Abstract
Structural maintenance of chromosome flexible domain containing 1 (Smchd1) is a chromatin regulatory gene for which mutations are associated with facioscapulohumeral muscular dystrophy and arhinia. The contribution of oocyte- and zygote-expressed SMCHD1 to early development was examined in mice (Mus musculus) using an siRNA knockdown approach. Smchd1 knockdown compromised long-term embryo viability, with reduced embryo nuclear volumes at the morula stage, reduced blastocyst cell number, formation and hatching, and reduced viability to term. RNAseq analysis of Smchd1 knockdown morulae revealed aberrant increases in expression of a small number of trophectoderm-related genes and reduced expression of cell proliferation genes, including S-phase kinase-associated protein 2 (Skp2). Smchd1 expression was elevated in embryos deficient for Caudal type homeobox transcription factor 2 (Cdx2, a key regulator of trophectoderm specification), indicating that Smchd1 is normally repressed by CDX2. These results indicate that Smchd1 plays an important role in the preimplantation embryo, regulating early gene expression and contributing to long-term embryo viability. These results extend the known functions of SMCHD1 to the preimplantation period and highlight important function for maternally expressed Smchd1 mRNA and protein.
Keywords: chromatin, nuclear programming, preimplantation embryo, pluripotency, epigenetic, transcription regulation
Introduction
Early mammalian embryos undergo extensive genome re-structuring as sperm and egg genomes are united and reprogrammed to totipotency. Changes in DNA methylation, histone modifications, and chromatin structure begin at fertilization and continue as embryos progress through cleavage to form blastocysts (Marcho et al. 2015). Correct activation and regulation of the embryonic genome and establishment of the early lineages are two essential processes that depend on correct chromatin remodeling and nuclear programming.
The mechanisms mediating this vast reprogramming are still being discovered. Maternal factors in the oocyte promote embryonic genome activation and expression of a 2-cell stage transiently expressed class of genes that contribute to early embryogenesis. Correct re-programming of the genome after this initial activation is crucial to development, including early cell lineage formation (Ko 2016). Lineage establishment begins early during cleavage with the formation of inner and outer cell populations, followed by differential cellular polarization and asymmetric cleavage. Major regulators (e.g., CHD1, CHD4, BRG1, SMYD3, HMGPI, POU5F1/OCT4, NANOG, CDX2, TEAD4, and HIPPO pathway genes) act sequentially to define early cellular identity (Carey et al. 2015; Cui and Mager 2018; Nishioka et al. 2009; O’Shaughnessy-Kirwan et al. 2015; Ralston et al. 2010; Sasaki 2015; Suzuki and Minami 2018; Suzuki et al. 2015; Wang et al. 2010; Yagi et al. 2007). Moreover, gene products that contribute to developmental reprogramming (e.g., INO80, SMARCA5) are encoded by maternally expressed mRNA in the oocyte (Miller and Hendrich 2018), as further examples of the importance of oocyte- and early embryo-expressed factors in long-term developmental programming.
Structural maintenance of chromosome flexible domain containing 1 (Smchd1) is a chromatin regulator that contributes to gene regulation throughout life. SMCHD1 possesses an N-terminal ATPase domain, a large central domain and a C-terminal SMC hinge domain. SMCHD1 functions in part as a transcriptional repressor that promotes DNA methylation of the X-chromosome and of autosomal loci, including imprinting control elements within imprinted domains and gene clusters such as the proto-cadherin cluster (Jansz et al. 2017). SMCHD1 heterodimerizes via its ATPase and hinge domains and with kleisin (e.g., cohesion) subunits forms a ring structure for binding to chromatin, and additionally interacts with other transcription regulators and chromatin proteins to bind to H3K9me3 and long noncoding RNAs to bind at sites of repression (Jansz et al. 2017). SMCHD1 binding sites overlap with CTCF binding sites indicating a role in controlling chromatin structure (Jansz et al. 2017). In most instances SMCHD1 deficiency leads to gene de-repression.
Defects in SMCHD1 contribute to facioscapulohumeral muscular dystrophy and congenital arhinia-microphthalmia, and cancer (Gordon et al. 2017; Jansz et al. 2017; Leong et al. 2013; Shaw et al. 2017). The first functions ascribed to SMCHD1 were in variegating transgene expression, its role as an epigenetic modifier, and then its role in X chromosome DNA methylation (Ashe et al. 2008; Blewitt et al. 2008; Gendrel et al. 2012; Gendrel et al. 2013). More recent studies found SMCHD1 also regulates many autosomal genes, imprinted gene clusters, and monoallelically expressed genes (Chen et al. 2015; Liu et al. 2016; Mason et al. 2017; Mould et al. 2013). Human mutations in SMCHD1 play a key role in facioscapulohumeral muscular dystrophy (FSHD) by releasing DUX4 gene repression and disrupting a muscle forming gene in cis, and also contribute to craniofacial abnormalities (Jansz et al. 2017; Lemmers et al. 2012) and cancer (Leong et al. 2013). As in mice, human SMCHD1 displays haploinsufficiency and variegating effects.
We report here that SMCHD1 has additional key roles in early embryos. Smchd1 is expressed both maternally in the oocyte, and embryonically after fertilization, siRNA knockdown reveals that reduction in both maternal and embryonic sources of SMCHD1 reduces blastocyst formation, hatching, embryo cell number, and term development. Transcriptome analysis reveals effects of siRNA knockdown on the expression of lineage-related genes and cell proliferation genes that support cell cycle progression and cellular specialization during preimplantation development. By eliminating both maternal and embryonic sources of SMCHD1, our studies reveal, for the first time, important roles for SMCHD1 in maintaining embryo quality and viability, with long-term effects of its preimplantation actions on term development. Additionally, we show that Cdx2 deficiency leads to increased Smchd1 expression consistent with differential regulation between inner cell mass and trophectoderm lineages. These results extend our understanding of early embryonic nuclear programming mechanisms and the potential consequences of mutations in a gene that is linked to health disorders in humans.
Materials and Methods
Embryo isolation, culture and transfer
All animal use was in accordance with guidelines of the Michigan State University Institutional Animal Care and Use Committee. Mice were maintained on 12 h light/dark cycles. For most studies, oocytes and embryos were obtained by superovulating 8-16 week old female mice (C57BL/6 × DBA/2)F1 (B6D2F1), The Jackson Laboratory, Bar Harbor, ME)], unmated or mated to B6D2F1 males. Oocytes were isolated at second meiotic metaphase (MII), and embryos were isolated at the 1-cell stage in M2 medium (Hogan et al., 1994) (Millipore, Billerica, MA). Cumulus cells were removed using hyaluronidase (Sigma-Aldrich, St. Louis, MO) as described (Chung et al. 2003). Embryos were cultured in KSOM medium (Erbach et al. 1994) in humidified atmospheres containing 5% O2 and 5% CO2 at 37°C.
The Cdx2tm1Fbe null allele (Chawengsaksophak et al. 1997) was maintained in an outbred (CD1) background. Mice were genotyped from ear punches by PCR (primers (5’-3’): ATATTGCTGAAGAGCTTGGCGGC, AGGGACTATTCAAACTACAGGAG and TAAAAGTCAACTGTGTTCGGATCC). Mice were maintained on 12 h light/dark cycles. ED3.5 and ED4.25 blastocysts were flushed from uteri with M2 medium after timed natural matings. We note that blastocysts lacking Cdx2 undergo pulsatile, failed attempts to expand their blastocoel, owing to loss of trophectoderm epithelial integrity (Strumpf et al. 2005), and hence morphological assessment of embryo quality was not feasible. All embryos were processed as described below.
For testing developmental potential, 10 to 20 embryos of either treatment group were transferred surgically at the 2-cell stage to each recipient as indicated, with embryos divided equally between the two uterine horns. The numbers of scrambled control and knockdown embryos transferred were kept the same in each experimental replicate as indicated. Recipients were euthanized 7 or 18 days after embryo transfer. The numbers of embryos and pups obtained from the two uteri were recorded for each treatment. For term development assessment, Caesarian section was performed 18.5 days after embryo transfer. For assessment of viability at ED8.5, uteri were dissected and implantation sites counted.
siRNA knockdown
For cytoplasmic microinjection, zygotes received approximately 10 pl of 100 μM Smchd1 siRNA or control scrambled siRNA. In some cases, a piezo pipette driver was used. siRNAs were obtained from GE Dharmacon (L-040501-01-0005, Lafayette, CO). The Smchd1 siRNA was a mixture of four oligonucleotides: GAAAUGAACUAAAAGCGU, GAAGGAGAAGGACGAGUUA, CUGUGAAAGAUGUCCGCUA, and CUAACAAGGUGGGAGCAUA. Confirmation of knockdown of Smchd1 mRNA and/or protein expression was obtained for each experiment by qRT-PCR (see below) or by immunofluorescence confocal microscopy (IFCM). Bioinformatics analysis revealed no other perfectly complementary mRNA targets of these siRNAs, and RNAseq results indicated no significant off-target effects on other mRNAs having partial sequence match (data not shown).
