ABSTRACT
The JAK/STAT pathway is a conserved metazoan signaling system that transduces cues from extracellular cytokines into transcriptional changes in the nucleus. JAK/STAT signaling is best known for its roles in immunity. However, recent work has demonstrated that it also regulates critical homeostatic processes in germline and somatic stem cells, as well as regenerative processes in several tissues, including the gonad, intestine and appendages. Here, we provide an overview of JAK/STAT signaling in stem cells and regeneration, focusing on Drosophila and highlighting JAK/STAT pathway functions in proliferation, survival and cell competition that are conserved between Drosophila and vertebrates.
KEY WORDS: Drosophila, Niche, Germline stem cells, Somatic stem cells, Testis, Intestine, Intestinal stem cells, Enterocytes, Wing disc, Cytokines, Proliferation, Survival, Differentiation, Cell competition, Niche competition, Inflammation, Regeneration, Cellular plasticity, Cell reprogramming
Summary: This Primer reviews the role of JAK/STAT signaling in stem cells and regeneration in Drosophila testis, intestine and appendages, and discusses the similarities between Drosophila and vertebrates.
Introduction
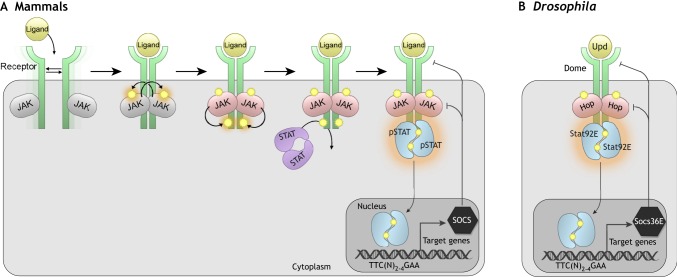
In the early 1990s, several research groups demonstrated that a family of Janus (JAK) tyrosine kinases and a family of latent cytosolic transcription factors, termed signal transducers and activator of transcription (STATs), mediated interferon signaling in cultured mammalian cells (Darnell, 1997). Subsequent work showed that numerous interleukins and growth factors also use JAKs and STATs to alter gene expression (Levy and Darnell, 2002). Since these early studies, a wealth of research on various model organisms and cell types has provided further insight into how JAKs and STATs function to translate a multitude of signals into developmental or homeostatic responses. JAK/STAT signal transduction involves the binding of extracellular ligands to transmembrane cytokine receptors, which results in the activation of cytosolic JAKs and then of STATs (Fig. 1A) (Bach et al., 1997; Darnell, 1997). Activated STAT dimers translocate to the nucleus, bind specific DNA sequences in target genes and alter gene expression (see Fig. 1 legend for details).
Fig. 1.
The JAK/STAT pathway. (A) The mammalian JAK/STAT pathway. An extracellular ligand (yellow) binds to a transmembrane cytokine receptor (green), which lacks intrinsic kinase activity and instead constitutively associates with JAKs (gray). Ligand binding to receptors causes a conformational switch, leading to JAK activation by trans-phosphorylation. The activated JAKs (pink) then phosphorylate the receptor on tyrosine residues in the cytoplasmic domain. Inactive STAT dimers (purple) are recruited to the receptor at those phospho-tyrosine sites and are phosphorylated by activated JAKs. The phosphorylated STAT (pSTAT) dimers (blue) assume an activated dimer conformation, translocate to the nucleus, bind specific DNA sequences in target genes [consensus TTC(N)2–4GAA] and alter gene expression. SOCS genes, which are targets of the JAK/STAT pathway, encode inhibitory proteins (black) that promote degradation of the cytokine receptor and JAKs, thereby providing a negative-feedback loop. Tyrosine phosphorylation events are indicated by orange halos. (B) The Drosophila JAK/STAT pathway consists of three Upd ligands, Upd, Upd2 and Upd3 (referred to here collectively as Upd), one receptor called Dome, one JAK called Hop, and one STAT, termed Stat92E. Socs36E is a Stat92E target gene that encodes a negative regulator of pathway activity.
STAT genes are widely conserved in metazoans, from the slime mold Dictyostelium discoideum, the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster to vertebrates (Hou et al., 1996; Kawata et al., 1997; Wang and Levy, 2006; Yan et al., 1996). In mammals, there are four JAK and seven STAT genes, and knockout mice have revealed expected roles in hematopoiesis and immunity, as well as unexpected roles in embryonic development (Levy, 1999). Most research on JAK/STAT signaling in non-mammalian species has been performed in Drosophila, where this pathway serves a myriad of developmental roles and cellular functions (Arbouzova and Zeidler, 2006). In Drosophila, three related ligands – Unpaired (Upd), Upd2 and Upd3 (Agaisse et al., 2003; Harrison et al., 1998; Hombria et al., 2005) – activate one receptor, which is called Domeless (Dome) (Brown et al., 2001; Chen et al., 2002). This leads to the activation of one JAK, termed Hopscotch (Hop) (Binari and Perrimon, 1994) and one STAT transcription factor, Stat92E (Hou et al., 1996; Yan et al., 1996) (Fig. 1B). Pathway activity is downregulated by Socs36E, a Suppressor of Cytokine Signaling (SOCS) protein, which is induced in a negative-feedback loop (Callus and Mathey-Prevot, 2002; Karsten et al., 2002). Core components of the Drosophila JAK/STAT pathway are homologous to interleukin 6 (IL-6), its receptor Gp130, the JAK Jak2 and STAT Stat3, which mediate inflammatory and proliferative responses in mammals (Rose-John, 2018).
JAK/STAT is one of a handful of conserved signal transduction pathways required for normal development and adult physiology, as well as for regenerative responses during infection and injury (Housden and Perrimon, 2014). In the past few years, numerous publications from many labs have revealed crucial roles for JAK/STAT signaling in conserved processes, ranging from stem cell self-renewal in homeostasis to proliferation and survival during regeneration. Additionally, JAK/STAT signaling orchestrates essential functions in cell competition and stem cell competition, which are also conserved processes. Of note, many of these findings have come from studies in Drosophila, which is amenable to powerful genetic tools. Given the relative simplicity of the pathway in flies, research on the roles of the JAK/STAT pathway in Drosophila stem cells and regeneration will likely have important ramifications for vertebrate model organisms.
Here, we review the functions of JAK/STAT signaling in stem cell biology and regeneration, focusing on three Drosophila tissues. First, we discuss how JAK/STAT signaling functions in the developing and adult testis, where cytokines constitutively produced by the stem cell niche control homeostatic functions such as self-renewal as well as regeneration after genetic ablation or irradiation. Second, we review the roles of the JAK/STAT pathway in the adult intestine, where cytokines produced by differentiated cells in response to infection or damage non-autonomously stimulate the proliferation and differentiation of tissue stem cells, thereby renewing the gut epithelium. Third, we discuss roles of JAK/STAT signaling in regenerating appendages, where cytokines produced after damage regulate cell division, survival and cellular plasticity. Finally, we discuss the parallels in JAK/STAT pathway function in stem cells and regeneration between Drosophila and vertebrates.
JAK/STAT signaling in stem cell homeostasis and regeneration in the Drosophila testis
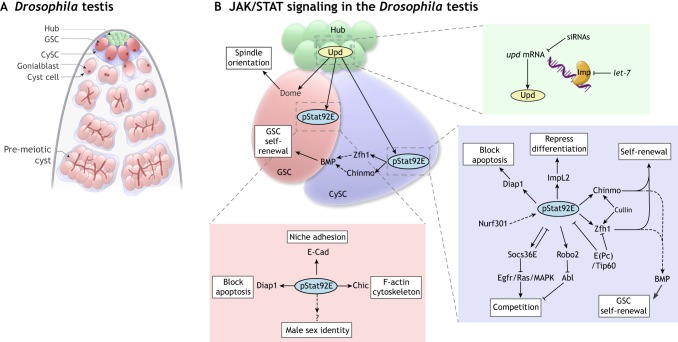
In the Drosophila testis, a group of quiescent somatic niche cells supports two resident stem cell populations (Fig. 2A): germline stem cells (GSCs) and somatic cyst stem cells (CySCs) (reviewed by Greenspan et al., 2015). The niche secretes short-range signals that promote the proliferation of these resident stem cells (Fig. 2B). GSCs proliferate and divide with oriented mitosis to produce a GSC daughter that remains in contact with the niche and another daughter that is displaced from the niche and differentiates into a spermatogonium and ultimately into individual spermatids (Fuller, 1998; Yamashita et al., 2003). CySCs divide to maintain the stem cell pool and to produce offspring that function as critical somatic support cells for the germline, akin to Sertoli cells in the mammalian testis (Gonczy and DiNardo, 1996; Oatley and Brinster, 2012). CySCs also provide essential support to GSCs as an extended niche (Leatherman and Dinardo, 2010). As we discuss below, studies have shown that JAK/STAT signaling regulates both GSCs and CySCs, from their initial development through to their functioning in the adult testis.
Fig. 2.
JAK/STAT signaling in homeostasis and regeneration in the Drosophila testis. (A) Schematic of the adult Drosophila testis. A group of quiescent somatic cells (green) forms the niche (also referred to as the ‘hub’) and secretes self-renewal cues for resident stem cells. GSCs (dark pink) and CySCs (dark blue) adhere to the niche. GSCs divide with oriented division to produce a gonialblast (light pink) that undergoes transit-amplifying divisions, resulting in a pre-meiotic cyst that gives rise to spermatids. The CySC divides to produce cyst cells (light blue) that become quiescent and ensheath the gonialblast. Two cyst cells continue to ensheath the associated spermatogonial cyst throughout spermatogenesis. (B) JAK/STAT signaling influences multiple processes in the testis; the insets detail events occurring in niche cells (green box), GSCs (pink box) and CySCs (blue box). Niche cells produce the cytokine Upd, which activates Stat92E (pStat92E) in GSCs and CySCs. Through its targets Zfh1 and Chinmo, pStat92E endows CySCs with the ability to support GSCs through BMP production. Upd can signal to Dome but not to Hop or Stat92E to promote spindle orientation during GSC division. In niche cells, upd mRNA is bound by Imp, which protects it from siRNAs, while Imp mRNA is a target of the let-7 microRNA. In GSCs, pStat92E regulates niche adhesion through E-Cad and the cytoskeleton via Chic. pStat92E also promotes male germline sexual identity through unknown targets and protects GSCs from irradiation-induced apoptosis through Diap1. In CySCs, pStat92E protects cells from irradiation-induced death through Diap1 and represses somatic differentiation through ImpL2. Chinmo and Zfh1 promote CySC self-renewal, and both are positively regulated by Cullin. pStat92E and Zfh1 are negatively regulated by E(Pc)/Tip60. Niche competition is controlled by the JAK/STAT target Socs36E, which represses Egfr/Ras/MAPK signaling. Niche competition is also regulated by another potential Stat92E target, Robo2, which represses the kinase Abl. pStat92E is positively regulated by Nurf301.
