Abstract
Background
Effective use of microbiology test results may positively influence patient outcomes and limit the use of broad-spectrum antibiotics. However, studies indicate that their potential is not fully utilized. We investigated microbiology test ordering practices and the use of test results for antibiotic decision-making in hospitals.
Methods
A multicentre cohort study was conducted during five months in 2014 in Medical departments across three hospitals in Western Norway. Patients treated with antibiotics for sepsis, urinary tract infections, skin and soft tissue infections, lower respiratory tract infections or acute exacerbations of chronic obstructive pulmonary disease were included in the analysis. Primary outcome measures were degree of microbiology test ordering, compliance with microbiology testing recommendations in the national antibiotic guideline and proportion of microbiology test results used to inform antibiotic treatment. Data was obtained from electronic- and paper medical records and charts and laboratory information systems.
Results
Of the 1731 patient admissions during the study period, mean compliance with microbiology testing recommendations in the antibiotic guideline was 89%, ranging from 81% in patients with acute exacerbations of chronic obstructive pulmonary disease to 95% in patients with sepsis. Substantial additional testing was performed beyond the recommendations with 298/606 (49%) of patients with lower respiratory tract infections having urine cultures and 42/194 (22%) of patients with urinary tract infections having respiratory tests. Microbiology test results from one of the hospitals showed that 18% (120/672) of patient admissions had applicable test results, but only half of them were used for therapy guidance, i.e. in total, 9% (63/672) of patient admissions had test results informing prescription of antibiotic therapy.
Conclusions
This study showed that despite a large number of microbiology test orders, only a limited number of tests informed antibiotic treatment. To ensure that microbiology tests are used optimally, there is a need to review the utility of existing microbiology tests, test ordering practices and use of test results through a more targeted and overarching approach.
Keywords: Microbiology testing, Antibiotic prescribing, Antimicrobial resistance, Hospitals, Cohort study
Background
Effective use of microbiology test results has been shown to influence patient outcomes, health care costs and appropriateness of antibiotic prescribing and – use [1–3]. Microbiology tests have also for years provided antimicrobial resistance (AMR) surveillance data, informing empiric antibiotic therapy guidelines. With increasing AMR rates globally, sensitive, specific and affordable microbiology tests could be important tools in providing targeted antibiotic treatment to patients. The tests may facilitate de-escalation of antibiotic therapy from broad- to narrow spectrum treatment, thereby limiting the selection of drug resistant bacteria.
However, several studies indicate that the potential of microbiology tests is not fully utilized. Firstly, clinicians feel they cannot make full use of microbiology tests due to prolonged turnaround times (TATs) [4, 5]. Secondly, although many guidelines provide microbiology test ordering recommendations and information on how to interpret and use test results [6, 7], studies show that microbiology test ordering and use of test results are substandard [8–11]. As diagnostic microbiology methods evolve and become more sophisticated, these inadequacies may increase and ultimately result in incorrect antibiotic treatment for patients, as well as inefficient use of human and laboratory resources [12].
There are some studies on yield and utility of blood cultures, skin and soft tissue (SST) cultures, urinary pneumococcal antigen (UPAg) and polymerase chain reaction (PCR) tests detecting respiratory pathogens, and evidence exists for excessive ordering of urine cultures in asymptomatic patients [13–17]. However, there is little knowledge on existing microbiology test ordering practices and clinical use of microbiology test results, which is needed to optimize use of the tests. The aim of our study was therefore to investigate microbiology test ordering practices in hospitals and how microbiology test results were used to inform antibiotic decision-making. Our hypotheses were that a majority of current microbiology test ordering practices did not adhere to recommendations in the national antibiotic guideline and that a minority of microbiology test results were used to guide antibiotic treatment.
Methods
Design, setting and study population
This study was a multicentre cohort study conducted in infectious diseases-, gastroenterology- and pulmonary medicine wards across three emergency care and teaching hospitals in Western Norway. Patient data were originally collected for a multicentre cluster randomized controlled intervention study, evaluating antibiotic stewardship interventions in hospital settings [18].
Hospital A and B were tertiary care hospitals with 1100 and 600 beds, respectively, offering a full range of microbiology testing services. Hospital C was a secondary care hospital with 160 beds, referring the majority of microbiology specimens to hospital A. Infectious diseases- and pulmonary medicine wards were selected as these specialties have the highest consumption of antibiotics and thus order a large proportion of microbiology tests. Gastroenterology was included since hospital B had a joint medication storage area for the pulmonary medicine- and gastroenterology wards.
