SUMMARY
Eukaryotic cells contain a large number of RNA–protein assemblies, generically referred to as ribonucleoprotein (RNP) granules. Such RNP granules include stress granules and P-bodies in the cytosol and the nucleolus, Cajal bodies, and paraspeckles in the nucleus. A variety of imaging approaches have been used to reveal different components, structural features, and dynamics of RNP granules. In this review, we discuss imaging approaches that have been used to study stress granules and the insights gained from these experiments. A general theme is that these approaches can be transferred to other RNP granules to examine similar aspects of their composition, ultrastructure, dynamics and control.
1. INTRODUCTION
Ribonucleoprotein (RNP) granules are higher-order assemblies composed of various RNAs and proteins, and they are ubiquitous in eukaryotes. RNP granules are diverse and vary in their components, regulation, and dynamics. Cytoplasmic RNP granules include stress granules, P-bodies, germ granules, and neuronal granules, whereas nuclear RNP granules include paraspeckles, the nucleolus, Cajal bodies, and RNA foci formed from repeat expansion RNAs in disease contexts (Gall 2000; Fox et al. 2002; Kiebler and Bassell 2006; Wojciechowska and Krzyzosiak 2011; Voronina et al. 2012; Buchan 2014). Different environmental or gene expression cues modulate each type of RNP granule. For example, although P-bodies are ubiquitously present at a basal level (Yang et al. 2004; Jain and Parker 2013), stress granules only form under conditions that disrupt translation initiation and lead to ribosomes running off the messenger RNA (mRNA) (Kedersha et al. 1999). Specialized RNP granules can also be found in specific cell types, such as neuronal granules and germ granules present in neurons and oocytes, respectively (Kiebler and Bassell 2006; Voronina et al. 2012).
Despite differences in the cellular context of assembly, RNP granules share a number of common characteristics. First, all RNP granules examined to date are nonmembrane-bound organelles. Second, by definition all RNP granules include a population of specific RNAs, which can vary between different RNP granules. For example, the nucleolus is dependent on pre-rRNA transcripts and Alu-containing RNAs for assembly, paraspeckles contain and are dependent on the NEAT1 long noncoding RNA (lncRNA), and cytoplasmic P-bodies and stress granules contain and are dependent on nontranslating mRNAs (Bond and Fox 2009; Protter and Parker 2016; Németh and Grummt 2018; Fox et al. 2018). Third, although RNP granules are often characterized and visualized by their specific components, many protein components can be found in more than one type of RNP granule, blurring the distinct lines that separate them. For example, several proteins, including the exoribonuclease Xrn1 and the RNA helicase and translational repressor Rck, are present in both P-bodies and stress granules and are also found in neuronal transport or germinal granules (Barbee et al. 2006; Buchan and Parker 2009; Buchan 2014). Fourth, RNP granules assemble through a combination of protein–protein, RNA–protein, and RNA–RNA interactions, leading to a networked assembly of RNPs, although the relative importance of these interactions can vary depending on the specific RNP granule (Kaiser et al. 2008; Jain and Vale 2017; Van Treeck et al. 2018).
RNP granules have been implicated in a number of biological processes. In some cases, RNP granules appear to be sites of localized activity, such as the nucleolus that functions as the site of ribosome biogenesis (Scheer et al. 1993) or Cajal bodies that play a role in small nuclear ribonucleoprotein (snRNP) assembly (Staněk and Neugebauer 2004; Strzelecka et al. 2010). Similarly, by concentrating components of the mRNA decay machinery, P-bodies have been proposed to increase the rates of mRNA decapping and/or 5′ to 3′ exonuclease decay (Sheth and Parker 2003). In contrast, germinal and neuronal RNP granules have been proposed to modulate translation rates through the storage of mRNAs until proper temporal or spatial release (Barbee et al. 2006; Kiebler and Bassell 2006; Strome and Updike 2015; Hubstenberger et al. 2017). In addition, the formation of stress granules, and possibly other RNP granules, can modulate signaling pathways (Kedersha et al. 2013).
Stress granules are particularly interesting RNP granules for a number of reasons. First, stress granules typically form during a variety of stress responses, including responses to heat shock, nutrient deprivation, hypo-osmotic conditions, and oxidative stress, all of which inhibit bulk translation initiation, leading to the release of most mRNAs from ribosomes (reviewed in Protter and Parker 2016). Subsets of these nontranslating mRNAs then coalesce into stress granules with their associated proteins (see below). Second, in mammalian cells in culture, a number of different genetic perturbations that limit stress granule formation lead to increased cell death during stress (Baguet et al. 2007; Eisinger-Mathason et al. 2008; Hofmann et al. 2012). Thus, stress granule formation seems to improve cell viability during the stress response, although the molecular mechanism for this effect is not clear. Finally, stress granules are very similar to germinal granules and RNP granules in neurons, the latter of which are important for at least some forms of synaptic plasticity (Barbee et al. 2006; McCann et al. 2011; Bakthavachalu et al. 2018).
Stress granules also have a number of connections to human diseases. For example, the ability to form stress granules can contribute to tumor progression, perhaps by helping tumor cells survive the insults of chemotherapeutic agents or transitions in the process of forming metastases (Anderson et al. 2015; Somasekharan et al. 2015; Grabocka and Bar-Sagi 2016). Stress granules also affect a number of viral infections; for example, the ability to form stress granules can enhance the innate immune response to dsRNA (reviewed in McCormick and Khaperskyy 2017). Moreover, mutations that affect stress granule disassembly or formation can be causative in degenerative diseases such as amyotrophic lateral sclerosis, frontotemporal lobar degeneration, and some muscle myopathies (Li et al. 2013; Ramaswami et al. 2013). One class of such mutations are dominant mutations in abundant RNA-binding proteins found in stress granules; the mutations increase the propensity of the proteins to form amyloid-like structures (Guo et al. 2011; Kim et al. 2013, Schwartz et al. 2015). This has led to the model in which stress granules create a high local concentration of these proteins, which then enhances their transition to a toxic amyloid-like state (Li et al. 2013; Ramaswami et al. 2013).
