Abstract
Proteins containing tyrosine kinase activity play critical roles in cancer signaling. Intracellular SRC-family kinases relay growth signals from numerous cell surface receptors and can be constitutively activated by oncogenic mutations, as can transmembrane growth factor receptors such as epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 2 (HER2) that signal via their tyrosine kinase activity. In this excerpt from his forthcoming book on the history of cancer research, Joe Lipsick looks back at the discovery of tyrosine kinases and the demonstration that the V-SRC protein encoded by Rous sarcoma virus was a tyrosine kinase.
NUCLEAR ENGINEERING
Cancer cells behave differently than normal cells in culture. Cancer cells do not respect their neighbors’ boundaries—they grow over and on top of one another. In contrast, normal cells display contact inhibition: they respect their neighbors’ boundaries and stop dividing once they touch. When dispersed in semisolid agar, individual cancer cells continue to proliferate and can form visible macroscopic colonies, whereas normal cells will not. These are the properties that allowed Howard Temin and Harry Rubin to develop a focus assay for the transformation of normal chicken embryonic fibroblasts by Rous sarcoma virus (RSV) in the late 1950s (see Lipsick 2019a).
Temin also found that RSV-transformed fibroblasts required much less serum to proliferate than did normal cells, displayed increased uptake of glucose and other nutrients, and used glycolysis rather than the far more efficient mitochondrial oxidative phosphorylation for energy production. Otto Warburg had first described this metabolic switch in the 1930s, and he believed it was the primary cause of cancer (see Lipsick 2019b). Temin’s results argued the opposite—the Warburg effect was a consequence rather than a cause of oncogenic transformation. RSV-transformed cells also differed in shape from normal cells. The transformed cells were less adherent and often rounded up, appearing more refractile when observed through a phase-contrast microscope. This morphologic change correlated with a disorganization of the cytoskeleton. Transformed cells also frequently displayed an increase in the size and/or number of nucleoli.
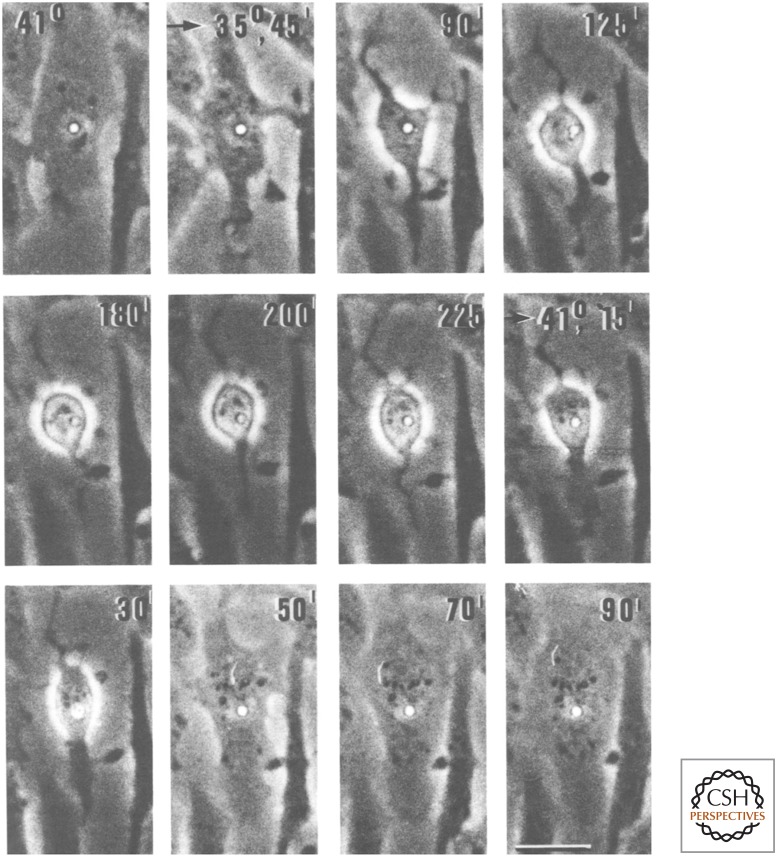
Some thought that constant changes in gene expression were required to maintain all these features of the transformed state. In 1978, Hartmut Beug and Thomas Graf performed a simple but elegant experiment to test this hypothesis (Fig. 1). They first transformed chicken embryo fibroblasts with a mutant of Rous sarcoma virus (ts-RSV) that caused temperature-sensitive oncogenic transformation. Steve Martin had previously shown that in such cells, the V-SRC oncogene was required for both the initiation and the maintenance of the transformed phenotype. These ts-RSV-transformed cells were then treated with cytochalasin, a reversible inhibitor of actin polymerization. Loss of cytoskeletal integrity resulted in the bulging of the nucleus at the top of cells grown on an adherent surface. Gentle centrifugation of these adherent, cytochalasin-treated cells caused enucleation, leaving behind adherent cytoplasts (enucleated cells with cytoplasm surrounded by an intact plasma membrane). Beug and Graf then observed cytoplasts from ts-RSV-transformed cells during shifts from the permissive temperature (35°C) to the nonpermissive temperature (41°C), and back again. Time-lapse videomicroscopy revealed that cytoplasts lacking a nucleus showed a transformed phenotype at the permissive temperature. These cytoplasts reverted to normal cell morphology when shifted to the nonpermissive temperature but could be “retransformed” by shifting them back to the permissive temperature. They also exhibited reversible changes in actin polymerization and in glucose transport. This experiment demonstrated that many aspects of oncogenic transformation do not require the cell nucleus or changes in gene expression.
Figure 1.
Reversible transformation of cytoplasts by temperature-sensitive Rous sarcoma virus, ts-RSV. Virus-infected fibroblasts were treated with cytochalasin, enucleated by gentle centrifugation, and incubated at two different temperatures. Incubation temperatures (41°C or 35°C) and time in minutes are indicated. The small hole in the center of the cell was the site of enucleation. (Modified from Beug H, et al. 1978. Cell 14: 843–856, with permission from Elsevier.)
SRC IT TO ME!
DNA makes RNA makes protein. And proteins do most of the work. The V-SRC protein was remarkable for its ability to affect so many different processes that resulted in the transformed phenotype, including cell shape, organization of the cytoskeleton, and nutrient transport. To understand how it exerted such a range of effects on cells, researchers needed to isolate V-SRC and determine its molecular activity(s). Different experimental approaches to this question were tried, but success ultimately came from a rabbit trick.
THE LOCK IS THE KEY
In 1890, Emil von Behring and Kitasato Shibasaburo found that rabbits infected with the bacteria that cause tetanus produced a substance in their blood that could protect naïve animals against the effects of tetanus toxin. Further experiments showed that these antitoxins were soluble and cell-free. Remarkably, each antitoxin was specific for the toxin that elicited it. Paul Ehrlich used the term “antibody” to describe these antitoxins. He reasoned that if two substances elicited different antibodies, then the two substances must differ in structure. He proposed that antibodies bound to these substances (antigens) very specifically, much as a lock requires a specific key. In the 1920s, Michael Heidelberger and Oswald Avery showed that antibodies could precipitate their cognate antigens, and that antibodies themselves were proteins. Antibodies then became powerful tools for identifying and purifying antigens.
Studies in the early 1960s showed that a DNA virus isolated from human adenoid tissue (adenovirus) and a monkey DNA virus that had contaminated poliovirus vaccines (simian virus 40, SV40) caused tumors in hamsters. While investigating oncogenic transformation by these viruses, Wallace Rowe and Robert Huebner discovered that virus-transformed cells contained proteins that appeared to be encoded by the viral genomes but were not present within infectious viral particles. Similar observations were made in cells transformed by polyoma, a DNA tumor virus affecting mice. One hypothesis to explain these observations was that some viral proteins required for replication early during the course of infection were not themselves incorporated into the mature virion. Remarkably, these virally encoded tumor antigens were present in oncogenically transformed cells even in the absence of viral replication.
Joan Brugge, who had studied oncogenic transformation by the SV40 virus as a graduate student with Janet Butel, joined Ray Erikson’s laboratory as a postdoctoral fellow. Following the logic of the discovery of SV40 tumor antigens, Brugge injected newborn rabbits subcutaneously with a strain of RSV that infected mammals in addition to birds. Two weeks later, tumors appeared—in some rabbits the tumors eventually regressed, whereas other rabbits developed metastatic disease. Brugge then used immunoprecipitation to test whether the sera from these rabbits could recognize a V-SRC protein in RSV-transformed cells.