Microbiology test ordering practices were analysed using data from patients discharged from the study wards between February 10th and July 11th 2014. Only data from patients receiving antibiotic treatment for sepsis, urinary tract infections (UTIs), skin and soft tissue infections (SSTIs), lower respiratory tract infections (LRTIs) or acute exacerbations of chronic obstructive pulmonary disease (AECOPD) was included in the analyses. Patients admitted for < 24 h, > 21 days and/or readmitted within 30 days were excluded. Clinical use of test results was analysed for patients at hospital A, as complete microbiology test results were available at this hospital.
Outcome measures
The primary outcome measures were microbiology test ordering practices and clinical use of microbiology test results. The secondary outcome measures were yield and TAT for the microbiology tests (Table 1).
Table 1.
Outcome measures
| Outcome | Description |
|---|---|
| Primary outcomes | |
| 1: Microbiology test ordering practices | Measured by a. Degree of compliance with test ordering recommendations in the Norwegian national antibiotic guideline, by diagnoses [6]. b. Degree of microbiology test ordering, i.e. the proportion of patients who had different specimens obtained within the first three days after initiation of antibiotic treatment, by diagnoses and hospital sites |
| 2: Clinical use of microbiology test results | The proportion of microbiology tests ordered on the day of admission used to guide antibiotic treatment. Use was assessed within the first two days after tests results were available to clinicians. For an antibiotic regime to be defined as adjusted in accordance with microbiology test result, it had to be susceptible to the identified pathogen and the regime least prone to drive antibiotic resistance. The evaluation took into account glomerular filtration rate and allergic reactions to antibiotics as recorded on admittance. |
| Secondary outcomes | |
| 1: Yield of microbiology tests | The proportion of patients for which a specific test was positive and identified the potential causative pathogen. Reported by test and diagnoses. |
| 2: Turnaround time for microbiology tests | Time in hours from the specimen was registered as received at the laboratories to final test results were available to clinicians in the electronic medical record. For blood cultures; time when gram stain results were made available to clinicians. |
Data collection
Patient data, including indication for antibiotic treatment, antibiotic treatment throughout the hospital stay, allergic reactions to antibiotics, glomerular filtration rate and number of days admitted were obtained from medical records and drug charts. Indications for antibiotic treatment were based on the treating physicians’ working diagnoses as recorded in patients’ medical records or drug charts on the day of initiation of antibiotic treatment. Laboratory data were collected from medical records to evaluate microbiology test ordering practices and yield, and from the laboratory information system to study clinical use of microbiology test results and TAT (Table 1). An overview of microbiology tests and test results are presented in Table 2. Bacterial cultures were identified by matrix assisted laser desorption ionization-time of light mass spectrometry (Maldi-Tof MS) and susceptibility testing was performed by disk diffusion tests or by minimum inhibitory concentration gradient tests. The PCR tests were developed in-house and the UPAg test was a lateral flow immunoassay.
Table 2.
Overview of microbiology tests and test results
| Microbiology tests | |
| Respiratory tests | Respiratory cultures Polymerase chain reaction (PCR) tests for viral and bacterial respiratory pathogens Urinary pneumococcal antigen tests |
| Skin and soft tissue cultures | Wound-, pus-, breastmilk- and/or tissue cultures |
| Blood cultures | |
| Urine cultures | |
| Test results | |
| Positive findings | Potential pathogen identified |
| a) Causative findings | Positive test results identifying causative pathogen |
| b) Non-causative findings | Positive test results reported as “contaminants”, “normal flora” or “mixed flora” |
| Negative findings | No pathogen identified |
Data analysis
Descriptive statistical analyses were performed on all outcome measures (Table 1). Chi-square test was applied to evaluate differences in microbiology testing practices between the hospitals, where the testing frequencies of each hospital were compared to the total test frequency of the two others. Fisher’s exact test was applied when numbers in one or more categories were < 5. Tests were two-sided and because of multiple testing, p-values < 0.01 were considered statistically significant. Statistical analyses were performed using the SPSS (Statistical Package for the Social Science) version 24.
Results
In total, 1731 patient admissions were included in the analyses of microbiology test ordering practices. The mean age was 68 years old (range 15–103 years), the female/male ratio was 0.48/0.52, mean length of stay was 6.8 days and the 30 day-mortality rate was 8% (142/1731). The distribution of diagnoses was as follows: LRTI 35%, AECOPD 24%, sepsis 18%, SSTI 12% and UTI 11% (Table 3). Of the total patient cohort, 48% were recruited from Hospital A, 27% from Hospital B and 25% from Hospital C.
Table 3.