Given their biological importance and connection to human disease, stress granules have become one of the most studied RNP granules. In this review, we take advantage of the diverse work on stress granules to illustrate how RNP granules can be studied using classical and modern microscopy-based approaches. These studies provide a wealth of information about the principles of stress granule formation and dynamics as well as highlight approaches that could be applied to other RNP granules.
2. PROTEIN COMPONENTS OF STRESS GRANULES
A starting point of analysis for any RNP granule is to identify its components. A variety of different approaches and methods have been used to compile the stress granule proteome (Table 1A). For many RNP granules, the discovery of the granule and an initial component occurred concurrently, by observing intracellular foci marked with a specific protein of interest. For example, stress granules were first defined by noticing that heat shock proteins and the predominantly nuclear mRNA-binding protein TIA1 formed cytoplasmic foci during heat shock and oxidative stress, respectively (Collier and Schlesinger 1986; Kedersha et al. 1999). Similarly, P-bodies were discovered by the observation that components of the mRNA decay machinery concentrated in cytoplasmic foci (Sheth and Parker 2003). Some RNP granules were first identified through contrast imaging, such as the discovery of Cajal bodies by silver staining (Cajal 1903), and although this provided information on their overall size and structure, identification of key protein components could only occur after the advent of molecular markers. Even now, punctate nuclear or cytoplasmic localization of a protein often leads to colocalization studies to determine if this protein is localized to a previously identified granule or is a member of a novel assembly.
Table 1.
Approaches to identify stress granule components
| A. Methods to identify protein components | ||||
|---|---|---|---|---|
| Method | Pros | Cons | Contribution | Reference |
| Immunofluorescence/tagging with fluorescent protein | Technically straightforward; can measure dynamics; can measure partition coefficients |
Inefficient for global analysis; examined protein must be enriched over background; need to avoid overexpression | Identification of multiple stress granule components; measured exchange rates of many components | Reviewed in Buchan and Parker 2009 |
| Purify stress granule cores followed by mass spec | Allows global analysis; able to identify proteins that are not enriched over the cytoplasm | Need to verify hits; may lose weakly associated factors during purification | Identified 228/317 components of yeast/mammalian stress granule proteomes | Jain et al. 2016 |
| APEX proximity labeling | Allows global analysis in vivo; able to identify proteins that are not enriched over the cytoplasm; able to identify transient interactors | Partitioning and dynamics of protein with stress granules increases noise and background | Identified ∼150 possible new stress granule components; showed differences in stress granule composition in different stresses, different cell types | Markmiller et al. 2018 |
| Bio-ID | Allows global analysis in vivo; able to identify proteins that are not enriched over the cytoplasm; able to identify transient interactors |
Partitioning and dynamics of protein with stress granules increases noise and background | Identified new components of stress granules; provided evidence for subassemblies within stress granules | Youn et al. 2018 |
| B. Methods to identify RNA components | ||||
|---|---|---|---|---|
| Method | Pros | Cons | Contribution | Reference(s) |
| FISH | Techincally straightforward; can be performed for bulk mRNA (oligodT) or target-specific RNAs |
Slow and expensive for high-throughput | mRNA enriched in stress granules; identified specific mRNAs enriched or depleted from stress granules |
Kedersha et al. 1999, 2002; Damgaard and Lykke-Andersen 2011; Zurla et al. 2011 |
| Purify stress granule cores and perform RNA-seq | Allows global analysis; coupled with smFISH can yield quantitative comparisons |
Need to verify hits by smFISH; may lose weakly associated RNAs during purification |
Length and translatability are key determinants for RNA targeting to stress granules; 10%–15% of total cytoplasmic mRNA is in stress granules | Khong et al. 2017 |
| Pellet followed by RNA-seq | Allows global analysis | Not only stress granules pellet; need to validate | AU-rich elements enriched in stress granules; identified differences in stress granules during different stresses | Namkoong et al. 2018 |
| smFISH | Validation is easy; allows quantification of numbers of mRNAs in stress granules | Expensive, low-throughput |
Validation of RNAs in sequencing data sets | Khong et al. 2017 |
FISH, fluoresence in situ hybridization; RNA-seq, RNA sequencing; smFISH, single-molecule FISH.
The discovery of protein components of stress granules has gone through two phases. In the initial experiments, multiple new components of stress granules were identified either by immunofluorescence (IF) using specific antibodies or by observing the accumulation of a given fluorescently tagged protein in stress granules (e.g., Kedersha et al. 2002; Tourrière et al. 2003; Antar et al. 2005; Nonhoff et al. 2007). This piecemeal approach led to the identification of approximately 75 different proteins or protein complexes that accumulate in stress granules (reviewed in Buchan and Parker 2009). The main types of stress granule components found in these targeted experiments were RNA-binding proteins and components of the translation machinery.
