Abstract
Nonsense-mediated mRNA decay (NMD) is arguably the best-studied eukaryotic messenger RNA (mRNA) surveillance pathway, yet fundamental questions concerning the molecular mechanism of target RNA selection remain unsolved. Besides degrading defective mRNAs harboring premature termination codons (PTCs), NMD also targets many mRNAs encoding functional full-length proteins. Thus, NMD impacts on a cell’s transcriptome and is implicated in a range of biological processes that affect a broad spectrum of cellular homeostasis. Here, we focus on the steps involved in the recognition of NMD targets and the activation of NMD. We summarize the accumulating evidence that tightly links NMD to translation termination and we further discuss the recruitment and activation of the mRNA degradation machinery and the regulation of this complex series of events. Finally, we review emerging ideas concerning the mechanistic details of NMD activation and the potential role of NMD as a general surveyor of translation efficacy.
Originally conceived as a quality control pathway that recognizes and specifically degrades aberrant messenger RNAs (mRNAs) with premature termination codons (PTCs), it has meanwhile become clear that nonsense-mediated mRNA decay (NMD) contributes to posttranscriptional gene regulation in a way that goes far beyond quality control. Exploring in which biological contexts NMD-mediated gene regulation plays an important role is a relatively new but rapidly expanding area of research that has been covered in recent reviews (Nasif et al. 2017; Nickless et al. 2017). Here, we summarize our current understanding regarding the molecular mechanism of NMD, with the focus on data obtained from mammalian systems. Despite more than 25 years of research and a wealth of biochemical data characterizing interactions between different NMD factors, their enzymatic functions and posttranslational modifications, the mechanism and criteria for selection of an mRNA for the NMD pathway are still not well understood. The slow progress in deciphering the mechanism of NMD can at least partially be attributed to the lack of a suitable in vitro system that faithfully recapitulates the key steps of NMD. Nevertheless, work from many laboratories during the last few years has provided compelling evidence that NMD is tightly coupled to the process of translation termination. During translation termination, it is decided whether the translated mRNA shall remain intact and serve as a template for additional rounds of translation or whether it shall be degraded by the NMD pathway (He and Jacobson 2015). In a nutshell, the current view is that NMD ensues when ribosomes at nonsense codons (hereafter called termination codon [TC]) fail to terminate correctly. Because of the tight link between NMD and translation termination, we begin this review with a brief overview of eukaryotic translation termination. For a more detailed review of the mechanism of translation termination, see Hellen (2018).
THE MECHANISM OF EUKARYOTIC TRANSLATION TERMINATION AND RIBOSOME RECYCLING
Translation termination is signaled by the presence of one of the three TCs in the A site of the ribosome. Canonical translation termination is marked by three key events: (1) proper recognition of the termination signal, (2) hydrolysis of the terminal peptidyl-tRNA bond and release of the nascent peptide, and (3) dissociation of the ribosome into its 60S and 40S subunits (Fig. 1A) (Dever and Green 2012; Simms et al. 2017). In comparison to the initiation and elongation steps, the step of translation termination is less well studied and accordingly much less is known regarding its molecular mechanism. Instead of cognate aminoacylated (aa) transfer RNAs (tRNAs) that get recruited to the A site of the ribosome during elongation, eukaryotic release factor 1 (eRF)1 binds the A site when it harbors one of the three TCs. On terminating ribosomes, eRF1 is found as a ternary complex with the GTPase eRF3 and GTP. After its recruitment to the A site, GTP hydrolysis by eRF3 stimulates a large conformational change in eRF1 that enhances polypeptide release by engaging the active site of the ribosome. Despite the extended conformational change of the middle and carboxy-terminal parts of eRF1, the amino-terminal part of the protein interacts stably with the TC throughout the process (Alkalaeva et al. 2006; Becker et al. 2012; Eyler et al. 2013; Brown et al. 2015; Shao et al. 2016).
Figure 1.
Schematic illustration of the sequential events taking place in normal translation termination and aberrant translation termination resulting in the activation of nonsense-mediated mRNA decay (NMD). (A) The presence of a termination codon (TC) in the A site of the ribosome provides the signal for translation termination that is marked by recognition of the TC by eukaryotic release factor (eRF)1 and eRF3, a conformational change of eRF1 that facilitates hydrolysis of the terminal peptidyl-transfer RNA (tRNA) bond and the release of the nascent peptide. Subsequently, release of the ribosome from the messenger RNA (mRNA) is facilitated by ABCE1. (B) When translation termination occurs in an unfavorable environment, UPF1 is activated by UPF2 and/or UPF3 that are free in the cytoplasm or positioned nearby on an exon junction complex (EJC). UPF1 activation involves its phosphorylation by SMG1, whose kinase activity is controlled by SMG8 and SMG9. The fate of the stalled ribosomes in the context of NMD activation remains unclear. Phosphorylated UPF1 (indicated by red dots) recruits the NMD-specific SMG6 endonuclease, which cleaves the targeted mRNA nearby the TC and the SMG5/SMG7 heterodimer that stimulates the exonucleolytic degradation of the mRNA by recruiting the CCR4/NOT complex. For clarity, a part of the NMD-activating complex (SMG1, UPF2, and UPF3) is depicted in gray to highlight the factors triggering mRNA degradation in the lower part of the panel.
Following GTP hydrolysis, eRF3 dissociates from the termination complex, allowing for the subsequent interaction of eRF1 with the ABC-type ATPase ABCE1 (Rli1 in yeast), a factor that stimulates the recycling of the ribosome by splitting the ribosomal subunits. ABCE1 contains two nucleotide-binding domains (NBDs) and a unique amino-terminal FeS cluster domain aligned by two diamagnetic [4Fe–4S]2+ clusters. ATP hydrolysis by ABCE1 causes extended conformational changes that provide the mechanical force leading to the dissociation of the 60S from the 40S ribosomal subunit (Pisarev et al. 2010; Becker et al. 2012). Structural studies showed that an initial closure of the NBDs positions the FeS cluster domain toward eRF1, exerting an immediate force that destabilizes the intersubunit interactions (Heuer et al. 2017). Cross-linking and mass spectrometry approaches showed that after ribosomal splitting, ABCE1 remains bound to the translational GTPase-binding site of the small ribosomal subunit, establishing major contacts with the S24e ribosomal protein mainly through NBD1 and FeS cluster domains. Structural data suggest that ABCE1 performs a tweezer-like movement that positions the FeS cluster domain in a cleft between S12 and rRNA on the small subunit (Kiosze-Becker et al. 2016). Additionally, ABCE1 associates with the 43S preinitiation complex (consisting of eIF1A, eIF2, eIF3, and initiator tRNA) but its role in translation initiation remains to be elucidated (Heuer et al. 2017). Biochemical data suggest that ABCE1 also accelerates the rate of the peptide release by eRF1 in an ATP-independent manner, providing a functional link between translation termination and ribosome recycling (Shoemaker and Green 2011).
Most of the information concerning translation termination originates from structural studies and in vitro assays. The isolation of translation termination intermediates can be achieved by using purified components in the presence of nucleotide analogs that trap specific steps (Shao et al. 2016) or mutated release factors (Alkalaeva et al. 2006; Brown et al. 2015). The biochemical dissection of translation termination was facilitated by the establishment of a reconstituted system with mammalian components containing purified ribosomal subunits, translation factors, and aminoacylated tRNAs. This in vitro translation system made possible the stepwise investigation of the configuration of termination complexes by using the toeprinting technique, which allows the identification of ribosomal positions on a reporter mRNA at single-nucleotide resolution (Alkalaeva et al. 2006). However, the establishment of similar approaches by using mammalian cell lysates, as well as studies of translation termination in living cells, remains a challenge in the field. The development of techniques that allow studies in living cells such as ribosome profiling and live cell imaging already provides valuable insight into the dynamics of translation termination (Wang et al. 2016; Shirokikh et al. 2017).