Antibodies and Immunofluorescence confocal microscopy
SMCDH1 expression was monitored using Rabbit-α-SMCHD1 polyclonal antibody from Abcam (ab122555; Cambridge, MA) diluted 1:100 for IFCM. The secondary antibody for detecting SMCHD1 was donkey-α-rabbit IgG Alexa Fluor 594 from Abcam (ab150076) diluted 1:300. Western blot analysis of embryonic stem cell (ESC) and fibroblast lysates confirmed reaction to a single band in ESCs of the expected size; a very minor secondary band was faintly seen in fibroblasts (Figure 1A). Antibodies for POU5F1/OCT4 and CDX2 were from Abcam (ab19857, 1:200 dilution) and BioGenex (AM392-5M, 1:20 dilution, San Ramone, CA) respectively, and detected using goat-α-mouse 488 (1:2000) and Donkey-α-Rabbit (1:2000) secondary antibodies from Life Technologies/Thermo Fisher (A11001, Carlsbad, CA) and Abcam (ab150076), respectively. Antibody against phospho- H2AX (Histone 2A family member X), conjugated with Fluorescein isothiocyanate (FITC), was from Millipore (16-202A) and was diluted 1:100. Embryos were fixed in 3.7% paraformaldehyde for 30 min, permeabilized with 0.5% Triton X-100 in phosphate buffered saline (PBS) for 30 min, and blocked in 2% bovine serum albumin for 45 min. Embryos were imaged using an Olympus FluoView 1000 Filter-based CLSM confocal microscope with a 60x oil immersion objective. Optical sections were collected at approximately 3 μm intervals and, where needed, compiled into Z-stacks. ICM cells were distinguished from TE cells in late blastocyst embryos and both cell populations quantified by staining embryos with antibodies to POU5F1 and CDX2. ICM cells were those that lacked CDX2 signals and displayed intense nuclear POU5F1 signals (average ~ 3-fold more intense than CDX2-positive TE cells), whereas TE cells were those with intense nuclear CDX2 signals. Significance of differences was determined by two-tailed t-test assuming equal variance.
Figure 1.
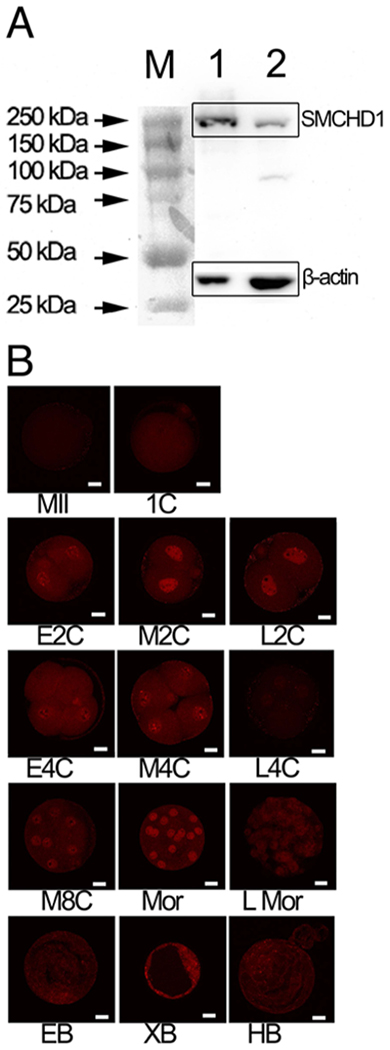
Detection of SMCHD1. A. Western blot testing of SMCHD1 antibody specificity. Blot was reacted withantibodies to mouse SMCHD1 and Actin. M, protein ladder; 1, embryo stem cells; 2, mouse embryonic fibroblasts. B. Expression of SMCHD1 in preimplantation embryos. Immunofluorescence confocal microscopy analysis of SMCHD1 expression during preimplantation development. Results were replicated across 2 independent collections for each stage. Embryos were fixed at the following times (hours post-hCG injection) during in vitro culture: 1C (n=16, 24 h), E2C (n=6, 32 h), M2C (n=8, 41 h), L2C (n=7, 44 h), E4C (n=16, 51 h), M4C (n=12, 57 h), L4C (n=21, 67 h), M8C (n=10, 76 h), LMor (n=6, 92 h), EB (n=10, 92 h), XB (n=7, 116 h), , and LB (n=6, 120 h). Note the periodic enrichment of SMCHD1 in nuclei at mid 2-cell (M2C), 4-cell (4C), 8-cell (8C) and morula (Mor) stages. E = early, M = mid, L = late. Mor = morula, EB = early blastocyst, XB = expanded blastocyst, HB = hatched blastocyst. Bar = 20 μm.
Western Blots
Mouse embryonic stem (ES) cell and mouse embryonic fibroblast cell cultures were lysed in RIPA buffer. Protein concentrations were measured using a Bradford assay, and lysates diluted to an equal concentration. Cell lysates were mixed 1:1 with Laemmli buffer, boiled for 10 min then immediately run on an 8% polyacrylamide gel with a 4% stacking layer for 50 min at 200V. Immobilon-P membrane (EMD Millipore) was activated by placing it in a sequence of 100% methanol (30 sec), deionized water (2 min), and then transfer buffer (20 min). Electroblotting was completed by submerging the transfer assembly in ice during transfer (1 h, 95V). Membranes were blocked for 1.5 h in 5% nonfat dry milk in transfer buffer (25 mM Tris, 192 mM Glycine). Antibodies (SMCHD1; Abcam ab122555, 1:1000 dilution; Actin Sigma A5441, 1:10,000 dilution) were incubated on the membrane overnight in the same solution. Chemiluminescent detection of proteins was completed using the SuperSignal mouse Immunoglobulin G (IgG) detection kit from Thermo (34081, Waltham, MA) and Imaged on a myECL (Thermo) imager following manufacturer instructions.
Response to ultraviolet light
SMCHD1 knockdown embryos and control embryos were exposed to UV-C radiation. In brief, 8-cell embryos 70 h post-hCG were exposed to UV-C radiation (50 J m−2) using a UV-Cross linker (SX-1000, Spectronic Corporation, Westbury, NY) in droplets of M2 medium. Embryos were transferred to KSOM medium for 1 h and then fixed in 3.7% paraformaldehyde for 30 min. Immunofluorescence detection was performed for phosphorylated histone H2AX (antibody 16-202A, Millipore, FITC conjugated). Embryos were imaged on an Olympus IX-71 microscope with a Retiga-6000 camera system (QImaging, Surrey, British Columbia, Canada). Image analysis was performed using NIS-Elements AR3.1 software, relative intensity units of nuclear fluorescence minus background fluorescence determined, and differences evaluated by two-tailed t-test assuming equal variance.
RNAseq transcriptome analysis
RNA was extracted from pools of 35 morulae (93 h post-hCG injection) that had been injected with either scrambled control or Smchd1 siRNA (5 pools each). Each pool of embryos was an independent biological replicate. RNA was extracted using the PicoPure RNA Isolation kit (Life Technologies, Grand Island, NY) with DNAse digestion to remove any contaminating DNA. Aliquots of approximately half of each sample were processed through the Ovation RNA-Seq System v2 using the Ribo-SPIA™ Technology (Nugen, San Carlos, CA), followed by fragmentation to 300 bp using a Covaris sonicator, and a brief S1 nuclease digestion as described (Head et al., 2011). After purification and end repair, cDNA was processed through the Ovation Ultralow DR Multiplex Systems 1-8 and 9-16 for end repair, barcoding and final library production.
Barcoded libraries were pooled, loaded on flow cells and sequenced with Illumina HiSeq 2500 in rapid run mode to generate 50 nucleotide single-end reads. To enhance effectiveness of cluster identification, samples were loaded at 65% of optimal loading concentration, along with PhiX Control library (Illumina, San Diego, CA) – adapter-ligated library obtained from randomly sheared PhiX DNA – added at 10% of loading concentration to increase read sequence complexity. The total numbers of PF (passed-filter) reads was 37.3 to 38.5 million (Table 1). Fraction of Q30 bases was 91% to 93% and average Q was 36.2 to 36.9.
Table 1.
Sequencing Quality Control and Alignment Statistics
| Treatment | Bio. Repl. # | Sequencing output (M of PF reads) | % of bases Q≥30 | Avg Q | Aligned to GRCm38 genome (M of reads) | Aligned to unique genes (M of reads) |
|---|---|---|---|---|---|---|
| scrambled siRNA | 1 | 38.5 | 93.0 | 36.93 | 33.8 | 14.6 |
| 2 | 37.9 | 91.7 | 36.40 | 32.0 | 13.8 | |
| 3 | 37.8 | 92.3 | 36.65 | 31.9 | 14.7 | |
| 4 | 38.4 | 92.1 | 36.57 | 32.7 | 13.7 | |
| 5 | 37.4 | 91.1 | 36.20 | 32.3 | 12.5 | |
| Smchd1 KD siRNA | 1 | 37.3 | 91.6 | 36.38 | 32.2 | 12.9 |
| 2 | 37.3 | 91.0 | 36.17 | 30.5 | 13.2 | |
| 3 | 37.5 | 91.2 | 36.22 | 32.0 | 13.3 | |
| 4 | 37.6 | 92.1 | 36.56 | 31.0 | 13.4 | |
| 5 | 37.8 | 91.7 | 36.38 | 31.7 | 12.7 |
Reads were aligned using TopHat2 (Kim et al. 2013) to mouse genome assembly GRCm38.p4. Reads aligned to ribosomal and transfer RNA (rRNA, tRNA) were removed. A total of 12.5M to 14.7M reads were aligned to unique non-rRNA/non-tRNA gene transcript sequences. Cuffdiff (Trapnell et al. 2013) was used for quantification and differential expression analyses; genes with q-value (false discovery rate) below 0.05 were considered differentially expressed (DE) genes (DEGs).
qRT-PCR
Embryos were stripped of zonae pellucidae using acidified Tyrode’s buffer and then washed in M2 medium. Embryos were counted, divided into groups, and then transferred clean microcentrifuge tubes where they were counted again. Embryos were extracted with 40 μl of PicoPure Extraction Buffer and incubated for 30 min at 42°C.