The identification of an initial set of stress granule localized proteins allowed further interrogation of stress granule assembly, disassembly, and modulation. Deletion or knockdown of some protein components has resulted in an inability of stress granules to form in certain cellular contexts. For example, cells lacking G3BP1 and G3BP2 no longer form stress granules in response to eIF2α phosphorylation or eIF4A inhibition but still assemble stress granules under heat shock or osmotic stress (Kedersha et al. 2016). Conversely, overexpression of certain RNA-binding proteins highly enriched in stress granules can lead to the accumulation of constitutive stress granules, which argues that these proteins play an assembly role in stress granule formation (Kedersha et al. 1999, 2005; Mazroui 2002; Wilczynska 2005; Baguet et al. 2007). The identification of robust protein markers for stress granules has also allowed genetic screens to identify modulators of stress granule assembly and disassembly globally (Ohn et al. 2008; Buchan et al. 2013; Yang et al. 2014).
In addition, analysis of stress granule components has permitted the elucidation of compositional heterogeneity in certain contexts. Stress granules are generally conserved in eukaryotes, as orthologs of multiple mammalian stress granule proteins are present in yeast and Drosophila stress granules (Buchan et al. 2008; Aguilera-Gomez and Rabouille 2017). However, stress granule composition can vary under different stresses (Buchan et al. 2008; 2010; Aulas et al. 2017). Specifically, stress granules induced in yeast cells from NaN3 treatment contain a number of initiation factors that are absent in stress granules induced from glucose deprivation (Buchan et al. 2010). Similarly, in mammalian cells, treatment with RocA, an eIF4A inhibitor, induces the formation of cytoplasmic foci that are positive for stress granule markers but lack accumulation of poly(A)-containing mRNAs, although exposure to ultraviolet (UV) forms noncanonical stress granules lacking eIF4G and eIF3 (Aulas et al. 2017).
The second phase of stress granule component discovery has been achieved through several systematic approaches. In one set of experiments, a stable substructure within stress granules (see below), referred to as a stress granule “core,” was purified and subjected to mass spectroscopy analysis (Jain et al. 2016). This analysis identified 228 and 317 proteins enriched in yeast and mammalian stress granules, respectively. Validation of a number of hits via microscopy extended the list of stress granule protein components (Jain et al. 2016). This unbiased approach identified expected stress granule components, such as RNA-binding proteins and translation components, and also unexpected components involved in metabolism and signaling (Jain et al. 2016). Similar results were seen in a systematic approach to identify yeast stress granule components based on screening the genome-wide library of green fluorescent protein (GFP)-tagged proteins to identify proteins that concentrate into cytoplasmic stress granules during extreme heat shock (Cherkasov et al. 2015).
Another systematic approach to identify components of stress granules has used proximity-based proteomics, such as APEX and BioID, which were originally applied to identifying protein components within membrane-bound compartments (Roux et al. 2012; Rhee et al. 2013). In these experiments, a stress granule component is expressed as a fusion protein with either ascorbate peroxidase or biotin ligase, which labels neighboring proteins within the cell. Subsequent purification and mass spectroscopy can identify the labeled proteins. Utilizing proximity ligation for proteomics of nonmembrane bound organelles has a unique set of challenges. First, even for a highly enriched protein in stress granules, the percentage of that protein in a granule can be relatively small, which reduces the specificity for biotin modification of proteins that are in the RNP granule. For example, only 18% of G3BP1 is estimated to accumulate within stress granules in U-2 OS cells during oxidative stress (Wheeler et al. 2017). A second issue for BioID in particular is that the reactive species generated has a half-life of ∼30 sec, which is similar to the exchange rate of many RNP granule proteins (Buchan and Parker 2009). Again, this will reduce the specificity of identifying strictly RNP granule components.
Despite these limitations, two recent studies have used BioID or APEX labeling to provide several new insights into the stress granule proteome (Markmiller et al. 2018; Youn et al. 2018). First, these experiments identified multiple new components interacting with stress granule components, some of which were verified to be localized within stress granules (Markmiller et al. 2018; Youn et al. 2018). Second, by examining stress granule formation in different cell types and under different stresses, it was observed that, although the majority of stress granule components are conserved, some protein components are stress-specific and cell type–specific (Markmiller et al. 2018). Third, by repeated BioID analysis of multiple RNP granule components, subgroups of proteins within stress granules that preferentially interacted with each other were identified (Youn et al. 2018). It will be of interest in future work to determine if these subgroups represent different regions within stress granules, different intermediates on the pathways of stress granule assembly or disassembly, or simply represent specific types of RNPs that can accumulate within stress granules.
Taken together, this compilation of work has led to a rich census of proteins enriched in stress granules. The components encompass a spectrum of protein functions including RNA-binding proteins, translation initiation factors, stress granule remodeling complexes such as protein chaperones and helicases (Jain et al. 2016), as well as unexpected proteins involved in metabolism and signaling (Kedersha et al. 2013). This provides a rich resource for the understanding of stress granule formation and disassembly, function, and regulation.
3. RNA COMPONENTS OF STRESS GRANULES
A second set of experiments has revealed the RNA content of stress granules (Table 1B). The first experiments on mRNAs within stress granules used oligo-dT probes to show that mRNAs were highly enriched in stress granules by in situ hybridization (Kedersha et al. 1999). Based on poly(A)-staining signals, anywhere from 5% to 50% of the total mRNA population was estimated to accumulate in stress granules (Kedersha et al. 1999; Anderson and Kedersha 2008; Mollet et al. 2008; Zurla et al. 2011). Similar to the work with proteins, initial experiments examining specific mRNAs by fluorescence in situ hybridization (FISH) showed that some mRNAs could be enriched in stress granules including 5′ terminal oligopyrimidine motif (5′ TOP) mRNAs (Damgaard and Lykke-Andersen 2011). In contrast, only a minor fraction of some mRNAs was observed to be present in stress granules (e.g., only 3% of β-actin mRNA was observed to be in stress granules during arsenite stress [Zurla et al. 2011]). Similarly, mRNAs encoding heat shock proteins were largely excluded from stress granules induced by heat shock (Kedersha and Anderson 2002). These observations suggested that the mRNA recruited to stress granules was a specific subset of the total mRNA, although the nature of that population remained to be determined.
