Abstract
The eukaryotic translation pathway has been studied for more than four decades, but the molecular mechanisms that regulate each stage of the pathway are not completely defined. This is in part because we have very little understanding of the kinetic framework for the assembly and disassembly of pathway intermediates. Steps of the pathway are thought to occur in the subsecond to second time frame, but most assays to monitor these events require minutes to hours to complete. Understanding translational control in sufficient detail will therefore require the development of assays that can precisely monitor the kinetics of the translation pathway in real time. Here, we describe the translation pathway from the perspective of its kinetic parameters, discuss advances that are helping us move toward the goal of a rigorous kinetic understanding, and highlight some of the challenges that remain.
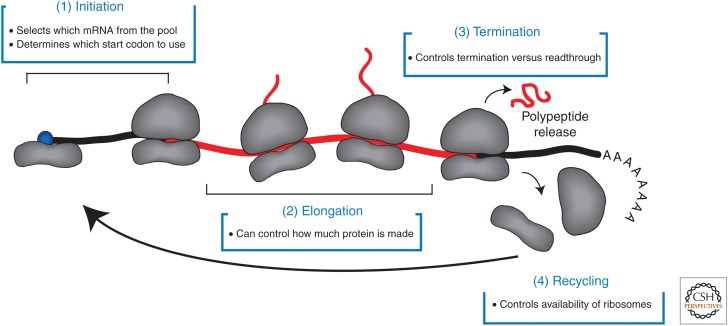
Determining how the process of translation is regulated in eukaryotic cells remains a central challenge in biology. Translation can be studied in vitro and in vivo using techniques that range from single-molecule analysis to genome-wide measurements. The latter are playing a key part in revealing the extent to which translational control plays a central role in regulating many aspects of cell physiology. The canonical pathway of translation can be depicted as four main stages (Fig. 1). The initiation stage determines which messenger RNA (mRNA) is recruited to the ribosome and which start codon is selected. During elongation, the ribosome translates the open reading frame (ORF) into a polypeptide chain. Termination occurs when the termination codon enters the aminoacyl (A) site of the ribosome, leading to the release of the polypeptide chain. Finally, recycling of the ribosome takes place so that it can enter another cycle of translation.
Figure 1.
Pathway of eukaryotic translation. The model depicts four main stages of translation. (1) The initiation stage includes the recruitment of the 40S subunit to the m7G cap structure, followed by its migration in a 3′ direction. Following start codon selection, the 60S subunit joins to form the 80S ribosome. This stage determines which messenger (mRNA) is selected from the pool and which start codon is chosen to initiate translation. (2) The elongation stage involves decoding the open reading frame (ORF) into a polypeptide chain. This stage can determine the rate at which protein is synthesized when initiation is very rapid (see main text). (3) Translation termination occurs when a termination codon is positioned in the A site of the ribosome, which results in the release of the polypeptide chain. The regulation of termination versus readthrough occurs at this step. (4) Following termination, the 40S and 60S subunits are recycled so that they can take part in another round of translation. For simplicity, the model depicts the recycling of the 40S subunit back onto the same mRNA. The rate of recycling can regulate the availability of ribosomal subunits for translation.
Translational efficiency (TE) relates the abundance of an mRNA to its rate of translation into protein. In yeast and mammals, the TE can vary between mRNAs by at least 20-fold, which provides a considerable range of translational control (Ingolia et al. 2009; Weinberg et al. 2016). Genome-wide analysis of the translatome indicates that the TE of a large number of different mRNAs is altered in response to changes in environmental conditions and events such as progression through stages of the cell cycle (Ingolia et al. 2011; Brar et al. 2012; Stumpf et al. 2013; Tanenbaum et al. 2015). A major challenge is to elucidate the molecular mechanisms that reprogram the translational machinery to determine: (1) what mRNA is selected for translation from the pool of mRNAs in the cell; (2) which start codon is used to specify the ORF that is translated; (3) how much protein is made from the selected mRNA; and (4) which termination codon is used.
As we discuss below, each stage of translation is generally believed to occur as a series of intermediate sequential substeps. Pathway intermediates are typically monitored at steady-state, which provides only limited information about how the pathway is regulated. To fully understand translational control, it will be essential to determine the rate at which intermediates are formed. This will require precise monitoring of the dynamics of the translational machinery in real time throughout the translation pathway in vitro and in vivo. In this review, we discuss the progress being made toward this goal and how we may achieve this with continued technological progress.
INITIATION
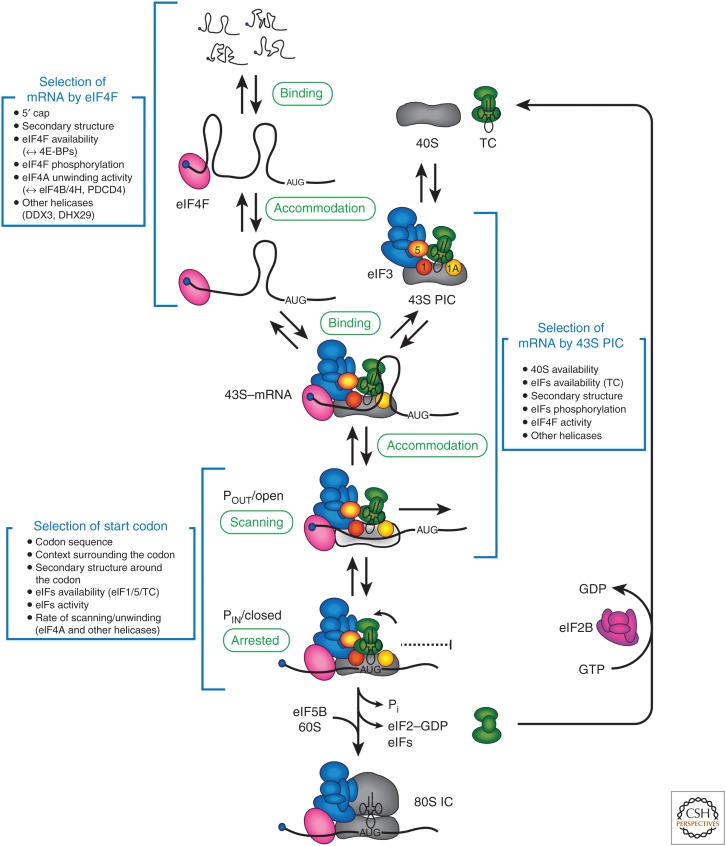
During initiation, an mRNA is recruited to the ribosome and a start codon is selected (for details of the initiation pathway, see Merrick and Pavitt 2018). Of all the stages of translation, initiation is the most different between the domains of life. This is highlighted by the fact that the combined molecular weight of the initiation factors is roughly an order of magnitude more in eukaryotes as compared to bacteria. Initiation can be separated into substeps, which are depicted in Figure 2. It is important to note, however, that the proposed substeps have generally been detected and characterized by virtue of their being relatively long-lived thermodynamically stable complexes. Being that many intermediates in the pathway are likely to exist on the subsecond or second timescale, new approaches for rapidly detecting pathway intermediates will be needed to verify the importance of these proposed intermediates. Moreover, whereas the initiation pathway is presented as a sequence of ordered steps, the order of binding and release of components during initiation is not known. It is also not clear whether different initiation factors exist in a free form, or are bound together in larger multifactor complexes. Two unanswered fundamental questions about the regulation of initiation are (1) how does the translation machinery discriminate between mRNAs for their recruitment to the ribosome; and (2) how is the selection of the ORF regulated? Here, we discuss how we can move toward a kinetic framework for the initiation stage and explain why this will be needed to fully answer these two questions.
Figure 2.
Cap-dependent translation initiation. The eukaryotic translation initiation pathway is depicted as a series of substeps, many of which are likely to be reversible (see main text for details). Potential sources of regulation for each substep are summarized in boxes and described in more detail in the main text. Selection of messenger RNA (mRNA) by eukaryotic initiation factor (eIF)4F: the mRNA is selected for translation by the binding and accommodation of the eIF4F complex. The accommodation step requires secondary structure in the mRNA to be unwound by the ATP-dependent helicase activity of eIF4A (secondary structure is depicted as “loops” in the 5′UTR). Selection of an mRNA by the 43S PIC: the 43S PIC (40S subunit, eIF1, eIF1A, ternary complex [TC], eIF3, and eIF5) is recruited to the 5′ end of the mRNA through its interaction with eIF4F. Productive recruitment occurs through initial binding and accommodation steps. The accommodation step requires a conformational change in the 40S subunit to a POUT/open conformation (described in the main text and depicted as a light gray-colored 40S subunit). Selection of the start codon: the base-pairing between the methionyl-transfer RNA (Met-tRNAi) and the start codon generates a scanning arrested 43S preinitiation complex (PIC). The arrested complex possesses a PIN/closed conformation. The final step of the pathway involves the recruitment of the 60S subunit to form the 80S initiation complex (IC). This step is stimulated by the GTPase activity of eIF5B. Following its release from the 40S subunit, the eIF2•GDP complex is recycled to eIF2•GTP by the guanine nucleotide exchange factor (GEF) activity of eIF2B. It should be noted that the possible function of poly(A)-binding protein (PABP) in circularization of the mRNA is not shown for clarity.
