Abstract
The super family of glutathione S-transferases (GSTs) is composed of multiple isozymes with significant evidence of functional polymorphic variation. Over the last three decades, data from cancer studies have linked aberrant expression of GST isozymes with the development and expression of resistance to a variety of chemicals, including cancer drugs. This review addresses how differences in the human GST isozyme expression patterns influence cancer susceptibility, prognosis and treatment. In addition to the well-characterized catalytic activity, recent evidence has shown that certain GST isozymes can regulate mitogen-activated protein kinases or can facilitate the addition of glutathione to cysteine residues in target proteins (S-glutathionylation). These multiple functionalities have contributed to the recent efforts to target GSTs with novel small molecule therapeutics. Presently, at least two drugs are in late-stage clinical testing. The evolving functions of GST and their divergent expression patterns in individuals make them an attractive target for drug discovery.
Keywords: GST, polymorphism, resistance, MAP kinase pathway
Introduction
A primary cause of cancer treatment failure and patient relapse is an acquired or intrinsic resistance to anticancer therapies. Acquisition of drug resistance can be attributed to various factors that include avoidance of apoptotic cell death, altered expression of multidrug resistance-associated proteins, altered drug metabolism or uptake, and/or overexpression of glutathione S-transferases (GSTs). GSTs are a family of Phase II detoxification enzymes that catalyse the conjugation of glutathione (GSH) to a wide variety of xenobiotics. This detoxification ability plays a role in cellular protection from environmental and oxidative stress, yet is also implicated in cellular resistance to drugs.
Chemotherapeutic-resistant tumor cell lines have been shown to overexpress GST isozymes. This overexpression leads to an accelerated detoxification of drug substrates and thus an acquired resistance. However, drug resistance is exhibited in cells expressing certain isoforms of GSTs even when that specific selecting drug is not an enzyme substrate. This anomaly may be explained by the ability of GSTs to act as ligand-binding proteins in the regulation of cell cycle components such as mitogen-activated protein kinases (MAPK) and extracellular-regulated kinases (ERK). This review will explore these various issues related to human GST gene expression as it correlates to cancer susceptibility, prognosis and treatment.
GST classification and human polymorphisms
Human GSTs are divided into three main families: cytosolic, mitochondrial and membrane-bound microsomal. Microsomal GSTs are designated as ‘membrane-associated proteins in eicosanoid and glutathione metabolism’ (MAPEGs). MAPEGs are structurally distinct from cytosolic GSTs but are functionally similar in the ability to catalyse the conjugation of GSH to electrophilic compounds (Hayes et al, 2005).
The mammalian cytosolic family of GSTs exist as monomers and are catalytically active in a homo- or heterodimeric state (Mannervik and Danielson, 1988). The cytosolic family is further divided into seven classes: Alpha, Mu, Omega, Pi, Sigma, Theta and Zeta. Classification is based on sharing greater than 60% identity within a class and focuses mainly on the more highly conserved N-terminal domain that contains a catalytically active tyrosine, cysteine or serine residue. The catalytic residue interacts with the thiol group of GSH (G-site), and crystal structure has confirmed a substrate binding site (H-site) that facilitates catalysis and is in proximity to the G-site (Dirr et al., 1994; Armstrong, 1997).
Alpha class
The GSTα isoform is mainly expressed in the liver and is encoded by a gene cluster localized on chromosome 6p12 (Table 1). This cluster contains five genes encoding proteins belonging to GSTA1–A5. Human tissues widely express transcripts for GSTA1, A2 and A4, whereas expression of GSTA3 is rare and GSTA5 has yet to be detected in human tissues. Epidemiological results show that aberrant expression of GSTα has been linked to an increased risk in colorectal cancer, ovarian cancer and clear cell renal cell carcinoma (Coles et al., 2001; Tetlow et al., 2004a; Chuang et al., 2005).
Table 1.
Cytosolic GSTs
| Class | Gene | Chromosome location |
|---|---|---|
| Alpha (α) | GSTA1 | 6p12 |
| GSTA2 | 6p12.2 | |
| GSTA3 | 6p12 | |
| GSTA4 | 6p12 | |
| GSTA5 | 6p12.1 | |
| Mu(μ) | GSTM1 | 1p13.3 |
| GSTM2 | 1p13 | |
| GSTM3 | 1p13.3 | |
| GSTM4 | 1p13.3 | |
| GSTM5 | 1p13.3 | |
| Omega (ω) | GSTO1 | 10q25.1 |
| GSTO2 | 10q25.1 | |
| Pi (π) | GSTP1 | 11q13-qter |
| Theta (θ) | GSTP1 | 22q11.2 |
| GSTT2 | 22q11.2 | |
| Zeta (ξ) | GSTZ1 | 14q24.3 |
The GSTA1 gene contains seven exons and is ~12 kb in length. A genetic polymorphism of GSTA1 is characterized by two alleles, GsTA1*A and GSTA1*B (Table 2). These alleles differ in promoter regions based on three linked single nucleotide substitutions at positions −567, −69 and −52. Specifically, the −52 substitution has been shown to increase promoter activity in GSTA1*A, thus making it more highly expressed. Coles et al. (2001) previously showed a correlation between GSTA1*B expression and an increased risk of colorectal cancer. However, as with many such published accounts, other groups have recently reported conflicting evidence finding no association between GST expression and susceptibility to colorectal cancer (van der Logt et al., 2004).
Table 2.
Genetic variations of GSTs
| Allele | Nucleotide variability |
|---|---|
| GSTA1*A | Wild-type |
| GSTA1*B | Promotor point mutation |
| GSTA2*A | Pro110;Ser112;Lys196;Glu210 |
| GSTA2*B | Pro110;Ser112;Lys196;ALa210 |
| GSTA2*C | Pro110;Thr112;Lys196;Glu210 |
| GSTA2*D | Pro110;Ser112;Asn196;Glu210 |
| GSTA2*E | Ser110;Ser112;Lys196;Glu210 |
| GSTO1*A | Ala140;Glu155 |
| GSTO1*B | Ala140;Glu155 deletion |
| GSTO1*C | Asp140;Glu155 |
| GSTO1*D | Asp140;Glu155 deletion |
| GSTO2*A | Asn142 |
| GSTO2*B | Asp142 |
| GSTZ1*A | Lys32;Arg42;Thr82 |
| GSTZ1*B | Lys32;Gly42;Thr82 |
| GSTZ1*C | Glu32;Gly42;Thr82 |
| GSTZ1*D | Glu32;Gly42;Met82 |
| GSTM1*A | Lys173 |
| GSTM1*B | Asn173 |
| GSTM1*0 | Deletion |
| GSTM1*1x2 | Duplication |
| GSTM3*A | Wild type |
| GSTM3*B | 3 base deletion, intron 6 |
| GSTM4*A | Tyr2517 |
| GSTM4*B | Cyt2517 |
| GSTT1*A | Thr104 |
| GSTT1*B | Pro104 |
| GSTT1*0 | Deletion |
| GSTT2*A | Met139 |
| GSTT2*B | I1e139 |
| GSTP1*A | I1e105;Ala114 |
| GSTP1*B | Val105;Ala114 |
| GSTP1*C | Val105;Val114 |
| GSTP1*D | I1e105;Val114 |
GSTA2 has not been extensively studied, but it is known to have several variants (GSTA2*A-E). Single nucleotide polymorphisms for GSTA2 are summarized in Table 2. The catalytic properties of GSTA2 variants A-D do not seem to differ; however, the novel variant GSTA2E shows reduced rates of catalysis when compared to A-D (Ning et al., 2004). This may be due to substitution of the highly conserved Pro residue (Ning et al., 2004).