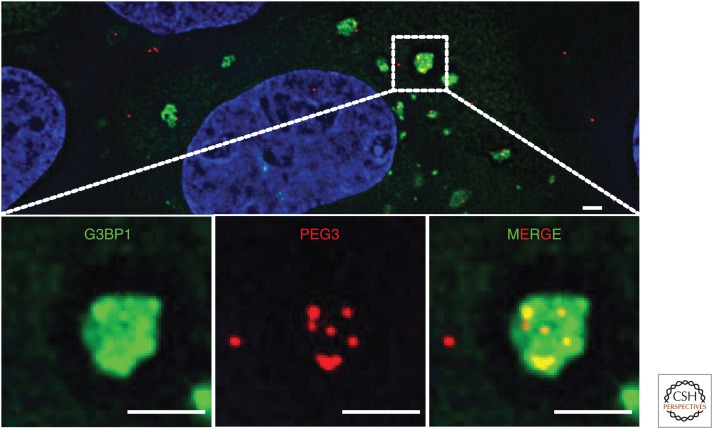
A more comprehensive analysis of the RNAs in both yeast and mammalian stress granules has come from purifying and sequencing stress granule cores (Khong et al. 2017). Importantly, by standardizing the RNA-seq analysis of stress granule cores to the fraction of a given mRNA accumulating in stress granules as determined by single-molecule FISH (smFISH) (Fig. 1), this work allowed a quantitative description of the stress granule transcriptome. This quantitative analysis revealed that ∼10–15% of cytoplasmic mRNA molecules accumulate within stress granules, and that this is a highly diverse population of RNAs with no single RNA accounting for >0.5% of the total stress granule RNA population (Khong et al. 2017). The stress granule transcriptome also revealed that ∼85% is mRNAs, with some lncRNAs also accumulating in stress granules (Khong et al. 2017). Similar results have been observed in the sequencing of mRNAs that pellet in response to stress (Namkoong et al. 2018), which is based on the premise that stress granules are large and pellet at moderate centrifugation. Thus, stress granules represent a diverse mixture of cellular RNAs, with essentially every mRNA being present in stress granules to some degree.
Figure 1.
Immunofluorescence (IF) and single-molecule fluorescence in situ hybridization (smFISH) reveal stress granule proteins and RNAs, respectively. IF of G3BP (green) delineates arsenite-induced stress granules. smFISH using probes to PEG3 (red) show robust localization of PEG3 RNAs to stress granules. Scale bar, 2 µm.
A striking observation was that individual mRNAs could vary from having 1% to 95% of their molecules in stress granules (Khong et al. 2017). Analysis of mRNA parameters revealed that long transcript length and poor translatability were the major determinants of recruitment to stress granules (Khong et al. 2017). In addition, mRNAs associated with the endoplasmic reticulum (ER) or mitochondrial membranes were generally excluded from stress granules, perhaps because they remain tethered to these membranes even after exiting translation. Additional sequence elements that correlate with increased mRNA accumulation in pellets during stress have been identified, such as AU-rich elements, but whether these directly target mRNAs into stress granules will require future experiments (Namkoong et al. 2018).
An emerging possibility is that long RNAs accumulate in stress granules simply because they can form more nonspecific RNA–RNA interactions in trans (Van Treeck et al. 2018). This possibility is first suggested by the limited correlation of the binding of stress granule proteins and RNA enrichment in stress granules (Khong et al. 2017). For example, on average, long mRNAs found in stress granules are not enriched in binding sites for stress granule proteins based on eCLIP analysis (Khong et al. 2017). Moreover, the NORAD lncRNA, which contains 19 binding sites for the stress granule component pumilio, accumulates in stress granules independent of pumilio (Namkoong et al. 2018). Evidence that RNA–RNA interactions could provide the basis for RNA enrichment in stress granules is that purified total yeast RNA will self-assemble in vitro under physiological conditions into large assemblies with the same transcript composition seen in the yeast stress granule transcriptome (Van Treeck et al. 2018). Thus, RNA–RNA interactions in trans are likely to play a contributing role to stress granule assembly in cells.
4. MICROSCOPIC ANALYSES OF STRESS GRANULES REVEAL INTERNAL ULTRASTRUCTURE
A variety of imaging approaches with higher resolution have been used to reveal that stress granules are irregular assemblies composed of dense “cores” surrounded by regions in which stress granule components are at lower concentrations. This organization was first suggested by examination of stress granules by electron microscopy (EM), which showed stress granules to be an electron-dense material with substructures of higher density (Souquere et al. 2009). Subsequent super-resolution microscopy using both STORM and SIM approaches showed that G3BP1, the poly(A)-binding protein PAB1, and mRNAs as assessed by oligo-dT FISH were all distributed nonuniformly in stress granules with clusters of “cores” surrounded by regions of lower density, referred to collectively as the “shell” (Jain et al. 2016). Refinement of stress granule core images by double-helix point spread function showed that cores have diameters of 175–190 nm on average, with G3BP and poly(A) mRNA concentrated ∼30× more in the core than the shell, and proteins concentrated ∼10× more in the shell than the bulk cytosol (Jain et al. 2016). Moreover, stress granule cores are stable in lysates and can be purified to identify their protein and RNA components (Jain et al. 2016; Khong et al. 2017). Additional evidence for cores within stress granules has come from superresolution imaging revealing that G3BP1 and IMP1 show restrained dynamics within 150–200-nm regions of the stress granule (Niewidok et al. 2018).
