 95.72 | 99.09 | 97.91 | 99.02 | 99.1 | 100 | ||||||||
| 7: B. vietnamiensis | 95.48 | 98.67 | 97.92 | 99.44 | 99.4 | 99.17 | 100 | |||||||
| 8: B. ambifaria | 95.76 | 99.23 | 97.92 | 99.02 | 99.12 | 99.45 | 99.32 | 100 | ||||||
| 9: B. anthina | 95.85 | 99.33 | 99.7 | 99.11 | 99.19 | 99.41 | 99.56 | 99.7 | 100 | |||||
| 10: B. humptydooensis | 94.86 | 97.54 | 96.38 | 97.55 | 97.59 | 97.86 | 98.02 | 98.23 | 98.22 | 100 | ||||
| 11: B. pseudomallei | 95.05 | 97.47 | 96.52 | 97.83 | 97.83 | 97.99 | 98.25 | 98.29 | 98.08 | 98.92 | 100 | |||
| 12: B. mallei | 95.07 | 97.4 | 96.45 | 97.76 | 97.79 | 97.93 | 98.22 | 98.23 | 98 | 98.89 | 99.87 | 100 | ||
| 13: B. cupriavidus | 91.5 | 90.99 | 91.29 | 91.29 | 91.21 | 91.14 | 91.14 | 91.21 | 91.56 | 91.49 | 91.63 | 91.63 | 100 | |
| 14: Ralstonia | 92.39 | 90.53 | 89.58 | 90.97 | 91.03 | 91.16 | 91.37 | 91.23 | 91.49 | 91.56 | 91.63 | 91.63 | 94.66 | 100 |
Whole genome sequences of B. cepacia
Due to recent developments in the molecular genetics of bacteria, usage of whole genome sequences (WGS) of the bacterial pathogens is gaining interest among clinical microbiologists. The WGS data helps in typing of the pathogens and in identifying the evolutionary pattern of the organism based on whole genome single nucleotide polymorphisms (SNPs). Till date 102 genome sequences have been deposited in NCBI for B. cepacia. This includes complete genomes and shotgun genome sequences. Further baseline data on B. cepacia WGS will help to identify region specific clones, which will be handy in identifying an outbreak situation.
Antimicrobial resistance mechanisms and therapy of Bcc
The mechanisms of antibiotic resistance of Bcc species have been intensively studied. Major resistance mechanism in Bcc is due to efflux pump overexpression mostly by members of the resistance-nodulation-division (RND) family [52–54]. B. cenocepacia strain J2315 was reported to encode 16 RND efflux systems [55, 56].
Ceftazidime and other extended-spectrum cephalosporins are the reliable treatment options for Bcc due to intrinsic resistance to many other classes of antimicrobials. Bcc were reported with class A β-lactamases conferring resistance to β-lactam antibiotics such as ceftazidime. This was first described in B. cepacia as the PenA-PenR system [57], which were later known as PenB and PenR (AmpR) [58]. Due to their complex role in activation of penB and ampC targets in the presence of an antibiotic susceptibilities to ceftazidime, cefotaxime, and meropenem were greatly reduced [58, 59].
Class A PenA β-lactamase was also reported in B. cenocepacia which is located on chromosome 2 and their genetic environment are similar to that of B. pseudomallei [60]. However, the B. cenocepacia enzyme has not yet been shown to be involved in β-lactam resistance. In a study by Hwang and Kim [58], a B. cenocepacia strain J2315 PenB β-lactamase had shown a Ser72Tyr substitution, due to which B. cenocepacia has intrinsic clavulanate resistance [58].
In addition, B. multivorans was reported to have a PenA enzyme (Bmul_3689 in B. multivorans ATCC 17616), that is closely related to PenB reported in Bcc [58, 61]. This PenB is also similar to KPC-2 a significant carbapenemase [62]. However, the B. multivorans enzyme is an inhibitor-resistant carbapenemase, unlike in B. pseudomallei which is an extended spectrum β-lactamase. Though the active role of PenA in clinical B. multivorans is it yet established. There were also reports on difference in efflux pump and outer membrane protein mediated resistance especially for colistin in Bcc and B. pseudomallei complex (Bpc) [63]. Due to these multiple difference in their resistance mechanisms, it is imperative to accurately identify Bcc from other Burkholderia spp for appropriate therapy.