Development of the testis involves the coalescence of primordial germ cells (PGCs), which become GSCs, and somatic gonadal precursors (SGPs), which give rise to niche cells and CySCs. Autonomous JAK/STAT pathway activation has been reported to regulate the migration of PGCs to the developing gonad (Brown et al., 2006). Once SGPs become niche cells, they induce expression of the cytokine Upd (Kitadate and Kobayashi, 2010; Okegbe and DiNardo, 2011). The reception of the Upd signal in PGCs leads to Stat92E-dependent acquisition of male sexual identity (Sheng et al., 2009b; Wawersik et al., 2005). The targets that regulate male germline sex are not known, but one possibility is the H3K4me2 reader PHD finger protein 7 (Phf7), which is induced in early male GSCs and required for their maintenance (Yang et al., 2012). JAK/STAT signaling also promotes the specification of CySCs from SGPs (Sinden et al., 2012).
upd continues to be produced by niche cells in the adult testis but declines with age (Kiger et al., 2001; Toledano et al., 2012; Tulina and Matunis, 2001). This drop in Upd is mediated by a decrease in IGF-II mRNA-binding protein (Imp), which protects the upd transcript from small interfering RNAs (Fig. 2B). Imp levels decline during aging due to increased expression of the heterochronic microRNA let-7, which targets Imp. The reduction in Upd at least partially accounts for the age-related decline of GSCs and spermatogenesis in older males (Boyle et al., 2007; Toledano et al., 2012). Males mutant for another upd gene (upd3) become infertile at a much younger age than wild-type controls (Wang et al., 2014). Little is known about upd3 in the testis, but it is tempting to speculate that, like upd, it is also expressed in niche cells and is subjected to a similar age-related decline.
Reception of the Upd signal is required autonomously in GSCs and CySCs for their residence in the niche (Issigonis et al., 2009; Kiger et al., 2001; Leatherman and Dinardo, 2008; Tulina and Matunis, 2001). In GSCs, JAK/STAT signaling does not induce self-renewal per se but regulates adhesion to the niche via the control of E-Cadherin (E-Cad) levels, thus tethering these cells to the source of self-renewal cues (Leatherman and Dinardo, 2010). In addition, Stat92E influences F-actin dynamics in GSCs through the Drosophila Profilin homolog Chickadee (Chic), and this may affect GSC-niche attachment (Shields et al., 2014). Stat92E levels in GSCs are regulated indirectly by the nucleosome-remodeling factor (NURF) complex and the histone demethylase Little imaginal discs (Lid) (Cherry and Matunis, 2010; Tarayrah et al., 2015). A recent study reported that Upd and its receptor Dome – but not Hop and Stat92E – control spindle orientation during GSC division, ensuring that one GSC daughter remains attached to the niche (Chen et al., 2018). How this partition of pathway signaling to include ligand and receptor but not kinase and transcription factor occurs in GSCs is not yet understood.
The GSC pool exhibits considerable plasticity, and this is dependent on JAK/STAT signaling. Indeed, dedifferentiation of spermatogonial cells into GSCs was first shown using a Stat92E temperature-sensitive allele; upon transient loss of Stat92E at the restrictive temperature, all GSCs are lost from the niche but, after returning to the permissive temperature, spermatogonia fragment and dedifferentiate into functional stem cells (Brawley and Matunis, 2004). Subsequent work showed that inhibiting Stat92E activity in dedifferentiating spermatogonia by mis-expressing Socs36E significantly decreases regeneration of the testis, presumably because these spermatogonia cannot effectively receive the Upd signal at the niche (Sheng et al., 2009a). Furthermore, although X-rays cause temporary infertility in Drosophila males due to the death of spermatogonial cells, GSCs (and CySCs) are resistant to death by ionizing radiation as a result of Stat92E-dependent regulation of the survival factor Death-associated inhibitor of apoptosis 1 (Diap1). As a consequence, the testis regenerates in about 15 days, and this is followed by the return of fertility (Betz et al., 2008; Hasan et al., 2015; Welshons and Russel, 1957).
In CySCs, JAK/STAT target genes regulate several crucial homeostatic activities, including self-renewal, support of GSCs, stem cell competition and inhibition of differentiation. Two key downstream Stat92E effectors in CySCs are zinc finger homeodomain 1 (zfh1) and chronologically inappropriate morphogenesis (chinmo), both of which encode transcriptional repressors. zfh1 and chinmo are upregulated by ectopic JAK/STAT signaling, suggesting they are direct target genes (Flaherty et al., 2010, 2009; Leatherman and Dinardo, 2008; Terry et al., 2006). Zfh1 and Chinmo are also positively regulated by Cullin-RING ubiquitin ligases, although the mechanism by which this occurs is not clear (Qian et al., 2015), whereas zfh1 and Stat92E are negatively regulated by the Enhancer of Polycomb (E(Pc))/Tip60 acetyl transferase complex (Feng et al., 2017).
Both Zfh1 and Chinmo are required autonomously for niche residence of CySCs; CySCs lacking either gene rapidly differentiate (Flaherty et al., 2010; Leatherman and Dinardo, 2008). The role of CySCs in supporting GSCs is also endowed by JAK/STAT signaling through zfh1 and chinmo, but not by other CySC self-renewal pathways (Albert et al., 2018; Amoyel et al., 2014a, 2016a, 2013; Flaherty et al., 2010; Leatherman and Dinardo, 2008, 2010). Ectopic activation of Stat92E or mis-expression of zfh1 or chinmo in CySCs is sufficient to expand both CySC and GSC pools. Mechanistically, Stat92E acts via Zfh1 and Chinmo to ultimately upregulate CySC secretion of bone morphogenetic proteins (BMPs), which are essential GSC self-renewal cues (Leatherman and Dinardo, 2010). This JAK/STAT-dependent production of BMPs by CySCs allows GSCs to proliferate and remain in an undifferentiated state even when they reside far from the endogenous niche.
CySCs also produce the secreted insulin inhibitor ImpL2, the Drosophila homolog of IGFBP7, in response to JAK/STAT signaling (Amoyel et al., 2016b; Terry et al., 2006). In the testis, local insulin cues induce Phosphoinositide 3-kinase/Tor activity in early CySC daughters, inducing their differentiation as well as non-autonomously promoting the differentiation of ensheathed spermatogonia (Amoyel et al., 2016b). Insulin action in CySCs is hindered within the testis stem cell niche by ImpL2. Thus, niche signals repress somatic differentiation in CySCs at least in part thorough Stat92E-dependent induction of ImpL2.
In numerous tissues and organisms, stem cells compete homeostatically for limited niche resources (Simons and Clevers, 2011). In the Drosophila testis, two types of stem cell competition (also termed ‘niche’ competition) have been reported: intra-lineage CySC-CySC competition and inter-lineage CySC-GSC competition. Clonal analyses of CySCs have revealed that they conform to neutral drift dynamics, a process in which stem cells are stochastically lost and replaced (Amoyel et al., 2014b; Klein and Simons, 2011). A CySC can gain clonal dominance if it obtains a competitive advantage, and this clone and its descendants can displace wild-type CySCs, which then differentiate. Once the CySC lineage is dominated by the competitive clone, CySCs can exert pressure on GSCs, leading to their ejection from the niche and subsequent differentiation. Stem cell competition in the testis was initially revealed by analyses of CySCs lacking the induced negative-feedback inhibitor Socs36E; these CySCs outcompete neighboring wild-type CySCs and then begin to force wild-type GSCs out of the niche (Amoyel et al., 2016a; Issigonis et al., 2009; Singh et al., 2010). One model for the increased competitiveness of Socs36E-mutant CySCs suggests that autonomous increases in JAK/STAT signaling cause Stat92E-dependent upregulation of integrin adhesion (Issigonis et al., 2009). However, this model is at odds with the aforementioned observation that increased JAK/STAT signaling in CySCs endows them with the ability to support (not outcompete) GSCs (Leatherman and Dinardo, 2010). Furthermore, it has been found that stem cell competition does not occur after clonal upregulation of JAK/STAT activity or of integrin adhesion (Amoyel et al., 2014b). Instead, the competitiveness of Socs36E-mutant CySCs was shown to result from increased Egfr/Ras/MAPK signaling (Amoyel et al., 2016a). Taken together, these results indicate that Socs36E is induced by JAK/STAT signaling in response to niche signals, and that Socs36E then dampens both JAK/STAT and MAPK signals to restrain competitive interactions. Socs36E-dependent CySC-GSC niche competition is also regulated by the NURF complex (Cherry and Matunis, 2010). Global reduction of Nurf301 partially rescues the loss of GSCs caused by competitive Socs36E-mutant CySCs, suggesting that the NURF complex positively regulates JAK/STAT signaling in CySCs. Finally, CySC-CySC competition is regulated by the axon guidance receptor Roundabout 2 (Robo2), another potential JAK/STAT target (Stine et al., 2014). Robo2 negatively regulates the Abelson (Abl) tyrosine kinase. CySCs mutant for Abl out-compete wild-type CySCs, but not wild-type GSCs, and their competitive behavior is inhibited by depletion of E-Cad.
Overall, this body of work demonstrates that JAK/STAT ligands from a dedicated stem cell niche promote proliferation and prevent differentiation of resident stem cells. Additionally, JAK/STAT signaling in GSCs is required for their adhesion to the niche and for optimal regeneration of the germline after ablation. CySCs are a crucial source of support for GSCs, and this property is endowed uniquely by JAK/STAT activity. Finally, in CySCs, the JAK/STAT target Socs36E restrains competitive interactions among CySCs and between CySCs and GSCs.
But how conserved are these functions for JAK/STAT signaling? In the mouse testis, Stat3 is not required for the self-renewal of spermatogonial stem cells but rather is needed for spermatogonial differentiation (Oatley et al., 2010). This suggests that the self-renewal function of STAT proteins in male GSCs may not be conserved. Furthermore, although transient-amplifying spermatogonia in the mouse testis can fragment and contribute to the spermatogonial stem cell pool (Nakagawa et al., 2010), whether JAK/STAT signaling is involved in this process during homeostasis or stress is not known. It has been shown that activated Stat3 in the mouse testis is concentrated in somatic Sertoli cells that support undifferentiated spermatogonial cells, consistent with the expression pattern of activated Stat92E in the Drosophila testis (Nagasawa et al., 2018). What activates Stat3 and its specific roles in Sertoli cells are not entirely clear. It is possible that the IL-6 family member leukemic inhibitory factor (LIF) may play a role. LIF activates Stat3 through the LIFR receptor (LIFR) binding chain, the Gp130 receptor signaling chain and Jak2 (Rose-John, 2018). Partial depletion of Lifr in Sertoli cells causes a reduction in adult testis volume and a disruption of the seminiferous epithelium (Curley et al., 2018), but the mechanistic details by which LIF regulates Sertoli cell function requires further investigation. Finally, LIF is crucial for the robust long-term self-renewal of pluripotent embryonic stem cells (ESCs) from various mouse strains and other rodent species (Buehr et al., 2008; Nichols et al., 2009; Smith et al., 1988). LIF promotes ESC proliferation and inhibits ESC differentiation (Ying and Smith, 2017), similar to the roles of Stat92E in stem cells in the Drosophila testis.
