Abstract
In mammalian hippocampus, new neurons are continuously produced from neural stem cells throughout life. This postnatal neurogenesis may contribute to information processing critical for cognition, adaptation, learning, and memory, and is implicated in numerous neurological disorders. During neurogenesis, the immature neuron stage defined by doublecortin (DCX) expression is the most sensitive to regulation by extrinsic factors. However, little is known about the dynamic biology within this critical interval that drives maturation and confers susceptibility to regulatory signals. This study aims to test the hypothesis that DCX-expressing immature neurons progress through developmental stages via activity of specific transcriptional networks. Using single-cell RNA-seq combined with a novel integrative bioinformatics approach, we discovered that individual immature neurons can be classified into distinct developmental subgroups based on characteristic gene expression profiles and subgroup-specific markers. Comparisons between immature and more mature subgroups revealed novel pathways involved in neuronal maturation. Genes enriched in less mature cells shared significant overlap with genes implicated in neurodegenerative diseases, while genes positively associated with neuronal maturation were enriched for autism-related gene sets. Our study thus discovers molecular signatures of individual immature neurons and unveils potential novel targets for therapeutic approaches to treat neurodevelopmental and neurological diseases.
Keywords: neurogenesis, neuronal maturation, single-cell RNA-seq, transcriptome, doublecortin
Introduction
In mammals, neuronal production ceases at birth in most brain regions. The dentate gyrus (DG) of the hippocampus is one of the few regions in mammalian brains and possibly the only region in the human brain that retains its ability to produce functional neurons throughout life. The precise function of this lifelong neurogenesis after early development, collectively called “adult neurogenesis,” remains to be clarified (Groves et al. 2013). However, extensive studies have shown that new neurons can integrate into existing hippocampal circuits and may contribute to information processing that is critical for cognition, adaptation, learning, and memory (Kempermann et al. 2015). Impaired adult neurogenesis is implicated in neuropsychiatric disorders, such as depression and schizophrenia, and in neurodegenerative diseases, such as Alzheimer's and Huntington's disease (Christian et al. 2014). Understanding the developmental processes and regulatory mechanisms of adult hippocampal neurogenesis will thus open up new approaches for treating brain diseases.
To develop into mature neurons, neural stem cells (NSCs) in the postnatal hippocampus undergo a multistep process that includes a precursor cell phase, an early survival phase, a post-mitotic maturation phase, and a late survival/integration phase (Kempermann et al. 2015). During this process, the newly generated cells are regulated by both intrinsic and extrinsic regulatory factors that affect proliferation, differentiation, survival, migration, axon extension, dendritic branching, spine formation, and integration. This process recapitulates, to some extent, neurogenesis in early development; however, certain regulatory mechanisms are unique to adult neurogenesis (Christian et al. 2014). Curiously, most newborn cells disappear within the first 2 weeks after cell division, but the few surviving cells develop into mature neurons and persist for a long time (Kempermann et al. 2015). Although extensive studies have focused on the precursor cell phase, the regulation of adult hippocampal neurogenesis seems to occur mostly during the survival and maturation phases of newborn cells (Kempermann et al. 2015); however, neither development nor regulation during this critical period is well understood.
Newborn cells between the neural progenitor phase and mature neuron phase, collectively called immature neurons, appear as early as Day 2 and persist to about 3 weeks after cell birth. Most, if not all of these, express polysialylated neural-cell adhesion molecule (PSA-CAM, product of Ncam1 gene) and the microtubule-associated protein doublecortin (DCX) in an overlapping manner. Therefore, DCX has been used widely to detect immature neurons in the adult hippocampus. However, interpreting DCX-expressing cells as a single population masks detailed developmental characteristics important for understanding biological functions. A limited number of markers have been used to further divide DCX-expressing cells into substages of development, but such markers are few and better markers are needed.
Attempts have been made to study gene expression in immature neurons during adult neurogenesis by using dissected DG tissue (Ma et al. 2009) or by laser capture micro-dissection of granule cells (Smrt et al. 2007). DCX-expressing cells have been isolated from adult Dcx-DsRed transgenic mice using fluorescence-activated sorting (FACS) followed by microarray analysis (Bracko et al. 2012). Although these studies provide important information about adult hippocampal neurogenesis, the characteristics of individual immature neuronal cells are not revealed. Single-cell transcriptome analysis allows researchers to characterize the cellular heterogeneity within in vivo organs with unprecedented resolution (Shapiro et al. 2013; Stegle et al. 2015). Recently, this method was used to study NSCs residing in the postnatal subventricular zone (Luo et al. 2015) and the DG of the hippocampus (Shin et al. 2015). These studies provide novel insight into the properties and regulatory mechanisms of adult NSCs. Until now, single-cell RNA-Seq has not been used for adult-born neurons, which is more challenging than studying NSCs, because not only do immature neurons constitute a minor population in the adult DG, but their processes are intertwined with other cells and the hippocampal structure.
In this study, we captured single cells from FACS-enriched immature neurons from the DG of 7 to 9-week-old Dcx-DsRed transgenic mice using an automated single-cell isolation and processing system (Pollen et al. 2014). Our RNA-sequencing transcriptome analyses showed that these DCX-DsRed+ single cells are highly similar to one another in their transcriptome and are enriched in genes involved in adult neurogenesis. Our further analysis classified these cells into 4 subgroups based on 3 clusters of genes with concordant expression patterns. One cell subgroup expressed astrocyte and stem cell genes that overlap with those enriched in NSCs, whereas another subgroup expressed genes enriched in mature neurons. We discovered several subgroup-specific markers that can define the developmental stages of immature neurons. A comparison between less mature and more mature subgroups revealed novel pathways involved in neuronal maturation. In addition, while the genes enriched in immature cells shared significant overlap with genes implicated in neurodegenerative diseases, the genes positively associated with the progression of neuronal maturation were robustly enriched for autism-related gene sets. Our single-cell expression study, therefore, provides new knowledge for understanding a critical developmental stage of new neurons born during postnatal neurogenesis at unprecedented resolution.
Materials and Methods
Mice
All animal procedures were performed according to protocols approved by the University of Wisconsin-Madison Care and Use Committee. The C57B/L6 mice (51–66 days old) used in this study were originally purchased from the Jackson Lab. Dcx-DsRed transgenic mice (51–66 days old) used in this study were created previously (Wang et al. 2007). A 3.7-kb DCX genomic DNA fragment covering the 5′ upstream region from the ClaI site to the translation start site was used to create DsRed mice.
Tissue Preparation, Immunohistochemistry, and Confocal Imaging
Brain tissue processing and histological analysis of mouse brains were performed as described in our publications (Wang et al. 2015). Refer to Supplementary Methods for more details. Z-stack confocal images were taken using Nikon A1 confocal microscope with 2 µm interstack intervals. The Z-stack images were analyzed using a Zeiss Apotome microscope equipped by StereoInvestigator and Microlucida software (MBF Biosciences, Inc.) and the marker positive cells in the DG of each animal were identified as described in our previous publications (Wang et al. 2015).
FACS Purification of DCX+ Cells Population From Postnatal Brains
Cell isolation from the DG by FACS was performed as described (Codega et al. 2014). DG tissue was microdissected from 51- to 66-day-old Dcx-DsRed mice as described previously (Guo et al. 2012) with modifications (Hagihara et al. 2009). All cell populations were isolated into single cells using a Becton Dickinson FACS Aria II. Ten thousand total alive or DCX-DsRed+ live cells were collected. Gates were set manually by using control samples derived from wild-type mice. Refer to Supplementary Methods for more details. Quantification of DsRed+ cells in FACS-isolated cells was performed by using a BX51 epifluorescence microscope (Olympus) with the assistance by the StereoInvestigator software (MicroBrightField) as described (Wang et al. 2015).
RNA Isolation, qPCR, Analyses of FACS-Isolated Cells
Total RNA was extracted from the FACS-isolated cell using the Direct-zol™ RNA MiniPrep Kit (Zymo Research Corporation, Irvine, CA, USA). The RNA was reverse transcribed by MessageBOOSTER™ Whole Transcriptome cDNA Synthesis Kit (MBWT80510, Illumina) for qPCR. Refer to Supplementary Methods for more details and PCR primers.
