Abstract
The conformational heterogeneity and dynamics of protein side chains contribute to function, but investigating exactly how is hindered by experimental challenges arising from the fast timescales involved and the spatial heterogeneity of protein structures. The potential of two-dimensional infrared (2D IR) spectroscopy for measuring conformational heterogeneity and dynamics with unprecedented spatial and temporal resolution has motivated extensive effort to develop amino acids with functional groups that have frequency-resolved absorptions to serve as probes of their protein microenvironments. We demonstrate the full advantage of the approach by selective incorporation of the probe p-cyanophenylalanine at six distinct sites in a Src homology 3 domain and the application of 2D IR spectroscopy to site-specifically characterize heterogeneity and dynamics and their contribution to cognate ligand binding. The approach revealed a wide range of microenvironments and distinct responses to ligand binding, including at the three adjacent, conserved aromatic residues that form the recognition surface of the protein. Molecular dynamics simulations performed for all the labeled proteins provide insight into the underlying heterogeneity and dynamics. Similar application of 2D IR spectroscopy and site-selective probe incorporation will allow for the characterization of heterogeneity and dynamics of other proteins, how heterogeneity and dynamics are affected by solvation and local structure, and how they might contribute to biological function.
Introduction
The now routine determination of structure has revolutionized our understanding of proteins and how structure contributes to function; however, proteins exist on a multi-tiered energy landscape with substates that interconvert on a wide range of timescales. Such conformational heterogeneity and dynamics are now also thought to be essential for function.1–6 A fundamental example is the entropic contribution to the thermodynamics of biological processes, which depends on the range of microstates accessible to protein and solvent, including states that interconvert rapidly, such as those of amino acid side chains.6 Additionally, proteins are complex structures, in which the side chains of their constituent amino acids create a wide range of microenvironments that may possess their own unique heterogeneity and dynamics. Thus a complete investigation of heterogeneity and dynamics to fully understand their role requires experimental approaches for measuring them with sufficient temporal and spatial resolution. While much has been learned, for example via NMR spectroscopy, IR spectroscopy provides an approach with an inherent picosecond timescale that makes possible direct detection of even the most rapidly interconverting states that could contribute to function, and its bond-specific spatial resolution allows for the characterization of different microenvironments. One dimensional (1D) IR spectroscopy is routinely used to characterize small molecules, as the frequency, linewidth, and number of distinct absorptions reflect the nature, heterogeneity, and number of distinct states populated. However, the interpretation of 1D spectra is complicated by the convolution of line broadening processes. This problem has motivated the application of two-dimensional (2D) techniques that not only deconvolute the contributions of these processes, but also allow for the elucidation of the underlying dynamics.7
Historically, IR studies of proteins have focused on the amide backbone vibrations, and thus backbone dynamics have been the focus of the majority of previous 2D IR studies.8,9 However, even in combination with isotopic labeling, the massive spectral congestion of the frequency region of the amide vibrations typically limits site-specific studies to peptides or very small proteins. Amide bonds also do not provide information about side chains, which is arguably as or more critical to our understanding of protein function and how it might be tailored by evolution. These issues have motivated intensive efforts to develop IR probes that may be incorporated into a protein at side chains and that possess environmentally sensitive absorptions within a “transparent” frequency window (1900–2300 cm−1) that is free of native protein absorptions.10–13 These probes are similar to small molecule ligands that also have absorptions in the transparent window, such as CO, which have been used for decades to characterize their local microenvironments within proteins that bind them.14–16 However, because the IR probes are incorporated into the protein via covalent attachment to an amino acid, they may be used to characterize any protein and at any position of interest. The sulfhydryl band of natively occurring cysteine had been exploited as a transparent window probe, but its weak signals thus far have limited its widespread use.17 The application of 2D IR spectroscopy with non-native transparent window probes was first demonstrated with p-cyanophenylalanine (CNF) to characterize the dynamics of the model 35 amino acid peptide HP35,18,19 and the approach was extended to measure the dynamics of a large, intact protein via the incorporation of azidophenylalanine into myoglobin.20 Additionally, the attachment of metal–carbonyl complexes and the installation of cyanocysteines have been used to introduce probes for 2D IR spectroscopy.21,22 However, thus far the approach has been applied almost exclusively for characterizing one position in a protein, not taking advantage of its potential high spatial resolution that enables study of different parts of the structure. One exception is a recent study that incorporated azidohomoalanine as a probe at six positions of a PDZ domain and obtained difference 2D IR spectra of the states before and after perturbation by photoinduced cis–trans isomerization of an azobenzene, which was covalently incorporated into the protein.23 Interestingly, only a change in absorption intensity was observed at one site, while none showed significant differences in the frequencies or linewidths. While this may suggest that the probes are not sensitive to changes in their local environment, the biological relevance of the two states characterized is uncertain. Additionally, the time evolution of different environments in a protein has yet to be measured, and so whether and how spatial variation in protein dynamics contributes to biological function has remained unexplored.
Src homology 3 (SH3) domains recognize proline-rich (PR) motifs to mediate eukaryotic protein–protein interactions in diverse cellular processes and serve as a model for the study of protein molecular recognition more broadly.24 SH3 domains are composed of a beta sheet core, a short 310 helix, and three loops: the RT loop, n-Src loop, and distal loop (Fig. 1). The domains recognize a linear sequence motif that contains a core consensus sequence, PxxP, with x typically a proline or a hydrophobic amino acid. In the complex, the PR ligand adopts a polyproline type II secondary structure and the two conserved proline side chains pack within grooves on the SH3 domain surface formed by a set of conserved tyrosine residues. Despite extensive study of SH3 domain-PR motif recognition by a vast range of approaches, many aspects of the process, such as the origins of specificity and underlying thermodynamics, remain poorly understood. The contributions of the dynamics of the SH3 domain, peptide ligands, and associated solvent to molecular recognition are likely to be important,25–32 but challenging to assess experimentally. While NMR spectroscopy has revealed binding-induced changes on the ps–ns and longer ms time-scales,25–27,32 few studies have characterized side chain dynamics,33,34 and none have explored their possible contribution to molecular recognition.
