Abstract
BAP1 is a tumor suppressor gene important to the development and prognosis of many cancers, especially uveal melanoma (UM). Its role in more common cancers such as breast and colon cancer is largely unknown. We collected the transcriptome profiling data sets from the TCGA uveal melanoma (TCGA-UVM), breast cancer (TCGA-BRCA), and colon cancer (TCGA-COAD) projects to analyze the expression of BAP1. We found that patients with UM and breast cancer, but not colon cancer, who died had a lower level of BAP1 gene expression compared to surviving patients. Importantly, in breast cancer patients, the lowest BAP1 expression levels corresponded to the dead young patients (age at diagnosis < 46). Since the number of cases in TCGA-BRCA was much higher than TCGA-UVM, we obtained highly correlated genes with BAP1 in invasive breast carcinomas. Then, we tested if these genes are also highly correlated with BAP1 in UM and colon cancer. We found that BAP1 is highly positively correlated with RBM15B and USP19 expression in invasive breast carcinoma, UM, and colon adenocarcinoma. All three genes are located in close proximity on the 3p21 tumor suppressor region that is commonly altered in many cancers.
Introduction
Both somatic and germline mutations in the BRCA1 associated protein-1 (BAP1) gene have been observed in many cancers. BAP1 tumor predisposition syndrome (BAP1 -TPDS) OMIM #614327 is associated with an increased risk for uveal melanoma (UM), malignant mesothelioma (MMe), cutaneous melanoma (CM), clear cell renal cell carcinoma (RCC) and potentially other cancers such as breast cancer [1–3].
UM is the most common and earliest presenting cancer in the BAP1-TPDS [1–3]. In addition, UM has the highest frequency of pathogenic somatic mutations in BAP1 [4, 5]. Although mutations in BAP1 are strongly associated with metastatic UMs (84%), it is not frequent in non-metastatic tumors (4%) [4]. Moreover, germline BAP1 mutations are associated with more aggressive UM [6]. BAP1 also plays a prognostic role in other cancers such as RCC and cholangiocarcinoma [7–9].
Although it is known that BAP1 is a tumor suppressor gene important to the development and prognosis of many cancers, especially UM, its role in more common cancers including breast and colon is largely unexplored.
Here, we collected RNA-seq data sets from TCGA-BRCA, -UVM, and -COAD projects at Genomic Data Commons Data Portal, and we employed computational techniques to investigate the role of BAP1 in UM and two common cancers, breast and colon. Furthermore, we evaluated how patient age, sex, and race may be associated with BAP1 gene expression and patient survival with these cancers. We hypothesized that BAP1 gene expression could be a potential signature for aggressive tumors. In addition we identified genes with expression that is highly correlated with BAP1. To determine the genes that may be critical to BAP1 functional pathways, we used the UM data set which is known to have a high level of BAP1 alteration to identify candidates, which were validated in the breast and colon cancer data sets.
We analyzed the data sets by applying two slightly different normalization methods. In the first method, we evaluated all genes and obtained z-scores to normalize the data. In the second method, we excluded the genes with values of zero for all patients in the data set, and then we obtained z-scores. Importantly, the results were independent of these two normalization strategies.
Results
Dead patients had low BAP1 expression compared to survivors in breast cancer and UM but not colon cancer
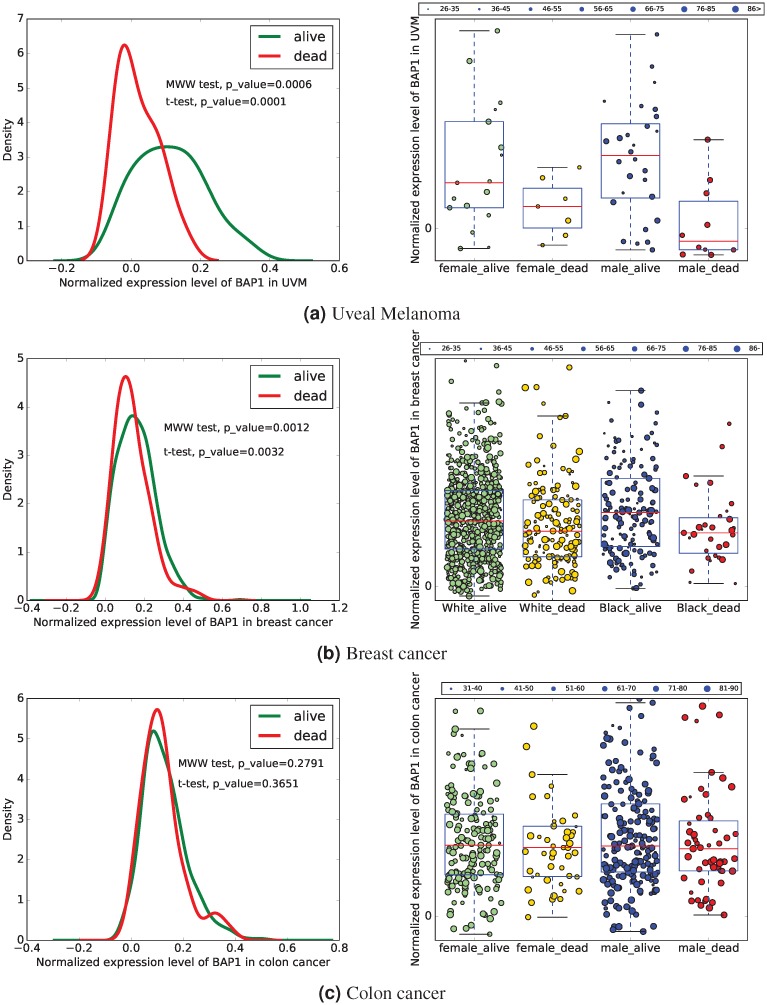
We divided UM patients in four sub-categories based on their gender and vital status: female-alive, female-dead, male-alive, and male-dead (Table 1). We also divided patients into sub-groups based on their age at diagnosis (grouped into intervals of 10 years starting from 26, which was the age of the youngest patient). The left panel of Fig 1(a) shows the normalized level of BAP1 for these patients, and the right sub-figure represent the density function for the values of BAP1 in dead and alive patients after excluding genes with zero values and normalizing the data set. This figure indicates that the expression level of BAP1 is lower in dead patients compared to the survivors regardless of their gender.