Cell Capture for RNA-seq
Cell capture and cDNA synthesis were performed as described (Pollen et al. 2014) with modifications. Immediately after FACS, collected cells were centrifuged and then resuspended in cell culture medium at a concentration of 500 cells/µL. The cell suspension was then mixed with C1 Cell Suspension Reagent (Fluidigm, 634833) at the recommended ratio of 3:2 immediately before loading 5 μL of this final mix on the C1 IFC. Immediately after capturing, the cells were visualized with a Leica TCS-LSI confocal microscope to inspect each capture site. Capture sites containing clean single cells were identified. If the capture site contained more than one cell or the cells had debris nearby, the port was marked to be removed from further analysis. Single-cell RNA extraction and mRNA amplification were performed on the C1 Single-Cell Auto Prep Integrated Fluidic Circuit (IFC) following the methods described in the protocol (PN 100-7168). The cDNA products were quantified using the Quant-iT PicoGreen double-stranded DNA (dsDNA) Assay Kit (Life Technologies) and high-sensitivity (HS) DNA chips (Agilent). All samples assayed had material present in the 1000–3000 bp range with concentrations varied from approximately 160 pg/μL to 1.5 ng/µL. The cDNA samples were diluted to <0.3 ng/μL, and 2 μL of diluted cDNA reaction products were then converted into libraries using the Nextera XT DNA Sample Preparation Kit (Illumina, FC-131-1096 and FC-131-1002) following the manufacturer's instruction. Briefly, 12 cycles of PCR amplification were used. After the PCR step, samples were pooled, cleaned with 0.9× Agencourt AMPure XP SPRI beads (Beckman Coulter lot no. 14669400 X2), eluted in Tris + EDTA buffer, and quantified using a HS DNA chip (Agilent). A cDNA negative control was included as negative control. Paired-end RNA-Sequencing was performed in 2 batches using Illumina Hiseq2500 system at UW-Madison Biotechnology Center, generating 100 bp length (first batch) and 150 bp length (second batch) reads.
RNA-Seq of DG Tissue
The DG tissue was microdissected from 3 (8–12 weeks old) C57BL6 male mice (biological triplicates) using our published method (Guo et al. 2012). The tissue was homogenized immediately in RLT buffer (Qiagen, RNeasy Micro Kit), and the tissue lysate was snap frozen and then stored in a −80 freezer until use. RNA isolation was performed using the RNeasy Micro Kit (Qiagen). The quality of RNA was assessed by Agilent before subjected to library synthesis using TruSeq mRNA and total RNA-stranded sample preparation kit (Illumina). The libraries were then subject to paired-end and 100 bp sequencing on an Illumina Hi-Seq2000 (Illumina).
Mapping and Expression Estimation
RSEM was used to align read pairs to the mm9 transcriptome and estimate gene expression levels (Li and Dewey 2011). The following command was used to execute alignment and quantification: rsem-calculate-expression—paired-end—bowtie2 -p 8 $sample_forward.fastq $sample_reverse.fastq $rsem_index $sample_output. We used a table of RSEM's “expected counts” as expression values in downstream analyses (see Supplementary Table 1). This table contains 64 columns of cells and 30 970 rows of transcripts. This set of transcripts also served as a “transcriptome background” for downstream calculations and analyses. To confirm that sorted Dcx-DsRed cells analyzed in this experiment were true DsRed-positive cells, read pairs from each sample were aligned to the DsRed sequence. The number of read pairs that successfully mapped to DsRed was normalized to the total number of read pairs generated for a given cell and expressed as a percentage (see Supplementary Fig. 2D).
Sample Clustering and Principal Component Analysis
For sample clustering with DCX-DsRed cells, whole DG tissue, and Nestin-CFP cells, the DESeq package was used in R. Fluidigm's SINGuLAR Analysis Toolset was used in R to perform a suite of analyses on DCX-DsRed single-cell expression data. First, “identifyOutliers()” was used to exclude 4 samples that displayed significantly aberrant expression profiles. The “autoAnalysis()” command was used to perform unsupervised clustering, principal component analysis, and expression heat mapping of the remaining 64 cells using the top 400 most variable genes as determined by ANOVA.
Enrichment Analyses
GO analysis was performed in identified gene clusters using PANTHER's statistical overrepresentation test with the “GO biological process complete” annotation set (http://pantherdb.org/) (Thomas et al. 2003). We curated sets of CNS cell type-specific genes using the website that accompanies the corresponding brain RNA-seq data (http://web.stanford.edu/group/barres_lab/brain_rnaseq.html) (Zhang et al. 2014). To build the neuron-specific gene list, for example, “Neuron” was selected and compared with all other cell types except “Neuron,” and the top 500 genes were used. Genes that are demethylated during development of the mammalian brain were provided by Lister et al. (2013) which represent the top 2 groups of genes indicated in Fig. 2 of the accompanying article. Genes linked to adult hippocampal neurogenesis were downloaded from the Mammalian Adult Neurogenesis Gene Ontology (MANGO) database (http://mango.adult-neurogenesis.de/documents/annotations?show=20&expression=true) (Overall et al. 2012). Disease-linked genes were curated from Phenocarta (http://gemma-doc.chibi.ubc.ca/phenocarta/), the DISEASES database (Pletscher-Frankild et al. 2015) (http://diseases.jensenlab.org/Search), and AutDB (Basu et al. 2009) (http://autism.mindspec.org/autdb/Welcome.do). Overrepresentation of gene sets was statistically assessed within identified gene clusters against the transcriptome background in R using the Modular Single-set Enrichment Test (Eisinger et al. 2013).
Figure 2.
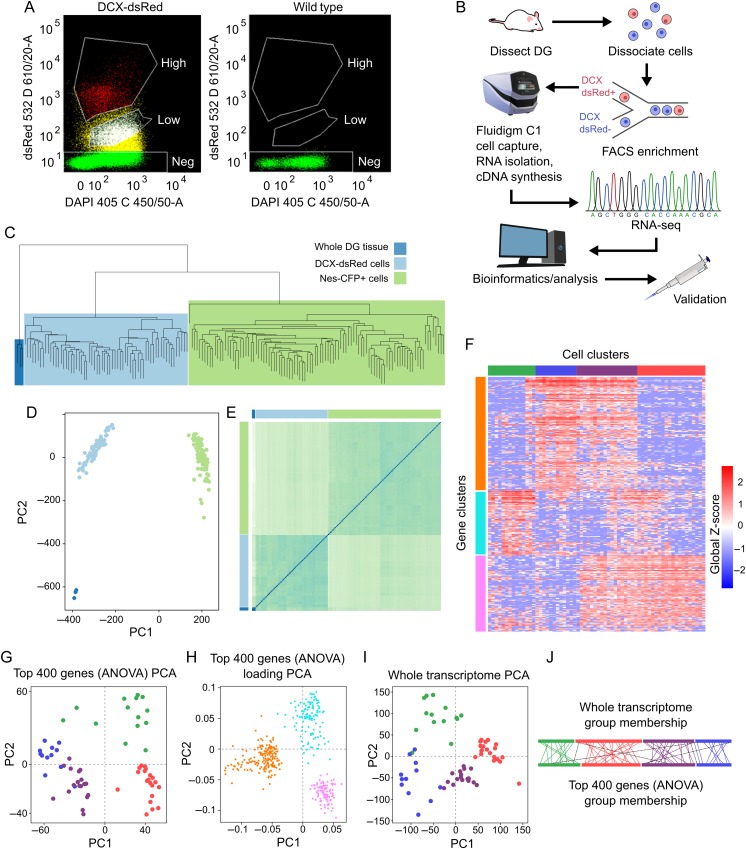
DCX-DsRed+ cells form a distinct, primary population but contains 4 subpopulations with distinct expression patterns. (A) Enrichment of DCX-DsRed+ cells through FACS. Sample fluorescence-side scatter dot plot graphs of dissociated single DG cells from a DCX-DsRed transgenic mouse (left) and a wild-type control mouse (right). Both DsRed High (red) and DsRed Low (white) populations in the transgenic mouse are absent from control mouse. We selected the DsRed-high population for subsequent analysis. (B) A schematic of the experimental design for single-cell transcriptome analysis. DG tissue was dissected from mice and dissociated into single cells, which were then subjected to FACS enrichment. DCX-DsRed+ cells were then loaded into the Fluidigm C1 system, which captures single cells, isolates RNA, and prepares cDNA within the capture chamber. Sequencing libraries were prepared outside of the C1 machine and RNA-sequencing was performed as described in Materials and Methods. Downstream analyses were carried out on expression data, and discoveries were validated histologically in brain sections. (C) Unsupervised clustering of whole-transcriptome RNA-seq data from individual DCX-DsRed+ cells (light blue) alongside individual Nestin-CFP+ cells (green, published by Shin et al (2015)) and whole DG tissue (dark blue). (D) PCA analysis of the same data. (E) Distance heatmap of the same data shows that DCX-DsRed+ cells tend to be more similar to DG tissue than Nestin-CFP+ cells. (F) Expression heatmap and clustering of cell samples and genes using the most variable 400 genes according to ANOVA significance. Each column represents a cell, and each row represents a gene. Red color reflects high expression, and blue represents low expression. (G) PCA plot of the DCX+ cell samples based on the 400 most variable genes. Samples are color-coded to match their group membership as shown in Figure 3A. (H) Loading PCA plot of genes used in Figure 3B. Genes are color-coded to match their group membership as shown in Figure 3A. (I) PCA analysis was repeated on cell samples using the entire transcriptome to confirm that DCX-DsRed+ subpopulations segregate according to their full expression profiles. (J) Sample group membership of cells when clustered by the whole transcriptome (top) and the most variable 400 genes (bottom). Colored lines show the relative position of each cell sample that belonged to the same group in both clustering methods, and black dotted lines indicate samples that were clustered into different groups.