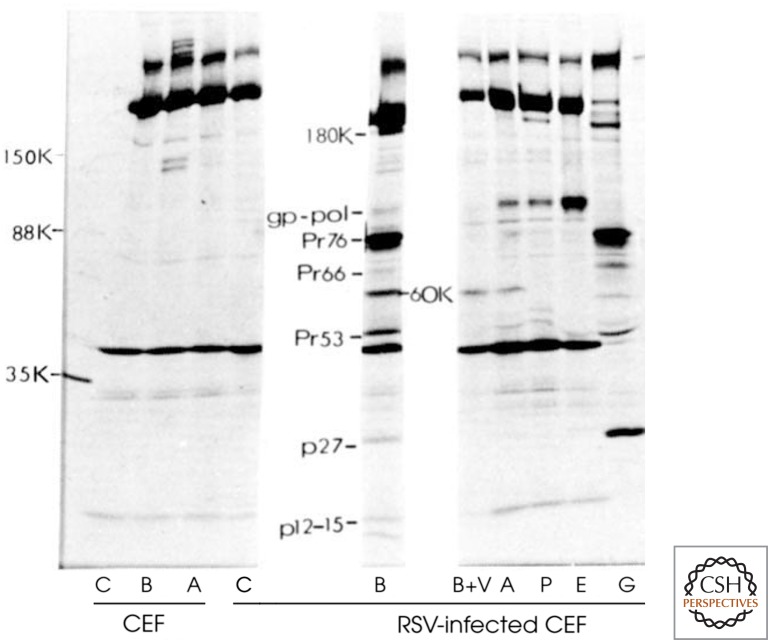
The key results published in 1977 are shown in Figure 2. Numerous controls are present, each directed at a different aspect of the experiment. Three different sera were used, one from normal uninfected rabbits (lanes labeled C), and two from RSV-infected tumor-bearing rabbits (lanes labeled A and B). Radioactive proteins immunoprecipitated from cell lysates by all of these sera were considered to be “background bands,” because they did not depend on the presence of RSV-induced tumors in the immunized animals. As another control, two different radioactively labeled cell lysates were used, one from uninfected chicken embryo fibroblasts, and one from RSV-infected oncogenically transformed chicken embryo fibroblasts. Radioactive proteins immunoprecipitated from RSV-infected but not from uninfected cells would be possible products of the viral V-SRC gene. However, the viral GAG, POL, and ENV genes also encoded protein products. Because tumor-bearing rabbit serum would also contain antibodies directed against these, Brugge used nonradioactively labeled viral particles (lane labeled B+V) to block the precipitation of their radiolabeled counterparts. What remained was a 60-kDa protein, the likely product of the V-SRC gene. The success of this experimental approach depended on the absence of the V-SRC protein within the viral particles and its presence within the transformed cells, as had been the case for the SV40 virus tumor antigen (T antigen).
Figure 2.
Identification of a 60-kDa V-SRC protein by radioimmunoprecipitation. (C) Control serum, (A and B) tumor-bearing rabbit sera, (B+V) Serum B blocked with virus, (P) anti-Pol serum, (E) anti-Env serum, (G) anti-Gag serum, (CEF) chicken embryo fibroblasts. (Adapted from Brugge and Erickson. 1977. Nature 269: 3346–3348, with permission from Springer Nature.)
Further evidence that the 60-kDa protein was the product of V-SRC was its absence in chicken fibroblasts infected by transformation-defective deletion mutants of RSV. Although it remained possible that the 60-kDa protein was the product of a cellular gene that was expressed at higher levels in RSV-transformed cells, the most likely hypothesis was that the 60-kDa protein was the product of the V-SRC oncogene. Important evidence that the 60-kDa protein was actually encoded by the RSV genome came from in vitro translation of RNA isolated from purified viral particles. In 1977, Anthony Peruchio in the Erikson laboratory had identified a 60-kDa protein that was translated from subgenomic RNA fragments of nondefective RNA, but not from subgenomic fragments of transformation-defective RSV mutants. This in vitro translated 60-kDa protein was then shown be identical to that of the protein immunoprecipitated from RSV-infected cells when analyzed by two-dimensional tryptic peptide mapping. The protein became known as p60v-src, for protein of 60 kDa encoded by V-SRC. Following the discovery of RNA splicing in adenovirus by the laboratories of Richard Roberts and Phillip Sharp in 1977, several laboratories showed that RSV used differential splicing to produce two different subgenomic messenger RNAs (mRNAs): one encoding the transmembrane ENV protein precursor and the other encoding p60v-src. These results provided an explanation for the observation that only subgenomic fragments of the full-length RSV genomic RNA could be translated in vitro to produce the V-SRC protein.
HAVE PROTEIN, NEED FUNCTION
The next big question was how one protein could cause such dramatic (and reversible) changes in cell morphology and proliferation. In the 1940s, Fritz Lipmann had proposed that ATP functioned within cells as the currency of energy transfer. By making and breaking phosphate-containing chemical bonds, enzymes could assist in the storage or the use of this energy. Studies of glycogen synthesis led Edwin Krebs and Edmond Fischer, in the 1950s, to discover that enzymes themselves could be regulated by the addition and removal of covalently bound phosphates. Enzymes that use the energy of ATP to add phosphates to other proteins became known as protein kinases from the Greek “kinein,” meaning “to move.” By the 1970s, a number of protein kinases had been shown to regulate cellular metabolism, in some cases acting as powerful intracellular effectors of the hormones that cells and tissues use to communicate with one another.
The rapid reversibility of the transformed state in fibroblasts infected with ts V-SRC suggested that a reversible protein modification, such as phosphorylation, might be involved in transformation. Marc Collett, another member of the Erikson laboratory, tested the hypothesis that p60v-src functioned as a protein kinase. He immunoprecipitated V-SRC protein from chicken embryo fibroblasts that had previously been infected with wild-type RSV. After washing to remove extraneous proteins, he added ATP containing a radioactive terminal (γ) phosphate group. If the radioactive terminal phosphate of ATP had been transferred to a protein, that protein would then become radioactive and would be detected by exposure of X-ray film.
Remarkably, Collett detected a radioactively labeled 50-kDa protein in immunoprecipitations of extracts of cells that had been infected by wild-type RSV, but neither in extracts of uninfected cells nor in extracts of cells infected by transformation-defective RSV. Furthermore, he could detect this activity using sera from rabbits bearing RSV-induced tumors but not from control rabbits. Moreover, when he tested cells infected with a ts-RSV maintained at either the permissive or nonpermissive temperature, only those immunoprecipitates prepared from ts-RSV-infected cells maintained at the permissive temperature contained the radioactive phosphoprotein of 50 kDa. Similar results were obtained by Arthur Levinson and J. Michael Bishop, who in their publication wrote, “We thank R. Erikson for telling us about rabbits and for candid discussions regarding work in progress.”
The simplest explanation was that V-SRC was indeed a protein kinase, and that it had phosphorylated itself (autophosphorylation). However, it turned out that the most abundant radiolabeled phosphoprotein was in fact the 50-kDa heavy chain of the antibody that had immunoprecipitated the V-SRC protein. The potential reversibility of protein phosphorylation by protein phosphatases provided an attractive explanation for the reversibility of oncogenic transformation by ts-RSV.
CHANCE FAVORS THE PREPARED MIND (AND THE LAZY BODY?)