GSTA3–3 is selectively expressed in steroidogenic tissues and plays a role in steroid hormone biosynthesis (Johansson and Mannervik, 2001). Three GSTA3 transcripts have been identified along with a p.I71L polymorphism found exclusively in African populations (Tetlow et al., 2004a). Polymorphisms in GSTA3 may affect steroidogenesis through altered protein levels or function, and it has recently been hypothesized that alterations in genes involved in steroidogenesis and sex steroid metabolism could potentiate risk factors for the development of ovarian cancer (Tiltman and Ali, 2001; Tetlow et al., 2004a).
Omega class
The Omega class of GSTs contains two members (GSTO1 and GSTO2) and a pseudogene (GSTO3p). Based on two defining features, the Omega class is both structurally and functionally distinct from other eukaryotic GSTs (Townsend and Tew, 2003). First, X-ray crystallography shows a unique 19-residue N-terminus extension that forms a structural unit quite unlike any found in other classes. At present, its function remains undefined (Board et al., 2000). Second, known substrates of other GSTs are not catalysed by GSTO.
GSTO1 is a single gene located on chromosome 10 that codes for proteins expressed abundantly in the liver, macrophages, glial and endocrine cells (Board et al., 2000; Yin et al., 2001). To date, four polymorphisms have been identified, GSTO1*A–D (see Table 2) (Board et al., 2000). Among the Australian, African and Chinese populations, GSTO1*A was the most prevalent haplotype with a frequency ranging from 0.6 to 0.9; whereas GSTO1*B*A was the least common, with a frequency of 0.01–0.05 (Whitbread et al., 2003). GSTO1*A demonstrated a GSH-dependent reduction of dehydroascorbate, a function characteristic of glutaredoxins rather than GSTs (Board et al., 2000). This allele was first described as the human monomethylarsenic acid reductase, (MMA[V]), and is the rate-limiting enzyme of inorganic arsenic metabolism (Zakharyan et al., 2001). Thioltransferase activity differs among the GSTO1*A-C polymorphisms and may contribute to an individual’s ability to metabolize arsenic (Tanaka-Kagawa et al., 2003).
GSTO2, while separated from GSTO1 by 7.5 kb on chromosome 10, shares 64% amino-acid identity (Whitbread et al., 2003). GSTO2, like GSTO1, is ubiquitously expressed and shares GSH-dependent dehydroascorbate reductase activity. However, GSTO2 has a high catalytic activity toward CDNB, and its overexpression induced apoptosis, suggesting a possible role in cell signaling (Wang et al., 2005).
Zeta class
The Zeta class (GSTZ1) is a single 10.9 kb gene located on chromosome 14 that codes for a 29 kDa protein (Table 1) (Blackburn et al., 1998). GSTZ1 was independently characterized as maleylacetoacetate isomerase (MAAI) and plays a putative isomerase role in the catabolic pathway of phenylalanine and tyrosine in addition to the GSH-dependent transformation of α- halogenated acids (Fernandez-Canon et al., 2002). GSTZ1 is preferentially expressed in hepatocytes and renal proximal tubule cells where phenylalanine and tyrosine are catabolized.
While the literature is limiting for this class, GSTZ1 polymorphisms have been identified (GSTZ1*A-D) (Table 2) (Blackburn et al., 1998, 2001). The isozyme GSTZ1*A has the highest catalytic activity toward dichloroacetic acid, an investigational drug for certain metabolic disorders and a nephrotoxic metabolite of industrial solvents (Board et al., 2000; Guo et al., 2005). In contrast, GSTZ1*D (p.T82M) has a reduced catalytic activity and has been associated with inborn errors in tyrosine metabolism, although the disorders have also been attributed to mutations in other enzymes. Rodent models deficient for GSTZ1 provide insight into its role in metabolic deficiencies. GSTZ1/MAAI converts maleylacetoacetate to fumarylacetoacetate (FAA). GSTZ1-deficient mice have an elevated urinary excretion of FAA and were subject to renal injury following phenylalanine and tyrosine overload (Fernandez-Canon et al., 2002). Four families have been identified that have GSTZ1-deficient members that have died within the first year of life. Whereas the clinical data for GSTZ1 are insufficient to deduce a role for GSTZ1 in inherited genetic disease, it is plausible that a perturbation in GSTZ1-mediated tyrosine metabolism is contributory to the described pathology.
Mu class
Five GST isoforms belonging to the mu class (GSTM1–5) have been described (Pearson et al., 1993). A gene cluster located on chromosome 1 encodes for GSTM1–5 (Table 1) (DeJong et al., 1991). The GSTM1 gene contains four different alleles allowing for several M1 class polymorphisms. GSTM1*A and GSTM1*B are functionally identical and only differ by one p.K173N amino-acid substitution (Table 2) (McLellan et al., 1997). The presence of the GSTM1*A allele has been associated with a decreased risk of bladder cancer with the implication that detoxification of possible bladder-specific carcinogens may occur in these individuals (Smith et al., 1995). In addition, McLellan et al. (1997) described a rapid enzyme activity phenotype in Saudi Arabian individuals attributed to a tandem M1 gene duplication resulting in two functional M1 genes. It was inferred that this rapid detoxification phenotype could have an increased protective effect against carcinogens (McLellan et al., 1997).
Loss of GSTM enzyme function is ascribed to a homozygous deletion of this gene resulting in the GSTM1*0 allele (Smith et al., 1994). It has been suggested that the mutation is a result of an unequal crossing over of the M1 and M2 loci (Xu et al., 1998), which are in close physical proximity and share 99% nucleotide sequence identity (Pearson et al., 1993). The frequency of GTSM1*0 individuals is approximately 67% in Australians, 50% in Caucasians and 22% in Nigerians (Smith et al., 1995). The GSTM null phenotype is associated with an increased risk of the lung, colon and bladder cancer and has also been associated with response rates to some chemotherapy (Smith et al., 1994; Lohmueller et al., 2003). For example, GSTM1*0 patients with acute myeloid leukemia (AML) have been shown to have a better response to adriamycin and cyclophosphamide treatment, although precise reasons for this correlation are not clear (Ambrosone et al., 2001).
Cytosolic prostaglandin E was purified from the human brain and characterized as GSTM2 (Beuckmann et al., 2000). The GSTM2*B allele has been shown to catalyse the conjugation of GSH to aminochrome, a redox cycling product of dopamine (Baez et al., 1997). Products formed during the redox cycling of catecholamines (such as dopamine) contribute to the processes involved in neurodegenerative diseases such as Parkinson’s disease and schizophrenia; therefore, GSTM2*B may play a cytoprotective role in neurodegeneration (Baez et al., 1997).