Similar microscopic approaches have revealed that other RNP granules also have an irregular shape with distinct substructures. For example, using lattice light sheet microscopy, the Meg-3 protein in C. elegans has been shown to form a dynamic domain that surrounds the bulk of the P-granule (Wang et al. 2014). Similarly, P-bodies in mammalian cells have been shown to have an irregular shape by EM tomography (Cougot et al. 2012) and to have subdomains with some fibrillar nature by EM (Yang 2004; Souquere et al. 2009). An interesting goal of future work will be to determine the nature of these subdomains and the assembly mechanisms that allow their formation and interchange of components.
5. IMAGING OF STRESS GRANULE ASSEMBLY AND DISASSEMBLY IN REAL TIME
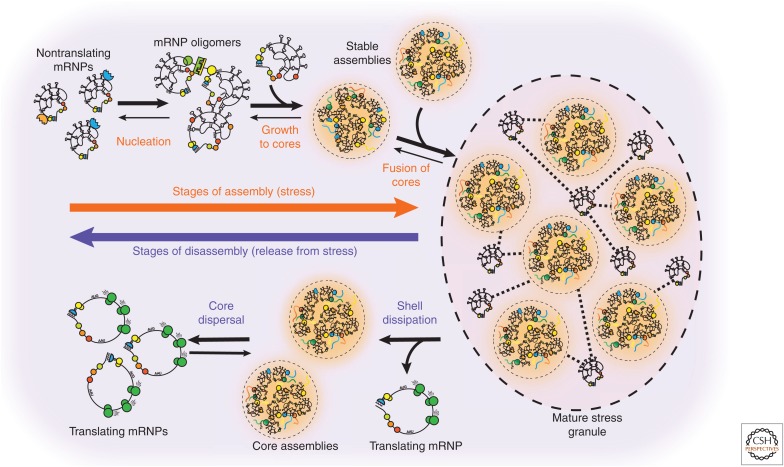
The use of fluorescently tagged stress granule proteins and imaging in real time has allowed the determination of the assembly and disassembly pathways of stress granules with multiple distinct steps (Fig. 2). When cells are first exposed to stress, there is a lag period of ∼10 min before the appearance of small stress granules (Kedersha et al. 2000; Ohshima et al. 2015; Wheeler et al. 2016). This lag may represent the time it takes for mRNAs to exit translation by ribosome runoff, as well as additional time-dependent events in the coalescing of nontranslating messenger ribonucleoproteins (mRNPs). The small stress granules that form early in the time course are biochemically stable in cell lysates, which is consistent with the first step in stress granule assembly being the coalescence of a group of nontranslating mRNPs into a stress granule core structure (Wheeler et al. 2016). Subsequently, the small stress granules increase in size both by accretion of new material and by fusion, resulting in a decrease of the total number of stress granules over time (Kedersha et al. 2000; Ohshima et al. 2015; Wheeler et al. 2016). During recovery from stress, stress granules disassemble in a reverse of this process, wherein large stress granules show a decrease in the concentration of components and fracture into smaller stress granules, which then disappear in a final step (Kedersha et al. 2000; Ohshima et al. 2015; Wheeler et al. 2016). An interesting area of current research is examining how perturbations to key stress granule components or remodeling machines affect the different stages of this assembly and disassembly pathway.
Figure 2.
Distinct steps of stress granule assembly and disassembly. Microscopy has revealed several distinct stages of stress granule assembly (orange, top) and disassembly (purple, bottom). In brief, during stress, translation is inhibited, and ribosomes run off their corresponding messenger ribonucleoproteins (mRNPs). These nontranslating mRNPs then oligomerize through either protein–protein, protein–RNA, or RNA–RNA interactions. As more RNAs enter the nontranslating pool, oligomers grow into stable core assemblies. Through fusion and RNP recruitment, a mature stress granule is produced with clear substructure. On stress release, mRNPs re-enter translation and stress granules begin to disassemble by a loss of the shell followed by core dispersal.
6. STRESS GRANULES ARE DYNAMIC ASSEMBLIES
6.1. Dynamics of Stress Granule Proteins
A variety of different approaches have been used to study the dynamics of proteins localized to stress granules (Table 2A). The most common has been fluorescence recovery after photobleaching (FRAP), in which a given component of stress granules is tagged with a fluorescent protein, bleached within a specific granule using a focused laser, and then the recovery of fluorescence is measured as a marker for exchange of bleached components for protein molecules that were originally outside the granule. Inverse FRAP (iFRAP) has also been used wherein proteins outside the granule are bleached and the rate of fluorescence loss from the granule gives a measure of protein exchange. A third approach has been to tag a stress granule protein with photoactivatable GFP, and then activate the fluorescence within an individual stress granule, which allows the experimenter to measure both the exchange of the protein from the activated granule and to determine if that protein transfers to other specific regions or granules within the cell.
Table 2.