RECRUITMENT OF mRNA TO THE 43S PREINITIATION COMPLEX (PIC)
In response to various physiological stimuli, the translation machinery is reprogrammed to change which mRNAs are preferentially selected for recruitment to the ribosome. A large amount of data indicate that reprogramming occurs in response to changes in the availability of at least two canonical initiation factors. These are the cap-binding protein, eukaryotic initiation factor (eIF)4E, which together with eIF4A and eIF4G forms the cap-binding complex, eIF4F; and the eIF2•GTP•Met-tRNAi ternary complex (TC). An increase in the availability of eIF4E (see Proud 2018) correlates with increased recruitment of growth-promoting mRNAs, whereas a decrease in its availability correlates with reduced recruitment of these mRNAs. In contrast, reduced availability of the TC more generally lowers global rates of translation, although the translation of some mRNAs containing upstream ORFs (uORFs) can be increased in response to reduced TC availability (see Wek 2018).
Regulation of 43S PIC Availability
The 43S PIC can be purified from eukaryotic cells as a stable complex containing the 40S subunit, eIF1, eIF1A, TC, eIF3, and eIF5. Thermodynamic frameworks for the yeast and human 43S PIC in the absence of mRNA have revealed that the stability of this complex is governed by a network of direct and indirect interactions among its components (reviewed in Fraser 2015). This complex can be formed on free ribosomal subunits, or on 40S subunits that are generated during ribosome recycling (see Hellen 2018). A kinetic framework for association/dissociation rates of eIF1, eIF1A, TC, eIF3, and eIF5 from the 40S subunit has not been determined and it is not known whether different kinetically favored complexes regulate the subsequent recruitment of specific mRNAs to the 43S PIC.
The availability of TC is one 43S PIC component that is regulated in response to changes in environmental conditions. In response to different cellular stresses, eIF2 is phosphorylated, which results in reduced TC availability for 40S binding by virtue of an altered interaction with its guanine nucleotide exchange factor, eIF2B (see Merrick and Pavitt 2018; Wek 2018). Initiation pathway models imply that binding of the TC to the 40S subunit is required before mRNA recruitment, suggesting that reduced TC levels would globally reduce translation. However, sucrose gradient, gel shift, and toeprinting assays that are generally used to monitor the mRNA•eIF4F•43S PIC complex (often described as the 48S PIC) rely on start codon recognition by the TC to stabilize the complex. Thus, it is unclear whether prior binding of the TC is required before mRNA recruitment to the 40S ribosomal subunit. To investigate this, we recently generated an equilibrium-binding assay that revealed a 10-fold reduction in the affinity of eIF3j for the eIF4F•43S PIC on mRNA accommodation into the mRNA entry channel (Sokabe and Fraser 2017). The TC was found to be absolutely required for the reduced eIF3j affinity in an mRNA-dependent, but AUG codon-independent manner. This is consistent with the recruitment of the TC being needed for mRNA to fully accommodate into the mRNA entry channel of the 40S subunit. What is currently not known is whether mRNA accommodation into the entry channel is absolutely required for the scanning stage of initiation, or whether scanning can occur on an mRNA that is partially accommodated into the entry channel of the 40S subunit. An m7G cap-proximal AUG codon is more frequently bypassed when TC availability is reduced in cells in response to increased eIF2 phosphorylation (Palam et al. 2011). A similar observation was made in cell-free extracts when mRNA concentration was increased (Dasso et al. 1990). These findings are consistent with TC not being absolutely required for mRNA recruitment to and accommodation into the decoding site of the 40S subunit, or the subsequent translocation of the 40S subunit. However, TC is of course absolutely necessary for the recognition of the initiation codon. More investigation will be needed to establish the precise role of TC in mRNA recruitment to the 43S PIC and whether its availability can influence mRNA selection in addition to start site selection. In addition to TC availability, it is possible that the amount of free 43S PICs for mRNA recruitment becomes limiting for translation when the majority of ribosomes are engaged in translation in actively growing cells. Under such conditions, the rate of ribosome recycling after termination would be expected to play a significant role in controlling translation (see below).
The Role of the eIF4F Complex in mRNA Selection
The molecular details of how eIF4F regulates the selective recruitment of different messages to the ribosome are complex. eIF4F comprises eIF4E, the m7G-cap-binding subunit; eIF4A, an RNA helicase protein; and eIF4G, a large scaffolding protein. It therefore is capable of numerous activities: binding to the m7G-cap; melting of RNA secondary structure; and interacting through eIF4G with other proteins such as the poly(A)-binding protein (PABP), eIF3, DDX3/ded1, and the protein kinase MNK1 (Lamphear et al. 1995; Imataka et al. 1998; Pyronnet et al. 1999; Korneeva et al. 2000; Hilliker et al. 2011; Soto-Rifo et al. 2012). Human eIF4F can be purified from cells as a stable trimeric complex, but in cells may also be present as a mixture of larger complexes containing the eIF4G-binding proteins and possessing different mRNA-binding properties (Merrick 2015). Given the dynamic aspects of eIF4F and its complexes, we will evaluate how the different substeps of mRNA selection are accomplished.
The first step in the interaction of eIF4F with mRNA is its binding to the m7G-cap through its eIF4E subunit. The rate of cap binding can be reduced if secondary structure in the mRNA “hides” the cap, sterically hindering its interaction with eIF4E (reviewed in Kozak 2005; Livingstone et al. 2010; Fraser 2015). Such weak RNA secondary structures are likely dynamic, being partially or completely unwound through thermal energy in the cell. When the m7G-cap is sufficiently exposed, eIF4E binds and forms an eIF4F•mRNA complex called the preaccommodated state. This complex is not thought to be very stable, although its rates of binding and dissociation are not well elucidated. It seems likely that mRNAs differ in how exposed their cap structures are. Under conditions of limiting eIF4F, the mRNAs must compete for binding to the factor. Those mRNAs that are best able to bind with well-exposed 5′ caps are often called “strong” mRNAs (reviewed in Gingras et al. 1999).
Thermodynamic frameworks have shown that eIF4E can bind to the m7G-cap in its free form, bound to eIF4G (and eIF4F associated factors), or associated with 4E-BPs. The binding of eIF4F to the cap is thermodynamically favored over that of uncomplexed eIF4E binding (Niedzwiecka et al. 2002), but this does not inform about the order in which these components bind to the cap structure. Heterogeneity in the order of binding is likely to occur, but whether this contributes to regulating specific mRNA recruitment is not known. The use of single-molecule analysis similar to that recently used to study the order of binding and release of components during bacterial initiation will likely help solve this question (Tsai et al. 2012; see Prabhakar et al. 2018).
The next step is the formation of an additional interaction involving the RNA-binding region of eIF4G and a cap-proximal portion of the mRNA that is unstructured. The resulting eIF4F•mRNA complex, called the accommodated state, is more stable, although again its rate of formation and dissociation are not known. Based on cross-linking experiments, it was shown that formation of the accommodated state is inhibited by cap-proximal secondary structure (Pelletier and Sonenberg 1985; Lawson et al. 1986). DEAD-box helicases, such as eIF4A, unwind duplex regions by local strand separation, whereby they can bind directly to the duplex region and destabilize it (Yang et al. 2007). Whether the opening of cap-proximal structures by thermal energy to allow eIF4A access to a duplex contributes to eIF4F accommodation is unknown. It is thought that eIF4F accommodation could function as a kinetic checkpoint for mRNA selection, as the accommodated state is more stable than the pre-accommodated state and therefore more likely to progress down the initiation pathway. Should the rate of accommodation of eIF4F not be suitably fast, it is possible that the entire eIF4F complex will dissociate from the mRNA, preventing its selection for translation. The accommodation model will need to be experimentally tested by determining detailed kinetic frameworks of eIF4F binding to different mRNAs. Obtaining high-resolution structures of mRNAs in the absence and presence of eIF4F (or other RNA-binding proteins) will provide the necessary structural information to explain how mRNAs are selected by eIF4F.
How can the mechanism of mRNA binding by eIF4F be regulated so that mRNA recruitment can be reprogrammed? It has long been proposed that mRNAs compete for limiting amounts of eIF4F in the cell (Gingras et al. 1999). The regulation of eIF4F availability is controlled by eIF4E-binding proteins (4E-BPs) acting as competitive inhibitors to prevent eIF4E binding to eIF4G (Richter and Sonenberg 2005). It is generally assumed that eIF4G (and associated proteins) cannot productively bind to the 5′ end of an mRNA and successfully recruit it to the ribosome in the absence of eIF4E. However, this can be accomplished in vitro by an eIF4G truncation lacking its eIF4E-binding domain (eIF4G682-1599 [Ali et al. 2001]). Whether full-length eIF4G can accomplish this has not been rigorously tested.