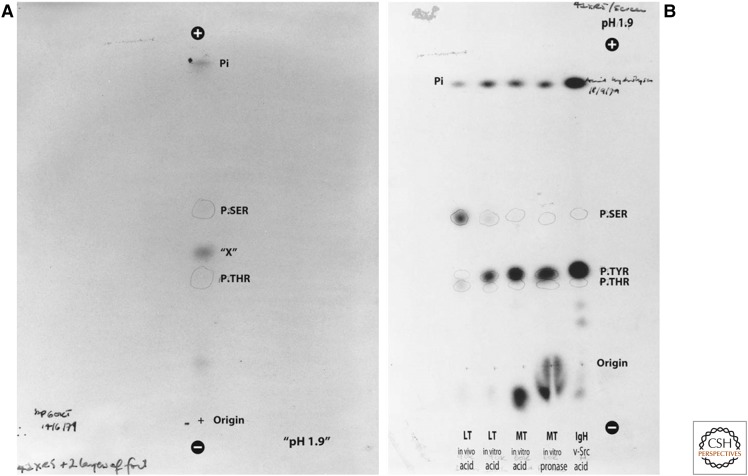
The discovery that V-SRC was a protein kinase caused many investigators to test whether their favorite oncoprotein might also function as a protein kinase. Tony Hunter and Walter Eckhart had been studying the three tumor antigens (small, middle, and large) encoded by murine polyoma virus, a small DNA tumor virus. In 1979, they used an antipolyoma virus tumor serum and the immune complex kinase assay described above to discover a protein kinase activity that coprecipitated with polyoma middle T antigen protein. In this case, the middle T antigen was phosphorylated. To determine whether it was phosphorylated on serine or threonine (the two amino acids known to be phosphorylated by most protein kinases in animal cells), Hunter hydrolyzed the protein to single amino acids and then separated them by electrophoresis on cellulose plates. To his surprise, he found a radioactive molecule that comigrated with neither phosphoserine nor phosphothreonine, but instead ran between them (Fig. 3). He reasoned that the new radioactively phosphorylated species might be phosphotyrosine, because tyrosine is the only other amino acid containing an accessible hydroxyl group. To test this hypothesis, he chemically synthesized a small amount of phosphotyrosine. He then found that the radioactive amino acid in the middle T-associated protein comigrated with the nonradioactive phosphotyrosine standard (detected by ninhydrin staining) in the thin-layer electrophoresis (TLE) used to separate phosphoamino acids.
Figure 3.
Discovery of protein tyrosine kinases. (A) Thin-layer electrophoresis revealed a phosphorylated “mystery” spot (“X”) in amino acid hydrolysis from immune-complex kinase assays of polyoma middle T antigen. (B) Phosphoamino acid analysis showed that V-SRC phosphorylated the immunoglobulin heavy chain (IgH) on tyrosine. Pronase hydrolysis of middle T gave a similar result, arguing against an artifact of acid hydrolysis. (Reprinted from Hunter T. 2015. Proc Natl Acad Sci 112: 7787–7882, with permission from the author.)
Meanwhile, Bart Sefton, Hunter’s neighbor at the Salk Institute, was studying transformation by RSV. Quite unexpectedly, the major phosphoamino acid in the antibody that had immunoprecipitated V-SRC also turned out to be tyrosine. In the course of repeating results before publication, the separation of phosphotyrosine from phosphothreonine in TLE stopped working. It turned out that the continued reuse of electrophoresis buffer had resulted in an increase in acidity from pH 1.9 to 1.7, thereby permitting the separation of phosphotyrosine and phosphothreonine in “old” buffer but not in the standard buffer. This observation also explained why Collett and Erikson had previously reported that V-SRC was a protein threonine kinase, when in reality it was a protein tyrosine kinase. Doing the experiment “right” (with fresh buffer) gave the “wrong” answer (phosphothreonine), whereas doing the experiment “wrong” (with old buffer) gave the “right” answer (phosphotyrosine). The latter required a happy coincidence of laziness (not making new buffer), followed by keen chemical insight and persistence (Fig. 4).
Figure 4.
A cartoon from a meeting held 10 years after the discovery of tyrosine kinases. (Drawn by Jamie Simon.)
Owen Witte and David Baltimore soon showed that the V-ABL protein encoded by the oncogene of Abelson murine leukemia virus also had tyrosine kinase activity. Within a few years, the V-FES/FPS, V-FGR, V-FMS, V-ROS, and V-YES oncogenes of avian and feline retroviruses were also shown to encode protein tyrosine kinases. Ironically, in 1983 Sara Courtneidge and Alan Smith discovered that the tyrosine kinase activity associated with polyoma middle T antigen was caused by the binding and activation of the normal C-SRC protein by middle T.
V-SRC: CLONE, SEQUENCE, REPEAT
The development of methods to create and propagate recombinant DNA molecules in the early 1970s in the laboratories of Paul Berg, Herbert Boyer, and Stanley Cohen led to concerns about potential biosafety hazards. Following a conference at Asilomar, California, in 1975, strict guidelines were established in the United States that included a prohibition on the molecular cloning of cancer-causing viruses. This prohibition was removed in 1978, and in 1980 the laboratories of J. Michael Bishop, Thomas Parsons, and Anna Marie Skalka reported the isolation of molecular clones of RSV. The Bishop laboratory also published a DNA sequence of V-SRC and a deduced protein sequence of p60v-src. The following year, Inder Verma’s laboratory determined the sequence of the V-MOS oncogene of Moloney murine sarcoma virus, and Russell Doolittle found that the deduced protein sequence had a region of similarity to p60v-src. Winona Barker and Margaret Dayhoff then reported significant homology between the carboxy-terminal region of the V-SRC protein and the recently published catalytic domain of cyclic AMP-dependent protein kinase (PKA). However, they noted that the V-SRC protein sequence deduced from the DNA sequence of the Schmidt-Ruppin A strain published by the Bishop laboratory differed by ∼40% from the V-SRC protein sequence deduced from the as yet unpublished sequence of the Prague C strain of RSV. Dennis Schwartz and Walter Gilbert had determined the latter directly from reverse-transcribed viral RNA.
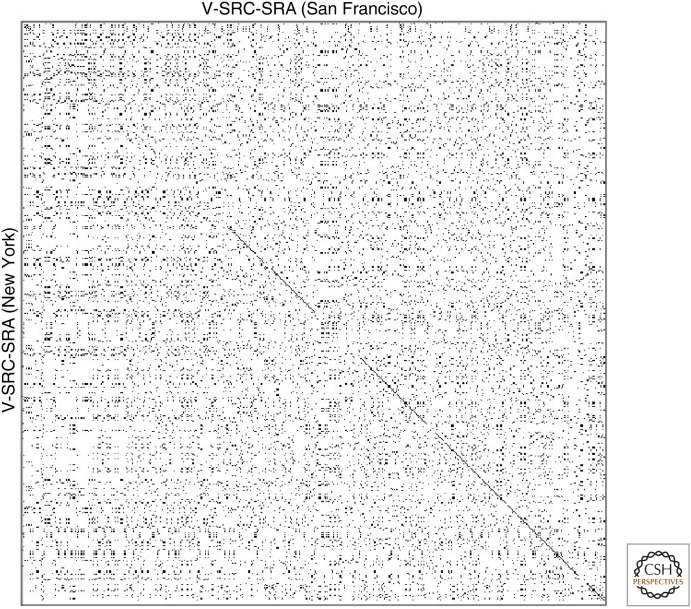
By 1982, the laboratory of Hidesaburo Hanafusa had also molecularly cloned and sequenced the Schmidt-Ruppin A strain. They found that their predicted V-SRC (New York) protein sequence was nearly identical to that of the Prague C strain determined by Schwartz and Gilbert. The Hanafusa laboratory then showed that two-dimensional peptide mapping of the p60v-src protein was consistent with their predicted protein sequence but not with the published V-SRC (San Francisco) protein sequence. The ∼40% discrepancy in predicted protein sequence was due to inaccuracies in the published DNA sequence, resulting in several frameshifts (Fig. 5). Rather fortuitously, this had not affected the conserved protein kinase domain near the carboxyl terminus used in the protein sequence alignments of Doolittle, Barker, and Dayhoff.
Figure 5.
A dot matrix plot showing amino acid identities between the two different deduced protein sequences of the V-SRC protein encoded by the Schmidt-Ruppin A (SRA) strain of Rous sarcoma virus. The broken diagonal line indicates regions of identity (∼60% of the overall sequence). Both sequences start in the upper left corner of the plot.