JAK/STAT signaling in homeostasis and regeneration in the Drosophila gut
The Drosophila digestive tract serves numerous functions, including digestion, absorption of nutrients and defense against pathogens (Lemaitre and Miguel-Aliaga, 2013). The midgut, which is the functional equivalent of the mammalian small intestine, is surrounded by visceral muscle (Fig. 3) and is maintained by a population of intestinal stem cells (ISCs) that give rise to all cell types within the midgut epithelium. Under normal conditions (Fig. 3A), ISCs divide asymmetrically to produce an ISC and an enteroblast (EB) that exits the cell cycle and differentiates directly into an absorptive enterocyte (EC). ISCs and EBs are collectively referred to as progenitor cells. Notch signaling in EBs then induces differentiation into ECs; accordingly, loss of Notch from the ISC lineage leads to tumors comprised primarily of ISCs (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). During homeostasis, ISCs divide slowly and the epithelium turns over every ∼2 weeks (Jiang et al., 2009; Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). However, following infection or damage the rate of ISC proliferation is substantially increased (Fig. 3B). During homeostasis, the proliferation of ISCs is regulated by several pathways, including the Egfr/Ras/MAPK pathway (Biteau and Jasper, 2011; Buchon et al., 2010; Jiang et al., 2011; Xu et al., 2011) and, as we discuss below, the JAK/STAT pathway.
Fig. 3.
JAK/STAT signaling in homeostasis and regeneration in the Drosophila intestine. (A) The intestinal epithelium resides on basal membrane and is surrounded by visceral muscle. Under normal conditions, intestinal stem cells (ISCs) within the epithelium divide slowly as a result of Egfr/Ras/MAPK signaling. ISCs divide asymmetrically to produce an ISC and an enteroblast (EB). Upd2 and Upd3 produced by different sources promote EB differentiation into enterocytes (ECs) through the activation of Stat92E (pStat92E) and its target Sox21a. (B) Upon damage or infection, several pathways, including the p38, JNK, Hpo/Wts and Msn/Wts pathways, activate AP-1 and Yki/Sd transcriptional complexes, which induce the upd3 gene. This results in ECs secreting high levels of Upd3, which acts on several cell types to promote regeneration. In visceral muscle, Upd3 activates Stat92E (pStat92E), which induces the expression of the EGF ligand Vein (Vn). In turn, Vn triggers Egfr/Ras/MAPK signaling in ISCs to promote their proliferation. In ISCs, Upd3 induces the expression of another EGF ligand, Spitz (Spi), that autonomously promotes ISC division through Egfr/Ras/MAPK. Autonomous JAK/STAT signaling in ISCs can also promote their proliferation (not shown). Upd3-mediated activation of Stat92E in EBs causes their rapid differentiation to ECs. After septic injury in the body cavity, macrophages produce Upd cytokines, and this remotely induces Stat92E activation in visceral muscle and ISCs, leading to autonomous and non-autonomous ISC proliferation and EB differentiation, as described above. Additionally, sustained JAK/STAT signaling in ISCs contributes to gut dysbiosis in aging intestines. Finally, JAK/STAT signaling in ISCs regulates the proliferation of ‘winning’ cells via the process of cell competition: the winners can be neoplastic cells outcompeting wild-type cells or wild-type cells eliminating suboptimal cells.
Under normal conditions, JAK/STAT ligands are produced at variable levels by several intestinal cell types. There is agreement that upd2 is expressed by ISCs/EBs/ECs and upd3 by ECs (Jiang et al., 2009; Liu et al., 2010; Osman et al., 2012; Zhou et al., 2013). Lineage tracing has shown that ISCs unable to transduce JAK/STAT signals can proliferate, indicating that this pathway is not required for ISC self-renewal under steady-state conditions (Beebe et al., 2010; Jiang et al., 2009). Under normal conditions, JAK/STAT activity is highest in EBs and, consistent with this, EBs deficient for Stat92E cannot differentiate into mature gut cells, leading to the formation of tumors composed of progenitor cells (Beebe et al., 2010; Jiang et al., 2009). Although Notch and Stat92E are required for EB differentiation into ECs, epistasis experiments to determine their relationship have yielded conflicting results. One group reported that the defective differentiation of Stat92E-mutant clones cannot be rescued by mis-expressing an activated form of Notch, suggesting that these pathways function in parallel (Beebe et al., 2010). However, another group reported the opposite (Liu et al., 2010). More recent work has shown that the transcription factor Sox21a acts downstream of Stat92E in EB differentiation (Zhai et al., 2017, 2015). Sox21a behaves as a positively regulated JAK/STAT target and, similar to loss of Stat92E, clonal or lineage-wide inactivation of Sox21a causes tumors composed of progenitor cells. Furthermore, ectopic expression of Sox21a rescues the differentiation defect in Stat92E-mutant clones and suppresses the tumors caused by Stat92E inactivation. These results have led to a model in which Stat92E acts upstream of Sox21a to control EC differentiation (Zhai et al., 2017).
The rate of ISC proliferation is significantly higher when homeostasis is disrupted, e.g. upon bacterial infection, or physical or chemical injury (Biteau et al., 2011). These insults result in the production of inflammatory cytokines – particularly Upd3 – by intestinal epithelial cells that drive epithelial regeneration by increasing ISC proliferation (Buchon et al., 2009a,b; Cronin et al., 2009; Jiang et al., 2009; Osman et al., 2012; Zhou et al., 2013). Recent work has identified a 4 kb enhancer that induces upd3 expression in EBs/ECs during bacterial infection or chemical injury (Houtz et al., 2017; Jiang et al., 2011; Zhou et al., 2013). During regeneration, this enhancer is upregulated by Yorkie (Yki)/Scalloped (Sd) and AP-1 (D-Jun and D-Fos) transcriptional complexes in response to several stress signals, including Jun N-terminal kinase (JNK), p38, Hippo (Hpo)/Warts (Wts) and Misshapen (Msn)/Wts (Houtz et al., 2017; Jiang et al., 2011; Karpowicz et al., 2010; Li et al., 2014; Ren et al., 2010; Shaw et al., 2010; Staley and Irvine, 2010).
The Upd ligands produced by stressed EBs and ECs activate JAK/STAT signaling in intestinal progenitor cells and in visceral muscle (Buchon et al., 2009b; Jiang et al., 2009; Zhou et al., 2013). Autonomous activation of the JAK/STAT pathway is sufficient to induce ISC proliferation and intestinal turnover (Beebe et al., 2010; Buchon et al., 2009a, 2010; Jiang et al., 2009; Lin et al., 2010). Stat92E target genes that are expressed in progenitors following damage or infection include windpipe, which encodes a leucine-rich repeat transmembrane protein that dampens pathway output by promoting degradation of the receptor Dome, thereby limiting ISC proliferation (Ren et al., 2015). The transcription factor Myc and the Argonaute protein Piwi are induced by pathway activation and are required for intestinal turnover in response to damage, but they are also upregulated by other regenerative pathways (Ren et al., 2013; Sousa-Victor et al., 2017).
Although autonomous Stat92E activation in ISCs is sufficient to induce intestinal renewal, work has suggested that the role of Stat92E activation in ISC proliferation is more complex and involves a feed-forward relay of EGF ligands. JAK/STAT signaling in progenitors and visceral muscle induces the production of the EGF ligands Spitz (Spi) and Vein (Vn), respectively. In autocrine and paracrine manners, these EGFs activate Egfr/Ras/MAPK in ISCs to accelerate proliferation and induce epithelial renewal (Biteau and Jasper, 2011; Buchon et al., 2010; Jiang et al., 2011; Zhou et al., 2013). Indeed, compensatory proliferation by ectopic Upd is blocked when ISCs are devoid of Egfr/Ras/MAPK signaling (Buchon et al., 2010; Jiang et al., 2011). These results indicate that JAK/STAT-dependent intestinal regeneration requires the upregulation of EGF ligands, which trigger MAPK-dependent proliferation in ISCs. Swift EB differentiation into ECs is also needed for epithelial renewal during infection/injury, and this occurs by Upd3-dependent activation of the JAK/STAT pathway in EBs (Buchon et al., 2009a; Jiang et al., 2009; Zhou et al., 2013). Interestingly, ISCs can be induced to proliferate as part of a systemic response to infection or wounding (Chakrabarti et al., 2016). After wounding and subsequent bacterial infection, for example, macrophages in the body cavity secrete Upd cytokines, resulting in remote JAK/STAT activation in intestinal progenitor cells and visceral muscle, subsequent induction of EGF ligands, and intestinal renewal. This response is necessary for survival from this type of injury.
JAK/STAT signaling has also been implicated in aging of the Drosophila intestine. Under homeostatic conditions, the intestinal epithelium of older flies deteriorates as a result of altered bacterial load (termed dysbiosis) and the subsequent triggering of JAK/STAT and other regenerative pathways (Biteau et al., 2008; Buchon et al., 2009b; Choi et al., 2008; Park et al., 2009). This deterioration is characterized by aberrant EC differentiation and a 10-fold increase in ISC proliferation, which impairs metabolism and shortens lifespan (Biteau et al., 2010). The excessive ISC proliferation in aged flies is significantly reduced in upd2 or upd3 single mutants, or in upd2, upd3 double mutants (Osman et al., 2012). This suggests that the protective regenerative response to damage or infection becomes dysregulated during aging and is mediated, at least in part, through sustained JAK/STAT signaling.
Upd cytokines also fuel the growth of intestinal neoplasia (Cordero et al., 2012; Patel et al., 2015). Aging ISCs sustain sporadic mutations, and ∼10% of aged male flies have inactivating mutations in the X-linked gene Notch, causing neoplastic ISC growths (Siudeja et al., 2015). When large Notch-mutant ISC tumors are induced, they compete against wild-type ECs for space in the epithelium (Patel et al., 2015), as has been observed for other neoplasia in the Drosophila gut (Suijkerbuijk et al., 2016). As the outcompeted ECs detach from the epithelium, they activate JNK and Yki, and secrete Upd2 and Upd3, and these cytokines accelerate the growth of the Notch-mutant tumors (Patel et al., 2015). JAK/STAT signaling also regulates another type of cell competition in the intestine in which wild-type epithelial cells compete against viable but less robust cells (e.g. those with reduced ribosome function). The ‘losing’ cells activate JNK and secrete Upd3, which non-autonomously increases ISC proliferation, thereby augmenting the growth of fitter wild-type cells (Kolahgar et al., 2015). In both competitive scenarios, the ‘losing’ cell secretes Upd cytokines, which increases the growth of the ‘winning’ cells.
In sum, JAK/STAT signaling is essential for ISC differentiation in the Drosophila gut and for intestinal regeneration after insults and infection. Damaged/stressed ECs secrete the inflammatory cytokine Upd3, which acts on neighboring cell types to induce production of EGF ligands that trigger Egfr/Ras/MAPK in ISCs. Although ectopic activation of JAK/STAT signaling in ISCs can induce proliferation, the feed-forward relay of EGF ligands is necessary for optimal regenerative responses. JAK/STAT signaling also controls competitive interactions between different cell types in the gut. Many of these findings are similar to those observed in mammals. For example, mouse ISCs also increase proliferation after damage or infection in response to IL-6, which promotes intestinal regeneration (McConnell et al., 2008; Rigby et al., 2007). Interestingly, a causal connection between inflammation and cancer in the intestine is well established, and IL-6 family members play important roles in colon cancer progression by regulating cell proliferation and cell survival (Taniguchi and Karin, 2014; West et al., 2015). Individuals with inflammatory bowel diseases are more likely to develop colorectal cancer, and studies have emphasized the role of IL-6, Gp130 and Stat3 in these cancers (Bollrath et al., 2009; Choi et al., 2017; Grivennikov et al., 2009; Putoczki et al., 2013). Competitive interactions between ISCs, similar to those observed in the midgut, also occur in the mouse intestine, and the success of dominant clones underlies tumor initiation when the intestine is damaged (Vermeulen et al., 2013). However, what roles cytokines and JAK/STAT signaling serve during this competitive process are not known.