For some studies, RNA was isolated using the Arcturus (Life Technologies) PicoPure RNA Isolation kit and reverse transcribed into cDNA using Quanta qScript cDNA Supermix (QuantaBio, Beverly, MA). Master mixes for each target were assembled using 5 μl of Taqman Universal qPCR mix and 0.5 μl of probe per well. Sample master mixes were prepared with a 1:5 dilution with nuclease free MilliQ water and samples were loaded onto a 384-well plate using an Eppendorf (Hauppauge, NY) epMotion M5073 liquid handler in 5 μl quantities. qRT-PCR was performed on a QuantStudio™ 7 Flex Real-Time PCR System (Life Technologies/Applied Biosystems) with standard Taqman chemistry. Primers and their efficiencies are described (Table 2). Data were preprocessed (baseline subtraction, threshold identification, outlier removal) and analyzed (relative gene expression ΔΔCt approach) with QuantStudio™ 6 & 7 Flex Real-Time PCR System software. Relative Quantity (RQ) values for control and SMCHD1 KD samples were compared using unpaired two-tailed t-test assuming equal variance to identify differentially expressed genes, and Benjamini-Hochberg procedure (Benjamini Y 1995) was applied due to multiple testing. The endogenous standard used was either Upstream binding transcription factor (Ubtf) or Ribosomal protein L18 (Rpl18). Ubtf was employed in initial studies to assess success of mRNA depletion with Smchd1 siRNA knockdown. Rpl18 was selected for later studies at the morula stage examining effects of siRNA knockdown on the expression of mRNAs identified as affected in the RNAseq analysis, as RNAseq data revealed it to be highly constant in expression level with minimal variance at that stage.
Table 2.
Primers for qPCR
| Target | Assay | Sequence/Catalog # | Primer Efficiency (%) |
|---|---|---|---|
| Arid3a | Taqman | Mm00492248_m1 | 96.6 |
| β-Actin | Syber Green | F-CTGAACCCTAAGGCCAACC R-CCAGAGGCATACAGGGACAG |
|
| Cdx2 | Syber Green | F-AGACAAATACCGGGTGGTGTA R-CCAGCTCACTTTTCCTCCTGA |
|
| Dppa1 | Taqman | Mm00626454_m1 | 92.9 |
| Eomes | Syber Green | F-TGTGACGGCCTACCAAAACA R-GCCGTGTACATGGAATCGTAG |
|
| Igf2r | Taqman | Mm00439576_m1 | 96.5 |
| Krt8 | Syber Green | F-CTTCATTGACAAGGTGCGCTT R-TTGCTCCTCGACGTCTTCTG |
|
| Nt5c3b | Taqman | Mm00518281_m1 | 92.4 |
| Peg10 | Taqman | Mm01167724_m1 | 100 |
| Rpl18 | Taqman | Mm02745785_g1 | 100 |
| Skp2 | Taqman | Mm00449925_m1 | 97.6 |
| Smchd1 | Taqman | Mm00512253_m1 | 89.3 |
| Smchd1 | Syber Green | F-TACTGACGGCGACGAAAGAAA R-CTCTGCAAGAGCAAATGGCAAA |
|
| Ubtf | Taqman | Mm00456972_m1 | 93.9 |
Other studies used embryos from Cdx2+/− inter se matings to test for effects of Cdx2 mutation on Smchd1 expression. Individual embryos were transferred to 20 μl of PicoPure Extraction Buffer (Arcturus Biosciences), and RNA isolation was performed according to the manufacturers protocol. Single-blastocyst qRT-PCR was performed to quantify Caudal-type homeobox transcription factor 2 (Cdx2), Eomesodermin (Eomes), Smchd1 and keratin 8 (Krt8) mRNAs as described (Blij et al., 2012; Ralston and Rossant, 2008; Strumpf et al., 2005). Cdx2−/− genotype was inferred based on lack of detectable Cdx2, and low expression of Eomes, and Krt8. Wild type and Cdx2−/+ genotypes were inferred by plotting expression values for Cdx2 versus the expression of the positively regulated CDX2 target genes, Eomes and Krt8. Embryos with high levels of Cdx2, Eomes and Krt8, were designated as wild type, and those with moderate Cdx2, Eomes and Krt8 expression were designated as Cdx2−/+. For analysis of Smchd1 knockdown effects, Cdx2+/+ and Cdx2+/− embryos were grouped together for comparison to Cdx2−/− embryos. Primers employed are described (Table 2). One-way ANOVA was performed to assess significance of differences between genotypes using GraphPad Prism version 7.0a for Mac (GraphPad Software, La Jolla CA).
Ingenuity Pathway Analysis
Differentially expressed genes (DEGs) identified by RNAseq analysis (q≤0.05) were uploaded into QIAGEN Ingenuity Pathway Analysis® (IPA, Redwood City, CA) and subjected to Core Analysis, in particular Canonical Pathway (CP) analysis, Disease and Functions (D/F) analysis, Upstream Regulator (UR) analysis, and Network analysis. For CP, D/F and UR analysis, IPA calculates overlap p-values, taking into account the number of DEGs and the number of all molecules in knowledge database that are implicated in that pathway (implicated in increase or decrease of disease/function; regulated by upstream regulator), as well as the total number of DEGs and the total number of molecules in knowledge database. In addition to overlap p-values, z-scores are calculated for CPs, D/Fs, and URs, which are based on how many DEG’s direction of change (upregulation or downregulation) is consistent with activation (z>0) or inhibition (z<0) of CPs and URs, and with increase (z>0) or decrease (z<0) of D/Fs. Since P(|z|>1.96)~0.05 for normal N(0,1) distribution, we consider CPs, URs and D/Fs with z>1.96 to be significantly activated or increased, and those with z<−1.96 to be significantly inhibited or decreased. Additionally, IPA uses a greedy algorithm to construct networks that incorporate DEGs – with some additional genes or other molecules where needed – in an attempt to reproduce possible mechanistic networks.
Data availability
RNAseq data from this study are available in the Gene Expression Omnibus (GEO accession number GSE92936) at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92936.
Results
Smchd1 protein and mRNA expression in oocytes and early embryos
Our first step in assessing the role of SMCHD1 in early embryogenesis was to evaluate temporal patterns of SMCHD1 expression by IFCM (Figure 1B). We observed periodicity in the intensity of nuclear SMCHD1 expression (Figure 1B), with peaks in mid 2-cell, 4-cell, and morula stages. Our previous study revealed Smchd1 mRNA expression mouse MII stage oocytes (Cheng et al. 2013). Microarray data for mouse oocytes and embryos (Zeng et al. 2004a; Zeng et al. 2004b) also revealed Smchd1 mRNA expression in germinal vesicle (GV) stage oocytes and at all stages of preimplantation development. Other published transcriptome data indicate a two-fold higher level of Smchd1 mRNA expression in ICM cells compared to whole blastocysts (Aksoy et al. 2013a; Aksoy et al. 2013b). The high level of expression in oocytes, comparable to expression throughout cleavage, indicates that maternal oocyte-mediated Smchd1 expression is significant, and may contribute to chromatin regulation during early cleavage stages.
Effects of siRNA knockdown on preimplantation development
We next assessed the impact of Smchd1 deficiency on preimplantation development by siRNA injection into fertilized zygotes followed by embryo culture. Because of the substantial contribution of oocyte-derived Smchd1 mRNA to expression in zygotes and early cleaving embryos, an siRNA knockdown approach was chosen, for its ability to reduce both maternal and embryonically expressed Smchd1 expression. Quantitative reverse transcription and polymerase chain reaction (qRT-PCR) and IFCM revealed 75-85% reduction in Smchd1 mRNA at the 4-cell, 8-cell and morula stages (Figure. 2A), and 63% reduction in SMCHD1 protein for the morula stage (Figure 2B), compared to controls that received the scrambled siRNA. In Smchd1 knockdown embryos, Smchd1 mRNA (Figure 2A) and protein (not shown) expression began to recover after the morula stage, consistent with the later burst of expression seen in normal embryos (Figure 1B).
Figure 2.
