Abstract
Nipah virus (NiV) is an emerging pathogen that, unlike other priority pathogens identified by WHO, is endemic to Southeast Asia. It is most commonly transmitted through exposure to saliva or excrement from the Pteropus fruit bat, or direct contact with intermediate animal hosts, such as pigs. NiV infection causes severe febrile encephalitic disease and/or respiratory disease; treatment options are limited to supportive care. A number of in-house diagnostic assays for NiV using serological and nucleic acid amplification techniques have been developed for NiV and are used in laboratory settings, including some early multiplex panels for differentiation of NiV infection from other febrile diseases. However, given the often rural and remote nature of NiV outbreak settings, there remains a need for rapid diagnostic tests that can be implemented at the point of care. Additionally, more reliable assays for surveillance of communities and livestock will be vital to achieving a better understanding of the ecology of the fruit bat host and transmission risk to other intermediate hosts, enabling implementation of a ‘One Health’ approach to outbreak prevention and the management of this zoonotic disease. An improved understanding of NiV viral diversity and infection kinetics or dynamics will be central to the development of new diagnostics, and access to clinical specimens must be improved to enable effective validation and external quality assessments. Target product profiles for NiV diagnostics should be refined to take into account these outstanding needs.
Keywords: nipah virus, NiV, henipavirus, in vitro diagnostics, outbreaks
Summary box.
Current diagnostics for Nipah virus (NiV), a WHO priority pathogen endemic to Southeast Asia, include in-house laboratory-based serological and nucleic acid amplification techniques.
This review identified a number of remaining diagnostic gaps, including lack of point-of-care testing, difficulties obtaining clinical specimens and limited understanding of viral diversity, infection kinetics or dynamics, and ecology of the wild life reservoir; target product profiles for NiV diagnostics should be refined accordingly.
Development of more reliable diagnostics will be key to the prevention and management of NiV disease, and to implementation of a ‘One Health’ approach comprising both human and animal surveillance and intervention.
Introduction
Nipah virus (NiV) infection is an emerging pathogen in Southeast Asia, first detected in the 1998–1999 outbreaks in Malaysia and Singapore, with seasonal outbreaks in Bangladesh and India since 2001, and a suspected 2014 outbreak in the Philippines.1–5 In the 1998–1999 outbreak, nearly 300 human cases with over 100 deaths were reported, and more than a million pigs were euthanised.6 Since 2001, NiV has infected hundreds with an average case fatality rate of 75%.5 Currently, there are no licensed vaccines or therapies, and only supportive treatment is available.
NiV has been identified as a high-priority pathogen by WHO due to the broad geographical distribution of the NiV host reservoir and its potential for zoonotic and human transmission, as well as limitations in treatment and prevention.7–9 However, there are few validated and regulated diagnostic tests available for NiV. This landscape analysis provides an overview of the current state of NiV diagnostics for screening, diagnosis and surveillance, highlighting further research and development needs.
Epidemiology
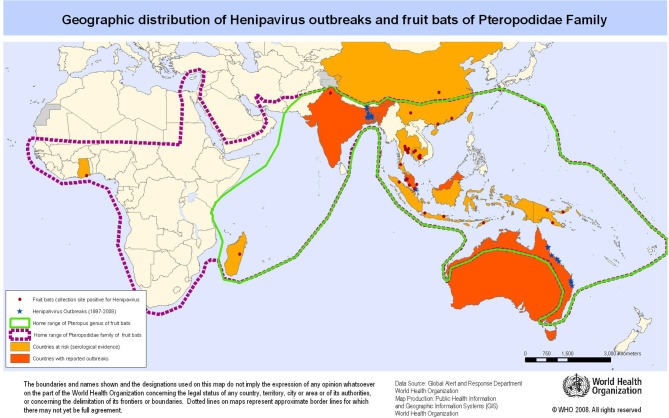
NiV is a member of the Paramyxoviridae family, sharing the genus H enipavirus because of its similarity and homology to Hendra virus (HeV) and the less pathogenic Cedar virus.10–12 Both NiV and HeV are carried by the Pteropus host endemic to South Asia and northern Australia.13–15 Infected bats shed NiV virus in their saliva, urine, semen and excreta, but are symptomless carriers. NiV surveillance has found evidence of infection in fruit bats across Southeast Asia,5 16 17 with seropositivity as far as West Africa18–20 and Brazil (figure 1).21 Sporadic outbreaks of human NiV infection have occurred in Malaysia, Singapore, India and Bangladesh since 1999.1 22–24
Figure 1.
Geographical distribution of henipavirus (Nipah and Hendra) outbreaks. Reprinted from Emergency preparedness and response—Nipah virus infection, WHO, Geographic distribution of henipavirus outbreaks and fruit bats of Pteropodidae family, copyright (2008).
Humans can be infected through direct exposure to Pteropus saliva or excrement, particularly through contaminated food.25 Date palm tree sap is a delicacy in Bengali culture, traditionally harvested by overnight collection which can be contaminated by opportune bats.14 26 27 Human NiV outbreaks show a strong seasonal pattern during winter and spring months, likely associated with the Pteropus breeding season and the date palm sap harvesting season.25
Pteropus bats may also infect intermediate hosts, where NiV typically presents as fever and severe respiratory distress.28 29 Direct contact with NiV-infected pigs was identified as the predominant mode of transmission in the 1998–1999 Malaysia–Singapore outbreaks, where 90% of the infected people were pig farmers.30 31 There is evidence of NiV seropositivity in other animals including cats, dogs, cattle and horses.28 30 32–37
Human-to-human transmission, while uncommon, appears to be possible through exposure to the body fluids of infected patients. Evidence of NiV human-to-human transmission was found in Bangladesh, where indications of respiratory distress were more common than in the Malaysian outbreak.38 39 Evidence indicated that human-to-human transmission resulted from direct contact with respiratory secretions of severely ill patients.39–42 NiV can persist on surfaces, posing further risk for fomite-borne NiV transmission.43
Clinical indications
NiV causes severe encephalitis in humans, characterised by vasculitis and necrosis in the central nervous system (CNS). The incubation period for NiV is typically 4–14 days.5 6 NiV primarily affects the CNS via endothelial, vascular, and parenchymal cell infection,28 with high rates of viral replication in neuronal cells.44
At the early stage, NiV infection typically presents as febrile encephalitis or pneumonia, and can be difficult to distinguish from other febrile illnesses. Respiratory distress was a hallmark in approximately 20% of cases in the Malaysia–Singapore outbreak and 70% of cases in Bangladesh–India. Depending on the severity, patients can also present with fever, malaise, headache, myalgia, nausea, vomiting, vertigo and disorientation.
