Abstract
The axolotl (Ambystoma mexicanum) provides critical models for studying regeneration, evolution, and development. However, its large genome (∼32 Gb) presents a formidable barrier to genetic analyses. Recent efforts have yielded genome assemblies consisting of thousands of unordered scaffolds that resolve gene structures, but do not yet permit large-scale analyses of genome structure and function. We adapted an established mapping approach to leverage dense SNP typing information and for the first time assemble the axolotl genome into 14 chromosomes. Moreover, we used fluorescence in situ hybridization to verify the structure of these 14 scaffolds and assign each to its corresponding physical chromosome. This new assembly covers 27.3 Gb and encompasses 94% of annotated gene models on chromosomal scaffolds. We show the assembly's utility by resolving genome-wide orthologies between the axolotl and other vertebrates, identifying the footprints of historical introgression events that occurred during the development of axolotl genetic stocks, and precisely mapping several phenotypes including a large deletion underlying the cardiac mutant. This chromosome-scale assembly will greatly facilitate studies of the axolotl in biological research.
In the late nineteenth century, the axolotl (Ambystoma mexicanum) emerged as a model for studying vertebrate development (Reiss et al. 2015) and has continued to serve as a uniquely informative model for studying diverse topics, including regeneration (Voss et al. 2009), evolution of germline (Evans et al. 2014), genome evolution (Smith et al. 2009; Voss et al. 2011; Keinath et al. 2015, 2017; Nowoshilow et al. 2018), and the origin of adaptive phenotypes (Voss and Shaffer 1997, 2000). Recent advances in sequencing, assembly, and genetic manipulation have led to a resurgence of this model and are beginning to surmount longstanding issues associated with the large size of the axolotl genome (∼32 Gb). The axolotl genome assembly has seen steady improvement over the last few years (Smith et al. 2005; Keinath et al. 2015; Evans et al. 2018; Nowoshilow et al. 2018). The latest version of the assembly harbors annotated models for a vast majority of axolotl genes on scaffolds that typically exceed a megabase in length (N50 ∼3 Mb) (Nowoshilow et al. 2018). Even with the advanced computational methods and extensive sampling of long reads brought to bear in the completion of the most recent assembly, the typical scaffold represents ∼1/1000th of a chromosome and contains on average 1–2 complete or fragmented genes and associated promotor/proximate regulatory sequences. Recognizing that greater contiguity will permit more in-depth analyses that leverage large-scale genomic signatures (i.e., identification of regulatory elements, genetic association studies, and genome evolution), we sought to improve the contiguity of the genome assembly toward the goal of achieving chromosome-scale scaffolding.
Initial attempts to develop a chromosome-scale assembly using a proximity ligation approach (DoveTail) (Putnam et al. 2016) revealed limitations of the approach and yielded few useful results, despite extensive sequencing/computational costs and time associated with analyses of the resulting data sets. This led us to adapt established meiotic mapping methods for Ambystoma as a tool to generate dense genome-wide scaffolding information. Hybrid crosses between A. mexicanum and A. tigrinum (and other species) have been generated and used to develop meiotic maps for the species and infer the positions of quantitative trait loci (QTL), sex, and Mendelian pigment mutants (Voss and Shaffer 1997, 2000; Voss et al. 2001; Voss and Smith 2005; Smith and Voss 2006, 2009; Page et al. 2013; Woodcock et al. 2017). For example, hybrid crosses have been used to dissect the genetic basis of paedomorphosis, a hallmark trait of salamanders (Voss and Shaffer 1997, 2000; Voss and Smith 2005). Paedomorphosis reflects the loss of metamorphic changes in a derived species relative to its ancestor. These studies have found that expression of paedomorphosis can largely be attributed to a single genomic region, known as the met QTL. In the last year, sparse meiotic mapping information was also used to identify the genes underlying two pigment mutants (Woodcock et al. 2017), however, the genes for many mutants remain unknown. Development of a chromosome-scale assembly would greatly accelerate gene discovery and make it possible to identify epigenetic changes associated with embryogenesis, post-embryonic development, and regeneration.
Here, we use low-coverage sequence data from 48 segregants of a hybrid A. mexicanum/A. tigrinum backcross to identify and localize 12.6 million SNPs and 27.3 Gb of corresponding A. mexicanum sequence on 14 chromosomal scaffolds, which corresponds to the known number of chromosome pairs. Additionally, we used fluorescent in situ hybridization (FISH) mapping of BAC clones to physically anchor scaffolds to specific chromosomes and verify that these scaffolds correspond to single (entire) chromosomes. We use this new assembly to improve resolution of orthologous regions between salamander and other vertebrates, identify regions of introgression that have persisted since the early establishment of axolotl genetic stocks, and precisely map the location of major effect loci, including sex, the met QTL, and a large deletion underlying cardiac, a classical Mendelian mutant.
Results
SNP typing and linkage analysis
Our analyses of low-coverage sequence data yielded 12.6 million A. mexicanum/A. tigrinum polymorphisms segregating in the mapping panel. These were distributed across 98,802 scaffolds with individual scaffolds containing between 1 and 12,108 polymorphisms. Although sequencing depth at any position was insufficient to reliably assess genotype, we were able to generate consensus genotypes (homozygous A. mexicanum: or heterozygous A. mexicanum/A. tigrinum) for 69,250 scaffolds by averaging across five or more adjacent SNPs. The vast majority of scaffolds were characterized by segregation ratios approximating 1:1, consistent with expectations from the backcross mapping panel. A small number of scaffolds with aberrant segregation ratios were flagged as potential assembly errors (Supplemental Table S1).