Pairwise Cell Cluster Differential Expression
The EBSeq package (Leng et al. 2013) was used in R to calculate fold changes and FDR-corrected P values using default parameters (fold-change thresholds of 0.7 and 1.4, FDR-adjusted P value of 0.05). Because EBSeq requires integer expression values, the table of expected counts produced by RSEM was rounded before testing. The percent of differentially expressed transcripts was calculated against the transcriptome background.
Data Deposit
Expression data for this experiment are available in Supplementary Table 3 and on the Gene Expression Omnibus (accession number GSE75901).
Results
Enrichment of DCX+ Immature Neurons Using FACS
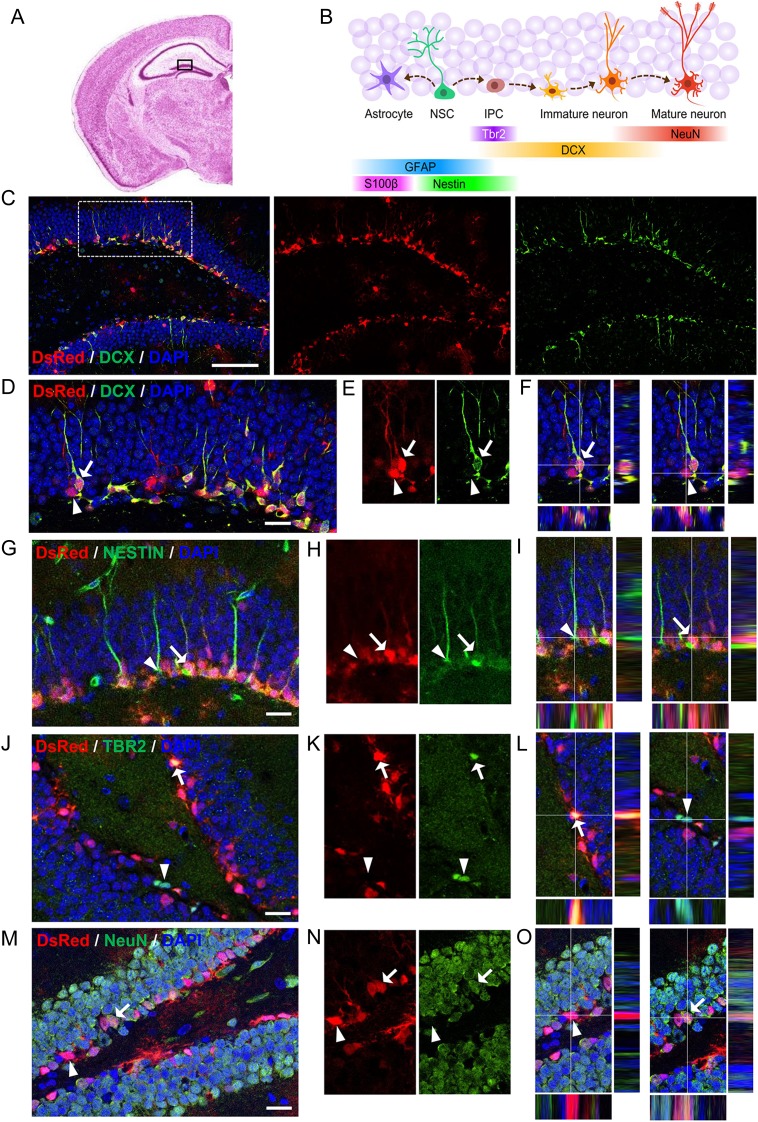
In the hippocampus of adult mammals, radial glia-like NSCs give rise to intermediate progenitors (IPCs), which differentiate into immature neurons and then mature neurons within a 4- to 6-week period of time (Fig. 1A,B). DCX is expressed as early as 2 days to about 3 weeks upon differentiation, thereby labeling a broad span in the life of immature neurons in the adult DG. Because most, if not all, adult-born new neurons in the DG transiently express DCX at some time during their development, we decided to characterize DCX-expressing cells in the DG for a better understanding of immature neurons during adult neurogenesis. It has been shown that, in a DCX-DsRed transgenic mouse line expressing DsRed under the promoter of DCX, there is nearly complete agreement between DsRed signal and DCX expression during embryonic brain development (Wang et al. 2007), and thus, they might be used to study DCX-expressing cells. Since this line of mice has not been evaluated after early development, we first characterized the DsRed-expressing (DsRed+) cells in the hippocampus of 51- to 66-day-old mice. We found that majority of the DsRed+ cells were localized in the subgranular zone (SGZ) of the DG, where DCX+ immature neurons reside (Fig. 1C,D). We further found that a majority (∼84%) of DsRed+ cells in the DG were positive for DCX, and almost all DCX+ cells were DsRed+ (Fig. 1C–F). We then used several known cell lineage markers to further define DsRed+ cells. We found that a small number of DsRed+ cells expressed the neural progenitor marker NESTIN (NESTIN+), but very few DsRed+ cells were GFAP+ (Fig. 1G–I; see Supplementary Fig. 1A). In addition, while DsRed+ cells exhibit significant overlap with TBR2, a transcription factor expressed in IPCs and immature neurons during both cortical development and adult neurogenesis (Fig. 1J–L), they were rarely labeled with astrocyte marker S100ß (see Supplementary Fig. 1B). Coexpression of DCX with the mature neuronal marker NeuN was shown before (Brown et al. 2003; Smrt et al. 2007). We found that, although some DsRed+ cells indeed expressed NeuN, significant numbers of the DsRed+ cells expressed DCX without NeuN (Fig. 1M–O). Therefore, in this DCX-DsRed transgenic line, DsRed primarily labels DCX+ immature neurons in the postnatal DG, making this line suitable for studying DCX-expressing cells in vivo.
Figure 1.
Characterization of DCX-DsRed transgenic mice for postnatal hippocampal neurogenesis. (A) An illustration of coronal section of Nissl-stained adult mouse brain (http://www.hms.harvard.edu/research/brain/atlas.html) showing the DG of the hippocampal region (boxed). (B) Schematic diagram showing stages of neurogenesis in the adult DG and cell lineage-specific markers. NSCs, neural stem cells; IPCs, immediate progenitor cells. (C–F) Confocal images showing that most of DsRed+ (red) cells in the DG express doublecortin (DCX, green). Scale bar in C, 100 μm. Scale bar in D–F, 20 μm. Arrow points to a DsRed+DCX+ cell. Arrowhead points to a DsRed+DCX− cells. (G–O) Confocal images showing colocalization (arrows) or a lack of colocalization (arrowheads), in the DG between DsRed+ cells and the cell lineage markers: Nestin (G–I), TBR2 (J–L), NeuN (M–O). Scale bars, 20 μm.
We dissected the DG tissues from 51- to 66-day-old Dcx-DsRed mice and dissociated the tissues into a single-cell suspension using a method optimized for adult DG NPC isolation (Guo et al. 2012). The single-cell suspension was then subjected to FACS enrichment of DsRed+ cells (Codega et al. 2014). We separated 2 DsRed+ populations based on the intensity of red fluorescence: the DsRed-high population, which is 5–8% of total live cells, and the DsRed-low population, which is 10–30% of total live cells (Fig. 2A). Since both the DsRed-high population had consistently high percentage of DsRed-positive cells among different FACS experiments, we decided to use the DsRed-high population as the DsRed+ cells for our study. We first assessed the purity of the FACS-purified DsRed+ population by acutely plating cells onto coverslips and fixing them. We found that most (> 95%) of the cells express high levels of DsRed+ (see Supplementary Fig. 2A,B). We then verified the enrichment of immature neurons in the sorted DsRed+ cells using quantitative PCR and confirmed that DsRed+ cells had higher Dcx and DsRed mRNA levels compared with total input cells (see Supplementary Fig. 2C). On the contrary, both NSC-enriched mRNAs, Gfap, Nestin and mature neuron-enriched mRNAs, NeuN, exhibited lower levels in DsRed+ cells compared with total input cells (see Supplementary Fig. 2C).