Severe cases include encephalitis with attending drowsiness and disorientation, which can rapidly progress to seizures and coma within 48 hours. Case fatality rates range from 40% (seen in the Malaysian outbreak7) to 75% (seen in the Bangladesh outbreak45) depending on severity, patient age and underlying health issues. Progression to encephalitis indicates a poor prognosis, with death within a median of 6 days after onset of symptoms.39
Approximately 20% of encephalitis survivors sustain neurological dysfunction including persistent seizures, disabling fatigue and behavioural abnormalities.46 NiV infection can persist as non-encephalitic or asymptomatic infection; late-onset encephalitis and relapse have been observed months following initial recovery.31 47
Disease management and prevention
Due to the high pathogenicity associated with henipavirus, NiV and HeV are classified as Biosafety Level-4 (BSL-4) agents.48 Safe handling of specimens requires physical infrastructure, personal protection equipment and strict operating procedures for both clinical and research operations.49 As BSL-4 facilities may be limited in many endemic settings, BSL-3 and BSL-2 facilities may be sufficient if the virus can be inactivated following specimen collection.50
At present, there are no antiviral drugs or human vaccines available for NiV. Treatment is limited to supportive care.5 6 Ribavirin has shown some evidence for a reduction in mortality, but its efficacy against NiV disease has not yet been established.51–53 Several vaccine candidates are in development that employ NiV glycoprotein (G) and fusion (F) proteins to stimulate a protective immune response in preclinical animal models. Some approaches target specific neutralising antibody responses; others have been evaluated for both immune response and efficacy.54–56 The Hendra G protein-targeted vaccine has been developed in Australia to protect horses against HeV and may offer protection against NiV as well.57 58
Phylogenetic diversity
The NiV genome consists of a single-stranded negative-sense RNA of approximately 18.2 kb and encodes for six major structural proteins: F, G, nucleocapsid (N), phosphoprotein (P), matrix protein (M) and RNA polymerase (L) and three accessory proteins (V, W and C).12 40 Genetic sequencing has shown that the NiV and HeV coding regions for the N, P, C, V, M, F and G proteins have 70%–90% genetic homology, with approximately 92%, 87%, 83% and 68% amino acid homology for the N, L, G and P proteins, respectively.10 12 40 59 60 Because of this homology, diagnostic tests can be cross-reactive for HeV and NiV, depending on the chosen RNA or antigen targets.
Genetic sequencing confirmed that the NiV strains circulating in Bangladesh and India (Clade I, or NiV-B) were different from the strains identified in Malaysia and Singapore (Clade II, or NiV-M),23 45 60–62 with >92% genetic and amino acid homology across NiV-M and NiV-B strains.7 10 44 45 61 63 64 Further analysis confirmed the presence of two distinct NiV-M subclade strains circulating in the north and south, likely arising from distinct hosts.64 65 Lineage diversity needs to be taken into consideration in order to identify strains specific to a particular region, as well as ongoing genetic mutations to assist detection of all circulating strains (including differentiation from HeV).
Nipah detection and diagnostic tests
Since the symptoms of NiV infection are similar to other febrile diseases, early diagnosis is critical for containment of an outbreak and to facilitate appropriate patient care. Laboratory testing for NiV includes nucleic acid amplification testing (NAAT, eg, PCR and sequencing), IgG/IgM/antigen ELISA, immunofluorescence assay, histopathology, and virus isolation and neutralisation.50 66 67 Table 1 describes the typical infrastructure, settings and resources required for the various test types.
Table 1.
Diagnostic capacity vs setting
| Test type | Infrastructure requirements (example) |
Training requirements (example) |
Test process time | NiV target population | Number of NiV in-house or LDTs* | Number of NiV commercial tests* |
| Virus isolation, neutralisation | HIGH/BSL-4 (reference laboratory) |
HIGH (advanced laboratory technician) |
5 days for cell culture | Human, animal |
>3 | 0 |
| NAAT | HIGH (reference laboratory) |
HIGH (advanced laboratory technician) |
2–3 hours (1–2 hours preparation) |
Human, animal |
>5 | 3 |
| NAAT POC | MODERATE (district hospital) |
MODERATE (laboratory technician) |
1–2 hours | Human, animal | 0 | 0 |
| ELISA, IFA | HIGH to MODERATE (regional laboratory, district hospital) |
MODERATE (laboratory technician) |
3–4 hours | Human, animal |
>5 | 1 (reagents only) |
| RDTs | LOW (clinic, health centre, field settings) |
LOW (nurse, healthcare worker) |
<30 min | Human, animal |
0 | 0 |
*In-house and LDTs described in sections below; commercial sources listed in online supplementary table S1.
BSL, biosafety level; IFA, immunofluorescence assay; LDT, laboratory-developed test; NAAT, nucleic acid amplification test; NiV, Nipah virus; POC, point of care; RDT, rapid diagnostic test.
bmjgh-2018-001118supp001.pdf (419.8KB, pdf)
Currently, the majority of international laboratories use in-house NiV assays; however, the degree of test validation varies widely.28 50 67 There are a small number of commercial PCR kits available and only one commercial source for reagents (not an assembled/validated kit) for ELISA testing (tables 2 and 3, and online supplementary table S1).
Table 2.
World Organisation for Animal Health test methods available for diagnosis of henipavirus and their purpose
| Method | Purpose | |||||
| Population freedom from infection | Individual animal freedom from infection prior to movement | Contribution to eradication policies | Confirmation of clinical cases | Prevalence of infection—surveillance (no clinical) | Immune status in individual animals or populations post-vaccination | |
| Agent identification* | ||||||
| Virus isolation | + | + | — | +++ | — | |
| RT-PCR and qRT-PCR | + | + | ++ | +++ | ++ | — |
| IHC | — | — | — | ++ | — | — |
| IFA | — | — | — | ++ | — | — |
| Detection of immune response† | ||||||
| ELISA | +++ | +++ | +++ | + | +++ | +++ |
| VNT | +++ | +++ | +++ | + | +++ | +++ |
| Bead assays | +++ | +++ | +++ | + | +++ | +++ |
+++, recommended method; ++, suitable method; +, may be used in some situations, but cost, reliability or other factors severely limit application; —, not appropriate for this purpose. Although not all of the tests listed as +++ or ++ have undergone formal validation, their routine nature and the fact that they have been used widely without dubious results make them acceptable.
*A combination of agent identification methods applied on the same clinical specimen is recommended.
†One of the listed serological tests is sufficient. Reproduced with permission from OIE Terrestrial Manual, 7th ed. (May 2015 update).70
IFA, immunofluorescence assay; IHC, immunohistochemistry; RT-PCR, reverse transcriptase PCR; VNT, virus neutralisation test; qRT-PCR, quantitative RT-PCR.
Table 3.
In-house NAAT tests for NiV*
| Assay type | Target | Reference laboratory | Reference |
| RT-PCR | NiV P, N genes | ICDDRB and IEDCR, CDC (USA) |
7 45 |
| Real-time RT-PCR | NiV N gene | Institut Pasteur (France) | 112 |
| SYBR qRT-PCR | NiV N gene | University of Malaya (Malaysia) | 113 |
| Duplex nested RT-PCR | NiV N gene | Chulalongkorn University Hospital (Thailand) | 114 |
| Nested RT-PCR | HeV/NiV P gene | Bernhard Nocht Institute (Germany), CDC (USA) | 18 115 |
| Nested RT-PCR | NiV L gene | CSIRO/AAHL, University of Cambridge, Zoological Society (UK) | 71 |
| SYBR qRT-PCR | NiV N gene | CSIRO/AAHL, University of Cambridge, Zoological Society (UK) | 71 |
| Real-time RT-PCR | NiV/HeV P, M, N genes | CSIRO/AAHL, University of Cambridge, Zoological Society (UK) | 71 |
| Multiplex bead-based real-time RT-PCR | NiV, HeV N and P genes |
CSIRO/AAHL (Australia) | 89 |
| Multiplex array card real-time RT-PCR | 26 multiplex panel including NiV N gene | CDC (Kenya, USA) | 90 |
| Multiplex array card real-time RT-PCR | 21 multiplex panel including NiV N gene | Kenya Medical Research Institute (Kenya), CDC (Kenya, USA) | 91 |
*Adapted with permission from Springer Customer Service Centre GmBH: Springer Nature Indian Journal of Virology,66 copyright 2013; Springer Customer Service Centre GmBH: Springer Nature Springer eBook,67 copyright 2012; and World Organisation for Animal Health (OIE).70
AAHL, Australian Animal Health Laboratory; CDC, Centers for Disease Control and Prevention; CSIRO, Commonwealth Scientific and Industrial Research Organisation; HeV, Hendra virus; ICDDRB, International Centre for Diarrhoeal Disease Research, Bangladesh; IEDCR, Institute of Epidemiology, Disease Control and Research; NiV, Nipah virus; RT-PCR, reverse transcriptase PCR; qRT-PCR, quantitative RT-PCR.