Consensus genotypes were used to infer the grouping and ordering of scaffolds within linkage groups. To avoid confounding effects of genotyping errors on the estimation of the recombination structure of the linkage map, we selected a subset of 8758 scaffolds with genotypes supported by more than 200 polymorphic sites to build a framework genetic map. After manual curation, these markers yielded 14 linkage groups spanning a total of 5364 cM (Fig. 1), which is consistent with microscopic observations of the A. mexicanum karyotype and chiasma frequencies (Callan 1966). The average intermarker distance within this framework map was <1 cM (0.6 cM), smaller than the minimum distance that can be estimated from 48 backcross offspring (∼2.1 cM). Notably, the met QTL interval was inflated relative to the rest of the map because recombinant individuals for this region were purposely included in the mapping panel (see below). After construction of the framework map, an additional 17,819 markers were placed on the meiotic map based on similarity of genotypes to the framework map. Additional scaffolding and orientation information from previously published RNA-seq studies (Evans et al. 2014; Bryant et al. 2017; Caballero-Pérez et al. 2018) and bulked-segregant analyses were used to tune internal ordering of scaffolds and add another 1097 previously unplaced scaffolds (grand total of 27,674). In total, 27.5 Gb of sequence was scaffolded onto 14 linkage groups (chromosomes), with the total length of scaffolded sequence per chromosome ranging from 3.14 to 0.66 Gb (Fig. 1; Supplemental Tables S2, S3). By comparison, the entire human genome is ∼3.2 Gb. Accordingly, our meiotic scaffolding approach resulted in an increase in scaffold N50 from 3.05 Mb to 2.46 Gb, a more than 800× increase in scaffold contiguity.
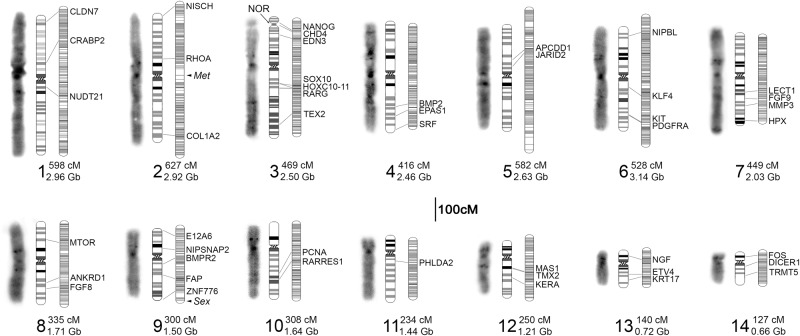
Figure 1.
Summary of assembled chromosomes from the A. mexicanum genome. Three images are shown for each chromosome (1–14): (left) microscopic image illustrating DAPI banding; (center) an idiogram summarizing banding patterns and relative sizes of chromosomes from several spreads (Supplemental Fig. S1); and (right) the corresponding linkage group. Groups of linked markers at a given position are represented by horizontal marks on linkage groups. The location of genes and hybridization signals from gene-anchored BACs are labeled with the corresponding gene ID and connecting lines. The length of each linkage group and assembled chromosome is given next to its numerical label. The locations of three additional features are highlighted: (met) a major quantitative trait locus controlling timing and incidence of metamorphosis; (sex) the sex determining locus; and (NOR) the nucleolar organizer, which harbors most ribosomal RNA copies.
To validate the large-scale structure of our scaffolded genome and more directly assign linkage groups to chromosomes, we performed FISH analyses using a collection of 46 gene-anchored BACs. These hybridizations confirmed that our linkage groups correspond to individual chromosomes, confirmed the large-scale ordering of scaffolds along the length of chromosomes, and allowed us to assign assembled scaffolds to karyotypically definable entities (Fig. 1; Supplemental Fig. S1).
Conservation of synteny and chromosome evolution
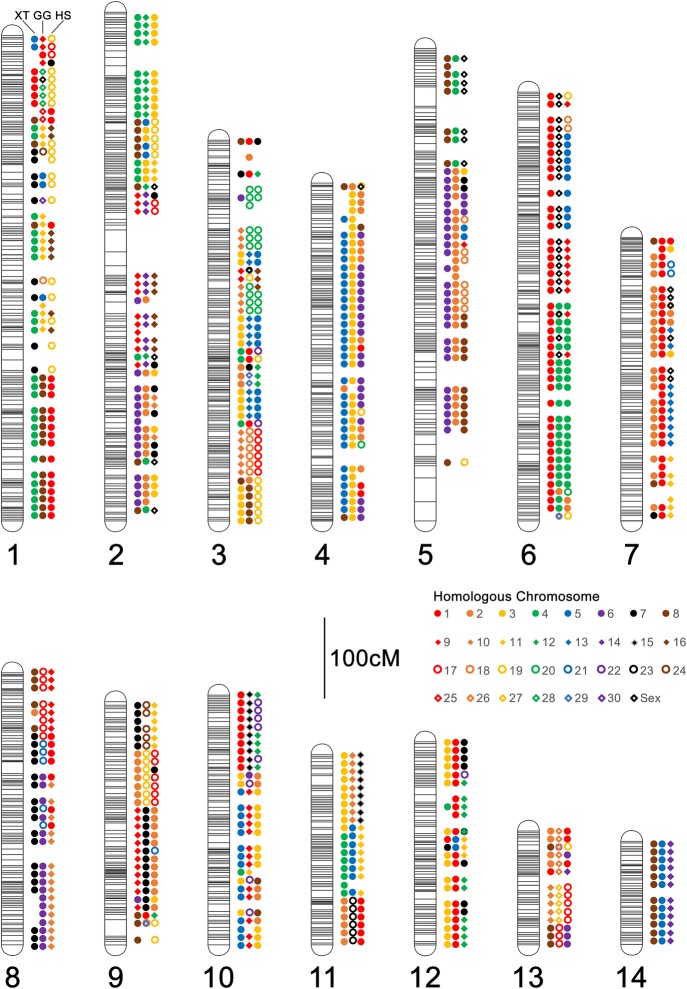
To further assess the degree to which our scaffolded genome recovers conserved gene orders across vertebrates, we aligned predicted A. mexicanum proteins to those annotated in the genomes of chicken, human, and frog (Xenopus tropicalis [XT]). These revealed broad-scale conservation of genome structure and fusion/fission events that underlie differences in chromosome number and content between taxa (Fig. 2; Supplemental Tables S4–S6). In general, these confirm patterns reported from earlier mapping studies, revealing that X. tropicalis and A. mexicanum (AM) genomes are composed of ancestral chromosomes and microchromosomes that were independently fused after the separation of frog and salamander lineages (Voss et al. 2011). These also confirm that conserved regions corresponding to chicken (GG) Chromosomes Z and 4 were linked in the ancestral amphibian (Smith and Voss 2006; Voss et al. 2011; Keinath et al. 2017) and identify two other conserved linkages (portions of GG8/11 homologous to AM1/XT4, and portions of GG6/21 homologous to AM8/XT7), which represent previously unreported fusion events that likely occurred in the common ancestral lineage of all extant amphibians. Moreover, the extensive conservation of synteny revealed by these analyses serves as additional support for the accuracy of the large-scale structure of the assembled chromosomes.