V-SRC VERSUS C-SRC
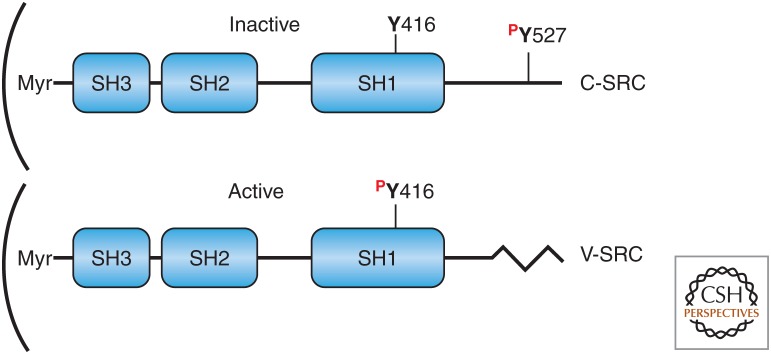
Some (but not all) RSV-induced tumor-bearing rabbit sera were able to immunoprecipitate the much less abundant normal C-SRC protein from uninfected normal cells. The molecular cloning and determination of the DNA sequences of V-SRC and C-SRC together with biochemical experiments in a number of laboratories led to a model in which V-SRC and C-SRC were both tyrosine kinases, but V-SRC had a much higher activity. V-SRC was phosphorylated on tyrosine residue Y416 within the kinase domain, whereas C-SRC was predominately phosphorylated on Y527. The transduction of the V-SRC oncogene by RSV resulted in a frameshift that caused a deletion of the carboxyl terminus of C-SRC that included Y527 and the surrounding amino acids (Fig. 6).
Figure 6.
Schematic diagrams of the C-SRC versus V-SRC proteins. Note that V-SRC has a carboxy-terminal truncation owing to a frameshift (jagged line) relative to C-SRC. This truncation removes Y527, the most abundant phosphotyrosine in C-SRC. Instead, the most abundant phosphotyrosine in V-SRC is Y416, a residue that is not highly phosphorylated in C-SRC. (SH) SRC homology domain, (Myr) myristoylation. Curved line, plasma membrane. (Adapted from Rosenberg NJ, Jolicoeur P. 1997. In Retroviruses (ed. Coffin JD, et al.), Chapter 10, Fig. 11, © Cold Spring Harbor Laboratory Press.)
The working hypothesis in the field was that phosphorylation of Y527 in C-SRC was inhibitory, and that phosphorylation of Y416 was required for the kinase activity of both V-SRC and C-SRC. The test of this hypothesis used what was then a new and very powerful technique, site-directed mutagenesis. Previously, experimental genetics had depended on the generation of random mutations (by chance, by X-rays, by chemicals, or by transposons) that caused detectable phenotypes (changes in visible and/or measurable traits). Considerable work was then required to identify the mutation (sometimes among many) that caused the phenotype. Michael Smith used techniques developed by Har Gobind Khorana to synthesize short pieces of DNA with predefined sequences. Smith then showed that one could introduce a defined mutation into a molecule of known DNA sequence by synthesizing DNA in vitro using an oligonucleotide “primer” containing the desired mutation. This technique rapidly became a workhorse of modern molecular genetics, permitting scientists to create and study “designer” mutations in a variety of molecules and organisms.
In 1987, the laboratories of Walter Eckhart, Thomas Roberts, and David Shalloway used this technology to substitute Y527 of C-SRC with phenylalanine (Y527F). The two amino acids are identical except for the absence of a phosphorylatable hydroxyl group in phenylalanine. As predicted, C-SRC(Y527F) had increased tyrosine kinase activity and high levels of phospho-Y416. Furthermore, C-SRC(Y527F) could oncogenically transform cells, whereas wild-type C-SRC could not. In addition, Y416F mutants of V-SRC or C-SRC(Y527F) had reduced tyrosine kinase activity and reduced transforming activity. These results indicated that C-SRC was indeed negatively regulated by phosphorylation at Y527. Further studies identified a C-SRC-specific kinase (CSK) responsible for the inhibitory phosphorylation of C-SRC at Y527, and several phosphotyrosine phosphatases (PTPs) that could remove this inhibitory phosphorylation. In normal cells, the phospho-Y527 “OFF” form predominates. The levels of the unphosphorylated Y527 “ON” form can be increased in response to various extracellular signals. V-SRC is oncogenic because it remains permanently stuck in the “ON” state.
V-SRC IS A LOUSY ENZYME
The ability of V-SRC to function as a protein tyrosine kinase was clear, but some aspects of its enzymatic activity were unusual. Most enzymes are efficient catalysts that have a high “turnover number.” They are capable of converting many molecules of raw material into many molecules of finished product in a short period of time. However, V-SRC had a low turnover number (i.e., it was very inefficient). Most enzymes are rather picky about the raw materials they will use (substrate specificity). However, V-SRC was not very picky and could phosphorylate many substrates, including some antibodies used in immune complex kinase assays. The regulation of SRC kinase activity was unraveled following the discovery of two new protein-binding domains.
SRC-HOMOLOGY DOMAINS
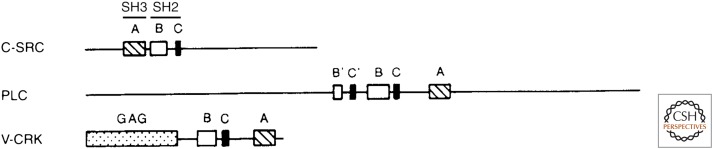
In 1984, Tony Pawson’s laboratory used recombinant DNA technology to create a panel of short in-frame insertion mutants of the SRC-related V-FPS oncogene. As expected, they found that some insertions within the conserved kinase domain abolished oncogenic transformation. Somewhat unexpectedly, a subset of insertional mutations upstream of the kinase domain also abolished oncogenic transformation but retained kinase activity. By comparing the amino acid sequences of several SRC-related tyrosine kinases (FPS/FES, SRC, YES, FGR, ABL, and LCK/TCK), the Pawson laboratory identified two discrete regions of sequence conservation that corresponded to the functionally important domains identified by insertional mutagenesis. In 1986, they termed the kinase domain “SH1” (SRC-homology domain 1), and the more amino-terminal region was called “SH2” (Fig. 6).
Meanwhile, Bruce Mayer in Hidesaburo Hanafusa’s laboratory had identified and sequenced a peculiar oncogene in the CT10 avian retrovirus (named for chicken tumor number 10) in 1988 that had been isolated by James Murphy and Albert Claude 50 years earlier. The oncogene in CT10 encoded an SH2 domain, but no SH1 (tyrosine kinase) domain. Comparison with sequences of other proteins revealed the presence of an additional conserved motif, termed SH3. Several SRC-related tyrosine kinases contained all three domains (SH1, SH2, and SH3), whereas the oncogene in CT10 contained only SH2 and SH3, as did the recently sequenced phospholipase C (PLC), an enzyme that cleaves phospholipids during intracellular signaling. However, the order of SH2 and SH3 domains differed among these proteins (Fig. 7). Furthermore, the FPS tyrosine kinase in which SH2 domains were first discovered lacked an SH3 domain. These observations indicated that SH2 and SH3 domains were modular and could function independently.
Figure 7.
SH2 and SH3 domains are present in SRC-family tyrosine kinases and in nonkinase proteins. (A) SH3 domain, (B and C) subdomains of the SRC-family SH2 domain also conserved in PLC. (Modified from Stahl ML, et al. 1988. Nature 332: 269–272, with permission from Springer Nature.)
Remarkably, immunoprecipitates of the GAG-fusion protein encoded by the oncogene in CT10 contained tyrosine kinase activity, although the virus itself did not encode a kinase domain. In addition, cells transformed by the CT10 virus contained elevated levels of phosphotyrosine in a number of cellular proteins. Mayer and Hanafusa called the new oncogene V-CRK, for CT10 regulator of kinase. Subsequent work in Masabumi Shabuya’s laboratory identified two normal C-CRK isoforms, one containing an additional carboxy-terminal SH3 domain. Only the isoform lacking this additional SH3 domain was oncogenic in cell culture, which was consistent with its absence in two independently isolated oncogenic retroviruses containing V-CRK.
In 1990, the Pawson laboratory showed that recombinant SH2 domains bound specifically to phosphotyrosine-containing proteins. Furthermore, SH2 domains were required in V-SRC and V-FPS proteins for efficient phosphorylation of some substrates. The laboratories of Stuart Schreiber and David Baltimore then showed that SH3 domains bound specifically to peptides containing proline-rich sequences. A relatively large number of proteins containing these domains have now been identified in the human genome database (approximately 110 proteins with SH2 domains; approximately 530 proteins with SH3 domains). Zhou Songyang and Lewis Cantley developed a powerful technology that used synthetic degenerate peptide libraries to determine the binding specificity of these protein–protein interaction domains. The discovery of SH2 and SH3 domains and their ligands provided a paradigm for the identification of other conserved regulatory domains and binding motifs in proteins involved in signal transduction.