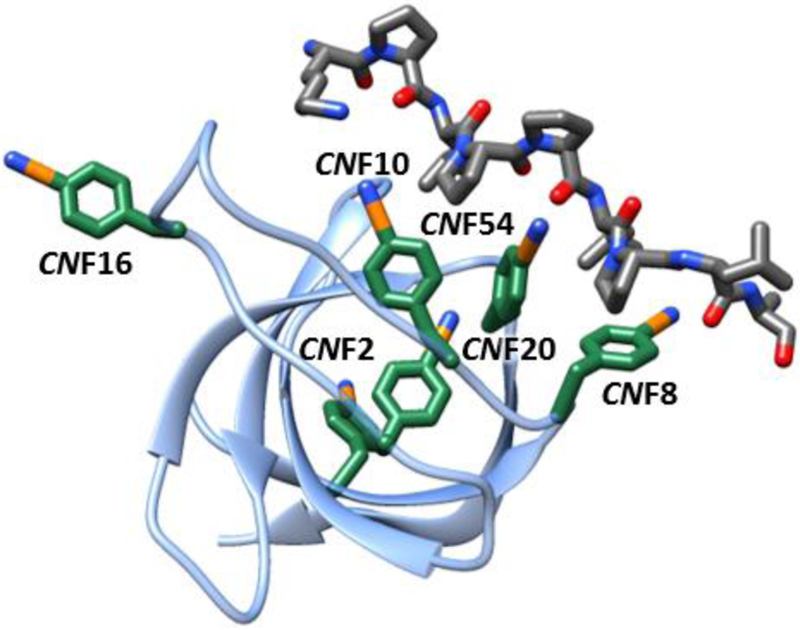
Fig. 1.
Structural model of the complex of SH3Sho1 and pPbs2 (2VKN) showing locations of CNF incorporation.
To demonstrate the full potential of 2D IR spectroscopy combined with site-specific labeling for measuring the heterogeneity and dynamics of proteins with high spatial and temporal resolution, and to characterize the involvement of side chain dynamics in molecular recognition, we incorporated CNF at six distinct sites of the SH3 domain from yeast protein Sho1 (SH3Sho1; Fig. 1) via amber suppression and characterized them both in the unligated state and when bound to its cognate peptide from Pbs2 (pPbs2). Specifically, CNF was introduced at each of the three conserved Tyr residues that line the binding surface (CNF8, CNF10, and CNF54), at a Tyr residue within the RT loop (CNF16), at a Phe residue in a more buried location (CNF20), and at a Tyr residue that is more distant from the ligand binding surface (CNF2) (Fig. 1). (Residue numbering is based on a standard numbering system for SH3 domains and the PR motif.35,36) 2D IR spectroscopy reveals site-specific heterogeneity and dynamics that depend significantly on ligand binding, including at adjacent conserved residues within the binding surface.
Experimental methods
CNF-labeled SH3Sho1 and pPbs2 were produced via amber suppression and Fmoc solid phase peptide synthesis, respectively. All procedures to express or synthesize, purify, and prepare the samples for analysis via FT IR, 2D IR, visible, circular dichroism, or fluorescence spectroscopy were performed as reported previously and are described more thoroughly in ESI.†37,38 Samples for 2D IR spectroscopy contained 4 mM SH3Sho1 for study of the unligated state or 4 mM SH3Sho1 and 4.8 mM pPbs2 for study of the complex in 50 mM sodium phosphate, pH 7.0, 100 mM NaCl, with the exception of CNF20, which was prepared with 5 mM protein and 6 mM pPbs2 due to lower labeling efficiency. 2D IR spectroscopy was performed in the conventional BOXCARS geometry as previously reported (see ESI†for a complete description). The FFCFs were determined via center line slope (CLS) analysis of the Tw-dependent 2D IR spectra in combination with fitting to the linear spectra.39,40 All experiments were performed in triplicate with independently prepared samples.
Preparation and execution of the MD simulations utilized computational resources of BigRed2 at Indiana University using Amber 2016.41 The charges for the CNF were derived via the R.E.D. Server.42 CNF was introduced into the crystal structural model of SH3Sho1 bound to pPbs2 (PDB ID 2VKN) at residues 10, 16, 20, 54 or 2 and 8 using Chimera (UCSF).43 The protein was solvated by a periodic 12 Å octahedron of TIP3 water, Na+ counter ions were added to neutralize the charge of the system, then additional Na+ and Cl− ions were added to make the system 150 mM in Na+ concentration according to the total number of water molecules. The particle mesh Ewald summation method with a non-bonded cut-off of 10 Å was employed for long-range interactions and the SHAKE procedure was used to constrain all bonds involving hydrogen atoms. Self-guided Langevin dynamics were run for 1 ns with a 2 ps local averaging time and target guiding temperature of 450 K. Ten frames from this trajectory (separated by 100 ps) were then extracted and used to start two sets of production MD simulations of 5 ns with 1 fs time steps, saving the coordinates and forces every 100 fs. Analysis of the MD simulations were performed using the cpptraj program of Amber16 and Matlab 18.0 (Mathworks). Calculation of the electric field (EF) along the CN to determine the EF time correlation function (TCF) was performed as described previously.44 Additional details about the MD simulation preparation and analysis are provided in ESI,† Experimental methods.
Results
Protein characterization
The IR probe, CNF, was successfully incorporated at each of the six locations in SH3Sho1 via amber suppression.45 While the CN group was chosen to be minimally perturbative compared to the other possible transparent window 2D IR probes,13 substitution of the native Tyr or Phe residues with CNF altered the size and hydrogen bonding potential of the side chains, introducing possible perturbation to structure or function. However, characterization of the variants via circular dichroism spectroscopy indicated that CNF incorporation resulted in no detectable perturbation in secondary structure, and fluorescence-based binding assays indicated that the probe incorporation led to at most two-fold change in the affinity for pPbs2 (Fig. S2 and S3, ESI†).
FT IR spectroscopy
The linear FT IR spectra were acquired first for the unligated proteins to investigate the spatial heterogeneity in the absence of the ligand. All spectra show an absorption band associated with the CN stretch around 2232.5–2236.3 cm−1 (Fig. S4, ESI†). For all absorptions, the frequency of maximum absorbance was the same within error of the first moment of the absorption, consistent with a single symmetrical band (Table S3, ESI†). Each spectrum was fit to a Gaussian function to determine the center frequency and linewidth (Table 1). The frequencies of the CNF incorporated at the protein surface, with the exception CNF10, were relatively high and similar to CNF in aqueous solution. In comparison, the absorptions for CNF10 and CNF20, which is positioned in the protein core, were found at lower frequencies (by 3–4 cm−1), indicating at these sites interactions with the local environments distinct from bulk solvent. The spectra also show small variation in the line-widths; the linewidth was the broadest for CNF16, whereas it was relatively narrow for CNF2, CNF8, and CNF54.