Figure 2.
Patterns of conserved synteny across assembled A. mexicanum chromosomes. Colored shapes represent the consensus homologous chromosomes (see inset legend) for each 10 cM interval. For each chromosome, homologs are shown in a consistent order from left to right: (left) X. tropicalis (XT); (center) chicken (Gallus gallus [GG]); (right) human (Homo sapiens [HS]). A. mexicanum chromosomes are designated by numerical labels corresponding to those in Figure 1.
Distribution of polymorphisms
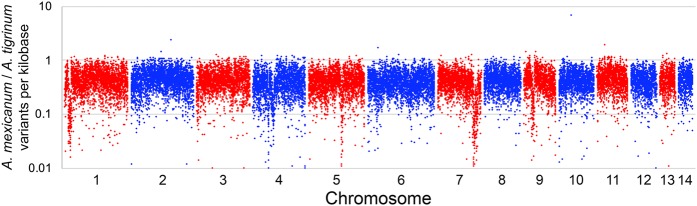
A recent study showed that the vast majority of A. mexicanum used in biological studies derive from an A. mexicanum/A. tigrinum mavortium cross that was performed in 1962 (Humphrey 1967; Woodcock et al. 2017). This cross was performed to introgress an albinism gene (tyr) into a captive population to create a new mutant strain. Analysis of SNPs across the axolotl genome reveal that the density of polymorphisms (in 1-Mb windows) is relatively consistent, averaging 0.43 polymorphisms per kilobase (SD = 0.20, N = 27,518 regions). However, the fingerprint of the tyr introgression is evident when examining density distributions for typed polymorphisms (Chromosome 7) (Fig. 3). The density of polymorphisms is lower at the position of tyr because more A. tigrinum-like sequences have remained in linkage disequilibrium within this region. Other regions show similar depression in SNP densities, including one region near the centromere of the sex chromosome and another large region on Chromosome 1. Notably, the kisspeptin-54 receptor (KISS1R) is located near the center of the presumptive A. tigrinum introgression on Chromosome 1. This gene is a key regulator of gonadotropin-releasing hormone signaling, and variation in this gene is known to be associated with the onset timing of sexual maturity in mammals (Seminara et al. 2003; Popa et al. 2008). We speculate that this and other introgressed A. tigrinum mavortium DNA sequences were retained because they confer fitness benefits in the context of laboratory culture and husbandry practices.
Figure 3.
The distribution of segregating variants within the mapping panel used to generate scaffolding information. The density of polymorphisms within 1-Mb intervals is relatively uniform; however, a few intervals show substantially reduced densities, including regions on Chromosomes 1, 5, 7, and 9.
The genetic basis of the cardiac mutant
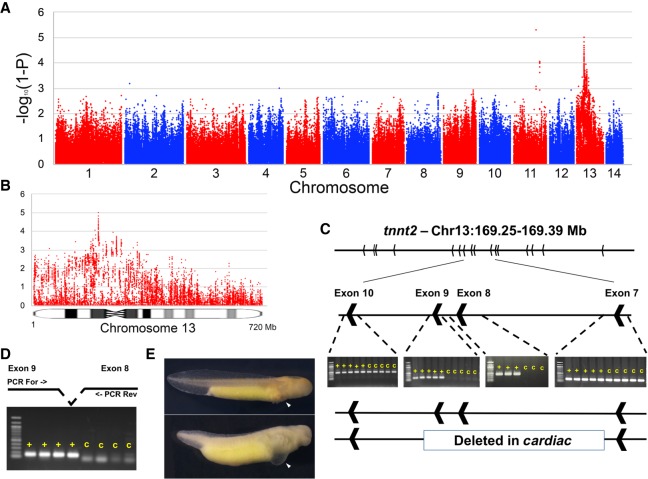
To further illustrate the utility of the assembly in performing genetic analyses in laboratory stocks of A. mexicanum, we sought to localize the mutation underlying the classical Mendelian mutant cardiac (c) (Humphrey 1972). Due to the large size of the genome and the cost associated with generating sufficient data to accurately assess genotypes from DNA sequencing data, we drew from successful efforts in newt (Notophthalmus viridescens) and used transcriptomic data to efficiently sample gene-anchored polymorphisms (Keinath et al. 2017). This sampling strategy effectively reduces the size of the genome and more deeply subsamples the ∼100 Mb of largely, gene-anchored sequences that are processed into mature transcripts. To further reduce costs associated with localizing the mutant, we used a bulked-segregant mapping strategy, sequencing RNAs from two pools of embryos that exhibited the c phenotype and two pools of their wild-type siblings (N = 10 per pool). Genotype/phenotype associations calculated from these data revealed a strong peak near the predicted location of the centromere of Chromosome 13 (Fig. 4). This is consistent with early gynogenetic diploid mapping experiments that predicted tight linkage between c and its centromere, but did not identify the specific chromosome carrying the c mutation (Armstrong 1984). Manual inspection of the region under this peak revealed that troponin 2 (tnnt2), a cardiac-specific troponin, was located under the association peak on Chromosome 13, although notably tnnt2 was not sampled at sufficient depth in our RNA-seq data to permit calculation of test statistics for the gene itself. To further assess this association, we performed genomic PCR using primers targeted to tnnt2 exons. These revealed that all exons could be amplified from wild-type individuals, as well as a specific and reproducible lack of amplification of exons 8 and 9 from cardiac homozygotes (Fig. 4C). We consider this strong evidence that this specific deletion within tnnt2 underlies the c mutation. Notably, mouse knockouts of the tnnt2 locus strongly phenocopy the axolotl c mutant, including failure of cardiac development, edema, and a lack of circulating red blood cells (Nishii et al. 2008).