Microbiology test ordering practices
| Microbiology test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Blood culture | Urine culture | SST culture1 | Resp. test2 | |||||||
| Diagnosis* | Hospital | % | p3 | % | p3 | % | p3 | % | p3 | |
| Sepsis | Hospital A | n = 205 | 95.1 | 0.64 | 70.2 | 0.02 | 10.7 | < 0.01 | 60.0 | < 0.01 |
| Hospital B | n = 42 | 88.1 | 0.06 | 76.2 | 0.81 | 2.4 | 0.34 | 78.6 | < 0.01 | |
| Hospital C | n = 73 | 97.3 | 0.38 | 86.3 | 0.01 | 1.4 | 0.02 | 17.8 | < 0.01 | |
| Total | n = 320 | 94.7 | 74.7 | 52.8 | 7.5 | |||||
| UTI | Hospital A | n = 67 | 59.7 | 0.77 | 91.0 | 0.64 | 3.0 | 0.27 | 31.3 | 0.02 |
| Hospital B | n = 50 | 70.0 | 0.05 | 90.0 | 0.54 | 2.0 | 1.00 | 36.0 | < 0.01 | |
| Hospital C | n = 77 | 49.4 | 0.04 | 94.8 | 0.28 | 0.0 | 0.28 | 3.9 | < 0.01 | |
| Total | n = 194 | 58.2 | 92.3 | 1.5 | 21.6 | |||||
| SSTI | Hospital A | n = 97 | 80.4 | 0.97 | 14.4 | < 0,01 | 68.0 | < 0.01 | 8.2 | 0.66 |
| Hospital B | n = 54 | 79.6 | 0.89 | 13.0 | 0.04 | 44.4 | 0.16 | 11.1 | 0.22 | |
| Hospital C | n = 52 | 80.8 | 0.92 | 50.0 | < 0,01 | 32.7 | < 0.01 | 1.9 | 0.12 | |
| Total | n = 203 | 80.3 | 23.2 | 52.7 | 7.4 | |||||
| LRTI | Hospital A | n = 287 | 80.8 | 0.07 | 48.4 | 0.73 | 1.0 | 0.51 | 68.3 | < 0.01 |
| Hospital B | n = 164 | 75.0 | 0.36 | 40.2 | 0.01 | 1.8 | 0.71 | 89.0 | < 0.01 | |
| Hospital C | n = 155 | 74.2 | 0.25 | 60.0 | < 0,01 | 1.9 | 0.70 | 20.0 | < 0.01 | |
| Total | n = 606 | 77.6 | 49.2 | 1.5 | 61.6 | |||||
| AECOPD | Hospital A | n = 172 | 74.4 | < 0.01 | 38.4 | 0.42 | 2.3 | 0.46 | 59.3 | 0.29 |
| Hospital B | n = 152 | 57.9 | 0.01 | 33.6 | 0.02 | 1.3 | 1.00 | 92.1 | < 0.01 | |
| Hospital C | n = 84 | 63.1 | 0.54 | 58.3 | < 0.01 | 1.2 | 1.00 | 14.3 | < 0.01 | |
| Total | n = 408 | 65.9 | 40.7 | 1.7 | 62.3 | |||||
| All | Total | n = 1731 | 76.1 | 53.7 | 8.7 | 49.3 | ||||
*AECOPD: acute exacerbation of chronic obstructive pulmonary disease; LRTI: lower respiratory infection; SSTI: skin and soft tissue infection; UTI: urinary tract infection
1SST culture: wound, pus, breastmilk or tissue culture
2Resp. test: respiratory culture, polymerase chain reaction (PCR) test for viral and bacterial respiratory pathogens and/or urinary pneumococcal antigen test
3p value for testing whether there is a significant difference between one hospital compared to the total frequencies of the two others by Chi-square test or by Fisher’s exact test when numbers in one or more categories were < 5
Guideline adherence
The degree of compliance with microbiology test ordering recommendations in the national antibiotic guideline was 89% across all diagnoses. Compliance was 95% in sepsis (blood culture), 92% in UTI (urine culture), 88% in LRTI (PCR test detecting respiratory pathogens, UPAg test, respiratory- or blood culture) and 81% in AECOPD (respiratory- and/or blood culture). There were no specific test ordering recommendations for SSTIs, however culture specimens were often obtained from the site of infection.
Testing practices by tests, diagnoses and hospital sites
Many patients had more than one microbiology sample collected regardless of diagnosis (Table 3). In the total cohort of patient admissions, the following microbiology tests were ordered: 76% blood cultures, 54% urine cultures, 49% respiratory tests and 9% skin or soft tissue cultures. Among the patients with LRTI and AECOPD, 49 and 41% had urine cultures taken, respectively. Concomitantly, 22% of the patients with UTI had respiratory tests performed. Test ordering practices varied between the three hospitals. Patients diagnosed with sepsis, LRTI and AECOPD had significantly more respiratory tests taken at hospital B than at the two other hospitals (p < 0.01), and the same groups of patients had significantly less respiratory tests taken at hospital C compared to the two other hospitals (p < 0.01).