A MOLECULAR JACKKNIFE
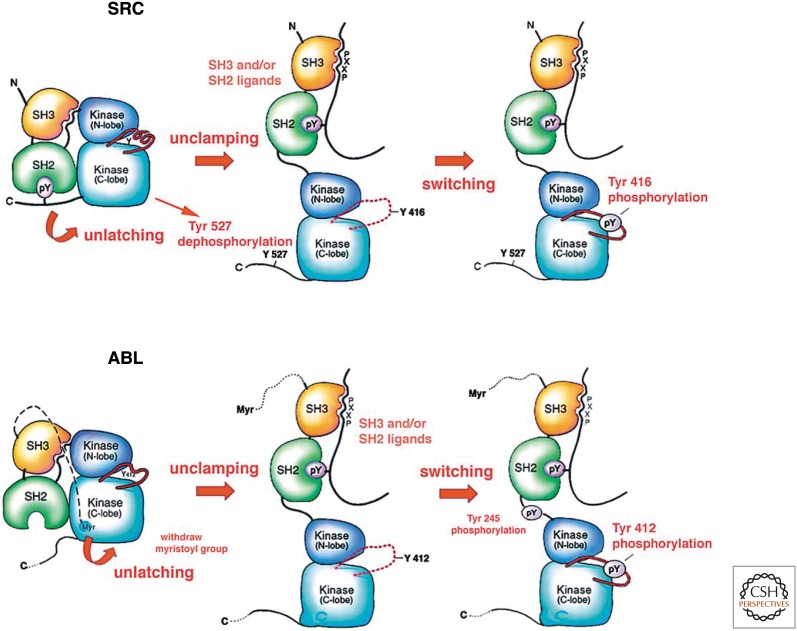
Structures of SRC-related proteins determined by X-ray crystallography in the laboratories of Michael Eck, Stephen Harrison, John Kuriyan, and Rik Wierenga in 1997 led to a very satisfying model consistent with many previous biochemical and genetic studies (Fig. 8). The regulation of C-SRC resembled a molecular jackknife. When it was tyrosine-phosphorylated at carboxy-terminal Y527, the phosphorylated residue interacted with a pocket within the SH2 domain and the C-SRC knife was closed. The binding of the amino-terminal SH3 domain to a proline-rich sequence that connects the kinase and SH2 domains helped ensure that the knife stayed closed. This interaction also caused a conformational change in the kinase domain that inhibited its activity. When C-SRC was dephosphorylated at Y527, the knife opened. A peptide loop that blocked the substrate binding site in the closed state was now tyrosine-phosphorylated (pY416) in the open state. This enabled the enzyme to actively phosphorylate other proteins on tyrosine residues. The model also explained the mechanism of oncogenic activation of V-SRC. During retroviral transduction of the V-SRC oncogene, a frameshift mutation occurred that resulted in the deletion of the carboxy-terminal tail containing Y527 (Fig. 6). Therefore, in the case of V-SRC, the jackknife was always open and the enzyme was always active. The oncogenic activation of C-SRC by a single amino acid substitution (Y527F) was also readily explained, because the mutant carboxy-terminal tail could not be phosphorylated.
Figure 8.
Intramolecular regulation of the C-SRC and C-ABL proteins by their SH2 and SH3 domains. (Adapted from Harrison SC. 2003. Cell 112: 737–740, with permission from Elsevier.)
The V-ABL oncogene of the Abelson murine leukemia virus was discovered in the laboratories of Stuart Aaronson and David Baltimore. As was the case for V-SRC, Owen Witte’s laboratory showed that V-ABL is a much more active tyrosine kinase than C-ABL. Jean Wang’s laboratory identified an inhibitory amino terminus in C-ABL with a covalently attached myristic acid group. Structural and biochemical studies from the Kuriyan laboratory showed that inactive C-ABL had a structure similar to C-SRC, but its amino terminus rather than a carboxy-terminal phosphorylation site functioned as the clasp that kept the jackknife closed. Interactions with other proteins via its SH2 and SH3 domains were thought to help stabilize the open kinase-active state of C-ABL during normal cellular responses to various signals. The amino-terminal clasp is deleted in the BCR-ABL fusion protein encoded by the Philadelphia chromosome t(9;22) that causes human chronic myelogenous leukemia (see Lipsick 2019c). As a result the BCR-ABL fusion protein is a constitutively active tyrosine kinase stuck in the open state. Among the most successful of the new targeted cancer therapies is a small molecule inhibitor (Gleevec/imatinib) of the BCR-ABL tyrosine kinase discovered by Brian Druker and Nicholas Lydon (see Lipsick 2019d).
LOCATION, LOCATION, LOCATION
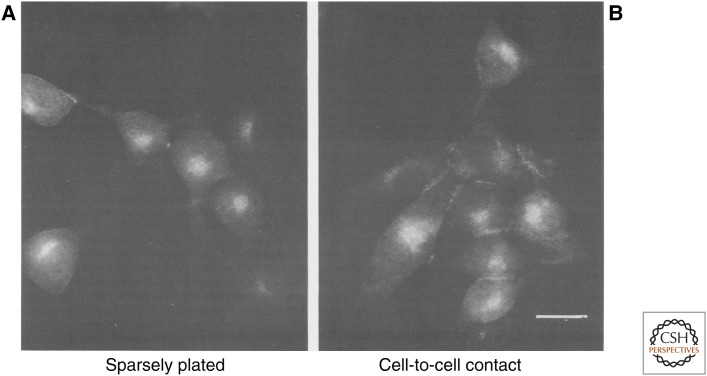
The location of a protein within the cell can be an important clue to its function. In 1979, Larry Rohrschneider used antibodies directed against the V-SRC protein to determine its intracellular location by immunofluorescence (Fig. 9). The V-SRC protein was primarily localized to the membranes of the cell, but was not apparent on the outer surface of living cells. Additional experiments showed that V-SRC was concentrated at cell–cell junctions and at focal adhesion plaques, structures used by cells to adhere to the extracellular matrix. Shortly thereafter, Sara Courtneidge in J. Michael Bishop’s laboratory showed, using biochemical fractionation, that the bulk of the V-SRC protein was associated with the plasma membrane.
Figure 9.
Localization of the C-SRC protein by indirect immunofluorescence. (Reprinted from Rohrschneider LR. 1979. Cell 16: 11–24, with permission from Elsevier.)
The corrected sequence of the V-SRC protein lacked a hydrophobic domain that might explain its membrane attachment. In 1984, Bart Sefton’s laboratory reported that myristic acid (a hydrophobic chain of 14 carbon atoms) was covalently attached to V-SRC (Fig. 6). This relatively rare protein modification had previously been reported to occur on a few other proteins, including the catalytic subunit of the signaling molecule cAMP-dependent protein kinase (PKA). The Sefton laboratory showed that this fatty acid was specifically attached to a glycine (G2) at the amino terminus of the processed protein. To test whether it was necessary and sufficient for the membrane localization of V-SRC, they used site-directed mutagenesis to replace this glycine either with alanine (G2A), a similarly hydrophobic amino acid, or with glutamic acid (G2E), a negatively charged amino acid. Neither the G2A nor the G2E mutant of V-SRC could oncogenically transform cells, nor were the mutant proteins localized to the membrane. However, these mutant proteins did remain highly active as tyrosine kinases. Levels of phosphotyrosine within infected cells were similarly elevated (from ∼0.04% to ∼0.20% of total phosphoamino acids) by the wild-type and mutant V-SRC proteins. These results indicated that membrane localization of V-SRC and, therefore, tyrosine phosphorylation of proteins at or near the membrane were required for oncogenic transformation.
The search for the critical proteins that must be phosphorylated by V-SRC to cause oncogenic transformation has been long and complicated—there are likely to be a number of targets. Two major strategies were used for the identification of physiologic substrates of V-SRC. Jonathan Cooper and Tony Hunter pioneered the use of two-dimensional gel electrophoresis of 32[P]-labeled phosphoproteins followed by alkali treatment to dephosphorylate serine and threonine residues. Thomas Parsons’s laboratory used anti-phosphotyrosine monoclonal antibodies very effectively to identify transformation-specific substrates, including focal adhesion kinase (FAK). The inhibition of phosphotyrosine phosphatases by vanadate greatly aided these efforts. Like V-SRC and C-SRC, FAK is localized at focal adhesions and mediates interactions between the intracellular actin cytoskeleton and the integrin transmembrane receptor proteins that bind to the extracellular matrix and regulate cell adhesion and migration. In 1994, Michael Schaller in the Parsons laboratory showed that FAK autophosphorylates on Y397, creating a binding site for the SH2 domain of C-SRC, which in turn results in additional tyrosine phosphorylation of FAK and other adjacent proteins. Recent studies have implicated increased levels/activity of C-SRC in the invasion and metastasis of some human cancers. The FAK-SRC axis may thus be important in these lethal alterations of cell adhesion and motility.
