Abstract
Ceramides, important players in signal transduction, interact with multiple cellular pathways, including p53 pathways. However, the relationship between ceramide and p53 is very complex, and mechanisms underlying their coregulation are diverse and not fully characterized. The role of p53, an important cellular regulator and a transcription factor, is linked to its tumor suppressor function. Ceramides are involved in the regulation of fundamental processes in cancer cells including cell death, proliferation, autophagy, and drug resistance. This regulation, however, can be pro-death or pro-survival depending on cancer type, the balance between ceramide species, the rate of their synthesis and utilization, and the availability of a specific array of downstream targets. This chapter highlights the central role of ceramide in sphingolipid metabolism, its role in cancer, specific effectors in ceramide pathways controlled by p53, and coregulation of ceramide and p53 signaling. We discuss the recent studies, which underscore the function of p53 in the regulation of ceramide pathways and the reciprocal regulation of p53 by ceramide. This complex relationship is based on several molecular mechanisms including the p53-dependent transcriptional regulation of enzymes in sphingolipid pathways, the activation of mutant p53 through ceramide-mediated alternative splicing, as well as modulation of the p53 function through direct and indirect effects on p53 coregulators and downstream targets. Further insight into the connections between ceramide and p53 will allow simultaneous targeting of the two pathways with a potential to yield more efficient anticancer therapeutics.
1. INTRODUCTION
Ceramides, members of the second largest class of membrane lipids, represent a family of more than 200 structurally related sphingolipids, compounds formed by a sphingoid base and a fatty acid linked via the amide bond (Hannun & Obeid, 2011, 2017). Over the past decades numerous studies have demonstrated that depending on their molecular structure, ceramides confer unique biophysical properties to the biological membranes (Castro, Prieto, & Silva, 2014). While the mechanisms of ceramide function in biological membranes and the biological consequences of ceramide alterations for cellular processes are still poorly understood, overwhelming data point to the role of these sphingolipids as important cellular regulators. Over the last 3 decades ceramides were established as second messengers regulating key cellular processes including cytoskeleton dynamics, endocytosis, protein transport and subcellular localization, cell cycle, autophagy, and apoptosis (Castro et al., 2014; Coant, Sakamoto, Mao, & Hannun, 2017; Dany & Ogretmen, 2015; Mullen, Hannun, & Obeid, 2012). Therefore, ceramides are indispensable for cellular homeostasis and control of fundamental functions, such as proliferation, migration, differentiation, adaptation to stress, survival, and senescence (Hannun & Obeid, 2008, 2017).
2. CERAMIDE SIGNALING—BIOLOGY
Ceramide, a central molecule in sphingolipid metabolism (Hannun & Obeid, 2008; Levy & Futerman, 2010), can be generated by the de novo pathway, which starts from the serine and palmitoyl-CoA condensation. It can be also produced by the salvage pathway, whereby sphingolipids are hydrolyzed in the lysosomes to sphingosine, which can be reacylated back to ceramide by one of the ceramide synthases (CerSs) (Coant et al., 2017). Finally, it can originate from the direct hydrolysis of complex sphingolipids such as sphingomyelin and glycosphingolipids (Hannun & Obeid, 2017). Overall, ceramide biosynthesis and the downstream function are interconnected with other lipid pathways, highly compartmentalized within the cell and also engaged in communication with organelle-specific metabolic pathways (Coant et al., 2017; Hannun & Obeid, 2011; Hoeferlin, Fekry, Ogretmen, Krupenko, & Krupenko, 2013). Collectively, these aspects of ceramide metabolism call for a “Many Ceramides” approach (Hannun & Obeid, 2011) for investigating ceramide effects and their significance for biological processes. Such an approach requires characterization of the temporal and spatial dynamics of different ceramide species, identification of pathways regulating such dynamics, and the close look at specific cellular compartments where the changes take place. Understanding of the complex aspects of ceramide biology will provide a comprehensive picture of the ceramide function, allow more precise predictions of the end point effects of ceramide alterations, and enable the development of therapeutic means to modulate ceramide-dependent processes (Hannun & Obeid, 2011). Ultimately, not only ceramides but also sphingosine 1-phosphate (S1P), as well as related complex sphingolipids, determine the overall cellular response to different stimuli, a concept represented by a “sphingolipid rheostat” mechanism, originally suggested in 1998 (Mandala et al., 1998). Importantly, increased ceramide metabolism by certain enzymes such as glucosylceramide synthase (GCS), sphingomyelin synthase, ceramide kinase, ceramidases, and sphingosine kinases (SPHKs) has been shown for cancer cells, underscoring the role of sphingolipid metabolism in cancer. Toward this end, the increase in sphingolipids with prosurvival function emerged as a novel target for anticancer therapy (Ogretmen, 2018).
3. CERAMIDE AND CANCER
Cancers are frequently characterized by altered levels of sphingolipids, including ceramide, but mechanisms by which these alterations result in tumor formation and progression are largely unknown. Ceramide-producing enzymes CerS2 (ceramide synthase 2) and CerS6 (ceramide synthase 6) were shown to be elevated in cancerous breast tissues as compared with normal breast tissues (Erez-Roman, Pienik, & Futerman, 2010). Endogenous C16-, C24-, and C24:1-ceramide levels were increased in human head and neck squamous cell carcinomas, compared to normal tissues levels (Karahatay et al., 2007). Importantly, C16- and C18-ceramides were demonstrated to play two opposing roles in human head and neck squamous cell carcinomas, prosurvival and proapoptotic, respectively (Senkal, Ponnusamy, Bielawski, Hannun, & Ogretmen, 2010). Over the years numerous studies presented evidence of the ceramide function in the signal transduction of cancer cells and in their response to nutrient and oxidative stress, chemotherapy, and radiation (Hannun & Obeid, 2017; Ogretmen & Hannun, 2004; Saddoughi et al., 2013). While initially ceramides were regarded as death-promoting signaling molecules, it is now believed that effect of the ceramide elevation depends on the specific ceramide structure and the cellular context (Mesicek et al., 2010; Schiffmann et al., 2009).
Ceramides also emerged as important biomarkers in cancer therapy. For example, increase of total plasma ceramide in the patients undergoing radiation therapy in combination with irinotecan chemotherapy was associated with favorable tumor response (Dubois et al., 2016). Analogously, CerS6 was identified as a member of the 19-gene-signature predicting survival of patients with colorectal cancer (Abdul Aziz et al., 2016). Mechanistic basis for the distinct functions of endogenous ceramides is not known; however, interaction of ceramides with specific proteins was suggested to be responsible for this, at least in part (Saddoughi et al., 2013).
Another important aspect of altered ceramide levels in cancer, specific elevation of ceramide-metabolizing enzymes, is connected to cancer resistance, a major obstacle in cancer therapy. Thus, elevation of SPHK1 and GCS in prostate cancer cells was found to be responsible for resistance to paclitaxel (Aoyama et al., 2017). Elevated GCS expression has been demonstrated to correlate with progression of breast, urinary, ovarian cancers, and leukemia (Liu, Hill, & Li, 2013), whereas upregulation of acid ceramidase on irradiation conferred prostate cancer resistance (Cheng et al., 2013). This opens an opportunity to target these enzymes for treatment of resistant tumors (Ogretmen, 2018).
4. P53: THE BEGINNING
The p53 transcription factor, originally discovered as an endogenous cellular protein bound to a simian virus 40 large T antigen (Lane & Crawford, 1979; Linzer & Levine, 1979), has been initially described as an oncogene. Later studies demonstrated that a normal p53 gene is not required for embryonic development, but its absence predisposes mice to neoplastic disease, and that the oncogenic mutant forms of p53 are not required for development of many types of cancer (Donehower et al., 1992). Thus, the p53 protein was established as tumor suppressor. It is also commonly referred to as the guardian of the genome, due to its strong ability to respond to DNA damage or to stress stimuli capable of the induction of such damage (Lane, 1992). TP53, the gene encoding for the p53 protein, is the most often mutated gene in human cancers with the mutation rate estimated to be above 50% across all cancer types (Shirole et al., 2016). Due to the fact that p53 protein does not easily respond to pharmacological intervention, it has been considered “undruggable,” and therapies capable of overcoming the oncogenic effects of mutant p53 have not been developed yet, almost 40 years after discovery of the protein function in cancer (Sabapathy & Lane, 2018). Nevertheless, the line of research evaluating p53 as a chemotherapeutic target tremendously expanded our understanding of the protein structure and function.