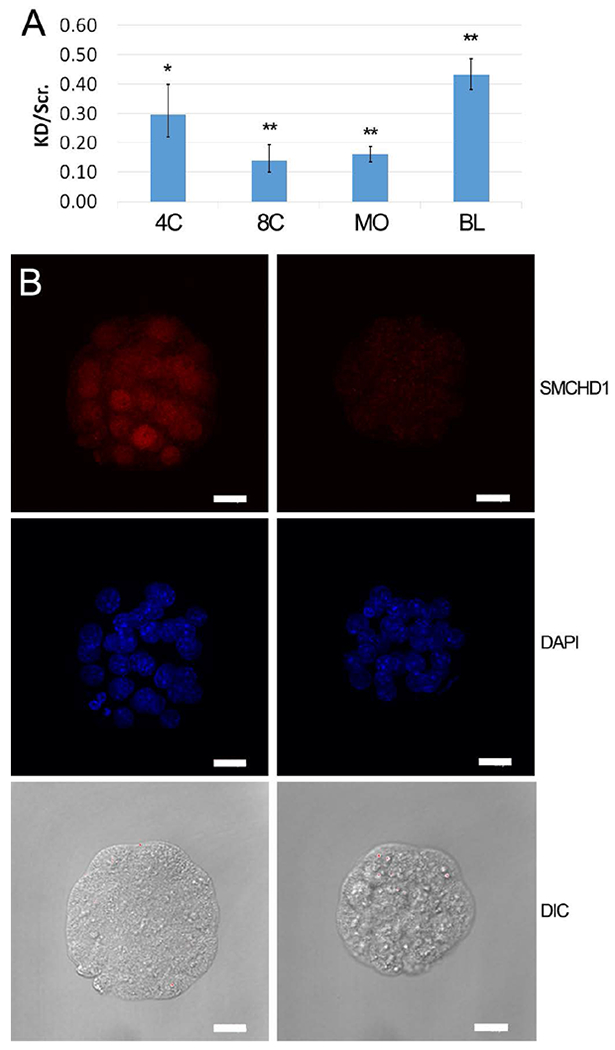
Confirmation of siRNA knockdown of SMCHD1. A: qRT-PCR analysis of Smchd1 mRNA fold expression ratio following zygotic microinjection of Smchd1 siRNA versus scrambled control siRNA. Data are the mean of the ratio (± s.e.m.) of knockdown: scrambled control injected embryos, calculated from 3 independent trials of 8-cell (n=20, 72 hphCG) and blastocyst (n=20,108 hphCG) stage embryos, and 5 independent trials of 4-cell (n=20, 66 hphCG) and morula (n=20, 92 hphCG) stage embryos. Statistical significance of difference between Smchd1 and scrambled control siRNA injected embryos was tested by two-tailed paired t-tests assuming equal variance and p-values were adjusted for multiple testing; (* indicates p <0.05; ** indicates p < 0.005). Relative expression values (RQs) were calculated using Ubtf as the internal reference gene. B: Immunofluorescence detection of SMCHD1 in morula stage embryos (92 hphCG) following zygotic microinjection of scrambled control and Smchd1 siRNA. Left panels, scrambled siRNA injection controls. Right panel, Smchd1 siRNA injected embryos. Representative images of embryos are shown. Knockdown was confirmed in seven independent replicate studies. Top row, SMCHD1 localization; Middle row, DAPI DNA staining; Bottom row, Differential interference microscopy images; Note also the smaller diameter, reduced nuclear dimensions, and cytoplasmic irregularities in the knockdown embryos (right) compared to scrambled siRNA controls (left). Note smaller embryo diameters and smaller nuclei in knockdown embryos (Table 4). Bar = 20 μm.
Smchd1 siRNA injection led to a significant increase in the number of morula arresting before the blastocyst stage (p < 0.05) and to a significant reduction in the number of blastocysts hatching by about one third (p < 0.01), compared to scrambled siRNA controls (Table 3). Examination of DAPI stained nuclei in morula stage embryos indicated possible reduction in nuclear volumes with siRNA knockdown (Figure 2B). Quantitative measurements of nuclear volumes in morulae confirmed a significant reduction in average nuclear volume at the morula stage (Table 4). Blastocyst total cell number was reduced by 12% (p < 0.02) (Table 5). The number of ICM cells in Smchd1 knockdown late stage blastocysts was reduced by 17% compared to control blastocysts (p < 0.001). The minor reduction in TE cell number was not statistically significant (p = 0.104) and there was no significant change in overall ICM:TE ratio (Table 5).
Table 3.
Effect of Smchd1 KD on preimplantation development
| siRNA | No. expts | Arrested (% of TOTAL) | Blastocysts (% of TOTAL) | Hatched (% of TOTAL) | Not hatched (% of TOTAL) | TOTAL |
|---|---|---|---|---|---|---|
| Scrambled | 8 | 13 (7) | 164 (93) | 94 (53) | 70 (40) | 177 |
| Smchd1 | 8 | 27 (15)* | 150 (85) | 61 (34) | 89 (50)** | 177 |
p < 0.05
p < 0.01 by chi-squared test
Table 4.
Effect of Smchd1 KD on morula nuclear diameter
| siRNA | no. expts | Nuclear Vol. (fL) avg (s.d.) | No. Nuclei | Nuclei/Morula avg |
|---|---|---|---|---|
| scrambled | 2 | 498 (105) | 48 | 29.6 |
| Smchd1 | 2 | 257 (24)** | 42 | 23.9 |
p < 0.0001
p < 0.001 by two tailed t-test assuming equal variance
Table 5.
Effect of Smchd1 KD on blastocyst cell number and allocation
| siRNA | ICM | TE | TOTAL | ICM:Total | ICM:TE | n | |
|---|---|---|---|---|---|---|---|
| no. expts | avg (s.d.) | avg (s.d.) | avg (s.d.) | avg (s.d.) | avg (s.d.) | ||
| Scrambled | 7 | 24(3.9) | 67(17) | 91(18) | 0.27(0.06) | 0.35(0.15) | 23 |
| Smchd1 | 7 | 20(2.9)** | 60(11) | 80(12)* | 0.25(0.03) | 0.33(0.07) | 23 |
p < 0.02
p < 0.001 by two-tailed t-test assuming equal variance
Term development effects
Because homozygous mutant Smchd1 females are not viable, and because Smchd1 is expressed in the oocyte, previous studies have not revealed whether early Smchd1 deficiency impairs embryo viability to term. Zygotes injected with scrambled control or Smchd1 siRNA were transferred to pseudopregnant foster mothers at the 2-cell stage (Table 6). Viability to term after Smchd1 knockdown was reduced from 76% to 47% of the embryos transferred (p < 0.005). There was no apparent deficiency in females (10 of 18) among knockdown pups that survived to 3 weeks of age. The reduction in viability of Smchd1 knockdown embryos to ED8.5 was less severe (88 vs. 72 %, p < 0.005). These observations indicate that Smchd1 expressed during preimplantation development is important for development to term.
Table 6.
Effect of Smchd1 KD on post-implantation development
| Term/2cell (%) | No. Expts (embryo transferred per side) | d 8.5/2cell (%) | No. Expts (embryo transferred per side) | |
|---|---|---|---|---|
| Control | 34/45 (76) | 3 (7/8,7/8,7/8) | 44/50 (88) | 3 (10,10,5) |
| siRNA | 21/45 (47)** | 3 (7/8,7/8,7/8) | 36/50 (72)* | 3 (10,10,5) |
p <0.05
p < 0.005 by Chi squared test
Response to ultraviolet light exposure
Because Smchd1 contributes to DNA repair (Coker and Brockdorff 2014) we tested whether a reduced DNA damage response could contribute to phenotypic abnormalities described above. DNA damage was assessed in embryos by IFCM for phosphorylated H2AX, which reveals DNA damage in cleaving mouse embryos, e.g., following zygotic H2O2 exposure (Qian et al. 2016). Zygotes were injected with Smchd1 or scrambled control siRNA and then cultured to the 8-cell stage, exposed to UV-C irradiation, and then fixed after 1 h of further culture. We observed no reduction in response to UV-C exposure, as measured by phospho-H2AX labeling of nuclei, with Smchd1 knockdown (Figure 3). Thus, a major deficiency of DNA damage response does not appear to be a significant factor in the effects of Smchd1 siRNA knockdown.
Figure 3.