V-ERBB ENCODES A TRANSMEMBRANE TYROSINE KINASE
Following the discovery of RSV in 1911, a number of other acutely transforming retroviruses that caused cancer were isolated from chickens. The avian erythroblastosis virus, first identified by Evan Stubbs and Jacob Furth in the 1930s, causes a rapidly fatal leukemia as a result of the proliferation of hematopoietic progenitor cells that have committed to differentiating into erythrocytes. Dominique Stehelin’s laboratory used nucleic acid hybridization to discover two different oncogenes of cellular origin, V-ERBA and V-ERBB, within some isolates of this virus. However, other isolates contained only the V-ERBB gene. Using a strategy similar to that which Joan Brugge used to identify V-SRC, Michael Hayman and his colleagues reported in 1983 that antisera from rats bearing AEV-induced tumors could immunoprecipitate the V-ERBB protein. This protein was associated with the plasma membrane of transformed cells, but, unlike V-SRC, at least a portion of V-ERBB was displayed on the outer surface of the cell. Several lines of evidence supported these conclusions—V-ERBB contained glycosyl modifications similar to those of cell surface and/or secreted proteins, the protein was associated with cell membranes in biochemical fractionation experiments, and it could be detected by immunofluorescence in living cells, which are impermeable to antibodies. Biochemical studies by Martin Privalsky in J. Michael Bishops’s laboratory, using antibodies raised in rabbits against a recombinant V-ERBB protein fragment produced in bacteria by Art Levinson’s group at Genentech, supported these findings.
C-ERBB IS THE EGF RECEPTOR—A REAL EYE-OPENER
Tadashi Yamamoto in Kumao Toyoshima’s laboratory deduced the protein sequence of V-ERBB from the proviral DNA sequence. Rather unexpectedly, the central region of V-ERBB was very similar to the tyrosine kinase domain of V-SRC. But near the amino terminus was a hydrophobic stretch of amino acids that looked like a transmembrane sequence. These results together with the biochemical experiments described above led to the conclusion that the amino terminus of the V-ERBB protein was displayed on the outer surface of the cell, whereas the tyrosine kinase domain and a carboxyl-terminal tail were located inside the cell.
In a seemingly unrelated line of experimentation, Stanley Cohen had been studying an epidermal growth factor (EGF) that he had discovered in the 1960s. This secreted protein from salivary gland extracts was purified using premature eyelid opening and tooth development in embryonic mice as a biological assay. The Cohen laboratory then used radioactively labeled EGF to identify a cell surface receptor (EGFR) for this growth factor. In 1980, they reported that EGFR was phosphorylated on tyrosine following the binding of EGF, rather than being phosphorylated on threonine as they had reported before the discovery of tyrosine kinases.
A race ensued to clone and sequence the EGFR gene. In 1984, the laboratories of Michael Waterfield, Joseph Schlessinger, and Axel Ullrich together used a monoclonal antibody to purify EGFR protein from the A431 human epidermoid cancer cell line, which produces 50-fold more receptor than most other cell lines. Remarkably, they found that short peptide sequences from fragments of the purified protein were homologous to the predicted amino acid sequence of the V-ERBB protein. A comparison of the complete cDNA sequence of the human EGFR revealed that it was indeed the human ortholog of chicken C-ERBB. Michael Rosenfeld’s laboratory independently came to the same conclusion by a different route. They used polyclonal anti-EGFR antibodies to screen a plasmid-based cDNA expression library from A431 cells. Both groups reported that A431 cells had amplified the EGFR gene, thereby accounting for the large number of receptor molecules.
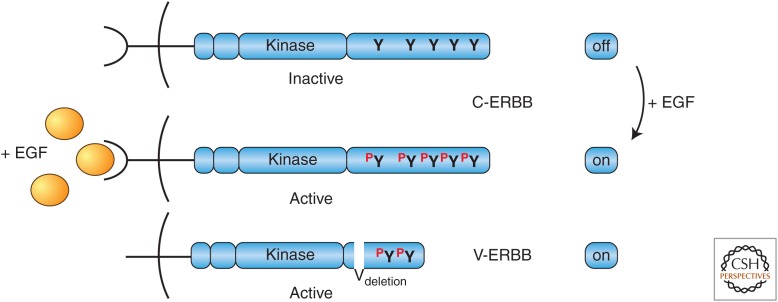
During retroviral transduction, V-ERBB had undergone a deletion of the sequences encoding most of the amino-terminal ligand-binding domain to which EGF normally binds. An additional deletion had removed some of the protein carboxy-terminal to the tyrosine kinase domain. Biochemical experiments showed that oncogenic V-ERBB was a constitutively active, ligand-independent form of the normally EGF-dependent receptor tyrosine kinase (C-ERBB/EGFR) (Fig. 10). Further experiments showed that the carboxyl terminus of EGFR was a major site of EGF-dependent tyrosine phosphorylation.
Figure 10.
The relationship between the retroviral V-ERB oncoprotein and its normal cellular homolog, C-ERB/EGFR. (Curved line) Plasma membrane. (Modified from Rosenberg N, Jolicoeur P. 1997. In Retroviruses (ed. Coffin JD, et al.), Chapter 10, Fig. 11, © Cold Spring Harbor Laboratory Press.)
The discovery that the EGFR was in fact C-ERBB added to the great excitement generated by another recently discovered connection between oncogenes and growth factors. One year earlier, two different laboratories had purified platelet-derived growth factor (PDGF), a protein that promotes fibroblast growth in culture. PDGF is stored in platelets, circulating fragments of specialized blood cells that participate in the formation of blood clots. Activated platelets release PDGF, thereby initiating wound healing by first attracting and then promoting the proliferation of quiescent tissue fibroblasts. Michael Waterfield’s group obtained peptide sequences of PDGF in collaboration with Carl-Henrik Heldin and Tom Deuel, as did Michael Hunkapiller and Leroy Hood in collaboration with Harry Antoniades. When they compared these sequences with a database of published protein sequences maintained by Russell Doolittle, it became apparent that the PDGF gene was homologous to the V-SIS oncogene of simian sarcoma virus that had recently been cloned and sequenced in Stuart Aaronson’s laboratory. Heldin’s laboratory had reported in 1982 that the PDGF receptor, like EGFR, was a ligand-dependent transmembrane receptor tyrosine kinase. In 1985, Carl Rettenmier and Charles Sherr, in collaboration with Richard Stanley’s laboratory, showed that the C-FMS proto-oncogene (initially discovered via its transduction in the McDonough feline sarcoma virus) encodes the receptor for CSF-1/M-CSF, a colony-stimulating factor that promotes the growth of macrophages. Together, the PDGF = SIS, EGFR = ERBB, and CSF1R = FMS identities made it clear that mutations of genes encoding normal cellular growth factors and their receptors could be the molecular basis of cancer.
IT IS A FAMILY AFFAIR
Robert Weinberg’s laboratory continued to use DNA transfection in 3T3 cells to identify new oncogenes (see Lipsick 2019c). Among other types of cancer, they investigated a series of tumors of the nervous system induced in the offspring of pregnant rats following treatment with the chemical carcinogen ethylnitrosourea (ENU). Four independently isolated oncogenes from these tumors, which were thought to be neuroblastomas, appeared to be the same gene. Therefore, they named the new oncogene NEU. Molecular cloning and DNA sequencing revealed that NEU was similar but not identical to EGFR/C-ERBB. Meanwhile, the laboratories of Stuart Aaronson, Axel Ulrich, and Tadashi Yamamoto had all discovered the same new EGFR/ERBB-related gene that turned out to be identical to NEU. Two additional EGFR/ERBB-related genes were then identified. These genes eventually became known as ERBB2/HER2, ERBB3/HER3, and ERBB4/HER4 (for human EGF receptor-like). In the ENU-induced rat tumors, NEU was activated by a point mutation that caused an unusual charged amino acid substitution within the hydrophobic transmembrane domain of the protein. This mutation greatly increased the tyrosine kinase activity of the oncogenic NEU protein by stabilizing protein dimers.