Table 1.
Parameters from fits of absorption spectra and FFCFs of CNF-labeled SH3Sho1
| Γ* (cm−1) | Δ1 (cm−1) | τ1 (ps) | Δs (cm−1) | Frequency (cm−1) | FWHMa (cm−1) | |
|---|---|---|---|---|---|---|
| CNF | 7.9 ± 0.2 | 2.7 ± 0.2 | 1.2 ± 0.2 | 1.5 ± 0.1 | 2236.7 ± 0.1 | 12.6 ± 0.1 |
| CNF2 | 4.9 ± 0.2 | 2.4 ± 0.1 | 1.9 ± 0.3 | 1.9 ± 0.1 | 2235.2 ± 0.1 | 9.4 ± 0.1 |
| CNF2-Pbs2 | 5.6 ± 0.1 | 2.9 ± 0.2 | 1.6 ± 0.5 | 1.9 ± 0.4 | 2235.4 ± 0.1 | 10.7 ± 0.1 |
| CNF8 | 7.4 ± 0.3 | 2.4 ± 0.2 | 1.6 ± 0.1 | 1.4 ± 0.1 | 2235.4 ± 0.1 | 9.5 ± 0.3 |
| CNF8-Pbs2 | 8.6 ± 0.2 | 2.7 ± 0.1 | 1.4 ± 0.1 | 1.6 ± 0.1 | 2236.1 ± 0.1 | 10.7 ± 0.2 |
| CNF10 | 6.2 ± 0.8 | 3.6 ± 0.2 | 5.8 ± 0.2 | 1.3 ± 0.2 | 2232.5 ± 0.1 | 11.3 ± 0.1 |
| CNF10-Pbs2 | 4.3 ± 0.1 | 2.3 ± 0.1 | 1.5 ± 0.4 | 3.7 ± 0.1 | 2233.3 ± 0.1 | 11.5 ± 0.1 |
| CNF16 | 10.2 ± 1.0 | 2.9 ± 0.5 | 1.7 ± 0.5 | 1.8 ± 0.2 | 2234.9 ± 0.1 | 12.8 ± 0.1 |
| CNF16-Pbs2 | 9.6 ± 0.1 | 3.1 ± 0.1 | 2.5 it 0.6 | 1.4 ± 0.3 | 2234.7 ± 0.1 | 12.6 ± 0.1 |
| CNF20 | 6.4 ± 0.1 | 2.9 ± 0.2 | 2.8 ± 0.6 | 1.8 ± 0.2 | 2233.6 ± 0.2 | 10.7 ± 0.1 |
| CNF20-Pbs2 | 7.2 ± 0.1 | 3.3 ± 0.1 | 0.7 ± 0.2 | 2.0 ± 0.2 | 2234.6 ± 0.1 | 11.0 ± 0.2 |
| CNF54 | 5.9 ± 0.2 | 2.2 ± 0.1 | 1.2 ± 0.2 | 2.0 ± 0.1 | 2236.3 ± 0.2 | 9.0 ± 0.2 |
| CNF54-Pbs2 | 5.9 ± 0.3 | 2.8 ± 0.3 | 2.9 ± 0.2 | 1.0 ± 0.5 | 2235.6 ± 0.1 | 9.8 ± 0.1 |
Full width at half maximum.
To assess the involvement of each CNF in the recognition of pPbs2, we next acquired the FT IR spectra of the SH3Sho1 variants in the ligand complex. As found for the unligated proteins, the spectra showed no evidence for multiple distinct states and were thus fit by a single Gaussian function. Ligand binding induced no significant change in the frequency for CNF2 or CNF16, located distant from the binding site and on the RT loop, respectively (Table 1). In contrast, for the three sites along the interaction surface (CNF8, CNF10, CNF54) and for the more buried residue (CNF20) the frequency was moderately sensitive to the binding of SH3Sho1 with pPbs2, shifting 0.6–1.0 cm−1. However, for CNF8, CNF10, and CNF20 the absorption shifted to higher frequency, whereas for CNF54 it uniquely shifted to lower frequency. Thus all these sites appear to be involved in ligand recognition, but how exactly they participate varies among them. Another result of ligand binding was the broadening of the absorptions of CNF2, CNF8 and CNF54. Line-broadening could arise from greater heterogeneity in the complexes that positions the CN probe in a variety of environments to engender a greater range of frequencies; however, inhomogeneous broadening is convoluted with line broadening due to other processes in 1D spectra, which limits interpretation.
2D IR spectroscopy
To better elucidate the differences in the inhomogeneity and dynamics among the CNF, we applied 2D IR spectroscopy.7 As frequency variation arises from interaction of the IR probe with the protein or solvent environment, the time evolution of the frequencies, spectral diffusion, reports on the dynamics of the surrounding protein or solvent. 2D IR spectroscopy was used to generate 2D correlation spectra that connect the frequencies of the CN probes before (horizontal axis) and after (vertical axis) a variable waiting time, Tw (Fig. 2 and Fig. S5, ESI†). For short Tw, the 2D spectra appear diagonally elongated, which indicates that most of the CN probes in the ensemble have the same initial and final frequencies. With increasing Tw, the 2D lineshapes became less elongated, reflecting that the frequencies have changed because the environment has changed during Tw. The Tw-dependent change in the 2D lineshape directly reports on the spectral diffusion of the CNF probe that results from the dynamics of its interaction with its environment.
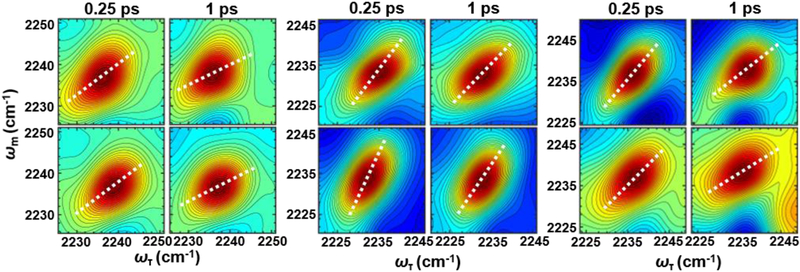
Fig. 2.