Following formation of the eIF4F•mRNA accommodated state, binding of the 43S PIC to the 5′ end of the mRNA occurs through an interaction between eIF4G and eIF3 (Lamphear et al. 1995; Korneeva et al. 2000; Morino et al. 2000; Villa et al. 2013). Binding and mRNA entry into the mRNA channel of the 40S ribosomal subunit requires an unstructured portion of the mRNA. To enable 43S PIC binding, a secondary structure near the cap-bound eIF4F complex is unwound by the helicase activity of its eIF4A subunit, together with eIF4B (or eIF4H) (Rozen et al. 1990; Rogers et al. 2001; Nielsen et al. 2011; Özeş et al. 2011; Harms et al. 2014; see Merrick and Pavitt 2018). The ability of eIF4F to unwind hairpins in vitro is thought to be limited to those with modest stability (∼ΔG −30 kcal/mol [Linder and Jankowsky 2011]). A limited unwinding potential of eIF4F may be caused by the fact that eIF4A, like other DEAD-box helicase proteins, functions as a nonprocessive helicase in the absence of eIF4G (Linder and Jankowsky 2011; García-García et al. 2015). However, using a force-generated, single-molecule assay, eIF4A was shown to behave as a processive helicase protein in the presence of eIF4G and eIF4B/4H (García-García et al. 2015). Single-molecule fluorescence resonance energy transfer (smFRET) has shown that eIF4B, eIF4H, and eIF4G promote unwinding by increasing the rate of cycling between “open” and “closed” conformations of eIF4A (Sun et al. 2012; Harms et al. 2014). It should be noted that the processivity mechanism used by eIF4A is still not clear. It is possible that, whereas eIF4A itself may not remain stably bound to an mRNA following ATP hydrolysis, cooperative interactions between eIF4A, eIF4G, eIF4B, and RNA may prevent eIF4A from dissociating from the eIF4F•eIF4B complex (thereby enabling eIF4A to take part in another round of unwinding). Monitoring rapid kinetics of the association and dissociation rates between eIF4F components during this process will explain this in greater detail. The interaction of Pdcd4 with eIF4A may function to inhibit the availability and/or unwinding activity of eIF4A, thereby decreasing 43S PIC binding on structured mRNAs (Yang et al. 2003). Finally, the recruitment of additional helicase proteins, such as DDX3/ded1p, to the eIF4F complex may regulate its ability to unwind secondary structure (Gao et al. 2016).
Accommodation of the 43S PIC onto mRNA
It is possible that the final stage of mRNA selection for translation is completed when the mRNA fully accommodates into the mRNA entry channel and decoding site of the 43S PIC (Fig. 2). However, it should be noted that the dissociation rate of a decoding site–accommodated mRNA has not been measured, so it is entirely possible that start codon recognition could serve as the final stage of mRNA selection. For mRNA accommodation into the entry channel and decoding site of the 40S subunit, the 43S PIC must adopt an open conformation of the decoding site. The binding of eIF1 and eIF1A to the 40S subunit generates this conformation (Pestova et al. 1998; Passmore et al. 2007; Hussain et al. 2014). However, other initiation components may contribute to stabilizing this conformation because structures of 40S subunit complexes that include eIF1 and eIF1A adopt a closed conformation (Hashem et al. 2013; Lomakin and Steitz 2013; Weisser et al. 2013). All structures to date have been determined in the absence of the eIF4F complex. It is therefore unknown whether this complex affects the conformation of the 40S subunit decoding site. In support of this possibility, eIF4F and eIF4B were recently shown to lower the affinity of eIF3j for the 43S PIC, but only in the presence of mRNA (Sokabe and Fraser 2017). This is likely caused by the fact that eIF3j binds to the mRNA entry channel and A site of the 43S PIC with a reduced affinity when mRNA is stabilized in the mRNA entry channel (Unbehaun et al. 2004; Fraser et al. 2007; Aylett et al. 2015; Sharifulin et al. 2016). Notably, an unstructured mRNA is required for the reduction of the affinity of eIF3j for the 43S PIC. This is consistent with the observation that secondary structure serves as a barrier to productive 48S PIC formation on mRNAs containing the iron responsive element ([IRE]; Muckenthaler et al. 1998). Thus, the 43S PIC likely binds to an mRNA first in a pre-accommodated state (with the mRNA not stably bound to the mRNA entry channel) and then in an accommodated state (with the mRNA stably bound to the mRNA entry channel). It is anticipated that high-resolution structures of eIF4F bound to the 43S PIC will help determine whether this affects the conformation of the 40S subunit decoding site to enable mRNA entry. Equally important is the need to generate kinetic assays to monitor the rate by which prebound mRNA accommodates into the decoding site of the 43S PIC or dissociates from the ribosome.
Generating a kinetic assay to monitor the rate of mRNA accommodation into the decoding site of the 43S PIC will help establish the extent to which the mRNA accommodation step can serve as a kinetic checkpoint to regulate mRNA selection. Reduced availability of the 43S PIC would be expected to regulate the pre-accommodation step if the 43S PIC becomes limiting for translation. This would also be expected to be the case when TC is limiting (when eIF2 is phosphorylated). Comparable to the regulation of eIF4F accommodation onto an mRNA, the rate at which secondary structure is unwound by eIF4A and other helicases could play a critical role in controlling such a checkpoint. In mammals, a direct interaction between eIF4G and eIF3 plays an important role in stabilizing the 43S PIC on an mRNA (Hinton et al. 2007; Villa et al. 2013). Immunoprecipitation assays have indicated that the affinity of eIF4G binding to eIF3 is enhanced by TORC1 activation by a yet-to-be-determined mechanism (Harris et al. 2006; Thoreen et al. 2012). Altering the lifetime of this interaction could regulate 48S PIC formation on different mRNAs. Should the rate of accommodation of mRNA into the decoding site of the 43S PIC not be suitably fast, it is entirely possible that the entire 43S PIC dissociates from the mRNA, preventing its selection for translation.
SCANNING AND START CODON SELECTION
Following accommodation of the mRNA into the entry channel and decoding site of the 43S PIC, the complex migrates in a 3′ direction until it selects a suitable codon with which to initiate protein synthesis. The scanning mechanism was first proposed by Kozak and Shatkin and has remained the primary mechanism to explain the process of start codon selection on the majority of eukaryotic cellular mRNAs (Kozak and Shatkin 1978; Kozak 1980). Progress toward elucidating the thermodynamic and kinetic frameworks of start site selection has been made, but how the 43S PIC scans/migrates in a 5′ to 3′ direction is less understood. This is largely caused by the fact that kinetic assays to monitor 43S PIC migration have not yet been developed. Thus, little is known about the extent to which the rate of scanning can contribute to the regulation of translation or selection of mRNA.
The Scanning Mechanism
The ability of the eIF4F•4B complex to migrate along an mRNA in a 5′ to 3′ processive manner is consistent with eIF4A serving as the translocation motor for the scanning 43S PIC. The unwinding step size of eIF4A is 11 ± 2 base pairs, which is the same as the major steps taken by the hepatitis C virus NS3 DEx(H/D) helicase (Dumont et al. 2006; García-García et al. 2015). This relatively large step size is remarkable given that the 43S PIC must inspect each nucleotide during the scanning process. Generating single-molecule assays that are able to monitor scanning of the 43S PIC in real time at single-nucleotide resolution will likely play an important role in determining how the step size of eIF4A helicase activity relates to the migration of the 43S PIC. In addition, the emergence of high-resolution structures of scanning intermediates will help provide a structural basis for scanning. The relationship between scanning and start site selection is discussed in more detail in the following section.
Kinetics of Start Codon Recognition
The fidelity of start site selection is primarily governed by mRNA cis-elements, such as the codon sequence, the context surrounding the codon (Kozak sequence), and the position and stability of secondary structure. An ORF starting with the AUG codon is >50-fold better translated than one with a near-AUG codon, such as UUG (Fekete et al. 2005). Similarly, an AUG in the optimal context can be >20-fold more efficiently selected than one in the weakest context (Kozak 1986). Start codon selection is one of the most well-characterized steps in the eukaryotic translation pathway (Lorsch and Dever 2010; Hinnebusch 2014, 2017). Recognition of the start codon requires concerted structural rearrangements of initiation factors, initiator Met-tRNAi, and the 40S subunit. This results in a stable, scanning-arrested, 43S PIC harboring Met-tRNAi base-paired with the start codon at the P site (see Jobe et al. 2018 and Merrick and Pavitt 2018 for details). Structurally, codon–anticodon base-pairing results in transition of the 40S subunit from an open to a closed conformation together with a movement of the Met-tRNAi from an unaccommodated (POUT) to an accommodated (PIN) state (Llácer et al. 2015; Hinnebusch 2017). The release of the γ phosphate from the hydrolyzed TC-bound GTP ensures that the process is irreversible. The kinetics of these events have been primarily identified and characterized using a purified reconstituted yeast system in conjunction with structural and genetic studies (reviewed in Hinnebusch 2017).