Single-Cell RNA-Seq of Immature Neurons From the Postnatal Mouse Dentate Gyrus
To ensure that we captured high-quality single DCX-DsRed+ immature neurons from the FACS-enriched population, we used the Fluidigm C1 IFC system, which has been successfully used to capture NSCs and neurons from developing human cortex for single-cell transcriptome analysis (Pollen et al. 2014). Immediately after cell capture, each capture site of the capture plate was evaluated under a microscope to ensure the presence of a single and intact cell (Fig. 2B). Capture sites with multiple cells or sites with cell debris were marked and later removed from analysis. The captured cells were then subjected to automated RNA isolation and cDNA synthesis through the Fluidigm C1 IFC system. The cDNA samples that have passed Agilent quality assessment were then subjected to library construction and RNA-sequencing.
In total, we sequenced 84 DCX-DsRed+ cells in 2 batches (see Supplementary Table 1). Sixteen cell samples were excluded from analysis for yielding small numbers of read pairs, and 4 samples were excluded after outlier analysis detected significantly aberrant expression profiles, indicating that they may not be true DCX-DsRed cells. Read alignment and expression estimation were performed with RSEM, and a table of RSEM expected counts were used for downstream analyses (see Supplementary Table 2 (Table_S2.csv)). For samples in the first sequencing run (21 cells), an average of 11.06 million sequencing read pairs were successfully aligned to the mouse transcriptome. For samples in the second run (43 cells), an average of 2.8 million read pairs were aligned. The discrepancy in sequencing depth had no effect on downstream analyses, as samples from Round 1 and Round 2 were randomly distributed in unsupervised clustering and PCA analysis. Mean Phred quality scores for reads ranged from 33.53 to 38.27, and 93.7% of all bases had Phred scores over 30. Although Dcx transcripts were detected in only 9 of the 64 sequenced cells, likely because expression is below the threshold of detection at this sequencing depth, DsRed transcripts were readily detectable in all cells across a range of abundance (see Supplementary Fig. 2D). In addition, we found that Ncam1, the gene that encodes the immature neuron marker PSA-NCAM, was expressed in all cells (see Supplementary Fig. 3D). Furthermore, reference genes, such as Actb, Gapdh, and Ubb, were expressed evenly at high levels across all samples (see Supplementary Fig. 3A–C). Therefore, we have indeed captured single immature neurons, and these single neurons yielded high-quality transcriptome data for further analyses.
DCX-DsRed+ Cells Are Highly Similar at the Whole Transcriptome Transcriptional Level
We first assessed the degree of similarity among sequenced DCX-DsRed+ cells at the whole-transcriptome level. To establish a baseline of comparison, we performed RNA-Seq on the DG tissue and obtained over 40 million mapped reads from each DG sample. We then compared the expression profiles of our DsRed+ single cells to those of whole DG tissue, as well as single-cell RNA-Seq data of NSCs from the DG of adult Nestin-CFP transgenic mice (Shin et al. 2015). In unsupervised clustering, our DsRed+ cells grouped together under a single level-2 branch (Fig. 2C). In PCA analysis, DsRed+ cells formed a tight cluster and were segregated from both NSCs and whole DG (Fig. 2D). The distance matrix of samples suggests that DG tissue is more similar to our DsRed+ cells than to NSCs (Fig. 2E), likely due to the relative abundance of neuronal cells present in the DsRed+ samples versus NSCs. These data suggest that DCX-DsRed marks a primary cell population with a relatively high degree of homogeneity at the whole-transcriptome level.
DCX-DsRed+ Cell Population Is Composed of Distinct Subpopulations
To investigate biological diversity within the DCX-DsRed+ cells, we performed unsupervised clustering of DCX-DsRed+ cells using the 400 most variable genes as determined by ANOVA. An expression heatmap was created, and 4-cell clusters were identified and assigned reference colors (Fig. 2F; green, blue, violet, and red). Cell clusters were distinguished by 3 groups of genes (orange, teal, and magenta), with concordant gene expression across sample clusters. PCA analysis of samples revealed that cell clusters were largely segregated (Fig. 2G). The PCA loading plot demonstrates the relationship between genes and cell clusters (Fig. 2H); red cells are localized to the lower right quadrant by virtue of high expression of magenta genes, while green cells are mapped to the upper right quadrant due to high expression of the teal gene group. Blue and violet cells are both located to the left by high expression of orange genes, while the violet cell group is shifted farther down because it also expresses magenta genes. To validate these results, we performed PCA analysis for the entire transcriptome. We found that the segregation and group membership were largely preserved (Fig. 3I,J), and we observed that red and violet cell clusters appear more similar to each other at the whole-transcript level. Collectively, these data demonstrate that the DsRed+ cell population is composed of several distinct subpopulations defined by significant gene expression patterns (Fig. 3A).
Figure 3.
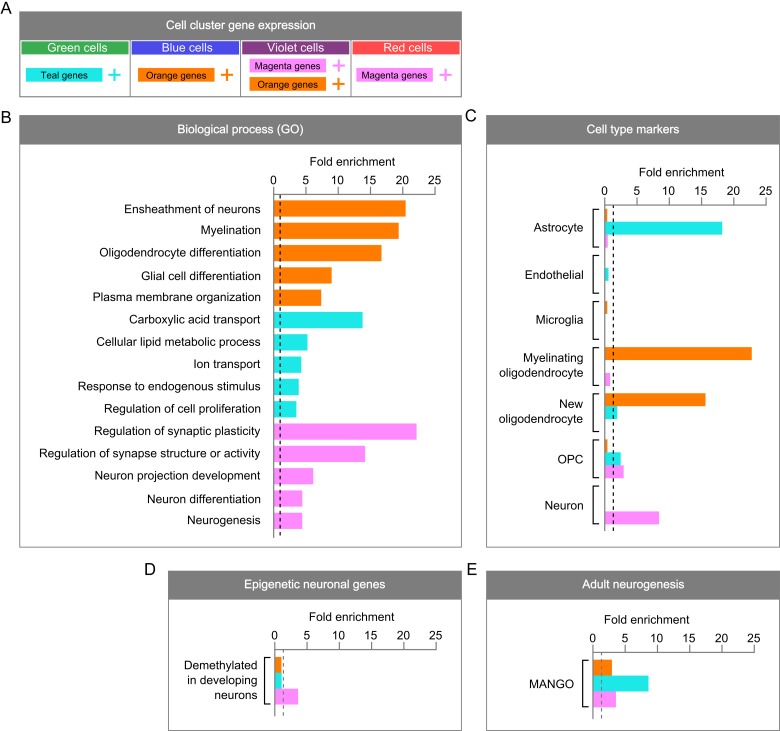
Characterization of the molecular signatures that define DCX-DsRed+ subpopulations. (A) A summary of DCX-DsRed+ subpopulations and the predominant gene clusters whose high expression defines them. (B) GO analysis of gene clusters. While enrichment was found for more GO terms, only 5 representative biological processes are shown. (C) Enrichment of CNS cell type-specific gene sets (500 genes in each each) within DCX-DsRed+ subpopulation gene clusters. For each enrichment test, fold enrichment is shown on the x-axis and a dotted line is shown at 1. (D) Enrichment of genes that are demethylated in neurons over developmental time within DCX-DsRed+ subpopulation gene clusters. (E) Enrichment of genes implicated in adult hippocampal neurogenesis within DCX-DsRed + subpopulation gene clusters.
DCX-DsRed+ Subpopulations Have Molecular Characteristics of Several Cell Lineages
We next sought to characterize the molecular signatures of subpopulations among DsRed+ cells. We first performed GO analysis on gene clusters whose differential expression defines the DsRed+ subpopulations (Fig. 3A,B). We found that orange genes were enriched for pathways related to myelination and oligodendrocyte differentiation, teal genes represented metabolic processes and cell proliferation, and the magenta gene cluster was strongly enriched for structure and function of the synapse and differentiation/development of neurons. Because gene networks that form the basis of diversity across DsRed+ subpopulations are enriched for functions that typify cell types like oligodendrocytes, neurons, and NSCs, it is possible that DCX+ cells in different stages of neurogenesis can be identified by molecular signatures characteristic of mature CNS cell types.