Figure 4.
Localization and characterization of the cardiac mutation. (A) The genome-wide distribution of P-values for tests of association between transcribed polymorphisms and the c mutant phenotype. (B) Distribution of P-values across Chromosome 13. (C) Identification and PCR validation of a large deletion that is associated with the c phenotype and removes exons 8 and 9 of tnnt2. (D) Verification that these same exons are expressed in wild-type individuals, but not in individuals with the c phenotype. (E) Example of two siblings from a cross segregating for the cardiac mutation: (top) wild-type; (bottom) cardiac homozygote. Note the thoracic edema and lack of erythrocytes in the developing heart (white arrows) of cardiac mutants (Supplemental Fig. S3).
Discussion
This study presents a relatively cost-effective approach to scaffolding the large axolotl genome and validates the resulting assembly using both FISH and analysis of synteny conservation across vertebrates. We also demonstrate the utility of this assembly in resolving patterns of genome-wide SNP variation that (1) reveal the footprints of a historical introgression event, and (2) resolve the genetic underpinnings of the classical mutant phenotype cardiac using a simple RNA-seq-based approach that efficiently navigates the large axolotl genome. Our work sets the stage for routine analyses at this scale, including not only SNP-based approaches, but other analyses that involve large-scale signatures such as epigenetic remodeling events and promotor–enhancer interactions that will be critical for dissecting the regulatory logic of A. mexicanum development and regeneration.
tnnt2 and cardiac
The cardiac mutation was first described 45 years ago (Humphrey 1972) and has been the subject of numerous descriptive, biochemical, and genetic analyses. Animals carrying homozygous recessive alleles for c lack organized cardiac myofibrils, do not generate a heartbeat, and die around the time of hatching (Humphrey 1972; Lemanski 1973). This mutation has been attributed to defects in the expression of tropomyosin (Zajdel et al. 1999), troponin (Sferrazza et al. 2007; Zhang et al. 2007), and noncoding RNA (Lemanski et al. 1996; Zhang et al. 2003; Kochegarov et al. 2013), and has been associated with a point mutation in the noncoding myofibril-inducing RNA (MIR) (Zhang et al. 2003). Our analyses provide strong evidence that the genetic basis of the mutation lies in a large deletion that resulted in the loss of internal tnnt2 exons. Notably, the interval does not contain the myofibril-inducing RNA, which lies on Chromosome 12 upstream of the Cholinergic Receptor Muscarinic 2 (CHRM2) gene. It seems plausible, however, that cis modulation of cardiac contractile activity by CHRM2 might underlie improvement in cardiac function observed in experiments dissecting the effects of myofibril-inducing RNA.
The verification of linkage between cardiac and the centromere highlights the potential of bulked-segregant analyses in axolotl and earlier approaches that were used to map genes prior to the advent of modern molecular methods. The availability of a chromosome-scale assembly will significantly facilitate forward genetic screens of additional mutants and allow for more accurate and precise design of gene-editing tools to empower reverse genetic experiments.
Localization of the major effect QTL: met
The mapping cross used for this study was originally designed to dissect the genetic basis of paedomorphosis in A. mexicanum, whereby this species reaches sexual maturity without undergoing metamorphic changes that are typical of most amphibian species. Many of the individuals selected for this study were known to possess recombinant genotypes near the met locus. As such, estimated recombination frequencies were inflated near the position of the met QTL (Chromosome 2) (Fig. 1). This inflation permitted us to more precisely define the limits of met and identify a relatively small (∼10 Mb) genomic region containing nine annotated genes, including homologs of NGFR, SETD2, KIF9, KLHL18, and five predicted loci (LOC101951429, LOC109139901, LOC102363594, LOC106731944, LOC102943813). It seems possible that variation in one or more of these genes may regulate the expression of metamorphic versus paedomorphic modes of development, or alternately, that a gene (present in other metamorphic species) may have been lost within this region of the A. mexicanum genome. Future efforts to sequence and assemble additional salamander genomes should shed light on the ancestral structure of this region and changes that are associated with life history evolution.
Broad utility in axolotl and other salamanders
We anticipate that the improved assembly presented here will facilitate diverse studies in urodeles by providing insights into genome structure that were not previously accessible in any salamander species. Long-range interactions are critical for regulating transcription, particularly in large genomes and even in the comparatively small human genome. Enhancer–promotor interactions may involve looping structures that arch over multiple intervening genes and span megabases (Whalen et al. 2016; Cao et al. 2017). Resolving such interactions will likely be critical to understanding the gene-regulatory basis of tissue regeneration. This assembly should also be useful for interpreting patterns from large-scale sampling of polymorphisms from species and natural populations. Thus far, it has been difficult to effectively control for linkage when estimating demographic parameters and genome-wide patterns of divergence (McCartney-Melstad et al. 2016; Rodríguez et al. 2017; Nunziata and Weisrock 2018), particularly given evidence for relatively strong conservation in genome structure between Ambystoma and newt (Keinath et al. 2017), which diverged ∼150 million years ago. Extension of similar approaches to species without existing methods for production of hybrid crosses should also be relatively straightforward, provided offspring or other meiotic products can be sampled from individuals with modest levels of heterozygosity.