The Lorsch laboratory has pioneered a reconstituted purified yeast system to monitor start site selection using fluorescently tagged components. What has emerged from this work is a model that describes a complex set of dynamic interactions between initiation components and the decoding site of the 40S subunit to ensure the fidelity of start site selection. Fast kinetic monitoring of a purified 40S subunit together with eIF1, eIF1A, and TC (40S•1•1A•TC) revealed that the rate of eIF5-dependent GTP hydrolysis is rapid (within a second) and somewhat insensitive to the presence of an mRNA (only a ∼1.5-fold stimulation was observed by adding an mRNA with an AUG codon) (Algire et al. 2005). In contrast to GTP hydrolysis, the rate of Pi release from 40S•1•1A•TC is sensitive to the presence of a prebound mRNA, but only if it possesses an AUG codon. The rate of Pi release was 0.04–0.06/sec (taking >40 sec to completion) in the absence of an AUG codon and was >100-fold accelerated in the presence of an mRNA with an AUG codon (6.7/sec). The rates of GTP hydrolysis and Pi release are similar in the presence of an mRNA containing an AUG codon, which is consistent with a model whereby GTP and GDP•Pi (possibly a hydrolysis transition state) are in rapid equilibrium within the 40S•1•1A•TC in the presence of eIF5 and independent of the codon in the P site. Upon start site selection, Pi release is the commitment step that makes GTP hydrolysis irreversible. The Lorsch laboratory also found that Pi release from 40S•1•1A•TC is >10-fold slower (0.54/sec) when saturating mRNA with an AUG codon was added together with eIF5 to trigger the reaction, instead of the mRNA being prebound to 40S•1•1A•TC (Algire et al. 2005). Because the rate of mRNA binding was not limiting for a saturated concentration, this 10-fold reduced rate is entirely consistent with Pi release being limited by a conformational change that occurs on placement of the AUG codon of an mRNA into the P site.
To characterize the conformational change of the 40S subunit on start site selection, a kinetic analysis was undertaken using a FRET signal between eIF1A and eIF1 to monitor the dissociation rate of eIF1 from 40S•1•1A•TC (Maag et al. 2005). The dissociation rate of eIF1 was accelerated 40-fold by the mRNA with an AUG codon, resulting in a dissociation rate (0.6/sec) that is the same as that of Pi release triggered by the same mRNA and eIF5 (Maag et al. 2005). This is consistent with eIF1 dissociation from the 40S•1•1A•TC complex acting as the rate-limiting step for Pi release. To verify this, when slowly or rapidly dissociating mutants of eIF1 (G107K or G107E) were used in this assay, the Pi release rate was appropriately reduced or increased (Algire et al. 2005; Nanda et al. 2013). It is worth noting that the rate of decrease in eIF1-eIF1A FRET in response to an mRNA containing an AUG codon would also be consistent with a rapid conformation change of eIF1 preceding its dissociation (with the rate of 9/sec). Such a conformational change may be similar to the recent structural model in which eIF1 adopts an altered conformation when Met-tRNAi adopts the accommodated conformation (PIN state described above) (Hussain et al. 2014; Llácer et al. 2015). It is important to note that the kinetic experiments were performed in the absence of eIF3 and eIF4F, so possible contributions of these components in this process could not be determined. Using a mammalian reconstituted system, both eIF3 and eIF4F (bound to the m7G cap) have been shown to inhibit eIF5-induced GTP hydrolysis in the absence of the AUG codon (Majumdar and Maitra 2005). This indicates that GTP hydrolysis is regulated by these initiation components, which were not present in the yeast work. It will be important in the future to rigorously explore these regulatory roles of eIF4F and eIF3 on GTP hydrolysis and Pi release using kinetic assays.
Upon start site selection, a conformational change occurs between the carboxy-terminal tail of eIF1A (1A-CTT) and the amino terminus of eIF5 (5-amino-terminal domain [NTD]). Specifically, the placement of the AUG codon at the P site induces a biphasic increase in a FRET signal between 1A-CTT and 5-NTD (24/sec and 0.4/sec in rates) (Nanda et al. 2013). This is consistent with a two-step conformational change of these proteins during start site selection. The rate of the slow phase in this process is once again very similar to the rate of AUG-dependent eIF1 dissociation. As expected, the slow kinetic phase can be reduced further, or increased, when more slowly or rapidly dissociating mutants of eIF1 were used (Nanda et al. 2013). Mutations in the 1A-CTT that reduce this slow phase (to 0.01–0.02/sec) further reduced the Pi release rate but did not alter the dissociation rate of eIF1. This implies that eIF1 dissociation is not in fact sufficient to induce Pi release, but is likely a key mediatory event in start site selection followed by the altered binding of eIF1A and eIF5.
How is the conformation of the 43S PIC altered on start codon recognition? Upon start codon recognition, the 43S PIC adopts a closed/PIN conformation, in which eIF1A binding is stabilized. This is shown in recent cryoelectron microscopy (cryo-EM) structures where eIF1A bridges the narrowed gap between head and body domains with its NTD interacting with the codon–anticodon duplex and +4 to +6 nucleotides of mRNA (Hussain et al. 2014; Llácer et al. 2015). The dissociation rate of fluorescently labeled eIF1A shows a biphasic pattern (with fast and slow phases) in the absence of an AUG codon (Maag et al. 2006). This is consistent with two distinct populations, each possessing a defined conformation of the 40S subunit. Importantly, AUG recognition increases the occupancy of the slow phase population from ∼30% to ∼90%. The 43S PIC may therefore be spontaneously switching back and forth between open and closed states even in the absence of mRNA, with AUG recognition shifting the equilibrium between these conformations toward the closed state. Thus, rather than a static model in which an open conformation 40S subunit slides along the mRNA during scanning, the scanning complex may dynamically sample the codon–anticodon interaction, whereas initiation factors function to help stabilize the conformation with stable base-pairing.
Start Codon Recognition during Scanning
The above kinetic studies provide precise information about the molecular steps and time frame for start codon recognition, but one wonders if the observed ∼twofold difference of the Pi release rates at AUG and near-AUG (UUG/AUU) codons is sufficient to enable a >50-fold difference in their translation efficiencies (Cheung et al. 2007; Nanda et al. 2009). As noted above, it is possible that the partially reconstituted system used in these studies (minus eIF4F and eIF3) possesses altered kinetics and/or reduced fidelity. This partial system allows for direct loading of an unstructured short mRNA to the 43S PIC without imposing migration of the 43S PIC driven by ATP hydrolysis, which is mediated by eIF4F. Lack of scanning may enable the prolonged lifetime (or iterative repositioning) of a near-cognate codon in the P site compared to a 43S PIC that migrates at a certain rate using an energy-dependent translocase (eIF4A). It will therefore be important to determine the rate of Pi release (i.e., fidelity) of the 43S PIC under more stringent physiologically relevant scanning conditions. Interestingly, placing a moderately stable hairpin (19 kcal/mol) ∼14 nt downstream of a weak AUG codon (near the leading edge of the 43S PIC) dramatically increases the likelihood of initiating translation at that codon (Kozak 1990). This is entirely consistent with improved recognition of a weak codon when the rate of scanning is reduced.
The rate of 43S PIC migration is generally believed not to be limiting for the overall rate of translation, as long as the 5′UTR is unstructured. This has been inferred from experiments that have shown that the extension of a 5′UTR does not result in decreased translation, and may even lead to increased translation (possibly caused by the increased capacity for loading multiple 43S PICs) (Kozak 1991). Nevertheless, secondary structure in the mRNA could reduce the kinetics of 43S PIC migration to a rate comparable to that needed to select a weak AUG codon. As mentioned earlier, the mechanism of codon recognition requires a dynamic set of interactions and conformational changes that likely reduces the rate of scanning. These include interactions of the Kozak sequence with eIFs and ribosomal proteins, codon–anticodon base-pairing, a closing of the mRNA decoding site conformation, and the generation of a PIN conformation. Consistent with this, in a cell-free extract, the closed/PIN scanning 43S PIC located at an optimum AUG (formed with nonhydrolyzable GTP) is stable at least for an hour, with marginal “sliding” to downstream start codons (Terenin et al. 2015). Presumably, the lifetime of the closed/PIN state formed at a weak/noncognate codon is much shorter to allow 43S PIC moving downstream more occasionally before Pi release.
Regulation of Start Site Selection
Early studies identified eIF1, eIF2, and eIF5 as the primary components that maintain the fidelity of start site selection in yeast (Donahue et al. 1988; Cigan et al. 1989; Yoon and Donahue 1992; Huang et al. 1997). Subsequent work characterizing additional yeast mutants has revealed that the fidelity of start site selection is maintained by coordinated interactions between the mRNA, the 40S subunit, and all initiation factors that make up the 43S PIC (eIF1, eIF1A, eIF2, eIF3, and eIF5 [reviewed in Hinnebusch 2014, 2017]). A conserved autoregulatory translation mechanism maintains the cellular concentrations of both eIF1 and eIF5, underscoring the importance of these components in maintaining fidelity (Ivanov et al. 2010; Loughran et al. 2012). What appears to be missing is a regulatory mechanism to control the fidelity of start site selection to reprogram the proteome in response to different cell stimuli. An attractive mechanism would be provided by regulated phosphorylation of fidelity-controlling components. Interestingly, Thr72 in eIF1 can be phosphorylated in response to arsenite stress, and this correlates with the upregulation of arsenite-inducible regulatory particle-associated protein (AIRAP) synthesis (Zach et al. 2014). This has been proposed to occur by increased leaky scanning of an inhibitory uORF that possesses a suboptimal AUG codon (Zach et al. 2014). The phosphorylation of eIF5 occurs at several sites in the carboxy-terminal domain in a cell-cycle-dependent manner (Homma et al. 2005). These phosphorylation sites are correlated with increased formation of the 43S PIC and cyclin B1 translation, although it is not clear whether they lead to selective or general translation activation. One anticipates that regulatory mechanisms will be identified that are able to precisely control start site selection. The ability to monitor the kinetics of start site selection using in vitro assays will undoubtedly help provide the necessary molecular insight into these mechanisms as they are found.