To more directly link DsRed+ subpopulations identified in our data to known cell lineages, we assessed overrepresentation of cell type-specific genes within underlying gene clusters (Fig. 3C). Using a published web-based tool (http://web.stanford.edu/group/barres_lab/brain_rnaseq.html) (Zhang et al. 2014), we curated sets of genes known to be enriched in various cell types in the brain relative to others as profiled in RNA-Seq experiments. Astrocytic genes were overrepresented within the teal gene cluster, while genes that distinguish new and myelinating oligodendrocytes predominated in the orange gene cluster. Neuronal genes were specifically enriched within the magenta gene cluster, and oligodendrocyte progenitor (OPC) genes exhibited mild enrichment in both teal and magenta genes. We then compared our 3 gene clusters with single-cell transcriptome data of Nestin-CFP cells (Shin et al. 2015) and found that only the astrocytic teal gene cluster showed significant enrichment, with expression levels decreasing over the pseudotime developmental trajectory (Shin et al. 2015; see Supplementary Fig. 4). In contrast, neither orange nor magenta genes show significant representation with the transcriptome of Nestin-CFP cells. Therefore, the teal gene cluster is more characteristic of adult DG NSCs compared with the other 2 clusters.
To further support the neuronal nature of magenta genes and evaluate their potential roles in immature neurons, we compared the magenta genes with those genes exhibiting dynamic changes in DNA methylation in neurons during the postnatal neuronal maturation period (Lister et al. 2013). We found that the cluster of magenta genes was specifically enriched for genes that are progressively demethylated in neurons during development (Lister et al. 2013) (Fig. 3D). Therefore, magenta cluster of genes may play important roles in the maturation of postnatal new neurons.
When gene clusters are viewed with respect to the cell clusters that express them, these data provide evidence that the green subpopulation of DsRed+ cells is more similar to neurogenic astrocytes and NSCs, while the red subpopulation is more similar to neurons. The blue subpopulation is relatively more similar to oligodendrocytes, while the violet cell cluster appears to express both neuronal and oligodendrocyte genes. However, all 3 gene clusters were enriched for genes linked to adult hippocampal neurogenesis by the Mammalian Adult Neurogenesis Gene Ontology (MANGO) database (Overall et al. 2012) (Fig. 3E). This, together with the similarity of transcriptional profiles among these subpopulations (Fig. 2), suggests that DsRed+ subpopulations are linked by their shared neurogenic nature and represent developing rather than mature cell types.
Single Gene Markers Define DCX-DsRed+ Subpopulations
DCX is commonly used as a marker for immature neurons, but DCX expression spans a wide range of 2–3 weeks in neuronal maturation (Kempermann et al. 2015). Therefore, it is of great interest to identify markers that can delineate stages of neuronal development that fall within the temporal range of DCX expression. Because single-cell gene expression is inherently stochastic, we developed an algorithm to declare cluster-specific genes that uses enrichment in both “mean expression level” among all cells in a cluster and “cluster coverage,” the proportion of cells in a cluster in which transcripts were detected. We used these criteria to identify genes that are enriched in each subgroup of cells. Using the most stringent criteria (fold change of 5 for both mean expression and coverage), we found 6 genes for the astrocyte-like DCX-DsRed+ subpopulation (green), 5 for the neuron-like cluster (red), 27 for the oligodendrocyte-like cluster (blue), and 2 for the neuron/oligodendrocyte-like cluster (violet) (see Supplementary Fig. 5). When we relaxed these criteria, more genes were identified in each subgroup, including some of the known cell lineage markers as expected. For example, Gfap, Sox2, Fabp7 (gene encoding Brain Lipid Binding Protein or BLBP) marked the astrocyte/stem cell-like (less mature) green group, while Calb1 (gene encoding Calbindin) and Rbfox3 (gene encoding NeuN) distinguished the more neuronal red and violet cell groups. Interestingly, while both Prox1 and Neurod1, genes that produce 2 transcription factors known to be enriched in granule neurons, were present in green, violet, and red groups of cells, only Prox1 was detected in the blue subgroups (see Supplementary Fig. 6). Since adult NSCs do not normally differentiate into oligodendrocytes, and because we were primarily interested in identifying novel markers in immature neurons, we decided to focus on the green, violet, and red groups for assessment of these markers at different developmental stages of immature neurons.
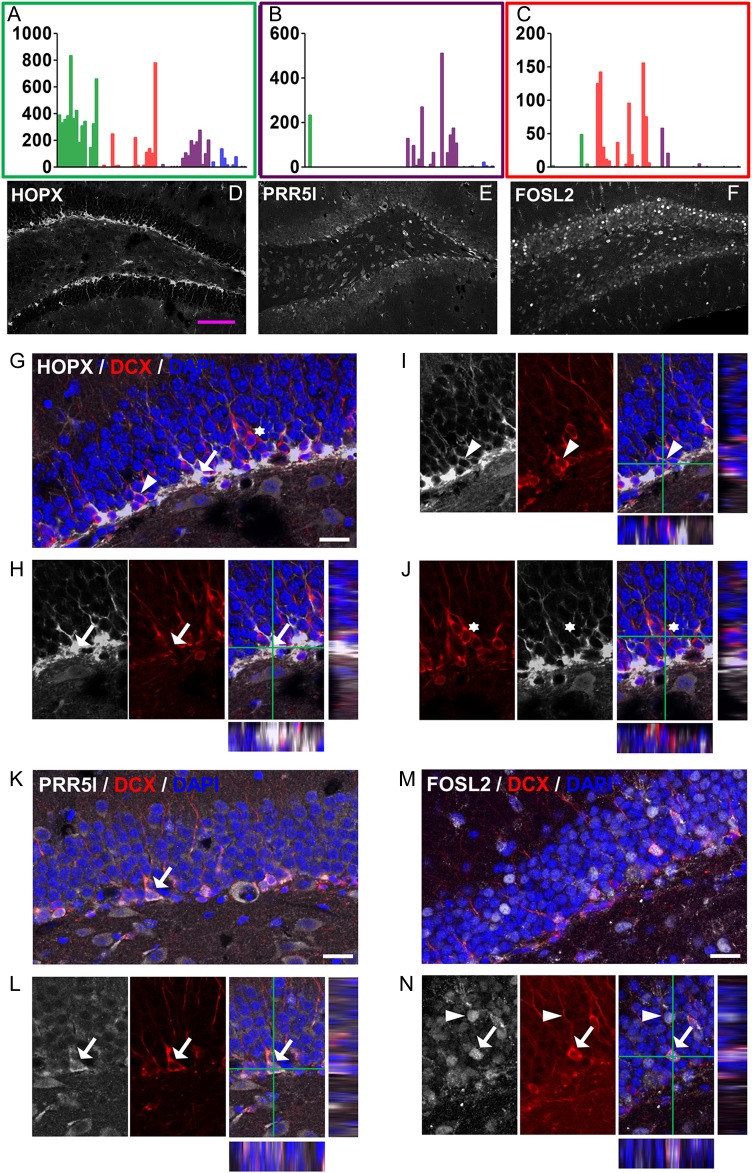
To ensure that subgroup-enriched mRNAs truly represent protein expression and to enhance the usefulness of our discovery, we screened commercially available antibodies for these markers that could be used for histological characterization of immature neurons. We identified antibodies for Homeodomain only protein (HOPX, green group), proline rich 5 like (PRR5L or Protor-2, Violet group), and Fos-related antigen 2 (FOSL2 or FRA2, red group). We found that both HOPX and PRR5L were expressed at higher levels in the SGZ compared with the rest of the DG, whereas FOSL2 seemed to be expressed at variable levels in most of the mature neurons (Fig. 4A–F). As expected, histological signals of all these antibodies show colocalization with DCX immuno reactivity (Fig. 4G–N). Therefore, these markers can be readily used to study immature neurons in the adult DG.
Figure 4.