5. P53 IS A TRANSCRIPTION FACTOR
Regardless of all the nicknames of the p53 protein, first and foremost, it is a transcription factor controlling expression of hundreds of targets (Fischer, 2017). At the transcriptional level, p53 acts exclusively as a direct transcriptional activator, whereas inhibitory effects on some targets are usually indirect and mediated by cyclin-dependent kinase inhibitor p21 (CDKN1A or Waf1) interfering with phosphorylation of the retinoblastoma-like proteins RBL1 (p107) and RBL2 (p130) (Quaas, Muller, & Engeland, 2012; Sullivan, Galbraith, Andrysik, & Espinosa, 2018). In turn, hypophosphorylated RBLs stabilize a multicomponent repressor DREAM (including dimerization proteins, RBLs, E2F4, and multivulval class B complex [MUVB]), which inhibits transcription by engaging E2F or cell cycle homology region sites and suppresses transcription (Fischer, 2017). In addition, p53 may indirectly suppress transcription by a variety of mechanisms, such as binding to and disabling specific transcription factors, outcompeting specific activators from promoters due to overlapping binding sites, recruiting chromatin-modifying complexes that block gene expression, and via regulation of both miRNAs and lincRNAs (Barsotti & Prives, 20101; Huarte et al., 2010).
The p53 protein is a highly labile and tightly controlled transcription factor present in nonstressed cells at low level due to its binding by E3 ubiquitin ligase MDM2, which enables p53 ubiquitination and proteasomal degradation (Hock & Vousden, 2014). On a variety of cellular cues, p53 escapes the control by MDM2, accumulates in the nuclei, and activates multiple genes to regulate diverse cellular processes including progression through the cell cycle, DNA damage repair, apoptosis, metabolic alterations, epigenomic modifications, shortening of telomeres, ferroptosis, etc (Haupt & Haupt, 2017). While numerous data indicate that p53 can regulate a wide array of biological processes, it should be noted that cell type, context, and specific stimuli define not just the different levels of gene expression but rather affect the transcription of very different sets of genes (Kastenhuber & Lowe, 2017). Thus, it is not surprising that depending on the cell type, p53 can generate opposite responses to the same stressor. Still, in many cases, mechanisms for such differences are not known. It has been suggested that context-dependent posttranslational modifications of p53 may dictate the interaction with a specific gene target (Kumari, Kohli, & Das, 2014). In addition, the kinetics of the p53 activation and the architecture of core promoters play a role in defining cellular response in a specific case: short p53 activation with rapid assembly of preinitiation complex and few rounds of RNA polymerase II reinitiation taking place at the p21 core promoter, and sustained activation with slow assembly of preinitiation complex that supports multiple rounds of transcription in case of Fas/APO1 and PUMA (Gomes & Espinosa, 2010; Morachis, Murawsky, & Emerson, 2010). The presence of epigenetic marks and other transcription factors at the target promoters may further attenuate the gene response to p53 activation (Itahana et al., 2016; Kastenhuber & Lowe, 2017). Thus, p53 functions in a complex network of regulators that support flexibility of cellular response to changing environment, depending on the cell type and state.
6. P53 AS A METABOLIC REGULATOR
Despite the canonical p53 definition as a tumor suppressor, one should keep in mind that ancestral members of the p53 gene family, found in unicellular protists and short-lived multicellular organisms (Drosophila) that do not develop cancers, function only in germ stem cells as protectors from DNA damage and uncontrolled proliferation (in the absence of DNA damage) (Wylie, Lu, D’Brot, Buszczak, & Abrams, 2014). These findings point to the cancer prevention by p53 as a later development of an ancient ability to prevent DNA damage and regulate growth of stem cells. It is also believed that the p53 regulatory network acquired diverse physiological roles before it was engaged in tumor suppression (Kastenhuber & Lowe, 2017). A few examples demonstrating the breadth of the p53 regulatory network include the response to various types of cellular stress, such as DNA damage (Williams & Schumacher, 2016), nutrient deprivation (Hoeferlin et al., 2013), hypoxia (Humpton & Vousden, 2016), and ribosomal stress (Deisenroth, Franklin, & Zhang, 2016). In addition, p53 regulates metabolic activities of the cell (Floter, Kaymak, & Schulze, 2017), the epigenome (Haupt & Haupt, 2017), and miRNAs (Liu, Zhang, Zhao, & Feng, 2017). Importantly, ceramide elevation is also observed in response to DNA damage, nutrient stress, senescence, autophagy, and apoptosis (Hannun & Obeid, 2017), pointing to a possible connection between the two pathways.
As a transcription factor, p53 regulates a variety of metabolic pathways, including glycolysis, pentose phosphate pathway, serine metabolism, mitochondrial metabolism, redox status, and lipid metabolism (Floter et al., 2017). By limiting NADPH production, p53 slows down many anabolic processes, including fatty acid biosynthesis. At the same time, wild-type (WT) p53 upregulates fatty acid oxidation by elevating expression of lipin 1 and guanidinoacetate methyltransferase (Liu, Zhang, Hu, & Feng, 2015). Moreover, via direct transcriptional control of such targets as sterol-regulatory-binding protein-1 (SREBP-1), acetyl-CoA dehydrogenase family member 11 (ACAD11), LIPIN1, malonyl-CoA decarboxylase (MCD), dehydrogenase/reductase 3 (DHRS3), carnitine palmitoyltransferase 1C (CPT1C) and caveolin 1, p53 controls multiple nodes in lipid metabolism (Parrales & Iwakuma, 2016).
Additional role of p53, different from the transcription activation, is known (Green & Kroemer, 2009; Vaseva & Moll, 2009). It is based on the ability of p53 to interact with myriads of cellular proteins thus affecting their functions. Importantly, this nontransactivation function of p53 can be fulfilled by WT protein and by its mutant variants. For example, while WT p53 suppresses the SREBP-1 levels via yet unidentified mechanisms, the R273H and R280K p53 mutants can bind to SREBP increasing its activity. Because p53 is frequently mutated in cancers, this further expands the processes controlled by p53. Another example includes the binding of mutant p53 to AMPK that inhibits this kinase activity (Zhou et al., 2014). The consequences of this effect can be profound taking into consideration that AMPK has recently emerged as a key regulator of cellular metabolism. Finally, it should be mentioned that p53 has yet another function associated with its translocation to mitochondria. This pathway involves the p53 interaction with members of the Bcl2 family and regulates apoptosis (Marchenko & Moll, 2014; Vaseva & Moll, 2009).
7. P53 IN TUMOR SUPPRESSION
Since the discovery of a tumor suppressor function of p53, it was believed to exert its cancer suppressor role by inducing senescence and apoptosis. Later it was demonstrated that p53 proficient mice, contrary to mice deficient in this protein, were completely suppressing development of thymic lymphomas even in the absence of p21, PUMA, and NOXA, critical players in p53-dependent senescence and programmed cell death (Valente et al., 2013). Moreover, mice expressing the p533KR mutant, deficient in apoptotic and growth arrest functions but proficient in noncanonical and metabolic-regulatory functions, were still protected from tumor formation (Wang et al., 2016). These studies underscore the significance of the p53-mediated transcription for tumor suppression. However, identification of the p53 target genes and responses contributing to tumor suppression is very challenging, and only a few of already identified mechanisms have been tested in vivo (Kaiser & Attardi, 2018).