The choice for antimicrobial therapy is usually chosen based on in vitro susceptibility, while duration of therapy be based upon clinical and microbiologic response. Use of combination regimen is commonly reported for Bcc [64]. However, it is still uncertain as the evidences were mostly limited to in vitro studies or small clinical experiences. For serious infection with susceptible strains, a two-drug combination of parenteral trimethoprim-sulfamethoxazole (5 mg/kg trimethoprim component every 6–12 h) plus a β-lactam (e.g., ceftazidime, piperacillin, meropenem) or a fluoroquinolone should be utilized [65]. For serious infection with trimethoprim-sulfamethoxazole-resistant strains or sulfa drug allergy, combination therapy guided by in vitro susceptibility results should be administered [66]. In a study by Blumer et al. [67], in 102 CF patients, meropenem/tobramycin and ceftazidime/tobramycin improved clinical status and reduced bacterial burden in 96 and 92% of treated patients, respectively.
Bonacorsi et al. had proven enhanced bactericidal activity of ciprofloxacin in combination with other agents [68]. Further, triple antimicrobial combination based on meropenem was suggested useful than double or single agents [69].
Macrolides in combination with other antimicrobials had shown moderate synergism [70], while specific combinations including fosfomycin/tobramycin exhibited poor activity against Bcc [71].
Conclusions
Conventional phenotypic methods could not discriminate Bcc and related genus, as there is an overlap in the biochemical characteristics. A single molecular target for differentiation of Bcc from non-Bcc and non-Burkholderia spp. is not reliable, while two or more molecular targets significantly improves the species level discrimination in Bcc. rpsU and/or 16S rRNA sequencing, VITEK2, VITEK MS and Bruker Biotyper could discriminate between Burkholderia spp. and non-Burkholderia spp. Whereas, Bcc complex level identification can be given by VITEK MS, Bruker Biotyper, 16S rRNA/rpsU/recA/hisA sequencing. For species level identification within Bcc hisA or recA sequencing are reliable. Recent advancements in genome sequencing using SNP phylogeny might help to accurately identify the clone of Bcc from non-Bcc and non-Burkholderia spp. Such identification is necessary to help in timely diagnosis of hospital acquired infections and to provide appropriate antimicrobial therapy.
Authors’ contributions
DRNK and VB conceptualized the article. VB and DRNK did the review of literature. The article was written, reviewed and finalised by VB and DRNK. Both authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
No funding was obtained for this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Henry DA, Mahenthiralingam E, Vandamme P, Coenye T, Speert DP. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:1073–1078. doi: 10.1128/JCM.39.3.1073-1078.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araque-Calderon Y, Miranda-Contreras L, Rodriguez-Lemoine V, Palacios-Pru EL. Antibiotic resistance patterns and SDS-PAGE protein profiles of Burkholderia cepacia complex isolates from nosocomial and environmental sources in Venezuela. Med Sci Monit. 2008;14(2):BR49–55. [PubMed] [Google Scholar]
- 3.Gautam V, Ray P, Vandamme P, Chatterjee SS, Das A, Sharma K, et al. Identification of lysine positive non-fermenting gram negative bacilli (Stenotrophomonas maltophilia and Burkholderia cepacia complex) Indian J Med Microbiol. 2009;27(2):128–133. doi: 10.4103/0255-0857.49425. [DOI] [PubMed] [Google Scholar]
- 4.Gautam V, Singh M, Singhal L, Kaur M, Kumar A, Ray P. Burkholderia cepacia complex in Indian cystic fibrosis patients. Indian J Med Res. 2012;136(5):882–883. [PMC free article] [PubMed] [Google Scholar]
- 5.Burkholder WH. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;40:115–117. [Google Scholar]