Yield
The total yield for blood-, urine-, respiratory- and SSTI cultures was 8, 29, 34 and 67%, respectively (Table 4). For blood cultures, the yield was 20% in sepsis- and 4% in LRTI patients. For LRTI patients, the yield of the PCR test detecting respiratory pathogens, UPAg test and respiratory cultures was 18, 9 and 33%, respectively. However, 52% of the respiratory cultures had non-causative findings.
Table 4.
Yield of microbiological specimen
| Diagnosis* | Test findings | Blood culture % | Urine culture % | SST culture1% | Respiratory culture % | RP-PCR2 % | UPAg3 % |
|---|---|---|---|---|---|---|---|
| Sepsis | Causative | 20 | 36 | 63 | 29 | 18 | 8 |
| Non-causative | 5 | 16 | 25 | 58 | |||
| Negative | 75 | 48 | 13 | 13 | 82 | 92 | |
| Total | 100 (n = 303) | 100 (n = 239) | 100 (n = 24) | 100 (n = 52) | 100 (n = 106) | 100 (n = 111) | |
| UTI | Causative | 17 | 54 | 33 | 0 | 0 | 0 |
| Non-causative | 4 | 17 | 67 | 80 | |||
| Negative | 79 | 28 | 0 | 20 | 100 | 100 | |
| Total | 100 (n = 113) | 100 (n = 179) | 100 (n = 3) | 100 (n = 15) | 100 (n = 27) | 100 (n = 21) | |
| SSTI | Causative | 4 | 30 | 70 | 40 | 0 | 0 |
| Non-causative | 4 | 15 | 23 | 60 | |||
| Negative | 92 | 55 | 7 | 0 | 100 | 100 | |
| Total | 100 (n = 163) | 100 (n = 47) | 100 (n = 107) | 100 (n = 5) | 100 (n = 11) | 100 (n = 4) | |
| LRTI | Causative | 4 | 16 | 67 | 33 | 17 | 9 |
| Non-causative | 2 | 20 | 22 | 52 | |||
| Negative | 94 | 63 | 11 | 15 | 83 | 91 | |
| Total | 100 (n = 470) | 100 (n = 298) | 100 (n = 9) | 100 (n = 185) | 100 (n = 240) | 100 (n = 196) | |
| AECOPD | Causative | 1 | 17 | 43 | 41 | 8 | 10 |
| Non-causative | 4 | 16 | 43 | 45 | |||
| Negative | 94 | 67 | 14 | 14 | 92 | 90 | |
| Total | 100 (n = 269) | 100 (n = 166) | 100 (n = 7) | 100 (n = 161) | 100 (n = 167) | 100 (n = 123) | |
| Total | Causative | 8 | 29 | 67 | 34 | 13 | 8 |
| Non-causative | 4 | 18 | 25 | 51 | |||
| Negative | 88 | 53 | 8 | 14 | 87 | 92 | |
| Total | 100 (n = 1318) | 100 (n = 929) | 100 (n = 150) | 100 n = (418) | 100 (n = 551) | 100 (n = 455) |
*AECOPD: acute exacerbation of chronic obstructive pulmonary disease; LRTI: lower respiratory infection; SSTI: skin and soft tissue infection; UTI: urinary tract infection
1SST culture: wound, pus, breastmilk or tissue culture; 2RP-PCR: Respiratory panel polymerase chain reaction test for viral and bacterial respiratory pathogens; 3UPAg: Urinary pneumococcal antigen tests
Turnaround time
Mean TAT was 25 h (95% CI, 22.4–27.7) for blood-, 37 h (95% CI, 31.2–42.6) for urine-, 56 h (95% CI, 49.5–63.0) for SST- and 80 h (95% CI, 60.5–99.6) for respiratory cultures.
Clinical use of test results
In hospital A, there were 828 patient admissions, of which 81 collected microbiology specimens at day > 1 after admission, leaving 747 cases eligible for inclusion in the analyses of clinical use of microbiology test results obtained on the day of admission. Of these, 672 (81%) had blood-, urine-, respiratory- and/or SST cultures taken and were included in the analyses of clinical use of test results (Fig. 1).
Fig. 1.