Methods
Sequencing of segregants for meiotic mapping
DNA was extracted from 48 individuals that were selected from a previously described mapping population that was generated by backcrossing F1 hybrid A. mexicanum/A. tigrinum to A. mexicanum (Voss and Smith 2005). Individuals were selected to include individuals of known sex and that were previously inferred to have inherited recombinant genotypes near the met locus (Voss and Smith 2005; Keinath et al. 2018). This number of individuals was also selected in an attempt to optimize trade-offs between our ability to identify and genotype informative SNPs and the cost/data volume associated with producing whole-genome shotgun data sets for large genomes. DNA was extracted via phenol-chloroform extraction (Sambrook and Russell 2001). Outsourced library prep and Illumina sequencing (HiSeq 2500 V4, 125 bp paired-end reads) was performed by HudsonAlpha Genome Services Laboratory. In total, these yielded about 14 billion reads, with each individual being sequenced to about 1× coverage.
SNP typing
Sequence data were aligned to the genome assembly (ambMex3) (Nowoshilow et al. 2018) using BWA-MEM with option -a (Li and Durbin 2009) and filtered by SAMtools view with option -F2308 (Li et al. 2009). Duplicates were marked with the bammarkduplicates2 tool from the biobambam package v.0.0.189 (Tischler and Leonard 2014), and alignments were processed by GATK V3.5 (McKenna et al. 2010; Van der Auwera et al. 2013) to identify variant sites and tabulate the number of reads supporting reference versus alternate alleles in each individual (using HaplotypeCaller -ploidy 2 --genotyping_mode DISCOVERY -stand_emit_conf 10 -stand_call_conf 30). Sites were then filtered to extract those that were homozygous for the reference base in a resequenced A. mexicanum (Ambystoma Genetic Stock Center ID# 130001.3, sequenced to 20× depth of coverage) and homozygous for the alternate base in a resequenced A. tigrinum (sequenced to 3× depth of coverage) in order to identify a subset of sites that are likely to differ consistently between the species. Prior to inferring genotypes, polymorphisms were filtered using VCFtools v0.1.13 (Danecek et al. 2011) to retain a subset of individual biallelic SNPs that were spaced at a minimum distance of 75 bp, characterized by observed A. tigrinum allele frequencies within the range expected for the backcross design (0.25 ± 0.15), and sampled in 18 or more individuals. Low-coverage SNP calls were accumulated across the length of each scaffold in order to infer whether each individual was homozygous A. mexicanum or heterozygous A. mexicanum/A. tigrinum by summing the number of reads that could be assigned to A. mexicanum or A. tigrinum and assessing the resulting ratios (A. tigrinum reads/A. tigrinum + A. mexicanum reads) to define a consensus genotype for the scaffold. Individuals with ratios reflecting no or low representation of A. tigrinum alleles (<0.20) were scored as homozygous A. mexicanum, and individuals with ratios approximating 50% A. tigrinum alleles (0.25–0.75) were scored as heterozygous A. mexicanum/A. tigrinum. Individuals with intermediate frequencies or low coverage (fewer than four reads) were scored as missing genotypes, as implemented by SparseGenotyping (https://github.com/timnat/SparseGenotyping).
Linkage analysis
A meiotic map was generated from a set of 26,577 genotyped scaffolds using JoinMap 4.1 (Van Ooijen 2006). A high-confidence backbone map was generated using a subset of 8758 scaffolds with genotypes supported by more than 200 polymorphic sites. This map was manually curated by examining all intervals >10 cM, recursively removing loci, and examining the effect of removal on map distance. Markers that increased map distance were considered erroneous genotypes and were removed from the backbone map. This backbone map was enriched with additional scaffolds with genotypes that were identical or highly similar (Jaccard distance <10%) to backbone scaffolds, without impacting the relative ordering or estimated recombinational distance between high-confidence markers on the backbone map.
Ordering and orientation with RNA-seq data
Paired-end RNA-seq data sets (all data from NCBI BioProject database under accession numbers PRJNA300706, PRJNA306100, PRJNA312389, and PRJNA354434) and bulked-segregant mapping studies (below) were aligned to genomic scaffolds and predicted gene models from ambMex3 using BWA-MEM (Li and Durbin 2009), and the resulting alignments were processed by Rascaf (Song et al. 2016) and AGOUTI (Zhang et al. 2016) to identify linkages that were supported by five or more read pairs (minimum mapping quality 20 with no mismatches for AGOUTI). The resulting linkage and orientation information was appended to the linkage map using ALLMAPS (Tang et al. 2015).
Karyotypic analyses and FISH mapping
Suspensions of mitotic cells for chromosome preparations were made according to a previously described protocol (Keinath et al. 2015). Cells were applied to a cold glass slide and immediately placed in a humid chamber at 60°C in order to spread chromosomes, spreads were then air-dried, aged overnight at 65°C, and held at −20°C in 100% ethanol until processing. To develop idiograms, DAPI-stained early metaphase chromosomes (two complete and well-resolved metaphases) were imaged, grayscaled, inverted, and contrasted using Adobe Photoshop (Demin et al. 2011). Chromosome images were straightened using ImageJ (https://imagej.nih.gov/ij/) and aligned for comparison. Two gradations of gray were used to depict reverse DAPI banding patterns: Strong DAPI-positive puncta (bands that do not necessarily span the breadth of large chromosomes) (Rudak and Callan 1976) are designated by black bars; other stronger and fainter bands are respectively designated by dark and light gray bands in idiograms (Supplemental Fig. S1). Patterns were further assessed by examining 10 additional spreads and larger numbers of spreads used for BAC hybridization.
For hybridization of individual genes, BAC libraries were screened by PCR using gene-specific primers as previously described (Smith et al. 2009) in order to identify a set of 46 BACs that correspond to known A. mexicanum genes (Supplemental Table S7). DNA from individual BAC clones was isolated using the QIAGEN Large Construct kit (QIAGEN Science) or via an outsourced purification service provided by Clemson University Genomics Institute. Labeling via nick translation, Cot2DNA suppression, and fluorescent in situ hybridizations were performed as previously described (Timoshevskiy et al. 2012). Two color fluorescence in situ hybridization was used to localize signals from BAC-derived markers on axolotl chromosomes. After localization of each new marker to a specific chromosome, according to size and DAPI banding patterns, markers on each particular chromosome were cohybridized to define their relative location and ordering.