- 6.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, et al. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia Palleroni and Holmes 1981 comb. nov. Microbiol Immunol. 1992;36(12):1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 7.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff, comb nov, Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 1973;1995(39):897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- 8.Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5(9):719–729. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 9.Coenye T, Vandamme P, Govan JR, Lipuma JJ. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39(10):3427–3436. doi: 10.1128/JCM.39.10.3427-3436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiska DL, Kerr A, Jones MC, Caracciolo JA, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan P. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative, nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–891. doi: 10.1128/jcm.34.4.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen GY, Stull TL, Burns JL. Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. J Clin Microbiol. 1993;31:788–792. doi: 10.1128/jcm.31.4.788-792.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu PYF, Dhi ZY, Lau YJ, Hu BS, Shyr JM, Tsai WS, Lin YH, Tseng CY. Comparison of different PCR approaches for characterization of Burkholderia (Pseudomonas) cepacia isolates. J Clin Microbiol. 1995;33:3304–3307. doi: 10.1128/jcm.33.12.3304-3307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpson IN, Finlay J, Winstanley DJ, Dehwurst N, Nelson JW, Butler SL, Govan JRW. Multi-resistance isolates possessing characteristics of both Burkholderia (Pseudomonas) cepacia and Burkholderia gladioli from patients with cystic fibrosis. J Antimicrob Chemother. 1994;34:353–361. doi: 10.1093/jac/34.3.353. [DOI] [PubMed] [Google Scholar]
- 14.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan JRW. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 15.Coenye T, LiPuma JJ, Henry D, Hoste B, Vandemeulebroucke K, Gillis M, Speert DP, Vandamme P. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int J Syst Evol Microbiol. 2001;51:271–279. doi: 10.1099/00207713-51-2-271. [DOI] [PubMed] [Google Scholar]
- 16.Coenye T, Mahenthiralingam E, Henry D, LiPuma JJ, Laevens S, Gillis M, Speert DP. Vandamme P. Burkholderia ambifaria sp. nov, a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int J Syst Evol Microbiol. 2001;51:1481–1490. doi: 10.1099/00207713-51-4-1481. [DOI] [PubMed] [Google Scholar]
- 17.Balandreau J, Viallard V, Cournoyer B, Coenye T, Laevens S, Vandamme P. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl Environ Microbiol. 2001;67:982–985. doi: 10.1128/AEM.67.2.982-985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coenye T, Holmes B, Kersters K, Govan JRW, Vandamme P. Burkholderia cocovenenans (van Damme et al. 1960) Gillis et al. 1995 and Burkholderia vandii Urakami et al. 1994 are junior subjective synonyms of Burkholderia gladioli (Severini 1913) Yabuuchi et al. 1993 and Burkholderia plantarii (Azegami et al. 1987) Urakami et al. 1994, respectively. Int J Syst Bacteriol. 1999;49:37–42. doi: 10.1099/00207713-49-1-37. [DOI] [PubMed] [Google Scholar]
- 19.Coenye T, Schouls LM, Govan JRW, Kersters K, Vandamme P. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int J Syst Bacteriol. 1999;49:1657–1666. doi: 10.1099/00207713-49-4-1657. [DOI] [PubMed] [Google Scholar]
- 20.Gillis M, Van Van T, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp nov for N2- fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. doi: 10.1099/00207713-45-2-274. [DOI] [Google Scholar]
- 21.Gilligan PH, Gage PA, Bradshaw LM, Schidlow DV, DeCicco BT. Isolation medium for the recovery of Pseudomonas cepacia from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 1985;22:5–8. doi: 10.1128/jcm.22.1.5-8.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch DF, Muszynski MJ, Pai CH, Marcon MJ, Hribar MM, Gilligan PH, Matsen JM, Ahlin PG, Hilman BC, Chartrand SA. Selective and differential medium for the recovery of Pseudomonas cepacia from the respiratory tract of patients with cystic fibrosis. J Clin Microbiol. 1987;25:1730–1734. doi: 10.1128/jcm.25.9.1730-1734.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry DA, Campbell ME, LiPuma JJ, Speert DP. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry D, Campbell M, McGimpsey C, Clarke A, Louden L, Burns JL, Roe MH, Vandamme P, Speert D. Comparison of isolation media for recovery of Burkholderia cepacia complex from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 1999;37:1004–1007. doi: 10.1128/jcm.37.4.1004-1007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehlberg LCC, Lucas Henrique Sales Andrade, Diego Magno Assis, Rosana Helena Vicente Pereira, Ana Cristina Gales, Elizabeth Andrade Marques. Performance of MALDI-ToF MS for species identification of Burkholderia cepacia complex clinical isolates. Diagn Microbiol Infect Dis. 2013;77:126–8. [DOI] [PubMed]