JAK/STAT signaling in regeneration of the Drosophila wing disc
The Drosophila wing imaginal disc is a larval tissue comprising pouch, hinge and notum regions that gives rise to the adult wing blade, wing hinge and body wall, respectively (Cohen, 1993). Wing discs exhibit plasticity and can be induced to regenerate after mechanical or genetic ablation (reviewed by Hariharan and Serras, 2017; Worley et al., 2012). Mechanical ablation involves cutting discs either in situ or ex vivo followed by culturing them in the abdomen of female hosts (Diaz-Garcia and Baonza, 2013). Genetic ablation techniques involve binary expression systems and thermo-sensitive repressors that can be used to deliver inducers of apoptosis, such as Reaper (Rpr), Head involution defective (Hid) or the JNK pathway ligand Eiger (Egr, which is homologous to TNFα), to specific subsets of wing cells in a time-controlled manner (Bergantinos et al., 2010; Herrera et al., 2013; Smith-Bolton et al., 2009). Regenerative processes in the wing disc can also be observed after widespread cell death following ionizing radiation (Jaklevic and Su, 2004). Using these various approaches, studies have shown that the JAK/STAT pathway plays a central role in controlling the response to tissue ablation and mediating wing disc regeneration.
Following ablation, apoptosis triggers the production of extracellular reactive oxygen species (ROS), which are detected both in the ablated cells and the surviving cells (Fig. 4) (Santabárbara-Ruiz et al., 2015). This burst of ROS release by dying cells activates autonomous JNK signaling to ensure commitment to death through a positive-feedback loop between JNK and hid or rpr (Santabárbara-Ruiz et al., 2015; Shlevkov and Morata, 2012). ROS production by dying cells also non-autonomously activates JNK and p38 in nearby surviving cells (Santabárbara-Ruiz et al., 2015). JNK activation may also be propagated non-autonomously by Egr/TNFα produced downstream of JNK in apoptotic cells (Pérez-Garijo et al., 2013). Overall, this JNK signaling is essential for appendage regeneration, as indicated by lineage-tracing studies showing that JNK-activated cells comprise a majority of the regenerated tissue (Bosch et al., 2008; Katsuyama et al., 2015).
Fig. 4.
JAK/STAT signaling during regeneration of the Drosophila wing disc. After genetic or mechanical ablation, ROS are produced by damaged cells (red), leading to autonomous JNK activation, which ensures commitment to apoptosis through a positive-feedback loop between JNK and Hid or Rpr. ROS produced by damaged cells also non-autonomously activate JNK and p38 in neighboring healthy, surviving cells (blue). JNK activation in dying cells may be propagated non-autonomously to surviving cells by Egr/TNFα. In the surviving cells, JNK signaling can induce proliferation. Additionally, JNK and p38 independently induce the expression of Upd, which subsequently activates Stat92E (pStat92E) in an autocrine and paracrine manner. pStat92E then increases the proliferation of surviving cells and inhibits apoptosis. pStat92E may also upregulate Dilp8 (dashed line), thereby promoting developmental delay. pStat92E in surviving cells is required for their plasticity and the ability to be ‘reprogrammed’ to other wing lineages. CtBP strongly inhibits JNK signaling and weakly inhibits JAK/STAT signaling cell-autonomously. Finally, pStat92E together with JNK activity downstream of Egr can trigger the formation of an ectopic wing blade (‘mis-specification’).
Activation of JNK and p38 in surviving cells induces Upd ligands, particularly Upd3 (Ahmed-de-Prado et al., 2018; Katsuyama et al., 2015; Klebes et al., 2005; La Fortezza et al., 2016; Pastor-Pareja et al., 2008; Santabárbara-Ruiz et al., 2015; Worley et al., 2018). Notably, one study found that the same 4 kb upd3 enhancer induced in the Drosophila gut in response to damage was also induced in the wing disc during regeneration (Worley et al., 2018), indicating that Upd production in response to inflammation is a common feature in intestinal and appendage regeneration. The Upd secreted by surviving cells acts in autocrine and paracrine manners (Ahmed-de-Prado et al., 2018; Katsuyama et al., 2015; La Fortezza et al., 2016; Santabárbara-Ruiz et al., 2015; Worley et al., 2018). In general, the induction of JAK/STAT signaling leads to proliferation in the regenerating tissue, consistent with established roles of Upd as a noted mitogen (Bach et al., 2003; Katsuyama et al., 2015; Tsai and Sun, 2004). Specifically, proliferation in response to ablation was shown to be reduced in hop-mutant discs (Katsuyama et al., 2015). However, another study reported that JAK/STAT signaling does not promote proliferation in regenerating tissue but instead promotes cell survival (La Fortezza et al., 2016). In fact, it was found that mitosis was increased in ablated hop heterozygous mutants. However, this study also reported that cell division in surviving cells within the wing disc is not altered in Stat92E heterozygous backgrounds (La Fortezza et al., 2016), despite the fact that Stat92E heterozygosity significantly inhibits cell proliferation in other growth contexts (Amoyel et al., 2014a). Therefore, the connection between JAK/STAT signaling and proliferation after ablation warrants further attention. Interestingly, JAK/STAT activity is required for proliferation of surviving cells during cricket limb regeneration (Bando et al., 2013).
Some studies have shown that Stat92E activation increases survival in regenerating cells (Ahmed-de-Prado et al., 2018; La Fortezza et al., 2016; Verghese and Su, 2016), which is consistent with known roles of the pathway. Cells with elevated JAK/STAT activity are at a competitive advantage: they compete with and eliminate nearby wild-type healthy cells (Rodrigues et al., 2012). During development, Stat92E signaling becomes restricted to hinge cells (Ayala-Camargo et al., 2013; Hatini et al., 2013; Johnstone et al., 2013), and the increased JAK/STAT activity in these cells makes them resistant to death by X-rays (Verghese and Su, 2016). The anti-apoptotic factor Diap1 acts downstream of Stat92E in maintaining the viability of posterior wing cells during development (Recasens-Alvarez et al., 2017). It has been shown that a diap1 regulatory element containing STAT binding sites can be upregulated by ectopic Stat92E in the wing disc, suggesting direct transcriptional control (Betz et al., 2008). However, hinge cells do not upregulate the diap1 gene after X-ray, indicating that other Stat92E targets are responsible for resistance to radiation (Verghese and Su, 2016). One possibility is zinc finger homeodomain 2 (zfh2), which is positively-regulated by Stat92E in hinge cells (Ayala-Camargo et al., 2013). Zfh2 is upregulated after genetic ablation and can repress JNK signaling and hid, thereby promoting the survival of JNK-activated cells (La Fortezza et al., 2016). These results were interpreted as JAK/STAT signaling downregulating JNK activity, which was also reported by another group (Ahmed-de-Prado et al., 2018). However, a separate study has suggested that JNK signaling does not increase in the absence of JAK/STAT in regenerating tissue (Katsuyama et al., 2015), so the precise links between JAK/STAT and JNK pathways in this context remain unclear.
JAK/STAT signaling in the regenerating wing disc may also have systemic consequences through upregulation of the Drosophila Insulin-like peptide 8 (Dilp8; Ilp8 – FlyBase). Dilp8 is produced by imaginal disc cells following mechanical or genetic ablation, and acts at a distance on neuroendocrine cells in the brain to delay the larval-to-pupal transition (Colombani et al., 2012; Garelli et al., 2012; Harris et al., 2016; Katsuyama et al., 2015; La Fortezza et al., 2016). It was reported that dilp8 induction is dependent on JAK/STAT activity and does not occur in wounded hop-mutant discs (Katsuyama et al., 2015). However, another study concluded that dilp8 is not transcriptionally regulated by JAK/STAT but rather by JNK signaling (La Fortezza et al., 2016). This latter result is consistent with other reports that dilp8 is a JNK target gene (Colombani et al., 2012; Harris et al., 2016).
During regenerative responses, surviving cells and their descendants often change their cell fate or identity to reconstruct the missing tissue/appendage. Imaginal disc cells possess such plasticity and can adopt other fates (Hariharan and Serras, 2017; Worley et al., 2012). For example, after ablation of the wing pouch, a large fraction of cells in the reconstructed pouch derive from hinge cells located outside of the ablation domain (Herrera et al., 2013; Smith-Bolton et al., 2009). After irradiation, hinge cells autonomously require Stat92E to participate in pouch reconstruction (Verghese and Su, 2016), indicating a crucial function of JAK/STAT signaling in cell reprogramming.
Regenerating organs need to have mechanisms to prevent aberrant reconstruction of the tissue. Imaginal discs have so-called ‘weak points’ where phenomena such as leg-to-wing trans-determination occur at low but reproducible rates (Maves and Schubiger, 1995; Schubiger, 1971). upd is strongly induced during this transdetermination event (Klebes et al., 2005), but the functional role of the JAK/STAT pathway in this process is not clear and warrants further investigation. Two recent papers have characterized one weak point in the dorsal posterior region of the wing disc where cells fated to give rise to the notum form ectopic wings after genetic ablation or irradiation (Verghese and Su, 2017; Worley et al., 2018). Although no ectopic wings are observed in wild-type regenerating wing discs after Egr-induced ablation, discs that are heterozygous for C-terminal binding protein (CtBP) display ectopic wings (in 20-40% of discs) emanating from the weak point (Worley et al., 2018). Based on this, it was suggested that damage to the normal pouch leads to the secretion of both Egr and Upd ligands that travel over many cells diameters and act on cells at the weak point. As CtBP autonomously represses JNK activity and, more weakly, JAK/STAT activity, cells at the weak point in a CtBP/+ background are more responsive to ectopic Egr and Upd (Worley et al., 2018). As such, Upd activates Stat92E in weak point cells, which represses notum identity genes and induces hinge fate (Worley et al., 2018), consistent with the developmental role of Stat92E (Ayala-Camargo et al., 2013; Hatini et al., 2013). In addition, Egr induces JNK signaling in weak point cells, which then upregulate wingless (wg) that promotes pouch identity in the ectopic wing (Harris et al., 2016; Worley et al., 2018). The formation of an ectopic appendage from the weak point has also been observed in ∼20% of regenerating wing discs after irradiation (Verghese and Su, 2017). Although the mechanisms of ectopic appendage formation in this context are less clear, it has been shown that Stat92E and Wg activity in weak point cells is also required, suggesting that similar mechanisms are at play (Verghese and Su, 2017).
Collectively, these studies have revealed that stress pathways in surviving cells induce the expression of Upd cytokines that regulate crucial parameters in regeneration. Upd ligands act non-autonomously to promote proliferation, inhibit apoptosis and mediate reprogramming of surviving cells. Additionally, these potent signals need to be constrained to prevent induction of an ectopic appendage after ablation or irradiation.