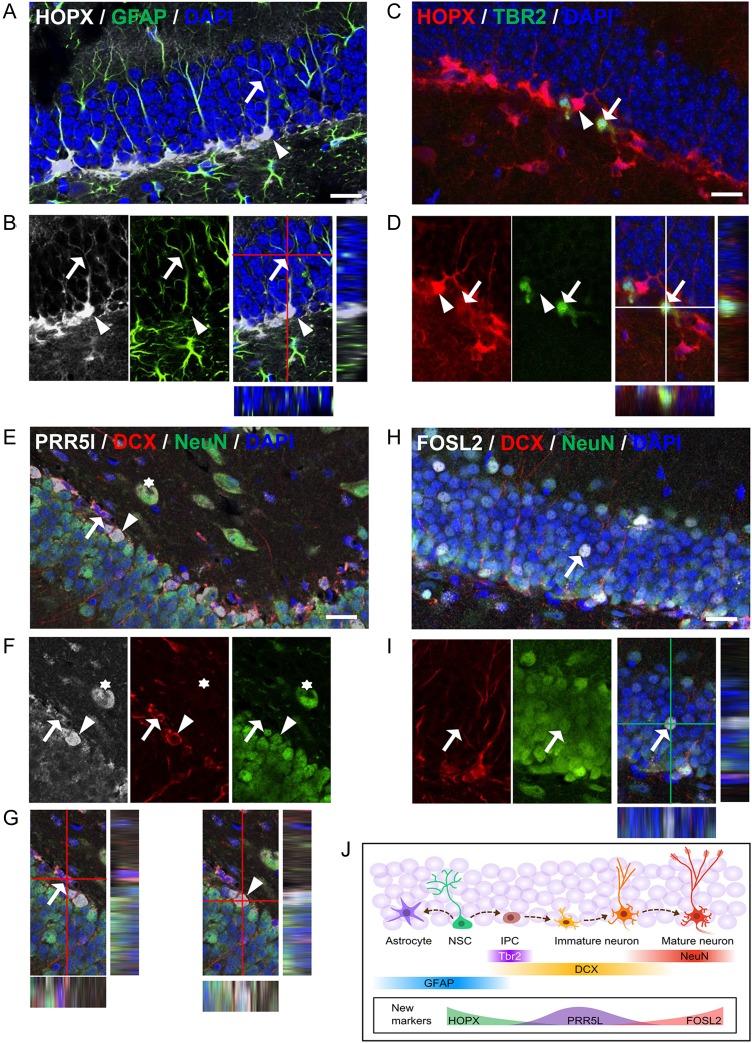
Genes enriched in specific DCX-DsRed subpopulations are expressed in DCX+ cells in the DG. (A–C) RNA-seq expression patterns of representative novel genes (A) Hopx, (B) Prr5l, and (C) Fosl2. (D–F) Confocal images showing expression patterns of the 3 genes in the DG. Scale bar, 100 μm. Both HOPX and PRR5L had higher expression levels in the SGZ compared with other areas of the DG. (G–J) Confocal images showing localization of HOPX in a significant number of DCX− cells (G, arrow) and some DCX+ cells (I, arrowhead), but not in other DCX+ cells (J, asterisk). (K–N) Confocal images showing localization of PRR5L (K,L) and FOSL2 (M,N) with DCX. Arrows and arrowheads indicate cells showing colocalization of markers. Scale bar in G–N, 20 μm.
DCX-DsRed+ Subpopulation Markers Represent Different Developmental Stages of Immature Neurons
DCX is used experimentally as a marker of developing neurons, and it is therefore possible that DCX-DsRed+ subpopulations represent stages of a single linear timeline as neurogenic cells differentiate and mature. Because adult NSCs are radial glia-like cells, it is likely that the astrocyte-like green cell cluster identified in the present experiment corresponds to the earliest developmental stage. Indeed, Nestin+ NSCs share the same pattern of relative gene cluster expression (high teal, low orange, and magenta) as the astrocyte-like DCX-DsRed+ subpopulation, which diminishes as NSCs differentiate into IPCs (see Supplementary Fig. 4). In contrast, the neuron-like red cell cluster is closest to a mature neuron endpoint. Supporting this conclusion, the mature neuron markers NeuN (Rbfox3) and Calbindin (Calb1) are most highly expressed in the red and violet cell clusters (see Supplementary Fig. 6A,B).
To further define the developmental stage of immature neurons that express these markers, we co-stained the above new markers with well-known cell lineage markers. We found that HOPX, known to be present in adult NSCs (De Toni et al. 2008; Shin et al. 2015), is expressed not only in a large number of GFAP+ radial glial-like NSCs (Fig 5A,B) but also a small number of TBR2-expressing cells (Fig 5C,D). However, HOPX is not expressed in any of the NeuN+ cells (see Supplementary Fig. 7A). Interestingly, HOPX exhibited distinct subcellular localization patterns in different cell types. In GFAP+ NSCs, where HOPX exhibited the highest expression levels, HOPX was localized in both the cytosol and the nuclei (Fig. 5A,B). In contrast, in DCX+ cells where HOPX was expressed at reduced levels, HOPX was localized only in the cytosol (Fig. 4I). A small number of TBR2+ cells expressed HOPX at low levels, but the pattern of expression resembled that of GFAP+ radial glia (Fig. 5C,D). Therefore, HOPX may mark the most immature population of the DCX+ cells, and its subcellular expression pattern may underlie its important function in adult neurogenesis.
Figure 5.
Genes enriched in specific DCX-DsRed subpopulations define different developmental stages of immature neurons. (A–D) Confocal images showing collocalization of HOPX with GFAP in radial glia-like neural stem cells (A,B, both arrow and arrowhead) and with TBR2 in neural progenitors (C,D, arrow indicates colocalization; Arrowheads point to a HOPX+TBR2− cell) in the DG. (E–G) Confocal images showing collocalization of PRR5L with DCX and NeuN in neurons. Arrow points to a DCX+NeuN− cell, whereas arrowhead indicates a DCX+NeuN+ cell. (H,I) Confocal images showing colocalization of FOSL2 with DCX and NeuN in neurons. Arrow points to a DCX−NeuN+ cell that expressed a high level of FOSL2. Scale bar in A–I, 20 μm. (J) Schematic diagram showing the expression patterns of both new and known markers during adult neurogenesis. NSC, neural stem cells; IPC, immediate progenitor cells.
PRR5L is a component of mammalian target of rapamycin complex 2 (mTORC2) that regulates actin cytoskeleton reorganization and cell migration (Gan et al. 2012). Its function in the brain is unknown. We found that PRR5L was expressed at 2 different levels in the DG. The DG cells expressing high levels of PRR5L (PRR5L-high cells) were all localized in the SGZ. PRR5L-high cells were positive for either DCX alone or both DCX and NeuN (DCX+NeuN+), but rarely NeuN alone (DCX-NeuN+, Fig 5E–G). PRR5L signal (high) was detected in TBR2+ cell in the SGZ (see Supplementary Fig. 7B). On the other hand, nearly all NeuN+ DG granule neurons outside of SGZ expressed low levels of PRR5L (PRR5L-low) (Figs 5E,F, 4D). Therefore, PRR5L-high marks immature neuron population (Fig. 4B) in the DG and its down regulation in mature neurons may underlie its functions in neuronal migration and maturation.
FOSL2 belongs to the AP-1 transcription factor family, which includes a number of isoforms of FOS and JUN (Wang et al. 2014). FOSL2 exhibited variable expression levels in DG granule neurons. The cells expressing the highest FOSL2 levels are all NeuN+ (DCX−) mature neurons (Fig. 5H,I). The sporadic distribution of FOSL2 expression patterns resembles that of cFOS, a neuronal activity-dependent immediate early gene (Clark et al. 2010). Some of the DCX+NeuN+ immature neurons also expressed FOSL2, but the expression levels were much lower than those in FOSL2-high neurons (Fig. 5H,I). Neither PRR5L nor FOSL2 showed strong signals in GFAP+ radial glia-like NSCs or TBR2+ progenitors (see Supplementary Fig. 7C,E). Therefore, FOSL2-high marks the most mature neuron population (red, Fig. 4C) in the DG, and its upregulation in mature neurons may point to a function in neuronal integration or synaptogenesis. In summary, therefore, the 3 markers HOPX, PRR5L, and FOSL2, representing the green, violet, and red groups of cells, can be used to identify DCX+ cells at different developmental stages.
Neuronal Maturation Is Mediated by Specific Molecular Pathways
Although DCX-DsRed+ cells are intrinsically similar (Fig. 2), subgroups of DsRed+ cells exhibited differential expression in subsets of genes (Fig. 6A; see Supplementary Fig. 8). We reasoned that dynamically regulated genes between DCX+ subgroups during the process of neuronal maturation might be important for driving the transition from a less mature to more mature phenotype. To investigate the underlying molecular features of the progression from the early, stem-like stage to the more mature neuronal stage of DCX-DsRed+ cells, we analyzed differentially expressed genes between the green and red groups of cells in a pairwise comparison. One thousand six hundred and sixty-five genes exhibited significant expression changes in this transition, with one-third upregulated in red compared with green, and two-thirds upregulated in green over red (Fig. 6A; see Supplementary Table 2 (Table_S2.csv)). We found that genes highly expressed in the less mature green subgroup of cells exhibited internal membrane localization, participated in lipid metabolism and poly(A) RNA binding, and were associated with the centrosome (Fig. 6B). Interestingly, enrichment was detected for 2 transcription factor-binding motifs that correspond to serum response factor (SRF), which is known to play important roles in cell proliferation. A full list of transcription factors and chromatin remodeling factors (Zhang et al. 2012) with differential expression between red and green cell clusters can be found in Supplementary Table 4, which may directly facilitate neuronal maturation via transcriptional regulation.