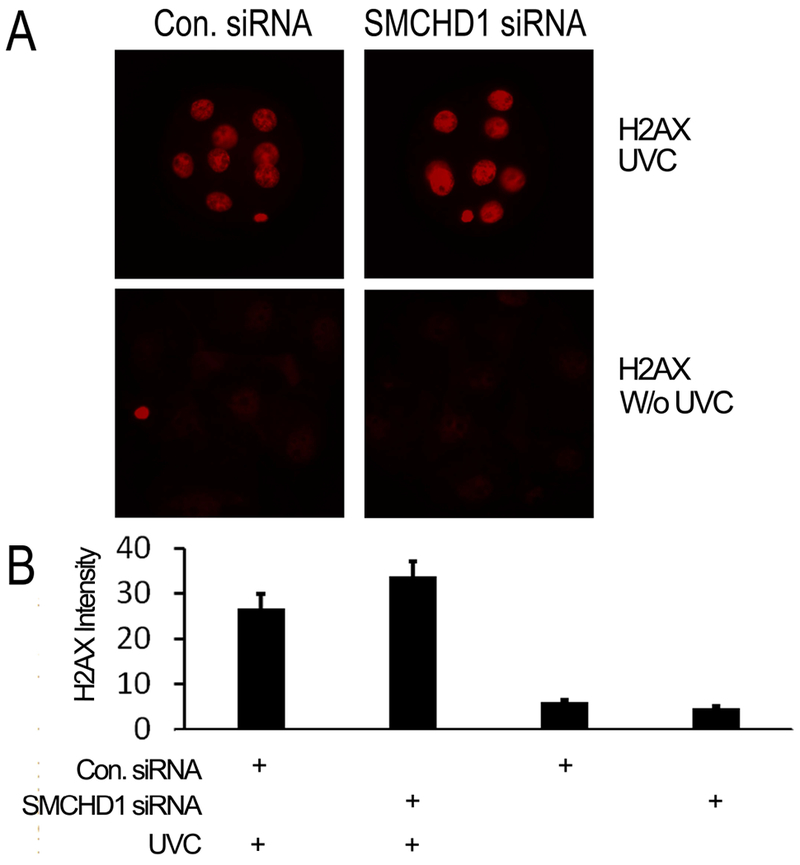
Smchd1 knockdown does not affect early embryonic DNA damage response. A: Representative images showing immunofluorescence confocal detection of histone H2AX in embryos with and without prior UV-C treatment. B. Quantitation of nuclear staining for histone H2AX in embryos with (9 control and 8 Smchd1 knockdown embryos) and without (3 control and 3 Smchd1 knockdown embryos) prior UV-C treatment. Bars show the mean (± s.e.m.) of relative intensity units. Mean values between scrambled control and Smchd1 siRNA knockdown embryos were not significantly different. UV-C treated control and Smchd1 knockdown embryos were significantly different from untreated embryos (p< 0.01 and p < 0.001, respectively). Embryos were imaged on an Olympus IX-71 microscope equipped with a Retiga-6000 camera system (Qimaging, Surrey, British Columbia, Canada). Image analysis was performed using NIS-Elements AR3.1 software and differences evaluated by two-tailed t-test assuming equal variance.
Effects of Smchd1 siRNA knockdown on embryonic gene expression
The results above revealed a significant effect of Smchd1 siRNA knockdown on morula and blastocyst development, associated with long-term effects on term development. Given that SMCHD1 regulates a large number of genes in other contexts, Smchd1 siRNA knockdown could affect a large number of genes in preimplantation embryos. To determine effects of siRNA knockdown on embryonic gene expression, we performed RNAseq comparisons of knockdown and control embryos (5 independent pools of each, 35 embryos per pool) at the morula stage, when knockdown is maximal. A total of 88 genes displayed significant (q < 0.05) differential expression. Of these, 25 exhibited 1.5-fold or greater increase in expression (4 at the level of 2-fold or greater), and 19 1.5-fold or greater decreases in expression (6 at the level of 2-fold or greater) (Table 7).
Table 7.
Genes differentially expressed in Smchd1 siRNA knockdown embryos
| Upregulated genes (fold-change ≥ 1.5) | |||
|---|---|---|---|
| Gene Symbol | Gene Name | q-value | Fold-Change KD/scrambled |
| Magea5 | melanoma antigen, family A, 5 | 1.22E-02 | 3.47 |
| Wfdc2 | WAP four-disulfide core domain 2 | 3.44E-02 | 2.52 |
| Prss42 | protease, serine 42 | 2.92E-02 | 2.50 |
| Dppa1 | developmental pluripotency associated 1 | 1.22E-02 | 2.07 |
| Spata13 | spermatogenesis associated 13 | 1.22E-02 | 1.98 |
| Peg10 | paternally expressed 10 | 2.92E-02 | 1.93 |
| Mbp | myelin basic protein | 1.22E-02 | 1.91 |
| Npm3-ps1 | nucleoplasmin 3, pseudogene 1 | 4.58E-02 | 1.86 |
| Tfap2a | transcription factor AP-2, alpha | 1.22E-02 | 1.76 |
| Ripply3 | ripply transcriptional repressor 3 | 3.44E-02 | 1.74 |
| Ank | progressive ankylosis | 1.22E-02 | 1.64 |
| Lcmt2 | leucine carboxyl methyltransferase 2 | 1.22E-02 | 1.64 |
| Ppp1r16b | protein phosphatase 1, regulatory (inhibitor) subunit 16B | 1.22E-02 | 1.59 |
| Arid3a | AT rich interactive domain 3A (BRIGHT-like) | 1.22E-02 | 1.58 |
| Tspan8 | tetraspanin 8 | 2.92E-02 | 1.57 |
| Stat5b | signal transducer and activator of transcription 5B | 1.22E-02 | 1.57 |
| Adprhl1 | ADP-ribosylhydrolase like 1 | 1.22E-02 | 1.56 |
| Hspb8 | heat shock protein 8 | 1.22E-02 | 1.55 |
| Tor4a | torsin family 4, member A | 1.22E-02 | 1.55 |
| Gm42372 | predicted gene, 42372 | 1.22E-02 | 1.53 |
| Oscp1 | organic solute carrier partner 1 | 1.22E-02 | 1.53 |
| Rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 | 3.99E-02 | 1.52 |
| Gm32113 | predicted gene, 32113 | 3.44E-02 | 1.52 |
| Gm30141 | predicted gene, 30141 | 1.22E-02 | 1.52 |
| Gm11525 | predicted gene 11525 | 1.22E-02 | 1.52 |
| Downregulated genes (fold-change ≥ 1.5) | |||
| Gene Symbol | Gene Name | q-value | Fold-Change scrambled/KD |
| Nt5c3b | 5’-nucleotidase, cytosolic IIIB | 1.22E-02 | 3.73 |
| Smchd1 | SMC hinge domain containing 1 | 1.22E-02 | 3.58 |
| Clu | clusterin | 3.99E-02 | 2.70 |
| Ccr4 | chemokine (C-C motif) receptor 4 | 1.22E-02 | 2.69 |
| Hspa1a | heat shock protein 1A | 2.92E-02 | 2.35 |
| Slc19a3 | solute carrier family 19, member 3 | 1.22E-02 | 2.02 |
| Rps4l | ribosomal protein S4-like | 1.22E-02 | 1.94 |
| Klrg2 | killer cell lectin-like receptor subfamily G, member 2 | 1.22E-02 | 1.75 |
| Gm7969 | predicted gene 7969 | 1.22E-02 | 1.72 |
| Skp2 | S-phase kinase-associated protein 2 (p45) | 1.22E-02 | 1.71 |
| Siglecg | sialic acid binding Ig-like lectin G | 2.92E-02 | 1.69 |
| Pfkfb3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 1.22E-02 | 1.69 |
| Enpp3 | ectonucleotide pyrophosphatase/phosphodiesterase 3 | 2.24E-02 | 1.68 |
| Psmc2 | proteasome (prosome, macropain) 26S subunit, ATPase 2 | 1.22E-02 | 1.67 |
| Ficd | FIC domain containing | 1.22E-02 | 1.56 |
| Lima1 | LIM domain and actin binding 1 | 1.22E-02 | 1.54 |
| Ptpn22 | protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | 3.99E-02 | 1.54 |
| Fam50a | family with sequence similarity 50, member A | 1.22E-02 | 1.53 |
| Gpkow | G patch domain and KOW motifs | 1.22E-02 | 1.50 |
The RNAseq analysis confirmed that the Smchd1 mRNA was reduced by more than 70%. Other genes displaying large reductions in expression and lowest q value (0.0122) included Nt5c3b (5’-nucleotidase, cytosolic IIIB; FC=3.7), Ccr4 (Chemokine (C-C motif) receptor 4; FC=2.7), Slc19a3 (solute carrier family 19, member 3; FC=2.0), Rps4l (Ribosomal protein S4 like, FC=1.9), Klrg2 (killer cell lectin-like receptor subfamily G, member 2; FC=1.8) and Skp2 (S-phase kinase-associated protein 2, a.k.a. p45; FC=1.7). Genes with the strongest increases and lowest q values included Magea5 (Melanoma antigen family A, protein 5, FC=3.5), Dppa1 (Developmental pluripotency associated 1; FC=2.1), the imprinted gene Peg10 (Paternally expressed gene 10; FC=1.9), Tfap2a (Transcription factor AP-2, alpha; FC=1.8). These latter genes are associated with TE lineage development, consistent with precocious and/or increased expression of at least a subset of TE-related genes in Smchd1 knockdown embryos.
We also assessed effects of Smchd1 knockdown on X-linked genes and on imprinted genes. There was no selective enrichment for X-linked genes among the DEGs identified by RNAseq. Aside from the imprinted gene Peg10, only one other imprinted gene (Igf2r, Insulin-like growth factor 2 receptor) was affected, with 1.36-fold lower expression. Thus, at the morula stage, Smchd1 deficiency did not exert a widespread effect on X-linked or imprinted genes.