Parallels in JAK/STAT signaling across tissues and across species
Taken together, these studies reveal proliferation, anti-apoptosis, regeneration, competition and positive regulation of Egfr/Ras/MAPK signaling as common functions of the Drosophila JAK/STAT pathway. Upd ligands act as mitogens for several stem cell populations, including ovarian follicle stem cells (FSCs) and neural stem cells, as well as CySCs and ISCs (Vied et al., 2012; Yasugi et al., 2008). JAK/STAT signaling induces expression of the anti-apoptotic factor diap1, which regulates cell survival of wing disc cells during development and of GSCs and CySCs after irradiation (Betz et al., 2008; Hasan et al., 2015; Recasens-Alvarez et al., 2017). This is reminiscent of the STAT-dependent transcriptional induction of pro-survival Bcl-2 family members in mouse cell lines (Catlett-Falcone et al., 1999; Fujio et al., 1997). Indeed, targeted disruption of JAK/STAT signaling in vivo in mice leads to reduced proliferation and decreased survival of multiple cell lineages, including the inner cell mass, and neuronal, hematopoietic and cardiac cells (Do et al., 2013; Kleppe et al., 2017; Levy and Lee, 2002; McLemore et al., 2001; Onishi and Zandstra, 2015; Takeda et al., 1997; Yoshida et al., 1996).
These studies also reveal that cytokine production and subsequent JAK/STAT activation represent a general mechanism for tissue regeneration across species. In Drosophila, pathway activation in surviving cells autonomously suppresses apoptosis, increases cell division and leads to systemic responses (Katsuyama et al., 2015; La Fortezza et al., 2016; Santabárbara-Ruiz et al., 2015; Verghese and Su, 2016; Worley et al., 2018). Consistent with this, mice deficient for il-6, gp130 or stat3 exhibit impaired liver regeneration (Cressman et al., 1996; Taub, 2004). IL-11, another IL-6 family member, is induced early in Xenopus tail regeneration and is required for this process (Tsujioka et al., 2017). JAK/STAT signaling is also crucial for the regeneration of diverse zebrafish cells and tissues, including cardiomyocytes, the retina and inner ear hair cells (Fang et al., 2013; Liang et al., 2012; Zhao et al., 2014).
JAK/STAT-dependent upregulation of proliferation and survival likely endows cells with the ability to prevail in competitive scenario: (1) in the midgut and in the wing disc, winning cells require elevated JAK/STAT signaling to eliminate losing cells and colonize the tissue (Kolahgar et al., 2015; Rodrigues et al., 2012); (2) CySCs lacking the JAK/STAT-induced inhibitor Socs36E eject wild-type CySCs (and GSCs) from the testis stem cell niche (Amoyel et al., 2016a; Issigonis et al., 2009; Singh et al., 2010); and (3) FSCs with increased JAK/STAT signaling outcompete wild-type FSCs for residence in the ovarian niche (Vied et al., 2012). It is not clear whether this last form of competition is linked to Socs36E-dependent repression of Egfr/Ras/MAPK, another FSC self-renewal pathway (Castanieto et al., 2014). Cell competition is also known to occur during mouse development, in mouse ESCs and ISCs, and in adult mouse tissues (Claveria et al., 2013; Martins et al., 2014; Ritsma et al., 2014; Sancho et al., 2013; Vermeulen et al., 2013), but the roles, if any, of the JAK/STAT pathway in these processes remains to be determined.
Conclusions
Despite the numerous discoveries about the roles of the JAK/STAT pathway in stem cells and regeneration, many fundamental questions remain. Although increased proliferation and survival are conserved functions of IL-6-like cytokines, the direct transcriptional STAT targets that regulate these processes are largely unknown. In the future, it will therefore be important to define such factors as they may represent new druggable targets for the treatment of hematological cancers, carcinoma and inflammatory diseases caused by aberrant JAK/STAT signaling (Calautti et al., 2018; Grivennikov et al., 2009; Koskela et al., 2012; Pilati et al., 2011; Tefferi, 2016; Yu et al., 2014). The roles of cell competition in mouse tumor initiation and cancer progression are only beginning to be unraveled (Claveria and Torres, 2016; Fernandez et al., 2016). Given the well-established roles of the Drosophila JAK/STAT pathway in fostering dominance in competition scenarios, future research should focus on the functions of the mammalian JAK/STAT pathway in competitive interactions underlying oncogenesis. Cellular plasticity is controlled by epigenetic remodeling of chromatin by histone modifying complexes, and this is essential for regenerative responses (Herrera and Morata, 2014; Lee et al., 2005). Components of the JAK/STAT pathway have been shown to influence epigenetic modifications and chromatin structure (Griffiths et al., 2011; Rui et al., 2016; Shi et al., 2006, 2008), and future work should focus on how the pathway regulates these processes in stem cells and during regeneration. Finally, it will be useful to investigate JAK/STAT signaling in new model genetic organisms. The regenerative properties of the planarian flatworm Schmidtea mediterranea are well established, and pluripotent stem cells called neoblasts can differentiate into all adult cell types (Adler and Sánchez Alvarado, 2015). A protein BLAST of the S. mediterranea genome (smedgd.stowers.org/) using Drosophila Stat92E as a query reveals SMU15029397 as a putative STAT homolog. In the future, it will be interesting to determine whether this potential STAT protein regulates stem cells, cellular plasticity and regeneration in this powerful model system.
Acknowledgements
We apologize to colleagues whose contributions to this field could not be cited owing to space constraints.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work in the Bach lab is supported by New York State Stem Cell Science contracts C32584GG and C32577GG, and by a National Institute of Child Health and Human Development grant (R03 HD090422). Deposited in PMC for release after 12 months.
References
- Adler C. E. and Sánchez Alvarado A. (2015). Types or states? cellular dynamics and regenerative potential. Trends Cell Biol. 25, 687-696. 10.1016/j.tcb.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaisse H., Petersen U. M., Boutros M., Mathey-Prevot B. and Perrimon N. (2003). Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5, 441-450. 10.1016/S1534-5807(03)00244-2 [DOI] [PubMed] [Google Scholar]
- Ahmed-de-Prado S., Diaz-Garcia S. and Baonza A. (2018). JNK and JAK/STAT signalling are required for inducing loss of cell fate specification during imaginal wing discs regeneration in Drosophila melanogaster. Dev. Biol. 441, 31-41. 10.1016/j.ydbio.2018.05.021 [DOI] [PubMed] [Google Scholar]
- Albert E. A., Puretskaia O. A., Terekhanova N. V., Labudina A. and Bokel C. (2018). Direct control of somatic stem cell proliferation factors by the Drosophila testis stem cell niche. Development 145, dev156315 10.1242/dev.156315 [DOI] [PubMed] [Google Scholar]
- Amoyel M., Sanny J., Burel M. and Bach E. A. (2013). Hedgehog is required for CySC self-renewal but does not contribute to the GSC niche in the Drosophila testis. Development 140, 56-65. 10.1242/dev.086413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M., Anderson A. M. and Bach E. A. (2014a). JAK/STAT pathway dysregulation in tumors: a Drosophila perspective. Semin. Cell Dev. Biol. 28, 96-103. 10.1016/j.semcdb.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M., Simons B. D. and Bach E. A. (2014b). Neutral competition of stem cells is skewed by proliferative changes downstream of Hh and Hpo. EMBO J. 33, 2295-2313. 10.15252/embj.201387500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M., Anderson J., Suisse A., Glasner J. and Bach E. A. (2016a). Socs36E controls niche competition by repressing MAPK signaling in the Drosophila testis. PLoS Genet. 12, e1005815 10.1371/journal.pgen.1005815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M., Hillion K. H., Margolis S. R. and Bach E. A. (2016b). Somatic stem cell differentiation is regulated by PI3K/Tor signaling in response to local cues. Development 143, 3914-3925. 10.1242/dev.139782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbouzova N. I. and Zeidler M. P. (2006). JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development 133, 2605-2616. 10.1242/dev.02411 [DOI] [PubMed] [Google Scholar]
- Ayala-Camargo A., Anderson A. M., Amoyel M., Rodrigues A. B., Flaherty M. S. and Bach E. A. (2013). JAK/STAT signaling is required for hinge growth and patterning in the Drosophila wing disc. Dev. Biol. 382, 413-426. 10.1016/j.ydbio.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach E. A., Aguet M. and Schreiber R. D. (1997). The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15, 563-591. 10.1146/annurev.immunol.15.1.563 [DOI] [PubMed] [Google Scholar]
- Bach E. A., Vincent S., Zeidler M. P. and Perrimon N. (2003). A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics 165, 1149-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando T., Ishimaru Y., Kida T., Hamada Y., Matsuoka Y., Nakamura T., Ohuchi H., Noji S. and Mito T. (2013). Analysis of RNA-Seq data reveals involvement of JAK/STAT signalling during leg regeneration in the cricket Gryllus bimaculatus. Development 140, 959-964. 10.1242/dev.084590 [DOI] [PubMed] [Google Scholar]
- Beebe K., Lee W.-C. and Micchelli C. A. (2010). JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev. Biol. 338, 28-37. 10.1016/j.ydbio.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Bergantinos C., Corominas M. and Serras F. (2010). Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 137, 1169-1179. 10.1242/dev.045559 [DOI] [PubMed] [Google Scholar]
- Betz A., Ryoo H. D., Steller H. and Darnell J. E. Jr. (2008). STAT92E is a positive regulator of Drosophila inhibitor of apoptosis 1 (DIAP/1) and protects against radiation-induced apoptosis. Proc. Natl. Acad. Sci. USA 105, 13805-13810. 10.1073/pnas.0806291105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binari R. and Perrimon N. (1994). Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 8, 300-312. 10.1101/gad.8.3.300 [DOI] [PubMed] [Google Scholar]
- Biteau B. and Jasper H. (2011). EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045-1055. 10.1242/dev.056671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C. E. and Jasper H. (2008). JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 3, 442-455. 10.1016/j.stem.2008.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Karpac J., Supoyo S., DeGennaro M., Lehmann R. and Jasper H. (2010). Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 6, e1001159 10.1371/journal.pgen.1001159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C. E. and Jasper H. (2011). Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9, 402-411. 10.1016/j.stem.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J., Phesse T. J., von Burstin V. A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S. et al. (2009). gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 15, 91-102. 10.1016/j.ccr.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Bosch M., Baguna J. and Serras F. (2008). Origin and proliferation of blastema cells during regeneration of Drosophila wing imaginal discs. Int. J. Dev. Biol. 52, 1043-1050. 10.1387/ijdb.082608mb [DOI] [PubMed] [Google Scholar]
- Boyle M., Wong C., Rocha M. and Jones D. L. (2007). Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell 1, 470-478. 10.1016/j.stem.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Brawley C. and Matunis E. (2004). Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 304, 1331-1334. 10.1126/science.1097676 [DOI] [PubMed] [Google Scholar]
- Brown S., Hu N. and Hombria J. C.-C. (2001). Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr. Biol. 11, 1700-1705. 10.1016/S0960-9822(01)00524-3 [DOI] [PubMed] [Google Scholar]