Example 2D IR spectra of CNF8 (left panel), CNF10 (center panel), and CNF54 (right panel) SH3Sho1 at Tw of 0.25 and 1 ps for the unligated protein (top row) and the pPbs2 complex (bottom row). Overlayed are the center lines from analysis of the lineshapes of the average spectra (white dotted line).
A useful quantity for describing spectral diffusion is the frequency–frequency correlation function (FFCF), which can be determined from analysis of the Tw-dependent 2D data via well-established methods.39,40 We applied a Kubo model46 that separates the dynamics into homogeneous and inhomogeneous contributions:
The latter two terms describe the dynamics among the inhomogeneous distribution of frequencies. The inhomogeneous dynamics are separated into two timescales, where Δ1 reflects the part of the frequency distribution sampled on the timescale τ1, and the static term Δs, reflects the part of the frequency distribution sampled slowly compared to the experimental time window (~5 ps, determined by the vibrational lifetime of the CN probe). The first term accounts for the homogeneous contribution to the FFCF. T1 is the vibrational lifetime, and the pure dephasing time, T2* = (Δ2τ)−1, describes very fast fluctuations in the motionally narrowed limit where the frequency amplitude and timescale cannot be separated (Δτ ≪ 1), which lead to a Lorentzian contribution to the line shape, Γ* = 1/πT2*. The center line slope (CLS) analysis yields a good approximation of the inhomogeneous part of the normalized FFCF (Fig. 2).39 Combined fitting of the CLS decays with the linear spectra was used to obtain the complete FFCFs (Table 1).40
The FFCFs of the CNFs of the unligated protein show variation in the line-broadening and underlying dynamics that reveal significant spatial heterogeneity in SH3Sho1 (Fig. 3 and Table 1). The FFCF for CNF10 was the most distinct, as it showed dynamics on a substantially slower timescale (τ1 of 5.8 ps) associated with relatively large inhomogeneous broadening (Δ1). For CNF20, the site most buried within the protein, τ1 also was slightly slower (2.8 ps). In comparison, the other sites at the protein surface showed faster dynamics similar to the amino acid in aqueous solution (1.2–1.7 ps).38 Another distinction among the sites revealed by the FFCFs was a large homogeneous component (Γ*) for CNF16, which actually underlay the larger linewidth observed in the 1D spectra. The homogeneous broadening is indicative of motion that is fast on the IR timescale and could be due to high orientational mobility of the side chain due to its location on a flexible loop, a contribution that was not specifically measured in this study. Consistent with this possibility, NMR order parameters determined for the amide backbone of the RT loop of SH3 domains generally indicate the backbone to be highly dynamic on the ps–ns timescale.25–27 Thus together the NMR and 2D IR data reflect the high overall flexibility of the RT loop, including both backbone and side chains.
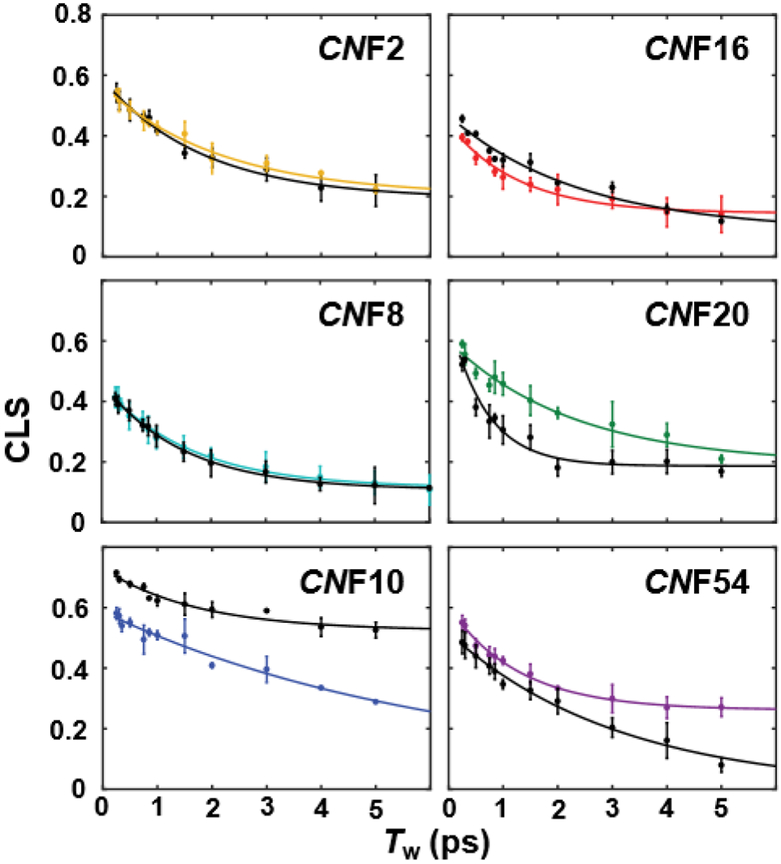
Fig. 3.
CLS decay curves (points) and fits (lines) for unligated SH3Sho1 (colored) and the pPbs2 complex (black).