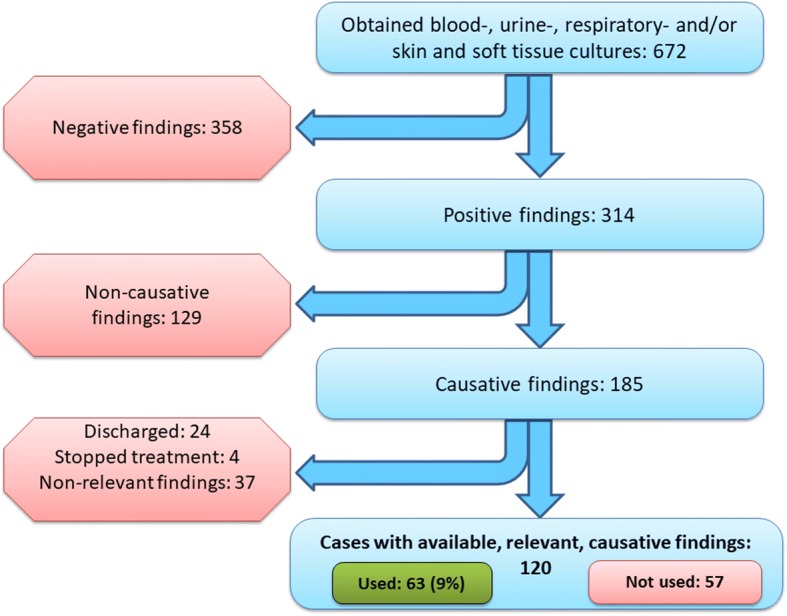
Patient admissions and use of microbiology test results
Of the 672 patient admissions, 358 (53%) had negative microbiology test results and 129 (19%) had non-causative findings. Among the remaining 185 cases, 37 had findings not relevant to their diagnoses, four had stopped antibiotic treatment and 24 were discharged when microbiology test results became available. Of the 120/672 (18%) inpatients with applicable findings, antibiotic treatment was adjusted according to test results only in 63 patients, i.e. 9% of the total number of patient admissions. Among the patients with the diagnoses SSTI and UTI, a majority had their antibiotic treatment adjusted in accordance with the test results, whereas treatment was adjusted only in a minority of the patients with AECOPD. As only 120 patient admissions had applicable test results, the number in each diagnostic group was low.
Discussion
The main finding of this study was that despite a large number of microbiology test orders, only a small fraction of test results informed antibiotic decision-making. We observed high compliance with test ordering recommendations in the national guideline, but excessive testing across diagnoses, contributing to a low yield. TATs were long and microbiology test results with causative pathogens were underused, both contributing to the low utilization of the tests.
To our knowledge, this is the first study reporting on microbiology test ordering practices and use of test results in clinical practice. A previous study on clinical laboratory- and imaging tests, reported that one third of tests were unnecessary and only half of relevant test results were used in patient follow up [11]. Studies investigating the yield of blood culture and UPAg tests, showed similar results to ours [13, 14, 19, 20]. The high rates of respiratory cultures with non-causative findings identified in our material are also in accordance with the literature, reporting that respiratory sampling procedures are challenging [21].
Our study shows that the existing microbiology tests, testing practices and use of test results are not in accordance with the objective of microbiology testing; only a minor fraction of patients benefitted from a test result and only a small proportion could be used to target therapy and minimize the use of broad spectrum antibiotics. This suggests that microbiology laboratory resources could be spent more efficiently than producing insignificant or negative results, although negative microbiology test results may be important for treatment in some infectious disease patients. Only half of the patients with test results identifying the causative pathogen had their antibiotic treatment tailored accordingly, meaning that antibiotic treatment was not optimized for the other half of the patients.
There are several explanations for these findings. Excessive testing and inadequate follow up of test results may be caused by clinician’s insufficient knowledge of microbiology [22, 23]. Diagnostic uncertainty and inadequate routines for microbiology testing in the emergency departments may also contribute to the large number of unnecessary test orders [24]. However, the inherent characteristics of the microbiology tests play a major role for the yield; although adhering to the guidelines when sampling specimens, the yield for several tests was low. However, in this study, we did not have information on the quality of sampling and transportation of specimens, which may also impact the yield [8, 25]. Long TATs associated with certain tests may reduce their utility. We observed particularly long TAT for respiratory cultures. This may partly be explained by the time-consuming challenge of identifying and separating respiratory pathogens from normal bacterial flora. One reason for the continuation of these practices, both in the microbiology laboratories and in the clinical units, may be a lack of communication between the two parties to improve microbiology testing practices [23].
Our findings show the need for a systematic review of the use of microbiology tests in clinical practice. Firstly, tests with low yield should be evaluated, particularly tests for respiratory infections as our and other studies show that microbiology test results are of little help to identify causative pathogens [14, 20, 21]. Thus, more specific and sensitive tests in the diagnostic work up of respiratory infections are needed. Secondly, there is a need to review the indications for microbiology tests. Obviously, restricting urine cultures to patients with possible UTIs may reduce unnecessary antibiotic treatment of asymptomatic bacteriuria [26]. Additionally, although the overall yield for blood cultures was low in our study, it varied significantly between patients suffering from respiratory infections in the lower end and sepsis patients in the higher end. This indicates that a stratification, prioritizing blood cultures for the more severely ill patients, may be appropriate and increase overall yield.