TRANSLATION-COUPLED RNA DEGRADATION PATHWAYS
Apart from synthesizing proteins, translation also acts as a quality-control checkpoint for template mRNAs, newly synthesized proteins, and the translation machinery itself. As discussed above, accurate translation termination occurs in the presence of an in-frame TC and requires the engagement of eRF1, which acts as a TC decoding factor, and of the translational GTPase eRF3 in the A site of the ribosome. For mRNAs lacking a TC (nonstop decay) and for those on which ribosomes stall inside an open reading frame (ORF) caused by strong secondary structures or the lack of a cognate tRNA (no-go decay), the rescue of otherwise trapped ribosomes is essential (see Heck and Wilusz 2018). In such cases, the roles of the decoding factor and the GTPase are performed by the HBS1-like protein (HBS1L; Uniprot Q9Y450) and protein Pelota homolog (PELO; Uniprot Q9BRX2; Dom34 in yeast), respectively. When the HBS1L/PELO complex binds a stalled ribosome, degradation of the corresponding mRNA is induced by an endonucleolytic cleavage. These ribosome rescue mechanisms may occur at any point during the translation cycle and require the recruitment of specific factors into the A site of the ribosome. Unlike no-go and nonstop mRNA decay that depend on specialized termination factors, NMD relies on the canonical termination factors eRF1 and eRF3 (Naeger et al. 1992; Dever and Green 2012; Simms et al. 2017).
NMD IS LINKED TO INEFFICIENT TRANSLATION TERMINATION
That NMD depends on translation is well documented (Carter et al. 1995; Thermann et al. 1998). It was initially suggested that only mRNAs associated with the nuclear cap-binding complex (CBC) were sensitive to NMD, leading to the idea that NMD can target mRNAs only during the first round of translation, the so-called pioneer round of translation (Maquat et al. 2010). However, it was later shown that NMD also targets eIF4E-bound mRNAs (Durand and Lykke-Andersen 2013; Rufener and Mühlemann 2013), consistent with data from Saccharomyces cerevisiae (Maderazo et al. 2003; Gao et al. 2005). This led to a revised, unified NMD model, according to which NMD can be triggered at any aberrant translation termination event during any round of translation (He and Jacobson 2015). This model posits that the likelihood for a ribosome to terminate correctly primarily depends on the availability of termination stimulating factors in the immediate vicinity of the TC. For classical NMD targets with a PTC located in a messenger ribonucleoprotein particle (mRNP) environment lacking such factors needed for proper translation termination, the first ribosome attempting to terminate at this PTC will most likely trigger NMD. However, the probability for a terminating ribosome to activate NMD at many other TCs located in more termination-friendly environments would be smaller and the NMD may therefore ensue only after several ribosomes have already terminated properly. This latter scenario might apply for most endogenous NMD targets and explain their mostly moderate NMD-dependent mRNA level reduction (Colombo et al. 2017).
In contrast to no-go and nonstop mRNA decay, which both use the specialized release factors HBS1L and PELO to free the stalled ribosomal subunits, translation termination on NMD-sensitive mRNAs appears to operate with the common release factors eRF1 and eRF3. What, then, is the difference between a termination event that activates NMD and one that does not? Evidence from S. cerevisiae and rabbit reticulocyte lysate indicates that ribosomes reside longer at the TC of NMD-sensitive compared with NMD-insensitive mRNAs (Amrani et al. 2004; Peixeiro et al. 2011). Based on these data, it was proposed that kinetically slow translation termination caused by the lack of termination-promoting factors may be the signal triggering NMD (reviewed in Kervestin and Jacobson 2012; He and Jacobson 2015). The so-far best-characterized termination-promoting factor is poly(A)-binding protein (PABP). Experiments using a fully reconstituted mammalian in vitro translation termination system showed that under limiting concentrations of release factors, PABP directly stimulates translation termination by enhancing the recruitment of eRF3 and eRF1 to the ribosome in a GTP- and peptide release-independent manner (Ivanov et al. 2016). Evidence from both yeast and mammalian cells suggests that NMD ensues when the termination occurs in an mRNP environment that is spatially distant from the PAPB. Accordingly, NMD can be antagonized when cytoplasmic PABP (Pab1p in yeast, PABPC1 in mammals) is artificially tethered into the vicinity of an otherwise NMD-triggering TC (Amrani et al. 2004; Behm-Ansmant et al. 2007; Eberle et al. 2008; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008) and the efficiency of suppression is correlated with the physical distance between the TC and the tethered PABP. Somewhat unexpectedly, given the reported interaction of eRF1 with the carboxy-terminal domain of PABP, it was further shown that the carboxyl terminus of PABPC1 is dispensable for suppression of NMD, whereas RRMs 1–3 are sufficient and RRMs 1–2 are necessary. Furthermore, abrogation of the PABPC1–eIF4G interaction reduces NMD suppression by PABPC1 and tethering of eIF4G downstream from an NMD-eliciting TC also suppresses NMD (Fatscher et al. 2014; Joncourt et al. 2014). Although the nature of the termination-promoting activity is still unknown, this finding suggests that it might lie in the formation of the circular structure that the mRNP is thought to adopt by eIF4G interacting with both the 5′ cap-bound eIF4E and the 3′ poly(A) tail-bound PABP (Wells et al. 1998).
UPF1, one of the key NMD factors, has been reported to associate with eRF1 and eRF3 (Czaplinski et al. 1998; Wang et al. 2001; Kashima et al. 2006), fueling the hypothesis that UPF1 antagonizes PABPC1 at termination events that trigger NMD (Kervestin and Jacobson 2012). There is evidence from coimmunoprecipitation experiments that UPF1 and PABPC1 compete for association with eRF3 (Singh et al. 2008). However, a very recent study using recombinant proteins in in vitro experiments showed that this interaction is most likely indirect, bridged by UPF3B (Neu-Yilik et al. 2017). In addition, interactions of eIF3 with hyperphosphorylated UPF1 (Isken et al. 2008) and with UPF2 (Morris et al. 2007) have also been detected, but their roles in NMD are not clear.
KEY EVENTS IN NMD ACTIVATION: THE PIVOTAL ROLE OF UPF1
NMD activation relies on the formation of dynamic protein complexes on the target mRNA that eventually lead to the degradation of the mRNA. The first NMD factors were identified by genetic screens in S. cerevisiae (Leeds et al. 1991, 1992) and Caenorhabditis elegans (Pulak and Anderson 1993), and mammalian homologs have been subsequently identified by homology searches (Applequist et al. 1997; Lykke-Andersen et al. 2000; Denning et al. 2001; Serin et al. 2001; Chiu et al. 2003; Reichenbach et al. 2003). More recently, additional screens in C. elegans and human cells have identified several additional proteins that appear to be needed for NMD (Longman et al. 2007; Anastasaki et al. 2011; Alexandrov et al. 2017; Hoque et al. 2017); but, apart from the RNA helicase DHX34 (Longman et al. 2013; Hug and Caceres 2014; Melero et al. 2016), their role in NMD is not yet known. Three of the initially identified NMD factors are conserved from yeast to humans: UPF1, UPF2, and UPF3 (of which mammals have two very similar genes, UPF3A and UPF3B) (Maquat 2004). SMG1, SMG5, SMG6, and SMG7, in contrast, appear to be present only in metazoans (Chen et al. 2008). As outlined below, some of the NMD factors are constituents of more than one of the biochemically characterized protein complexes that form along the pathway of NMD and also have multiple functions in NMD, which complicates the determination of the temporal order of events and ultimately the understanding the molecular mechanism of NMD.