While PCR is recommended as the most sensitive method for diagnosis of active NiV infection, NiV-specific IgM ELISA is an alternative serological approach where PCR is not available.25 Histopathology (immunohistochemistry) is used post mortem to confirm NiV diagnosis in fatal cases, and virus isolation and neutralisation techniques are generally used for confirmation and restricted to BSL-4 facilities with stringent safety precautions.
Specimen collection should be performed in the early stages of disease, typically from throat and nasal swabs, cerebrospinal fluid, urine and blood.6 In animals, NiV has been detected in respiratory secretions, blood, urine, faeces and organ tissues,28 37 66 with evidence that NiV initially infects the respiratory system, followed by later stage dissemination into the nervous system.50 68 There are limited data available for viral kinetics, but throat swabs have indicated 103–104 genome copies of NiV in severe cases and 106 in acute cases in humans.46 Similar results have been seen in animal blood and serum,22 69 with up to 1010 copies detected in animal brain tissue.56
Specifically for animal health, the World Organisation for Animal Health (OIE) has developed the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals to provide internationally agreed diagnostic laboratory methods and requirements for the production and control of vaccines and other biological products, towards the goal of facilitating international trade in animals and animal products and to contribute to the improvement of animal health services worldwide.70 Details of the specific tests and parameters are described in table 2.
Nucleic acid tests
NAATs such as PCR are often the preferred method for detection of active viral infection as they are highly sensitive (detecting as few as 20 viral genomes); however, the infrastructure required may not be feasible in all endemic settings. Further, these tests can lose sensitivity if the viral genome undergoes significant genetic mutation or if new strains differ in the regions of probe design. To ensure coverage for genetic variants, probes are typically designed for highly conserved (resistant to mutation) sequences present in all known strain sequences for the pathogen of interest.71 Full genome sequencing has been performed on the NiV-B and NiV-M strains (and substrains) along with the Cambodian and Philippine variants.7 10 44 45 60 63 64 72
Reverse-transcriptase PCR (RT-PCR) tests for NiV have targeted the conserved N, M or P genome segments (table 3). Several types of PCR tests for NiV have been developed, including conventional RT-PCR, nested RT-PCR and real-time RT-PCR (also known as quantitative PCR or qPCR). Real-time PCR has been shown to be 1000 times more sensitive as conventional PCR, and is now used almost exclusively.67
In addition, next-generation sequencing and deep sequencing enable a direct read of the viral genome, allowing virus and clade identification without prior knowledge of the composition.73 Complex and expensive, this approach is not practical for screening larger numbers of samples in a diagnostic context; however, this technique provides a comprehensive and unbiased approach to viral analysis. Used in retrospect, it can identify the phylogenetic evolution of viral isolates over time and geography.74 75
Serological assays
Serological tests can directly detect NiV antigens, as well as IgM and IgG antibodies raised against NiV antigens. In general, serological tests are less sensitive to minor genetic variation and more broadly cross-reactive within subtypes and strains.45 Antigen and IgM tests can be used to detect active infection; detection of anti-NiV IgM in serum peaks after 9 days of illness (based on hospital admittance) and can persist for at least 3 months. Since IgG can persist long after convalescence, IgG tests are primarily used for epidemiological studies and surveillance; detection of anti-NiV IgG peaks after 17 days of illness and can persist for more than 8 months.76 Existing in-house serological tests for NiV are shown in table 4.
Table 4.
In-house serological tests for NiV*
| Assay type | Target | Reference laboratory | Reference |
| ELISA | NiV IgG, IgM | ICDDRB and IEDCR (Bangladesh), CDC (USA) | 7 |
| ELISA | NiV neutralising Ab | DVS (Malaysia) | 116 |
| ELISA | NiV G protein | Universiti Putra (Malaysia) | 117 118 |
| Western blot, ELISA | NiV N, M proteins | Canadian Science Center for Human and Animal Health (Canada) | 119 |
| ELISA | NiV N protein | Chinese National Diagnostic Center for Animal Diseases (China) | 120 |
| ELISA | NiV N protein | Institute of Tropical Medicine (Japan) | 121 |
| ELISA | NiV/HeV P proteins | CSIRO/AAHL (Australia), University of Malaya (Malaysia), China Epidemiology Center (China) | 122 |
| ELISA | NiV/HeV N, HeV P proteins | CDC (USA) | 78 |
| ELISA | NiV/HeV G protein | National Institute for Infectious Diseases (Japan) | 80 |
| ELISA | NiV M protein | MARDI, Universiti Putra Malaysia, Monash University Malaysia | 123 |
| ELISA | NiV N protein | National Institute of High Security Animal Diseases/ICAR (India) | 79 |
| Multiplex bead-based antibody capture | NiV G, HeV G proteins | CSIRO/AAHL (Australia), Uniformed Services University of Health Sciences (USA) | 94 |
*Adapted with permission from Springer Customer Service Centre GmBH: Springer Nature Indian Journal of Virology,66 copyright 2013; and Springer Customer Service Centre GmBH: Springer Nature Springer eBook,67 copyright 2012.
AAHL, Australian Animal Health Laboratory; CDC, Centers for Disease Control and Prevention; CSIRO, Commonwealth Scientific and Industrial Research Organisation; DVS, Department of Veterinary Services; HeV, Hendra virus; ICAR, Indian Council of Agricultural Research; ICDDRB, International Centre for Diarrhoeal Disease Research, Bangladesh; IEDCR, Institute of Epidemiology, Disease Control and Research; MARDI, Malaysian Agricultural Research and Development Institute; NiV, Nipah virus.
IgM ELISA is typically the first-line NiV serological diagnostic test, followed by serum neutralisation or PCR as a confirmatory test.1 50 67 For increased specificity, recombinant proteins, monoclonal or polyclonal antibodies are used as capture agents. Recombinant NiV M protein has been shown to be highly reactive against antibodies in porcine sera.77 Taking advantage of the >90% homology between HeV and NiV N and G proteins but ~30% difference between their P proteins, polyclonal antibodies have been developed to capture both NiV and HeV N antigens,78 79 preferential detection of HeV P antigens,78 and capture of both NiV and HeV G antigens including live virus.80 ELISA combinations of these polyclonal antibodies can enable pan-henipavirus (NiV and HeV) detection, as well as differentiation between NiV and HeV. However, it is difficult to compare assay sensitivity due to differences in sample preparation and units of quantitation (pfu/mL vs infectious units vs TCID50).