The GSTM3 locus contains two alleles, A and B. The GSTM3*B allele has a three base pair deletion in intron 6 that introduces a recognition motif for the transcription factor YY1 (Inskip et al., 1995). GSTM3*A and *B are expressed in the brain; yet, there appears to be no direct relationship between GSTM3 expression and the incidence of astrocytomas (Hand et al., 1996). The GSTM3*AA genotype is associated with an increased risk for laryngeal squamous cell carcinoma, whereas GSTM3*BB was putatively protective (Matthias et al., 1998). In addition, GSTM3*AA was shown to occur more frequently in patients with multiple cutaneous basal cell carcinoma than GSTM3*BB (Yengi et al., 1996).
Recently, two new polymorphisms have been detected in the GSTM3 gene, a rare p.G147W substitution and a more common p.V224I substitution. These two polymorphisms can combine to form four different isoforms (Tetlow et al., 2004b). The p.W147 variant seems to exhibit decreased catalytic and specific activity, whereas the p.I224 variant has shown increased catalytic and specific activity (Tetlow et al., 2004b).
Theta class
The Theta class of GSTs consists of two different subfamilies: GSTT1 and GSTT2. Genes encoding both proteins are colocalized on chromosome 22 and are separated by 50 kb (Pemble et al., 1994; Tan et al., 1995). Polymorphisms exist within both genes including a null phenotype (GSTT1*0) that exhibits decreased catalytic activity and has been associated with an increased risk of cancers of the head, neck and oral cavity (Strange and Fryer, 1999).
A single nucleotide polymorphism in exon 3 results in two different variants, GSTT1*A and GSTT1*B (Table 2). GSTT1*A has a threonine at residue 104, which is substituted to a proline in the GSTT1*B allele (Alexandrie et al., 2002). The GSTT1*B allele shows a decreased catalytic activity when compared to the GSTT1*A allele, which could be attributed to the conformational change induced by the proline substitution (Alexandrie et al., 2002).
The presence of a G to A transition at intron 2 has given rise to a pseudogene (GSTT2P) of GSTT2. Evidence points to transcription of this gene, although the gene product is thought to be inactive. An allele identified in GSTT2 involves a rare p.M139L substitution (Coggan et al., 1998). A phenotype relating to this substitution has yet to be identified, although there is no clear indication that this substitution affects enzyme function (Coggan et al., 1998).
GST null phenotype and cancer prognosis
Deletion of the GSTM1 and GSTT1 genes results in a ‘null’ genotype characterized by a general deficit in enzymatic activity. Individuals homozygous for these deletions are thought to be at increased risk for malignancies as a consequence of a decreased capacity to detoxify possible carcinogens. Where this phenotype has been studied as a predictive factor for cancer prognosis or response to therapy, the results have been contingent upon tumor type. Patients with GSTM1*0 or GSTT1*0 showed a better survival rate after chemotherapeutic treatment for invasive ovarian cancer compared to other patients (Howells et al., 2001). However, results were contrary for ovarian cancer patients who were double null for GSTM1 and GSTT1. When compared to patients with the wild-type GSTM1 or GSTT1 allele, the null genotype patients exhibited a diminished response to chemotherapy resulting in both a poorer prognosis as well as a decrease in remission rates (Howells et al., 1998).
The occurrence of acute myelogenous leukemia (AML) is associated with exposure to carcinogenic chemicals, particularly benzene and its derivatives. This association of AML occurrence with carcinogen exposure provides a rationale for examining chemical detoxification enzymes, such as GSTs, in patients with AML (Ye and Song, 2005). Specifically, GSTM1 and GSTT1 phenotypes were examined in patients with AML, and it was shown that patients null for GSTT1 exhibited an increased toxicity and reduced survival rate following chemotherapeutic treatment (Davies et al., 2001). In contrast to this lower survival rate, patients null for GSTM1 or GSTT1 showed a twofold reduction in cancer relapse during remission (Stanulla et al., 2000).
GST null genotypes have also been examined when studying patients with chronic myeloid leukemia (CML). A reciprocal chromosomal translocation of t(9;22) results in a BCR-ABL fusion protein that causes a disruption of normal signaling pathways (Mondal et al., 2005). It is this pathway disruption that is thought to cause the initiation of CML. When comparing CML patients to a control population, a strong correlation was found between CML incidence and the presence of the GSTT1*0 genotype (Mondal et al., 2005). The frequency of GSTT1*0 in the CML patients varied greatly from the control group, whereas the frequency of the GSTM1*0 genotype did not (Mondal et al., 2005).
Epidemiological assessment
Small-scale population studies, particularly with cancer patients, are prone to significant limitations. There are numerous correlative reports that fall into this category, and deriving firm conclusions from such data are difficult. What is clear is that a single variable, such as is represented by a GST polymorphism, is not likely to afford a straightforward risk factor for analysis of cancer susceptibility or response to chemotherapy. Multivariables such as carcinogen exposure, pharmacokinetic/pharmacodynamic polymorphisms and population diversities must also be taken into account. This consideration only serves to further complicate the ability to draw consistent conclusions.
The literature is replete with correlative epidemiological reports where small sample size precludes definitive conclusions of cause and effect between GST expression and cancer incidence. As a phase II detoxification enzyme, GST is unlikely to act in isolation. As such, in the absence of multivariant analysis of other metabolic enzymes, any statistical correlations are likely to be oversimplistic. Moreover, the ligand-binding properties of GST (particularly those identified recently for GSTπ) will also serve to enhance the level of complexity for any correlative analysis. Collectively, present studies serve to show that levels of GST expression contribute to disease progression and prognosis; however, at best, the data are confusing and sometimes conflicting.
GST Pi
The GST Pi class is encoded by a single gene spanning approximately 3 kb and located on chromosome 11 (Cowell et al., 1988). Four active, functionally different polymorphisms (GSTP1*A-D) have been identified (Ali-Osman et al., 1997). The GSTP1 genotype has been associated with differences in chemotherapeutic response and cancer susceptibility and is overexpressed in a wide variety of tumors including ovarian, NSCLC, breast, colon, pancreas and lymphoma (Tew, 1994).
GSTP1*A has been reported to play a role in the acquisition of resistance to cisplatin via formation of platinum–GSH conjugates (Goto et al., 1999). Patients expressing homozygosity for the GSTP1*B allele have a diminished capacity to detoxify platinum-based anticancer agents, thus making this phenotype favorable for response rates (Stoehlmacher et al., 2002). An p.I105V substitution in the GSTP1*B allele results in a substantial reduction in catalytic activity and diminished detoxification capacity in individuals possessing the p.V105 as compared to individuals possessing the p.I105 allele (Watson et al., 1998). GSTp1*C, an allelic variant that is predominant in malignant glioma cells, differs from other GSTP1 variants by two transitions resulting in p.I104V and p.A113V (Lo and Ali-Osman, 1998). Individuals positive for GSTp1*C seem to have a lower incidence of breast cancer (Maugard et al., 2001). However, the precise relevance of this variant to disease occurrence or progression is not yet established.