UPF1, an ATP-Dependent RNA Helicase, Is at the Heart of NMD
UPF1 is an ATP-dependent RNA helicase of the helicase superfamily 1 (Fairman-Williams et al. 2010) and a key factor in NMD. UPF1 seems to play a central role in substrate selection and it forms the hub for assembling other NMD factors and recruiting the RNA decay factors. Its helicase activity is tightly regulated and essential for NMD in both yeast and humans (Weng et al. 1998; Franks et al. 2010). ATP binding to the ATP-binding pocket located between the two RecA domains (Rec1A and Rec2A) reduces the affinity of UPF1 for RNA and induces the catalytically active conformation (Bhattacharya et al. 2000; Cheng et al. 2007). Mammalian UPF1 contains three domains that regulate its helicase activity: an amino-terminal domain rich in cysteines and histidines (CH domain); 1B and 1C subdomains embedded within the Rec1A domain; and a low-structured carboxy-terminal SQ-rich domain (Cheng et al. 2007; Chakrabarti et al. 2011; Fiorini et al. 2013). Additionally, UPF1 is regulated by phosphorylation of multiple SQ and TQ motifs located in the amino- and carboxy-terminal parts of the protein and this phosphorylation/dephosphorylation cycle is essential for NMD in metazoans (Yamashita et al. 2001; Ohnishi et al. 2003; Okada-Katsuhata et al. 2012). UPF1 phosphorylation is catalyzed by SMG1, a phosphatidylinositol 3-kinase-related protein kinase (Yamashita 2013).
Is UPF1 Binding Sufficient to Mark mRNAs for Degradation?
The questions of when and where UPF1 binds mRNAs and whether mere binding of UPF1 is sufficient to mark RNAs for degradation have long been investigated and have given rise to contradictory findings. In support of specific UPF1 recruitment to NMD-targeted transcripts, UPF1 has been found significantly enriched on PTC-containing mRNAs compared with PTC-less mRNAs at steady state (Johansson et al. 2007; Johns et al. 2007; Silva et al. 2008; Hwang et al. 2010; Kurosaki et al. 2014; Lee et al. 2015). On the other hand, CLIP-seq and immunoprecipitation experiments in human and mouse cells showed that UPF1 is considerably enriched on the 3′ untranslated region (UTR) of mRNAs independently of whether or not they are targeted by NMD (Hurt et al. 2013; Zund et al. 2013), suggesting that UPF1 binding to RNA is not the discriminatory step in NMD target identification. Furthermore, it appears that UPF1 associates with mRNA before translation starts at seemingly random positions on the RNA and that translating ribosomes subsequently displace UPF1 from the ORF, resulting in the majority of UPF1 being detected on 3′UTRs at steady state (Fig. 2) (Hogg and Goff 2010; Hurt et al. 2013; Zund et al. 2013). Based on the available data, it is possible that translation affects UPF1-RNA association in two independent ways: (1) translation removes UPF1 from the CDS; and (2) UPF1 gets recruited by release factors (Ivanov et al. 2008) and in yeast cells also by ribosomal proteins (Min et al. 2013) to the 3′UTR of NMD targets. The ATPase activity of UPF1 was shown to be required for releasing UPF1 from nontarget mRNAs (Lee et al. 2015), and there is evidence that UPF1 phosphorylation occurs specifically on NMD targets (Kurosaki et al. 2014). Thus, a plausible scenario is that UPF1 initially binds all mRNAs and that improper/delayed translation termination triggers phosphorylation of the remaining RNA-bound UPF1, eventually activating NMD, whereas ATP hydrolysis results in the release of UPF1 from mRNAs that undergo proper translation termination. In this working model, UPF1 functions as a time bomb that destroys the mRNA if not removed quickly enough, an idea that was postulated some time ago (Hilleren and Parker 1999).
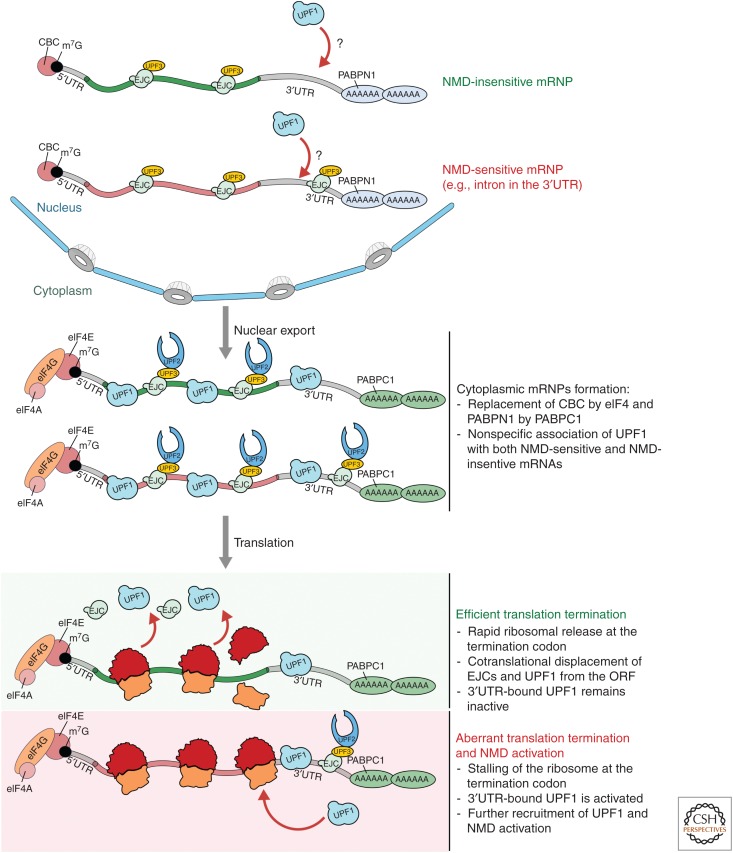
Figure 2.
Distribution and molecular architecture of factors that affect nonsense-mediated mRNA decay (NMD) on NMD-sensitive and NMD-insensitive messenger RNAs (mRNAs) at various stages. NMD-insensitive (green) and NMD-sensitive (red) messenger ribonucleoprotein particles (mRNPs) are loaded with UPF3 bound to exon junction complexes (EJCs) before export to the cytoplasm, and it is possible (although not known) that some UPF1 also associates with mRNA in the nucleus. In the cytoplasm, UPF1 is bound to all mRNAs independent of their sensitivity to NMD. UPF2 interacts with UPF3 and the nuclear cap-binding complex (CBC) and the nuclear poly(A)-binding protein (PABPN)1 are replaced by their cytoplasmic counterparts eIF4F (composed of eIF4E, G, and A) and PABPC1, respectively. During translation, UPF1 and EJCs are displaced from the coding regions of the mRNA by elongating ribosomes, while they remain associated on the 3′ untranslated region (UTR). In the context of normal translation termination, ribosome release is very efficient, and the corresponding mRNAs persist. When translation termination is not efficient, however, NMD ensues by UPF1 activation and/or recruitment of additional UPF1 molecules to the site of termination. The presence of an EJC downstream from the termination codon (TC) increases the concentration of UPF2 and UPF3 in the vicinity of UPF1, which is thought to facilitate the activation of NMD.