Figure 6.
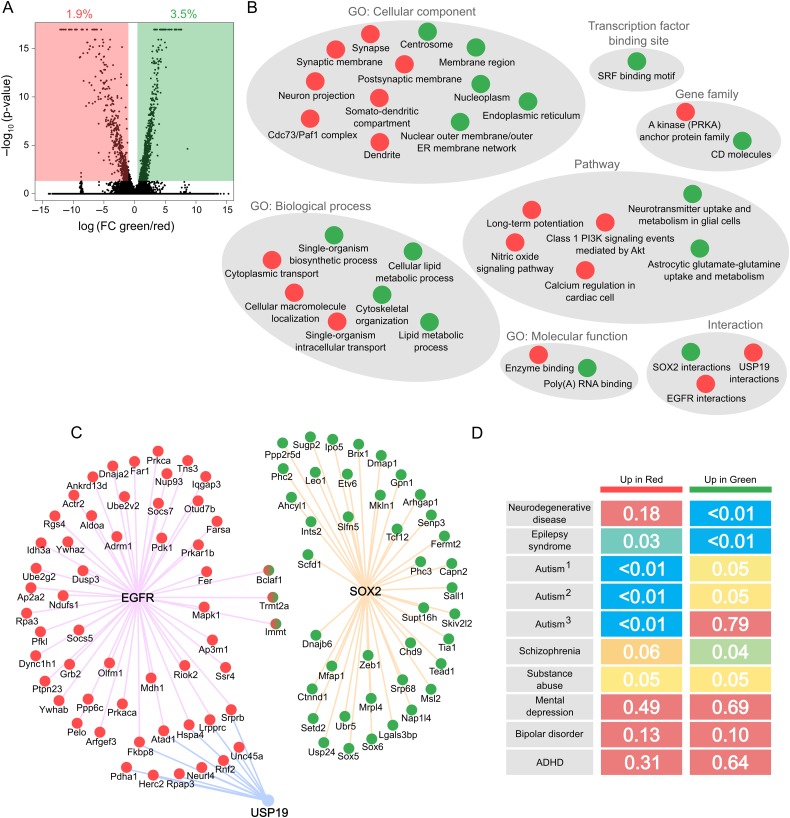
Gene expression changes from Stem-like DCX-DsRed+ cells to neuron-like DCX-DsRed+ cells. (A) Volcano plot showing log2 fold changes of individual genes on the x-axis and –log10P values on the y-axis. 1.9% of genes were differentially expressed at higher levels in red cells than in green, while 3.5% of genes were differentially expressed at higher levels in green cells relative to red. (B) Pathway analysis of genes upregulated in green and red cell clusters. Gray circles include categories of the same type (e.g., “GO: Cellular component”). Categories enriched in genes expressed higher in green cells are shown as green circles, and enriched categories within genes expressed higher in red cells are shown as red circles. (C) Individual genes that comprise enriched interactions among differentially expressed genes in red versus green cell clusters. The color of each circle signifies whether it is more highly expressed in red or in green DCX-DsRed cell groups, while 3 genes (Bclaf1, Trmt2a, and Immt) are shown as both red and green, because they possess transcript variants that are differentially expressed in opposite directions. Interactions are shown as colored lines connecting genes to EGFR (pink), USP19 (blue), and SOX2 (orange). (D) Enrichment heatmap for disease-related genes in differentially expressed genes. Diseases are represented by rows and upregulated genes in red and green cell clusters are represented by columns. P values are shown in each cell and are color coded from 0 (blue) to 0.1 (red). The Autism1 gene set was curated by Phenocarta, Autism2 by AutDB, and Autism3 by the DISEASES database. All other gene sets are from Phenocarta.
Genes preferentially expressed in the green group of immature cells were also enriched for interactions with SOX2, a known transcription factor enriched in NSCs. In contrast, genes characteristic of the red subgroup of neuron-like cells were enriched for synapse and dendrite localization, cellular transport, nitric oxide signaling and long-term potentiation. Genes with increased expression in the red cell cluster were also enriched for interactions with EGFR, which is known to regulate neuronal development and signaling (Oyagi and Hara 2012), and for interactions with the deubiquitinating enzyme USP19 that has recently been shown to regulate Hes1 stability and neuronal differentiation in ES cells (Kobayashi et al. 2015). Individual interacting genes for SOX2, EGFR, and USP19 are shown in Figure 6C and include numerous novel candidates for involvement in neurogenesis at the stage of new neuron survival and development. Therefore, gene networks that are differentially expressed between green and red dsRed+ subgroups unveil expected molecular hallmarks as well as novel genes and pathways involved in the progressive maturation of immature neurons.
We then assessed disease associations of the genes differentially expressed between the red and green subgroups using Phenocarta, DISEASES, and AutDB (Basu et al. 2009; Pletscher-Frankild et al. 2015). We found that genes implicated in neurodegenerative disease shared significant overlap with those enriched in the green cell cluster, suggesting that defects in immature cells or early stages of neural differentiation, rather than neuronal maturation, may be the primary contributor to the etiology of degenerative disorders such as Alzheimer's disease (Fig. 6D). In contrast, genes positively associated with the progression from green to red cell clusters were robustly enriched for autism-related gene sets, suggesting that the development of autism may be driven by dysregulation of candidate genes and networks initialized within the maturation process of immature neurons, when neurogenesis is critically susceptible to environmental influences. Genes linked to epilepsy were overrepresented in both up and downregulated genes during this period, reflecting a complex basis for the disease at multiple stages. Genes associated with other mental health diseases thought to have developmental components, such as depression, bipolar disorder, and ADHD, showed no enrichment among differentially expressed genes, while schizophrenia and substance abuse gene sets displayed borderline significance. Candidate genes identified in this study therefore represent potentially valuable targets for therapies designed to treat neurodevelopmental diseases.
Discussion
Using a combination of FACS enrichment, automated single-cell capturing and processing, and RNA-sequencing, we characterized molecular signatures of immature neurons born in the postnatal hippocampus. Our data provide the first transcriptome analysis of immature new neurons at single-cell resolution.
Single-cell transcriptome analysis provides a powerful tool for understanding cellular diversity and developmental processes of complex tissues, especially in mammalian brains. Two recent studies have used single-cell transcriptome analysis to characterize neural stem and progenitors directly isolated from the postnatal SVZ (Luo et al. 2015) and the DG (Shin et al. 2015). These studies provided novel information that had not been shown previously by analyzing bulk populations of cells even after FACS enrichment, demonstrating the power and utility of single-cell analysis. Despite extensive interest in adult neurogenesis and the awareness of immature neurons as a key regulatory point, little has been done to characterize immature neurons. Using similar FACS enrichment of DsRed+ cells from the same DCX-DsRed transgenic mouse line we used, Bracko et al. (2012) determined gene expression profiles through microarray analysis. Their study demonstrated that DsRed expression-based cell sorting can be used to isolate immature neurons with differential gene expression profiles from those more immature cells isolated from SOX2-GFP transgenic mice. We obtained a similar result when we compared our single-cell data from DCX-DsRed mice with recently published single-cell RNA-Seq data of CFP+ cells isolated from Nestin-CFP transgenic mice. However, compared with Bracko et al., not only does our study have the advantage of depth and richness of RNA-sequencing, but we also have single-cell resolution. During FACS procedure, we also observed a DsRed-low population, which we found to have high proportion of cells with no or lower than detectable levels of DsRed, upon further microscopic analysis. It is unclear whether these cells belong to a separate category of cells or an artifact due to low-level, nonspecific activities. Overall, our data provide much needed complementary information that extends recent publication on NSCs to immature neurons.