NEU/ERBB2/HER2 became of even greater interest in 1987 when Dennis Slamon reported that this gene was amplified in ∼25% of a series of human breast cancers, and that HER2 amplification correlated with poor prognosis. Mark Greene’s laboratory had shown that monoclonal antibodies directed against the extracellular domain of the rat NEU protein could reverse the transformed phenotype of fibroblasts transfected with the activated NEU oncogene. This reversion correlated with decreased levels of the protein at the cell surface and within the cell. These experiments indicated that monoclonal antibodies might be useful as therapeutic agents for the treatment of human cancers with mutations and/or increased expression of cell surface receptor proteins. Over the next several years, Dennis Slamon pushed for the development of anti-HER2 antibodies for the treatment of women whose breast cancers had amplification of HER2. Eventually, scientists at Genentech developed Herceptin/trastuzumab, a “humanized” mouse monoclonal antibody directed against the human HER2 extracellular domain that, when combined with chemotherapy, has been quite successful as a treatment for patients with “HER2-positive” breast cancer.
IT TAKES TWO TO TANGO
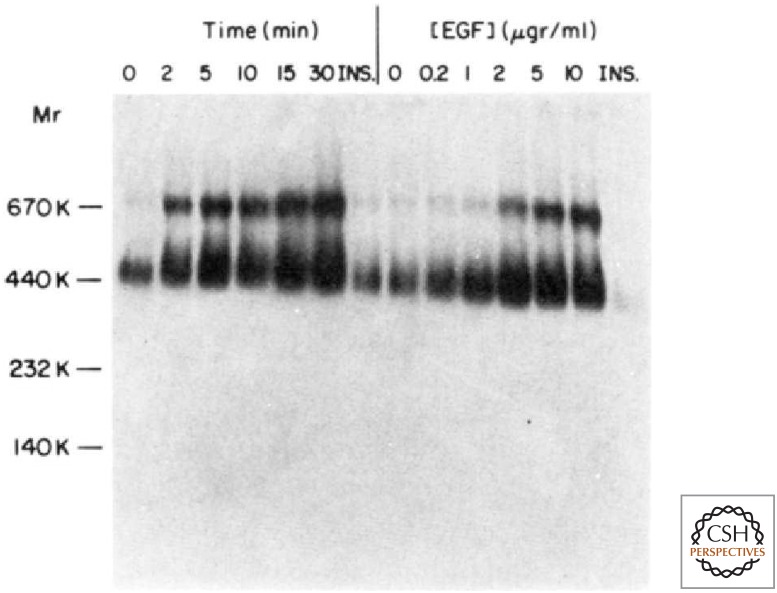
Biochemical studies of EGF binding to the EGFR had implied a 1:1 ligand–receptor interaction. However, careful analysis by Scatchard plots provided evidence for two EGF binding sites with different affinities. These data were consistent with a model of negative cooperativity in which binding of the first EGF molecule reduced the affinity of binding by a second EGF molecule. Joseph Schlessinger’s laboratory produced monoclonal antibodies against EGFR following the immunization of mice with A431 cells. In 1981, they reported that these anti-EGFR antibodies could block EGF binding, but could themselves cause EGF-like responses in living cells. Importantly, the normally bivalent antibody had both of these properties, but a monovalent Fab fragment could block EGF binding but not cause EGF-like responses. In 1987, Yosef Yarden and Schlessinger provided biochemical evidence for a rapid and reversible dimerization of purified EGFR following the binding of EGF (Fig. 11). Furthermore, the increased phosphorylation of the receptor on tyrosine correlated with this dimerization. Based on these results, they proposed that EGF functioned by inducing dimerization of an otherwise monomeric EGFR.
Figure 11.
EGF-induced dimerization of purified EGFR. (Left) A time course of phosphorylation following addition of EGF and analysis by nondenaturing gel electrophoresis. (Right) The dependence of the reaction up on the exposure to different concentrations of EGF for 15 minutes. Insulin (INS) was used as a negative control. (From Yarden Y, Schlessinger J. 1987. Biochemistry 26: 1443–1451, with permission from American Chemical Society.)
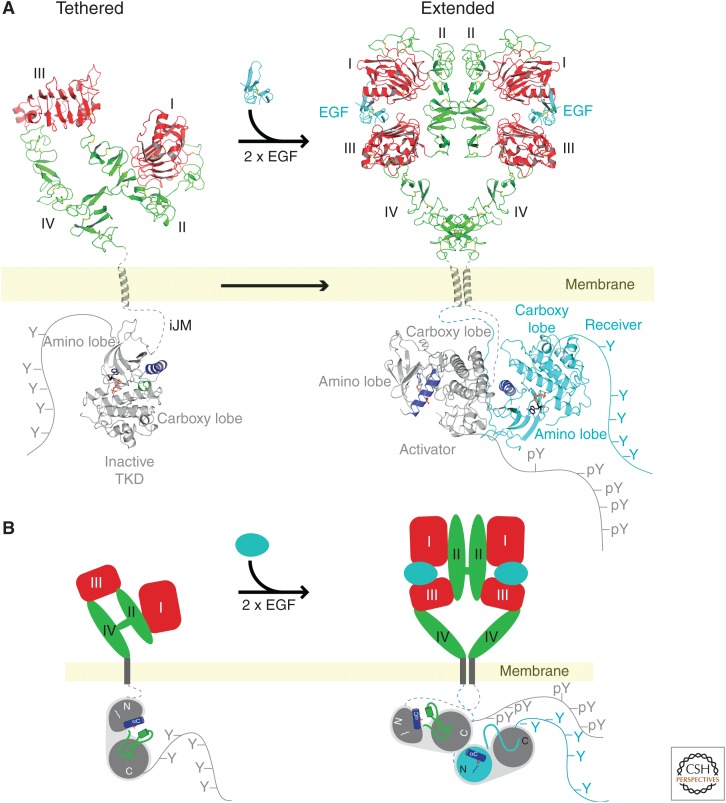
Schlessinger’s laboratory then used affinity labeling with EGF, deletion mutants of EGFR, and chicken/human EGFR chimeras to show that two noncontiguous extracellular domains (called I and III) were both required for high-affinity EGF binding. It was generally assumed (and many textbooks showed) that a dimer of EGF was responsible for dimerization of the activated EGFR. However, a comparison of the structures of the “free” and “bound” EGFR ligand-binding domains determined by X-ray crystallography in the laboratories of Anthony Burgess and Colin Ward, Daniel Leahy, Mark Lemmon, and Shigeyuki Yokoyama told quite a different story (Fig. 12). In the absence of EGF, the EGFR ligand-binding domain folded up on itself and was “auto-inhibited” (i.e., the jackknife was “closed”). On EGF binding, the ligand-binding domain unfolded and extended a finger-like projection from the surface opposite to the bound EGF. The fingers of two adjacent ligand-binding domains then clasped one another to form the ligand-activated EGFR dimer.
Figure 12.
A summary of structural studies of EGF receptor binding and tyrosine kinase activation. (A) Known molecular structures. (B) Simplified cartoon showing changes upon EGF binding. (Reprinted from Lemmon MA, et al. 2014. Cold Spring Harbor Perspect Biol 6: a020768, © Cold Spring Harbor Laboratory Press.)
Biochemical studies had suggested that HER2/NEU might be different from other members of the EGFR family. Researchers identified ligands for EGFR, HER3, and HER4, but not for HER2/NEU. Nevertheless, HER2/NEU was capable of forming heterodimers with the other three receptors. A comparison of the structure of all four receptor ligand-binding domains proved informative. In the absence of ligand, EGFR, HER3, and HER4 had very similar “auto-inhibited” structures. In each case, the fickle finger of dimerization was buried within. In contrast, HER2/NEU adopted an “open” conformation in the absence of ligand—it was always ready for a dimerization handshake. These results may help to explain why amplification of HER2/NEU (but not other members of the HER family) is frequently found in human breast cancers. Only HER2/NEU is likely to undergo frequent dimerization and tyrosine kinase activation in the absence of extracellular ligands.