8. MUTANT P53 FUNCTION
The variety and frequency of p53 mutations in cancer deserve special consideration. Simplistically, the 393-amino-acid protein is organized in several functional domains, the amino terminal transactivation domain (residues 1–92), the DNA-binding domain (DBD, residues 101–306), the oligomerization domain (residues 307–355), and the carboxy-terminal domain (residues 356–393). Of note, tetramerization of p53 is crucial for its activity as a transcription factor (Lomax, Barnes, Hupp, Picksley, & Camplejohn, 1998). While the high rate of the TP53 mutations affecting more than 300 amino acid residues creates an impediment for development of the p53-targeting therapeutics, a novel paradigm of “a rainbow of mutants” may lead to the development of efficient, mutant type—specific therapeutics (Sabapathy & Lane, 2018). The idea of the p53 mutants possessing functional differences suggests differential approach for targeting tumors with specific types of the protein mutations. Thus, mutations in the amino terminal domain often result in the utilization of an alternative start codon and expression of a shorter version of the protein, p47, defective in induction of the cell cycle arrest, but retaining selective transactivation function and ability to induce apoptotic genes (Phang et al., 2015). These mutations predispose organisms to cancer but predict better survival on therapeutic intervention. Distinct from the amino terminal domain, mutations in oligomerization domain result in the loss of function by p53 proteins, inability to form homotetramers, limited or absent transactivation capability, and absent tumor suppressor activity (Giacomazzi et al., 2014). Importantly, approximately 85% of all TP53 mutations are found in the central 205-amino-acid DBD (Bouaoun et al., 2016). Most of the p53 DBD mutations lead to the loss of the ability to activate genes targeted by nonmutated p53 (Lee et al., 2012). Indeed, the rate of tumorigenesis is similar in TP53‒/‒ mice and mice homozygous for mutant TP53, confirming the loss of tumor suppression function by the mutant. Interestingly, the dominant-negative effect of the p53 mutations makes WT allele dysfunctional when the cells are stressed by chemotherapeutics or irradiation (Lee et al., 2012; Willis, Jung, Wakefield, & Chen, 2004). So, an approach to downregulate only mutant p53 keeps promise for improvement of the tumor sensitivity to cancer drugs.
In the past decades numerous studies confirmed the “gain of function” hypothesis, which proposed that some DBD mutants may not just have lost the WT p53 activity but rather acquired an oncogenic activity (gained function), independent of their effects on the WT protein (Freed-Pastor & Prives, 2012). The addiction of tumors to mutant p53 was demonstrated in the mutant TP53 knock-in mice, as the tamoxifen-induced mutant p53 ablation resulted in reduction of tumor growth, induction of apoptosis, and tumor regression, leading to 37% increase in animal survival (Alexandrova et al., 2015). It is believed that “gain of function” characteristics of the mutant p53 could be achieved via one of the two major mechanisms: a mutation conferring ability to transactivate the completely new set of genes (possibly via interaction with other transcription factors and chromatin remodeling proteins) and/or the interaction with other cellular proteins/signaling pathways (Freed-Pastor & Prives, 2012; Kim & Lozano, 2018). Precise details of both of these mechanisms will be dictated by the cell type and context and may be rather dynamic. Not all but only some of the p53 mutants display “gain of function” properties, and these properties seem to manifest in cancer cells only (Sabapathy & Lane, 2018).
Thus, accumulating evidence points to a potential benefit of therapeutic approaches targeting p53, as long as they take into account the cancer type, p53 status, and cellular context.
9. P53 AND CERAMIDE PATHWAYS
Cancer research in the past decades consistently implicated both the p53 and ceramide pathways in the regulation of cell growth, cell cycle arrest, senescence, and apoptosis. Therefore, it is not surprising that the connection between the two has been regularly reviewed (Carroll, Donaldson, & Obeid, 2015; Hage-Sleiman, Esmerian, Kobeissy, & Dbaibo, 2013; Heffernan-Stroud & Obeid, 2011).
Even the first study, which discovered a dose- and time-dependent elevation of the endogenous ceramide following the p53 activation by actinomycin D or gamma irradiation, observed a complex relationship between p53 and ceramide. The authors concluded that both p53-dependent and p53-independent stress response pathways could be activated by DNA damage, and ceramide accumulation could be a common feature in both of them (Dbaibo et al., 1998). Since then, numerous reports have demonstrated ceramide function both, downstream of p53 activation, upstream of p53, or independent of p53, in different cell types and under different stimuli (Heffernan-Stroud & Obeid, 2011). Interestingly, in Caenorhabditis elegans, radiation-induced apoptosis of germ cells was induced by the parallel pathways of CEP-1 (worm homologue of p53)-mediated elevation of the BH3-domain protein EGL-1, and CerS-mediated ceramide accumulation that converged at the mitochondrial membrane (Deng et al., 2008), thus representing yet another, synergistic, relationship between the p53 and ceramide.
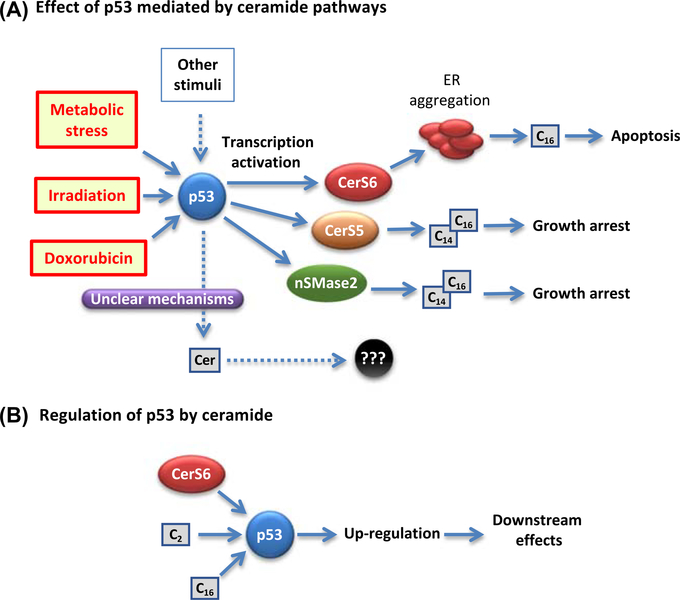
Several sphingolipid-metabolizing enzymes, which produce or degrade ceramide, are regulated by p53 (Fig. 1). Thus, activation of the DNA damage pathway by doxorubicin resulted in the increase of neutral sphingomyelinase 2 (nSMase 2) activity and elevation of ceramide levels, as well as in induction of cell growth arrest. Activation of the nSMase was a result of direct transcriptional activation by p53 at a novel start site upstream of exon 3 (Shamseddine et al., 2015). In contrast, there is practically no evidence on acid sphingomyelinase (aSMase) activation by p53. In one of the recent studies, a derivative of PRIMA-1 (p53 reactivation and induction of massive apoptosis, compound 8b) was found to activate aSMase in H1299 cells (null for p53). PRIMA-1 is a synthetic compound that forms adducts with the p53 thiol groups and facilitates proper protein folding, thus correcting effects of p53-disabling mutations (Soans, Evans, Cipolla, & Fernandes, 2014). However, when the activated targets in H1299 p53+/+ and H1299 cells were compared, this led to a conclusion that the compound demonstrated an off-target effect and activated the TNF receptor superfamily pathway in these cells, arguing against p53 involvement. In a different study, p53 was shown to activate the nSMase but not aSMase (Sawada et al., 2002). Studies in different cell lines, leukemic and colon carcinoma, revealed that, on p53 upregulation, the de novo pathway is a major contributor to ceramide accumulation. Interestingly, the increase in ceramide was associated with the transcriptional upregulation of CerS5 (ceramide synthase 5) and not with change in serine palmitoyltransferase (Panjarian et al., 2008).
Figure 1. Circuits connecting the p53 function and ceramide signaling.
(A) The induction of p53 has been associated with transcriptional activation of several enzymes of ceramide metabolism including CerS6 (in response to folate-mediated metabolic stress); CerS5 (in response to γ-irradiation); and nSMase (in response to doxorubicin). In all cases, C14- and/or C16-ceramides accumulated that caused antiproliferative effects (growth arrest or apoptosis). Other stress stimuli, which induce p53, can cause accumulation of certain ceramides, thus implicating p53 in this outcome, though precise underlying mechanisms remain unclear. (B). As part of signal transduction, ceramides themselves could activate p53 as a downstream target. Such effect has been demonstrated for C2- and C16-ceramides as well as for CerS6 (perhaps through the C16 generation as an intermediate step).