Thirdly, there is a need to reduce TATs and increase the proportion of microbiology test results available at an early stage of patient treatment. Potential measures are expansion of molecular diagnostics, rapid and point-of-care test services, as well as revision of testing processes within the microbiology laboratories, shown to reduce TAT significantly [27, 28]. These measures, promoting rapid delivery of microbiology test results with better performance characteristics, are even more important in settings with higher rates of AMR than Norway [29]; In such settings, the identification of causative pathogens and their susceptibility to antibiotic agents, is crucial for appropriate and targeted antibiotic treatment. Furthermore, clinicians need to increase their knowledge of different microbiology tests; when to order them and how to apply the test results. Systematic measures such as providing education, audit with feedback on microbiology test ordering and use of test results, as well as establishing decision support for microbiology testing in computerized provider order entry systems, may be useful [30]. In order to accommodate all these challenges adequately, there is a need for clinical- and microbiology laboratory staff to work in partnership. Moreover, to develop sustainable and efficient solutions, there is a need for a targeted and overarching approach.
An improved utilisation of microbiology services is vital both for the individual infectious disease patients in need of optimised antibiotic therapy and for the containment of AMR. Microbiology tests can contribute to reduced use of broad spectrum antibiotics and antibiotics in general, thereby limiting the impetus for development and selection of drug resistant bacteria. With improved availability of microbiology test results and increased test accuracy, treatment can be more targeted and broad spectrum antibiotics saved [31]. Additionally, rapid access to microbiology test results differentiating viral and bacterial infections, may reduce unnecessary use of antibiotics [32].
In summary, this study raises several questions regarding the future of microbiology testing. How can we utilize microbiology testing and the laboratory resources more efficiently? Which diagnostic tests do we need to develop? And how can we improve interdisciplinary collaboration around the infectious disease patient? Thus, more research is needed on how to optimize the collection of microbiology samples, how to develop and implement new diagnostic methods and how to reduce TAT for microbiology tests, taking into account the potential impact on patient outcomes, antibiotic prescribing and development of AMR, as well as on use of human and laboratory resources.
The study has some limitations. Microbiology test results were mainly based on traditional culturing and Maldi-Tof MS. Use of novel technology such as molecular diagnostics could have decreased TAT and increased the proportion of test results used to inform antibiotic treatment [27]. Patient data used for analysis in this study were originally collected for an interventional study on antibiotic prescribing in hospitals [18]. However, we supplemented with microbiology data to accommodate the needs of this study. Data collection was limited to departments of internal medicine in Western Norway, potentially reducing the external validity. This is however a relatively large, multicentre study, applying an extensive amount of different data and covering a wide range of clinical scenarios.
Conclusion
This study identified high compliance with microbiology testing recommendations in the national guideline. There was however extensive ordering of additional tests, many tests had low yield and only a small proportion of test results informed antibiotic decision-making. This highlights that the current use of microbiology laboratory services is suboptimal. There is a need both for tests with better performance characteristics and improved test ordering practices. Furthermore, use of microbiology test results to inform antibiotic decision-making needs to be optimized in order to ensure adequate patient treatment and more targeted therapy. To fill these gaps there is a need for an overarching approach with a clear call to fulfil the objective of microbiology testing; to provide rapid, sensitive test results to individual patients, but also to facilitate prudent use of antibiotics.
Acknowledgements
We would like to thank the hospitals and study wards for participating in the study.
Funding
This work was supported by the Norwegian Advisory Unit for Antibiotic Use in Hospitals at Haukeland University Hospital. Author E.C.’s work was funded by Economic and Social Science Research Council (ESRC) as part of the Antimicrobial Cross Council initiative [ES/P008313/1] supported by the seven UK research councils, and the Global Challenges Research Fund and the National Institute for Health Research, UK Department of Health [HPRU-2012-10047] in partnership with Public Health England.
Availability of data and materials
The datasets generated and/or analysed during the current study regarding individual patient data are not publicly available in concordance with the approval from the Data Protection Officer (2013/9352), but are available from the corresponding author on reasonable request.
Abbreviations
- AECOPD
Acute exacerbation of chronic obstructive pulmonary disease
- AMR
Antimicrobial resistance
- LRTI
Lower respiratory tract infection
- Maldi-Tof MS
Matrix assisted laser desorption ionization-time of light mass spectrometry
- PCR
Polymerase chain reaction
- SST(I)
Skin and soft tissue (infection)
- TAT
Turnaround time
- UPAg
Urinary pneumococcal antigen tests
- UTI
Urinary tract infection
Authors’ contributions
Study design: BS, JSW, IS. Data collection and validation: BS, JSW, LKSK, PCL, IS. Data analysis: BS, JSW, RMN, IS. Interpretation of data: BS, JSW, PCL, SH, RMN, EC, HS, BRK, LKSK, IS. Writing of manuscript: BS, IS. Critical assessment and approval of manuscript: BS, JSW, PCL, SH, RMN, EC, HS, BRK, LKSK, IS. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Western Regional Committee for Medical and Health Research Ethics in Norway assessed the study, and concluded that no ethical approval was needed. The Committee approved the waiver of informed consent on the condition that all patients received written information about the study, with opportunity to withdraw from it (2013/1305). Data collection was also approved by The Data Protection Officer at all three hospitals (2013/9352).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Brita Skodvin, Phone: +47 91 55 87 15, Email: brita.skodvin@helse-bergen.no.