ELONGATION
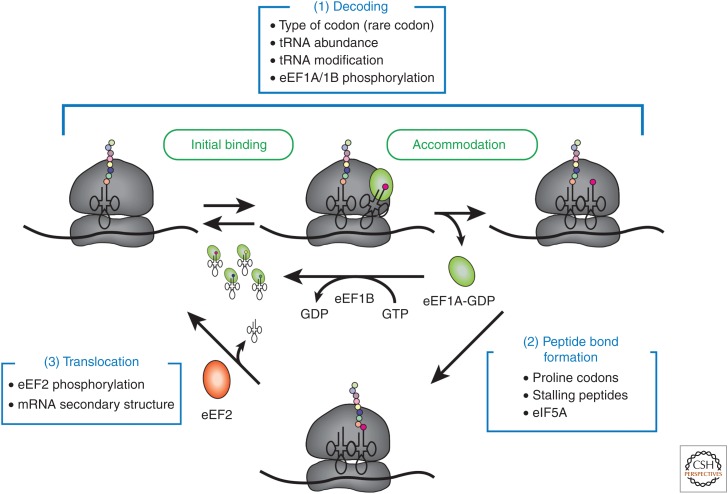
During elongation, the 80S ribosome decodes the ORF of the mRNA into protein (for a review of the elongation pathway in eukaryotes, see Dever et al. 2018). The process of elongation can be regulated at all three substeps: (1) decoding of the codon, (2) peptide bond formation, and (3) translocation of the reading frame (Fig. 3). The rate-limiting step of the elongation process is almost always the decoding of the codon, which is governed by the rate of delivery and accommodation of the cognate aminoacyl transfer RNA (aa-tRNA) to the A site of the ribosome (reviewed in Wohlgemuth et al. 2011). Importantly, the peptidyl-transferase center (PTC) of the ribosome is one of its most highly conserved regions, highlighting the fact that the fundamental mechanism of peptide bond formation is conserved across all organisms (reviewed in Rodnina and Wintermeyer 2009; Simonovic and Steitz 2009). Maintaining the speed and accuracy of translation requires precise coordination between the ribosome, mRNA, tRNA, and elongation factors (reviewed in Rodnina et al. 2017). The rate of elongation can in some instances control protein abundance, which implies that it can serve as the rate-limiting step of translation. However, this almost certainly depends on the mRNA that is being studied and the growth state of the cell. Recent analysis of yeast ribosome profiling data together with methods to monitor translation in real time have indicated that initiation rates in yeast likely vary between 4 and 240 sec (∼60-fold) on different mRNAs, whereas elongation rates appear to vary between three and five amino acids per second (∼twofold) (Shah et al. 2013; Wang et al. 2016; Yan et al. 2016). However, direct measurements of translation on mRNAs containing different rare codons in vitro have indicated that elongation rates can in fact vary by as much as ∼20-fold (reviewed in Rodnina 2016). This suggests that elongation can provide a mechanism with considerable potential to regulate protein synthesis, especially on mRNAs for which initiation is not rate limiting. It is anticipated that more precise methods to measure initiation and elongation in real time will help to fully understand the precise contributions that initiation and elongation stages can have on regulating translation on different mRNAs.
Figure 3.
Model of eukaryotic elongation. Elongation is depicted as three main stages: (1) During the decoding step, eEF1A•GTP recruits the appropriate aminoacyl-transfer RNA (aa-tRNA) to the A site of the ribosome. Productive recruitment occurs through initial binding and accommodation steps. The accommodation step requires the hydrolysis of eEF1A-bound GTP, followed by release of eEF1A•GDP. The rate of decoding can be reduced by the presence of rare codons. Following its release, eEF1A•GDP is recycled by the guanine nucleotide exchange factor (GEF) activity of eEF1B. (2) Peptide bond formation between the aa-tRNA in the A site and the peptidyl-tRNA in the P site is catalyzed by the ribosomal RNA (rRNA) through different substeps (not shown). The rate of peptide bond formation is affected by recruitment of eIF5A. (3) The translocation step moves the mRNA relative to the ribosome by one codon. This step requires the recruitment of eEF2•GTP, followed by GTP hydrolysis and release of eEF2•GDP. Potential sources of regulation for each step are summarized in boxes and described in more detail in the main text.
Regulation of Elongation by Codon Optimality
The rate of elongation in all organisms can be regulated by the degree of codon optimality of an mRNA. Optimal codons correspond to tRNA species that are abundant and rapidly accommodated into the ribosome during elongation, whereas nonoptimal codons (rare codons) interact with tRNAs that are low in abundance and therefore more slowly accommodated into the A site of the ribosome during elongation (reviewed in Chamary et al. 2006; Plotkin and Kudla 2011; Koutmou et al. 2015; Rodnina 2016; Hanson and Coller 2018). The introduction of ribosome profiling (see Ingolia et al. 2018) and other techniques to monitor translation in real time in living cells is helping to reveal the extent to which codon optimality contributes to the regulation of translation by influencing ribosome pausing at these codons (Gardin et al. 2014; Lareau et al. 2014; Weinberg et al. 2016; Yan et al. 2016). It has been estimated that codon optimality can account for about 30% of the variation that exists between mRNA abundance and protein abundance in human cells (Vogel et al. 2010; Hanson and Coller 2018). A region of mRNA where rare codons have a particularly strong effect on translation is that directly following the start codon. This is likely caused by the fact that slowing ribosomes in this region of the mRNA further decreases the rate of initiation by slowing the rate that ribosomes clear the start codon (Mitarai et al. 2008; Potapov et al. 2012; Chu et al. 2014). How can the regulation of elongation through rare codons be regulated in response to changes in cell stimuli? One solution would be to alter the concentration of tRNAs that decode rare codons. Accordingly, tRNA concentrations can be increased or decreased in cells by altering both transcription and turnover rates in response to different cell stimuli (Kirchner and Ignatova 2015; Wilusz 2015). This would enable control of the translation of mRNAs that contain a disproportionate number of rare codons in their ORF.
An additional function of rare codons is to slow down ribosomes in mRNA sequences that encode low complexity regions found in between folded domains. This ensures that complex domains are given suitable time to correctly fold (reviewed in Komar 2009). This function can be augmented by structural features in an mRNA that slow the rate of elongation at the translocation step (Fig. 3) (Nackley et al. 2006; Kudla et al. 2009). Importantly, in addition to regulating the rate at which a protein is synthesized from its mRNA, the regulation of elongation by rare codons also contributes to the regulation of translation fidelity, protein folding, and mRNA stability (reviewed in Hanson and Coller 2018).
Regulation of Translation by Elongation Factors
The elongation step of translation in all organisms requires two GTP-binding factors (reviewed in Dever et al. 2018). In eukaryotes, eEF1A binds to an aa-tRNA and recruits it to the ribosome in a GTP-dependent manner. Following the accommodation of the aa-tRNA in the A site of the ribosome, the GTP is hydrolyzed and eEF1A•GDP is released. The GDP-bound form of eEF1A is then recycled back to eEF1A•GTP by the guanine nucleotide exchange factor (GEF), eEF1B. Both eEF1A and eEF1B can be differentially phosphorylated in cells, with evidence suggesting that phosphorylation enhances the rate of GEF activity (reviewed in Traugh 2001; Browne and Proud 2002). This could increase the rate at which eEF1A can rebind to an aa-tRNA following each round of elongation and therefore increase the overall rate of aa-tRNA recruitment and translation.
More attention has been given to the regulated phosphorylation of eEF2 in cells, in part because the eEF2 kinase (eEF2K) is an attractive drug target in cancer (reviewed in Browne and Proud 2002; Liu and Proud 2016). Similar to EF-G in bacteria, eEF2 promotes the translocation step of eukaryotic elongation. Importantly, the phosphorylation of eEF2 inhibits its activity by reducing its affinity for the ribosome by roughly an order of magnitude (Carlberg et al. 1990). Inhibiting elongation (and therefore translation) by eEF2 phosphorylation therefore enables cells to survive nutrient deprivation (Leprivier et al. 2013). Although it has not been rigorously explored, one would expect that stalling translation by eEF2 phosphorylation would lead to mRNA decay in a similar way to that observed for other mechanisms of elongation stalling (reviewed in Hanson and Coller 2018; Heck and Wilusz 2018). It will be interesting in the future to determine on a genome-wide basis whether this kind of regulation of elongation functions to preferentially target specific mRNAs. Generating this information will help design additional experiments on an mRNA-by-mRNA basis to fully understand how this mechanism of elongation regulation functions to control translation in cells.