Another ceramide synthase, CerS6, was recently found to be directly transcriptionally activated by p53 via a noncanonical binding site, 11 nucleotides upstream of transcription start site (Fekry, Jeffries, et al., 2016). In contrast to the upregulation of ceramide synthases, SPHK1, a sphingolipid enzyme balancing the ceramide/S1P rheostat (Baran et al., 2007), was shown to be downregulated by p53 (Heffernan-Stroud et al., 2012; Taha et al., 2004). This regulation involves both canonical effector caspases of the mitochondrial apoptotic pathway and other, noncaspase/lysosomal proteases, such as cathepsins. More recent data revealed that p53-mediated activation of caspase-2 was required for SPHK1 proteolysis. In the triple-negative breast cancer cells with mutated p53, caspase-2 was not activated by doxorubicin treatment, and SPHK1 activity was preserved. However, suppression of the CHK1 was able to activate both caspase-2 and SPHK1 degradation (Carroll, Bonica, Shamseddine, Hannun, & Obeid, 2018).
Overall, the information regarding the regulatory effects of p53 on sphingolipid enzymes and the mechanisms involved in this regulation is rather scarce.
10. ACTIVATION OF P53 BY CERAMIDE
The relationship between ceramide and p53 is often interrogated by treatment of cells with exogenous ceramides. Because natural ceramides are highly hydrophobic and are not soluble in aqueous media, either short-chain analogs (C2- or C6-ceramides) or soluble derivatives of the ceramide carrying pyridinium group have been used in these studies (Hou et al., 2011). For example, C2-ceramide was shown to induce SKN-SH cell death via elevation of p53, subsequent increase in Bax/Bcl-2 ratio and caspase activation (Kim et al., 2002). Similarly, in murine myeloid NSF.H7 cells, C2-ceramide activated the protein phosphatase 2A (PP2A) with a consequent dephosphorylation of the Bcl-2, that allowed for direct binding of Bcl-2 and p53, thus preventing Bcl-2 from disabling the proapoptotic Bad and Bax proteins and leading to apoptosis (Deng, Gao, & May, 2009). In this case, p53 function was not dependent on ceramide but contributed to its downstream mechanisms. On the other hand, treatment of HCT116 adenocarcinoma cells with exogenous C16-ceramide upregulated expression of p53 and, additionally, modulated expression of 50 cellular proteins (Renert et al., 2009). Experiments in which mouse fibroblasts were treated by alkylating agent MNNG (1-methyl-3-nitro-1-nitrosoguanidine) demonstrated that DNA damage response triggered in these cells was mediated by ceramide independently of p53 (Yang & Duerksen-Hughes, 2001). Nevertheless, the authors concluded that although the pathways were separable, they were interacting with each other, as the effects of ceramide on p53 levels and stability were noted. Another study concluded, based on the differences in morphology and DNA staining, as well as on the timing of the apoptotic death, that radiation-induced apoptosis was p53 dependent and that ceramide-induced apoptosis was not (Shi, Wuergler, Blattmann, & Crompton, 2001). Furthermore, a complex relationship between p53 and ceramide was also found on investigation of the cellular response to folate stress (Hoeferlin et al., 2013). Stressing A549 cells by either depletion of folate from culture media or by ectopic expression of a major folate-regulatory enzyme caused p53-dependent activation of de novo ceramide biosynthesis and CerS6-dependent C16-ceramide elevation followed by apoptosis. Furthermore, overexpression of CerS6 by transient transfection, without any additional stress, resulted in strong p53 elevation and apoptosis, suggesting a feedback mechanism between C16-ceramide and the tumor suppressor (Fig. 1).
Connection between p53 and ceramide pathways is often exploited in search for the combinational therapeutics. Coadministration of C6-ceramide with chemotherapeutic drug vincristine caused necrosis and apoptosis in multiple cell lines. Significant activation of AMPK followed by p53 activation, inactivation of mTORC1, and downregulation of Bcl-1/Hif-1α were observed in these cells. Activated p53 translocated to mitochondria and bound to cyclophilin D, which led to mitochondrial permeability pore opening and cell necrosis. The authors suggested that p53 activation by these synergistic drugs activates both necroptosis and apoptosis (Chen et al., 2015). Although the number of studies demonstrating ceramide effects on p53 is steadily increasing, our knowledge of the mechanisms of ceramide effects on p53 levels and activity is still rudimentary.
11. ACTIVATION OF OTHER GENES BY CERAMIDE
Ceramide has been reported to regulate expression of several genes. Early investigations of the ceramide function identified several genes regulated by ceramide, such as cyclooxygenase (Ballou, Chao, Holness, Barker, & Raghow, 1992) and IL-6 (Laulederkind, Bielawska, Raghow, Hannun, & Ballou, 1995). MMP-1 expression was shown to be induced by endogenous or short-chain exogenous ceramides via activation of three distinct MAPK pathways in fibroblasts (Reunanen et al., 1998). Ceramide treatment of human hepatocarcinoma cells induced p53-independent elevation of the p21 mRNA and protein, as well as G1 cell cycle arrest (Kim, Kang, Kim, & Choi, 2000). Furthermore, both exogenous and endogenous ceramides were shown to reduce the hTERT (human telomerase reverse transcriptase) promoter activity (Wooten & Ogretmen, 2005). The mechanism of TERT inhibition involved reduction of acetylation of the Sp3 transcription factor, increasing the ratio of Sp3/Sp1 occupying the promoter and reducing the recruitment of RNA polymerase II to the promoter. Interestingly, a sphingolipid enzyme GCS implicated in the development of resistance to cancer chemotherapy was found to be transcriptionally upregulated by the ceramide, with the Sp1 transcription factor being essential for this regulation (Liu et al., 2008). In a different study, cannabinoid treatment—induced de novo ceramide generation activated transcription and increased protein levels of the stress-regulated protein 8 (p8) and its downstream targets Atf-4 and TRB3, leading to apoptosis of the pancreatic cancer cells (Carracedo et al., 2006). As in the case of p53, the mechanisms by which ceramide affects expression of its numerous targets are not fully understood.
12. CERAMIDE SIGNALING: MECHANISMS
Over the years, extensive research presented compelling evidence that changes in cellular ceramide levels are followed by activation of downstream effectors, which results in cell cycle arrest and adaptation, senescence, or programmed cell death.
Because ceramide is a hydrophobic molecule synthesized in and localized to the membranes, one of the plausible mechanisms of its effects on the downstream signaling molecules could be the changes in the biophysical properties of the membranes (lipid rafts formation, fluidity, etc.) where the changes in ceramide occur. These changes affect both the arrangement and properties of the membrane proteins, such as receptors, transporters, and signaling complexes that will transmit the signal further down the line (Edmond et al., 2015; Lockshon et al., 2012). In addition, both short- and long-chain ceramides were able to form stable ceramide channels in planar membranes in vitro, though the experimental evidence for this process in vivo is still missing (Hernandez-Corbacho, Salama, Canals, Senkal, & Obeid, 2017; Siskind & Colombini, 2000). Alternatively, ceramide elevation resulting from treatment of A549 cells with methotrexate caused ER (endoplasmic reticulum) membrane aggregation leading to apoptosis (Fekry, Esmaeilniakooshkghazi, Krupenko, & Krupenko, 2016).
Another mechanism could be the direct binding of ceramide to the components of a signaling pathway that changes their activity. Direct binding of ceramide has been shown for protein kinases PKCα, PKCδ, PKCz, KSR, and c-Raf. PKCα was shown to be activated on ceramide binding, and its translocation from cytosol to the membrane was delayed, whereas PKCδ showed no change in activity on binding but rather decreased autophosphorylation (Huwiler, Fabbro, & Pfeilschifter, 1998). PKCz interaction with ceramide via the cysteine-rich domain was found to elevate its activity and induce the interaction with stress-activated protein kinase (SAPK) complex, resulting in growth suppression (Bourbon, Yun, & Kester, 2000). Similarly, both endogenous and exogenous ceramides were able to bind to the KSR cysteine-rich domain C1 and activate it via autophosphorylation that promoted binding to and transactivation of Raf-1 (Zhang et al., 1997) (Yin et al., 2009). Curiously, using the photoactivatable iodine-labeled ceramide, c-Raf itself was identified as a ceramide-binding protein, which was activated on ligand binding and further activated MAPK1 and MAPK3 signaling cascade (Huwiler et al., 1996). Importantly, the regulation by direct physical binding to its target is not limited to ceramide. Recently, S1P was demonstrated to function as a cofactor of TRAF2 (TNF receptor—associated factor 2), which, on binding, stimulated the ubiquitin-ligase activity of the protein toward RIP1 (Alvarez et al., 2010).