Table 1. Data set for UVM.
| categories | number of cases | number of files |
| female | 40 | 40 |
| male | 48 | 48 |
| sub-groups | number of cases | number of files |
| female-alive | 31 | 31 |
| female-dead | 9 | 9 |
| male-alive | 32 | 32 |
| male-dead | 16 | 16 |
Fig 1. BAP1 expression.
The left and right sub-figures respectively represent the density function and the values of BAP1 after normalizing the data set excluding genes with zero values. The size of circles represents the age at diagnosis.
We also divided breast cancer female patients in four sub-categories based on their race and vital status: black-alive, black-dead, white-alive, and white-dead (Table 2). The left sub-figure of Fig 1(b) shows the normalized (z-score) value of BAP1 for survival categories of black and white female breast cancer patients, and the right sub-figure compares the BAP1 expression in survivors with the BAP1 expression in dead patients. In this figure, we also divided breast cancer patients into sub-groups based on their age at diagnosis (grouped into intervals of 10 years starting from 26, which was the age of the youngest patient). We found that dead patients, regardless of their race, have a lower level of BAP1 expression compared with living patients. Importantly, the smallest level of BAP1 corresponds to the young (age at diagnosis < 46 years old) patients who died.
Table 2. Data set for BRCA.
| categories | number of cases | number of files |
| female | 1084 | 1208 |
| male | 12 | 13 |
| sub-groups-females | number of cases | number of files |
| black-alive | 151 | 156 |
| black-dead | 29 | 31 |
| white-alive | 636 | 705 |
| white-dead | 112 | 159 |
Additionally, we investigated the expression level of BAP1 in colon cancer. We divided patients with colon cancer in four sub-categories based on their gender and vital status: female-alive, female-dead, male-alive, and male-dead (Table 3). The left sub-figure of Fig 1(c) shows the normalized level of BAP1 for these patients, and the right sub-figure represents the difference between the BAP1 expression in dead and alive patients. We also divided colon cancer patients into sub-groups based on their age at diagnosis (grouped into intervals of 10 years starting from 31, which was the age of the youngest patient). This figure indicates that the expression level of BAP1 in patients with colon cancer is not correlated with vital status or gender.
Table 3. Data set for COAD.
| categories | number of cases | number of files |
| female | 214 | 224 |
| male | 240 | 252 |
| sub-groups | number of cases | number of files |
| female-alive | 168 | 177 |
| female-dead | 46 | 47 |
| male-alive | 184 | 196 |
| male-dead | 56 | 56 |
BAP1 is highly positively correlated with RBM15B and USP19
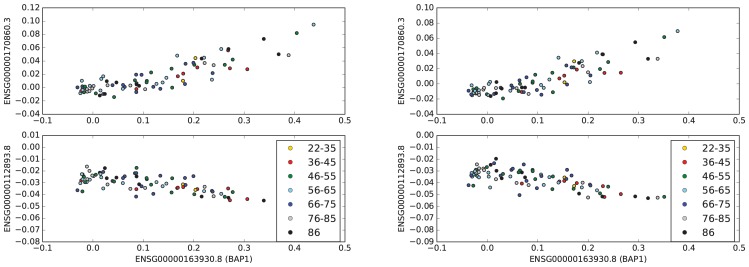
We found the genes that were highly correlated (absolute value of correlation coefficient > 0.8) with BAP1 in the entire TCGA-UVM dataset. Although there were 24 genes with a high positive correlation with BAP1, there were no genes with a high negative correlation with BAP1. Table 4 shows the genes that are highly positively correlated with BAP1 in the entire data set of UM. Top panels (and bottom panels) of Fig 2 show the scatter plot of values of BAP1 and the genes with the highest positive and negative correlation coefficients with BAP1 in UM.