ELISA test reagents can be translated into a lateral flow format to create a rapid diagnostic test (RDT) that can be implemented in a non-laboratory environment, typically paired with minimal specimen processing (blood, plasma, swabs).81 The lateral flow format enables a faster time to result (10–30 min), although with a lower detection sensitivity than their ELISA counterparts.82 Currently, there are no NiV (or HeV) RDTs available for humans or animals.
Serological confirmation of NiV infection is typically performed by gold-standard seroneutralisation assays using live NiV, which requires a high-containment BSL-4 facility.50 To circumvent the need for BSL-4 containment, non-infectious pseudotyped virus particles can be used in neutralisation assays under a BSL-2 environment.67 NiV pseudoparticles have been derived from vesicular stomatitis virus or lentivirus (HIV), providing a more convenient laboratory method for diagnostic confirmation.83–86
Multiplex panels
As the symptoms of NiV infection can mimic other febrile diseases, severe cases are at risk for fatality with misdiagnosis and delayed treatment. A syndromic approach involves testing for pathogens based on a syndrome such as febrile or encephalitic disease; a shift from individual tests to multiplex panels to quickly identify or eliminate likely pathogens from a single specimen. Consideration must be given to the cost of additional reagents and more sophisticated instrumentation; however, a rapid and efficient diagnosis scheme that impacts infection control may be cost-saving overall.87 88 As NiV outbreaks can be both regional and seasonal, region-specific panels may be more cost-effective and can be deployed ‘as needed’ for case detection as well as surveillance of risk factors across patient populations.
Multiplex PCR assays have been developed in the laboratory for differential detection of multiple pathogens or clades, using microarray, microcard or microsphere technologies. Microsphere array (Luminex) technology has been developed for detection and differentiation of HeV and NiV isolates.89 These microsphere assays achieved differentiation of HeV and NiV, with the sensitivity of HeV detection comparable with individual qPCR. Multiplex TaqMan array card technology, typically a 384-well microfluidic card for simultaneous PCR reactions, has been developed for differential detection of acute febrile illness, including NiV, as 26-member and 21-member real-time RT-PCR panels.90 91 The clinical performance exhibited an overall 86%–88% sensitivity and 97%–99% specificity compared with the individual real-time PCR assays. The 26-member panel includes 15 viruses, 8 bacteria and 3 protozoa. The 21-pathogen CNS multiple-pathogen assay includes two parasites, six bacterial pathogens and 13 viruses. Microarray technology has been developed for simultaneous interrogation of respiratory and gastrointestinal disease, including as global fever and biothreat agents92 93; such automated platforms enable ‘sample in, result out’ processing, but no panel includes NiV at the present date.
Multiplex immunoassay platforms (Luminex) for differential diagnosis employing microspheres (beads) coated with soluble HeV and NiV G proteins, respectively, have been developed.94 Two different test formats were evaluated: a binding assay to differentiate between HeV and NiV antibodies, and a blocking or inhibition assay for detection and differentiation of HeV and NiV neutralising antibodies. By comparison, conventional ELISA tests were incapable of antibody differentiation. These bead-based Luminex assays have also been used to detect henipavirus antibodies in fruit bats and pigs.17 19 95–98 More recently, the Luminex assay was used to assess HeV infection mice,99 humans,100 and horses and dogs.101
Challenges for NiV diagnostics
Viral diversity and infection kinetics and dynamics
The ideal NiV diagnostic test would detect all NiV lineages across all regions with equal sensitivity; however, many of the early PCR tests were developed from Bangladesh–India (NiV-B) or Malaysia–Singapore (NiV-M) strains. Thus, NiV diagnostics could benefit from a consensus-driven pan-NiV probe set. Additionally, further information is needed on viral and immune kinetics of NiV to identify the pathogenesis of NiV in blood and non-blood samples. Understanding the course of NiV infection at different stages of the disease would help to identify the window for effective intervention, and to better monitor transmission and recovery.
Clinical validation
For diagnostic test developers, clinical specimens are key to the validation process; however, validation for NiV tests has been limited due to the lack of NiV-positive sera available.67 78 While animal samples may be used to investigate the performance characteristics of a test, there is a risk that the antigens found in specific animal-sourced NiV infections are not the same as those found in humans or other animals.78 The WHO International Biological Reference Preparations serve as reference sources for ensuring the reliability of in vitro biological diagnostic procedures and are distributed by the WHO custodian laboratories. As new diagnostic tests are developed, these agencies could perform routine external quality assessment (EQA) monitoring of tests using up-to-date clinical specimen panels and reference standards. This role will be particularly important when test developers have little incentive to seek international regulatory approval.
Point-of-care testing
Given the often rural and remote NiV outbreak settings, sensitive and accurate diagnostics for NiV must be deployable under a range of circumstances, particularly at the point of care (POC). Existing POC and ‘near-POC’ NAAT platforms have lower infrastructure requirements than laboratory-based diagnostics, with automated sample preparation resulting in fewer training requirements for healthcare workers and cartridge-based formats allowing tests to be self-contained. They can therefore be more easily implemented in decentralised laboratories or field-based settings.102–104 Given the range of assays already developed for these commercial NAAT platforms, current NiV PCR assays could likely be readily adapted to the POC format.
RDTs are ideal for field testing and low infrastructure settings such as the clinic or home.81 RDTs have been developed to effectively screen and triage suspected high-risk cases such as Lassa, Ebola and Dengue.105–107 Similar to Ebola, the pathogenicity of NiV makes specimen processing a challenge in a non-laboratory setting, thus preliminary work in antigen ELISA development78–80 could serve as a starting point for an antigen NiV RDT.
Surveillance
Education campaigns may be helpful to increase awareness of the risk for NiV infection in the community and promote appropriate care-seeking behaviour, as well as to maintain vigilance for case identification by healthcare personnel.108–110 Early detection of NiV outbreaks should trigger a coordinated mobilisation of resources for infection control, as well as patient isolation, care and contact tracing.
Surveillance tools for NiV should include reliable laboratory assays for early detection of disease in communities and livestock.6 70 A One Health approach is key to understanding the fruit bat ecology, NiV disease seasonality and the transmission risk of other intermediate hosts. Without understanding the wildlife reservoir, the risk of reintroduction into animal or human populations cannot be managed.58 111
Conclusion
In addition to playing a central role in the recognition and control of outbreaks, diagnostic tests can enable a more nuanced understanding of the window of positivity and duration of infection, transmission risk and risk factors for severity for NiV, one of the most widespread agents of febrile encephalitic disease. In particular, diagnostics to support early detection will be critical for ‘hot spot’ interventions and containment (online supplementary table S2). However, several of the gaps identified in the 2016 WHO R&D Blueprint remain, including a lack of routine EQA, understanding of NiV viral and antibody kinetics, well-characterised and up-to-date proficiency panels, and accurate surveillance data.
bmjgh-2018-001118supp002.pdf (520.2KB, pdf)
Target product profiles for NiV should be refined to include the need to identify all known lineages of NiV, and the benefits of RDT POC diagnostics and syndromic panels. As diagnostics are a key element to achieving the goals of the R&D blueprint, WHO is coordinating research and funding through product development partnerships with groups such as FIND to ensure the development, evaluation and delivery of high-quality, affordable diagnostics for NiV.