Recent searches for ceramide-binding proteins using bifunctional lipids with a photoactivatable diazirine group as a bait revealed 20 candidates among cellular cytosolic proteins that were cross-linked to the labeled ceramide (Bockelmann et al., 2018). Identified targets are involved in a variety of cellular functions, including ceramide transport, DNA damage response (DNA damage binding protein 1, and DnaJ homologue DnaJc7), protein ubiquitination (COP9 signalosome subunit 3), protein sorting (sorting nexin-1 and 2), trafficking (unc-119 A and B), and metabolism (GLO1, ALDH7A1, ADO, PTGR2, HMGCS2, AKR1C13, CTPS1, PAOX). This underscores the broad spectrum of processes regulated by ceramide (Bockelmann et al., 2018). However, none of the ceramide-metabolizing enzymes have been identified in this work; this is likely due to exclusion of all cellular membranes from the experiments.
Another plausible mechanism for regulation of gene expression by ceramide could be the activation of protein phosphatase 1 (PP1) and PP2A by ceramide. While PP1 was shown to bind ceramide on its own both in vitro and in vivo (Sumanasekera et al., 2012), the activation of PP2A was mediated by the direct binding of ceramide to the inhibitor of PP2A (IPP2A/SET (SET nuclear protoco-oncogene)), as demonstrated by pull-down of SET on biotinylated C6-ceramide and avidin column (Mukhopadhyay et al., 2009). Moreover, specificity toward C18-ceramide, and not to C16-ceramide, was described for the PP2A inhibitor. Ceramide binding by PP1 inhibited the enzyme, whereas binding of ceramide by SET resulted in activation of PP2A accompanied by proteasomal degradation of c-Myc and tumor suppression (Mukhopadhyay et al., 2009; Oaks & Ogretmen, 2014). Change in the activity of either major cellular phosphatase may also affect phosphorylation status of multiple targets, such as transcription factors, signaling proteins, etc., and contribute to regulation of gene activity as well.
In line with these findings, it has been shown that in HEK293 cells, natural C16- and C24-ceramides bind and inhibit PP1, thus preserving phosphorylation of a group of splicing-regulatory proteins containing the evolutionarily conserved motif RVxF recognized by PP1 (Sumanasekera et al., 2012). On the other hand, in multidrug resistant NCI/ADR-RES ovarian cancer cells expressing mutant p53 (five-amino-acid deletion in exon 5), the synthetic pyridinium C6-ceramide activated PP1 thus causing dephosphorylation of serine/arginine-rich splicing-factor 1 and inducing its translocation to the nucleus (Patwardhan et al., 2014). This enabled the alternative splicing of the pre-mRNA encoding the p53 deletion mutant and restoration of the mRNA splice variant translated into WT p53. In a similar manner, recovery of the WT p53 mRNA expression on GCS knockdown and ceramide accumulation in OVCAR-8 and NCI/ADR-RES cells (both heterozygous for the p53 exon 5 deletion) led to the elevation of the p53 transcriptional targets p21, Bax, and PUMA, as well as induction of apoptosis (Liu et al., 2011). The same group has also reported that the inhibition of GCS, accompanied by the elevation of intracellular ceramide, restored the WT p53 activity in the cancer cells heterozygous for a missense R273H mutant. This further sensitized cells toward doxorubicin and increased apoptosis (Hosain et al., 2016). Ceramide-mediated alternative splicing by the function of the splice factor SRp30a was also shown for Bcl-x and caspase 9 (Chalfant et al., 2002; Massiello & Chalfant, 2006).
Thus the number of ceramide-binding protein targets is increasing as we learn more about the signaling functions of ceramide, yet we still do not understand how ceramide is delivered to specific targets (especially to cytosolic proteins), how specificity of the binding is achieved, and what mechanisms modulate the targets activity. The downstream effectors of ceramide-binding proteins activated/inhibited by ceramide are also often unknown.
13. CONCLUSIONS
Ceramides emerged as important novel signaling entities in the early 90s (Hannun, Obeid, & Wolff, 1993), and the past 25 years of research have revealed significant structural variability of ceramides, their involvement in vast array of biological processes, as well as very complex and intertwined relationships with their targets. An important role for ceramide in cancer development and response to therapeutic interventions has been also established, although our understanding of precise mechanisms guiding the cell type—specific and context-depending responses generated by ceramide alterations is still limited.
Tumor suppressor protein p53, discovered about 15 years earlier, is now, perhaps, the most studied protein, with the number of publications considering it approaching 90,000. These studies revealed many details about the protein structure, regulation, biological function, and its critical role for cancer suppression and therapy. Interestingly, similar to ceramide, the mechanisms of the p53 function are also cell type—specific and stimulus-specific, and highly context-dependent. Despite the years of intensive research, our understanding of these mechanisms is still incomplete.
In recent years, numerous studies indicated connections between the ceramide and p53 pathways (Fig. 1). Indeed, both of the pathways have been implicated in the critical cell fate decision processes, such as cell cycle arrest, senescence, and apoptosis. In these processes, ceramide and p53 can function upstream or downstream of each other, making the overall relationship quite complex. In this regard, due to cell type—dependent and context- and stimulus-dependent nature of both pathways, their intersection increases the variability of the response exponentially, making the repertoire of adaptive changes significantly larger. Accordingly, the task of examining these adaptations is extremely complex. Further investigation of the sphingolipid enzyme structures and regulation, identification of ceramide-specific protein targets, and changes that ceramide binding confers, are necessary to significantly improve our ability to untangle the mixed and sometimes contradictory data regarding the connection between p53 and ceramide. As our knowledge of both the ceramide and p53 function expands, it will reveal the regulatory nodes that can be efficiently targeted for therapeutic intervention in cancers.