The 4 subgroups of cells in our analysis are categorized by 3 sets of genes. Only teal genes show high expression in Nestin-CFP cells (Shin et al. 2015), but expression of these genes declines along the developmental pseudo-timeline proposed by Shin et al. In addition, HOPX, the representative marker of the teal gene cluster, labels mostly NSCs, progenitors, and some immature neurons, but not mature neurons. These data support our hypothesis that these teal genes define the green group of cells as the more progenitor-like type of cells among the DCX-DsRed population. The neuronal (magenta) genes are not only enriched for adult neurogenesis regulators found in the MANGO database (Overall et al. 2012), but also for genes that have been shown to undergo dynamic DNA demethylation during the active neuronal maturation period of postnatal forebrain development (Lister et al. 2013). Therefore, these genes are likely important for development and maturation of immature neurons. Our comparison between the green and red groups of cells yielded both known and novel information. The overrepresentation of genes highly expressed in the green cluster of cells for SRF-binding elements suggests the involvement of SRF in the regulation of immature stem cells. Interestingly, HOPX has been shown to interact with SRF and inhibit the activities of SRF in proliferating cells (De Toni et al. 2008). The distinct subcellular localization patterns of HOPX in NSCs versus immature neurons may play a critical role in HOPX interaction with SRF and subsequent regulation of SRF signaling and downstream target genes. Another surprise is the increased EGFR pathway in the red group of cells versus the green group of cells. Although EGF and its receptor EGFR are known to be critical for stem cell proliferation, the function of EGFR in neurons is less well-known (Ayuso-Sacido et al. 2006). A number of studies have demonstrated important roles for EGFR in neuronal signaling, survival, axonal growth, and stress responses (Oyagi and Hara 2012). However, the function of EGFR in maturation of adult-born new neurons remains unexplored. Enrichment of genes implicated in human brain pathologies within genes found to be dynamically regulated during maturation of adult-born mouse neurons suggests that this rodent model may be a valuable translational tool for uncovering new targets for the development of treatments in human disease.
Curiously, one of the 3 clusters of genes identified in this study is enriched specifically for oligodendrocyte (orange) genes. The orange genes are highly expressed in 2 subgroups of DsRed+ cells: the blue group, which is characterized by high expression of oligodendrocyte genes alone, and the violet group, which expresses both neuronal (magenta) and orange genes. Studies have shown that Nestin-expressing NSCs in the adult DG do not differentiate into the oligodendrocyte lineage. It is therefore possible that the presence of these genes is a result of nonspecific activities of the DCX promoter. Because the violet cell cluster shares expression of neuronal genes with the red cluster while retaining activity of genes related to other cell types, it may represent a more immature neuron stage. The fact that PRR5L, the representative marker of the violet group, exhibited highest expression levels in the SGZ and in immature neurons support this notion. It may therefore be possible that oligodendrocyte genes are transiently activated in developing neurons before their neuronal fate is decided. In fact, it has been shown that exogenously expressed AsclI/Math1 in adult DG neuroprogenitors pushes them into the oligodendrocyte lineage (Jessberger et al. 2008). This provocative hypothesis remains to be tested.
Finally, adult neurogenesis is a dynamic process that changes with age. Although the function of adult neurogenesis remains a discussion (Groves et al. 2013; Kempermann et al. 2015), we hope that our analysis leads the way for further analysis of disease models and older mice that are more relevant to human disease conditions.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by grants from the National Institutes of Health (grant number R01MH080434, R01MH078972, and R21NS095632 to X.Z.), the International Rett Syndrome Foundation (3007 to X.Z.), an NIH Molecular Biosciences Training grant (MBTG: T32 GM07215 to E.M.J), a Center grant from the National Institutes of Health to the Waisman Center (P30HD03352), a grant from the Chinese Ministry of Science and Technology (2014CB942801 to F.W.), and a grant from the Natural Science Foundation of China (31270027 to F.W.).
Supplementary Material
Notes
We thank Cheryl T. Strauss for editing, Sandra Splinter-BonDurant, Andrew O'Guin, and Yina Xing for technical assistance, Alex Pollen, Tom Nowakowski, and Aaron Diaz for advice in single-cell analysis, and the Zhao lab members for helpful discussion. Conflict of Interest: None declared.
References
- Ayuso-Sacido A, Graham C, Greenfield JP, Boockvar JA. 2006. The duality of epidermal growth factor receptor (EGFR) signaling and neural stem cell phenotype: cell enhancer or cell transformer? Curr Stem Cell Res Ther. 1:387–394. [DOI] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. 2009. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 37:D832–D836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracko O, Singer T, Aigner S, Knobloch M, Winner B, Ray J, Clemenson GD Jr, Suh H, Couillard-Despres S, Aigner L et al. . 2012. Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J Neurosci. 32:3376–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JP, Couillard-Despres S, Cooper-Kuhn CM, Winkler J, Aigner L, Kuhn HG. 2003. Transient expression of doublecortin during adult neurogenesis. J Comp Neurol. 467:1–10. [DOI] [PubMed] [Google Scholar]
- Christian KM, Song HJ, Ming GL. 2014. Functions and dysfunctions of adult hippocampal neurogenesis. Annu Rev Neurosci. 37:243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PJ, Kohman RA, Miller DS, Bhattacharya TK, Haferkamp EH, Rhodes JS. 2010. Adult hippocampal neurogenesis and c-Fos induction during escalation of voluntary wheel running in C57BL/6J mice. Behav Brain Res. 213:246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. 2014. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 82:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Toni A, Zbinden M, Epstein JA, Ruiz i Altaba A, Prochiantz A, Caille I. 2008. Regulation of survival in adult hippocampal and glioblastoma stem cell lineages by the homeodomain-only protein HOP. Neural Dev. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger BE, Saul MC, Driessen TM, Gammie SC. 2013. Development of a versatile enrichment analysis tool reveals associations between the maternal brain and mental health disorders, including autism. BMC Neurosci. 14:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Wang J, Wang C, Sommer E, Kozasa T, Srinivasula S, Alessi D, Offermanns S, Simon MI, Wu D. 2012. PRR5L degradation promotes mTORC2-mediated PKC-delta phosphorylation and cell migration downstream of Galpha12. Nat Cell Biol. 14:686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves JO, Leslie I, Huang GJ, McHugh SB, Taylor A, Mott R, Munafo M, Bannerman DM, Flint J. 2013. Ablating adult neurogenesis in the rat has no effect on spatial processing: evidence from a novel pharmacogenetic model. PLoS Genet. 9:e1003718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Patzlaff NE, Jobe EM, Zhao X. 2012. Isolation of multipotent neural stem or progenitor cells from both the dentate gyrus and subventricular zone of a single adult mouse. Nat Protoc. 7:2005–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara H, Toyama K, Yamasaki N, Miyakawa T. 2009. Dissection of hippocampal dentate gyrus from adult mouse. J Vis Exp. 33:e1543 10.3791/1543. [DOI] [PMC free article] [PubMed]
- Jessberger S, Toni N, Clemenson GD Jr, Ray J, Gage FH. 2008. Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat Neurosci. 11:888–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Song H, Gage FH. 2015. Neurogenesis in the Adult Hippocampus. Cold Spring Harbor Perspect Biol. 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Iwamoto Y, Takashima K, Isomura A, Kosodo Y, Kawakami K, Nishioka T, Kaibuchi K, Kageyama R. 2015. Deubiquitinating enzymes regulate Hes1 stability and neuronal differentiation. FEBS J. 282:2475–2487. [DOI] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. 2013. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 29:1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD et al. . 2013. Global epigenomic reconfiguration during mammalian brain development. Science. 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Coskun V, Liang A, Yu J, Cheng L, Ge W, Shi Z, Zhang K, Li C, Cui Y et al. . 2015. Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell. 161:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. 2009. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 323:1074–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall RW, Paszkowski-Rogacz M, Kempermann G. 2012. The mammalian adult neurogenesis gene ontology (MANGO) provides a structural framework for published information on genes regulating adult hippocampal neurogenesis. PLoS ONE. 7:e48527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyagi A, Hara H. 2012. Essential roles of heparin-binding epidermal growth factor-like growth factor in the brain. CNS Neurosci Therap. 18:803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletscher-Frankild S, Palleja A, Tsafou K, Binder JX, Jensen LJ. 2015. DISEASES: text mining and data integration of disease-gene associations. Methods. 74:83–89. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P et al. . 2014. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 32:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. 2013. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 14:618–630. [DOI] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL et al. . 2015. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 173:360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Eaves-Egenes J, Barkho BZ, Santistevan NJ, Zhao C, Aimone JB, Gage FH, Zhao X. 2007. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis. 27:77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Teichmann SA, Marioni JC. 2015. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 16:133–145. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13:2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Tidei JJ, Polich ED, Gao Y, Zhao H, Perrone-Bizzozero NI, Guo W, Zhao X. 2015. Positive feedback between RNA-binding protein HuD and transcription factor SATB1 promotes neurogenesis. Proc Natl Acad Sci USA. 112:E4995–E5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun D, Wang Y, Ren F, Pang S, Wang D, Xu S. 2014. FOSL2 positively regulates TGF-beta1 signalling in non-small cell lung cancer. PLoS ONE. 9:e112150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Qiu R, Tsark W, Lu Q. 2007. Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res. 85:3567–3573. [DOI] [PubMed] [Google Scholar]
- Zhang HM, Chen H, Liu W, Liu H, Gong J, Wang H, Guo AY. 2012. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 40:D144–D149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N et al. . 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.