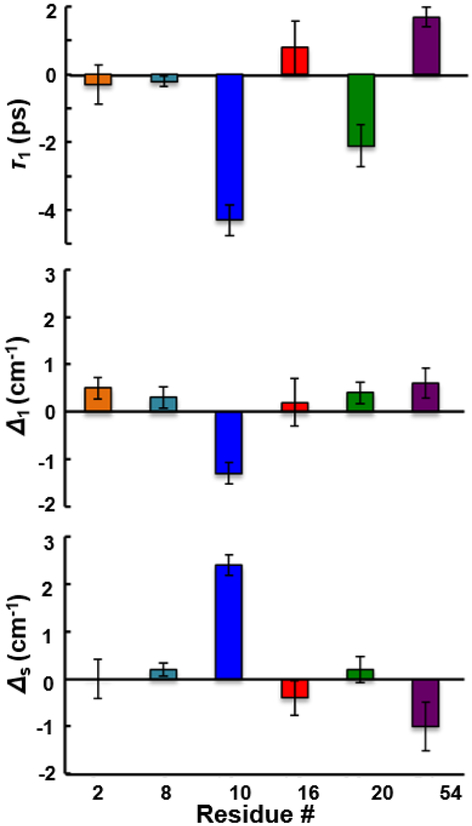
To next investigate how the heterogeneity and dynamics of the microenvironment of each CNF are affected by ligand binding, we characterized the variants in the complex with pPbs2. Interestingly, in comparison to the linear spectra, the spectral dynamics showed more substantial, and also site-specific, changes in response to ligand binding. The FFCFs for CNF2 and CNF16 did not differ significantly between the unligated protein and complex (Fig. 4), similarly insensitive to binding as the linear spectra. Surprisingly, unlike the linear spectra, the FFCF for CNF8 also was not significantly affected by pPbs2 binding, despite the probe’s location at the recognition surface. In contrast, the dynamics for CNF10, CNF20, and CNF54 were affected by pPbs2 binding, and how they changed differed among the sites. For CNF20 and CNF54, the CLS decayed more rapidly in the complexes, whereas for CNF10 it decayed more slowly (Fig. 3). The parameters from fitting the FFCF to the two inhomogeneous timescales indicate that the slower overall decay for CNF10 in the complex arises from an increase in inhomogeneity associated with slowly interconverting states (Δs), along with a decrease associated with rapidly inter-converting states (Δ1) (Fig. 4 and Table 1). In addition, the uniquely slow timescale of dynamics among the frequency distribution Δ1 exhibited for CNF10 of the free protein, was faster in timescale in the complex, becoming similar to the other CNF at the protein surface. Compared to CNF10, the FFCF fit parameters for CNF54 showed opposite changes, a reduction of inhomogeneity from slowly interconverting states (Δs) and a small increase from rapidly interconverting states (Δ1), while the timescale τ1 became slower. In contrast, for CNF20 the more rapid decay of the CLS in the complex arose from the faster timescale of dynamics, with little change in inhomogeneity. Remarkably, the sensitivity of the FFCFs for the CNF at the three, adjacent, conserved aromatic residues along the binding interface differed dramatically, with one (CNF8) showing no effect from pPbs2 binding, and the other two (CNF10 and CNF54) showing significant but opposite effects.
Fig. 4.
Binding-induced changes in the timescale of dynamics (upper panel) and the inhomogeneous distribution of frequencies sampled rapidly (middle panel) and slowly (bottom panel) reflected by the FFCFs. Error bars are standard deviations from three sets of experiments with independently prepared samples.
Molecular dynamics simulations
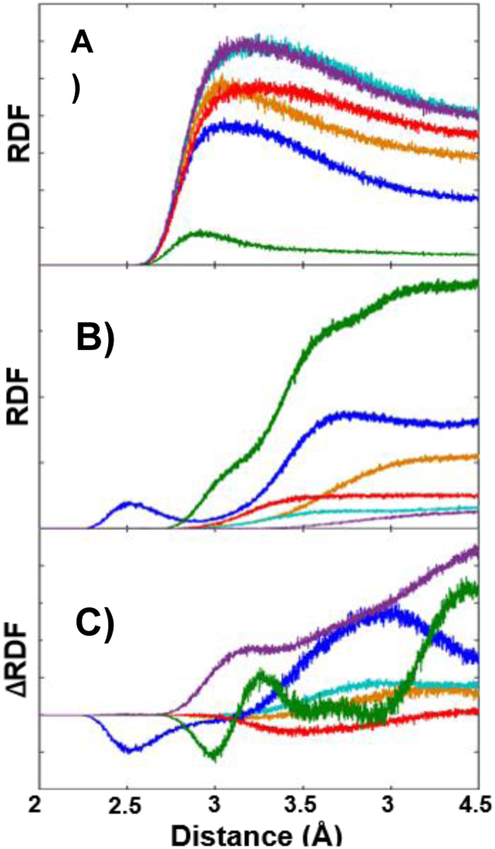
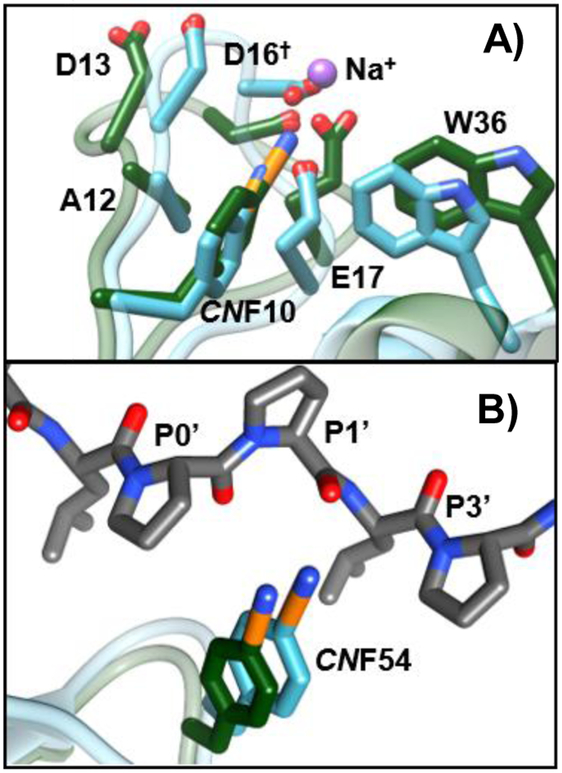
Clearly the spectral data for the CNF variants indicate complex, distinct, and importantly, site-specific changes in local environments and dynamics with ligand binding. To gain insight into the origins of the differences, we performed molecular dynamics (MD) simulations of SH3Sho1 labeled with CNF at each site for the unligated protein and the pPbs2 complex. We sought to identify the parts of the protein or ligand that influence each CN probe by determining the radial distribution functions (RDFs) for the distance of the cyano nitrogen to different parts of the protein to assess how often they approach each other and how this is changed by ligand binding (Fig. 5 and Fig. S9, ESI†). For the unligated protein, CNF20 and CNF10 were in close contact more frequently with surrounding protein and less so with solvent water relative to the other sites. In addition, in the unligated protein but not the pPbs2 complex, CNF10 frequently closely approached (~2.5 Å) a sodium ion, which appears to stabilize the charge density of the highly acidic RT loop (Fig. 6A). These distinct molecular environments are a possible contributor to the slower timescale τ1 found for CNF10 and CNF20 in the unligated protein, as well as the large change in the faster FFCF component of CNF10 in the complex (Table 1). Spectral dynamics on such fast timescales are likely to reflect, at least in part, the surrounding solvent fluctuations. Interactions of water with ions are found by previous studies to decrease hydrogen bond switching rates by a factor of three to four,47 in line with slower dynamics reported by the FFCF of CNF10.