REFERENCES
- Abdul Aziz NA, Mokhtar NM, Harun R, Mollah MM, Mohamed Rose I, Sagap I, et al. (2016). A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Medical Genomics, 9(1), 58 10.1186/s12920-016-0218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA, et al. (2015). Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature, 523(7560), 352–356. 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, et al. (2010). Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature, 465(7301), 1084–1088. 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama Y, Sobue S, Mizutani N, Inoue C, Kawamoto Y, Nishizawa Y, et al. (2017). Modulation of the sphingolipid rheostat is involved in paclitaxel resistance of the human prostate cancer cell line PC3-PR. Biochemical and Biophysical Research Communications, 486(2), 551–557. 10.1016/j.bbrc.2017.03.084. [DOI] [PubMed] [Google Scholar]
- Ballou LR, Chao CP, Holness MA, Barker SC, & Raghow R (1992). Interleukin-1-mediated PGE2 production and sphingomyelin metabolism. Evidence for the regulation of cyclooxygenase gene expression by sphingosine and ceramide. Journal of Biological Chemistry, 267(28), 20044–20050. [PubMed] [Google Scholar]
- Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, et al. (2007). Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. Journal of Biological Chemistry, 282(15), 10922–10934. 10.1074/jbc.M610157200. [DOI] [PubMed] [Google Scholar]
- Barsotti AM, & Prives C (2010). Noncoding RNAs: The missing “linc” in p53-mediated repression. Cell, 142(3), 358–360. 10.1016/j.cell.2010.07.029. [DOI] [PubMed] [Google Scholar]
- Bockelmann S, Mina JGM, Korneev S, Hassan DG, Mueller D, Hilderink A, et al. (2018). A search for ceramide binding proteins using bifunctional lipid analogs yields CERT-related protein StarD7. The Journal of Lipid Research. 10.1194/jlr.M082354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. (2016). TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics data. Human Mutation, 37(9), 865–876. 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- Bourbon NA, Yun J, & Kester M (2000). Ceramide directly activates protein kinase C zeta to regulate a stress-activated protein kinase signaling complex. Journal of Biological Chemistry, 275(45), 35617–35623. 10.1074/jbc.M007346200. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Gironella M, Lorente M, Garcia S, Guzman M, Velasco G, et al. (2006). Cannabinoids induce apoptosis of pancreatic tumor cells via endoplasmic reticulum stress-related genes. Cancer Research, 66(13), 6748–6755. 10.1158/0008-5472.CAN-06-0169. [DOI] [PubMed] [Google Scholar]
- Carroll B, Donaldson JC, & Obeid L (2015). Sphingolipids in the DNA damage response. Advances in Biological Regulation, 58, 38e52 10.1016/j.jbior.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll BL, Bonica J, Shamseddine AA, Hannun YA, & Obeid LM (2018). A role for caspase-2 in sphingosine kinase 1 proteolysis in response to doxorubicin in breast cancer cells - implications for the CHK1-suppressed pathway. FEBS Open Biology, 8(1), 27–40. 10.1002/2211-5463.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro BM, Prieto M, & Silva LC (2014). Ceramide: A simple sphingolipid with unique biophysical properties. Progress in Lipid Research, 54, 53–67. 10.1016/j.plipres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Chalfant CE, Rathman K, Pinkerman RL, Wood RE, Obeid LM, Ogretmen B, et al. (2002). De novo ceramide regulates the alternative splicing of caspase 9 and Bcl-x in A549 lung adenocarcinoma cells. Dependence on protein phosphatase-1. Journal of Biological Chemistry, 277(15), 12587–12595. 10.1074/jbc.M112010200. [DOI] [PubMed] [Google Scholar]
- Chen MB, Jiang Q, Liu YY, Zhang Y, He BS, Wei MX, et al. (2015). C6 ceramide dramatically increases vincristine sensitivity both in vivo and in vitro, involving AMP-activated protein kinase-p53 signaling. Carcinogenesis, 36(9), 1061–1070. 10.1093/carcin/bgv094. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Bai A, Beckham TH, Marrison ST, Yount CL, Young K, et al. (2013). Radiation-induced acid ceramidase confers prostate cancer resistance and tumor relapse. Journal of Clinical Investigation, 123(10), 4344–4358. 10.1172/JCI64791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coant N, Sakamoto W, Mao C, & Hannun YA (2017). Ceramidases, roles in sphingolipid metabolism and in health and disease. Advances in Biological Regulation, 63, 122–131. 10.1016/j.jbior.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dany M, & Ogretmen B (2015). Ceramide induced mitophagy and tumor suppression. Biochimica et Biophysica Acta, 1853(10 Pt B), 2834–2845. 10.1016/j.bbamcr.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dbaibo GS, Pushkareva MY, Rachid RA, Alter N, Smyth MJ, Obeid LM, et al. (1998). p53-dependent ceramide response to genotoxic stress. Journal of Clinical Investigation, 102(2), 329–339. 10.1172/JCI1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Franklin DA, & Zhang Y (2016). The evolution of the ribosomal protein-MDM2-p53 pathway. Cold Spring Harbor Perspectives in Medicine, 6(12). 10.1101/cshperspect.a026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Gao F, & May WS (2009). Protein phosphatase 2A inactivates Bcl2’s antiapoptotic function by dephosphorylation and up-regulation of Bcl2-p53 binding. Blood, 113(2), 422–428. 10.1182/blood-2008-06-165134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Yin X, Allan R, Lu DD, Maurer CW, Haimovitz-Friedman A, et al. (2008). Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science, 322(5898), 110–115. 10.1126/science.1158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr., Butel JS, et al. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 356(6366), 215–221. 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dubois N, Rio E, Ripoche N, Ferchaud-Roucher V, Gaugler MH, Campion L, et al. (2016). Plasma ceramide, a real-time predictive marker of pulmonary and hepatic metastases response to stereotactic body radiation therapy combined with irinotecan. Radiotherapy and Oncology: Journal of the European Society for Therapeutic Radiology and Oncology, 119(2), 229–235. 10.1016/j.radonc.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Edmond V, Dufour F, Poiroux G, Shoji K, Malleter M, Fouque A, et al. (2015). Downregulation of ceramide synthase-6 during epithelial-to-mesenchymal transition reduces plasma membrane fluidity and cancer cell motility. Oncogene, 34(8), 996–1005. 10.1038/onc.2014.55. [DOI] [PubMed] [Google Scholar]
- Erez-Roman R, Pienik R, & Futerman AH (2010). Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochemical and Biophysical Research Communications, 391(1), 219–223. 10.1016/j.bbrc.2009.11.035. [DOI] [PubMed] [Google Scholar]
- Fekry B, Esmaeilniakooshkghazi A, Krupenko SA, & Krupenko NI (2016). Ceramide synthase 6 is a novel target of methotrexate mediating its antiproliferative effect in a p53-dependent manner. PLoS One, 11(1), e0146618 10.1371/journal.pone.0146618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekry B, Jeffries KA, Esmaeilniakooshkghazi A, Ogretmen B, Krupenko SA, & Krupenko NI (2016). CerS6 is a novel transcriptional target of p53 protein activated by non-genotoxic stress. Journal of Biological Chemistry, 291(32), 16586e16596 10.1074/jbc.M116.716902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M (2017). Census and evaluation of p53 target genes. Oncogene, 36(28), 3943–3956. 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floter J, Kaymak I, & Schulze A (2017). Regulation of metabolic activity by p53. Metabolites, 7(2). 10.3390/metabo7020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, & Prives C (2012). Mutant p53: One name, many proteins. Genes & Development, 26(12), 1268–1286. 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomazzi J, Graudenz MS, Osorio CA, Koehler-Santos P, Palmero EI, Zagonel-Oliveira M, et al. (2014). Prevalence of the TP53 p.R337H mutation in breast cancer patients in Brazil. PLoS One, 9(6), e99893 10.1371/journal.pone.0099893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NP, & Espinosa JM (2010). Disparate chromatin landscapes and kinetics of inactivation impact differential regulation of p53 target genes. Cell Cycle, 9(17), 3428–3437. 10.4161/cc.9.17.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, & Kroemer G (2009). Cytoplasmic functions of the tumour suppressor p53. Nature, 458(7242), 1127–1130. 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage-Sleiman R, Esmerian MO, Kobeissy H, & Dbaibo G (2013). p53 and ceramide as collaborators in the stress response. International Journal of Molecular Sciences, 14(3), 4982–5012. 10.3390/ijms14034982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, & Obeid LM (2008). Principles of bioactive lipid signalling: Lessons from sphingolipids. Nature Reviews. Molecular Cell Biology, 9(2), 139–150. 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Hannun YA, & Obeid LM (2011). Many ceramides. Journal of Biological Chemistry, 286(32), 27855–27862. 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, & Obeid LM (2017). Sphingolipids and their metabolism in physiology and disease. Nature Reviews. Molecular Cell Biology. 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM, & Wolff RA (1993). The novel second messenger ceramide: Identification, mechanism of action, and cellular activity. Advances in Lipid Research, 25, 43–64. [PubMed] [Google Scholar]