Following selection and accommodation of aa-tRNA into the A site of the ribosome, the intrinsic rate of peptide bond formation is generally not limiting for elongation (Wohlgemuth et al. 2010). However, this can in fact be limiting for certain combinations of peptidyl-tRNAs located in the P site and aa-tRNAs in the A site. In particular, the amino acid proline (Pro) is a poor acceptor and donor for peptide bond formation in bacteria (Pavlov et al. 2009). Consistent with this, stretches of Pro codons in bacteria and eukaryotes appear to stall the elongating ribosome by virtue of slowing down the rate of peptidyl transfer. This stalling can be overcome by the addition of EF-P and eIF5A, respectively, in bacterial and eukaryotic translation assays (Doerfel et al. 2013; Gutierrez et al. 2013; Ude et al. 2013; Schuller et al. 2017; Dever et al. 2018; Rodnina 2018). Further characterization of eIF5A has indicated that it may have a more general role in accelerating the rate of peptide bond formation during every round of elongation no matter what the sequence (Schuller et al. 2017). Importantly, the ability of EF-P to limit the rate of translation depends on the rate of initiation being fast enough to enable elongation to act as the rate-limiting step (Hersch et al. 2014). Consistent with this, reporters used to study the regulation of elongation by eIF5A using in vitro and in vivo assays have generally contained very short unstructured 5′UTRs to avoid initiation becoming rate limiting for translation. No mechanism has yet been identified that naturally regulates the availability and/or activity of eIF5A, although eIF5A abundance is reduced in response to drugs that inhibit its modification (reviewed in Mathews and Hershey 2015). While eIF5A is phosphorylated in yeast (Kang et al. 1993), this modification does not appear to regulate its function. It will therefore be important in the future to determine whether the function of this important protein is regulated in response to different cell stimuli.
The Regulation of Elongation
A detailed kinetic framework for the process of elongation has been made by using a reconstituted purified system from bacteria (reviewed in Wohlgemuth et al. 2011; Rodnina 2016). Similar work is only just beginning to use purified eukaryotic systems and it is anticipated that equally detailed kinetic frameworks will eventually be generated. It has become clear that the regulation of elongation is complex and can have profound outcomes affecting the overall rate of translation, frameshifting, protein folding, and mRNA stability. Nevertheless, separating these outcomes can be difficult, especially in intact cells. A challenge for the future will therefore be to generate assays that can precisely monitor all these events in real time so that a more complete understanding of how regulation of the rate of elongation controls translation and the proteome.
TERMINATION AND RECYCLING
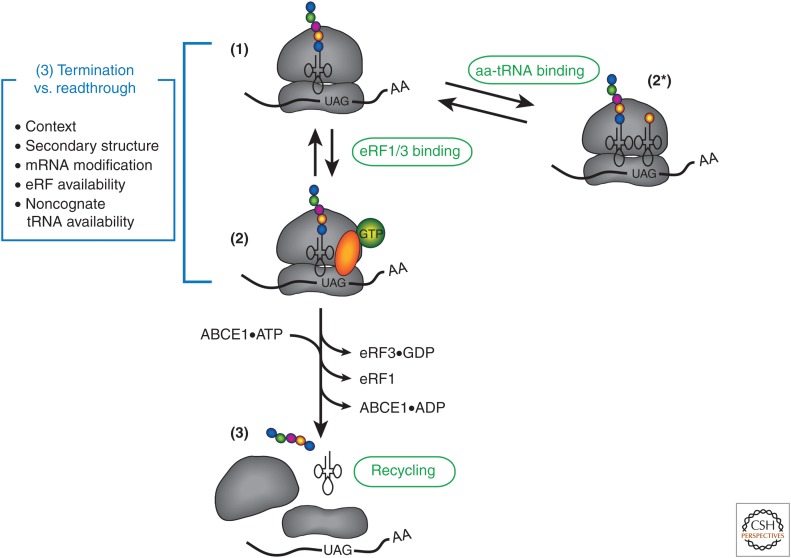
The stages of termination and recycling in eukaryotes (see Hellen 2018) appear to be rather different from those described in bacteria. A fundamental difference between the systems is that the steps occur independently in bacteria, but are tightly coupled in eukaryotic cells. This was only possible to determine through the use of reconstituted purified systems and a careful kinetic analysis of the process (reviewed in Dever and Green 2012; Jackson et al. 2012). A kinetic checkpoint exists at the point when a termination codon enters the A site of the ribosome (Fig. 4). At this checkpoint, one of two outcomes is possible: (1) the eRF1/eRF3•GTP complex can be recruited, resulting in the termination of polypeptide synthesis; or (2) a noncognate aa-tRNA can be recruited to enable the process of elongation to continue. A precise kinetic framework to explain how this decision is made is not yet clear, but some mechanistic details have emerged as to the steps that are taken during each of these two alternative outcomes. Understanding how this checkpoint works is a major goal for translation regulation, in part because of the fact that many genetic diseases can be attributed to the generation of a premature termination codon being introduced into the ORF of various genes (Mort et al. 2008; Keeling et al. 2014).
Figure 4.
Translation termination and ribosome recycling. (1) A pre-termination complex is shown to contain a stop codon in the A site of the ribosome. Two possible pathways for subsequent steps are depicted. (2) The recruitment of an eRF1/eRF3•GTP complex to the ribosome in the first step of the termination pathway. (2*) An alternative possibility is that a noncognate aminoacyl transfer RNA (aa-tRNA) is recruited to the A site of the ribosome to continue the elongation stage of translation (stop codon readthrough). Potential sources of regulation for determining which pathway is followed are summarized in the box and described in more detail in the main text. (3) Following the recruitment of eRF1/eRF3•GTP to the ribosome, a number of steps are undertaken to complete translation termination and are described in detail in the main text. Briefly, these steps involve the release of eRF3•GDP, recruitment of ABCE1•ATP, and the release of the polypeptide chain. Finally, the 60S ribosomal subunit is dissociated by the hydrolysis of the ATP bound to ABCE1. As mentioned in the main text, the precise pathway of 40S subunit recycling following termination is poorly understood.
Termination Codon Recognition
A cartoon that depicts the generally accepted termination pathway is shown in Figure 4 (see also Hellen 2018). In the first step of termination, a pre-termination complex (pre-TC) is generated with a termination codon in the A site and a peptidyl-tRNA in the P site of the ribosome. The empty A site of the ribosome allows for a trimeric complex (eRF1/eRF3•GTP) to be stably recruited. Recruitment to the ribosome stimulates the hydrolysis rate of GTP bound to eRF3, promoting its release from the ribosome (Frolova et al. 1996). The release of eRF3 enables eRF1 to correctly position its GGQ loop into the PTC of the 60S subunit to help promote peptide release (Alkalaeva et al. 2006; Shoemaker and Green 2011). Unexpectedly, it was discovered that the binding of the ATP-binding cassette protein E1 (ABCE1 in mammals and Rli1 in Saccharomyces cerevisiae) together with eRF3 synergistically accelerates peptide release (Shoemaker and Green 2011). The important thing to note in making this discovery is that it was only possible through the kinetic analysis of the termination process. This kinetic assay further revealed that eIF5A binding to the ribosome further stimulates the rate of peptide release by ∼17-fold (Schuller et al. 2017). This is likely caused by eIF5A binding to the exit site (E site) of the ribosome following release of the deacylated tRNA, where it stabilizes the peptidyl-tRNA. While the ATPase activity of ABCE1/Rli1 is not required for promoting peptide release, it is required for promoting the subsequent release of the 60S subunit (Pisarev et al. 2010; Barthelme et al. 2011; Shoemaker and Green 2011). Importantly, this function of ABCE1/Rli1 was subsequently verified in live cells by using ribosome profiling to monitor the presence of ribosomes around termination codons and the 3′UTR of mRNAs (Young et al. 2015).
Ribosome Recycling
Following the release of the 60S subunit, the separated 40S ribosomal subunits are bound by initiation factors so that they can take part in another round of translation (Pisarev et al. 2007). The binding of initiation factors to posttermination complexes is often depicted as an ordered process, but it should be noted that the order of release and binding of components from the separated ribosomal subunits is still not known. This is because these events have been monitored exclusively by using sucrose gradients, which do not allow for the order of binding to be determined (Pisarev et al. 2007). Importantly, the expected time for the entire process of termination and recycling is likely to be on the order of seconds. It will therefore be necessary in the future to generate a kinetic assay that can monitor the process of termination and ribosome recycling in real time to solve this question. Recent studies have suggested that eIF3 can play an active role in the termination process in vivo (Beznoskova et al. 2013, 2015). However, these studies were limited by the fact that cross-linking agents were necessary to stabilize complexes before sucrose gradient analysis. A kinetic assay will therefore be needed to verify whether eIF3 plays a role in termination, or whether it functions only in the recycling of the 40S subunit (as suggested from reconstitution assays).