Fig. 5.
RDFs for distance of the cyano nitrogen to (A) water oxygen atoms, (B) all heavy atoms excluding solvent and CNF side chain, and (C) change in the RDFs for these heavy atoms upon ligand binding for CNF2 (orange), CNF8 (teal), CNF10 (blue), CNF16 (red), CNF20 (green), and CNF54 (purple) SH3Sho1.
Fig. 6.
Overlay of average structures from MD simulations showing side chains of residues in local environment of (A) CNF10 and (B) CNF54 when unligated (green) and bound to pPbs2 (teal).
Comparison of the MD simulations for the unligated protein to pPbs2 complex indicate substantial changes to the environments at CNF10, CNF20, and CNF54 – those residues with FFCFs sensitive to binding – and much less so for the other sites (Fig. 5C). For CNF10, the change in RDFs indicate that the CN experiences an overall increase in the frequency of closely approaching atoms (<4 Å). These contacts involve a variety of residues, especially Asp13 and Glu17 of the RT loop, but also Ala12, Trp36, and Asp16‡ (Fig. 6A and Fig. S10, S11, ESI†). The enhanced packing of CNF10 within such a heterogeneous environment accompanies conformational adjustment of the RT loop upon pPbs2 binding. In comparison, the RDFs for CNF54 similarly indicated an overall increase in the proximity of atoms in the pPbs2 complex, but also increased amplitude at much closer distance (~3 Å). In addition, whereas the CN of CNF54 was rarely in close proximity to any specific protein atoms in the unligated state, in the pPbs2 complex it packed between the conserved P0′ and P3′ side chains of the ligand, placing it near the carbonyl oxygen of P1′ during almost the entire simulation (within 4 Å in 95% of frames) (Fig. 6B and Table S6, ESI†). Notably, in native SH3 domains Tyr54 forms a hydrogen bond with the backbone carbonyl of P1′.36 Thus, the CN probe appears to retain sensitivity to a native interaction with the ligand in the pPbs2 complex. For CNF20 the RDFs showed an increase in the amplitude at longer distances (>4 Å), but at close distance no net change, rather only a shift in amplitude. In contrast, the MD simulations showed little change in their environment or dynamics for CNF8, CNF2, and CNF16 when SH3Sho1 binds pPbs2.
In attempt to better understand the solvatochromism and spectral diffusion of the CN probes, we assessed whether the spectral data could be described by a vibrational Stark effect, the frequency dependence on the electric field projected onto the transition dipole of the vibration. The temporal autocorrelation functions of the electric field (EF TCF) along the CN probes showed dynamics on the several ps and longer timescales, similarly to the FFCFs determined for the CN probes. Additionally, in agreement with the FFCFs, the EF TCFs indicated greatest perturbation upon pPbs2 binding to the dynamics at CNF10. However, the Stark effect did not fully account for all the differences among the sites and changes due to pPbs2 binding in either the FFCFs or 1D absorptions (Fig. S6 and S7, ESI†). As noted previously, the environment surrounding an IR probe in a protein is non-uniform, and particularly for the CN probe, specific, local interactions including hydrogen bonding and repulsive interactions due to close packing are likely to also contribute substantially to the solvatochromism.12,48,49 In the MD simulations water molecules appeared within hydrogen bonding (HB) distance (3 Å) to the CN for ~10% of the frames for all the surface-exposed residues (i.e. all but CNF20), and the occurrence unexpectedly was slightly more frequent in the pPbs2 complex (Table S5, ESI†), consistent with the blue-shift of many absorptions. The hydrogen bonding time correlation functions (HB TCFs) showed timescales of dynamics similar to the FFCFs and better agreement than did the EF TCFs, but also did not quantitatively correlate with all the spectral data, particularly for CNF54 (Fig. S8, ESI†). More extensive simulations that more completely sample conformational space might lead to better agreement. In addition, short-range repulsive interactions not captured by the simple analysis also likely contribute substantially to the solvatochromism, as has been found in previous theoretical studies of the small molecule model system p-tolunitrile.50 Further work to apply more complex modelling clearly is needed to fully describe the microscopic interactions underlying the solvatochromism and spectral diffusion of the CN probe in the complex environments experienced in proteins.
Discussion
Altogether, the experimental data and simulations provide a spatially detailed picture of the molecular changes of SH3Sho1 upon binding to its ligand pPbs2. At CNF10, pPbs2 binding is accompanied by an increase in interaction with slowly fluctuating, heterogeneous environment from the RT loop and other parts of the protein, which is associated with an increase in inhomogeneity of slowly interconverting states. In contrast, when SH3Sho1 binds pPbs2, CNF54 experiences a relatively uniform environment where it interacts with specific parts of the ligand, which is associated with smaller inhomogeneity of slowly interconverting states. CNF20 shows no net change in the frequency of closely approaching atoms, and no substantial changes are found in the inhomogeneity reflected by the FFCF. However, the timescale of dynamics was sensitive to ligand binding. Thus, ligand binding affects not only the surface-exposed residues that are directly involved in the interaction, but its impact also is transmitted farther into the core of the protein. Similar binding-induced perturbations to local side chain conformations and dynamics have been found for other proteins and evoked in mechanisms of allostery.2–4
The lack of sensitivity of the FFCF of CNF8 to pPbs2 binding, while the average frequency shifts by a magnitude similar to those of CNF10 and CNF54, was unexpected. In the native protein CNF8 is a conserved Tyr positioned within the binding surface, and the side chain shows significant contact with pPbs2 in the crystal structure (2VKN). However, in agreement with the experimental results, the MD simulations showed little change in the dynamics at CNF8 induced by ligand binding. For comparison, we also analyzed the proximity of water and protein to the three native Tyr residues at the recognition surface by determining the RDFs for the distance to the hydroxyl oxygens, and the variable sensitivity to pPbs2 binding among the sites was in agreement with that found for the CNF (Fig. S12, ESI†), supporting that the heterogeneity and dynamics reported by the probes are comparable to the native protein.