More recent structural studies of a Drosophila EGFR from Mark Lemmon’s laboratory indicated the active form of this and other EGFR family members may be an asymmetric dimer with only one tightly bound EGF molecule, thus providing an explanation for the puzzling phenomenon of negative cooperativity. In addition, studies of the nematode EGFR (LET-23) have shown that it is a constitutive dimer that still requires EGF (LIN-3) binding for its activation—this is similar to the activation of the human insulin receptor family.
Studies by John Kuriyan’s group have provided further insights about EGFR dimerization and activation. As is the case for SRC and ABL, the inactive EGFR kinase domain is auto-inhibited by an “activation” loop. This loop switches into the “open” position in an asymmetric kinase dimer. The “head” of one EGFR kinase domain bumps the “tail” of another EGFR kinase domain, resulting in a conformational change in which one of the kinase domains becomes activated (Fig. 12). A detailed understanding of the mechanism of activation of the EGFR family is important because of frequent mutation and amplification of these receptors in human malignancies, including cancers of the brain, breast, and lung cancer. This knowledge is critical for development of drugs that target them.
INDUCIBLE BUILDING BLOCKS
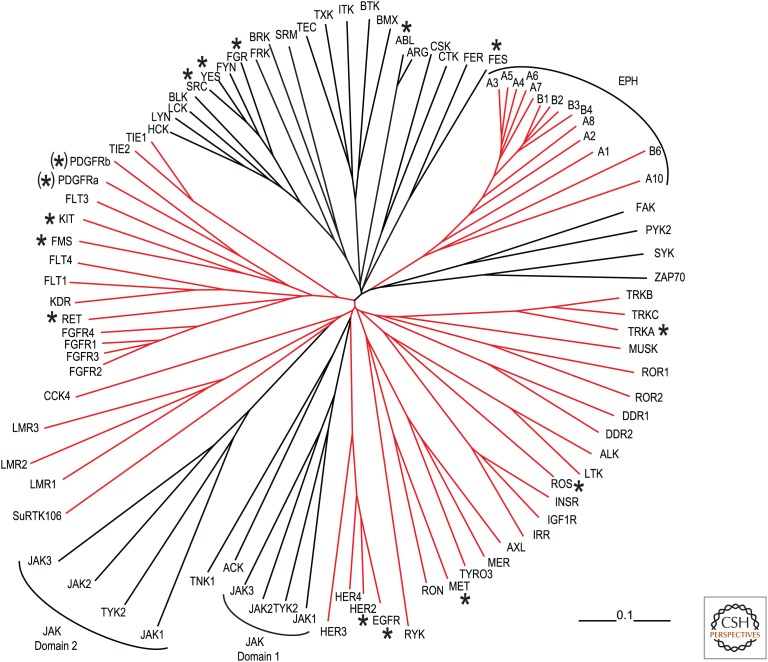
The human genome is predicted to encode 556 different protein kinases. By comparing the amino acid sequences, one can deduce evolutionary relationships and predict protein functions (Fig. 13). Of the 90 predicted protein tyrosine kinases in humans, including several discovered as oncoproteins, 58 appear to be transmembrane receptor tyrosine kinases.
Figure 13.
A phylogenetic tree of human protein tyrosine kinases. Those first discovered as oncoproteins are indicated by asterisks. The parentheses indicate two receptors for the ligand encoded by the V-SIS oncogene (PDGF). The JAK (for just another kinase, later renamed Janus kinase) family protein kinases each have two kinase domains. (Red lines) Transmembrane tyrosine kinases. (Alignment from kinase.com/static/colt/data/human/kinome/groups/tk.aln.)
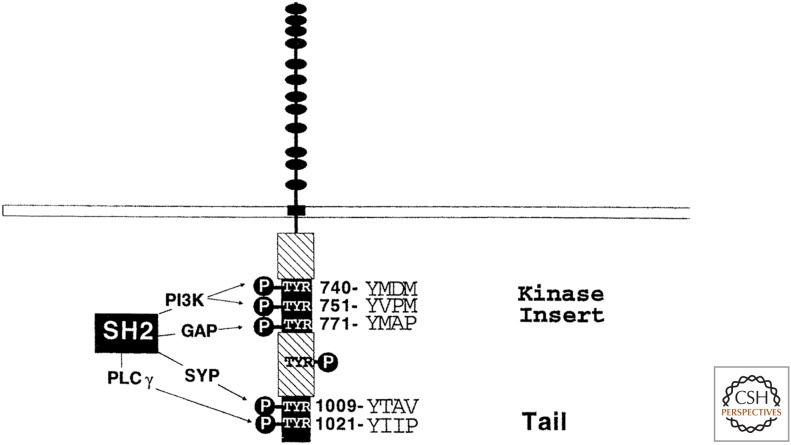
The major substrates that are phosphorylated by EGFR and by other receptor tyrosine kinases are the receptors themselves (“autophosphorylation,” which is often actually transphosphorylation by a dimeric partner). Carboxy-terminal to, or sometimes within, the kinase domain of each receptor is a tyrosine-rich region. When phosphorylated, these tyrosines act as docking sites for other proteins containing phosphotyrosine-binding domains. Direct biochemical purification, functional screening of bacterial expression libraries, and molecular cloning of genes encoding SH2 domain-containing proteins have identified numerous activation-dependent receptor-binding proteins. These include the p85 subunit of phosphatidylinositide 3-kinase (PI3K), phospholipase C, GRB2 (growth factor receptor bound protein), GAB1 (GRB2-associated binding protein), and SHC (SRC homology 2 domain-containing protein). Mutational analyses have shown that specific phosphotyrosines within the tails of activated receptors are specific for different receptor-binding proteins (Fig. 14). Therefore, one of the main functions of tyrosine phosphorylation is to provide an inducible scaffold up on which various intracellular signaling proteins can reversibly assemble at the plasma membrane. The specificity of this assembly is determined by the affinity of different SH2 (and other) domains for their cognate peptide ligands.
Figure 14.
Schematic diagram of the interaction of SH2 domains of different signaling proteins with specific phosphotyrosine motifs on PDGF receptor tyrosine kinase. (Adapted from Pawson T, et al. 1993. Philos Trans R Soc Lond B Biol Sci 340: 279–285, with permission from The Royal Society.)
The discovery and detailed understanding of tyrosine kinases has led to the development of a large number of new cancer drugs including small-molecule inhibitors of kinase domains and monoclonal antibodies directed against the extracellular domains of receptor tyrosine kinases. The first major therapeutic successes based on our knowledge of oncogene activation in human cancer, Herceptin and Gleevec, are the fruits of these long labors.
Footnotes
From the forthcoming volume Stalking the Enemy Within: A History of Cancer Research, by Joseph Lipsick
Additional Perspectives on A History of Cancer Research available at www.cshperspectives.org
SUGGESTED READING
*Article is in this collection.
- Brugge JS, Erikson RL. 1977. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature 269: 346–348. [DOI] [PubMed] [Google Scholar]
- Cohen S. 2008. Origins of growth factors: NGF and EGF. J Biol Chem 283: 33793–33797. 10.1074/jbc.X800008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. 2003. Variation on a Src-like theme. Cell 112: 737–740. [DOI] [PubMed] [Google Scholar]
- Hunter T. 2014. The genesis of tyrosine phosphorylation. Cold Spring Harb Perspect Biol 6: a020644 10.1101/cshperspect.a020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. 2015. Discovering the first tyrosine kinase. Proc Natl Acad Sci 112: 7877–7882. 10.1073/pnas.1508223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J, Ferguson KM. 2014. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol 6: a020768 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lipsick J. 2019a. A history of cancer research: tumor viruses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lipsick J. 2019b. A history of cancer research: cell growth and metabolism. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lipsick J. 2019c. A history of cancer research: human proto-oncogenes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lipsick J. 2019d. A history of cancer research: genetically targeted therapies. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JT. 2003. Focal adhesion kinase: the first ten years. J Cell Sci 116: 1409–1416. [DOI] [PubMed] [Google Scholar]
- Pawson T. 2004. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116: 191–203. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. 2014. Receptor tyrosine kinases: legacy of the first two decades. Cold Spring Harb Perspect Biol 6: a008912 10.1101/cshperspect.a008912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MJ, Stacey MM, Liu BA, Pawson T. 2013. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb Perspect Biol 5: a008987 10.1101/cshperspect.a008987. [DOI] [PMC free article] [PubMed] [Google Scholar]