SMG1 Complex Regulates UPF1 Phosphorylation
In mammalian cells, the SMG1-mediated UPF1 phosphorylation is essential for NMD (Yamashita et al. 2001; Okada-Katsuhata et al. 2012) and appears to occur in a translation-dependent manner (Yamashita et al. 2001, 2009; Yamashita 2013). At steady state, most UPF1 of an immortalized cell is hypophosphorylated (Chiu et al. 2003; Okada-Katsuhata et al. 2012). The kinase activity of SMG1 is regulated by SMG8 and SMG9, two proteins forming a complex with SMG1 that keeps SMG1 in a kinase-inactive conformation (Yamashita et al. 2009; Arias-Palomo et al. 2011; Fernandez et al. 2011; Melero et al. 2014). SMG8 inhibits SMG1 activity in vitro but enhances UPF1 phosphorylation in vivo (Yamashita et al. 2009), indicating that in addition to regulating the kinase activity of SMG1, SMG8 also functions in recruiting the SMG1 complex to UPF1. UPF1 can be coimmunoprecipitated from human cells in a complex with SMG1, eRF1, and eRF3 (the so-called SURF complex [Kashima et al. 2006]). These interactions support the view that UPF1 phosphorylation occurs in the context of translation termination (Fig. 1B) (Kashima et al. 2006; Kurosaki et al. 2014). UPF2 (see below) and DHX34 (Hug and Caceres 2014) contribute to the activation of SMG1-catalyzed UPF1 phosphorylation.
UPF2 and UPF3 Are Involved in NMD Activation
UPF2 is conserved in eukaryotes and functions as a ring-like adaptor protein between UPF1 and UPF3 (Serin et al. 2001; Chamieh et al. 2008; Melero et al. 2012). It consists of three MIF4G (middle fragment of eIF4G) domains, with the first two domains providing structural support (Clerici et al. 2009) and the third domain interacting with UPF3B (Kadlec et al. 2004). UPF2’s interaction with the CH domain of UPF1 induces a large conformational change of this domain that triggers the helicase activity of UPF1 by switching it from an RNA-clamping to an RNA-unwinding mode (Fig. 1B) (Chakrabarti et al. 2011). In addition to its interaction with UPF1 and UPF3B, UPF2 has been reported to bind to the DEAD-box helicase Dbp6 in yeast (DDX51 in Homo sapiens) (Fourati et al. 2014) and to SMG1 and ribosomal proteins of both subunits (Lopez-Perrote et al. 2016). UPF2 also directly interacts with eRF3 through a carboxy-terminal region that is essential for UPF3B binding but does not interfere with UPF1 binding (Lopez-Perrote et al. 2016). Furthermore, UPF2 forms a complex with SURF, allowing its direct association to ribosomes in an exon junction complex (EJC)-independent manner (Lopez-Perrote et al. 2016). The fact that UPF2 appears to associate with some NMD-related components simultaneously and with others in a mutually exclusive way implies an important role for UPF2 in orchestrating the order of events leading to NMD activation (Lopez-Perrote et al. 2016). Somewhat surprisingly, given its central role in NMD, degradation of a subset of NMD targets has been reported to occur independently of UPF2 (Gehring et al. 2005).
UPF3 is the least conserved of the three core NMD factors (Culbertson and Leeds 2003). At steady state, it predominately resides in the nucleus (Serin et al. 2001), but it is thought to shuttle with the mRNP to the cytoplasm in which NMD takes place (Trcek et al. 2013). Rather than binding RNA, the conserved amino-terminal putative RNA recognition motif (RRM) appears to interact with UPF2 (Kadlec et al. 2004). In addition, the three EJC core factors eIF4A3, MAGOH, and Y14 interact through a composite binding site with a short motif located at the carboxyl terminus of UPF3 (Chamieh et al. 2008). Thus, UPF3 and UPF2 bridge UPF1 to the EJC, providing a molecular link between NMD and the EJC (Chamieh et al. 2008). UPF3 and UPF2 together were also shown to stimulate the helicase activity of UPF1 (Chamieh et al. 2008). Mammals contain two UPF3 paralogs, UPF3A and UPF3B (UPF3B is located on the X chromosome and is also called UPF3X). UPF3A and UPF3B are similar in their amino acid sequence and both associate with other NMD factors but UPF3A is less efficient in destabilizing NMD reporter mRNAs in tethering assays (Lykke-Andersen et al. 2000; Kunz et al. 2006; Chan et al. 2009; Hug and Caceres 2014). In mice, the conditional knockout of UPF3A even enhanced the NMD response of a fraction of endogenous NMD targets, suggesting that UPF3A may function as an antagonist of UPF3B by sequestering UPF2 from the NMD machinery (Shum et al. 2016).
Degradation of NMD-Targeted mRNAs
As mentioned above, UPF1 phosphorylation seems to be a prerequisite for the direct or indirect recruitment of nucleases that degrade the targeted mRNA (Lejeune et al. 2003; Isken et al. 2008). Endonucleolytic cleavage in the vicinity of the NMD-eliciting TC is mediated by SMG6 and the SMG5/SMG7 heterodimer recruits exonucleolytic decay activities (Mühlemann and Lykke-Andersen 2010). SMG6 is a metazoan-specific NMD factor that confers its endonuclease activity through a triad of catalytically active Asp residues in its carboxy-terminal PIN domain (Huntzinger et al. 2008; Eberle et al. 2009). In immortalized human cells (HeLa and HEK-293), the bulk of NMD targets appears to be degraded by SMG6 (Boehm et al. 2014; Lykke-Andersen et al. 2014; Schmidt et al. 2014). The unprotected RNA ends generated by the endonucleolytic cleavage are then subjected to exonucleolytic degradation by the 5′–3′exonuclease XRN1 and by 3′–5′ endonucleases (Gatfield and Izaurralde 2004; Huntzinger et al. 2008; Eberle et al. 2009; Franks et al. 2010). Notably, SMG6 interacts with phosphorylated T28 of UPF1 (Okada-Katsuhata et al. 2012) and additionally in a phosphorylation-independent manner with the unique stalk protruding from the Rec1A domain of UPF1 (Chakrabarti et al. 2014; Nicholson et al. 2014). In tethering experiments, SMG6 requires UPF1 and SMG1 to degrade a reporter transcript, implying that the SMG6 endonuclease might adopt its active conformation only when bound to phosphorylated UPF1 (Nicholson et al. 2014).
SMG5 and SMG7 form a highly stable heterodimer (Jonas et al. 2013) that binds to phosphorylated S1096 of UPF1 in vivo (Ohnishi et al. 2003; Okada-Katsuhata et al. 2012) and triggers RNA degradation when tethered to reporter transcripts (Unterholzner and Izaurralde 2004; Cho et al. 2013). This activity requires the carboxy-terminal proline-rich region of SMG7, which was shown to interact with CNOT8 (POP2), the catalytic subunit of the CCR4-NOT deadenylase complex (Loh et al. 2013). Tethered SMG7 has further been reported to induce reporter transcript decay by DCP2-mediated decapping and XRN1-mediated 5′-to-3′ exonucleolytic degradation (Unterholzner and Izaurralde 2004). These factors are presumably recruited indirectly via the CCR4-NOT complex. There is ample evidence that SMG5/SMG7 and SMG6 RNA degradation pathways act in, at least partially, a redundant way. Codepletion of SMG6 with SMG5 or SMG7 inhibits NMD much more effectively than depletion of each of the factors alone (Luke et al. 2007; Jonas et al. 2013; Metze et al. 2013; Colombo et al. 2017). A transcriptome-wide approach revealed that SMG6 and SMG7 target essentially the same sets of transcripts (Colombo et al. 2017), even though individual mRNAs may have preferences for either SMG6- or SMG7-mediated decay (Ottens et al. 2017).