Although it is well established that GSTπ is overexpressed in a wide variety of solid tumors, prostate cancer is the only example in which the absence or reduced expression of GSTπ is associated with tumor incidence. GSTπ is widely expressed in normal prostate tissue; however, its presence is undetectable in malignant cells (Nelson et al., 2001). Studies examining the absence of GSTπ in human prostate cancer show that hypermethylation of the GSTπ regulatory region is the most common somatic alteration identified (Lin et al., 2001). This alteration results in the loss of GSTπ expression and is proposed to occur during pathogenesis of the disease (Lee et al., 1994). Recently, a methyl-CpG binding domain protein has been identified that mediates hypermethylation of the GSTπ regulatory region (Bakker et al., 2002). These findings provide a possible target for restoration of GSTp activity. GST expression (and/or activity) of specific isoforms is lost in some individuals with allelic variation.
Recently, Dang et al. (2005) examined the effect of GSTP1 on the survival and proliferation of human colon cancer cells. GSTP1 wild type and deficient colon cancer cells were grown under serum deprivation and low-density seeding conditions, and cellular apoptosis, oxidative stress and kinase signaling were examined. Lack of GSTP1 expression resulted in an increase in cellular oxidative stress and resultant apoptosis when cells were cultured under growth-limiting conditions (Dang et al., 2005). In addition, the presence of GSTP1 was essential for the mediation of MAPK kinase and ERK kinase signaling (Dang et al., 2005). Subsequent in vivo data correlated with the in vitro findings showing that GSTP1 plays a crucial role in early tumorigenesis (Dang et al., 2005). This study illustrates that GSTP1 is important for human colon cancer cell survival and proliferation, and overall these results show that GSTP1 plays a critical role in protection from cell cycle arrest and oxidative stress under growth-limiting conditions (Dang et al., 2005).
GSTs and kinase regulation
It has been speculated that the absence or decreased expression of GSTπ results in a reduced detoxification of possible carcinogens that may be causal to malignant transformation and disease progression. In addition, the GST-mediated conjugation of GSH to a number of anticancer drug substrates has long been linked to anticancer drug resistance in a variety of tumors. A disparity of this is that GSTπ has a weak affinity for the majority of anticancer drugs, although its increased expression is highly correlated with multidrug resistance. From this it can be inferred that the capacity of GSTs to regulate kinase-dependent proliferation pathways, especially in the case of GSTπ, may be of more consequence than its catalytic properties alone.
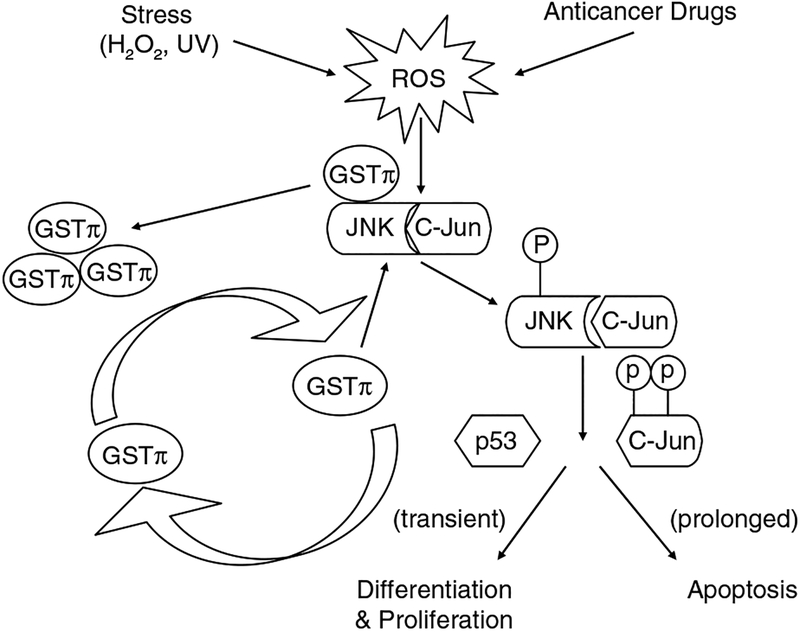
GSTs play a regulatory role in cellular signaling by forming protein:protein interactions with critical kinases involved in controlling stress response, apoptosis and proliferation. The ligand-binding capacity of GST results in the negative regulation of signaling pathways through sequestration of signaling kinases. The first example of GST-mediated kinase regulation is the characterization of GSTπ as a Jun kinase (JNK) inhibitor (Adler et al., 1999). JNK has been implicated in proapoptotic signaling and may be required for the induced cytotoxicity of a variety of chemotherapy agents (Yin et al., 2000). Phosphorylation of c-Jun activates JNK resulting in subsequent activation of downstream effectors. In non-stressed cells, low JNK1 catalytic activity is orchestrated and maintained through its sequestration within the protein complex that includes at least GSTπ and JNK (Adler et al., 1999). However, under conditions of oxidative or chemical stress, a dissociation of the GSTπ:JNK complex occurs releasing GSTπ for oligomerization, and activation of released JNK allows for the subsequent induction of apoptosis (Figure 1) (Adler et al., 1999; Davis et al., 2001). The high levels of GSTπ in many tumors may be a consequence of an acquired dependence on the protein. Because of the proliferative nature of tumor cells, many kinase pathways are dysregulated, and as a consequence, tumor cells may attempt to compensate by enhancing expression of GSTπ in an attempt to control kinase activity.
Figure 1.
Under non-stressed conditions, GSTπ inhibits JNK phosphorylation by sequestering JNK/c-Jun. Exposure to anticancer drugs or oxidative stress can alter the redox potential of the cell resulting in the oligomerization of GSTπ and the dissociation of the GSTπ:JNK complex. JNK can then become phosphorylated and subsequently activate downstream kinases and transcription factors. In some cases, transient or low exposure to stress can induce cell proliferation. During prolonged or high exposure, apoptosis can be induced.
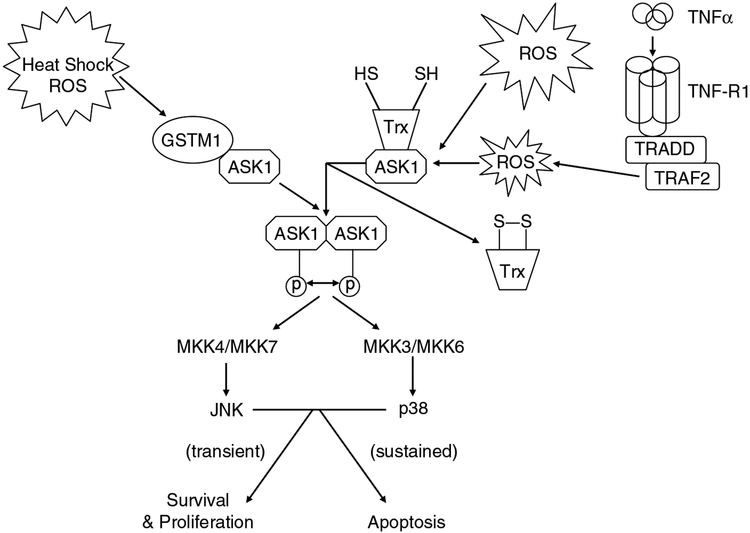
Another example of GST-mediated kinase regulation is evidence that GSTM1 binds to and inhibits the activity of ASK1 (Cho et al., 2001). ASK1 is an MAP kinase kinase kinase that activates the JNK and p38 pathways leading to cytokine- and stress-induced apoptosis (Ichijo et al., 1997). Under normal conditions, ASK1 exhibits low activity because of its sequestration via GSTM1. This protein:protein interaction forms a GSTM1:ASK1 complex, which is dissociated under stressful conditions leading to the release and activation of ASK1 (Figure 2) (Saitoh et al., 1998; Dorion et al., 2002). This mechanism is similar to the one proposed for GSTπ:JNK. In conditions such as oxidative stress or heat shock, GSTM1 oligomerizes allowing for the release of ASK1 and subsequent induction of apoptosis (Dorion et al., 2002). Impaired clinical response to therapy in a variety of tumor types has been associated with an altered expression of GSTM1. Thus, any enzymatic influence GSTM1 plays in anticancer drug resistance is only further augmented by its role in kinase regulation.