To extend the analysis of Smchd1-regulated genes and the effects of Smchd1 siRNA knockdown on their expression, qRT-PCR was applied to an expanded set of samples of morula and blastocyst stage embryos (up to 7 biological replicates of morula stage and 3 replicates of blastocyst stage embryos) (Table 8). For six of the eight mRNAs successfully tested, qRT-PCR confirmed effects of Smchd1 knockdown. The Peg10 mRNA was poorly detected at the morula stage by qRT-PCR, but additional qRT-PCR analyses of blastocyst stage embryos confirmed changes in Peg10 mRNA expression (Table 8). Effects on the expression of AT-rich interacting domain-containing protein 3A (Arid3a) mRNA were not confirmed by qRT-PCR for morula or blastocyst stage embryos. The Tfap2a mRNA tended to be increased in expression at the morula stage, but its expression was highly variable in morula samples, and no statistically significant difference was seen at the blastocyst stage. It is noted that developmental profiles of these mRNAs indicate peaks of expression at different stages (Zeng et al. 2004b), which could have affected reproducibility and sensitivity of measurements by qRT-PCR.
Table 8.
Quantitative RT-PCR analysis of mRNA expression changes with Smchd1 siRNA knockdown at morula and blastocyst stages.
| Morula | Blastocyst | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | FC1 | p-value2 | q-value3 | n4 | FC1 | p-value2 | q-value3 | n4 |
| Arid3a** | −1.034 | 0.867 | 0.867 | 5 | N/A | N/A | N/A | |
| Dppa1* | 3.619 | 0.002 | 0.014 | 6 | N/A | N/A | N/A | |
| Igf2r* | −1.383 | 0.028 | 0.039 | 6 | N/A | N/A | N/A | |
| Nt5c3b* | −3.480 | 0.011 | 0.039 | 5 | N/A | N/A | N/A | |
| Nt5c3b** | −4.247 | 0.00007 | 0.001 | 7 | N/A | N/A | N/A | |
| Peg10** | −1.055 | 0.431 | 0.464 | 4 | 1.461 | 0.023 | 0.036 | 3 |
| Skp2* | −1.619 | 0.018 | 0.032 | 5 | N/A | N/A | N/A | |
| Skp2** | −1.441 | 0.002 | 0.009 | 7 | N/A | N/A | N/A | |
| Smchd1* | −6.441 | 0.014 | 0.033 | 5 | N/A | N/A | N/A | |
| Smchd1** | −4.842 | 0.012 | 0.034 | 5 | −1.830 | 0.017 | 3 | |
| Tfap2a* | 1.402 | 0.416 | 0.529 | 5 | 1.074 | 0.428 | 3 | |
Fold-change (positive: upregulated by Smchd1 siRNA knockdown, negative: downregulated); relative expression values (RQs) were calculated using Rpl18 as the internal reference gene.
T-test p-value.
q = p-value adjusted by Benjamini-Hochberg procedure
Number of biological replicates per group.
SPIA amplified cDNA as template
Unamplified cDNA as template, N/A, not analyzed.
Ingenuity Pathway Analysis
Ingenuity Pathways Analysis (IPA) was employed to identify biological pathways and processes affected by siRNA-mediated Smchd1 deficiency. One biological function (cell death) displayed a significant positive z-score (z > 1.96) indicating increased activity. Cell proliferation was also significantly affected (multiple entries p = 4.9 × 10−2 to 1.8 × 10−4) with several entries having negative z-scores below the level of significance consistent with possible inhibition. These results suggest that the negative effect of Smchd1 siRNA on embryo cell number could reflect a combination of increased cell death and reduced cell division. Affected canonical pathways included protein ubiquitination, which encompasses a known function for Skp2, one of the genes affected by Smchd1 siRNA knockdown.
CDX2 represses Smchd1 expression
Our observations suggest a possible role for SMCHD1 regulating genes differentially associated with the TE and ICM lineages. To investigate Smchd1 regulation further in these two lineages, we examined Smchd1 expression in embryos lacking CDX2, a transcription factor that is essential for promoting TE gene expression and repressing ICM genes within the developing TE (Strumpf et al., 2005), and which binds to the Smchd1 gene (Nishiyama et al. 2009). We harvested ED3.5 and ED4.25 blastocysts from Cdx2−/+ inter se matings (Figure 4) and examined the expression of Cdx2 and its targets, Krt8 and Eomes (Ralston et al., 2008) by qPCR. As expected, Cdx2 and Krt8 expression was significantly decreased in Cdx2−/− embryos compared to non-mutant (p=0.0018 and p<0.0001, respectively) and ED3.5 and ED4.25. Eomes expression followed this trend, but differences were not significant at ED3.5 (p=0.128). We next examined expression of Smchd1 in Cdx2 null and control blastocysts. Loss of Cdx2 resulted in a significant increase in Smchd1 expression in Cdx2−/− embryos compared to non-mutant controls at ED3.5 (~2-fold overall, p=0.0003), attributable to a large increase in Smchd1 mRNA expression in a subset of embryos (~5-fold in three of the embryos assayed). At ED4.25, elevation of Smchd1 mRNA expression was more prominent, with 3 of 4 embryos displaying notably elevated expression compared to non-mutant controls (p=0.0019). At both stages, the increase in Smchd1 mRNA expression brought on by Cdx2 deficiency displays a notable degree of inter-embryo variation. Overall, these results indicate that CDX2 represses Smchd1 expression, and may enrich Smchd1 expression in the ICM and thereby inhibit TE-related gene expression.
Figure 4.
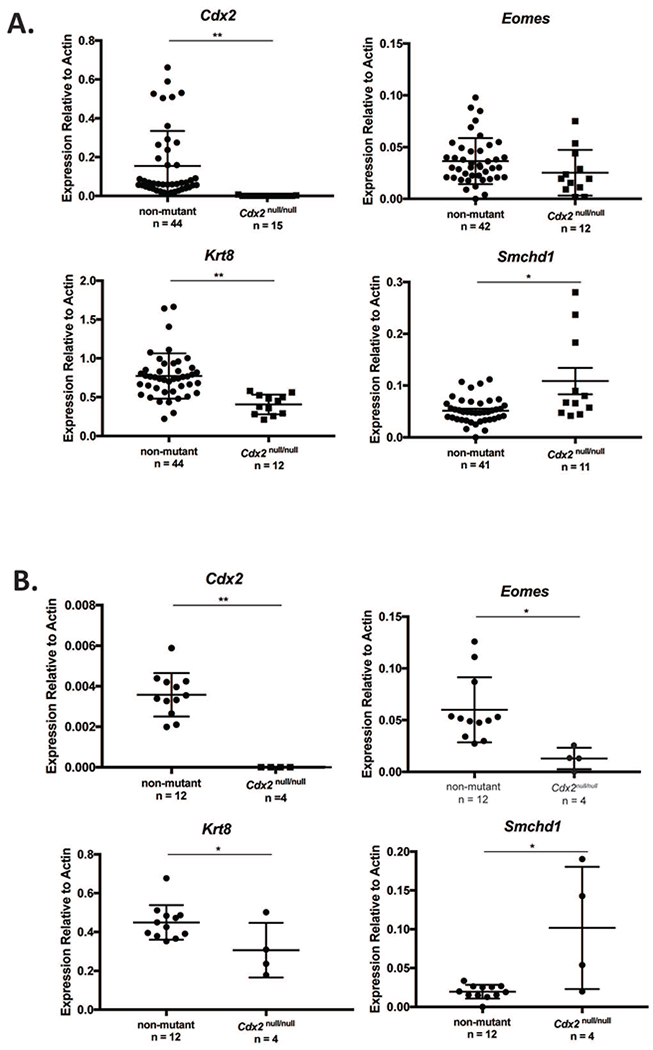
Effect of Cdx2 gene mutation on Smchd1 mRNA expression. Smchd1 mRNA is expressed more highly in Cdx2−/− embryos as compared to homozygous and heterozygous embryos (ED3.5). A: Expression of genes in ED3.5 blastocysts. Cdx2, Eomes, Krt8 and Smchd1 mRNAs were quantified relative to β-actin in wild type, Cdx2−/+ and Cdx2−/− blastocysts. Individual dots represent single blastocysts. B: Expression of genes in ED4.25 blastocysts. Mean expression values (± s.e.m.) for each genotype, normalized to β-actin, is indicated. Genotypes of embryos were inferred as described in Methods. One-way ANOVA was performed to assess significance of gene expression.
Discussion
Our results reveal important, novel roles for SMCHD1 during preimplantation development, modulating genes that are differentially expressed between the two primary lineages, and promoting cell survival and proliferation. These crucial early preimplantation functions for SMCHD1 were not detected previously in embryos from heterozygous Smchd1 mutant crosses, most likely because such crosses do not eliminate the maternal supply of Smchd1 mRNA and protein present in the oocyte. These preimplantation functions contribute to long-term embryo viability and successful pregnancy. SMCHD1 deficiency in the preimplantation embryo, mediated by siRNA knockdown, impairs blastocyst formation and hatching, reduces cell number, and impedes cell cycle progression as evidenced by reduced nuclear volume at the morula stage. Viability to term was more severely impacted than viability to ED8.5, further indicating persistent, long-term effects of early preimplantation SMCHD1 deficiency. These results reveal preimplantation roles for SMCHD1 in early programming of the embryonic genome that support long-term development.