Table 4. Highly correlated genes with BAP1 in the entire UVM dataset.
| transcript (gene) | (cor, p-value) | location |
|---|---|---|
| ENSG00000114316.11 (USP4) | (0.84,1.4e-24) | 3p21.31 |
| ENSG00000177352.9 (CCDC71) | (0.832,1.0e-23) | 3p21.31 |
| ENSG00000259956.1 (RBM15B) | (0.838,2.4e-24) | 3p21.2 |
| ENSG00000178149.15 (DALRD3) | (0.808,1.8e-21) | 3p21.31 |
| ENSG00000161202.16 (DVL3) | (0.854,4.1e-26) | 3q27.1 |
| ENSG00000114767.6 (RRP9) | (0.824,5.7e-23) | 3p21.2 |
| ENSG00000114650.17 (SCAP) | (0.84,1.3e-24) | 3p21.31 |
| ENSG00000171135.12 (JAGN1) | (0.802,6.5e-21) | 3p25.3 |
| ENSG00000172046.17 (USP19) | (0.838,2.2e-24) | 3p21.31 |
| ENSG00000175792.10 (RUVBL1) | (0.806,2.7e-21) | 3q21.3 |
| ENSG00000164062.11 (APEH) | (0.842,8.0e-25) | 3p21.31 |
| ENSG00000183624.12 (HMCES) | (0.804,4.2e-21) | 3q21.3 |
| ENSG00000176095.10 (IP6K1) | (0.824,5.9e-23) | 3p21.31 |
| ENSG00000114738.9 (MAPKAPK3) | (0.815,4.5e-22) | 3p21.2 |
| ENSG00000163932.12 (PRKCD) | (0.824,6.3e-23) | 3p21.1 |
| ENSG00000187091.12 (PLCD1) | (0.816,3.7e-22) | 3p22.2 |
| ENSG00000163719.17 (MTMR14) | (0.848,2.1e-25) | 3p25.3 |
| ENSG00000213672.6 (NCKIPSD) | (0.824,6.4e-23) | 3p21.31 |
| ENSG00000114544.14 (SLC41A3) | (0.819,1.8e-22) | 3q21.3 |
| ENSG00000270170.1 (NCBP2-AS2) | (0.851,9.2e-26) | 3q29 |
| ENSG00000144659.9 (SLC25A38) | (0.857,1.8e-26) | 3p22.1 |
| ENSG00000170860.3 (LSM3) | (0.876,6.3e-29) | 3p25.1 |
| ENSG00000132153.13 (DHX30) | (0.824,6.9e-23) | 3p21.31 |
| ENSG00000163930.8 (BAP1) | (1.0,0.0) | 3p21.1 |
| ENSG00000145191.10 (EIF2B5) | (0.822,9.8e-23) | 3q27.1 |
Fig 2. Highly correlated genes with BAP1 in UVM.
The top sub-figure shows the scatter plot of the values of BAP1 and the gene, which has the maximum positive correlation with BAP1 in the entire UVM data set. The bottom sub-figure shows the scatter plot of the values of BAP1 and the gene, which has the maximum negative correlation with BAP1 in all patients with UVM. The right panel shows the exact same scatter plot as the left panel using n-normalization.
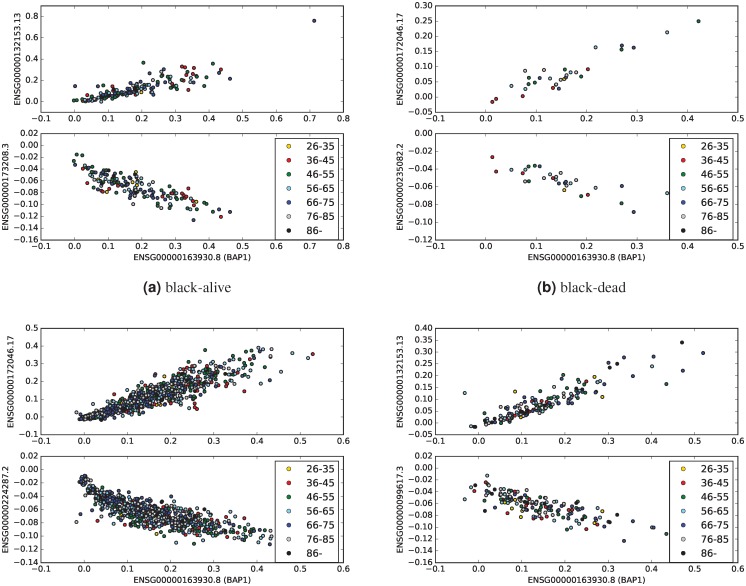
In patients with breast invasive carcinomas, BAP1 is highly positively correlated with NSG00000132153.13 (DHX30), ENSG00000259956.1 (RBM15B), and ENSG00000172046.17 (USP19), in all four categories (Table 5). Importantly, all these three genes were also highly positive correlated with BAP1 in UM (Table 1). DHX30 has the highest positive correlation with BAP1 in black-alive and white-dead categories, while USP19 has the maximum positive correlation with BAP1 in black-dead and white-alive breast cancer categories. Fig 3 presents scatter plots of values of BAP1 and the genes with the highest positive and negative correlation coefficient with BAP1 in all four breast patients categories.