Figure 2.
GSTμ and thioredoxin (Trx) can act as inhibitors of ASK1. Stresses such as heat shock or reactive oxygen species can result in the release of ASK1 from the GSTμ:ASK1 or TRX:ASK1 complex (respectively). ASK1 oligomerizes and is activated through autophosphorylation, which in turn activates downstream kinases such as MKK4/MKK7, MKK3/MKK6, JNK and p38. The fate of the cell (either proliferation or apoptosis) is dependent upon the time/concentration exposure to the stress.
In addition to acting as a negative regulator of kinase activation, GSTπ has also been shown to play a necessary role in the glutathionylation of 1-cys peroxiredoxin (1-cysPrx). Oxidation of the catalytic cysteine of 1-cysPrx has been associated with its loss of peroxidase activity (Manevich and Fisher, 2005). Recently, it was shown that heterodimerization of 1-cysPrx with GSTπ mediates the glutathionylation of the previously oxidized cysteine thus restoring its peroxidase activity (Manevich and Fisher, 2005). From this study, it was concluded that the glutathionylation and subsequent GSH-mediated reduction of 1-cysPrx requires heterodimerization with GSTπ (Manevich and Fisher, 2005). This provides the first example in which GSTπ functions in the glutathionylation of oxidized cysteine residues. In addition, our lab has found that cells deficient in GSTP1–1 and/or GSTP2–2 have a reduced capacity to respond to oxidative or nitrosative stress by enacting glutathionylation of a select group of target proteins (Townsend et al., 2005). These findings imply that GSTπ may play a direct role in control of post-translational glutathionylation reactions.
Pharmacogenetics and GSTs
Epidemiological evidence shows the existence of significant variations in individual patient response to cancer chemotherapy. These variations include altered drug metabolism, genetic polymorphisms and their functional significance, varied exposure to carcinogens, and population diversities such as race and ethnicity. When attempting to design an effective therapeutic regime, it is necessary to consider the different susceptibility and pharmacogenetic factors involved. The complex genetics and expression patterns of GSTs should provide a variable platform for rational drug design and serve to make GSTs an applicable target for anticancer therapy.
In tumors where GST overexpression has an adverse effect on therapeutic response, distinct strategies have been adopted to target GSTs. One pharmacological approach is the design of GST inhibitors to increase the efficacy of chemotherapeutics that would otherwise be detoxified by GSTs. Broadly, GST inhibitors can be classified as competitive or non-competitive. Two of the best-characterized inhibitors are ethacrynic acid (EA) and the GSH analog TLK199 [γ-glutamyl-S-(benzyl)-cysteinyl-R-phenyl glycine diethyl ester]. EA acts as a non-competitive inhibitor of GSH for GST binding as well as depleting the GSH cofactor by forming an EA-GSH conjugate (Ploemen et al., 1994). The binding to both GST and GSH serves to inhibit enzyme activity. Our earlier studies have shown this to be effective in sensitizing tumors that otherwise exhibited chemotherapy resistance (Tew et al., 1998). TLK199 is a selective inhibitor of GSTπ and can also act as a chemosensitizer potentiating the toxicity of numerous anticancer agents in different tumor cell lines. Sensitivity to a range of anticancer drugs, including nitrogen mustards, was enhanced in xenograft models with elevated GST levels (Morgan et al., 1996). Because of the structural similarities to GSH, TLK199 has also been shown to effectively inhibit the multidrug resistance-associated protein 1 (MRP-1), reversing the resistance of a variety of agents in NlH3T3 cells transfected with MRP-1 (O’Brien et al., 1999).
Interestingly enough, TLK199 exhibited myeloproliferative effects in mice during preclinical studies (Ruscoe et al., 2001). In vitro hematopoiesis experiments showed that mice deficient in GSTπ exhibit an increase in myeloid cell differentiation and proliferation (Gate et al., 2004). The molecular basis for this effect is possibly due to an increase in bone marrow progenitor cells proliferating and differentiating thus increasing production of circulating mature blood cells (Gate et al., 2004). In addition, a probable cause of these myeloproliferative effects is the ability of TLK199 to disrupt the protein:protein interaction of the JNK:GSTp complex (Adler et al., 1999; Ruscoe et al., 2001; Wang et al., 2001). As a consequence of this effect, JNK activity is enhanced and, in the case of the bone marrow compartment, this has been causally associated with the increase in cellular proliferation (Gate et al., 2004).
A new class of 7-nitro-2,1,3,-benzoxadiazole derivatives have been designed as suicide inhibitors of GSTs. These NBDs thioethers have micromolar to nanomolar affinities for binding to the H-site of GSTA1–1, GSTP1–1 and GSTM2–2 (Ricci et al., 2005). NDBs have been shown to induce tumor cell death by dissociating the JNK:GSTπ complex thus initiating apoptosis (Turella et al., 2005). When compared to the earlier example of bone marrow, this serves to illustrate the different role of JNK in transformed cells. A representative model of these NBD derivatives is 6-(7-nitro-2,1,3,-benzoxadia-zole-4-ylthio)hexanol or NBDHEX (Ricci et al., 2005). Their utility as possible anticancer drugs has yet to be established; however, mechanistically their impact upon kinase pathways does conform to literature predictions.
Another newly designed GSTA1–1-specific inhibitor is the GSH conjugate L-γ-glutamyl-(S-9-fluorenylmethyl)-L-cysteinyl-glycine (Cacciatore et al., 2005). This inhibitor possesses a fluorenylmethyl group as a cysteine S-derivatized moiety. It is thought that the H-site of GSTA1–1 will favor the binding of this isoform with a greater affinity, whereas the fluorescent properties will allow for easier monitoring during biological tests (Cacciatore et al., 2005). L-γ-glutamyl-(S-9-fluorenyl-methyl)-L-cysteinyl-glycine was shown to be effective in inhibiting GSTA1–1, GSTP1–1 and GSTM2–2 with the greatest inhibition shown against the alpha isoform (Cacciatore et al., 2005). Again, any practical utility as an anticancer drug has yet to be established.