The site-specific information about the contribution of different residues in a SH3 domain to ligand recognition reveals the variable involvement of the three conserved aromatic residues at the binding interface – the dynamics of two of these residues, CNF10 and CNF54, were sensitive to ligand binding, whereas those of CNF8 were unchanged. In agreement with these results, our previous study of SH3Sho1 binding to pPbs2 labeled with carbon–deuterium (C–D) transparent window IR probes revealed multiple bands for C–D bonds incorporated at P3′,51 indicating conformational heterogeneity at the proline residue of the ligand closest to CNF8. This observation is consistent with the insensitivity to ligand binding of the FFCF of CNF8, suggesting that at this position the protein and ligand do not as tightly interact. Also in line with this picture, mutation of Tyr8 to alanine results in a ~17-fold decrease in binding affinity, whereas the same mutation at Tyr54 results in ~350-fold decrease.52 Thus, while not apparent in the crystal structure, the IR data indicate site-dependent engagement of the SH3 domain with the ligand. Additional support for this conclusion is provided by previous NMR studies of the complexes of the Apb1 SH3 domain and a number of PR peptides where linear chemical shift variation among the complexes provided evidence for rapid dynamics that depended on the position probed.32
A longstanding question about SH3 domain recognition regards the thermodynamics that typically include large unfavorable entropy contributions, unexpected for the highly hydrophobic interaction with the PR motif in which solvent should be displaced. Toward explaining the thermodynamics, the protein, ligand, and, more recently, water dynamics have been suggested to play a critical role.25–31 Reduction in conformational freedom of the loops and restriction of interfacial water molecules upon complexation would engender unfavorable entropy changes. Notably, side chain dynamics play a key role in entropy changes of molecular recognition by other proteins.1,6 Similarly, we expect that the site-specific changes in the heterogeneity and dynamics uncovered in this study of SH3Sho1–pPbs2 recognition are important to the binding thermodynamics. In the complex with pPbs2, CNF10 appears to have increased interaction with a heterogeneous protein environment, whereas CNF54 experiences a more homogeneous environment upon intercalation between the conserved proline residues of the ligand. In contrast, CNF8 is sensitive to ligand binding, as evident from the induced shift in the average absorption frequency, but binding does not alter the inhomogeneity or dynamics of its environment. Thus, our study indicates how part of the recognition surface, specifically around the region of CNF54, could contribute more substantially to unfavorable binding entropy than other parts of the protein, such as probed by CNF10, and particularly CNF8. Furthermore, the data underscores how generating a complete molecular description of protein recognition and other aspects of function will require the ability to measure conformational heterogeneity and dynamics with residue-specific precision.
Conclusions
This study demonstrated how 2D IR spectroscopy in combination with the introduction of frequency-resolved IR probes may be used to reveal site-specific changes in the heterogeneity and dynamics of protein microenvironments, and the application of the approach toward developing a more complete molecular description of the recognition of SH3 domains. Importantly, the observed spectral changes were induced by cognate ligand binding, the biological function of the protein domain, which in turn suggests that the observed dynamics may be biologically significant. As expected, molecular recognition is clearly complex, but additional studies involving more probes in different proteins and contexts will help to elucidate the process. Further development of the experimental methods will advance our ability to characterize protein heterogeneity and dynamics to better enable investigation into their contribution to molecular recognition, catalysis, and other biochemical processes.
Supplementary Material
Acknowledgements
This material is based upon work supported by the National Science Foundation under Grant no. MCB-1552996. S. R. and R. E. H. were partially supported by the Indiana University Chemical and Quantitative Biology Program Training Grant (T32 GM109825). Computational aspects of this research were supported in part by Lilly Endowment, Inc., through its support for the Indiana University Pervasive Technology Institute, and in part by the Indiana METACyt Initiative. The Indiana METACyt Initiative at IU was also supported in part by Lilly Endowment, Inc.
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cp06146g
Unique, consecutive numbering according to the consensus sequence of SH3 domains is not possible for Asp16 and Tyr16/CNF16 due to a two-residue insertion at this site in the sequence of SH3Sho1 and other yeast SH3 domains.53
Notes and references
- 1.Palmer AG III, Annu. Rev. Biophys. Biomol. Struct, 2001, 30, 129–155. [DOI] [PubMed] [Google Scholar]
- 2.Popovych N, Sun S, Ebright RH and Kalodimos CG, Nat. Struct. Mol. Biol, 2006, 13, 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai CJ, del Sol A and Nussinov R, J. Mol. Biol, 2008, 378, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petit CM, Sapienza PJ, Fuentes EJ and Lee AL, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 18249–18254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zafra Ruano A, Cilia E, Couceiro JR, Ruiz Sanz J, Schymkowitz J, Rousseau F, Luque I and Lenaerts T, PLoS Comput. Biol, 2016, 12, e1004938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wand AJ and Sharp KA, Annu. Rev. Biophys, 2018, 47, 41–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamm P and Zanni M, Concepts and Methods of 2D Infrared Spectroscopy, Cambridge University Press, New York, 2011. [Google Scholar]