Comparative synteny analyses
Annotated genes were aligned with BLASTP, using default alignment parameters (Altschul et al. 1990; Camacho et al. 2009) to Human (Homo sapiens RefSeq proteins from hg38: UCSC Genome Browser annotations [http://hgdownload.cse.ucsc.edu/goldenpath] translated refGene downloaded November 27, 2017), Chicken (Gallus gallus Ensembl proteins from galGal4: UCSC Genome Browser annotations [http://hgdownload.cse.ucsc.edu/goldenpath] Enpep downloaded November 27, 2017) and Xenopus tropicalis (v9.0: Xenbase [https://www.xenbase.org/other/static/ftpDatafiles.jsp] gene model peptides downloaded April 24, 2018) proteins. All genes with a bitscore ≥100 and ≥99% of the best match were considered orthologs and integrated into comparative maps. Orthologous segments were defined across the map by tabulating the corresponding chromosomal location of genes encoding presumptively orthologous proteins and calculating the consensus across all 10 cM bins.
Sequencing of bulked segregants
Preparation and sequencing of RNA pools
A cross (Ambystoma Genetic Stock Center [AGSC - RRID:SCR_006372] spawn ID: 14427) was generated between two heterozygous cardiac carriers (AGSC IDs: 13911.2 [male] and 13970.1 [female]), and embryos were raised in 10% Holtfretter's solution (Armstrong et al. 1989) at 20°C until completion of early cardiac development (for wild-type embryos: RRID:AGSC_100E). A total of 20 cardiac mutants (RRID:AGSC_104E) and 20 wild-type embryos were sampled for RNA extraction. Individual embryos were dissociated with 23- and 26-gauge needles, and RNA was isolated with TRIzol and then further purified using a QIAGEN RNeasy Mini Kit with DNase treatment. RNA extracts were pooled in groups of 10 (two pools of wild-type and two pools of cardiac homozygotes) in equimolar ratios. These RNAs were used to generate outsourced poly(A) RNA-seq libraries and were sequenced on an Illumina HiSeq 2500 (V4 sequencing chemistry, 125 bp paired-end reads) by HudsonAlpha Genome Services Laboratory.
Analysis of RNA-seq data sets
Reads from each pool of cardiac homozygotes or wild-type individuals were mapped to the complete set of annotated genomic gene models (Nowoshilow et al. 2018) (downloaded from https://www.axolotl-omics.org/assemblies) using BWA-MEM (Li and Durbin 2009). Alignments were post-processed to remove duplicate reads using picard-tools-1.97 (http://broadinstitute.github.io/picard) using default parameters recommended via GATK best practices, and raw SNPs were identified for the complete set of transcriptome sequences with a putative human or chicken homolog and all genomic gene models using GATK V3.5 (McKenna et al. 2010; Van der Auwera et al. 2013) using the following parameters: HaplotypeCaller -ploidy 2 --genotyping_mode DISCOVERY -stand_emit_conf 10 -stand_call_conf 30. SNPs were filtered to identify sites that segregated two alleles in all four pools within each cross, and that were supported by 20 or more independent reads.
Statistical assessment of association
Association between mutant phenotypes and segregating SNPs was expressed as the absolute difference in the frequencies of the nonreference allele in pools of cardiac homozygotes versus wild-type individuals (Δ). SNPs that are not associated with a mutation should segregate randomly, whereas SNPs that are tightly linked to a mutation should be homozygous in affected individuals (cardiac homozygotes) and heterozygous in two-thirds of unaffected (wild-type) individuals. This value has a theoretical range between zero and one, with an expected value of for a perfectly associated SNP. Probability values for association statistics were calculated based on the normalized (transformed) distribution of observed Δ values (Supplemental Fig. S2).
Data access
Sequence data for this study have been submitted to the NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject) under accession numbers PRJNA477812 (hybrid cross) and PRJNA509654 (Ambystoma tigrinum). Polymorphism data have been submitted to the European Variation Archive (EVA; https://www.ebi.ac.uk/eva/) under accession number PRJEB30506. The Genome assembly is deposited to the GenBank assembly (https://www.ncbi.nlm.nih.gov/assembly) under accession number GCA_002915635.2 and as an assembly hub accessible through the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway?genome=Amex_PQ.v4&hubUrl=http://salamander.uky.edu/hubExamples/hubAssembly/Amex_PQ.v4.HUB/hub.txt). The source code of the SparseGenotyping software can be found as Supplemental Code and is available on GitHub, https://github.com/timnat/SparseGenotyping.
Supplementary Material
Acknowledgments
This work was funded by grants from the National Institutes of Health (NIH) (R24OD010435) and Department of Defense (DOD) (W911NF1110475) to S.R.V. Animals used in this study were provided by the Ambystoma Genetic Stock Center, which is currently funded by the NIH (P40OD019794) and previously by the National Science Foundation (NSF) (DBI-0951484) to S.R.V. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of NIH, DOD, or NSF. Partial computational support was provided by The University of Kentucky High Performance Computing complex.