The up-regulation of such lineage-related marker genes at the morula stage following Smchd1 knockdown, including some that are upregulated later in development of the TE lineage and placenta, is reminiscent of other studies (e.g., CHD4 ablation) in which multiple lineage-appropriate marker genes display precocious and/or continued expression leading to indeterminate lineage specification (O’Shaughnessy-Kirwan et al. 2015). Deficiency or Smchd1 may thus lead to precocious and/or prolonged expression of certain lineage-related genes, resulting in the establishment of intermediate cell states with a mixed marker gene expression profile and inadequate specialization, compromising long-term viability. Such a subtle mode of gene modulation leading to intermediate cell states would account for the lack of a major shift in ICM:TE ratio, but nevertheless a significant quantitative negative effect on embryo viability owing to disruptions in cellular specialization coupled with impact on pluripotency maintenance.
There was no selective effect of Smchd1 deficiency on X-linked gene expression. This was not surprising, because X chromosome inactivation (XCI) is very limited in the preimplantation embryo. Although embryonic Xist transcripts can be detected as early as the two-cell stage, XCI appears limited to genes near the X chromosome inactivation center during most of preimplantation development, with evidence of spreading of the inactivation as embryos reach the blastocyst stage (Latham 1996). Indeed, gynogenetic and parthenogenetic embryos, which do not possess imprinted Xist alleles and do not undergo early XCI nevertheless form blastocysts. Thus, although Smchd1 deficiency impacts XCI in postimplantation stage embryos, this is not a significant feature of the preimplantation effects of siRNA knockdown.
The effect of Smchd1 siRNA knockdown on cell number and nuclear volume could occur via SKP2. SKP2 is an E3 ligase and substrate recognition component of the SKP1-Cullin-Fbox (SCF) protein complex, which controls the transition from G1 to S phase of the cell cycle (Nakayama et al., 2000). SKP2 also associates with E1A binding protein/p300 to inhibit TP53-mediated apoptosis (Kitagawa et al. 2008), another function consistent with effects highlighted in our IPA results. Reduced cell division and increased cell death due to diminished SKP2 expression could thus account for reduced blastocyst cell number, while slower progression from G1 to S phase could account for reduced nuclear volumes in morulae following Smchd1 siRNA knockdown.
By regulating the CIP/KIP class of cyclin-dependent kinase inhibitors, SKP2 also modulates signaling through RHO-ROCK (Ras homolog gene family member a, - RHO-associated coiled-coil containing protein kinase 1), affecting cytoskeleton formation and cellular polarity (Besson et al., 2004), which is known to impact Hippo pathway signaling (Kono et al. 2014; Mihajlovic and Bruce 2016). Thus, Smchd1 may, through Skp2, promote proliferation, survival, and polarization required for the emergence of the TE lineage. SMCHD1 knockdown could thus impact both early lineages, reducing numbers of both cell types, with minimal change in ICM:TE ratio.
RNAseq and qRT-PCR analyses revealed reduced expression of the Nt5c3b mRNA. NT5C3B mediates the hydrolysis of 7-methyl GMP to 7 methyl guanosine, which can modulate mRNA translation. This deficiency could modulate the timely translation of maternal mRNAs during early cleavage stages.
Another striking aspect of these results is that significant phenotypic perturbations in blastocyst stage embryos are achieved despite partial recovery of Smchd1 expression in blastocysts, with a subsequent diminishment in survival to birth, and a more severe effect on term development as compared to development to ED8.5. Whether reduced term development is solely due to compromised blastocyst viability, or is also partly due to gene programming abnormalities arising during cleavage stages with SMCHD1 deficiency is unknown. In humans, SMCHD1 mutations display autosomal dominance, partial penetrance, and variegated effects (Daxinger et al. 2015; Lemmers et al. 2012; van den Boogaard et al. 2016). While studies of effects of Smchd1 mutations in mice reveal lethal developmental defects and effects on DNA methylation across the genome at later stages, more subtle effects have not been explored. Our data indicate that early SMCHD1 deficiency, even if transient, may inhibit essential epigenetic changes at small subsets of target genes, leading to significant defects in developmental programming of embryonic genomes during cleavage that disrupt later events and long-term development.
These results extend the known functions of SMCHD1 to the preimplantation period. Our RNAseq analysis identified novel effects of SMCHD1 on gene expression and IPA analysis provides new insight into additional biological pathways and functions impacted by SMCHD1. The phenotype following Smchd1 knockdown indicates crucial roles for SMCHD1 in promoting cleavage, inhibiting apoptosis, supporting blastocyst formation and hatching, and contributing to the establishment of cell lineage identities. Additional roles may include modulating mRNA translation, cell shape, and cell adhesion during cleavage, and early embryonic genome developmental programming processes. Further studies of SMCHD1 function in the early embryo could reveal additional effects of Smchd1 mutations on reproduction, fertility, and organogenesis, and improve our understanding of how SMCHD1 mutations may affect human development health.
Acknowledgments
Funding
This work was supported in part by grants from the National Institutes of Health, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the National Institute of General Medical Science (R01HD043092, R01HD075903, and T32HD087166 to KEL and R01GM104009 to AR), by MSU AgBioResearch, and by the Michigan State University. ALS was supported by T32HD087166. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
Authors declare no competing interests related to these studies.
Data availability
RNAseq data from this study are available in the Gene Expression Omnibus (GEO accession number GSE92936) at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92936.
References
- Aksoy I, Jauch R, Chen J, Dyla M, DIvakar U, Bogu GK, Teo R, Leng Ng CK, Herath W, Lili S, Hutchins AP, Robson P, Kolatkar PR, Stanton JA (2013a). RNA-sequencing technology identifies specific genes expressed in the ICM of mouse embryos. Gene Expression Omnibus accession no. GSE44553.
- Aksoy I, Jauch R, Chen J, Dyla M, Divakar U, Bogu GK, Teo R, Leng Ng CK, Herath W, Lili S, Hutchins AP, Robson P, Kolatkar PR, Stanton LW (2013b). Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J, 32(7), 938–953. 10.1038/emboj.2013.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, Butterfield NC, Wicking C, Blewitt ME, Wilkins SJ, Anderson GJ, Cox TC, Whitelaw E (2008). A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol, 9(12), R182 10.1186/gb-2008-9-12-r182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y HY (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of Royal Statistical Society, 57 289–300 [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, Kay GF, Whitelaw E (2008). SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet, 40(5), 663–669. 10.1038/ng.142 [DOI] [PubMed] [Google Scholar]
- Carey TS, Cao Z, Choi I, Ganguly A, Wilson CA, Paul S, Knott JG (2015). BRG1 Governs Nanog Transcription in Early Mouse Embryos and Embryonic Stem Cells via Antagonism of Histone H3 Lysine 9/14 Acetylation. Mol Cell Biol, 35(24), 4158–4169. 10.1128/MCB.00546-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F (1997). Homeosis and intestinal tumours in Cdx2 mutant mice. Nature, 386(6620), 84–87. 10.1038/386084a0 [DOI] [PubMed] [Google Scholar]
- Chen K, Hu J, Moore DL, Liu R, Kessans SA, Breslin K, Lucet IS, Keniry A, Leong HS, Parish CL, Hilton DJ, Lemmers RJ, van der Maarel SM, Czabotar PE, Dobson RC, Ritchie ME, Kay GF, Murphy JM, Blewitt ME (2015). Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc Natl Acad Sci U S A, 112(27), E3535–3544. 10.1073/pnas.1504232112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Gaughan J, Midic U, Han Z, Liang CG, Patel BG, Latham KE (2013). Systems genetics implicates cytoskeletal genes in oocyte control of cloned embryo quality. Genetics, 193(3), 877–896. 10.1534/genetics.112.148866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YG, Ratnam S, Chaillet JR, Latham KE (2003). Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol Reprod, 69(1), 146–153. 10.1095/biolreprod.102.014076 biolreprod.102.014076[pii] [DOI] [PubMed] [Google Scholar]