Table 5. Highly correlated genes with BAP1 in BRCA data set.
| transcript (gene) | black-alive (cor, p-value) |
black-dead (cor, p-value) |
white-alive (cor, p-value) |
white-dead (cor, p-value) |
all-data (cor, p-value) |
location |
|---|---|---|---|---|---|---|
| ENSG00000132153.13 (DHX30) | (0.839,8.1e-42) | (0.905,2.8e-12) | (0.87,6.4e-219) | (0.895,1.3e-56) | (0.87,1.3e-321) | 3p21.31 |
| ENSG00000163930.8 (BAP1) | (1.0,0.0e+00) | (1.0,0.0e+00) | (1.0,0.0e+00) | (1.0,0.0e+00) | (1.0,0.00e+00) | 3p21.1 |
| ENSG00000198218.9 (QRICH1) | (0.732,6.0e-27) | (0.827,9.8e-09) | (0.853,2.3e-201) | (0.841,1.6e-43) | (0.82,8.7e-255) | 3p21.31 |
| ENSG00000126062.3 (TMEM115) | (0.726,2.6e-26) | (0.806,4.6e-08) | (0.828,6.7e-180) | (0.848,8.1e-45) | (0.81,1.9e-248) | 3p21.31 |
| ENSG00000259956.1 (RBM15B) | (0.834,8.0e-41) | (0.911,1.1e-12) | (0.86,1.6e-208) | (0.86,1.5e-48) | (0.85,4.6e-302) | 3p21.2 |
| ENSG00000178252.16 (WDR6) | (0.752,4.1e-29) | (0.896,9.2e-12) | (0.848,3.2e-197) | (0.858,4.3e-47) | (0.83,4.7e-274) | 3p21.31 |
| ENSG00000176095.10 (IP6K1) | (0.733,4.8e-27) | (0.823,1.3e-08) | (0.861,7.9e-210) | (0.87,1.0e-49) | (0.83,9.9e-266) | 3p21.31 |
| ENSG00000172046.17 (USP19) | (0.802,1.3e-35) | (0.925,1.1e-13) | (0.893,2.1e-246) | (0.87,8.8e-51) | (0.87,0.0e+00) | 3p21.31 |
| ENSG00000076201.13 (PTPN23) | (0.773,1.2e-31) | (0.856,8.6e-10) | (0.848,1.2e-196) | (0.846,1.5e-44) | (0.83,3.6e-274) | 3p21.31 |
Fig 3. Highly correlated genes with BAP1 in BRCA sub-categories.
In each panel, the top sub-figures show the scatter plot of the values of BAP1 and the gene, which has the maximum positive correlation with BAP1 in the given BRCA sub-category. The bottom sub-figures shows the scatter plot of the values of BAP1 and the gene, which has the maximum negative correlation with BAP1 in the given sub-category.
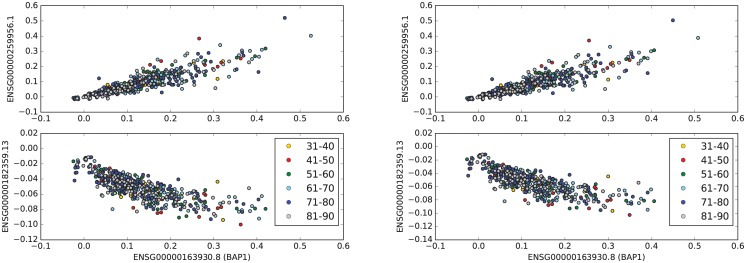
We also obtained the genes that were highly correlated with BAP1 in samples obtained from primary tumor of patients with colon adenocarcinoma (See Table 6 and Fig 4). Interestingly, USP19 and RBM15B were also highly correlated with BAP1 expression in this data set.
Table 6. Highly correlated genes with BAP1 in the entire COAD dataset.
| transcript (gene) | (cor, p-value) | location |
|---|---|---|
| ENSG00000144028.13 (SNRNP200) | (0.802, 2.23e-107) | 2q11.2 |
| ENSG00000126062.3 (TMEM115) | (0.838, 5.45e-126) | 3p21.31 |
| ENSG00000259956.1 (RBM15B) | (0.875, 8.43e-150) | 3p21.2 |
| ENSG00000172046.17 (USP19) | (0.854, 2.15e-135) | 3p21.31 |
| ENSG00000070413.17 (DGCR2) | (0.823, 1.65e-117) | 22q11.21 |
| ENSG00000168066.19 (SF1) | (0.802, 3.84e-107) | 11q13.1 |
| ENSG00000114867.18 (EIF4G1) | (0.803, 1.74e-107) | 3q27.1 |
| ENSG00000163930.8 (BAP1) | (1.0, 0.00e+00) | 3p21.1 |
| ENSG00000169180.10 (XPO6) | (0.81, 4.12e-111) | 16p12.1 |
| ENSG00000076201.13 (PTPN23) | (0.838, 1.84e-125) | 3p21.31 |
| ENSG00000176095.10 (IP6K1) | (0.837, 3.53e-125) | 3p21.31 |
Fig 4. Highly correlated genes with BAP1 in COAD.