Acknowledgments
We gratefully acknowledge all those who attended the WHO R&D Blueprint Roadmaps: Consultation on Nipah in March 2018. Editorial assistance for later drafts was provided by Rachel Wright, PhD, funded by FIND, according to Good Publication Practice guidelines (http://annals.org/aim/fullarticle/2424869/good-publication-practice-communicating-company-sponsored-medical-research-gpp3). Publication of this article was funded by FIND.
Footnotes
Handling editor: Seye Abimbola
Contributors: LTM contributed to drafting the manuscript and provided background research for the manuscript. CK-C contributed insight into diagnostic needs for outbreak pathogens. All authors reviewed, edited and approved the final version of the manuscript.
Funding: FIND was funded for this work by UK aid from the UK Government.
Disclaimer: The opinions expressed in this article are those of the authors and do not necessarily reflect those of the institutions or organisations with which they are affiliated.
Competing interests: None declared.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1. Sherrini BA, Chong TT. Nipah encephalitis—an update. Med J Malaysia 2014;69(Suppl A:1):03–11. [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention (CDC) Update: outbreak of Nipah virus—Malaysia and Singapore, 1999. MMWR Morb Mortal Wkly Rep 1999;48:335–7. [PubMed] [Google Scholar]
- 3. Ching PK, de los Reyes VC, Sucaldito MN, et al. . Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis 2015;21:328–31. 10.3201/eid2102.141433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chua KB. Nipah virus outbreak in Malaysia. J Clin Virol 2003;26:265–75. 10.1016/S1386-6532(02)00268-8 [DOI] [PubMed] [Google Scholar]
- 5. WHO , 2018. Nipah Virus (NiV) Infection. http://www.who.int/csr/disease/nipah/en/ (accessed 25 Feb 2018).
- 6. CDC , 2018. Nipah Virus (NiV). https://www.cdc.gov/vhf/nipah/index.html (accessed 25 Feb 2018).
- 7. Chua KB, Bellini WJ, Rota PA, et al. . Nipah virus: a recently emergent deadly paramyxovirus. Science 2000;288:1432–5. 10.1126/science.288.5470.1432 [DOI] [PubMed] [Google Scholar]
- 8. Epstein JH, Field HE, Luby S, et al. . Nipah virus: impact, origins, and causes of emergence. Curr Infect Dis Rep 2006;8:59–65. 10.1007/s11908-006-0036-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. WHO , 2017. R&D Blueprint for action to prevent epidemics. WHO RD Bluepr. http://www.who.int/blueprint/en/ (accessed 12 Sep 2017).
- 10. Harcourt BH, Tamin A, Ksiazek TG, et al. . Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 2000;271:334–49. 10.1006/viro.2000.0340 [DOI] [PubMed] [Google Scholar]
- 11. Marsh GA, de Jong C, Barr JA, et al. . Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog 2012;8:e1002836 10.1371/journal.ppat.1002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang L, Harcourt BH, Yu M, et al. . Molecular biology of Hendra and Nipah viruses. Microbes Infect 2001;3:279–87. 10.1016/S1286-4579(01)01381-8 [DOI] [PubMed] [Google Scholar]
- 13. Clayton BA, Wang LF, Marsh GA. Henipaviruses: an updated review focusing on the pteropid reservoir and features of transmission. Zoonoses Public Health 2013;60:69–83. 10.1111/j.1863-2378.2012.01501.x [DOI] [PubMed] [Google Scholar]
- 14. Luby SP, Rahman M, Hossain MJ, et al. . Foodborne transmission of Nipah virus, Bangladesh. Emerg Infect Dis 2006;12:1888–94. 10.3201/eid1212.060732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yob JM, Field H, Rashdi AM, et al. . Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg Infect Dis 2001;7:439–41. 10.3201/eid0703.017312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sendow I, Field HE, Adjid A, et al. . Screening for Nipah virus infection in West Kalimantan province, Indonesia. Zoonoses Public Health 2010;57:499–503. 10.1111/j.1863-2378.2009.01252.x [DOI] [PubMed] [Google Scholar]
- 17. Sendow I, Ratnawati A, Taylor T, et al. . Nipah virus in the fruit bat Pteropus vampyrus in Sumatera, Indonesia. PLoS One 2013;8:e69544 10.1371/journal.pone.0069544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drexler JF, Corman VM, Gloza-Rausch F, et al. . Henipavirus RNA in African bats. PLoS One 2009;4:e6367 10.1371/journal.pone.0006367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Field HE, Mackenzie JS, Daszak P. Henipaviruses: emerging paramyxoviruses associated with fruit bats. Curr Top Microbiol Immunol 2007;315:133–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayman DTS, Suu-Ire R, Breed AC, et al. . Evidence of henipavirus infection in West African fruit bats. PLoS One 2008;3:e2739 10.1371/journal.pone.0002739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Araujo J, Lo MK, Tamin A, et al. . Antibodies against Henipa-like viruses in Brazilian bats. Vector-Borne and Zoonotic Diseases 2017;17:271–4. 10.1089/vbz.2016.2051 [DOI] [PubMed] [Google Scholar]
- 22. Clayton BA, Middleton D, Arkinstall R, et al. . The nature of exposure drives transmission of Nipah viruses from Malaysia and Bangladesh in ferrets. PLoS Negl Trop Dis 2016;10:e0004775 10.1371/journal.pntd.0004775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lo Presti A, Cella E, Giovanetti M, et al. . Origin and evolution of Nipah virus. J Med Virol 2016;88:380–8. 10.1002/jmv.24345 [DOI] [PubMed] [Google Scholar]
- 24. Mire CE, Cross RW, Geisbert JB, et al. . Human-monoclonal-antibody therapy protects nonhuman primates against advanced Lassa fever. Nat Med 2017;23:1146–9. 10.1038/nm.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. WHO , 2018. Nipah Virus Infection factsheet. http://www.who.int/mediacentre/factsheets/fs262/en/ (accessed 25 Jan 2018).