Author contributions: J.J.S. and S.R.V. conceived of the study. N.T., J.J.S., V.A.T., S.R.V., D.H., and M.C.K. contributed analyses. J.J.S. and S.R.V. authored the manuscript with the assistance of N.T. and V.A.T. D.H. and M.C.K. provided additional edits.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.241901.118.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Armstrong JB. 1984. Genetic mapping in the Mexican axolotl, Ambystoma mexicanum. Can J Genet Cytol 26: 1–6. 10.1139/g84-001 [DOI] [PubMed] [Google Scholar]
- Armstrong JB, Duhon ST, Malacinski GM. 1989. Raising the axolotl in captivity. In Developmental biology of the axolotl (ed. Armstrong JB, Malacinski GM), pp. 220–227. Oxford University Press, New York. [Google Scholar]
- Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, Lee TJ, Leigh ND, Kuo TH, Davis FG, et al. 2017. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep 18: 762–776. 10.1016/j.celrep.2016.12.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Pérez J, Espinal-Centeno A, Falcon F, García-Ortega LF, Curiel-Quesada E, Cruz-Hernández A, Bako L, Chen X, Martinez O, Alberto Arteaga-Vázquez M, et al. 2018. Transcriptional landscapes of Axolotl (Ambystoma mexicanum). Dev Biol 433: 227–239. 10.1016/j.ydbio.2017.08.022 [DOI] [PubMed] [Google Scholar]
- Callan HG. 1966. Chromosomes and nucleoli of the axolotl, Ambystoma mexicanum. J Cell Sci 1: 85–108. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Anyansi C, Hu X, Xu L, Xiong L, Tang W, Mok MTS, Cheng C, Fan X, Gerstein M, et al. 2017. Reconstruction of enhancer-target networks in 935 samples of human primary cells, tissues and cell lines. Nat Genet 49: 1428–1436. 10.1038/ng.3950 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demin S, Pleskach N, Svetlova M, Solovjeva L. 2011. High-resolution mapping of interstitial telomeric repeats in Syrian hamster metaphase chromosomes. Cytogenet Genome Res 132: 151–155. 10.1159/000321676 [DOI] [PubMed] [Google Scholar]
- Evans T, Wade CM, Chapman FA, Johnson AD, Loose M. 2014. Acquisition of germ plasm accelerates vertebrate evolution. Science 344: 200–203. 10.1126/science.1249325 [DOI] [PubMed] [Google Scholar]
- Evans T, Johnson AD, Loose M. 2018. Virtual Genome Walking across the 32 Gb Ambystoma mexicanum genome; assembling gene models and intronic sequence. Sci Rep 8: 618 10.1038/s41598-017-19128-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey RR. 1967. Albino axolotls from an albino tiger salamander through hybridization. J Hered 58: 95–101. 10.1093/oxfordjournals.jhered.a107572 [DOI] [PubMed] [Google Scholar]
- Humphrey RR. 1972. Genetic and experimental studies on a mutant gene (c) determining absence of heart action in embryos of the Mexican axolotl (Ambystoma mexicanum). Dev Biol 27: 365–375. 10.1016/0012-1606(72)90175-3 [DOI] [PubMed] [Google Scholar]
- Keinath MC, Timoshevskiy VA, Timoshevskaya NY, Tsonis PA, Voss SR, Smith JJ. 2015. Initial characterization of the large genome of the salamander Ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Sci Rep 5: 16413 10.1038/srep16413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath MC, Voss SR, Tsonis PA, Smith JJ. 2017. A linkage map for the Newt Notophthalmus viridescens: insights in vertebrate genome and chromosome evolution. Dev Biol 426: 211–218. 10.1016/j.ydbio.2016.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath MC, Timoshevskaya N, Timoshevskiy VA, Voss SR, Smith JJ. 2018. Miniscule differences between sex chromosomes in the giant genome of a salamander. Sci Rep 14: 17882 10.1038/s41598-018-36209-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochegarov A, Moses A, Lian W, Meyer J, Hanna MC, Lemanski LF. 2013. A new unique form of microRNA from human heart, microRNA-499c, promotes myofibril formation and rescues cardiac development in mutant axolotl embryos. J Biomed Sci 20: 20 10.1186/1423-0127-20-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanski LF. 1973. Morphology of developing heart in cardiac lethal mutant Mexican axolotls, Ambystoma mexicanum. Dev Biol 33: 312–333. 10.1016/0012-1606(73)90140-1 [DOI] [PubMed] [Google Scholar]
- Lemanski LF, Nakatsugawa M, Bhatia R, Erginel-Unaltuna N, Spinner BJ, Dube DK. 1996. A specific synthetic RNA promotes cardiac myofibrillogenesis in the Mexican axolotl. Biochem Biophys Res Commun 229: 974–981. 10.1006/bbrc.1996.1910 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney-Melstad E, Mount GG, Shaffer HB. 2016. Exon capture optimization in amphibians with large genomes. Mol Ecol Resour 16: 1084–1094. 10.1111/1755-0998.12538 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii K, Morimoto S, Minakami R, Miyano Y, Hashizume K, Ohta M, Zhan DY, Lu QW, Shibata Y. 2008. Targeted disruption of the cardiac troponin T gene causes sarcomere disassembly and defects in heartbeat within the early mouse embryo. Dev Biol 322: 65–73. 10.1016/j.ydbio.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, Pippel M, Winkler S, Hastie AR, Young G, Roscito JG, et al. 2018. The axolotl genome and the evolution of key tissue formation regulators. Nature 554: 50–55. 10.1038/nature25458 [DOI] [PubMed] [Google Scholar]
- Nunziata SO, Weisrock DW. 2018. Estimation of contemporary effective population size and population declines using RAD sequence data. Heredity 120: 196–207. 10.1038/s41437-017-0037-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RB, Boley MA, Kump DK, Voss SR. 2013. Genomics of a metamorphic timing QTL: met1 maps to a unique genomic position and regulates morph and species-specific patterns of brain transcription. Genome Biol Evol 5: 1716–1730. 10.1093/gbe/evt123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa SM, Clifton DK, Steiner RA. 2008. The role of kisspeptins and GPR54 in the neuroendocrine regulation of reproduction. Annu Rev Physiol 70: 213–238. 10.1146/annurev.physiol.70.113006.100540 [DOI] [PubMed] [Google Scholar]
- Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res 26: 342–350. 10.1101/gr.193474.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiß C, Olsson L, Hossfeld U. 2015. The history of the oldest self-sustaining laboratory animal: 150 years of axolotl research. J Exp Zool B Mol Dev Evol 324: 393–404. 10.1002/jez.b.22617 [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Burgon JD, Lyra M, Irisarri I, Baurain D, Blaustein L, Gocmen B, Künzel S, Mable BK, Nolte AW, et al. 2017. Inferring the shallow phylogeny of true salamanders (Salamandra) by multiple phylogenomic approaches. Mol Phylogenet Evol 115: 16–26. 10.1016/j.ympev.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Rudak E, Callan HG. 1976. Differential staining and chromatin packing of the mitotic chromosomes of the newt Triturus cristatus. Chromosoma 56: 349–362. 10.1007/BF00292955 [DOI] [Google Scholar]
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, et al. 2003. The GPR54 gene as a regulator of puberty. N Engl J Med 349: 1614–1627. 10.1056/NEJMoa035322 [DOI] [PubMed] [Google Scholar]
- Sferrazza GF, Zhang C, Jia P, Lemanski SL, Athauda G, Stassi A, Halager K, Maier JA, Rueda-de-Leon E, Gupta A, et al. 2007. Role of myofibril-inducing RNA in cardiac TnT expression in developing Mexican axolotl. Biochem Biophys Res Commun 357: 32–37. 10.1016/j.bbrc.2007.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Voss SR. 2006. Gene order data from a model amphibian (Ambystoma): new perspectives on vertebrate genome structure and evolution. BMC Genomics 7: 219 10.1186/1471-2164-7-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Voss SR. 2009. Amphibian sex determination: segregation and linkage analysis using members of the tiger salamander species complex (Ambystoma mexicanum and A. t. tigrinum). Heredity 102: 542–548. 10.1038/hdy.2009.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Putta S, Walker JA, Kump DK, Samuels AK, Monaghan JR, Weisrock DW, Staben C, Voss SR. 2005. Sal-Site: integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics 6: 181 10.1186/1471-2164-6-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Putta S, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR. 2009. Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics 10: 19 10.1186/1471-2164-10-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Shankar DS, Florea L. 2016. Rascaf: improving genome assembly with RNA sequencing data. Plant Genome 9: 1–12. 10.3835/plantgenome2016.03.0027 [DOI] [PubMed] [Google Scholar]
- Tang H, Zhang X, Miao C, Zhang J, Ming R, Schnable JC, Schnable PS, Lyons E, Lu J. 2015. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol 16: 3 10.1186/s13059-014-0573-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoshevskiy VA, Sharma A, Sharakhov IV, Sharakhova MV. 2012. Fluorescent in situ hybridization on mitotic chromosomes of mosquitoes. J Vis Exp 67: e4215 10.3791/4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler G, Leonard S. 2014. biobambam: tools for read pair collation based algorithms on BAM files. Source Code Biol Med 9: 13 10.1186/1751-0473-9-13 [DOI] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11 10 11–33 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. 2006. JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands. [Google Scholar]
- Voss SR, Shaffer HB. 1997. Adaptive evolution via a major gene effect: paedomorphosis in the Mexican axolotl. Proc Natl Acad Sci 94: 14185–14189. 10.1073/pnas.94.25.14185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss SR, Shaffer HB. 2000. Evolutionary genetics of metamorphic failure using wild-caught vs. laboratory axolotls (Ambystoma mexicanum). Mol Ecol 9: 1401–1407. 10.1046/j.1365-294x.2000.01025.x [DOI] [PubMed] [Google Scholar]
- Voss SR, Smith JJ. 2005. Evolution of salamander life cycles: A major-effect quantitative trait locus contributes to discrete and continuous variation for metamorphic timing. Genetics 170: 275–281. 10.1534/genetics.104.038273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss SR, Smith JJ, Gardiner DM, Parichy DM. 2001. Conserved vertebrate chromosome segments in the large salamander genome. Genetics 158: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss SR, Epperlein HH, Tanaka EM. 2009. Ambystoma mexicanum, the axolotl: a versatile amphibian model for regeneration, development, and evolution studies. Cold Spring Harb Protoc 2009: pdb emo128 10.1101/pdb.emo128 [DOI] [PubMed] [Google Scholar]
- Voss SR, Kump DK, Putta S, Pauly N, Reynolds A, Henry RJ, Basa S, Walker JA, Smith JJ. 2011. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res 21: 1306–1312. 10.1101/gr.116491.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen S, Truty RM, Pollard KS. 2016. Enhancer–promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet 48: 488–496. 10.1038/ng.3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock MR, Vaughn-Wolfe J, Elias A, Kump DK, Kendall KD, Timoshevskaya N, Timoshevskiy V, Perry DW, Smith JJ, Spiewak JE, et al. 2017. Identification of mutant genes and introgressed tiger salamander DNA in the laboratory axolotl, Ambystoma mexicanum. Sci Rep 7: 6 10.1038/s41598-017-00059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajdel RW, Dube DK, Lemanski LF. 1999. The cardiac mutant Mexican axolotl is a unique animal model for evaluation of cardiac myofibrillogenesis. Exp Cell Res 248: 557–566. 10.1006/excr.1999.4419 [DOI] [PubMed] [Google Scholar]
- Zhang C, Dube DK, Huang X, Zajdel RW, Bhatia R, Foster D, Lemanski SL, Lemanski LF. 2003. A point mutation in bioactive RNA results in the failure of mutant heart correction in Mexican axolotls. Anat Embryol (Berl) 206: 495–506. 10.1007/s00429-003-0315-8 [DOI] [PubMed] [Google Scholar]
- Zhang C, Pietras KM, Sferrazza GF, Jia P, Athauda G, Rueda-de-Leon E, Maier JA, Dube DK, Lemanski SL, Lemanski LF. 2007. Molecular and immunohistochemical analyses of cardiac troponin T during cardiac development in the Mexican axolotl, Ambystoma mexicanum. J Cell Biochem 100: 1–15. 10.1002/jcb.20918 [DOI] [PubMed] [Google Scholar]
- Zhang SV, Zhuo L, Hahn MW. 2016. AGOUTI: improving genome assembly and annotation using transcriptome data. GigaScience 5: 31 10.1186/s13742-016-0136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.