- 8.Ganim Z, Chung HS, Smith AW, DeFlores LP, Jones KC and Tokmakoff A, Acc. Chem. Res, 2007, 41, 432–441. [DOI] [PubMed] [Google Scholar]
- 9.Moran SD, Woys AM, Buchanan LE, Bixby E, Decatur SM and Zanni MT, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 3329–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin JK, Jimenez R and Romesberg FE, J. Am. Chem. Soc, 2001, 123, 2426–2427. [DOI] [PubMed] [Google Scholar]
- 11.Suydam IT and Boxer SG, Biochemistry, 2003, 42, 12050–12055. [DOI] [PubMed] [Google Scholar]
- 12.Kim H and Cho M, Chem. Rev, 2013, 113, 5817–5847. [DOI] [PubMed] [Google Scholar]
- 13.Adhikary R, Zimmermann J and Romesberg FE, Chem. Rev, 2017, 117, 1927–1969. [DOI] [PubMed] [Google Scholar]
- 14.Rella CW, Rector KD, Kwok A, Hill JR, Schwettman HA, Dlott DD and Fayer MD, J. Phys. Chem, 1996, 100, 15620–15629. [DOI] [PubMed] [Google Scholar]
- 15.Bandaria JN, Dutta S, Hill SE, Kohen A and Cheatum CM, J. Am. Chem. Soc, 2008, 130, 22–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiro TG, Soldatova AV and Balakrishnan G, Coord. Chem. Rev, 2013, 257, 511–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozinski M, Garrett-Roe S and Hamm P, J. Phys. Chem. B, 2008, 112, 7645–7650. [DOI] [PubMed] [Google Scholar]
- 18.Urbanek DC, Vorobyev DY, Serrano AL, Gai F and Hochstrasser RM, J. Phys. Chem. Lett, 2010, 1, 3311–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung JK, Thielges MC and Fayer MD, Proc. Natl. Acad. Sci. U. S. A, 2011, 108, 3578–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thielges MC, Axup JY, Wong D, Lee HS, Chung JK, Schultz PG and Fayer MD, J. Phys. Chem. B, 2011, 115, 11294–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woys AM, Mukherjee SS, Skoff DR, Moran SD and Zanni MT, J. Phys. Chem. B, 2013, 117, 5009–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King JT and Kubarych KJ, J. Am. Chem. Soc, 2012, 134, 18705–18712. [DOI] [PubMed] [Google Scholar]
- 23.Stucki-Buchli B, Johnson PJM, Bozovic O, Zanobini C, Koziol KL, Hamm P, Gulzar A, Wolf S, Buchenberg S and Stock G, J. Phys. Chem. A, 2017, 121, 9435–9445. [DOI] [PubMed] [Google Scholar]
- 24.Zarrinpar A, Bhattacharyya RP and Lim WA, Sci. STKE, 2003, 2003, Re8. [DOI] [PubMed] [Google Scholar]
- 25.Arold S, O’Brien R, Franken P, Strub MP, Hoh F, Dumas C and Ladbury JE, Biochemistry, 1998, 37, 14683–14691. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Pawley NH and Nicholson LK, J. Mol. Biol, 2001, 313, 873–887. [DOI] [PubMed] [Google Scholar]
- 27.Ferreon JC, Volk DE, Luxon BA, Gorenstein DG and Hilser VJ, Biochemistry, 2003, 42, 5582–5591. [DOI] [PubMed] [Google Scholar]
- 28.Ferreon JC and Hilser VJ, Biochemistry, 2004, 43, 7787–7797. [DOI] [PubMed] [Google Scholar]
- 29.Palencia A, Cobos ES, Mateo PL, Martínez JC and Luque I, J. Mol. Biol, 2004, 336, 527–537. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Garcia JM, Ruiz-Sanz J and Luque I, Biochem. J, 2012, 442, 443–451. [DOI] [PubMed] [Google Scholar]
- 31.Zafra-Ruano A and Luque I, FEBS Lett, 2012, 586, 2619–2630. [DOI] [PubMed] [Google Scholar]
- 32.Stollar EJ, Lin H, Davidson AR and Forman-Kay JD, PLoS One, 2012, 7, e51282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittermaier A and Kay LE, Protein Sci, 2004, 13, 1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reif B, Xue Y, Agarwal V, Pavlova MS, Hologne M, Diehl A, Ryabov YE and Skrynnikov NR, J. Am. Chem. Soc, 2006, 128, 12354–12355. [DOI] [PubMed] [Google Scholar]
- 35.Larson SM and Davidson AR, Protein Sci, 2000, 9, 2170–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW and Schreiber SL, Cell, 1994, 76, 933–945. [DOI] [PubMed] [Google Scholar]
- 37.Horness RE, Basom EJ and Thielges MC, Anal. Methods, 2015, 7, 7234–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramos S, Scott KJ, Horness RE, Le Sueur AL and Thielges MC, Phys. Chem. Chem. Phys, 2017, 19, 10081–10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwak K, Park S, Finkelstein IJ and Fayer MD, J. Chem. Phys, 2007, 127, 124503. [DOI] [PubMed] [Google Scholar]
- 40.Park S, Kwak K and Fayer MD, Laser Phys. Lett, 2007, 4, 704–718. [Google Scholar]
- 41.Case DA, Betz RM, Cerutti DS, Cheatham ITE, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L and Kollman PA, AMBER, University of California, San Francisco, 2016. [Google Scholar]
- 42.Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P and Dupradeau FY, Nucleic Acids Res, 2011, 39, W511–W517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE, J. Comput. Chem, 2004, 13, 1605–1612. [DOI] [PubMed] [Google Scholar]
- 44.Fried SD, Wang LP, Boxer SG, Ren P and Pande VS, J. Phys. Chem. B, 2013, 117, 16236–16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R and Schultz PG, J. Am. Chem. Soc, 2006, 128, 13984–13985. [DOI] [PubMed] [Google Scholar]
- 46.Kubo R, Adv. Chem. Phys, 1969, 15, 101–127. [Google Scholar]
- 47.Fayer MD, Wong D, Rosenfeld DE, Fenn EE and Park S, Acc. Chem. Res, 2009, 42, 1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fafarman AT, Sigala PA, Herschiag D and Boxer SG, J. Am. Chem. Soc, 2010, 132, 12811–12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blasiak B, Ritchie AW, Webb LJ and Cho M, Phys. Chem. Chem. Phys, 2016, 18, 18094–18111. [DOI] [PubMed] [Google Scholar]
- 50.Choi JH and Cho M, J. Chem. Phys, 2011, 134, 154513. [DOI] [PubMed] [Google Scholar]
- 51.Horness RE, Basom EJ, Mayer JP and Thielges MC, J. Am. Chem. Soc, 2016, 138, 1130–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marles JA, Dahesh S, Haynes J, Andrews BJ and Davidson AR, Mol. Cell, 2004, 14, 813–823. [DOI] [PubMed] [Google Scholar]
- 53.Brown T, Brown N and Stollar EJ, PLoS One, 2018, 13, e0193128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.