Prodrugs have been designed as inactive agents that are converted to active cytotoxics upon exposure to tumor tissues exhibiting high expression of activating enzymes. This targeting strategy minimizes toxicity towards normal tissues while allowing for an increased delivery of active agent to the tumor tissue. Enhanced expression of GSTs in many tumors makes GSTs a promising target for prodrug therapy. One approach has been to design prodrugs exploiting the ability of GSTs to mediate cleavage of sulfonamides by promoting a β- elimination reaction. A logical extension of this concept is the synthesis of drugs as inactive compounds via GSH conjugation through a sulfone linkage. TLK286, [γ-glutamyl-α-amino-β-(2-ethyl-N,N,N’,N’-tetrakis (2-chloroethyl) phosphorodiamidate)-sulfonyl-propionyl-(R)-(—) phenylglycine], is the lead candidate from a novel class of latent drugs activated in cancer cells by GSTP1*A (Lyttle et al., 1994). TLK286 is cleaved by a GSTP1*A-promoted β-elimination reaction into a nitrogen mustard and GSH analog that can then alkylate cellular nucleophiles (Morgan et al., 1998). GSTP1–1 expression is correlated to drug sensitivity both in cell culture and in animal models (Morgan et al., 1998; Rosario et al., 2000). In addition, GSTP1*A was reported to be adaptively downregulated at both the protein and transcript level following chronic exposure to TLK286 (Rosario et al., 2000). Collectively, these results serve to support the concept that TKL286 will selectively target tumors expressing high levels of GSTP1*A thus preferentially exerting its cytotoxic effect on these tissues.
A series of sulfonylhydrazine prodrugs have been synthesized that utilize the reductive environment of hypoxic tumors to generate cytodestructive species. 1,2-Bis(methylsufonyl)-1-(2-chloroethyl)2-[[1-(4-nitropheny-l)ethoxy]carbonyl]hydrazine (KS119) and 1,2-bis (methylsufonyl)-1-(2-chloroethyl)2-[[1-(4-nitrobenzylox-yl)ethoxy]carbonyl]hydrazine (PNBC) are activated under hypoxic conditions through the reduction of their nitroaromatic groups (Seow et al., 2005). Upon reductive activation, these drugs release the cytotoxic alkylating species 1,2-bis(methylsulfonyl)-1-(2-chloroethyl)hydrazine (90CE) (Seow et al., 2005). KS119 requires reduction of its nitro group to activate the release of 90CE, yet PNBC can also be activated through nucleophilic attack by thiols such as GSH and GST. In vitro studies demonstrated the ability of KS199 and PNBC to kill cells from solid tumors while maintaining minimal toxicity to normal cells under ordinary aerobic conditions (Seow et al., 2005). This exhibits hypoxia-selective activity against tumor tissue while remaining relatively non-toxic to normal tissue.
A group of nitric oxide releasing prodrugs have been designed to target GST-associated drug resistance. Two NO-releasing diazeniumdiolates, [O2-(2,4-dinitrophenyl) 1-[(4-ethoxycarbonyl)piperazin-1-yl]diazen-1-ium-1,2-diolate] (JS-K) and [O2-(2,4-dinitrophenyl) 1-[4-(N,N-diethylcarboxamido)piperazin-1-yl]diazen-1–1,2-ium-diolate] (CB-3–100), have been shown to effectively inhibit GSTp while increasing arsenic and cisplatin toxicity in previously resistant tumor cells (Liu et al., 2004). This GSTπ inhibition by JS-K and CB-3–100 also leads to an enhanced activation of JNK and ERK of the MAPK pathway supporting previous studies showing the role of GSTs in MAPK inhibition. Enhanced MAPK pathway activation in conjunction with increased accumulation of metallochemotherapeutics make these prodrugs candidates for cancer therapy.
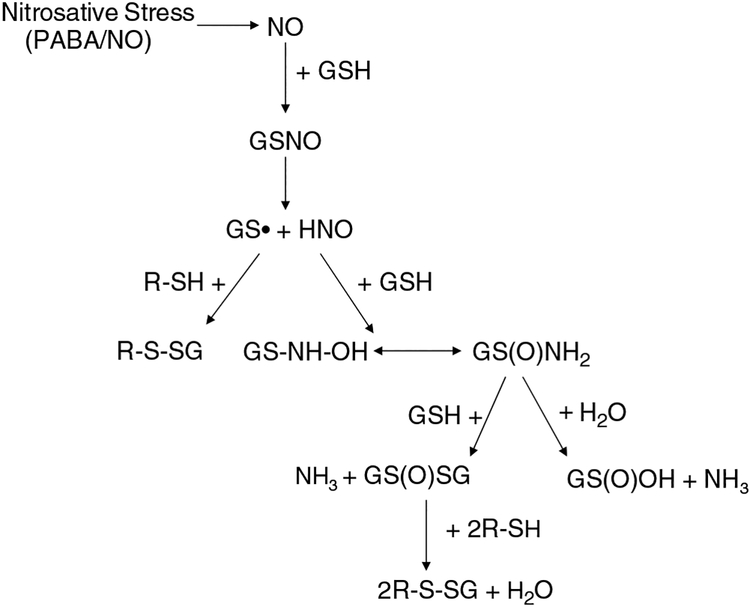
Another nitric oxide releasing drug is O(2)-[2,4-dinitro-5-(N-methyl-N-4-carboxyphenylamino) phenyl] 1-N,N-dimethylamino)diazen-1-ium-1,2-diolate (PABA/NO). GSTπ efficiently metabolizes PABA/NO to nitric oxide and has a preferential selectivity to GSTπ over GSTα (Findlay et al., 2004). In addition to releasing nitric oxide, treatment with PABA/NO has been shown to induce activation of p38, JNK and ERK (Findlay et al., 2004). It has not yet been shown whether kinase activation is caused by direct interaction with NO or interaction with the drug and/or its metabolites; however, this ability to activate stress-related kinases is significant due to the observed function of GSTπ as a negative regulatory switch for these kinase pathways. Figure 3 shows the possible S-thiolation intermediates following treatment with nitric oxide releasing drugs such as PABA/NO.
Figure 3.
Nitric oxide (NO) can react with GSH to form GSNO and subsequently a glutathionyl radical GS· and nitroxyl (HNO). This can react with GSH to give N-hydroxysulfenamide (GS-NHOH), which can rearrange to generate a sulfinamide (GS(O)NH2). Reaction of GS(O)NH2 with GSH forms GSH disulfide-S-oxide (GS(O)SG) or can be further oxidized to sulfinic acid (GS(O)OH) and NH3. The key intermediates leading to the synthesis of GS(O)SG are the sulfinamides (GS(O)NH2 and GS(O)-NH-SG). The reaction of GS(O)SG or GS· with a reduced protein thiol (R-SH) leads to the formation of a mixed disulfide.
Conclusions and perspectives
Many studies have provided epidemiological linkage of polymorphic expression of GSTs with cancer incidence and prognosis although, at times, the data are fragmented and inconsistent. These inconsistencies serve to emphasize the importance of considering the limitations and variables of each study when attempting to draw any definitive conclusions. GSTs have also been shown to play critical roles in kinase signaling as well as glutathionylation, indicating that their function extends beyond their endogenous catalytic properties. These functions in control of cell signaling and post-translational modification pathways provide novel avenues for exploration of new drugs. It is unlikely that 10 years ago, a myeloproliferative function would have been ascribed to GSTπ inhibition. The fact that the importance of S-glutathionylation has yet to be fully appreciated gives credence to the possibility that antagonists/agonists of this pathway may have biological consequences. In addition, the GSTπ activated prodrug Telcyta (TLK286) is presently in late-stage clinical development. The results of Phase III trials should be available in 2006, and these will influence eventual FDA registration of the drug. Obviously, development of such drugs will influence future drug discovery efforts.