- 26. Nahar N, Sultana R, Gurley ES, et al. . Date palm sap collection: exploring opportunities to prevent Nipah transmission. Ecohealth 2010;7:196–203. 10.1007/s10393-010-0320-3 [DOI] [PubMed] [Google Scholar]
- 27. Nahar N, Paul RC, Sultana R, et al. . Raw sap consumption habits and its association with knowledge of Nipah virus in two endemic districts in Bangladesh. PLoS One 2015;10:e0142292 10.1371/journal.pone.0142292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hooper P, Zaki S, Daniels P, et al. . Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect 2001;3:315–22. 10.1016/S1286-4579(01)01385-5 [DOI] [PubMed] [Google Scholar]
- 29. Sauerhering L, Zickler M, Elvert M, et al. . Species-specific and individual differences in Nipah virus replication in porcine and human airway epithelial cells. J Gen Virol 2016;97:1511–9. [DOI] [PubMed] [Google Scholar]
- 30. Chua KB, Goh KJ, Wong KT, et al. . Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 1999;354:1257–9. 10.1016/S0140-6736(99)04299-3 [DOI] [PubMed] [Google Scholar]
- 31. Goh KJ, Tan CT, Chew NK, et al. . Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med Overseas Ed 2000;342:1229–35. 10.1056/NEJM200004273421701 [DOI] [PubMed] [Google Scholar]
- 32. Chowdhury S, Khan SU, Crameri G, et al. . Serological evidence of henipavirus exposure in cattle, goats and pigs in Bangladesh. PLoS Negl Trop Dis 2014;8:e3302 10.1371/journal.pntd.0003302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Epstein JH, Rahman SA, Zambriski JA, et al. . Feral cats and risk for Nipah virus transmission. Emerg Infect Dis 2006;12:1178–9. 10.3201/eid1207.050799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Middleton DJ, Weingartl HM. Henipaviruses in their natural animal hosts. Curr Top Microbiol Immunol 2012;359:105–21. [DOI] [PubMed] [Google Scholar]
- 35. Middleton WH, Morrissy C. Experimental transmission of Nipah virus infection to pigs and cats : Nature reviews. 39 Sydney, Australia: Drug Discovery, 1999. [Google Scholar]
- 36. Mills JN, Alim AN, Bunning ML, et al. . Nipah virus infection in dogs, Malaysia, 1999. Emerg Infect Dis 2009;15:950–2. 10.3201/eid1506.080453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parashar UD, Sunn LM, Ong F, et al. . Case–control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998–1999 outbreak of severe encephalitis in Malaysia. J Infect Dis 2000;181:1755–9. 10.1086/315457 [DOI] [PubMed] [Google Scholar]
- 38. Hossain MJ, Gurley ES, Montgomery JM, et al. . Clinical presentation of nipah virus infection in Bangladesh. Clin Infect Dis 2008;46:977–84. 10.1086/529147 [DOI] [PubMed] [Google Scholar]
- 39. Hsu VP, Hossain MJ, Parashar UD, et al. . Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis 2004;10:2082–7. 10.3201/eid1012.040701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan YP, Chua KB, Koh CL, et al. . Complete nucleotide sequences of Nipah virus isolates from Malaysia. J Gen Virol 2001;82(Pt 9):2151–5. 10.1099/0022-1317-82-9-2151 [DOI] [PubMed] [Google Scholar]
- 41. Gurley ES, Montgomery JM, Hossain MJ, et al. . Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 2007;13:1031–7. 10.3201/eid1307.061128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luby SP, Hossain MJ, Gurley ES, et al. . Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg Infect Dis 2009;15:1229–35. 10.3201/eid1508.081237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hassan MZ, Sazzad HMS, Luby SP, et al. . Nipah virus contamination of hospital surfaces during outbreaks, Bangladesh, 2013–2014. Emerg Infect Dis 2018;24:15–21. 10.3201/eid2401.161758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. MK L, Miller D, Aljofan M, et al. . Characterization of the antiviral and inflammatory responses against Nipah virus in endothelial cells and neurons. Virology 2010;404:78–88. [DOI] [PubMed] [Google Scholar]
- 45. MK L, Lowe L, Hummel KB, et al. . Characterization of Nipah virus from outbreaks in Bangladesh, 2008–2010. Emerg Infect Dis 2012;18:248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sejvar JJ, Hossain J, Saha SK, et al. . Long-term neurological and functional outcome in Nipah virus infection. Ann Neurol 2007;62:235–42. 10.1002/ana.21178 [DOI] [PubMed] [Google Scholar]
- 47. Tan CT, Goh KJ, Wong KT, et al. . Relapsed and late-onset Nipah encephalitis. Ann Neurol 2002;51:703–8. 10.1002/ana.10212 [DOI] [PubMed] [Google Scholar]
- 48. CDC , 2018. Biosafety in Microbiological and Biomedical Laboratories (BMBL) 5th Edition—CDC. https://www.cdc.gov/biosafety/publications/bmbl5/ (accessed 12 Feb 2018).
- 49. WHO , 2014. Infection prevention and control (IPC) guidance summary. http://www.who.int/csr/disease/ebola/evd-guidance-summary/en/ (accessed 1 Feb 2018).
- 50. Daniels P, Ksiazek T, Eaton BT. Laboratory diagnosis of Nipah and Hendra virus infections. Microbes Infect 2001;3:289–95. 10.1016/S1286-4579(01)01382-X [DOI] [PubMed] [Google Scholar]
- 51. Chong H-T, Kamarulzaman A, Tan C-T, et al. . Treatment of acute Nipah encephalitis with ribavirin. Ann Neurol 2001;49:810–3. 10.1002/ana.1062 [DOI] [PubMed] [Google Scholar]
- 52. Freiberg AN, Worthy MN, Lee B, et al. . Combined chloroquine and ribavirin treatment does not prevent death in a hamster model of Nipah and Hendra virus infection. J Gen Virol 2010;91:765–72. 10.1099/vir.0.017269-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Georges-Courbot MC, Contamin H, Faure C, et al. . Poly(I)-poly(C12U) but not ribavirin prevents death in a hamster model of Nipah virus infection. Antimicrob Agents Chemother 2006;50:1768–72. 10.1128/AAC.50.5.1768-1772.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Broder CC, Weir DL, Reid PA. Hendra virus and Nipah virus animal vaccines. Vaccine 2016;34:3525–34. 10.1016/j.vaccine.2016.03.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Satterfield BA, Dawes BE, Milligan GN. Status of vaccine research and development of vaccines for Nipah virus. Vaccine 2016;34:2971–5. 10.1016/j.vaccine.2015.12.075 [DOI] [PubMed] [Google Scholar]
- 56. Walpita P, Cong Y, Jahrling PB, et al. . A VLP-based vaccine provides complete protection against Nipah virus challenge following multiple-dose or single-dose vaccination schedules in a hamster model. NPJ Vaccines 2017;2:21 10.1038/s41541-017-0023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Broder CC, Xu K, Nikolov DB, et al. . A treatment for and vaccine against the deadly Hendra and Nipah viruses. Antiviral Res 2013;100:8–13. 10.1016/j.antiviral.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Middleton D, Pallister J, Klein R, et al. . Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg Infect Dis 2014;20:372–9. 10.3201/eid2003.131159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chan YP, Koh CL, Lam SK, et al. . Mapping of domains responsible for nucleocapsid protein–phosphoprotein interaction of Henipaviruses. J Gen Virol 2004;85(Pt 6):1675–84. 10.1099/vir.0.19752-0 [DOI] [PubMed] [Google Scholar]
- 60. Halpin K, Mungall BA. Recent progress in henipavirus research. Comp Immunol Microbiol Infect Dis 2007;30:287–307. 10.1016/j.cimid.2007.05.008 [DOI] [PubMed] [Google Scholar]
- 61. Arankalle VA, Bandyopadhyay BT, Ramdasi AY, et al. . Genomic characterization of Nipah virus, West Bengal, India. Emerg Infect Dis 2011;17:907–9. 10.3201/eid1705.100968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yadav PD, Raut CG, Shete AM, et al. . Detection of Nipah virus RNA in fruit bat (Pteropus giganteus) from India. Am J Trop Med Hyg 2012;87:576–8. 10.4269/ajtmh.2012.11-0416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chadha MS, Comer JA, Lowe L, et al. . Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 2006;12:235–40. 10.3201/eid1202.051247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Harcourt BH, Lowe L, Tamin A, et al. . Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg Infect Dis 2005;11:1594–7. 10.3201/eid1110.050513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. AbuBakar S, Chang LY, Ali AR, et al. . Isolation and molecular identification of Nipah virus from pigs. Emerg Infect Dis 2004;10:2228–30. 10.3201/eid1012.040452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kulkarni DD, Tosh C, Venkatesh G, et al. . Nipah virus infection: current scenario. Indian J Virol 2013;24:398–408. 10.1007/s13337-013-0171-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang LF, Daniels P. Diagnosis of henipavirus infection: current capabilities and future directions. Curr Top Microbiol Immunol 2012;359:179–96. 10.1007/82_2012_215 [DOI] [PubMed] [Google Scholar]
- 68. Baseler L, Scott DP, Saturday G, et al. . Identifying early target cells of Nipah virus infection in Syrian hamsters. PLoS Negl Trop Dis 2016;10:e0005120 10.1371/journal.pntd.0005120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cong Y, Lentz MR, Lara A, et al. . Loss in lung volume and changes in the immune response demonstrate disease progression in African green monkeys infected by small-particle aerosol and intratracheal exposure to Nipah virus. PLoS Negl Trop Dis 2017;11:e0005532 10.1371/journal.pntd.0005532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. World Organisation for Animal Health (OIE) , 2015. Manual of diagnostic tests and vaccines for terrestrial animals. Chapter 2114 Nipah Hendra Virus Dis. http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.01.14_NIPAH_HENDRA.pdf (accessed 25 Feb 2018).