Human UPF1 also interacts with mRNA decay factors independently of SMG5/SMG7, namely, with the decapping factors DCP1, DCP2, and PNRC2 (Lykke-Andersen 2002; Lejeune et al. 2003; Fenger-Gron et al. 2005; Isken et al. 2008; Cho et al. 2009; Lai et al. 2012). PNRC2 binds directly to DCP1 and UPF1 (Lai et al. 2012; Cho et al. 2013). The published data regarding whether or not SMG5 interacts with PNRC2 and is involved in the PNRC2-dependent decay pathway are contradictory and further investigation is needed (Cho et al. 2013; Loh et al. 2013). It is noteworthy that many NMD factors colocalize with decapping and 5′–3′ exonucleolytic degradation components in processing bodies (P-bodies) (Fukuhara et al. 2005; Durand et al. 2007; Franks et al. 2010) but the formation of detectable P-bodies in cells is not required for NMD (Stalder and Mühlemann 2009).
NMD-INDUCING FEATURES
As outlined above, the current model posits that NMD is stimulated when the TC occurs in a microenvironment of the mRNP that is unfavorable for translation termination. Assuming that aberrant mRNAs with PTCs are degraded by the same mechanism as endogenous NMD targets with full-length ORFs, one would expect to find common features that render mRNAs susceptible to NMD. Transcriptome-wide approaches to identify NMD targets in cells of different species revealed that the majority of NMD-sensitive transcripts do not contain PTCs but are ordinary mRNAs coding for seemingly full-length functional proteins (Rehwinkel et al. 2006; Lykke-Andersen and Jensen 2015; Colombo et al. 2017). Despite the fact that in most of these studies it was not possible to distinguish between direct and indirect targets, there is ample evidence that NMD can target both normal and erroneous transcripts. Among the NMD-inducing features, the presence of the 3′-most exon–exon junction >50 nt downstream from the TC is the feature with the strongest predictive value for NMD susceptibility (Colombo et al. 2017). PTCs resulting from mutations in the ORF or from aberrant or alternative splicing, as well as genes with an intron in the 3′UTR mostly belong to this class of NMD targets. In addition, long 3′UTRs (>1000 nt in mammalian cells) (Buhler et al. 2006), the presence of actively translated short upstream ORFs (uORFs) (Hurt et al. 2013), or selenocysteine codons (UGA) in cells grown in the absence of selenium (Moriarty et al. 1998) are also features that can—but not always do—trigger NMD (Fig. 3). uORF translation often inhibits translation of the main ORF, either constitutively or in response to stress (Young and Wek 2016). Under such circumstances, ribosomes terminate at the TC of the uORF with usually several EJCs remaining bound further downstream on the mRNA, which creates an NMD-promoting translation termination environment. Despite all these empirically determined features, it has so far remained impossible to computationally predict NMD targets with high confidence. Based on the aforementioned ability of PABP to suppress NMD, it has been suggested that the physical distance between the TC and the poly(A) tail might be the crucial feature determining NMD sensitivity (Stalder and Mühlemann 2008). Because such information about the 3D architecture of mRNPs is currently not available, this TC-to-poly(A) tail distance is virtually impossible to measure and predict.
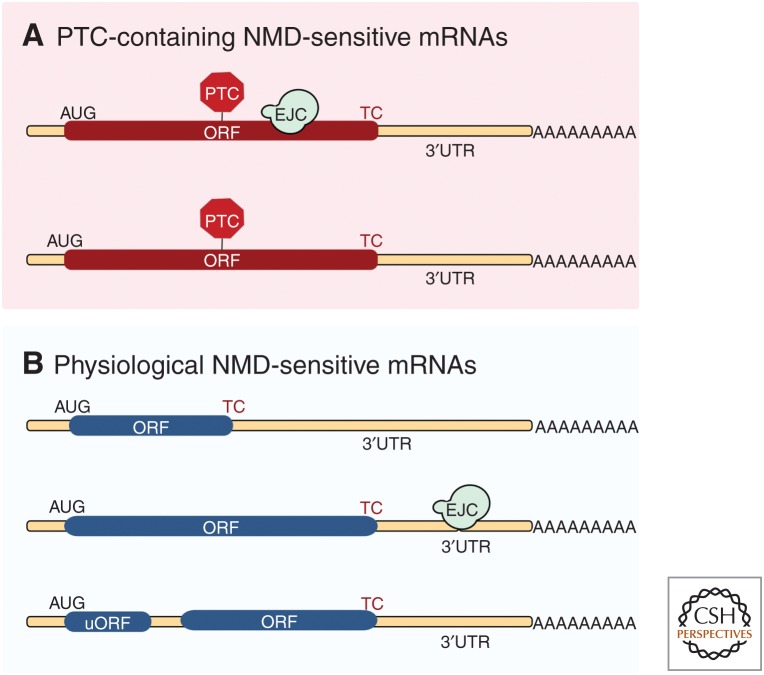
Figure 3.
Features that can render aberrant and physiological mRNAs sensitive to nonsense-mediated mRNA decay (NMD). (A) Messenger RNAs (mRNAs) in which the open reading frame (ORF) is truncated caused by the presence of a premature termination codon (PTC) are typically sensitive to NMD. The presence of one or several exon junction complexes (EJCs) downstream from the PTC is an important NMD-enhancing feature. (B) In mRNAs with an intact ORF coding for a full-length protein, long 3′ untranslated regions (UTRs), EJCs located in the 3′UTR, or short upstream ORFs (uORFs) can also trigger NMD.
The EJC as an NMD Enhancer
EJCs are multiprotein complexes organized around a central core of four proteins: eIF4A3, MLN51, and MAGOH/Y14, a heterodimer that interacts with UPF3B (Buchwald et al. 2010). The EJC core binds tightly to RNA without apparent sequence specificity, generally ∼24 nt upstream of spliced exon–exon junctions during the course of splicing (Le Hir et al. 2000a,b). However, a transcriptome-wide mapping of eIF4A3 in human cells indicated that not all splicing events result in the deposition of an EJC and a significant fraction of eIF4A3 was detected on mRNA in noncanonical positions (Sauliere et al. 2012; Singh et al. 2012). EJCs are deposited on mRNA in the nucleus during splicing and accompany the mature mRNAs to the cytoplasm, in which those EJCs located in ORFs are thought to be removed by the first ribosome translating the mRNA (Fig. 2). EJCs located in the 3′UTR of an mRNP >30 nt downstream from the TC in contrast will remain bound to the mRNA (Le Hir et al. 2016). The presence of EJCs in mRNPs was shown to activate translation mediated by an interaction of MLN51 with eIF3 subunits a and d (Chazal et al. 2013). Those EJCs that remain bound to the mRNA after translation has begun (i.e., those bound >30 nt downstream of the TC) strongly promote the activation of NMD on the corresponding mRNAs (Buhler et al. 2006; Metze et al. 2013) by recruiting NMD factors UPF3, UPF2, and/or SMG6 to the mRNP (Le Hir et al. 2001; Gehring et al. 2005; Kashima et al. 2010). A similar NMD-promoting effect was observed by tethering EJC components to the 3′UTR of reporter transcripts (Lykke-Andersen et al. 2000; Gehring et al. 2003; Palacios et al. 2004). The presence of EJCs in 3′UTRs was reported to enhance the efficiency of SMG6-mediated endonucleolysis without altering the cleavage site (Boehm et al. 2014). This enhanced SMG6-mediated NMD activity can be attributed to the interaction of SMG6 with the EJC via two conserved EJC-binding motifs (EBMs) in the amino-terminal portion of SMG6 (Kashima et al. 2010). Interestingly, the SMG6 EBMs interact with the same surface of the EJC as UPF3B (Kashima et al. 2010), suggesting that these interactions are mutually exclusive and may therefore occur sequentially.