- 26.Jones AM, Dodd ME, Govan JR, Barcus V, Doherty CJ, Morris J, et al. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax. 2004;59(11):948–951. doi: 10.1136/thx.2003.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antony B, Cherian EV, Boloor R, Shenoy KV. A sporadic outbreak of Burkholderia cepacia complex bacteremia in pediatric intensive care unit of a tertiary care hospital in coastal Karnataka. South India. Indian J Pathol Microbiol. 2016;59:197–199. doi: 10.4103/0377-4929.182010. [DOI] [PubMed] [Google Scholar]
- 28.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 29.Reik R, Spilker T, Lipuma JJ. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J Clin Microbiol. 2005;43:2926–2928. doi: 10.1128/JCM.43.6.2926-2928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marko DC, Saffert RT, Cunningham SA, Hyman J, Walsh J, Arbefeville S, et al. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of nonfermenting gram-negative bacilli isolated from cultures from cystic fibrosis patients. J Clin Microbiol. 2012;50(6):2034–2039. doi: 10.1128/JCM.00330-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alby K, Gilligan PH, Miller MB. Comparison of Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) Mass Spectrometry Platforms for the Identification of Gram-Negative Rods from Patients with Cystic Fibrosis. J Clin Microbiol. 2013;51(11):3852–3854. doi: 10.1128/JCM.01618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manji R, Bythrow M, Branda JA, Burnham CA, Ferraro MJ, Garner OB, et al. Multi-center evaluation of the VITEK® MS system for mass spectrometric identification of non-Enterobacteriaceae Gram-negative bacilli. Eur J Clin Microbiol Infect Dis. 2014;33(3):337–346. doi: 10.1007/s10096-013-1961-2. [DOI] [PubMed] [Google Scholar]
- 33.Plongla R, Panagea T, Pincus DH, Jones MC, Gilligan PH. Identification of Burkholderia and uncommon glucose non-fermenting Gram-negative Bacilli isolated from patients with cystic fibrosis using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) J Clin Microbiol. 2016;54(12):3071–3072. doi: 10.1128/JCM.01623-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautam V, Sharma M, Singhal L, Kumar S, Kaur P, Tiwari R, Ray P. MALDI-TOF mass spectrometry: an emerging tool for unequivocal identification of non-fermenting Gram-negative bacilli. Indian J Med Res. 2017;145(5):665–672. doi: 10.4103/ijmr.IJMR_1105_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stryjewski ME, LiPuma JJ, Messier RH, Reller LB, Alexander BD. Sepsis, multiple organ failure, and death due to Pandoraea pnomenusa infection after lung transplantation. J Clin Microbiol. 2003;41(5):2255–2257. doi: 10.1128/JCM.41.5.2255-2257.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jørgensen IM, Johansen HK, Frederiksen B, Pressler T, Hansen A, Vandamme P, et al. Epidemic spread of Pandoraea apista, a new pathogen causing severe lung disease in cystic fibrosis patients. Pediatr Pulmonol. 2003;36:439–446. doi: 10.1002/ppul.10383. [DOI] [PubMed] [Google Scholar]
- 37.Atkinson RM, LiPuma JJ, Rosenbluth DB, Dunne WM. Chronic colonization with Pandoraea apista in cystic fibrosis patients determined by repetitive-element-sequence PCR. J Clin Microbiol. 2006;44:833–836. doi: 10.1128/JCM.44.3.833-836.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pimentel JD, MacLeod C. Misidentification of Pandoraea sputorum isolated from sputum of a patient with cystic fibrosis and review of Pandoraea species infections in transplant patients. J Clin Microbiol. 2008;46(9):3165–3168. doi: 10.1128/JCM.00855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caraher E, Collins J, Herbert G, Murphy PG, Gallagher CG, Crowe MJ, et al. Evaluation of in vitro virulence characteristics of the genus Pandoraea in lung epithelial cells. J Med Microbiol. 2008;57:15–20. doi: 10.1099/jmm.0.47544-0. [DOI] [PubMed] [Google Scholar]