- 71. Feldman KS, Foord A, Heine HG, et al. . Design and evaluation of consensus PCR assays for henipaviruses. J Virol Methods 2009;161:52–7. 10.1016/j.jviromet.2009.05.014 [DOI] [PubMed] [Google Scholar]
- 72. Reynes J-M, Counor D, Ong S, et al. . Nipah virus in Lyle's flying foxes, Cambodia. Emerg Infect Dis 2005;11:1042–7. 10.3201/eid1107.041350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Matranga CB, Gladden-Young A, Qu J, et al. . Unbiased deep sequencing of RNA viruses from clinical samples. J Vis Exp 2016;113 10.3791/54117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Matranga CB, Andersen KG, Winnicki S, et al. . Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol 2014;15:519 10.1186/s13059-014-0519-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Carroll MW, Matthews DA, Hiscox JA, et al. . Temporal and spatial analysis of the 2014–2015 Ebola virus outbreak in West Africa. Nature 2015;524:97–101. 10.1038/nature14594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ramasundrum V, Tan C, Chua K, et al. . Kinetics of IgM and IgG seroconversion in Nipah virus infection. Neurol J Southeast Asia 2014;5:23–8. [Google Scholar]
- 77. Masoomi Dezfooli S, Tan WS, Tey BT, et al. . Expression and purification of the matrix protein of Nipah virus in baculovirus insect cell system. Biotechnol Prog 2016;32:171–7. 10.1002/btpr.2192 [DOI] [PubMed] [Google Scholar]
- 78. Chiang C-F, Lo MK, Rota PA, et al. . Use of monoclonal antibodies against Hendra and Nipah viruses in an antigen capture ELISA. Virol J 2010;7:115 10.1186/1743-422X-7-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kulkarni DD, Venkatesh G, Tosh C, et al. . Development and evaluation of recombinant nucleocapsid protein based diagnostic ELISA for detection of Nipah virus infection in pigs. J Immunoassay Immunochem 2016;37:154–66. 10.1080/15321819.2015.1074922 [DOI] [PubMed] [Google Scholar]
- 80. Kaku Y, Noguchi A, Marsh GA, et al. . Antigen capture ELISA system for henipaviruses using polyclonal antibodies obtained by DNA immunization. Arch Virol 2012;157:1605–9. 10.1007/s00705-012-1338-3 [DOI] [PubMed] [Google Scholar]
- 81. Koczula KM, Gallotta A, assays Lflow. Lateral flow assays. Essays Biochem 2016;60:111–20. 10.1042/EBC20150012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen Y, Chan K-H, Hong C, et al. . A highly specific rapid antigen detection assay for on-site diagnosis of MERS. J Infect 2016;73:82–4. 10.1016/j.jinf.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tamin A, Harcourt BH, Lo MK, et al. . Development of a neutralization assay for Nipah virus using pseudotype particles. J Virol Methods 2009;160:1–6. 10.1016/j.jviromet.2009.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kaku Y, Noguchi A, Marsh GA, et al. . A neutralization test for specific detection of Nipah virus antibodies using pseudotyped vesicular stomatitis virus expressing green fluorescent protein. J Virol Methods 2009;160:7–13. 10.1016/j.jviromet.2009.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Khetawat D, Broder CC. A functional henipavirus envelope glycoprotein pseudotyped lentivirus assay system. Virol J 2010;7:312 10.1186/1743-422X-7-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pernet O, Schneider BS, Beaty SM, et al. . Evidence for henipavirus spillover into human populations in Africa. Nat Commun 2014;5 10.1038/ncomms6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Demby AH, Chamberlain J, Brown DW, et al. . Early diagnosis of Lassa fever by reverse transcription-PCR. J Clin Microbiol 1994;32:2898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Perkins MD, Kessel M. What Ebola tells us about outbreak diagnostic readiness. Nat Biotechnol 2015;33:464–9. 10.1038/nbt.3215 [DOI] [PubMed] [Google Scholar]
- 89. Foord AJ, White JR, Colling A, et al. . Microsphere suspension array assays for detection and differentiation of Hendra and Nipah viruses. Biomed Res Int 2013;2013:289295–8 10.1155/2013/289295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu J, Ochieng C, Wiersma S, et al. . Development of a TaqMan array card for acute-febrile-illness outbreak investigation and surveillance of emerging pathogens, including Ebola virus. J Clin Microbiol 2016;54:49–58. 10.1128/JCM.02257-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Onyango CO, Loparev V, Lidechi S, et al. . Evaluation of a TaqMan array card for detection of central nervous system infections. J Clin Microbiol 2017;55:2035–44. 10.1128/JCM.02469-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Southern TR, Racsa LD, Albariño CG, et al. . Comparison of film array and quantitative real-time reverse transcriptase PCR for detection of Zaire ebolavirus from contrived and clinical specimens. J Clin Microbiol 2015;53:2956–60. 10.1128/JCM.01317-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. BioFire Defense , 2018. BioFire Defense. FilmArray® Test Kits. http://biofiredefense.com/biosurveillance-systems/filmarray-test-kits/ (accessed 17 Sep 2019).