The top sub-figure shows the scatter plot of the values of BAP1 and the gene, which has the maximum positive correlation with BAP1 in the entire colon cancer data set. The bottom sub-figure shows the scatter plot of the values of BAP1 and the gene, which has the maximum negative correlation with BAP1 in all patients with colon adenocarcinoma. The right panel shows the exact same scatter plot as the left panel using n-normalization.
The genes that were highly correlated with BAP1 in the entire TCGA-BRCA, -UVM, and -COAD datasets have been presented in Tables 4, 5, and 6. There are three genes other than BAP1, ENSG00000259956.1 (RBM15B), ENSG00000172046.17 (USP19), and ENSG00000176095.10 (IP6K1), that belong to all three tables. Note that IP6K1 was not in the list of highly correlated genes in the all four categories of breast cancer, because its correlation coefficient with BAP1 in the black-alive category was 0.73, which was less than the threshold 0.8. However, its correlation coefficient with BAP1 in the other three categories of breast cancer patients was greater than 0.8.
No genes are highly negatively correlated with BAP1
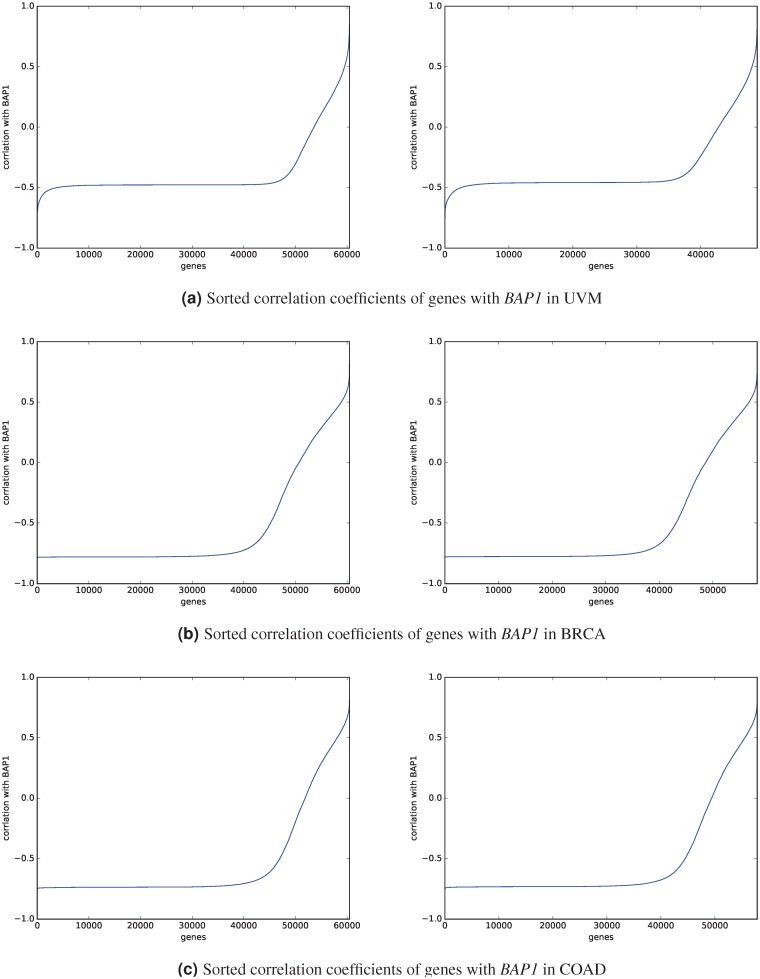
Fig 5 shows the sorted correlation coefficients of all genes with BAP1 in UM, breast, and colon cancer patients. Although most of genes are negatively correlated with BAP1, there is no gene, which is highly negatively correlated (correlation coefficient < −0.8) with BAP1 in all data sets. Each category of breast cancer patients has some genes that are highly negatively correlated with BAP1, but there is no gene that is highly negatively correlated with BAP1 in at least two categories. Furthermore, there are also no genes in the TCGA-UVM data set and also in the TCGA-COAD data set with correlation coefficient less than −0.8 with BAP1.
Fig 5. Sorted correlation coefficients of genes with BAP1.
The left sub-figure shows the sorted correlation of all genes with BAP1 in the dataset including genes with zero-values. The right sub-figure shows the results after using n-normalization (excluding genes with zero-values in the entire dataset).
Excluding genes with zero values did not change the results
We applied two normalization methods: 1) obtaining z-scores for each patient without excluding any genes and 2) eliminating genes with values of zero for all patients, then obtaining z-scores for each patient (for more details see Methods). These two normalization methods led to the same results.