Methods used to study dynamics of stress granule components
| A. Methods to study stress granule protein dynamics | |||
|---|---|---|---|
| Method | Component | Finding | Reference |
| FRAP/iFRAP | Many | Most proteins have a fast exchange rate, with t1/2 of recovery of <30 sec; specific proteins can have fast exchanging and slow exchanging populations; some proteins are very slow | Reviewed in Buchan and Parker 2009 |
| Photoactivatable GFP, fluorescence decay after photoactivation (FDAP) | PAGFP-G3BP1 and PAGFP-IMP1 | Allows measurement of exchange rates; can measure exchange between different stress granules |
Niewidok et al. 2018 |
| Tracking and localization microscopy (TALM) | IMP1 and G3BP1 | Both IMP1 and G3BP1 have two pools within stress granules: a dynamic fraction, and a relatively immobile fraction termed a nanocore | Niewidok et al. 2018 |
| B. Methods to study stress granule RNA dynamics | |||
|---|---|---|---|
| Method | Component | Finding | Reference(s) |
| FRAP/iFRAP | MS2-GFP tagged β-Gal mRNA; fluorescent poly(U)22 for mRNA detection |
Mostly mobile, short residence time in stress granules; ∼30% of mRNA in stress granules is immobile, mobile fraction has two exchange rates (40 sec and 275 sec) | Mollet et al. 2008; Zhang et al. 2011 |
| Single-molecule RNA tracking | Reporter RNA with MS2 stem loops to visualize | Two interaction types: brief and long-lived; larger stress granules hold RNAs longer; reporter RNAs relatively immobile within granules |
Moon et al. 2018 |
Examination of numerous stress granule components by these approaches has revealed several principles of their dynamics. First, many protein components are highly dynamic and exchange with the surrounding cytosol with t1/2 exchange rates on the order of 1–30 sec (reviewed in depth in Buchan and Parker 2009). For example, FRAP analysis of GFP-TIA-1 revealed a residence half-life of 2 sec (Kedersha et al. 2000). Second, by using photoactivatable GFP it can be shown that proteins that exchange from one granule can rapidly re-enter other stress granules (Niewidok et al. 2018). Third, some proteins exist in two pools within stress granules, with one pool showing rapid recovery in FRAP experiments, and the other failing to recover, indicative of a stably associated fraction of a given protein within stress granules (Buchan and Parker 2009). The difference between the stable and dynamic pool of proteins within stress granules could be the presence of different posttranslational modifications, different exchange rates based on being in the core or shell region of the stress granule, or differences in engaged individual protein–protein interactions.
Although many protein components exchange quickly, some show dramatically slower exchange rates. For example, the stress granule proteins FAST and Pab1 show slower exchange rates from stress granules than that of TIA1 or TTP, with FAST being extremely slow at exchanging (Kedersha et al. 2005). In principle, a slow exchange rate can occur through multiple molecular mechanisms. First, often in large assemblies, an increased number of interactions (e.g., higher valency) can lead to a slower off-rate simply because the off-rate requiring the loss of all interactions (Banani et al. 2016). Alternatively, a slow off-rate can occur if there are multiple binding sites within the stress granule, such that the probability of local rebinding is very high before exchange. Finally, a slow off-rate can also occur by formation of a single very stable interaction. An interesting area of future work will be in examining how different exchange rates of stress granule components are altered by specific protein–protein and/or protein–RNA interactions.
Although poorly understood at this time, exchange rates of proteins in stress granules do not just represent the biochemical on–off rates of proteins with their binding partners within stress granules; we should expect that these dynamics will be influenced by protein modification enzymes and energy-dependent stress granule remodeling complexes. For example, stress granule assembly and disassembly are known to be affected by a number of protein modification enzymes (e.g., Hilliker and Parker 2008), as well as protein chaperones such as the VCP or Hsp70/40 complexes, and RNA helicases, which can enhance or promote stress granule stability (Walters et al. 2015; Jain et al. 2016). Consistent with ATP-driven machines affecting stress granule dynamics, depletion of ATP from cells reduces the dynamics of G3BP1 protein exchange (Jain et al. 2016). Similarly, although the RHAU helicase exchanges from stress granules within 10 sec, a point mutation disrupting ATP hydrolysis by RHAU leads to a much slower exchange rate (Challupníková et al. 2008). This is consistent with the properties of DEAD box helicases, in which ATP hydrolysis generally leads to decreased RNA binding. An important area of future research will be to understand how different stress granule remodeling complexes affect the exchange rates of specific components.
One powerful approach for studying the dynamics of stress granules will be to use single-particle tracking and localization microscopy (TALM) (Appelhans et al. 2012). In this approach, proteins are tagged with protein modules such as the Halo or SNAP tags (Juillerat et al. 2003; Kogure et al. 2006) that can be labeled with exogenously provided chemical dyes. When the chemical dyes are provided at substoichiometric levels the limited number of molecules that are fluorescently labeled allows the experimenter to track the behavior of individual molecules. An initial application of TALM to stress granules illustrates its ability to yield new insights. Specifically, this approach shows that individual molecules of both G3BP1 and IMP1 show prolonged residence times within subregions of stress granules, referred to as nanocores (Niewidok et al. 2018). Such nanocores were measured to be ∼150–200 nm in diameter, similar to the size of cores defined by superresolution microscopy (Jain et al. 2016), and provide evidence from living cells for an internal core structure of stress granules. The power of tracking individual proteins also provided evidence for different diffusion rates of the mobile fraction of G3BP1 within stress granules and the cytosol, providing evidence for a different viscosity within stress granules (Niewidok et al. 2018).
The real power of TALM for the analysis of stress granules and other RNP assemblies will come from the analysis of how perturbations of the system affect the behavior of individual molecules. For example, deletion of an amino-terminal dimerization and caprin-interacting domain of G3BP1 led to a higher fraction of G3BP1 molecules in the bound nanocore region, although the molecular basis for this difference remains to be determined (Niewidok et al. 2018). Future experiments applying TALM to individual proteins and mRNAs and determining how their dynamics are altered by various perturbations to stress granule components, modifying enzymes, and stress granule remodeling complexes promises to be a rich area of investigation.