Jannicke S. Wathne, Email: jannicke.slettli.wathne@sav.no
P. Christoffer Lindemann, Email: paul.christoffer.lindemann@helse-bergen.no.
Stig Harthug, Email: stig.harthug@helse-bergen.no.
Roy M. Nilsen, Email: Roy.Miodini.Nilsen@hvl.no
Esmita Charani, Email: e.charani@imperial.ac.uk.
Heidi Syre, Email: heidi.syre@sus.no.
Baard R. Kittang, Email: bard.reiakvam.kittang@haraldsplass.no
Lars K. S. Kleppe, Email: lars.kare.selland.kleppe@sus.no
Ingrid Smith, Email: ismith@who.int.
References
- 1.Huang AM, Newton D, Kunapuli A, Gandhi TN, Washer LL, Isip J, et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. CID. 2013;57(9):1237–1245. doi: 10.1093/cid/cit498. [DOI] [PubMed] [Google Scholar]
- 2.Ly T, Gulia J, Pyrgos V, Waga M, Shoham S. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther Clin Risk Manag. 2008;4(3):637–640. doi: 10.2147/tcrm.s2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perez KK, Olsen RJ, Musick WL, Cernoch PL, Davis JR, Land GA, et al. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med. 2013;137(9):1247–1254. doi: 10.5858/arpa.2012-0651-OA. [DOI] [PubMed] [Google Scholar]
- 4.McCall SJ, Souers RJ, Blond B, Massie L. Physician satisfaction with clinical laboratory services: a College of American Pathologists Q-probes study of 81 institutions. Arch Pathol Lab Med. 2016;140(10):1098–1103. doi: 10.5858/arpa.2015-0486-CP. [DOI] [PubMed] [Google Scholar]
- 5.Skodvin B, Aase K, Charani E, Holmes A, Smith I. An antimicrobial stewardship program initiative: a qualitative study on prescribing practices among hospital doctors. Antimicrob Resist Infect Control. 2015;4:24. doi: 10.1186/s13756-015-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norwegian Directorate of Health. National guideline on antimicrobial use in Norwegian hospitals 2013; [Available from: https://helsedirektoratet.no/retningslinjer/antibiotika-i-sykehus.
- 7.Infectious Diseases Society of America. IDSA Practice Guidelines [Available from: https://www.idsociety.org/practice-guidelines/search-homepage/#/score/DESC/0/+//.
- 8.Hawkins R. Managing the pre- and post-analytical phases of the total testing process. Ann Lab Med. 2012;32(1):5–16. doi: 10.3343/alm.2012.32.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiff GD, Hasan O, Kim S, Abrams R, Cosby K, Lambert BL, et al. Diagnostic error in medicine: analysis of 583 physician-reported errors. Arch Intern Med. 2009;169(20):1881–1887. doi: 10.1001/archinternmed.2009.333. [DOI] [PubMed] [Google Scholar]
- 10.Callen J, Georgiou A, Li J, Westbrook JI. The safety implications of missed test results for hospitalised patients: a systematic review. BMJ Qual Saf. 2011;20(2):194–199. doi: 10.1136/bmjqs.2010.044339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch C, Roberts K, Petruccelli C, Morgan DJ. The frequency of unnecessary testing in hospitalized patients. Am J Med. 2018;131(5):500–503. doi: 10.1016/j.amjmed.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014;27(4):783–822. doi: 10.1128/CMR.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long B, Koyfman A. Best clinical practice: blood culture utility in the emergency department. J Emerg Med. 2016;51(5):529–539. doi: 10.1016/j.jemermed.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Dinh A, Duran C, Davido B, Lagrange A, Sivadon-Tardy V, Bouchand F, et al. Cost effectiveness of pneumococcal urinary antigen in emergency department: a pragmatic real-life study. Intern Emerg Med. 2018;13(1):69–73. doi: 10.1007/s11739-016-1586-4. [DOI] [PubMed] [Google Scholar]
- 15.Holter JC, Muller F, Bjorang O, Samdal HH, Marthinsen JB, Jenum PA, et al. Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: a 3-year prospective study in Norway. BMC Infect Dis. 2015;15(1):64. doi: 10.1186/s12879-015-0803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens DL, Bisno AL, Chambers HF, Everett ED, Dellinger P, Goldstein EJ, et al. Practice guidelines for the diagnosis and management of skin and soft-tissue infections. Clin Infect Dis. 2005;41(10):1373–1406. doi: 10.1086/497143. [DOI] [PubMed] [Google Scholar]