Discussion
Herein, we analyzed publicly available data sets from the TCGA and showed that BAP1 plays a prognostic role in both UM and breast cancer. In contrast, survival was not associated with BAP1 expression in patients with colon cancer. There is a study showing that mutation of BAP1 only significantly affected overall survival in female patients with RCC [10]. In this project, which is the first study to our knowledge that evaluated BAP1 associations with race and gender as well as age, we found that the BAP1 expression does not correlate with race or gender in these data sets. However, the dead UM and breast cancer patients had lower levels of BAP1 expression compared with surviving patients. Saliently, young dead patients with breast cancer had the lowest level of BAP1. This finding suggests that the level of BAP1 could be a signature of aggressive tumors in UM and breast cancer, especially for young breast cancer patients. In contrast, survival was not associated with BAP1 expression in patients with colon cancer.
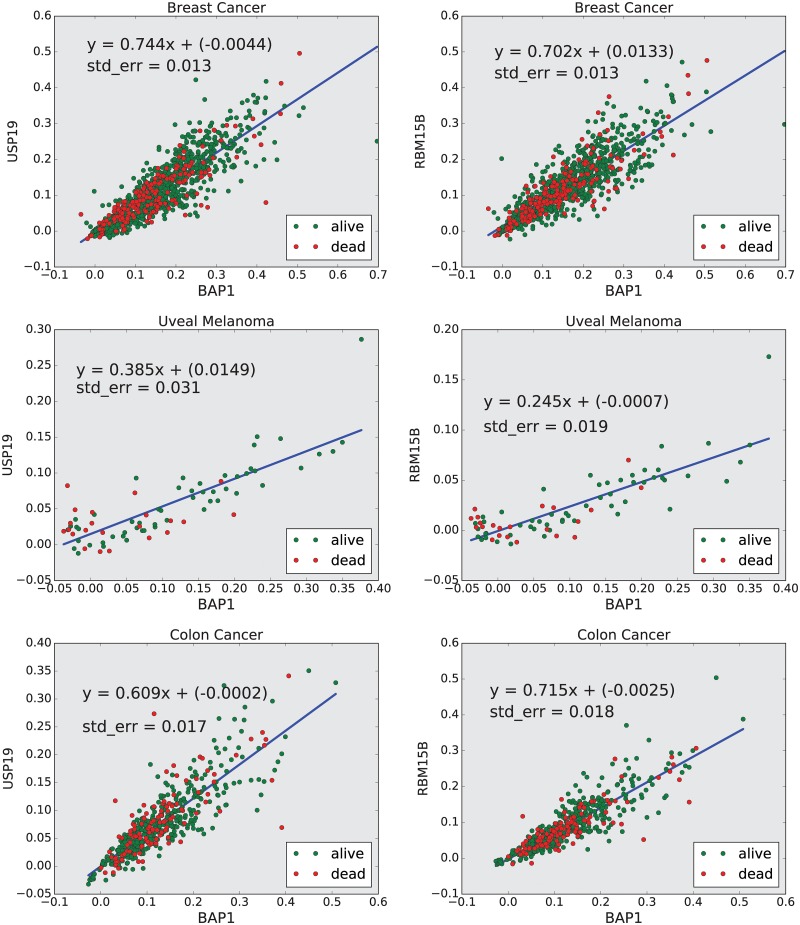
We discovered that there are three genes (DHX30, USP19, and RBM15B), which are highly positively correlated with BAP1 in both black and white breast cancer patients. Among these three genes, two, USP19 and RBM15B are also highly positively correlated with BAP1 in UM. Importantly, USP19 and RBM15B were also among 10 genes with a high positive correlation with BAP1 expression in colon adenocarcinoma. To further learn these relationships, we performed a regression analysis to obtain the linear functions that predict the levels of USP19 and RBM15B from the level of BAP1. We found that the slopes of the lines modeling these linear relationships were higher in breast and colon cancers compared to uveal melanoma (See Fig 6).
Fig 6. Regression analysis.
This figure shows the linear function that predicts the normalized expression value of USP19 and RBM15B from the normalized level of BAP1.
The role of BAP1 in breast cancer is still not clear. BAP1 is located in the 3p21 chromosomal region, which is commonly deleted in breast cancer patients [11]. This suggests that BAP1 could be an important driver for tumorigenesis in breast cancer. However, according to cBioPortal.org, there is a low frequency of BAP1 somatic mutation (1.8%) in breast cancer. Also, a recent study shows that biallelic inactivation of BAP1 is rare in breast cancer [12]. Our data show that BAP1 expression is decreased in a subset of UM and breast cancer patients. This suggests that haploinsufficiency of BAP1, likely as part of the 3p21 chromosomal deletion, could be still important in the pathogenesis of breast cancers.
BAP1, which is a nuclear-localized tumor suppressor protein, has an ubiquitin carboxy-terminal hydrolase (UCH) domain resulting in its deubiquitinase activity. Although earlier studies suggested that BAP1 function is through modulation of the BRCA1, later studies showed that it is an independent tumor suppressor gene [10]. It has been shown that the deubiquitinating activity and nuclear localization are both required for BAP1-mediated tumor suppression [13]. The tumor suppressor function of BAP1 is through its impact on cell cycle regulation, chromatin remodeling and DNA damage. BAP1, which is expressed in a temporal and spatial pattern during breast development and remodeling, binds to the BRCA1 RING finger motif, and pathogenic mutations in that domain negatively impact BAP1 binding [14]. Additionally, a decrease in the level of BRCA1 protein has been observed in the malignant mesothelioma cells with BAP1 deletion, while transduction of the mutants as well as WT BAP1 results in an increase in the level of BRCA1 protein [15]. BAP1 enhances BRCA1-mediated suppression of cell proliferation through stabilizing BRCA1 [14]. However, BAP1 function is not through deubiquitination of the BRCA1/BARD1 complex.