It is likely that some initiation factors can bind to a 40S subunit before the release of mRNA. This is implied by the fact that a post-termination 40S subunit can resume scanning in a 3′ direction following the translation of a short uORF. The scanning 40S subunit must recruit a new TC before it can select a start codon for another round of translation on the same mRNA. This process is termed reinitiation and has been reviewed in detail elsewhere (Hinnebusch 2011; Jackson et al. 2012). The fact that the TC is not required to resume scanning supports a mechanism by which this complex is not needed for stabilizing mRNA in the decoding site of the 40S subunit. However, this may only be the case for a 40S subunit that is already bound to an mRNA following the termination event. Importantly, the resumption of scanning following termination is restricted to uORFs that are translated in a short amount of time (Kozak 2001). It has been suggested that this limitation is caused by the dissociation rate of eIF4G from eIF3, or possibly the dissociation rate of eIF3 from the 40S subunit (Pöyry et al. 2004; Mohammad et al. 2017). As mentioned above, the affinity of eIF4G for eIF3 is regulated in a TORC1-dependent manner (Harris et al. 2006; Thoreen et al. 2012). It will be interesting to determine whether this change in affinity corresponds to a change in the dissociation rate, which could conceivably alter the efficiency of reinitiation.
Reading through Termination Codons
When paused with a termination codon in its A site, there is a possibility that an aa-tRNA will bind to the ribosome to decode the codon and continue the elongation cycle (a process called stop codon readthrough). Because the fidelity of termination is high, the competition between noncognate, or cognate, aa-tRNA binding and eRF1/eRF3•GTP binding must favor eRF1/eRF3•GTP binding. However, recent genome-wide data sets have unexpectedly indicated that readthrough can be regulated and is more pervasive than previously thought (although still comparatively rare) (Dunn et al. 2013). A number of factors have been shown to alter the likelihood of readthrough in cells. These include the sequence of the termination codon, the context surrounding the codon, secondary structure in the mRNA, mRNA modifications, and the concentration of endogenous suppressor aa-tRNAs (Cassan and Rousset 2001; Chao et al. 2003; Firth et al. 2011; Karijolich and Yu 2011). Often, various combinations of these and other factors regulate readthrough, making it difficult to precisely understand the mechanism(s) by which readthrough is regulated. Some organisms even use cognate aa-tRNAs to decode termination codons by default, and terminate at these codons only if a poly(A) tail is in close proximity (Heaphy et al. 2016; Swart et al. 2016; Zahonova et al. 2016). Context-dependent readthrough regulation is also apparent in the process of nonsense-mediated mRNA decay (NMD) in eukaryotes (see Karousis and Mühlemann 2018). For NMD, an mRNA is degraded when a termination codon is present within the ORF of an mRNA, but not when it is in the correct position (reviewed in Popp and Maquat 2013; Kurosaki and Maquat 2016; Celik et al. 2017). The ability of cells to differentiate between termination codons in a context-dependent manner raises a fundamental question of how termination codons are recognized by the translation machinery. As mentioned above, the kinetics of termination has begun to be elucidated, but the mechanism of regulated readthrough is still poorly defined. To address this, it is anticipated that genome-wide data sets from cells that show regulated readthrough will continue to provide a list of mRNAs for further study. Biochemical approaches will then be needed to determine the mechanism by which the decision is made to read through versus terminate translation. One goal will be to reconstitute the process of readthrough using a purified system. Although this is an ambitious challenge, the ability to monitor this checkpoint in real time will enable a complete kinetic framework to be developed. Both bulk assays and single-molecule approaches will be needed for developing these frameworks. This information will ultimately help toward development of small-molecule drugs that can precisely alter this process to promote readthrough of premature termination codons that cause a number of genetic diseases.
FUTURE DIRECTIONS
A goal of future work will be to precisely determine how the translation pathway can be reprogrammed to control what mRNA is selected for translation, which initiation/termination codons are used, and how much protein is synthesized from individual mRNAs. This will require the identification of where specific regulation checkpoints in the pathway are located. Using reconstituted purified systems for real-time bulk and single-molecule assays will establish a kinetic framework for the pathway. Using these assays, one can precisely determine how variables such as the concentration of components can alter progress of different mRNAs through the pathway. Nevertheless, reconstituted systems also suffer from limitations that must be considered. For example, the kinetics of these systems is unlikely to approach that found in intact cells, which may mean that the rate-limiting step of the pathway in vitro is different from that found in vivo. Reconstituted systems typically use in vitro transcribed mRNAs, so possible contributions of other mRNA-binding proteins that are present in cells (forming various mRNPs) and RNA modifications are not easy to test. Therefore, whereas reconstituted systems will be important in generating in vitro models for the pathway of translation, it is essential that these models be tested in intact cells. The power of this combination has been highlighted by the successful test of start site selection models in yeast (reviewed in Hinnebusch and Lorsch 2012; Hinnebusch 2014, 2017). It is anticipated that the use of new genome engineering methods will further enhance the ability to test in vitro models for the mammalian translation pathway. Continued improvements to imaging techniques will also make it increasingly possible to precisely monitor translation regulation on single mRNAs in live cells (see Biswas et al. 2018). It is hoped that once the regulation of translation is understood in sufficient detail, it will be possible to understand how the dysregulation of translation can lead to the generation and maintenance of different disease states.
ACKNOWLEDGMENTS
We thank John Hershey for many thought-provoking conversations about the regulation of eukaryotic translation. We are grateful to the Fraser laboratory for stimulating discussion and comments on the manuscript. We gratefully acknowledge support for this work from National Institutes of Health (NIH) Grant R01 GM092927.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ali IK, McKendrick L, Morley SJ, Jackson RJ. 2001. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J 20: 4233–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Lorsch JR. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262. [DOI] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136. [DOI] [PubMed] [Google Scholar]
- Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N. 2015. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol 22: 269–271. [DOI] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. 2011. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci 108: 3228–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznoskova P, Cuchalova L, Wagner S, Shoemaker CJ, Gunisova S, von der Haar T, Valasek LS. 2013. Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read-through in yeast cells. PLoS Genet 9: e1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beznoskova P, Wagner S, Jansen ME, von der Haar T, Valasek LS. 2015. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res 43: 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Biswas J, Liu Y, Singer RH, Wu B. 2018. Fluorescence imaging methods to investigate translation in single cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. 2002. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem 269: 5360–5368. [DOI] [PubMed] [Google Scholar]
- Carlberg U, Nilsson A, Nygard O. 1990. Functional properties of phosphorylated elongation factor 2. Eur J Biochem 191: 639–645. [DOI] [PubMed] [Google Scholar]
- Cassan M, Rousset JP. 2001. UAG read through in mammalian cells: Effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol Biol 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik A, He F, Jacobson A. 2017. NMD monitors translational fidelity 24/7. Curr Genet 63: 1007–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. 2006. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat Rev Genet 7: 98–108. [DOI] [PubMed] [Google Scholar]
- Chao AT, Dierick HA, Addy TM, Bejsovec A. 2003. Mutations in eukaryotic release factors 1 and 3 act as general nonsense suppressors in Drosophila. Genetics 165: 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev 21: 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T. 2014. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J 33: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan AM, Pabich EK, Feng L, Donahue TF. 1989. Yeast translation initiation suppressor sui2 encodes the α subunit of eukaryotic initiation factor 2 and shares sequence identity with the human α subunit. Proc Natl Acad Sci 86: 2784–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso MC, Milburn SC, Hershey JW, Jackson RJ. 1990. Selection of the 5′-proximal translation initiation site is influenced by mRNA and eIF-2 concentrations. Eur J Biochem 187: 361–371. [DOI] [PubMed] [Google Scholar]
- Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4: a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88. [DOI] [PubMed] [Google Scholar]
- Donahue TF, Cigan AM, Pabich EK, Valavicius BC. 1988. Mutations at a Zn(II) finger motif in the yeast eIF-2 β gene alter ribosomal start-site selection during the scanning process. Cell 54: 621–632. [DOI] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I Jr, Pyle AM, Bustamante C. 2006. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. 2013. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2: e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. 2005. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J 24: 3588–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Wills NM, Gesteland RF, Atkins JF. 2011. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res 39: 6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS. 2015. Quantitative studies of mRNA recruitment to the eukaryotic ribosome. Biochimie 114: 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA. 2007. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26: 811–819. [DOI] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, Kisselev L. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2: 334–341. [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Putnam AA, Bowers HA, Guenther UP, Ye X, Kindsfather A, Hilliker AK, Jankowsky E. 2016. Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. eLife 5: e16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García C, Frieda KL, Feoktistova K, Fraser CS, Block SM. 2015. RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science 348: 1486–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardin J, Yeasmin R, Yurovsky A, Cai Y, Skiena S, Futcher B. 2014. Measurement of average decoding rates of the 61 sense codons in vivo. eLife 3: e03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. 2013. eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson G, Coller J. 2018. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms U, Andreou AZ, Gubaev A, Klostermeier D. 2014. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res 42: 7911–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC Jr. 2006. mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 25: 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. 2013. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153: 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy SM, Mariotti M, Gladyshev VN, Atkins JF, Baranov PV. 2016. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in Condylostoma magnum. Mol Biol Evol 33: 2885–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Heck AM, Wilusz J. 2018. The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SJ, Elgamal S, Katz A, Ibba M, Navarre WW. 2014. Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J Biol Chem 289: 28160–28171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R. 2011. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell 43: 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83: 779–812. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42: 589–611. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. 2012. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb Perspect Biol 4: a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton TM, Coldwell MJ, Carpenter GA, Morley SJ, Pain VM. 2007. Functional analysis of individual binding activities of the scaffold protein eIF4G. J Biol Chem 282: 1695–1708. [DOI] [PubMed] [Google Scholar]