- Brown S., Zeidler M. P. and Hombria J. E. C.-G. (2006). JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev. Dyn. 235, 958-966. 10.1002/dvdy.20709 [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Chakrabarti S. and Lemaitre B. (2009a). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333-2344. 10.1101/gad.1827009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Poidevin M., Pradervand S. and Lemaitre B. (2009b). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200-211. 10.1016/j.chom.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Kuraishi T. and Lemaitre B. (2010). Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 8, 152 10.1186/1741-7007-8-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q.-L. and Smith A. (2008). Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287-1298. 10.1016/j.cell.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Calautti E., Avalle L. and Poli V. (2018). Psoriasis: A STAT3-centric view. Int. J. Mol. Sci. 19, E171 10.3390/ijms19010171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callus B. A. and Mathey-Prevot B. (2002). SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene 21, 4812-4821. 10.1038/sj.onc.1205618 [DOI] [PubMed] [Google Scholar]
- Castanieto A., Johnston M. J. and Nystul T. G. (2014). EGFR signaling promotes self-renewal through the establishment of cell polarity in Drosophila follicle stem cells. eLife 3, e04437 10.7554/eLife.04437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett-Falcone R., Landowski T. H., Oshiro M. M., Turkson J., Levitzki A., Savino R., Ciliberto G., Moscinski L., Fernández-Luna J. L., Nuñez G. et al. (1999). Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10, 105-115. 10.1016/S1074-7613(00)80011-4 [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Dudzic J. P., Li X., Collas E. J., Boquete J.-P. and Lemaitre B. (2016). Remote control of intestinal stem cell activity by haemocytes in Drosophila. PLoS Genet. 12, e1006089 10.1371/journal.pgen.1006089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-W., Chen X., Oh S. W., Marinissen M. J., Gutkind J. S. and Hou S. X. (2002). mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 16, 388-398. 10.1101/gad.955202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Cummings R., Mordovanakis A., Hunt A. J., Mayer M., Sept D. and Yamashita Y. M. (2018). Cytokine receptor-Eb1 interaction couples cell polarity and fate during asymmetric cell division. eLife 7, e33685 10.7554/eLife.33685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry C. M. and Matunis E. L. (2010). Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell 6, 557-567. 10.1016/j.stem.2010.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N.-H., Kim J.-G., Yang D.-J., Kim Y.-S. and Yoo M.-A. (2008). Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging Cell 7, 318-334. 10.1111/j.1474-9726.2008.00380.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. R., Bakir I. A., Hart A. L. and Graham T. A. (2017). Clonal evolution of colorectal cancer in IBD. Nat. Rev. Gastroenterol. Hepatol. 14, 218-229. 10.1038/nrgastro.2017.1 [DOI] [PubMed] [Google Scholar]
- Claveria C. and Torres M. (2016). Cell competition: mechanisms and physiological roles. Annu. Rev. Cell Dev. Biol. 32, 411-439. 10.1146/annurev-cellbio-111315-125142 [DOI] [PubMed] [Google Scholar]
- Claveria C., Giovinazzo G., Sierra R. and Torres M. (2013). Myc-driven endogenous cell competition in the early mammalian embryo. Nature 500, 39-44. 10.1038/nature12389 [DOI] [PubMed] [Google Scholar]
- Cohen S. M. (1993). Imaginal disc development. Cold Spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- Colombani J., Andersen D. S. and Leopold P. (2012). Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336, 582-585. 10.1126/science.1216689 [DOI] [PubMed] [Google Scholar]
- Cordero J. B., Stefanatos R. K., Myant K., Vidal M. and Sansom O. J. (2012). Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut. Development 139, 4524-4535. 10.1242/dev.078261 [DOI] [PubMed] [Google Scholar]
- Cressman D. E., Greenbaum L. E., DeAngelis R. A., Ciliberto G., Furth E. E., Poli V. and Taub R. (1996). Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 274, 1379-1383. 10.1126/science.274.5291.1379 [DOI] [PubMed] [Google Scholar]
- Cronin S. J. F., Nehme N. T., Limmer S., Liegeois S., Pospisilik J. A., Schramek D., Leibbrandt A., Simoes R. M., Gruber S., Puc U. et al. (2009). Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340-343. 10.1126/science.1173164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curley M., Milne L., Smith S., Atanassova N., Rebourcet D., Darbey A., Hadoke P. W. F., Wells S. and Smith L. B. (2018). Leukemia inhibitory factor-receptor is dispensable for prenatal testis development but is required in sertoli cells for normal spermatogenesis in mice. Sci. Rep. 8, 11532 10.1038/s41598-018-30011-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E., Jr (1997). STATs and gene regulation. Science 277, 1630-1635. 10.1126/science.277.5332.1630 [DOI] [PubMed] [Google Scholar]
- Diaz-Garcia S. and Baonza A. (2013). Pattern reorganization occurs independently of cell division during Drosophila wing disc regeneration in situ. Proc. Natl. Acad. Sci. USA 110, 13032-13037. 10.1073/pnas.1220543110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do D. V., Ueda J., Messerschmidt D. M., Lorthongpanich C., Zhou Y., Feng B., Guo G., Lin P. J., Hossain M. Z., Zhang W. et al. (2013). A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 27, 1378-1390. 10.1101/gad.221176.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Gupta V., Karra R., Holdway J. E., Kikuchi K. and Poss K. D. (2013). Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 110, 13416-13421. 10.1073/pnas.1309810110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Shi Z. and Chen X. (2017). Enhancer of polycomb coordinates multiple signaling pathways to promote both cyst and germline stem cell differentiation in the Drosophila adult testis. PLoS Genet. 13, e1006571 10.1371/journal.pgen.1006571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L. C., Torres M. and Real F. X. (2016). Somatic mosaicism: on the road to cancer. Nat. Rev. Cancer 16, 43-55. 10.1038/nrc.2015.1 [DOI] [PubMed] [Google Scholar]
- Flaherty M. S., Zavadil J., Ekas L. A. and Bach E. A. (2009). Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev. Dyn. 238, 2235-2253. 10.1002/dvdy.21989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty M. S., Salis P., Evans C. J., Ekas L. A., Marouf A., Zavadil J., Banerjee U. and Bach E. A. (2010). chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev. Cell 18, 556-568. 10.1016/j.devcel.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujio Y., Kunisada K., Hirota H., Yamauchi-Takihara K. and Kishimoto T. (1997). Signals through gp130 upregulate bcl-x gene expression via STAT1-binding cis-element in cardiac myocytes. J. Clin. Invest. 99, 2898-2905. 10.1172/JCI119484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M. T. (1998). Genetic control of cell proliferation and differentiation in Drosophila spermatogenesis. Semin. Cell Dev. Biol. 9, 433-444. 10.1006/scdb.1998.0227 [DOI] [PubMed] [Google Scholar]
- Garelli A., Gontijo A. M., Miguela V., Caparros E. and Dominguez M. (2012). Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336, 579-582. 10.1126/science.1216735 [DOI] [PubMed] [Google Scholar]
- Gonczy P. and DiNardo S. (1996). The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development 122, 2437-2447. [DOI] [PubMed] [Google Scholar]
- Greenspan L. J., de Cuevas M. and Matunis E. (2015). Genetics of gonadal stem cell renewal. Annu. Rev. Cell Dev. Biol. 31, 291-315. 10.1146/annurev-cellbio-100913-013344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths D. S., Li J., Dawson M. A., Trotter M. W. B., Cheng Y.-H., Smith A. M., Mansfield W., Liu P., Kouzarides T., Nichols J. et al. (2011). LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease. Nat. Cell Biol. 13, 13-21. 10.1038/ncb2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.-Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L. et al. (2009). IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103-113. 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan I. K. and Serras F. (2017). Imaginal disc regeneration takes flight. Curr. Opin. Cell Biol. 48, 10-16. 10.1016/j.ceb.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. E., Setiawan L., Saul J. and Hariharan I. K. (2016). Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. eLife 5, e11588 10.7554/eLife.11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D. A., McCoon P. E., Binari R., Gilman M. and Perrimon N. (1998). Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 12, 3252-3263. 10.1101/gad.12.20.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S., Hétié P. and Matunis E. L. (2015). Niche signaling promotes stem cell survival in the Drosophila testis via the JAK-STAT target DIAP1. Dev. Biol. 404, 27-39. 10.1016/j.ydbio.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatini V., Kula-Eversole E., Nusinow D. and Del Signore S. J. (2013). Essential roles for stat92E in expanding and patterning the proximodistal axis of the Drosophila wing imaginal disc. Dev. Biol. 378, 38-50. 10.1016/j.ydbio.2013.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera S. C. and Morata G. (2014). Transgressions of compartment boundaries and cell reprogramming during regeneration in Drosophila. eLife 3, e01831 10.7554/eLife.01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera S. C., Martin R. and Morata G. (2013). Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet. 9, e1003446 10.1371/journal.pgen.1003446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombria J. C.-G., Brown S., Häder S. and Zeidler M. P. (2005). Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev. Biol. 288, 420-433. 10.1016/j.ydbio.2005.09.040 [DOI] [PubMed] [Google Scholar]
- Hou X. S., Melnick M. B. and Perrimon N. (1996). Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell 84, 411-419. 10.1016/S0092-8674(00)81286-6 [DOI] [PubMed] [Google Scholar]
- Housden B. E. and Perrimon N. (2014). Spatial and temporal organization of signaling pathways. Trends Biochem. Sci. 39, 457-464. 10.1016/j.tibs.2014.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtz P., Bonfini A., Liu X., Revah J., Guillou A., Poidevin M., Hens K., Huang H.-Y., Deplancke B., Tsai Y.-C. et al. (2017). Hippo, TGF-beta, and Src-MAPK pathways regulate transcription of the upd3 cytokine in Drosophila enterocytes upon bacterial infection. PLoS Genet. 13, e1007091 10.1371/journal.pgen.1007091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issigonis M., Tulina N., de Cuevas M., Brawley C., Sandler L. and Matunis E. (2009). JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science 326, 153-156. 10.1126/science.1176817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklevic B. R. and Su T. T. (2004). Relative contribution of DNA repair, cell cycle checkpoints, and cell death to survival after DNA damage in Drosophila larvae. Curr. Biol. 14, 23-32. 10.1016/j.cub.2003.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G. and Edgar B. A. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343-1355. 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Grenley M. O., Bravo M.-J., Blumhagen R. Z. and Edgar B. A. (2011). EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84-95. 10.1016/j.stem.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone K., Wells R. E., Strutt D. and Zeidler M. P. (2013). Localised JAK/STAT pathway activation is required for Drosophila wing hinge development. PloS ONE 8, e65076 10.1371/journal.pone.0065076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P., Perez J. and Perrimon N. (2010). The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 137, 4135-4145. 10.1242/dev.060483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten P., Häder S. and Zeidler M. P. (2002). Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech. Dev. 117, 343-346. 10.1016/S0925-4773(02)00216-2 [DOI] [PubMed] [Google Scholar]