- 40.Martina PF, Martínez M, Frada G, Alvarez F, Leguizamón L, Prieto C, et al. First time identification of Pandoraea sputorum from a patient with cystic fibrosis in Argentina: a case report. BMC Pulm Med. 2017;17:33. doi: 10.1186/s12890-017-0373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryan MP, Pembroke JT, Adley CC. Ralstonia pickettii: a persistent Gram-negative nosocomial infectious organism. J Hosp Infect. 2006;62:278–284. doi: 10.1016/j.jhin.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Waugh JB, Granger WM, Gaggar A. Incidence, relevance and response for Ralstonia respiratory infections. Clin Lab Sci. 2010;23(2):99–106. [PMC free article] [PubMed] [Google Scholar]
- 43.Orme J, Rivera-Bonilla T, Loli A, Blattman NN. Native valve endocarditis due to Ralstonia pickettii: a case report and literature review. Case Rep Infect Dis. 2015;2015:324675. doi: 10.1155/2015/324675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lai HW, Shen YH, Chien LJ, Tseng SH, Mu JJ, Chan YJ, et al. Outbreak of Ralstonia pickettii bacteremia caused by contaminated saline solution in Taiwan. Am J Infect Control. 2016;44(10):1191–1192. doi: 10.1016/j.ajic.2016.03.074. [DOI] [PubMed] [Google Scholar]
- 45.Papaleo MC, Perrin E, Maida I, Fondi M, Fani R, Vandamme P. Identification of species of the Burkholderia cepacia complex by sequence analysis of the hisA gene. J Med Microbiol. 2010;59(10):1163–1170. doi: 10.1099/jmm.0.019844-0. [DOI] [PubMed] [Google Scholar]
- 46.Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, et al. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38:3165–3173. doi: 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, et al. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 48.Frickmann H, Neubauer H, Loderstaedt U, Derschum H, Hagen RM. rpsU-based discrimination within the genus Burkholderia. Eur J Microbiol Immunol (Bp). 2014;4(2):106–116. doi: 10.1556/EuJMI.4.2014.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ginther JL, Mayo M, Warrington SD, Kaestli M, Mullins T, Wagner DM, et al. Identification of Burkholderia pseudomallei near-neighbor species in the northern territory of Australia. PLoS Negl Trop Dis. 2000;9(6):e0003892. doi: 10.1371/journal.pntd.0003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suarez-Moreno ZR, Devescovi G, Myers M, Hallack L, Mendonca-Previato L, Caballero-Mellado J, et al. Commonalities and differences in regulation of N-acyl homoserine lactone quorum sensing in the beneficial plant-associated Burkholderia species cluster. Appl Environ Microbiol. 2010;76(13):4302–4317. doi: 10.1128/AEM.03086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guglierame P, Pasca MR, De Rossi E, Buroni S, Arrigo P, Manina G, Riccardi G. Efflux pump genes of the resistance-nodulation-division family in Burkholderia cenocepacia genome. BMC Microbiol. 2006;6:66. doi: 10.1186/1471-2180-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tseng SP, Tsai WC, Liang CY, Lin YS, Huang JW, Chang CY, Tyan YC, Lu PL. The contribution of antibiotic resistance mechanisms in clinical Burkholderia cepacia complex isolates: an emphasis on efflux pump activity. PLoS ONE. 2014;9:e104986. doi: 10.1371/journal.pone.0104986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Podnecky NL, Rhodes KA, Schweizer HP. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol. 2015;6:305. doi: 10.3389/fmicb.2015.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, Fondi M, Perrin E, Decorosi F, Viti C, Giovannetti L, Leoni L, Fani R, Riccardi G, Mahenthiralingam E, Buroni S. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS ONE. 2011;6:e18902. doi: 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buroni S, Matthijs N, Spadaro F, Van Acker H, Scoffone VC, Pasca MR, Riccardi G, Coenye T. Differential roles of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob Agents Chemother. 