- 94. Bossart KN, McEachern JA, Hickey AC, et al. . Neutralization assays for differential henipavirus serology using Bio-Plex Protein Array Systems. J Virol Methods 2007;142:29–40. 10.1016/j.jviromet.2007.01.003 [DOI] [PubMed] [Google Scholar]
- 95. Breed AC, Yu M, Barr JA, et al. . Prevalence of henipavirus and rubulavirus antibodies in pteropid bats, Papua New Guinea. Emerg Infect Dis 2010;16:1997–9. 10.3201/eid1612.100879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hayman DTS, Wang L-F, Barr J, et al. . Antibodies to henipavirus or henipa-like viruses in domestic pigs in Ghana, West Africa. PLoS One 2011;6:e25256 10.1371/journal.pone.0025256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Peel AJ, Baker KS, Crameri G, et al. . Henipavirus neutralising antibodies in an isolated island population of African fruit bats. PLoS One 2012;7:e30346 10.1371/journal.pone.0030346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Peel AJ, McKinley TJ, Baker KS, et al. . Use of cross-reactive serological assays for detecting novel pathogens in wildlife: assessing an appropriate cutoff for henipavirus assays in African bats. J Virol Methods 2013;193:295–303. 10.1016/j.jviromet.2013.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Dups J, Middleton D, Yamada M, et al. . A new model for Hendra virus encephalitis in the mouse. PLoS One 2012;7:e40308 10.1371/journal.pone.0040308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Playford EG, McCall B, Smith G, et al. . Human Hendra virus encephalitis associated with equine outbreak, Australia, 2008. Emerg Infect Dis 2010;16:219–23. 10.3201/eid1602.090552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. McNabb L, Barr J, Crameri G, et al. . Henipavirus microsphere immuno-assays for detection of antibodies against Hendra virus. J Virol Methods 2014;200:22–8. 10.1016/j.jviromet.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Goel N, Ritchie AV, Mtapuri-Zinyowera S, et al. . Performance of the SAMBA I and II HIV-1 Semi-Q Tests for viral load monitoring at the point-of-care. J Virol Methods 2017;244:39–45. 10.1016/j.jviromet.2017.03.001 [DOI] [PubMed] [Google Scholar]
- 103. Jani IV, Meggi B, Vubil A, et al. . Evaluation of the whole-blood Alere Q NAT point-of-care RNA assay for HIV-1 viral load monitoring in a primary health care setting in Mozambique. J Clin Microbiol 2016;54:2104–8. 10.1128/JCM.00362-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Moyo S, Mohammed T, Wirth KE, et al. . Point-of-care Cepheid Xpert HIV-1 viral load test in rural African communities is feasible and reliable. J Clin Microbiol 2016;54:3050–5. 10.1128/JCM.01594-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Boisen ML, Cross RW, Hartnett JN, et al. . Field validation of the ReEBOV antigen rapid test for point-of-care diagnosis of ebola virus infection. J Infect Dis 2016;214(suppl 3):S203–S209. 10.1093/infdis/jiw261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Shaffer JG, Grant DS, Schieffelin JS, et al. . Lassa fever in post-conflict Sierra Leone. PLoS Negl Trop Dis 2014;8:e2748 10.1371/journal.pntd.0002748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shukla MK, Singh N, Sharma RK, et al. . Utility of dengue ns1 antigen rapid diagnostic test for use in difficult to reach areas and its comparison with dengue NS1 ELISA and qRT-PCR. J Med Virol 2017;89:1146–50. 10.1002/jmv.24764 [DOI] [PubMed] [Google Scholar]
- 108. Dhillon J, Banerjee A. Controlling Nipah virus encephalitis in Bangladesh: policy options. J Public Health Policy 2015;36:270–82. 10.1057/jphp.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nahar N, Asaduzzaman M, Sultana R, et al. . A large-scale behavior change intervention to prevent Nipah transmission in Bangladesh: components and costs. BMC Res Notes 2017;10:225 10.1186/s13104-017-2549-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Naser AM, Hossain MJ, Sazzad HM, et al. . Integrated cluster- and case-based surveillance for detecting stage III zoonotic pathogens: an example of Nipah virus surveillance in Bangladesh. Epidemiol Infect 2015;143:1922–30. 10.1017/S0950268814002635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Forum on Microbial Threats, Board on Global Health, Institute of Medicine , 2015. Emerging viral diseases: the one health connection: workshop Summary. Washington (DC) : National Academies Press (US). http://www.ncbi.nlm.nih.gov/books/NBK280057 (accessed 25 Feb 2018). [PubMed]
- 112. Guillaume V, Lefeuvre A, Faure C, et al. . Specific detection of Nipah virus using real-time RT-PCR (TaqMan). J Virol Methods 2004;120:229–37. 10.1016/j.jviromet.2004.05.018 [DOI] [PubMed] [Google Scholar]
- 113. Chang LY, Ali AR, Hassan SS, et al. . Quantitative estimation of Nipah virus replication kinetics in vitro. Virol J 2006;3:47 10.1186/1743-422X-3-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wacharapluesadee S, Hemachudha T. Duplex nested RT-PCR for detection of Nipah virus RNA from urine specimens of bats. J Virol Methods 2007;141:97–101. 10.1016/j.jviromet.2006.11.023 [DOI] [PubMed] [Google Scholar]
- 115. Tong S, Chern SW, Li Y, et al. . Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J Clin Microbiol 2008;46:2652–8. 10.1128/JCM.00192-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kashiwazaki Y, Na YN, Tanimura N, et al. . A solid-phase blocking ELISA for detection of antibodies to Nipah virus. J Virol Methods 2004;121:259–61. 10.1016/j.jviromet.2004.06.015 [DOI] [PubMed] [Google Scholar]
- 117. Eshaghi M, Tan WS, Mohidin TB, et al. . Nipah virus glycoprotein: production in baculovirus and application in diagnosis. Virus Res 2004;106:71–6. 10.1016/j.virusres.2004.06.011 [DOI] [PubMed] [Google Scholar]
- 118. Eshaghi M, Tan WS, Chin WK, et al. . Purification of the extra-cellular domain of Nipah virus glycoprotein produced in Escherichia coli and possible application in diagnosis. J Biotechnol 2005;116:221–6. 10.1016/j.jbiotec.2004.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Berhane Y, Berry JD, Ranadheera C, et al. . Production and characterization of monoclonal antibodies against binary ethylenimine inactivated Nipah virus. J Virol Methods 2006;132:59–68. 10.1016/j.jviromet.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 120. Chen JM, Yu M, Morrissy C, et al. . A comparative indirect ELISA for the detection of henipavirus antibodies based on a recombinant nucleocapsid protein expressed in Escherichia coli. J Virol Methods 2006;136:273–6. 10.1016/j.jviromet.2006.05.003 [DOI] [PubMed] [Google Scholar]
- 121. Yu F, Khairullah NS, Inoue S, et al. . Serodiagnosis using recombinant Nipah virus nucleocapsid protein expressed in Escherichia coli. J Clin Microbiol 2006;44:3134–8. 10.1128/JCM.00693-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chen JM, Yaiw KC, Yu M, et al. . Expression of truncated phosphoproteins of Nipah virus and Hendra virus in Escherichia coli for the differentiation of henipavirus infections. Biotechnol Lett 2007;29:871–5. 10.1007/s10529-007-9323-8 [DOI] [PubMed] [Google Scholar]
- 123. Joseph NM, Ho KL, Tey BT, et al. . Production of the virus-like particles of Nipah virus matrix protein in Pichia pastoris as diagnostic reagents. Biotechnol Prog 2016;32:1038–45. 10.1002/btpr.2279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjgh-2018-001118supp001.pdf (419.8KB, pdf)
bmjgh-2018-001118supp002.pdf (520.2KB, pdf)