6.2. Dynamics of RNAs in Stress Granules
To date, few experiments have addressed the dynamics of mRNAs within stress granules (Table 2B). Given the large size of mRNAs, we should expect an mRNP within stress granules to have multiple interaction sites with other stress granule mRNPs, either through multivalent mRNA-binding proteins (Protter and Parker 2016), or through direct RNA–RNA interactions between mRNAs (Van Treeck et al. 2018). A high valency of interaction for a specific mRNA within stress granules would be expected to give a slow rate of exchange (Banani et al. 2016). Some observations consistent with this expectation have come from examining the exchange rates of bulk poly(A)+ mRNAs within stress granules by the injection of fluorescent oligo(U) probes into COS7 cells. Specifically, these experiments revealed that ∼30% of the mRNAs within stress granules are immobile and exchange only very slowly, if at all (Zhang et al. 2011). Additionally, two exchanging populations of mRNAs were observed. Approximately 35% of the mRNAs exchanged with half-times of ∼30 sec, whereas a second population (∼30% of mRNAs) exchanged with a half-time of ∼275 sec (Zhang et al. 2011). These results show that mRNAs can interact with stress granules in different modes and that overall, the exchange rates of mRNAs from stress granules are significantly slower than those of most stress granule proteins. Multiple modes of mRNA interaction with stress granules were also observed in the FRAP analysis of a β-Gal reporter mRNA tagged with multiple MS2-GFP fusion proteins, although the half-times for exchange, 2 and 58 sec, were much faster (Mollet et al. 2008). Whether these faster rates are caused by the specific mRNA being examined or differences in the experimental setup remain to be determined.
7. PERSPECTIVES AND FUTURE DIRECTIONS
Much progress has been made since the discovery of stress granules approximately 30 years ago. The ultrastructure of stress granules, as well as the protein and RNAs that accumulate in stress granules, are now known. Specific protein–protein and RNA–RNA interactions that contribute to their assembly have been defined and provide an intellectual framework for understanding perturbations that affect stress granule assembly. In addition, a variety of modification enzymes and stress granule remodeling complexes are known to alter assembly and disassembly. Finally, a number of mutations in stress granule components that lead to human disease have been identified.
Many of the outstanding key issues in understanding stress granules can be addressed by imaging approaches. For example, understanding how specific stress granule components are affected by alterations in stress granule remodeling complexes can be achieved using TALM and chemical or genetic inhibition of stress granule remodeling complexes. Similarly, understanding the dynamics of mRNA interaction with stress granules, and how that interaction is modulated, will be achieved with live single-molecule imaging of mRNAs and stress granules over time. Furthermore, understanding how pathogenic mutations affect the behavior of key components of stress granules may yield insight into how perturbations of stress granules can influence human disease.
Finally, it should be noted that all of the approaches discussed in this review are applicable to other RNP granules, and can be used to understand their composition, ultrastructure, dynamics, and regulation. All of these areas promise rich investigation in the near future.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Aguilera-Gomez A, Rabouille C. 2017. Membrane-bound organelles versus membrane-less compartments and their control of anabolic pathways in Drosophila. Dev Biol 428: 310–317. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P. 2015. Stress granules, P-bodies and cancer. Biochimica et Biophysica Acta 1849: 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. 2005. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav 4: 350–359. [DOI] [PubMed] [Google Scholar]
- Appelhans T, Richter CP, Wilkens V, Hess ST, Piehler J, Busch KB. 2012. Nanoscale organization of mitochondrial microcompartments revealed by combined tracking and localization microscopy. Nano Lett 12: 610–616. [DOI] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P. 2017. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130: 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet A, Degot S, Cougot N, Bertrand E, Chenard MP, Wendling C, Kessler P, Le Hir H, Rio MC, Tomasetto C. 2007. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci 120: 2774–2784. [DOI] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. 2016. Compositional control of phase-separated cellular bodies. Cell 166: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko A-M, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavachalu B, Huelsmeier J, Sudhakaran IP, Hillebrand J, Singh A, Petrauskas A, Thiagarajan D, Sankaranarayanan M, Mizoue L, Anderson EN, et al. 2018. RNP-granule assembly via Ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98: 754–766. [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH. 2009. Paraspeckles: Nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. 2014. mRNP granules: Assembly, function, and connections with disease. RNA Biol 11: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. 2009. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 36: 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon JH, Parker R. 2010. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R. 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153: 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SR. 1903. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en lo diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Investig Biol Univ Madr 2: 129–221. [Google Scholar]
- Challupníková K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. 2008. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem 283: 35186–35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Grousl T, Theer P, Vainshtein Y, Gläßer C, Mongis C, Kramer G, Stoecklin G, Knop M, Mogk A, et al. 2015. Systemic control of protein synthesis through sequestration of translation and ribosome biogenesis factors during severe heat stress. FEBS Lett 589: 3654–3664. [DOI] [PubMed] [Google Scholar]
- Collier NC, Schlesinger MJ. 1986. The dynamic state of heat shock proteins in chicken embryo fibroblasts. J Cell Biol 103: 1495–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Cavalier A, Thomas D, Gillet R. 2012. The dual organization of P-bodies revealed by immunoelectron microscopy and electron tomography. J Mol Biol 420: 17–28. [DOI] [PubMed] [Google Scholar]
- Damgaard CK, Lykke-Andersen J. 2011. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev 25: 2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason TSK, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shavanowitz J, Hunt DF, Macara IG, et al. 2008. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell 31: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AKL, Lyon CE, Andersen J, Mann M, Lamond AI. 2002. Paraspeckles: A novel nuclear domain. Curr Biol 12: 13–25. [DOI] [PubMed] [Google Scholar]
- Fox AH, Nakagawa S, Hirose T, Bond CS. 2018. Paraspeckles: Where long noncoding RNA meets phase separation. Trends Biochem Sci 43: 124–135. [DOI] [PubMed] [Google Scholar]
- Gall JG. 2000. Cajal bodies: The first 100 years. Annu Rev Cell Dev Biol 16: 273–300. [DOI] [PubMed] [Google Scholar]