- Katsuyama T., Comoglio F., Seimiya M., Cabuy E. and Paro R. (2015). During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc. Natl. Acad. Sci. USA 112, E2327-E2336. 10.1073/pnas.1423074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata T., Shevchenko A., Fukuzawa M., Jermyn K. A., Totty N. F., Zhukovskaya N. V., Sterling A. E., Mann M. and Williams J. G. (1997). SH2 signaling in a lower eukaryote: a STAT protein that regulates stalk cell differentiation in dictyostelium. Cell 89, 909-916. 10.1016/S0092-8674(00)80276-7 [DOI] [PubMed] [Google Scholar]
- Kiger A. A., Jones D. L., Schulz C., Rogers M. B. and Fuller M. T. (2001). Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542-2545. 10.1126/science.1066707 [DOI] [PubMed] [Google Scholar]
- Kitadate Y. and Kobayashi S. (2010). Notch and Egfr signaling act antagonistically to regulate germ-line stem cell niche formation in Drosophila male embryonic gonads. Proc. Natl. Acad. Sci. USA 107, 14241-14246. 10.1073/pnas.1003462107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebes A., Sustar A., Kechris K., Li H., Schubiger G. and Kornberg T. B. (2005). Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development 132, 3753-3765. 10.1242/dev.01927 [DOI] [PubMed] [Google Scholar]
- Klein A. M. and Simons B. D. (2011). Universal patterns of stem cell fate in cycling adult tissues. Development 138, 3103-3111. 10.1242/dev.060103 [DOI] [PubMed] [Google Scholar]
- Kleppe M., Spitzer M. H., Li S., Hill C. E., Dong L., Papalexi E., De Groote S., Bowman R. L., Keller M., Koppikar P. et al. (2017). Jak1 integrates cytokine sensing to regulate hematopoietic stem cell function and stress hematopoiesis. Cell Stem Cell 21, 489-501 e487. 10.1016/j.stem.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolahgar G., Suijkerbuijk S. J., Kucinski I., Poirier E. Z., Mansour S., Simons B. D. and Piddini E. (2015). Cell competition modifies adult stem cell and tissue population dynamics in a JAK-STAT-dependent manner. Dev. Cell 34, 297-309. 10.1016/j.devcel.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela H. L., Eldfors S., Ellonen P., van Adrichem A. J., Kuusanmäki H., Andersson E. I., Lagström S., Clemente M. J., Olson T., Jalkanen S. E. et al. (2012). Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl. J. Med. 366, 1905-1913. 10.1056/NEJMoa1114885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fortezza M., Schenk M., Cosolo A., Kolybaba A., Grass I. and Classen A. K. (2016). JAK/STAT signalling mediates cell survival in response to tissue stress. Development 143, 2907-2919. 10.1242/dev.132340 [DOI] [PubMed] [Google Scholar]
- Leatherman J. L. and Dinardo S. (2008). Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 3, 44-54. 10.1016/j.stem.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherman J. L. and Dinardo S. (2010). Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat. Cell Biol. 12, 806-811. 10.1038/ncb2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Maurange C., Ringrose L. and Paro R. (2005). Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature 438, 234-237. 10.1038/nature04120 [DOI] [PubMed] [Google Scholar]
- Lemaitre B. and Miguel-Aliaga I. (2013). The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 47, 377-404. 10.1146/annurev-genet-111212-133343 [DOI] [PubMed] [Google Scholar]
- Levy D. E. (1999). Physiological significance of STAT proteins: investigations through gene disruption in vivo. Cell. Mol. Life Sci. 55, 1559-1567. 10.1007/s000180050395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. E. and Darnell J. E. Jr. (2002). Stats: transcriptional control and biological impact. Nat Rev. Mol. Cell Biol. 3, 651-662. 10.1038/nrm909 [DOI] [PubMed] [Google Scholar]
- Levy D. E. and Lee C.-K. (2002). What does Stat3 do? J. Clin. Invest. 109, 1143-1148. 10.1172/JCI0215650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Li S., Mana-Capelli S., Roth Flach R. J., Danai L. V., Amcheslavsky A., Nie Y., Kaneko S., Yao X., Chen X. et al. (2014). The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev. Cell 31, 291-304. 10.1016/j.devcel.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wang D., Renaud G., Wolfsberg T. G., Wilson A. F. and Burgess S. M. (2012). The stat3/socs3a pathway is a key regulator of hair cell regeneration in zebrafish. [corrected]. J. Neurosci. 32, 10662-10673. 10.1523/JNEUROSCI.5785-10.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Xu N. and Xi R. (2010). Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of drosophila intestinal stem cells. J. Mol. Cell Biol. 2, 37-49. 10.1093/jmcb/mjp028 [DOI] [PubMed] [Google Scholar]
- Liu W., Singh S. R. and Hou S. X. (2010). JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J. Cell. Biochem. 109, 992-999. 10.1002/jcb.22482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins V. C., Busch K., Juraeva D., Blum C., Ludwig C., Rasche V., Lasitschka F., Mastitsky S. E., Brors B., Hielscher T. et al. (2014). Cell competition is a tumour suppressor mechanism in the thymus. Nature 509, 465-470. 10.1038/nature13317 [DOI] [PubMed] [Google Scholar]
- Maves L. and Schubiger G. (1995). Wingless induces transdetermination in developing Drosophila imaginal discs. Development 121, 1263-1272. [DOI] [PubMed] [Google Scholar]
- McConnell B. B., Klapproth J. M., Sasaki M., Nandan M. O. and Yang V. W. (2008). Krüppel-like factor 5 mediates transmissible murine colonic hyperplasia caused by Citrobacter rodentium infection. Gastroenterology 134, 1007-1016. 10.1053/j.gastro.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLemore M. L., Grewal S., Liu F., Archambault A., Poursine-Laurent J., Haug J. and Link D. C. (2001). STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity 14, 193-204. 10.1016/S1074-7613(01)00101-7 [DOI] [PubMed] [Google Scholar]
- Micchelli C. A. and Perrimon N. (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475-479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Nagasawa K., Imura-Kishi K., Uchida A., Hiramatsu R., Kurohmaru M. and Kanai Y. (2018). Regionally distinct patterns of STAT3 phosphorylation in the seminiferous epithelia of mouse testes. Mol. Reprod. Dev. 85, 262-270. 10.1002/mrd.22962 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Sharma M., Nabeshima Y., Braun R. E. and Yoshida S. (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62-67. 10.1126/science.1182868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Jones K., Phillips J. M., Newland S. A., Roode M., Mansfield W., Smith A. and Cooke A. (2009). Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat. Med. 15, 814-818. 10.1038/nm.1996 [DOI] [PubMed] [Google Scholar]
- Oatley J. M. and Brinster R. L. (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev. 92, 577-595. 10.1152/physrev.00025.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Kaucher A. V., Avarbock M. R. and Brinster R. L. (2010). Regulation of mouse spermatogonial stem cell differentiation by STAT3 signaling. Biol. Reprod. 83, 427-433. 10.1095/biolreprod.109.083352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B. and Spradling A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470-474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Okegbe T. C. and DiNardo S. (2011). The endoderm specifies the mesodermal niche for the germline in Drosophila via Delta-Notch signaling. Development 138, 1259-1267. 10.1242/dev.056994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi K. and Zandstra P. W. (2015). LIF signaling in stem cells and development. Development 142, 2230-2236. 10.1242/dev.117598 [DOI] [PubMed] [Google Scholar]
- Osman D., Buchon N., Chakrabarti S., Huang Y.-T., Su W.-C., Poidevin M., Tsai Y.-C. and Lemaitre B. (2012). Autocrine and paracrine unpaired signaling regulate intestinal stem cell maintenance and division. J. Cell Sci. 125, 5944-5949. 10.1242/jcs.113100 [DOI] [PubMed] [Google Scholar]
- Park J.-S., Kim Y.-S. and Yoo M.-A. (2009). The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging 1, 637-651. 10.18632/aging.100054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja J. C., Wu M. and Xu T. (2008). An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis. Model. Mech. 1, 144-154. 10.1242/dmm.000950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. H., Dutta D. and Edgar B. A. (2015). Niche appropriation by Drosophila intestinal stem cell tumours. Nat. Cell Biol. 17, 1182-1192. 10.1038/ncb3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Garijo A., Fuchs Y. and Steller H. (2013). Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. eLife 2, e01004 10.7554/eLife.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati C., Amessou M., Bihl M. P., Balabaud C., Nhieu J. T., Paradis V., Nault J. C., Izard T., Bioulac-Sage P., Couchy G. et al. (2011). Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J. Exp. Med. 208, 1359-1366. 10.1084/jem.20110283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putoczki T. L., Thiem S., Loving A., Busuttil R. A., Wilson N. J., Ziegler P. K., Nguyen P. M., Preaudet A., Farid R., Edwards K. M. et al. (2013). Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 24, 257-271. 10.1016/j.ccr.2013.06.017 [DOI] [PubMed] [Google Scholar]
- Qian Y., Ng C. L. and Schulz C. (2015). CSN maintains the germline cellular microenvironment and controls the level of stem cell genes via distinct CRLs in testes of Drosophila melanogaster. Dev. Biol. 398, 68-79. 10.1016/j.ydbio.2014.11.014 [DOI] [PubMed] [Google Scholar]
- Recasens-Alvarez C., Ferreira A. and Milán M. (2017). JAK/STAT controls organ size and fate specification by regulating morphogen production and signalling. Nat. Commun. 8, 13815 10.1038/ncomms13815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Wang B., Yue T., Yun E.-Y., Ip Y. T. and Jiang J. (2010). Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl. Acad. Sci. USA 107, 21064-21069. 10.1073/pnas.1012759107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Shi Q., Chen Y., Jiang A., Ip Y. T., Jiang H. and Jiang J. (2013). Drosophila Myc integrates multiple signaling pathways to regulate intestinal stem cell proliferation during midgut regeneration. Cell Res. 23, 1133-1146. 10.1038/cr.2013.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Zhang Y., Li M., Wu L., Wang G., Baeg G.-H., You J., Li Z. and Lin X. (2015). Windpipe controls Drosophila intestinal homeostasis by regulating JAK/STAT pathway via promoting receptor endocytosis and lysosomal degradation. PLoS Genet. 11, e1005180 10.1371/journal.pgen.1005180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby R. J., Simmons J. G., Greenhalgh C. J., Alexander W. S. and Lund P. K. (2007). Suppressor of cytokine signaling 3 (SOCS3) limits damage-induced crypt hyper-proliferation and inflammation-associated tumorigenesis in the colon. Oncogene 26, 4833-4841. 10.1038/sj.onc.1210286 [DOI] [PubMed] [Google Scholar]
- Ritsma L., Ellenbroek S. I., Zomer A., Snippert H. J., de Sauvage F. J., Simons B. D., Clevers H. and van Rheenen J. (2014). Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362-365. 10.1038/nature12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A. B., Zoranovic T., Ayala-Camargo A., Grewal S., Reyes-Robles T., Krasny M., Wu D. C., Johnston L. A. and Bach E. A. (2012). Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development 139, 4051-4061. 10.1242/dev.076760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose-John S. (2018). Interleukin-6 family cytokines. Cold Spring Harbor Perspect. Biol. 10, a028415 10.1101/cshperspect.a028415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L., Drennan A. C., Ceribelli M., Zhu F., Wright G. W., Huang D. W., Xiao W., Li Y., Grindle K. M., Lu L. et al. (2016). Epigenetic gene regulation by Janus kinase 1 in diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 113, E7260-E7267. 10.1073/pnas.1610970113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho M., Di-Gregorio A., George N., Pozzi S., Sánchez J. M., Pernaute B. and Rodríguez T. A. (2013). Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev. Cell 26, 19-30. 10.1016/j.devcel.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santabárbara-Ruiz P., Lopez-Santillan M., Martinez-Rodriguez I., Binagui-Casas A., Perez L., Milan M., Corominas M. and Serras F. (2015). ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during drosophila regeneration. PLoS Genet. 11, e1005595 10.1371/journal.pgen.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger G. (1971). Regeneration, duplication and transdetermination in fragments of the leg disc of Drosophila melanogaster. Dev. Biol. 26, 277-295. 10.1016/0012-1606(71)90127-8 [DOI] [PubMed] [Google Scholar]