Acknowledgements
This publication was made possible by grant number NCI CA53783 from the National Institutes of Health and grant number T32 ES012878 from the National Institutes of Environmental Health Sciences, NIH.
References
- Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD et al. (1999). EMBO J 18: 1321–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrie AK, Rannug A, Juronen E, Tasa G, Warholm M. (2002). Pharmacogenetics 12: 613–619. [DOI] [PubMed] [Google Scholar]
- Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J. (1997). J Biol Chem 272: 10004–10012. [DOI] [PubMed] [Google Scholar]
- Ambrosone CB, Sweeney C, Coles BF, Thompson PA, McClure GY, Korourian S et al. (2001). Cancer Res 61: 7130–7135. [PubMed] [Google Scholar]
- Armstrong RN. (1997). Chem Res Toxicol 10: 2–18. [DOI] [PubMed] [Google Scholar]
- Baez S, Segura-Aguilar J, Widersten M, Johansson AS, Mannervik B. (1997). Biochem J 324(Part 1): 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker J, Lin X, Nelson WG. (2002). J Biol Chem 277: 22573–22580. [DOI] [PubMed] [Google Scholar]
- Beuckmann CT, Fujimori K, Urade Y, Hayaishi O. (2000). Neurochem Res 25: 733–738. [DOI] [PubMed] [Google Scholar]
- Blackburn AC, Coggan M, Tzeng HF, Lantum H, Polekhina G, Parker MW et al. (2001). Pharmacogenetics 11: 671–678. [DOI] [PubMed] [Google Scholar]
- Blackburn AC, Woollatt E, Sutherland GR, Board PG. (1998). Cytogenet Cell Genet 83: 109–114. [DOI] [PubMed] [Google Scholar]
- Board PG, Coggan M, Chelvanayagam G, Easteal S, Jermiin LS, Schulte GK et al. (2000). J Biol Chem 275: 24798–24806. [DOI] [PubMed] [Google Scholar]
- Cacciatore I, Caccuri AM, Cocco A, De Maria F, Di Stefano A, Luisi G et al. (2005). Amino Acids 29: 255–261. [DOI] [PubMed] [Google Scholar]
- Cho SG, Lee YH, Park HS, Ryoo K, Kang KW, Park J et al. (2001). J Biol Chem 276: 12749–12755. [DOI] [PubMed] [Google Scholar]
- Chuang ST, Chu P, Sugimura J, Tretiakova MS, Papavero V, Wang K et al. (2005). Am J Clin Pathol 123: 421–429. [DOI] [PubMed] [Google Scholar]
- Coggan M, Whitbread L, Whittington A, Board P. (1998). Biochem J 334(Part 3): 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles B, Nowell SA, MacLeod SL, Sweeney C, Lang NP, Kadlubar FF. (2001). Mutat Res 482: 3–10. [DOI] [PubMed] [Google Scholar]
- Cowell IG, Dixon KH, Pemble SE, Ketterer B, Taylor JB. (1988). Biochem J 255: 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang DT, Chen F, Kohli M, Rago C, Cummins JM, Dang LH. (2005). Cancer Res 65: 9485–9494. [DOI] [PubMed] [Google Scholar]
- Davies SM, Robison LL, Buckley JD, Tjoa T, Woods WG, Radloff GA et al. (2001). J Clin Oncol 19: 1279–1287. [DOI] [PubMed] [Google Scholar]
- Davis W Jr, Ronai Z, Tew KD. (2001). J Pharmacol Exp Ther 296: 1–6. [PubMed] [Google Scholar]
- DeJong JL, Mohandas T, Tu CP. (1991). Biochem Biophys Res Commun 180: 15–22. [DOI] [PubMed] [Google Scholar]
- Dirr H, Reinemer P, Huber R. (1994). Eur J Biochem 220: 645–661. [DOI] [PubMed] [Google Scholar]
- Dorion S, Lambert H, Landry J. (2002). J Biol Chem 277: 30792–30797. [DOI] [PubMed] [Google Scholar]
- Fernandez-Canon JM, Baetscher MW, Finegold M, Burlingame T, Gibson KM, Grompe M. (2002). Mol Cell Biol 22: 4943–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay VJ, Townsend DM, Saavedra JE, Buzard GS, Citro ML, Keefer LK et al. (2004). Mol Pharmacol 65: 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gate L, Majumdar RS, Lunk A, Tew KD. (2004). J Biol Chem 279: 8608–8616. [DOI] [PubMed] [Google Scholar]
- Goto S, Iida T, Cho S, Oka M, Kohno S, Kondo T. (1999). Free Radic Res 31: 549–558. [DOI] [PubMed] [Google Scholar]
- Guo X, Dixit VS, Liu HP, Shroads AL, Henderson GN, James MO et al. (2005). Drug Metab Dispos: Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Hand PA, Inskip A, Gilford J, Alldersea J, Elexpuru-Camiruaga J, Hayes JD et al. (1996). Carcinogenesis 17: 1919–1922. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. (2005). Annu Rev Pharmacol Toxicol 45: 51–88. [DOI] [PubMed] [Google Scholar]
- Howells RE, Holland T, Dhar KK, Redman CW, Hand P, Hoban PR et al. (2001). Int J Gynecol Cancer 11: 107–112. [DOI] [PubMed] [Google Scholar]
- Howells RE, Redman CW, Dhar KK, Sarhanis P, Musgrove C, Jones PW et al. (1998). Clin Cancer Res 4: 2439–2445. [PubMed] [Google Scholar]
- Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T et al. (1997). Science 275: 90–94. [DOI] [PubMed] [Google Scholar]
- Inskip A, Elexperu-Camiruaga J, Buxton N, Dias PS, MacIntosh J, Campbell D et al. (1995). Biochem J 312(Part 3): 713–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson AS, Mannervik B. (2001). J Biol Chem 276: 3306133065. [DOI] [PubMed] [Google Scholar]
- Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS et al. (1994). Proc Natl Acad Sci USA 91: 11733–11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Tascilar M, Lee WH, Vles WJ, Lee BH, Veeraswamy R et al. (2001). Am J Pathol 159: 1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li C, Qu W, Leslie E, Bonifant CL, Buzard GS et al. (2004). Mol Cancer Ther 3: 709–714. [PubMed] [Google Scholar]
- Lo HW, Ali-Osman F. (1998). Chem Biol Interact 111–112: 91–102. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. (2003). Nat Genet 33: 177–182. [DOI] [PubMed] [Google Scholar]
- Lyttle MH, Satyam A, Hocker MD, Bauer KE, Caldwell CG, Hui HC et al. (1994). J Med Chem 37: 1501–1507. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Fisher AB. (2005). Free Radic Biol Med 38: 1422–1432. [DOI] [PubMed] [Google Scholar]