Long 3′UTRs
That mRNAs with a long 3′UTR can be targeted by NMD has been observed in yeast, plants, and animals (Pulak and Anderson 1993; Muhlrad and Parker 1999; Buhler et al. 2006; Kertesz et al. 2006; Eberle et al. 2008; Singh et al. 2008; Hogg and Goff 2010; Hurt et al. 2013; Boehm et al. 2014; Colombo et al. 2017) and, accordingly, insertion of long 3′UTRs into reporter constructs can render them NMD-sensitive (Huang et al. 2011; Yepiskoposyan et al. 2011). Because the physical distance between TC and PABP strongly affects the half-life of NMD reporter transcripts (Eberle et al. 2008), it is very likely that those long 3′UTRs that render mRNAs sensitive to NMD adopt an extended 3′UTR structure in which the poly(A) tail is too far away from the TC to promote proper translation termination. Consistent with this idea, intramolecular secondary structures that bring a distal poly(A) tail into close proximity with an otherwise NMD-triggering TC effectively antagonize NMD, even in the context of EJC-enhanced NMD (Eberle et al. 2008). Although the NMD-suppressing effect of PABP is well documented (Amrani et al. 2004; Behm-Ansmant et al. 2007; Eberle et al. 2008; Ivanov et al. 2008; Silva et al. 2008; Singh et al. 2008), the underlying mechanism remains elusive. Interestingly, several endogenous mRNAs with long 3′UTRs were found to be immune to NMD (Weil and Beemon 2006; Hogg and Goff 2010; Yepiskoposyan et al. 2011; Hurt et al. 2013). A subset of these NMD-insensitive mRNAs with long 3′UTRs contains AU-rich elements within the first 200 nt downstream from the TC, which were shown to be necessary and sufficient to inhibit NMD (Toma et al. 2015). However, the mechanism by which this NMD protection is conferred is not known. Another well-studied example of an NMD-resistant transcript with a long 3′UTR is the unspliced viral RNA of Rous sarcoma virus (RSV). RSV harbors a 400-nt RNA stability element (RSE) located immediately downstream from the gag TC, which is required to confer NMD immunity on the viral RNA (Weil and Beemon 2006). It has been found that the polypyrimidine tract-binding protein 1 (PTBP1) binds to the RSE and thereby blocks UPF1 association with this region (Ge et al. 2016). Insertion of PTBP1-binding sites into NMD reporter constructs downstream of the TC rendered these transcripts NMD-insensitive (Ge et al. 2016).
OPEN QUESTIONS
Despite the fact that NMD has been an area of intensive research for the past two decades, many points still remain unclear. A long-standing open question is whether all eukaryotes have NMD. Trypanosomes have homologs of UPF1 and UPF2 but UPF1 depletion showed no effect on the RNA level of genes that were predicted NMD targets, leading the investigators to conclude that a classical NMD pathway might be absent in Trypanosoma brucei (Delhi et al. 2011). In contrast, NMD appears to function in Giardia lamblia, another parasitic protist, despite the lack of recognizable homologs of UPF2 and UPF3 (Chen et al. 2008). Although UPF3 seems to be absent from most protists, a UPF3 homolog has been found in Paramecium tetraurelia and it was shown to play a role in NMD (Contreras et al. 2014). Interestingly, however, this UPF3 variant lacks the motifs required for interacting with the EJC, indicating that in these early-branched eukaryotes, NMD might function independently of EJCs, similar to S. cerevisiae, which lacks EJCs altogether. Consistently, NMD in Tetrahymena thermophila has recently been shown to be EJC-independent (Tian et al. 2017). In addition, this study identified a novel protozoa-specific nuclease that is responsible for degrading many NMD targets, a Smg6-like bacterial YacP nuclease (NYN) domain-containing protein (Smg6L). These recent findings corroborate the view that NMD in different species can occur by several different RNA decay pathways and that the evolutionarily conserved part may be confined to the substrate RNA selection, in which UPF1 plays a key function.
Recent ribosome profiling data in S. cerevisiae revealed that NMD-targeted mRNAs that lack PTCs according to their annotated sequences tend to show elevated rates of out-of-frame translation (Celik et al. 2017a). Such out-of-frame translation would lead to premature translation termination, which could explain why these mRNAs are NMD targets. It has therefore been proposed that NMD should be considered as a probabilistic quality-control pathway that continually detects errors throughout the translation process (Celik et al. 2017b). This is an interesting novel concept that needs to be further investigated.
Apart from understanding the mechanistic details that govern NMD activation in the context of inefficient translation termination, it would also be interesting to assess whether interconnections between NMD and other translation-dependent mRNA degradation mechanisms exist. Preliminary evidence supports the notion of such cross talk at least with regard to the mRNA degradation machinery involved. When endonucleolytic cleavage occurs upstream of a PTC, the resulting 5′RNA fragment was stabilized on depletion of nonstop decay components Pelota and Hbs1 in Drosophila cells (Hashimoto et al. 2017). Along the same lines, it was shown in C. elegans that the degradation of PTC-containing mRNAs as well as endogenous NMD targets is coupled to nonstop decay (Arribere and Fire 2018). This implies that the same mRNA could be targeted by more than one translation-dependent RNA surveillance pathway, allowing for more efficient RNA degradation.
It is evident from this review that many fundamental questions regarding the NMD phenomenon are still unsolved. Notably, it is currently not even clear whether what is commonly described as NMD constitutes one defined biochemical pathway or maybe several different pathways with requirements for an overlapping set of proteins. Thus, we are still far away from a comprehensive understanding of the biological functions of NMD, its underpinning molecular mechanism(s), and the evolutionary forces that led to the emergence of NMD.