- Shaw R. L., Kohlmaier A., Polesello C., Veelken C., Edgar B. A. and Tapon N. (2010). The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147-4158. 10.1242/dev.052506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X. R., Brawley C. M. and Matunis E. L. (2009a). Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell 5, 191-203. 10.1016/j.stem.2009.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng X. R., Posenau T., Gumulak-Smith J. J., Matunis E., Van Doren M. and Wawersik M. (2009b). Jak-STAT regulation of male germline stem cell establishment during Drosophila embryogenesis. Dev. Biol. 334, 335-344. 10.1016/j.ydbio.2009.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Calhoun H. C., Xia F., Li J., Le L. and Li W. X. (2006). JAK signaling globally counteracts heterochromatic gene silencing. Nat. Genet. 38, 1071-1076. 10.1038/ng1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Larson K., Guo D., Lim S. J., Dutta P., Yan S.-J. and Li W. X. (2008). Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat. Cell Biol. 10, 489-496. 10.1038/ncb1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields A. R., Spence A. C., Yamashita Y. M., Davies E. L. and Fuller M. T. (2014). The actin-binding protein profilin is required for germline stem cell maintenance and germ cell enclosure by somatic cyst cells. Development 141, 73-82. 10.1242/dev.101931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov E. and Morata G. (2012). A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death Differ. 19, 451-460. 10.1038/cdd.2011.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons B. D. and Clevers H. (2011). Strategies for homeostatic stem cell self-renewal in adult tissues. Cell 145, 851-862. 10.1016/j.cell.2011.05.033 [DOI] [PubMed] [Google Scholar]
- Sinden D., Badgett M., Fry J., Jones T., Palmen R., Sheng X., Simmons A., Matunis E. and Wawersik M. (2012). Jak-STAT regulation of cyst stem cell development in the Drosophila testis. Dev. Biol. 372, 5-16. 10.1016/j.ydbio.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. R., Zheng Z., Wang H., Oh S. W., Chen X. and Hou S. X. (2010). Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J. Cell. Physiol. 223, 500-510. 10.1002/jcp.22073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siudeja K., Nassari S., Gervais L., Skorski P., Lameiras S., Stolfa D., Zande M., Bernard V., Frio T. R. and Bardin A. J. (2015). Frequent somatic mutation in adult intestinal stem cells drives neoplasia and genetic mosaicism during aging. Cell Stem Cell 17, 663-674. 10.1016/j.stem.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M. and Rogers D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688-690. 10.1038/336688a0 [DOI] [PubMed] [Google Scholar]
- Smith-Bolton R. K., Worley M. I., Kanda H. and Hariharan I. K. (2009). Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell 16, 797-809. 10.1016/j.devcel.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Victor P., Ayyaz A., Hayashi R., Qi Y., Madden D. T., Lunyak V. V. and Jasper H. (2017). Piwi is required to limit exhaustion of aging somatic stem cells. Cell Reports 20, 2527-2537. 10.1016/j.celrep.2017.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley B. K. and Irvine K. D. (2010). Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 20, 1580-1587. 10.1016/j.cub.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine R. R., Greenspan L. J., Ramachandran K. V. and Matunis E. L. (2014). Coordinate regulation of stem cell competition by Slit-Robo and JAK-STAT signaling in the Drosophila testis. PLoS Genet. 10, e1004713 10.1371/journal.pgen.1004713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S. J., Kolahgar G., Kucinski I. and Piddini E. (2016). Cell competition drives the growth of intestinal adenomas in drosophila. Curr. Biol. 26, 428-438. 10.1016/j.cub.2015.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Noguchi K., Shi W., Tanaka T., Matsumoto M., Yoshida N., Kishimoto T. and Akira S. (1997). Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94, 3801-3804. 10.1073/pnas.94.8.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K. and Karin M. (2014). IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin. Immunol. 26, 54-74. 10.1016/j.smim.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Tarayrah L., Li Y., Gan Q. and Chen X. (2015). Epigenetic regulator Lid maintains germline stem cells through regulating JAK-STAT signaling pathway activity. Biol. Open 4, 1518-1527. 10.1242/bio.013961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R. (2004). Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 5, 836-847. 10.1038/nrm1489 [DOI] [PubMed] [Google Scholar]
- Tefferi A. (2016). Myeloproliferative neoplasms: a decade of discoveries and treatment advances. Am. J. Hematol. 91, 50-58. 10.1002/ajh.24221 [DOI] [PubMed] [Google Scholar]
- Terry N. A., Tulina N., Matunis E. and DiNardo S. (2006). Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev. Biol. 294, 246-257. 10.1016/j.ydbio.2006.02.048 [DOI] [PubMed] [Google Scholar]
- Toledano H., D'Alterio C., Czech B., Levine E. and Jones D. L. (2012). The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 485, 605-610. 10.1038/nature11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-C. and Sun Y. H. (2004). Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis 39, 141-153. 10.1002/gene.20035 [DOI] [PubMed] [Google Scholar]
- Tsujioka H., Kunieda T., Katou Y., Shirahige K., Fukazawa T. and Kubo T. (2017). interleukin-11 induces and maintains progenitors of different cell lineages during Xenopus tadpole tail regeneration. Nat. Commun. 8, 495 10.1038/s41467-017-00594-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulina N. and Matunis E. (2001). Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science 294, 2546-2549. 10.1126/science.1066700 [DOI] [PubMed] [Google Scholar]
- Verghese S. and Su T. T. (2016). Drosophila Wnt and STAT Define Apoptosis-Resistant Epithelial Cells for Tissue Regeneration after Irradiation. PLoS Biol. 14, e1002536 10.1371/journal.pbio.1002536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese S. and Su T. T. (2017). STAT, Wingless, and Nurf-38 determine the accuracy of regeneration after radiation damage in Drosophila. PLoS Genet. 13, e1007055 10.1371/journal.pgen.1007055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., Morrissey E., van der Heijden M., Nicholson A. M., Sottoriva A., Buczacki S., Kemp R., Tavare S. and Winton D. J. (2013). Defining stem cell dynamics in models of intestinal tumor initiation. Science 342, 995-998. 10.1126/science.1243148 [DOI] [PubMed] [Google Scholar]
- Vied C., Reilein A., Field N. S. and Kalderon D. (2012). Regulation of stem cells by intersecting gradients of long-range niche signals. Dev. Cell 23, 836-848. 10.1016/j.devcel.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. and Levy D. E. (2006). C. elegans STAT cooperates with DAF-7/TGF-beta signaling to repress dauer formation. Curr. Biol. 16, 89-94. 10.1016/j.cub.2005.11.061 [DOI] [PubMed] [Google Scholar]
- Wang L., Sexton T. R., Venard C., Giedt M., Guo Q., Chen Q. and Harrison D. A. (2014). Pleiotropy of the Drosophila JAK pathway cytokine Unpaired 3 in development and aging. Dev. Biol. 395, 218-231. 10.1016/j.ydbio.2014.09.015 [DOI] [PubMed] [Google Scholar]
- Wawersik M., Milutinovich A., Casper A. L., Matunis E., Williams B. and Van Doren M. (2005). Somatic control of germline sexual development is mediated by the JAK/STAT pathway. Nature 436, 563-567. 10.1038/nature03849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshons W. J. and Russel W. L. (1957). The effect of x-rays on the drosophila testis and a method for obtaining spermatogonial mutation rates. Proc. Natl. Acad. Sci. USA 43, 608-613. 10.1073/pnas.43.7.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West N. R., McCuaig S., Franchini F. and Powrie F. (2015). Emerging cytokine networks in colorectal cancer. Nat. Rev. 15, 615-629. 10.1038/nri3896 [DOI] [PubMed] [Google Scholar]
- Worley M. I., Setiawan L. and Hariharan I. K. (2012). Regeneration and transdetermination in Drosophila imaginal discs. Annu. Rev. Genet. 46, 289-310. 10.1146/annurev-genet-110711-155637 [DOI] [PubMed] [Google Scholar]
- Worley M. I., Alexander L. A. and Hariharan I. K. (2018). CtBP impedes JNK- and Upd/STAT-driven cell fate misspecifications in regenerating Drosophila imaginal discs. eLife 7, e30391 10.7554/eLife.30391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Wang S. Q., Tan D., Gao Y., Lin G. and Xi R. (2011). EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Dev. Biol. 354, 31-43. 10.1016/j.ydbio.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Yamashita Y. M., Jones D. L. and Fuller M. T. (2003). Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547-1550. 10.1126/science.1087795 [DOI] [PubMed] [Google Scholar]
- Yan R., Small S., Desplan C., Dearolf C. R. and Darnell J. E. Jr (1996). Identification of a Stat gene that functions in Drosophila development. Cell 84, 421-430. 10.1016/S0092-8674(00)81287-8 [DOI] [PubMed] [Google Scholar]
- Yang S. Y., Baxter E. M. and Van Doren M. (2012). Phf7 controls male sex determination in the Drosophila germline. Dev. Cell 22, 1041-1051. 10.1016/j.devcel.2012.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T., Umetsu D., Murakami S., Sato M. and Tabata T. (2008). Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development 135, 1471-1480. 10.1242/dev.019117 [DOI] [PubMed] [Google Scholar]
- Ying Q.-L. and Smith A. (2017). The art of capturing pluripotency: creating the right culture. Stem Cell Reports 8, 1457-1464. 10.1016/j.stemcr.2017.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Taga T., Saito M., Suematsu S., Kumanogoh A., Tanaka T., Fujiwara H., Hirata M., Yamagami T., Nakahata T. et al. (1996). Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc. Natl. Acad. Sci. USA 93, 407-411. 10.1073/pnas.93.1.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Lee H., Herrmann A., Buettner R. and Jove R. (2014). Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer 14, 736-746. 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- Zhai Z., Kondo S., Ha N., Boquete J.-P., Brunner M., Ueda R. and Lemaitre B. (2015). Accumulation of differentiating intestinal stem cell progenies drives tumorigenesis. Nat. Commun. 6, 10219 10.1038/ncomms10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Boquete J.-P. and Lemaitre B. (2017). A genetic framework controlling the differentiation of intestinal stem cells during regeneration in Drosophila. PLoS Genet. 13, e1006854 10.1371/journal.pgen.1006854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.-F., Wan J., Powell C., Ramachandran R., Myers M. G. Jr and Goldman D. (2014). Leptin and IL-6 family cytokines synergize to stimulate Muller glia reprogramming and retina regeneration. Cell Reports 9, 272-284. 10.1016/j.celrep.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Rasmussen A., Lee S. and Agaisse H. (2013). The UPD3 cytokine couples environmental challenge and intestinal stem cell division through modulation of JAK/STAT signaling in the stem cell microenvironment. Dev. Biol. 373, 383-393. 10.1016/j.ydbio.2012.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]