- Coker H, Brockdorff N (2014). SMCHD1 accumulates at DNA damage sites and facilitates the repair of DNA double-strand breaks. J Cell Sci, 127(Pt 9), 1869–1874. 10.1242/jcs.140020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Mager J (2018). Transcriptional Regulation and Genes Involved in First Lineage Specification During Preimplantation Development. Adv Anat Embryol Cell Biol, 229 31–46. 10.1007/978-3-319-63187-5_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Tapscott SJ, van der Maarel SM (2015). Genetic and epigenetic contributors to FSHD. Curr Opin Genet Dev, 33 56–61. 10.1016/j.gde.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbach GT, Lawitts JA, Papaioannou VE, Biggers JD (1994). Differential growth of the mouse preimplantation embryo in chemically defined media. Biol Reprod, 50(5), 1027–1033 [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Apedaile A, Coker H, Termanis A, Zvetkova I, Godwin J, Tang YA, Huntley D, Montana G, Taylor S, Giannoulatou E, Heard E, Stancheva I, Brockdorff N (2012). Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive x chromosome. Dev Cell, 23(2), 265–279. 10.1016/j.devcel.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Tang YA, Suzuki M, Godwin J, Nesterova TB, Greally JM, Heard E, Brockdorff N (2013). Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol Cell Biol, 33(16), 3150–3165. 10.1128/MCB.00145-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CT, Xue S, Yigit G, Filali H, Chen K, Rosin N, Yoshiura KI, Oufadem M, Beck TJ, McGowan R, Magee AC, Altmuller J, Dion C, Thiele H, Gurzau AD, Nurnberg P, Meschede D, Muhlbauer W, Okamoto N, Varghese V, Irving R, Sigaudy S, Williams D, Ahmed SF, Bonnard C, Kong MK, Ratbi I, Fejjal N, Fikri M, Elalaoui SC, Reigstad H, Bole-Feysot C, Nitschke P, Ragge N, Levy N, Tuncbilek G, Teo AS, Cunningham ML, Sefiani A, Kayserili H, Murphy JM, Chatdokmaiprai C, Hillmer AM, Wattanasirichaigoon D, Lyonnet S, Magdinier F, Javed A, Blewitt ME, Amiel J, Wollnik B, Reversade B (2017). De novo mutations in SMCHD1 cause Bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat Genet, 49(2), 249–255. 10.1038/ng.3765 [DOI] [PubMed] [Google Scholar]
- Jansz N, Chen K, Murphy JM, Blewitt ME (2017). The Epigenetic Regulator SMCHD1 in Development and Disease. Trends Genet, 33(4), 233–243. 10.1016/j.tig.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol, 14(4), R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Lee SH, McCormick F (2008). Skp2 suppresses p53-dependent apoptosis by inhibiting p300. Mol Cell, 29(2), 217–231. 10.1016/j.molcel.2007.11.036 [DOI] [PubMed] [Google Scholar]
- Ko MS (2016). Zygotic Genome Activation Revisited: Looking Through the Expression and Function of Zscan4. Curr Top Dev Biol, 120 103–124. 10.1016/bs.ctdb.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Kono K, Tamashiro DA, Alarcon VB (2014). Inhibition of RHO-ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev Biol, 394(1), 142–155. 10.1016/j.ydbio.2014.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE (1996). X chromosome imprinting and inactivation in the early mammalian embryo. Trends Genet, 12(4), 134–138. 0168952596100172 [pii] [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, Krom YD, Klooster R, Sun Y, den Dunnen JT, Helmer Q, Donlin-Smith CM, Padberg GW, van Engelen BG, de Greef JC, Aartsma-Rus AM, Frants RR, de Visser M, Desnuelle C, Sacconi S, Filippova GN, Bakker B, Bamshad MJ, Tapscott SJ, Miller DG, van der Maarel SM (2012). Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet, 44(12), 1370–1374. 10.1038/ng.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong HS, Chen K, Hu Y, Lee S, Corbin J, Pakusch M, Murphy JM, Majewski IJ, Smyth GK, Alexander WS, Hilton DJ, Blewitt ME (2013). Epigenetic regulator Smchd1 functions as a tumor suppressor. Cancer Res, 73(5), 1591–1599. 10.1158/0008-5472.CAN-12-3019 [DOI] [PubMed] [Google Scholar]
- Liu R, Chen K, Jansz N, Blewitt ME, Ritchie ME (2016). Transcriptional profiling of the epigenetic regulator Smchd1. Genom Data, 7 144–147. 10.1016/j.gdata.2015.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcho C, Cui W, Mager J (2015). Epigenetic dynamics during preimplantation development. Reproduction, 150(3), R109–120. 10.1530/REP-15-0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AG, Slieker RC, Balog J, Lemmers R, Wong CJ, Yao Z, Lim JW, Filippova GN, Ne E, Tawil R, Heijmans BT, Tapscott SJ, van der Maarel SM (2017). SMCHD1 regulates a limited set of gene clusters on autosomal chromosomes. Skelet Muscle, 7(1), 12 10.1186/s13395-017-0129-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihajlovic AI, Bruce AW (2016). Rho-associated protein kinase regulates subcellular localisation of Angiomotin and Hippo-signalling during preimplantation mouse embryo development. Reprod Biomed Online, 33(3), 381–390. 10.1016/j.rbmo.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Miller A, Hendrich B (2018). Chromatin Remodelling Proteins and Cell Fate Decisions in Mammalian Preimplantation Development. Adv Anat Embryol Cell Biol, 229 3–14. 10.1007/978-3-319-63187-5_2 [DOI] [PubMed] [Google Scholar]
- Mould AW, Pang Z, Pakusch M, Tonks ID, Stark M, Carrie D, Mukhopadhyay P, Seidel A, Ellis JJ, Deakin J, Wakefield MJ, Krause L, Blewitt ME, Kay GF (2013). Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin, 6(1), 19 10.1186/1756-8935-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H (2009). The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell, 16(3), 398–410. 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, Stagg C, Sharova L, Correa-Cerro LS, Bassey U, Hoang H, Kim E, Tapnio R, Qian Y, Dudekula D, Zalzman M, Li M, Falco G, Yang HT, Lee SL, Monti M, Stanghellini I, Islam MN, Nagaraja R, Goldberg I, Wang W, Longo DL, Schlessinger D, Ko MS (2009). Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell, 5(4), 420–433. 10.1016/j.stem.2009.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy-Kirwan A, Signolet J, Costello I, Gharbi S, Hendrich B (2015). Constraint of gene expression by the chromatin remodelling protein CHD4 facilitates lineage specification. Development, 142(15), 2586–2597. 10.1242/dev.125450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Li Z, Zhang Y, Huang Y, Wu Q, Ru G, Chen M, Wang B (2016). Response of Mouse Zygotes Treated with Mild Hydrogen Peroxide as a Model to Reveal Novel Mechanisms of Oxidative Stress-Induced Injury in Early Embryos. Oxid Med Cell Longev, 2016 1521428 10.1155/2016/1521428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J (2010). Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development, 137(3), 395–403. 10.1242/dev.038828 [DOI] [PubMed] [Google Scholar]
- Sasaki H (2015). Position- and polarity-dependent Hippo signaling regulates cell fates in preimplantation mouse embryos. Semin Cell Dev Biol, 47-48 80–87. 10.1016/j.semcdb.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Shaw ND, Brand H, Kupchinsky ZA, Bengani H, Plummer L, Jones TI, Erdin S, Williamson KA, Rainger J, Stortchevoi A, Samocha K, Currall BB, Dunican DS, Collins RL, Willer JR, Lek A, Lek M, Nassan M, Pereira S, Kammin T, Lucente D, Silva A, Seabra CM, Chiang C, An Y, Ansari M, Rainger JK, Joss S, Smith JC, Lippincott MF, Singh SS, Patel N, Jing JW, Law JR, Ferraro N, Verloes A, Rauch A, Steindl K, Zweier M, Scheer I, Sato D, Okamoto N, Jacobsen C, Tryggestad J, Chernausek S, Schimmenti LA, Brasseur B, Cesaretti C, Garcia-Ortiz JE, Buitrago TP, Silva OP, Hoffman JD, Muhlbauer W, Ruprecht KW, Loeys BL, Shino M, Kaindl AM, Cho CH, Morton CC, Meehan RR, van Heyningen V, Liao EC, Balasubramanian R, Hall JE, Seminara SB, Macarthur D, Moore SA, Yoshiura KI, Gusella JF, Marsh JA, Graham JM Jr., Lin AE, Katsanis N, Jones PL, Crowley WF Jr., Davis EE, FitzPatrick DR, Talkowski ME (2017). SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat Genet, 49(2), 238–248. 10.1038/ng.3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development, 132(9), 2093–2102. 10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Minami N (2018). CHD1 Controls Cell Lineage Specification Through Zygotic Genome Activation. Adv Anat Embryol Cell Biol, 229 15–30. 10.1007/978-3-319-63187-5_3 [DOI] [PubMed] [Google Scholar]
- Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T, Manabe I, Imai H, Minami N (2015). CHD1 acts via the Hmgpi pathway to regulate mouse early embryogenesis. Development, 142(13), 2375–2384. 10.1242/dev.120493 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol, 31(1), 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard ML, Jfl Lemmers R, Camano P, van der Vliet PJ, Voermans N, van Engelen BG, Lopez de Munain A, Tapscott SJ, van der Stoep N, Tawil R, van der Maarel SM (2016). Double SMCHD1 variants in FSHD2: the synergistic effect of two SMCHD1 variants on D4Z4 hypomethylation and disease penetrance in FSHD2. Eur J Hum Genet, 24(1), 78–85. 10.1038/ejhg.2015.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Sengupta S, Magnani L, Wilson CA, Henry RW, Knott JG (2010). Brg1 is required for Cdx2-mediated repression of Oct4 expression in mouse blastocysts. PLoS One, 5(5), e10622 10.1371/journal.pone.0010622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A (2007). Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development, 134(21), 3827–3836. dev.010223 [pii]10.1242/dev.010223 [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM 2004a. Preimplantation embryo development (MOE430A). Gene Expression Omnibus acession no. GSE1749. [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM (2004b). Transcript profiling during preimplantation mouse development. Dev Biol, 272(2), 483–496. 10.1016/j.ydbio.2004.05.018S0012160604003628 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNAseq data from this study are available in the Gene Expression Omnibus (GEO accession number GSE92936) at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92936.