ACKNOWLEDGMENTS
The research in the laboratory of O.M. is supported by the National Center of Competence in Research (NCCR) RNA & Disease funded by the Swiss National Science Foundation (SNSF), by the SNSF Grant 31003A-162986, and by the canton of Bern.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alexandrov A, Shu MD, Steitz JA. 2017. Fluorescence amplification method for forward genetic discovery of factors in human mRNA degradation. Mol Cell 65: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136. [DOI] [PubMed] [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118. [DOI] [PubMed] [Google Scholar]
- Anastasaki C, Longman D, Capper A, Patton EE, Caceres JF. 2011. Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res 39: 3686–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist SE, Selg M, Raman C, Jack HM. 1997. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res 25: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias-Palomo E, Yamashita A, Fernandez IS, Nunez-Ramirez R, Bamba Y, Izumi N, Ohno S, Llorca O. 2011. The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev 25: 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Fire AZ. 2018. Nonsense mRNA suppression via nonstop decay. eLife 7: e33292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. 2007. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 26: 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm V, Haberman N, Ottens F, Ule J, Gehring NH. 2014. 3′UTR length and messenger ribonucleoprotein composition determine endocleavage efficiencies at termination codons. Cell Rep 9: 555–568. [DOI] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. 2015. Structural basis for stop codon recognition in eukaryotes. Nature 524: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E. 2010. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc Natl Acad Sci 107: 10050–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. 2006. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′UTR length. Nat Struct Mol Biol 13: 462–464. [DOI] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 270: 28995–29003. [DOI] [PubMed] [Google Scholar]
- Celik A, Baker R, He F, Jacobson A. 2017a. High-resolution profiling of NMD targets in yeast reveals translational fidelity as a basis for substrate selection. RNA 23: 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik A, He F, Jacobson A. 2017b. NMD monitors translational fidelity 24/7. Curr Genet 63: 1007–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41: 693–703. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Bonneau F, Schussler S, Eppinger E, Conti E. 2014. Phospho-dependent and phospho-independent interactions of the helicase UPF1 with the NMD factors SMG5-SMG7 and SMG6. Nucleic Acids Res 42: 9447–9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh H, Ballut L, Bonneau F, Le Hir H. 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 15: 85–93. [DOI] [PubMed] [Google Scholar]
- Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. 2009. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol 16: 747–753. [DOI] [PubMed] [Google Scholar]
- Chazal PE, Daguenet E, Wendling C, Ulryck N, Tomasetto C, Sargueil B, Le Hir H. 2013. EJC core component MLN51 interacts with eIF3 and activates translation. Proc Natl Acad Sci 110: 5903–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Su LH, Sun CH. 2008. Incomplete nonsense-mediated mRNA decay in Giardia lamblia. Int J Parasitol 38: 1305–1317. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. 2007. Structural and functional insights into the human Upf1 helicase core. EMBO J 26: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE. 2003. Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA 9: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim KM, Kim YK. 2009. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell 33: 75–86. [DOI] [PubMed] [Google Scholar]
- Cho H, Han S, Choe J, Park SG, Choi SS, Kim YK. 2013. SMG5-PNRC2 is functionally dominant compared with SMG5-SMG7 in mammalian nonsense-mediated mRNA decay. Nucleic Acids Res 41: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. 2009. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J 28: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Karousis ED, Bourquin J, Bruggmann R, Mühlemann O. 2017. Transcriptome-wide identification of NMD-targeted human mRNAs reveals extensive redundancy between SMG6- and SMG7-mediated degradation pathways. RNA 23: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J, Begley V, Macias S, Villalobo E. 2014. An UPF3-based nonsense-mediated decay in Paramecium. Res Microbiol 165: 841–846. [DOI] [PubMed] [Google Scholar]
- Culbertson MR, Leeds PF. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev 13: 207–214. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhi P, Queiroz R, Inchaustegui D, Carrington M, Clayton C. 2011. Is there a classical nonsense-mediated decay pathway in trypanosomes? PLoS ONE 6: e25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. 2001. Cloning of a novel phosphatidylinositol kinase-related kinase: Characterization of the human SMG-1 RNA surveillance protein. J Biol Chem 276: 22709–22714. [DOI] [PubMed] [Google Scholar]
- Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4: a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Lykke-Andersen J. 2013. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol 20: 702–709. [DOI] [PubMed] [Google Scholar]
- Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F. 2007. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol 178: 1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Stalder L, Mathys H, Orozco RZ, Mühlemann O. 2008. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol 6: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55. [DOI] [PubMed] [Google Scholar]
- Eyler DE, Wehner KA, Green R. 2013. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem 288: 29530–29538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. 2010. SF1 and SF2 helicases: Family matters. Curr Opin Struct Biol 20: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatscher T, Boehm V, Weiche B, Gehring NH. 2014. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA 20: 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915. [DOI] [PubMed] [Google Scholar]
- Fernandez IS, Yamashita A, Arias-Palomo E, Bamba Y, Bartolome RA, Canales MA, Teixido J, Ohno S, Llorca O. 2011. Characterization of SMG-9, an essential component of the nonsense-mediated mRNA decay SMG1C complex. Nucleic Acids Res 39: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini F, Boudvillain M, Le Hir H. 2013. Tight intramolecular regulation of the human Upf1 helicase by its N- and C-terminal domains. Nucleic Acids Res 41: 2404–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourati Z, Roy B, Millan C, Coureux PD, Kervestin S, van Tilbeurgh H, He F, Uson I, Jacobson A, Graille M. 2014. A highly conserved region essential for NMD in the Upf2 N-terminal domain. J Mol Biol 426: 3689–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 143: 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. 2005. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547. [DOI] [PubMed] [Google Scholar]
- Gao Q, Das B, Sherman F, Maquat LE. 2005. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc Natl Acad Sci 102: 4258–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. 2004. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429: 575–578. [DOI] [PubMed] [Google Scholar]
- Ge Z, Quek BL, Beemon KL, Hogg JR. 2016. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. eLife 5: e11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. 2003. Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. 2005. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell 20: 65–75. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Takahashi M, Sakota E, Nakamura Y. 2017. Nonstop-mRNA decay machinery is involved in the clearance of mRNA 5′-fragments produced by RNAi and NMD in Drosophila melanogaster cells. Biochem Biophys Res Commun 484: 1–7. [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. 2015. Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu Rev Genet 49: 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Heck AM, Wilusz J. 2018. The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kotter P, Berninghausen O, Becker T, Beckmann R, Tampe R. 2017. Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat Struct Mol Biol 24: 453–460. [DOI] [PubMed] [Google Scholar]
- Hilleren P, Parker R. 1999. mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA 5: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. 2010. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell 143: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Park JY, Chang YJ, Luchessi AD, Cambiaghi TD, Shamanna R, Hanauske-Abel HM, Holland B, Pe'ery T, Tian B, et al. 2017. Regulation of gene expression by translation factor eIF5A: Hypusine-modified eIF5A enhances nonsense-mediated mRNA decay in human cells. Translation (Austin) 5: e1366294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF. 2011. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell 43: 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N, Caceres JF. 2014. The RNA helicase DHX34 activates NMD by promoting a transition from the surveillance to the decay-inducing complex. Cell Rep 8: 1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. 2010. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell 39: 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. 2008. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Mikhailova T, Eliseev B, Yeramala L, Sokolova E, Susorov D, Shuvalov A, Schaffitzel C, Alkalaeva E. 2016. PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res 44: 7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, He F, Spatrick P, Li C, Jacobson A. 2007. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci 104: 20872–20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. 2007. Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons. Mol Cell Biol 27: 5630–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Weichenrieder O, Izaurralde E. 2013. An unusual arrangement of two 14-3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev 27: 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncourt R, Eberle AB, Rufener SC, Mühlemann O. 2014. Eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay by two genetically separable mechanisms. PLoS ONE 9: e104391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11: 330–337. [DOI] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. 2006. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. 2010. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev 24: 2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D. 2006. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 34: 6147–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S, Jacobson A. 2012. NMD: A multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiosze-Becker K, Ori A, Gerovac M, Heuer A, Nurenberg-Goloub E, Rashid UJ, Becker T, Beckmann R, Beck M, Tampe R. 2016. Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry. Nat Commun 7: 13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. 2006. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA 12: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE. 2014. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 28: 1900–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Cho H, Liu Z, Bowler MW, Piao S, Parker R, Kim YK, Song H. 2012. Structural basis of the PNRC2-mediated link between mRNA surveillance and decapping. Structure 20: 2025–2037. [DOI] [PubMed] [Google Scholar]
- Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke-Andersen J. 2015. Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol Cell 59: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5: 2303–2314. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, Culbertson MR. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 12: 2165–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. 2000a. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. 2000b. Pre-mRNA splicing alters mRNP composition: Evidence for stable association of proteins at exon–exon junctions. Genes Dev 14: 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Sauliere J, Wang Z. 2016. The exon junction complex as a node of post-transcriptional networks. Nat Rev Mol Cell Biol 17: 41–54. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell 12: 675–687. [DOI] [PubMed] [Google Scholar]
- Loh B, Jonas S, Izaurralde E. 2013. The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev 27: 2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF. 2007. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev 21: 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D, Hug N, Keith M, Anastasaki C, Patton EE, Grimes G, Caceres JF. 2013. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res 41: 8319–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Perrote A, Castano R, Melero R, Zamarro T, Kurosawa H, Ohnishi T, Uchiyama A, Aoyagi K, Buchwald G, Kataoka N, et al. 2016. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res 44: 1909–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B, Azzalin CM, Hug N, Deplazes A, Peter M, Lingner J. 2007. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res 35: 7688–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. 2015. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 16: 665–677. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. 2000. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Chen Y, Ardal BR, Lilje B, Waage J, Sandelin A, Jensen TH. 2014. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev 28: 2498–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo AB, Belk JP, He F, Jacobson A. 2003. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol Cell Biol 23: 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. 2004. Nonsense-mediated mRNA decay: A comparative analysis of different species. Current Genomics 5: 175–190. [Google Scholar]
- Maquat LE, Tarn WY, Isken O. 2010. The pioneer round of translation: Features and functions. Cell 142: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero R, Buchwald G, Castano R, Raabe M, Gil D, Lazaro M, Urlaub H, Conti E, Llorca O. 2012. The cryo-EM structure of the UPF–EJC complex shows UPF1 poised toward the RNA 3′ end. Nat Struct Mol Biol 19: 498–505. [DOI] [PubMed] [Google Scholar]
- Melero R, Uchiyama A, Castano R, Kataoka N, Kurosawa H, Ohno S, Yamashita A, Llorca O. 2014. Structures of SMG1–UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure 22: 1105–1119. [DOI] [PubMed] [Google Scholar]
- Melero R, Hug N, Lopez-Perrote A, Yamashita A, Caceres JF, Llorca O. 2016. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat Commun 7: 10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metze S, Herzog VA, Ruepp MD, Mühlemann O. 2013. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 19: 1432–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min EE, Roy B, Amrani N, He F, Jacobson A. 2013. Yeast Upf1 CH domain interacts with Rps26 of the 40S ribosomal subunit. RNA 19: 1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE. 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol 18: 2932–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C, Wittmann J, Jack HM, Jalinot P. 2007. Human INT6/eIF3e is required for nonsense-mediated mRNA decay. EMBO Rep 8: 596–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann O, Lykke-Andersen J. 2010. How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol 7: 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. 1999. Aberrant mRNAs with extended 3′UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5: 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeger LK, Schoborg RV, Zhao Q, Tullis GE, Pintel DJ. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev 6: 1107–1119. [DOI] [PubMed] [Google Scholar]
- Nasif S, Contu L, Mühlemann O. 2017. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin Cell Dev Biol 75: 78–87. [DOI] [PubMed] [Google Scholar]
- Neu-Yilik G, Raimondeau E, Eliseev B, Yeramala L, Amthor B, Deniaud A, Huard K, Kerschgens K, Hentze MW, Schaffitzel C, et al. 2017. Dual function of UPF3B in early and late translation termination. EMBO J 36: 2968–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Josi C, Kurosawa H, Yamashita A, Mühlemann O. 2014. A novel phosphorylation-independent interaction between SMG6 and UPF1 is essential for human NMD. Nucleic Acids Res 42: 9217–9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickless A, Bailis JM, You Z. 2017. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. 2003. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell 12: 1187–1200. [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. 2012. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res 40: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottens F, Boehm V, Sibley CR, Ule J, Gehring NH. 2017. Transcript-specific characteristics determine the contribution of endo- and exonucleolytic decay pathways during the degradation of nonsense-mediated decay substrates. RNA 23: 1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E. 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757. [DOI] [PubMed] [Google Scholar]
- Peixeiro I, Inacio A, Barbosa C, Silva AL, Liebhaber SA, Romao L. 2011. Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res 40: 1160–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, Anderson P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 7: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Raes J, Izaurralde E. 2006. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem Sci 31: 639–646. [DOI] [PubMed] [Google Scholar]
- Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. 2003. A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574. [DOI] [PubMed] [Google Scholar]
- Rufener SC, Mühlemann O. 2013. eIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 20: 710–717. [DOI] [PubMed] [Google Scholar]
- Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonqueze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H. 2012. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol 19: 1124–1131. [DOI] [PubMed] [Google Scholar]
- Schmidt SA, Foley PL, Jeong DH, Rymarquis LA, Doyle F, Tenenbaum SA, Belasco JG, Green PJ. 2014. Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res 43: 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. 2001. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol 21: 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Murray J, Brown A, Taunton J, Ramakrishnan V, Hegde RS. 2016. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell 167: 1229–1240.e1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh NE, Archer SK, Beilharz TH, Powell D, Preiss T. 2017. Translation complex profile sequencing to study the in vivo dynamics of mRNA-ribosome interactions during translation initiation, elongation and termination. Nat Protoc 12: 697–731. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci 108: E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum EY, Jones SH, Shao A, Dumdie J, Krause MD, Chan WK, Lou CH, Espinoza JL, Song HW, Phan MH, et al. 2016. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell 165: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. 2008. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Thomas EN, Zaher HS. 2017. Ribosome-based quality control of mRNA and nascent peptides. Wiley Interdiscip Rev RNA 10.1002/wrna.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Rebbapragada I, Lykke-Andersen J. 2008. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 6: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Mühlemann O. 2008. The meaning of nonsense. Trends Cell Biol 18: 315–321. [DOI] [PubMed] [Google Scholar]
- Stalder L, Mühlemann O. 2009. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA 15: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. 1998. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J 17: 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Yang W, Zhang J, Dang H, Lu X, Fu C, Miao W. 2017. Nonsense-mediated mRNA decay in Tetrahymena is EJC independent and requires a protozoa-specific nuclease. Nucleic Acids Res 45: 6848–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma KG, Rebbapragada I, Durand S, Lykke-Andersen J. 2015. Identification of elements in human long 3′UTRs that inhibit nonsense-mediated decay. RNA 21: 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Sato H, Singer RH, Maquat LE. 2013. Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev 27: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596. [DOI] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW. 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20: 880–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X. 2016. Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165: 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil JE, Beemon KL. 2006. A 3′UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus. RNA 12: 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW. 1998. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4: 205–214. [PMC free article] [PubMed] [Google Scholar]
- Yamashita A. 2013. Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay. Genes Cells 18: 161–175. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev 15: 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Mühlemann O. 2011. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 17: 2108–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Wek RC. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291: 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zund D, Gruber AR, Zavolan M, Mühlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′UTRs. Nat Struct Mol Biol 20: 936–943. [DOI] [PubMed] [Google Scholar]