USP19, which has been mostly studied in skeletal muscle atrophy, is a deubiquitinating enzyme [16]. It has been observed that the depletion of USP19, which is located in close proximity to BAP1 at 3p21.3 region, results in defects in cell cycle progression; USP19-depleted cells over-express KPC1 and have reduced rates of growth [17]. It has been also reported that USP19 inhibits TNFα-induced apoptosis via the stabilization of c-IAPs, which are ubiquitin ligases that regulate the stability of a variety of apoptotic and non-apoptotic proteins [18]. Importantly, a mutation in USP19 and LAMB2 has been observed in 12 patients with UM at high-risk of metastasis [19].
The RNA binding motif protein RBM15B (OTT3), which also located in the same chromosomal region 3p21, has been identified as a binding partner of the Epstein-Barr virus mRNA export factor [20] and found to be a cofactor of the nuclear export receptor NXF1 [21]. Recently, it has been shown that RBM15B is located in a nuclear macromolecular complex through its direct binding to CDK11p110-cyclin L complexes and the splicing factor 9G8 [22]. Moreover, RBM15B is a functional competitor of the SR proteins, capable of inhibiting the formation of the spliceosomal E complex and antagonizing the positive effect of the CDK11p110-cyclin L2α complex on splicing [22]. Note that expression of the large CDK11p110 protein kinase isoforms is ubiquitous and constant throughout the cell cycle [23].
Of note, all highly positively correlated genes with BAP1 in UM and breast cancer are located in chromosome 3. In metastatic UM, monosomy of chromosome 3 has been associated with short survival, compared to metastases with disomy or partial change in chromosome 3 [24]. Chromosome 3 has also been reported to play a role in prognosis of some other cancers, including in colon cancer and AML [25, 26]. Additionally, loss of chromosome 3 is well known as a poor prognostic factor in primary UM [5]. Importantly, allelic loss of several distinct regions on chromosome 3p including 3p25, 3p21.22, 3p21.3, 3p12.13 and 3p14 is one of the earliest and most frequent genomic abnormalities involved in a wide spectrum of major epithelial cancers including breast and colon [11, 27]. Chromosome 3p deletions occur in more than 80% of breast carcinomas [27]. This finding suggests that USP19 and RBM15B could be in a functional pathway of BAP1 or could be altered based on regional loss of chromosome 3p21.
In summary, our findings suggest that BAP1 expression has prognostic significance in both UM and breast cancer. Future work to analyze BAP1 expression in other cancers will be performed. An effort to develop targeted therapeutics for cells deficient in BAP1 expression will be of great translational interest. The role of genes RBM15B and USP19 may be important in BAP1 function and give further insight into the mechanism of BAP1 tumor suppression and additional targets for therapy.
Materials and methods
We downloaded the TCGA transcriptome profiling data (HTSeq-FPKM-UQ files) of patients with UM, invasive breast carcinoma, and colon adenocarcinoma, from the GDC data portal (TCGA-UVM, TCGA-BRCA, and TCGA-CODA). For each of these cancer types, we downloaded the data sets data sets based on the age at diagnosis; we downloaded the data sets in the batches of 10 years interval starting from the minimum age at diagnosis. We divided the breast cancer data set into 2 main categories females and males. Since there were only 12 male patients, we only analyzed the data of female patients. We divided the female breast cancer patients to four sub-groups: black-alive (151 cases, 156 HTSeq-FPKM-UQ files), black-dead (29 cases, 31 HTSeq-FPKM-UQ files), white-alive (636 cases, 705 HTSeq-FPKM-UQ files), white-dead (112 cases, 159 HTSeq-FPKM-UQ files) (Table 2).
We divided UM and colon cancer patients into four sub-categories female-alive, female-dead, male-alive, and male-dead. Since the race was unknown for approximately half of these data sets, we only divided these two datasets based on gender, vital status, and age at diagnosis to several sub-categorizes. The TCGA-UVM data set includes 88 cases: 40 females and 44 males (Table 1). Since the number of cases in each category was small, we obtained the correlations with BAP1 in the entire UVM data set rather than in each category. We collected the gene expression data from primary tumors of 456 patients with colon adenocarcinoma from TCGA-COAD project. This data set includes 478 HTSeq-FPKM-UQ files (Table 3).
In a recent study, it has been shown that gene expression profiles of patients have similar distributions, while the distribution of the expression value of each gene is different [28]. Furthermore, the genes’ expression level is a representation of interaction network of genes, if we normalize the value of each genes separately then we loose information. Thus, the best way to normalize the gene expression data sets from the primary tumors is normalizing each file (transcriptome profiling data of each patient), separately. Therefore, we standardized the gene expression profile of each patient separately to normalize the data. We applied two normalization methods: 1) obtaining z-scores for each patient without excluding any genes and 2) eliminating genes with values of zero for all patients, then obtaining z-scores for each patient. We called the second method n-normalization.