- Haupt S, & Haupt Y (2017). P53 at the start of the 21st century: Lessons from elephants. F1000Research, 6, 2041 10.12688/f1000research.12682.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan-Stroud LA, Helke KL, Jenkins RW, De Costa AM, Hannun YA, & Obeid LM (2012). Defining a role for sphingosine kinase 1 in p53-dependent tumors. Oncogene, 31(9), 1166–1175. 10.1038/onc.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan-Stroud LA, & Obeid LM (2011). p53 and regulation of bioactive sphingolipids. Advances in Enzyme Regulation, 51(1), 219–228. 10.1016/j.advenzreg.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Corbacho MJ, Salama MF, Canals D, Senkal CE, & Obeid LM (2017). Sphingolipids in mitochondria. Biochimica et Biophysica Acta, 1862(1), 56–68. 10.1016/j.bbalip.2016.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock AK, & Vousden KH (2014). The role of ubiquitin modification in the regulation of p53. Biochimica et Biophysica Acta, 1843(1), 137–149. 10.1016/j.bbamcr.2013.05.022. [DOI] [PubMed] [Google Scholar]
- Hoeferlin LA, Fekry B, Ogretmen B, Krupenko SA, & Krupenko NI (2013). Folate stress induces apoptosis via p53-dependent de novo ceramide synthesis and up-regulation of ceramide synthase 6. Journal of Biological Chemistry, 288(18), 12880–12890. 10.1074/jbc.M113.461798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosain SB, Khiste SK, Uddin MB, Vorubindi V, Ingram C, Zhang S, et al. (2016). Inhibition of glucosylceramide synthase eliminates the oncogenic function of p53 R273H mutant in the epithelial-mesenchymal transition and induced pluripotency of colon cancer cells. Oncotarget, 7(37), 60575–60592. 10.18632/oncotarget.11169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Jin J, Zhou H, Novgorodov SA, Bielawska A, Szulc ZM, et al. (2011). Mitochondrially targeted ceramides preferentially promote autophagy, retard cell growth, and induce apoptosis. The Journal of Lipid Research, 52(2), 278–288. 10.1194/jlr.M012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell, 142(3), 409–419. 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpton TJ, & Vousden KH (2016). Regulation of cellular metabolism and hypoxia by p53. Cold Spring Harbor Perspectives in Medicine, 6(7). 10.1101/cshperspect.a026146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huwiler A, Brunner J, Hummel R, Vervoordeldonk M, Stabel S, van den Bosch H, et al. (1996). Ceramide-binding and activation defines protein kinase c-Raf as a ceramide-activated protein kinase. Proceedings of the National Academy of Sciences of the United States of America, 93(14), 6959–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huwiler A, Fabbro D, & Pfeilschifter J (1998). Selective ceramide binding to protein kinase C-alpha and -delta isoenzymes in renal mesangial cells. Biochemistry, 37(41), 14556–14562. 10.1021/bi981401i. [DOI] [PubMed] [Google Scholar]
- Itahana Y, Zhang J, Goke J, Vardy LA, Han R, Iwamoto K, et al. (2016). Histone modifications and p53 binding poise the p21 promoter for activation in human embryonic stem cells. Scientific Reports, 6, 28112 10.1038/srep28112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser AM, & Attardi LD (2018). Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death and Differentiation, 25(1), 93–103. 10.1038/cdd.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, et al. (2007). Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): Attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Letters, 256(1), 101–111. 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber ER, & Lowe SW (2017). Putting p53 in context. Cell, 170(6), 1062–1078. 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MP, & Lozano G (2018). Mutant p53 partners in crime. Cell Death and Differentiation, 25(1), 161–168. 10.1038/cdd.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Chae HS, Bach JH, Lee MW, Kim KY, Lee WB, et al. (2002). P53 mediates ceramide-induced apoptosis in SKN-SH cells. Oncogene, 21(13), 2020–2028. 10.1038/sj.onc.1205037. [DOI] [PubMed] [Google Scholar]
- Kim WH, Kang KH, Kim MY, & Choi KH (2000). Induction of p53-independent p21 during ceramide-induced G1 arrest in human hepatocarcinoma cells. Biochemistry and Cell Biology, 78(2), 127–135. [PubMed] [Google Scholar]
- Kumari R, Kohli S, & Das S (2014). p53 regulation upon genotoxic stress: intricacies and complexities. Molecular & Cellular Oncology, 1(3), e969653 10.4161/23723548.2014.969653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP (1992). Cancer. p53, guardian of the genome. Nature, 358(6381), 15–16. 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Lane DP, & Crawford LV (1979). T antigen is bound to a host protein in SV40-transformed cells. Nature, 278(5701), 261–263. [DOI] [PubMed] [Google Scholar]
- Laulederkind SJ, Bielawska A, Raghow R, Hannun YA, & Ballou LR (1995). Ceramide induces interleukin 6 gene expression in human fibroblasts. The Journal of Experimental Medicine, 182(2), 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Teoh WW, Phang BH, Tong WM, Wang ZQ, & Sabapathy K (2012). Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell, 22(6), 751–764. 10.1016/j.ccr.2012.10.022. [DOI] [PubMed] [Google Scholar]
- Levy M, & Futerman AH (2010). Mammalian ceramide synthases. IUBMB Life, 62(5), 347–356. 10.1002/iub.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer DI, & Levine AJ (1979). Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell, 17(1), 43–52. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang C, Hu W, & Feng Z (2015). Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Letters, 356(2 Pt A), 197–203. 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang C, Zhao Y, & Feng Z (2017). MicroRNA control of p53. Journal of Cellular Biochemistry, 118(1), 7–14. 10.1002/jcb.25609. [DOI] [PubMed] [Google Scholar]
- Liu YY, Hill RA, & Li YT (2013). Ceramide glycosylation catalyzed by glucosylceramide synthase and cancer drug resistance. Advances in Cancer Research, 117, 59–89. 10.1016/B978-0-12-394274-6.00003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Patwardhan GA, Bhinge K, Gupta V, Gu X, & Jazwinski SM (2011). Suppression of glucosylceramide synthase restores p53-dependent apoptosis in mutant p53 cancer cells. Cancer Research, 71(6), 2276–2285. 10.1158/0008-5472.CAN-10-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Yu JY, Yin D, Patwardhan GA, Gupta V, Hirabayashi Y, et al. (2008). A role for ceramide in driving cancer cell resistance to doxorubicin. FASEB Journal, 22(7), 2541–2551. 10.1096/fj.07-092981. [DOI] [PubMed] [Google Scholar]
- Lockshon D, Olsen CP, Brett CL, Chertov A, Merz AJ, Lorenz DA, et al. (2012). Rho signaling participates in membrane fluidity homeostasis. PLoS One, 7(10), e45049 10.1371/journal.pone.0045049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax ME, Barnes DM, Hupp TR, Picksley SM, & Camplejohn RS (1998). Characterization of p53 oligomerization domain mutations isolated from Li-Fraumeni and Li-Fraumeni like family members. Oncogene, 17(5), 643–649. 10.1038/sj.onc.1201974. [DOI] [PubMed] [Google Scholar]
- Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, et al. (1998). Sphingoid base 1-phosphate phosphatase: A key regulator of sphingolipid metabolism and stress response. Proceedings of the National Academy of Sciences of the United States of America, 95(1), 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, & Moll UM (2014). Mitochondrial death functions of p53. Molecular & Cellular Oncology, 1(2), e955995 10.1080/23723548.2014.955995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiello A, & Chalfant CE (2006). SRp30a (ASF/SF2) regulates the alternative splicing of caspase-9 pre-mRNA and is required for ceramide-responsiveness. The Journal of Lipid Research, 47(5), 892–897. 10.1194/jlr.C600003-JLR200. [DOI] [PubMed] [Google Scholar]
- Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, et al. (2010). Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cellular Signalling, 22(9), 1300–1307. 10.1016/j.cellsig.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morachis JM, Murawsky CM, & Emerson BM (2010). Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes & Development, 24(2), 135–147. 10.1101/gad.1856710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Saddoughi SA, Song P, Sultan I, Ponnusamy S, Senkal CE, et al. (2009). Direct interaction between the inhibitor 2 and ceramide via sphingolipid-protein binding is involved in the regulation of protein phosphatase 2A activity and signaling. FASEB Journal, 23(3), 751–763. 10.1096/fj.08-120550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TD, Hannun YA, & Obeid LM (2012). Ceramide synthases at the centre of sphingolipid metabolism and biology. The Biochemical Journal, 441(3), 789–802. 10.1042/BJ20111626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oaks J, & Ogretmen B (2014). Regulation of PP2A by sphingolipid metabolism and signaling. Frontiers in Oncology, 4, 388 10.3389/fonc.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogretmen B (2018). Sphingolipid metabolism in cancer signalling and therapy. Nature Reviews Cancer, 18(1), 33–50. 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogretmen B, & Hannun YA (2004). Biologically active sphingolipids in cancer pathogenesis and treatment. Nature Reviews Cancer, 4(8), 604–616. [DOI] [PubMed] [Google Scholar]
- Panjarian S, Kozhaya L, Arayssi S, Yehia M, Bielawski J, Bielawska A, et al. (2008). De novo N-palmitoylsphingosine synthesis is the major biochemical mechanism of ceramide accumulation following p53 up-regulation. Prostaglandins & Other Lipid Mediators, 86(1—4), 41–48. 10.1016/j.prostaglandins.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Parrales A, & Iwakuma T (2016). p53 as a regulator of lipid metabolism in cancer. International Journal of Molecular Sciences, 17(12). 10.3390/ijms17122074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan GA, Hosain SB, Liu DX, Khiste SK, Zhao Y, Bielawski J, et al. (2014). Ceramide modulates pre-mRNA splicing to restore the expression of wild-type tumor suppressor p53 in deletion-mutant cancer cells. Biochimica et Biophysica Acta, 1841(11), 1571–1580. 10.1016/j.bbalip.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang BH, Othman R, Bougeard G, Chia RH, Frebourg T, Tang CL, et al. (2015). Amino-terminal p53 mutations lead to expression of apoptosis proficient p47 and prognosticate better survival, but predispose to tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America, 112(46), E6349–E6358. 10.1073/pnas.1510043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaas M, Muller GA, & Engeland K (2012). p53 can repress transcription of cell cycle genes through a p21(WAF1/CIP1)-dependent switch from MMB to DREAM protein complex binding at CHR promoter elements. Cell Cycle, 11(24), 4661–4672. 10.4161/cc.22917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renert AF, Leprince P, Dieu M, Renaut J, Raes M, Bours V, et al. (2009). The proapoptotic C16-ceramide-dependent pathway requires the death-promoting factor Btf in colon adenocarcinoma cells. Journal of Proteome Research, 8(10), 4810–4822. 10.1021/pr9005316. [DOI] [PubMed] [Google Scholar]
- Reunanen N, Westermarck J, Hakkinen L, Holmstrom TH, Elo I, Eriksson JE, et al. (1998). Enhancement of fibroblast collagenase (matrix metalloproteinase-1) gene expression by ceramide is mediated by extracellular signal-regulated and stress-activated protein kinase pathways. Journal of Biological Chemistry, 273(9), 5137–5145. [DOI] [PubMed] [Google Scholar]
- Sabapathy K, & Lane DP (2018). Therapeutic targeting of p53: All mutants are equal, but some mutants are more equal than others. Nature Reviews. Clinical Oncology, 15(1), 13–30. 10.1038/nrclinonc.2017.151. [DOI] [PubMed] [Google Scholar]
- Saddoughi SA, Gencer S, Peterson YK, Ward KE, Mukhopadhyay A, Oaks J, et al. (2013). Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Molecular Medicine, 5(1), 105–121. 10.1002/emmm.201201283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada M, Nakashima S, Kiyono T, Yamada J, Hara S, Nakagawa M, et al. (2002). Acid sphingomyelinase activation requires caspase-8 but not p53 nor reactive oxygen species during Fas-induced apoptosis in human glioma cells. Experimental Cell Research, 273(2), 157–168. 10.1006/excr.2001.5437. [DOI] [PubMed] [Google Scholar]
- Schiffmann S, Sandner J, Birod K, Wobst I, Angioni C, Ruckhaberle E, et al. (2009). Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis, 30(5), 745–752. 10.1093/carcin/bgp061. [DOI] [PubMed] [Google Scholar]
- Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, & Ogretmen B (2010). Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB Journal, 24(1), 296–308. 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseddine AA, Clarke CJ, Carroll B, Airola MV, Mohammed S, Rella A, et al. (2015). P53-dependent upregulation of neutral sphingomyelinase-2: Role in doxorubicin-induced growth arrest. Cell Death & Disease, 6, e1947 10.1038/cddis.2015.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirole NH, Pal D, Kastenhuber ER, Senturk S, Boroda J, Pisterzi P, et al. (2016). TP53 exon-6 truncating mutations produce separation of function isoforms with protumorigenic functions. Elife, 5 10.7554/eLife.17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YQ, Wuergler FE, Blattmann H, & Crompton NE (2001). Distinct apoptotic phenotypes induced by radiation and ceramide in both p53-wild-type and p53-mutated lymphoblastoid cells. Radiation and Environmental Biophysics, 40(4), 301–308. [DOI] [PubMed] [Google Scholar]
- Siskind LJ, & Colombini M (2000). The lipids C2- and C16-ceramide form large stable channels. Implications for apoptosis. Journal of Biological Chemistry, 275(49), 38640–38644. 10.1074/jbc.C000587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soans E, Evans SC, Cipolla C, & Fernandes E (2014). Characterizing the sphingomyelinase pathway triggered by PRIMA-1 derivatives in lung cancer cells with differing p53 status. Anticancer Research, 34(7), 3271–3283. [PubMed] [Google Scholar]
- Sullivan KD, Galbraith MD, Andrysik Z, & Espinosa JM (2018). Mechanisms of transcriptional regulation by p53. Cell Death and Differentiation, 25(1), 133–143. 10.1038/cdd.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanasekera C, Kelemen O, Beullens M, Aubol BE, Adams JA, Sunkara M, et al. (2012). C6 pyridinium ceramide influences alternative pre-mRNA splicing by inhibiting protein phosphatase-1. Nucleic Acids Research, 40(9), 4025–4039. 10.1093/nar/gkr1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha TA, Osta W, Kozhaya L, Bielawski J, Johnson KR, Gillanders WE, et al. (2004). Down-regulation of sphingosine kinase-1 by DNA damage: Dependence on proteases and p53. Journal of Biological Chemistry, 279(19), 20546–20554. 10.1074/jbc.M401259200. [DOI] [PubMed] [Google Scholar]
- Valente LJ, Gray DH, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, et al. (2013). p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Reports, 3(5), 1339–1345. 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, & Moll UM (2009). The mitochondrial p53 pathway. Biochimica et Biophysica Acta, 1787(5), 414–420. 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SJ, Li D, Ou Y, Jiang L, Chen Y, Zhao Y, et al. (2016). Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Reports, 17(2), 366–373. 10.1016/j.celrep.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AB, & Schumacher B (2016). p53 in the DNA-Damage-Repair Process. Cold Spring Harbor Perspectives in Medicine, 6(5). 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A, Jung EJ, Wakefield T, & Chen X (2004). Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene, 23(13), 2330–2338. 10.1038/sj.onc.1207396. [DOI] [PubMed] [Google Scholar]
- Wooten LG, & Ogretmen B (2005). Sp1/Sp3-dependent regulation of human telomerase reverse transcriptase promoter activity by the bioactive sphingolipid ceramide. Journal of Biological Chemistry, 280(32), 28867–28876. 10.1074/jbc.M413444200. [DOI] [PubMed] [Google Scholar]
- Wylie A, Lu WJ, D’Brot A, Buszczak M, & Abrams JM (2014). p53 activity is selectively licensed in the Drosophila stem cell compartment. Elife, 3, e01530 10.7554/eLife.01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, & Duerksen-Hughes PJ (2001). Activation of a p53-independent, sphingolipid-mediated cytolytic pathway in p53-negative mouse fibroblast cells treated with N-methyl-N-nitro-N-nitrosoguanidine. Journal of Biological Chemistry, 276(29), 27129–27135. 10.1074/jbc.M100729200. [DOI] [PubMed] [Google Scholar]
- Yin X, Zafrullah M, Lee H, Haimovitz-Friedman A, Fuks Z, & Kolesnick R (2009). A ceramide-binding C1 domain mediates kinase suppressor of ras membrane translocation. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology, 24(3—4), 219–230. 10.1159/000233248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yao B, Delikat S, Bayoumy S, Lin XH, Basu S, et al. (1997). Kinase suppressor of Ras is ceramide-activated protein kinase. Cell, 89(1), 63–72. [DOI] [PubMed] [Google Scholar]
- Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, et al. (2014). Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Molecular Cell, 54(6), 960–974. 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]