- Grabocka E, Bar-Sagi D. 2016. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell 167: 1803–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Chen Y, Zhou X, Kar A, Ray P, Chen X, Rao EJ, Yang M, Ye H, Zhu L, et al. 2011. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nat Struct Mol Biol 18: 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Parker R. 2008. Stressed out? Make some modifications! Nat Cell Biol 10: 1129–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G. 2012. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell 23: 3775–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot J-B, Munier A, Fradet M, et al. 2017. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 68: 1–14. [DOI] [PubMed] [Google Scholar]
- Jain S, Parker R. 2013. The discovery and analysis of P bodies. Adv Exp Med Biol 768: 23–44. [DOI] [PubMed] [Google Scholar]
- Jain A, Vale RD. 2017. RNA phase transitions in repeat expansion disorders. Nature 546: 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. 2016. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K. 2003. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol 10: 313–317. [DOI] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. 2008. De novo formation of a subnuclear body. Science 322: 1713–1717. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. 2002. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30: 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of Tia-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. 2002. Evidence that ternary complex (eIF2–GTP–tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13: 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. 2013. Stress granules and cell signaling: More than just a passing phase? Trends Biochem Sci 38: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. 2016. G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212: 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. 2017. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68: 808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. 2006. Neuronal RNA granules: Movers and makers. Neuron 51: 685–690. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure T, Karasawa S, Araki T, Saito K, Kinjo M, Miyawaki A. 2006. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat Biotechnol 24: 577–581. [DOI] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. 2013. Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201: 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo E-C, Krach F, Yang D, Sen A, et al. 2018. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172: 590–598.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R. 2002. Trapping of messenger RNA by fragile X mental retardation protein into cytoplasmic granules induces translation repression. Hum Mol Gen 11: 3007–3017. [DOI] [PubMed] [Google Scholar]
- McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. 2011. The ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci 108: E655–E662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Khaperskyy DA. 2017. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 17: 647–660. [DOI] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell 19: 4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, Lyon K, Parker R, Stasevich TJ. 2018. Imaging of single mRNA translation repression reveals diverse interactions with mRNP granules. bioRxiv 10.1101/332692. [DOI] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, Lee JH. 2018. Systematic characterization of stress-induced RNA granulation. Mol Cell 70: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A, Grummt I. 2018. Dynamic regulation of nucleolar architecture. Curr Opin Cell Biol 52: 105–111. [DOI] [PubMed] [Google Scholar]
- Niewidok B, Igaev M, Pereira da Graca A, Strassner A, Lenzen C, Richter CP, Piehler J, Kurre R, Brandt R. 2018. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J Cell Biol 217: 1303–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo M-L, Lehrach H, Krobitsch S. 2007. Ataxin-2 interacts with the DEAD/H-Box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 18: 1385–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 10: 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima D, Arimoto-Matsuzaki K, Tomida T, Takekawa M, Ichikawa K. 2015. Spatio-temporal dynamics and mechanisms of stress granule assembly. PLoS Comput Biol 11: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R. 2016. Principles and properties of stress granules. Trends Cell Biol 26: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. 2013. Altered ribostasis: RNA–protein granules in degenerative disorders. Cell 154: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. 2013. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339: 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. 2012. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196: 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Thiry M, Goessens G. 1993. Structure, function and assembly of the nucleolus. Trends Cell Biol 3: 236–241. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Cech TR, Parker RR. 2015. Biochemical properties and biological functions of FET proteins. Annu Rev Biochem 84: 355–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TGP, Zhang F, Ng T, Delattre O, Evdokimova V, et al. 2015. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol 208: 913–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. 2009. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci 122: 3619–3626. [DOI] [PubMed] [Google Scholar]
- Staněk D, Neugebauer KM. 2004. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 166: 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Updike D. 2015. Specifying and protecting germ cell fate. Nat Rev Mol Cell Biol 16: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. 2010. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 17: 403–409. [DOI] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci 115: 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Paix A, Seydoux G. 2012. The P granule component PGL-1 promotes the localization and silencing activity of the PUF protein FBF-2 in germline stem cells. Development 139: 3732–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RW, Muhlrad D, Garcia J, Parker R. 2015. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA 21: 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JT, Smith J, Chen B-C, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G. 2014. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3: e04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. 2016. Distinct stages in stress granule assembly and disassembly. eLife 5: e18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Jain S, Khong A, Parker R. 2017. Isolation of yeast and mammalian stress granule cores. Methods 126: 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A. 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 118: 981–992. [DOI] [PubMed] [Google Scholar]
- Wojciechowska M, Krzyzosiak WJ. 2011. Cellular toxicity of expanded RNA repeats: Focus on RNA foci. Hum Mol Genet 20: 3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EKL. 2004. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci 117: 5567–5578. [DOI] [PubMed] [Google Scholar]
- Yang X, Shen Y, Garre E, Hao X, Krumlinde D, Cvijović M, Arens C, Nyström T, Liu B, Sunnerhagen P. 2014. Stress granule-defective mutants deregulate stress responsive transcripts. PLoS Genet 10: e1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J-Y, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. 2018. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell 69: 517–532.e11. [DOI] [PubMed] [Google Scholar]
- Zhang J, Okabe K, Tani T, Funatsu T. 2011. Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J Cell Sci 124: 4087–4095. [DOI] [PubMed] [Google Scholar]
- Zurla C, Lifland AW, Santangelo PJ. 2011. Characterizing mRNA interactions with RNA granules during translation initiation inhibition. PLoS ONE 6: e19727. [DOI] [PMC free article] [PubMed] [Google Scholar]