- 17.Flokas ME, Andreatos N, Alevizakos M, Kalbasi A, Onur P, Mylonakis E. Inappropriate Management of Asymptomatic Patients with Positive Urine Cultures: a systematic review and meta-analysis. Open Forum Infect Dis. 2017;4(4):ofx207. doi: 10.1093/ofid/ofx207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wathne JS, Kleppe LKS, Harthug S, Blix HS, Nilsen RM, Charani E, et al. The effect of antibiotic stewardship interventions with stakeholder involvement in hospital settings: a multicentre, cluster randomized controlled intervention study. Antimicrob Resist Infect Control. 2018;7:109. doi: 10.1186/s13756-018-0400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabak YP, Vankeepuram L, Ye G, Jeffers K, Gupta V, Murray PR. Blood culture turnaround time in US acute care hospitals and implications for laboratory process optimization. J Clin Microbiol. 2018;56:e00500-18. [DOI] [PMC free article] [PubMed]
- 20.Piso RJ, Iven-Koller D, Koller MT, Bassetti S. The routine use of urinary pneumococcal antigen test in hospitalised patients with community acquired pneumonia has limited impact for adjustment of antibiotic treatment. Swiss Med Wkly. 2012;142:w13679. doi: 10.4414/smw.2012.13679. [DOI] [PubMed] [Google Scholar]
- 21.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–S72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaschke AJ, Hersh AL, Beekmann SE, Ince D, Polgreen PM, Hanson KE. Unmet diagnostic needs in infectious disease. Diagn Microbiol Infect Dis. 2015;81(1):57–59. doi: 10.1016/j.diagmicrobio.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skodvin B, Aase K, Brekken AL, Charani E, Lindemann PC, Smith I. Addressing the key communication barriers between microbiology laboratories and clinical units: a qualitative study. J Antimicrob Chemother. 2017;72(9):2666–2672. doi: 10.1093/jac/dkx163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leis JA, Rebick GW, Daneman N, Gold WL, Poutanen SM, Lo P, et al. Reducing antimicrobial therapy for asymptomatic bacteriuria among noncatheterized inpatients: a proof-of-concept study. Clin Infect Dis. 2014;58(7):980–983. doi: 10.1093/cid/ciu010. [DOI] [PubMed] [Google Scholar]
- 25.Sloan CE, Bernard S, Nachamkin I. Appropriateness of expectorated sputum cultures in the hospital setting. Diagn Microbiol Infect Dis. 2015;83(1):74–76. doi: 10.1016/j.diagmicrobio.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Stagg A, Lutz H, Kirpalaney S, Matelski JJ, Kaufman A, Leis J, et al. Impact of two-step urine culture ordering in the emergency department: a time series analysis. BMJ Qual Saf. 2017;27:140-47. [DOI] [PMC free article] [PubMed]
- 27.Kothari A, Morgan M, Haake DA. Emerging technologies for rapid identification of bloodstream pathogens. Clin Infect Dis. 2014;59(2):272–278. doi: 10.1093/cid/ciu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kommedal O, Aasen JL, Lindemann PC. Genetic antimicrobial susceptibility testing in gram-negative sepsis - impact on time to results in a routine laboratory. APMIS. 2016;124(7):603–610. doi: 10.1111/apm.12549. [DOI] [PubMed] [Google Scholar]
- 29.NORM/NORM-VET. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway 2017 [Available from:https://unn.no/fag-og-forskning/norm-norsk-overvakingssystem-for-antibiotikaresistens-hos-mikrober#rapporter
- 30.Baron JM, Dighe AS. Computerized provider order entry in the clinical laboratory. J Pathol Inform. 2011;2:35. doi: 10.4103/2153-3539.83740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buehler SS, Madison B, Snyder SR, Derzon JH, Cornish NE, Saubolle MA, et al. Effectiveness of practices to increase timeliness of providing targeted therapy for inpatients with bloodstream infections: a laboratory medicine best practices systematic review and meta-analysis. Clin Microbiol Rev. 2016;29(1):59–103. doi: 10.1128/CMR.00053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egilmezer E, Walker GJ, Bakthavathsalam P, Peterson JR, Gooding JJ, Rawlinson W, et al. Systematic review of the impact of point-of-care testing for influenza on the outcomes of patients with acute respiratory tract infection. Rev Med Virol. 2018;28(5):e1995. doi: 10.1002/rmv.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study regarding individual patient data are not publicly available in concordance with the approval from the Data Protection Officer (2013/9352), but are available from the corresponding author on reasonable request.