2014;58:7424–7429. doi: 10.1128/AAC.03800-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trepanier S, Prince A, Huletzky A. Characterization of the penA and penR genes of Burkholderia cepacia 249 which encode the chromosomal class A penicillinase and its LysR-type transcriptional regulator. Antimicrob Agents Chemother. 1997;41:2399–2405. doi: 10.1128/AAC.41.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hwang J, Kim HS. Cell wall recycling-linked coregulation of AmpC and PenB beta-lactamases through ampD mutations in Burkholderia cenocepacia. Antimicrob Agents Chemother. 2015;59:7602–7610. doi: 10.1128/AAC.01068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vadlamani G, Thomas MD, Patel TR, Donald LJ, Reeve TM, Stetefeld J, Standing KG, Vocadlo DJ, Mark BL. The beta-lactamase gene regulator AmpR is a tetramer that recognizes and binds the D-Ala-D-Ala motif of its repressor UDP-N-acetylmuramic acid (MurNAc)-pentapeptide. J Biol Chem. 2015;290:2630–2643. doi: 10.1074/jbc.M114.618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FSL. The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poirel L, Rodriguez-Martinez JM, Plesiat P, Nordmann P. Naturally occurring Class A ss-lactamases from the Burkholderia cepacia complex. Antimicrob Agents Chemother. 2009;53:876–882. doi: 10.1128/AAC.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papp-Wallace KM, Taracila MA, Gatta JA, Ohuchi N, Bonomo RA, Nukaga M. Insights into beta-lactamases from Burkholderia species, two phylogenetically related yet distinct resistance determinants. J Biol Chem. 2013;288:19090–19102. doi: 10.1074/jbc.M113.458315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rhodes Katherine A, Schweizer Herbert P. Antibiotic resistance in Burkholderia species. Drug Resist Updat. 2016;28:82–90. doi: 10.1016/j.drup.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gautam V, Shafiq N, Singh M, Ray P, Singhal L, Jaiswal NP, Prasad A, Singh S, Agarwal A. Clinical and in vitro evidence for the antimicrobial therapy in Burkholderia cepacia complex infections. Expert Rev Anti Infect Ther. 2015;13(5):629–663. doi: 10.1586/14787210.2015.1025056. [DOI] [PubMed] [Google Scholar]
- 65.Pegues DA. Burkholderia cepacia complex. http://www.antimicrobe.org/b19.asp.
- 66.Nimri L, Sulaiman M, Hani OB. Community-acquired urinary tract infections caused by Burkholderia cepacia complex in patients with no underlying risk factor. JMM Case Rep. 2017;4(1):e005081. doi: 10.1099/jmmcr.0.005081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blumer JL, Saiman L, Konstan MW, Melnick D. The efficacy and safety of meropenem and tobramycin vs ceftazidime and tobramycin in the treatment of acute pulmonary exacerbations in patients with cystic fibrosis. Chest. 2005;128(4):2336–2346. doi: 10.1378/chest.128.4.2336. [DOI] [PubMed] [Google Scholar]
- 68.Bonacorsi S, Fitoussi F, Lhopital S, Bingen E. Comparative in vitro activities of meropenem, imipenem, temocillin, piperacillin, and ceftazidime in combination with tobramycin, rifampin, or ciprofloxacin against Burkholderia cepacia isolates from patients with cystic fibrosis. Antimicrob Agents Chemother. 1999;43(2):213–217. doi: 10.1128/AAC.43.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aaron SD, Ferris W, Henry DA, et al. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am J Respir Crit Care Med. 2000;161(4 Pt 1):1206–1212. doi: 10.1164/ajrccm.161.4.9907147. [DOI] [PubMed] [Google Scholar]
- 70.Saiman L, Chen Y, Gabriel PS, Knirsch C. Synergistic activities of macrolide antibiotics against Pseudomonas aeruginosa, Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. Antimicrob Agents Chemother. 2002;46(4):1105–1107. doi: 10.1128/AAC.46.4.1105-1107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacLeod DL, Barker LM, Sutherland JL, et al. Antibacterial activities of a fosfomycin/tobramycin combination: a novel inhaled antibiotic for bronchiectasis. J Antimicrob Chemother. 2009;64(4):829–836. doi: 10.1093/jac/dkp282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.