- Mannervik B, Danielson UH. (1988). CRC Crit Rev Biochem 23: 283–337. [DOI] [PubMed] [Google Scholar]
- Matthias C, Bockmuhl U, Jahnke V, Jones PW, Hayes JD, Alldersea J et al. (1998). Pharmacogenetics 8: 91–100. [PubMed] [Google Scholar]
- Maugard CM, Charrier J, Pitard A, Campion L, Akande O, Pleasants L et al. (2001). Int J Cancer 91: 334–339. [DOI] [PubMed] [Google Scholar]
- McLellan RA, Oscarson M, Alexandrie AK, Seidegard J, Evans DA, Rannug A et al. (1997). Mol Pharmacol 52: 958–965. [DOI] [PubMed] [Google Scholar]
- Mondal BC, Paria N, Majumdar S, Chandra S, Mukhopadhyay A, Chaudhuri U et al. (2005). Eur J Cancer Prev 14: 281–284. [DOI] [PubMed] [Google Scholar]
- Morgan AS, Ciaccio PJ, Tew KD, Kauvar LM. (1996). Cancer Chemother Pharmacol 37: 363–370. [DOI] [PubMed] [Google Scholar]
- Morgan AS, Sanderson PE, Borch RF, Tew KD, Niitsu Y, Takayama T et al. (1998). Cancer Res 58: 2568–2575. [PubMed] [Google Scholar]
- Nelson WG, De Marzo AM, Deweese TL, Lin X, Brooks JD, Putzi MJ et al. (2001). Ann NY Acad Sci 952: 135–144. [DOI] [PubMed] [Google Scholar]
- Ning B, Wang C, Morel F, Nowell S, Ratnasinghe DL, Carter W et al. (2004). Pharmacogenetics 14: 35–44. [DOI] [PubMed] [Google Scholar]
- O’Brien mL, Vulevic B, Freer S, Boyd J, Shen H, Tew KD. (1999). J Pharmacol Exp Ther 291: 1348–1355. [PubMed] [Google Scholar]
- Pearson WR, Vorachek WR, Xu SJ, Berger R, Hart I, Vannais D et al. (1993). Am J Hum Genet 53: 220–233. [PMC free article] [PubMed] [Google Scholar]
- Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM et al. (1994). Biochem J 300(Part 1): 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploemen JH, Van Schanke A, Van Ommen B, Van Bladeren PJ. (1994). Cancer Res 54: 915–919. [PubMed] [Google Scholar]
- Ricci G, De Maria F, Antonini G, Turella P, Bullo A, Stella L et al. (2005). J Biol Chem 280: 26397–26405. [DOI] [PubMed] [Google Scholar]
- Rosario LA, O’Brien ML, Henderson CJ, Wolf CR, Tew KD. (2000). Mol Pharmacol 58: 167–174. [DOI] [PubMed] [Google Scholar]
- Ruscoe JE, Rosario LA, Wang T, Gate L, Arifoglu P, Wolf CR et al. (2001). J Pharmacol Exp Ther 298: 339–345. [PubMed] [Google Scholar]
- Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y et al. (1998). EMBO J 17: 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow HA, Penketh PG, Shyam K, Rockwell S, Sartorelli AC. (2005). Proc Natl Acad Sci USA 102: 9282–9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cm Smith, Kelsey KT Wiencke JK, Leyden K, Levin S, Christiani DC. (1994). Cancer Epidemiol Biomarkers Prev 3: 471–477. [PubMed] [Google Scholar]
- Smith G, Stanley LA, Sim E, Strange RC, Wolf CR. (1995). Cancer Surv 25: 27–65. [PubMed] [Google Scholar]
- Stanulla M, Schrappe M, Brechlin AM, Zimmermann M, Welte K. (2000). Blood 95: 1222–1228. [PubMed] [Google Scholar]
- Stoehlmacher J, Park DJ, Zhang W, Groshen S, Tsao-Wei DD, Yu MC et al. (2002). J Natl Cancer Inst 94: 936–942. [DOI] [PubMed] [Google Scholar]
- Strange RC, Fryer AA. (1999). IARC Sci Publ, 231–249. [PubMed] [Google Scholar]
- Tan KL, Webb GC, Baker RT, Board PG. (1995). Genomics 25: 381–387. [DOI] [PubMed] [Google Scholar]
- Tanaka-Kagawa T, Jinno H, Hasegawa T, Makino Y, Seko Y, Hanioka N, Ando M. (2003). Biochem Biophys Res Commun 301: 516–520. [DOI] [PubMed] [Google Scholar]
- Tetlow N, Coggan M, Casarotto MG, Board PG. (2004a). Pharmacogenetics 14: 657–663. [DOI] [PubMed] [Google Scholar]
- Tetlow N, Robinson A, Mantle T, Board P. (2004b). Pharmacogenetics 14: 359–368. [DOI] [PubMed] [Google Scholar]
- Tew KD. (1994). Cancer Res 54: 4313–4320. [PubMed] [Google Scholar]
- Tew KD, O’Brien M, Laing NM, Shen H. (1998). Chem Biol Interact 111–112: 199–211. [DOI] [PubMed] [Google Scholar]
- Tiltman AJ, Ali H. (2001). Histopathology 39: 266–272. [DOI] [PubMed] [Google Scholar]
- Townsend D, Tew K. (2003). Am J Pharmacogenomics 3: 157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend DM, Findlay VJ, Fazilev F, Ogle M, Fraser J, Saavedra J et al. (2005). Mol Pharmacol: Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turella P, Cerella C, Filomeni G, Bullo A, De Maria F, Ghibelli L et al. (2005). Cancer Res 65: 3751–3761. [DOI] [PubMed] [Google Scholar]
- van der Logt EM, Bergevoet SM, Roelofs HM, van Hooijdonk Z, te Morsche RH, Wobbes T et al. (2004). Carcinogenesis 25: 2407–2415. [DOI] [PubMed] [Google Scholar]
- Wang L, Xu J, Ji C, Gu S, Lv Y, Li S et al. (2005). Int J Mol Med 16: 19–27. [PubMed] [Google Scholar]
- Wang T, Arifoglu P, Ronai Z, Tew KD. (2001). J Biol Chem 276: 20999–21003. [DOI] [PubMed] [Google Scholar]
- Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. (1998). Carcinogenesis 19: 275–280. [DOI] [PubMed] [Google Scholar]
- Whitbread AK, Tetlow N, Eyre HJ, Sutherland GR, Board PG. (2003). Pharmacogenetics 13: 131–144. [DOI] [PubMed] [Google Scholar]
- Xu S, Wang Y, Roe B, Pearson WR. (1998). J Biol Chem 273: 3517–3527. [DOI] [PubMed] [Google Scholar]
- Ye Z, Song H. (2005). Eur J Cancer 41: 980–989. [DOI] [PubMed] [Google Scholar]
- Yengi L, Inskip A, Gilford J, Alldersea J, Bailey L, Smith A et al. (1996). Cancer Res 56: 1974–1977. [PubMed] [Google Scholar]
- Yin Z, Ivanov VN, Habelhah H, Tew K, Ronai Z. (2000). Cancer Res 60: 4053–4057. [PubMed] [Google Scholar]
- Yin ZL, Dahlstrom JE, Le Couteur DG, Board PG. (2001). J Histochem Cytochem 49: 983–987. [DOI] [PubMed] [Google Scholar]
- Zakharyan RA, Sampayo-Reyes A, Healy SM, Tsaprailis G, Board PG, Liebler DC et al. (2001). Chem Res Toxicol 14: 1051–1057. [DOI] [PubMed] [Google Scholar]