- Homma MK, Wada I, Suzuki T, Yamaki J, Krebs EG, Homma Y. 2005. CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cycle progression. Proc Natl Acad Sci 102: 15688–15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HK, Yoon H, Hannig EM, Donahue TF. 1997. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev 11: 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llácer JL, Fernández IS, Munoz A, Martin-Marcos P, Savva Christos G, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Gradi A, Sonenberg N. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17: 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci 107: 18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2012. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 86: 45–93. [DOI] [PubMed] [Google Scholar]
- *.Jobe A, Liu Z, Gutierrez-Vargas C, Frank J. 2018. New insights into ribosome structure and function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HA, Schwelberger HG, Hershey JW. 1993. Translation initiation factor eIF-5A, the hypusine-containing protein, is phosphorylated on serine in Saccharomyces cerevisiae. J Biol Chem 268: 14750–14756. [PubMed] [Google Scholar]
- Karijolich J, Yu YT. 2011. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474: 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Karousis ED, Mühlemann O. 2018. Nonsense-mediated mRNA decay begins where translation ends. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Xue X, Gunn G, Bedwell DM. 2014. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet 15: 371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S, Ignatova Z. 2015. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 16: 98–112. [DOI] [PubMed] [Google Scholar]
- Komar AA. 2009. A pause for thought along the co-translational folding pathway. Trends Biochem Sci 34: 16–24. [DOI] [PubMed] [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE. 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF)3 and eIF4A to human eIF4G-1. J Biol Chem 275: 41369–41376. [DOI] [PubMed] [Google Scholar]
- Koutmou KS, Radhakrishnan A, Green R. 2015. Synthesis at the speed of codons. Trends Biochem Sci 40: 717–718. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1980. Evaluation of the “scanning model” for initiation of protein synthesis in eukaryotes. Cell 22: 7–8. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1991. Effects of long 5′ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr 1: 117–125. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 2001. Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29: 5226–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37. [DOI] [PubMed] [Google Scholar]
- Kozak M, Shatkin AJ. 1978. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J Biol Chem 253: 6568–6577. [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. 2016. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129: 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 270: 21975–21983. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3: e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson TG, Ray BK, Dodds JT, Grifo JA, Abramson RD, Merrick WC, Betsch DF, Weith HL, Thach RE. 1986. Influence of 5′ proximal secondary structure on the translational efficiency of eukaryotic mRNAs and on their interaction with initiation factors. J Biol Chem 261: 13979–13989. [PubMed] [Google Scholar]
- Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, et al. 2013. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153: 1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. 2011. From unwinding to clamping—The DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12: 505–516. [DOI] [PubMed] [Google Scholar]
- Liu R, Proud CG. 2016. Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol Sin 37: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone M, Atas E, Meller A, Sonenberg N. 2010. Mechanisms governing the control of mRNA translation. Phys Biol 7: 021001. [DOI] [PubMed] [Google Scholar]
- Llácer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Steitz TA. 2013. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 500: 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE. 2010. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem 285: 21203–21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR. 2005. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275. [DOI] [PubMed] [Google Scholar]
- Maag D, Algire MA, Lorsch JR. 2006. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol 356: 724–737. [DOI] [PubMed] [Google Scholar]
- Majumdar R, Maitra U. 2005. Regulation of GTP hydrolysis prior to ribosomal AUG selection during eukaryotic translation initiation. EMBO J 24: 3737–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Hershey JW. 2015. The translation factor eIF5A and human cancer. Biochim Biophys Acta 1849: 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC. 2015. eIF4F: A retrospective. J Biol Chem 290: 24091–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitarai N, Sneppen K, Pedersen S. 2008. Ribosome collisions and translation efficiency: Optimization by codon usage and mRNA destabilization. J Mol Biol 382: 236–245. [DOI] [PubMed] [Google Scholar]
- Mohammad MP, Munzarova Pondelickova V, Zeman J, Gunisova S, Valasek LS. 2017. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res 45: 2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol 20: 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort M, Ivanov D, Cooper DN, Chuzhanova NA. 2008. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 29: 1037–1047. [DOI] [PubMed] [Google Scholar]
- Muckenthaler M, Gray NK, Hentze MW. 1998. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell 2: 383–388. [DOI] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. 2006. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 314: 1930–1933. [DOI] [PubMed] [Google Scholar]
- Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. 2009. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol 394: 268–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Saini AK, Munoz AM, Hinnebusch AG, Lorsch JR. 2013. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J Biol Chem 288: 5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, et al. 2002. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Behrens MA, He Y, Oliveira CL, Jensen LS, Hoffmann SV, Pedersen JS, Andersen GR. 2011. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res 39: 2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özeş AR, Feoktistova K, Avanzino BC, Fraser CS. 2011. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J Mol Biol 412: 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palam LR, Baird TD, Wek RC. 2011. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem 286: 10939–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50. [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. 2009. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci 106: 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1985. Photochemical cross-linking of cap binding proteins to eukaryotic mRNAs: Effect of mRNA 5′ secondary structure. Mol Cell Biol 5: 3222–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. 2011. Synonymous but not the same: The causes and consequences of codon bias. Nat Rev Genet 12: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp MW, Maquat LE. 2013. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov I, Makela J, Yli-Harja O, Ribeiro AS. 2012. Effects of codon sequence on the dynamics of genetic networks. J Theor Biol 315: 17–25. [DOI] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ. 2004. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Prabhakar A, Puglisi EV, Puglisi JD. 2018. Single-molecule fluorescence applied to translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Proud CG. 2018. Regulation of translation by cap-binding proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033050. [DOI] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480. [DOI] [PubMed] [Google Scholar]
- Rodnina MV. 2016. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci 25: 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rodnina MV. 2018. Translation in prokaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. 2009. Recent mechanistic insights into eukaryotic ribosomes. Curr Opin Cell Biol 21: 435–443. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fischer N, Maracci C, Stark H. 2017. Ribosome dynamics during decoding. Philos Trans R Soc Lond B Biol Sci 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr, Richter NJ, Lima WF, Merrick WC. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 276: 30914–30922. [DOI] [PubMed] [Google Scholar]
- Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. 1990. Bidirectional RNA helicase activity of eukaryotic translation initiation factors 4A and 4F. Mol Cell Biol 10: 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. 2017. eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. 2013. Rate-limiting steps in yeast protein translation. Cell 153: 1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifulin DE, Bartuli YS, Meschaninova MI, Ven’yaminova AG, Graifer DM, Karpova GG. 2016. Exploring accessibility of structural elements of the mammalian 40S ribosomal mRNA entry channel at various steps of translation initiation. Biochim Biophys Acta 1864: 1328–1338. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci 108: E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Steitz TA. 2009. A structural view on the mechanism of the ribosome-catalyzed peptide bond formation. Biochim Biophys Acta 1789: 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS. 2017. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc Natl Acad Sci 114: 6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T. 2012. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J 31: 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. 2013. The translational landscape of the mammalian cell cycle. Mol Cell 52: 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Atas E, Lindqvist L, Sonenberg N, Pelletier J, Meller A. 2012. The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucleic Acids Res 40: 6199–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Serra V, Petroni G, Nowacki M. 2016. Genetic codes with no dedicated stop codon: Context-dependent translation termination. Cell 166: 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD. 2015. Regulation of mRNA translation during mitosis. eLife 4: e07957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Akulich KA, Andreev DE, Polyanskaya SA, Shatsky IN, Dmitriev SE. 2016. Sliding of a 43S ribosomal complex from the recognized AUG codon triggered by a delay in eIF2-bound GTP hydrolysis. Nucleic Acids Res 44: 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traugh JA. 2001. Insulin, phorbol ester and serum regulate the elongation phase of protein synthesis. Prog Mol Subcell Biol 26: 33–48. [DOI] [PubMed] [Google Scholar]
- Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. 2012. Heterogeneous pathways and timing of factor departure during translation initiation. Nature 487: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85. [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. 2004. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N, Do A, Hershey JW, Fraser CS. 2013. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 288: 32932–32940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Abreu Rde S, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. 2010. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X. 2016. Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165: 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, Bartel DP. 2016. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep 14: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. 2013. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat Struct Mol Biol 20: 1015–1017. [DOI] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. 2015. Controlling translation via modulation of tRNA levels. Wiley Interdiscip Rev RNA 6: 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Rodnina MV. 2010. Optimization of speed and accuracy of decoding in translation. EMBO J 29: 3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV. 2011. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos Trans R Soc Lond B Biol Sci 366: 2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME. 2016. Dynamics of translation of single mRNA molecules in vivo. Cell 165: 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH. 2003. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. 2007. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell 28: 253–263. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Donahue TF. 1992. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol 12: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. 2015. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell 162: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zach L, Braunstein I, Stanhill A. 2014. Stress-induced start codon fidelity regulates arsenite-inducible regulatory particle-associated protein (AIRAP) translation. J Biol Chem 289: 20706–20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahonova K, Kostygov AY, Sevcikova T, Yurchenko V, Elias M. 2016. An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr Biol 26: 2364–2369. [DOI] [PubMed] [Google Scholar]