We performed several statistical analyses to see if the expression value of BAP1 is different in alive patients versus the dead ones. We applied the Mann-Whitney-Wilcoxon (MWW), also known as Mann-Whitney U-test, which is a nonparametric test of the null hypothesis that it is equally likely that a randomly selected value from one sample will be less than or greater than a randomly selected value from a second sample [29]. We have also used t-test to investigate whether the means of expression level of BAP1 in two groups of alive and dead patients are statistically different from each other.
In order to obtain the genes, which are highly correlated with BAP1, first we normalized data by obtaining z-scores for each file independently. Then, we calculated the Pearson correlation between BAP1 and all other genes. Specifically, we have a data set [p1, ⋯, pn], where n is the number of patients. Each pi is a list of gene expression values, i.e. pi = [g1⋯, gm], where each gj is the expression value of transcript (gene) j, and m is the total number of transcripts (m = 60483). That means the data set is an n × m-dimensional matrix D = [gij], where gij is the expression of transcript j in file (patient) i.
To normalize the data set, we obtained the z-score for each pi. Thus, we got a new matrix , where ’s are normalized values. Then, for each transcript j, we obtained Pearson correlation between the list and the corresponding list for BAP1 ().
Data Availability
The data underlying the results presented in the study are available from the TCGA-UVM, TCGA-BRCA, and TCGA-COAD projects at the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov).
Funding Statement
This research has been supported in part by the Mathematical Biosciences Institute and the National Science Foundation under grant DMS 1440386, and by National Eye Institute award K08EY022672. Additional funds were provided by the Ohio Lions Eye Research Foundation, the Patti Blow Fund in Ophthalmology, and The Ohio State University Department of Ophthalmology and Visual Science. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Online Mendelian Inheritance in Man, OMIM. Baltimore, MD: Johns Hopkins University; 2014. Available from: http://www.omim.org/entry/614327#1.
- 2. Pilarski R, Rai K, Cebulla C, Abdel-Rahman M. BAP1 Tumor Predisposition Syndrome; 1993. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27748099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abdel-Rahman MH, Pilarski R, Cebulla CM, Massengill JB, Christopher BN, Boru G, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. Journal of medical genetics. 2011;48(12):856–9. 10.1136/jmedgenet-2011-100156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harbour JW, Onken MD, Roberson EDO, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science (New York, NY). 2010;330(6009):1410–3. 10.1126/science.1194472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell. 2017;32(2):204–220.e15. 10.1016/j.ccell.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Njauw CNJ, Kim I, Piris A, Gabree M, Taylor M, Lane AM, et al. Germline BAP1 Inactivation Is Preferentially Associated with Metastatic Ocular Melanoma and Cutaneous-Ocular Melanoma Families. PLoS ONE. 2012;7(4):e35295 10.1371/journal.pone.0035295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Piva F, Giulietti M, Occhipinti G, Santoni M, Massari F, Sotte V, et al. Computational analysis of the mutations in BAP1, PBRM1 and SETD2 genes reveals the impaired molecular processes in renal cell carcinoma. Oncotarget. 2015;6(31):32161–32168. 10.18632/oncotarget.5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peña-Llopis S, Vega-Rubín-de Celis S, Liao A, Leng N, Pavía-Jiménez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature Genetics. 2012;44(7):751–759. 10.1038/ng.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rai K, Pilarski R, Cebulla CM, Abdel-Rahman MH. Comprehensive review of BAP1 tumor predisposition syndrome with report of two new cases. Clinical Genetics. 2016;89(3):285–294. 10.1111/cge.12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ricketts CJ, Linehan WM. Gender Specific Mutation Incidence and Survival Associations in Clear Cell Renal Cell Carcinoma (CCRCC). PLOS ONE. 2015;10(10):e0140257 10.1371/journal.pone.0140257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene. 2007;26(52):7283–301. 10.1038/sj.onc.1210547 [DOI] [PubMed] [Google Scholar]
- 12. Chui J, Singh A, Gill AJ. Loss of BAP1 expression is very rare in breast carcinoma. Pathology. 2017;49(5):557–560. 10.1016/j.pathol.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 13. Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, et al. BRCA1-Associated Protein-1 Is a Tumor Suppressor that Requires Deubiquitinating Activity and Nuclear Localization. Cancer Research. 2008;68(17):6953–6962. 10.1158/0008-5472.CAN-08-0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh La, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16(9):1097–112. 10.1038/sj.onc.1201861 [DOI] [PubMed] [Google Scholar]
- 15. Hakiri S, Osada H, Ishiguro F, Murakami H, Murakami-Tonami Y, Yokoi K, et al. Functional differences between wild-type and mutant-type BRCA1-associated protein 1 tumor suppressor against malignant mesothelioma cells. Cancer Science. 2015;106(8):990–999. 10.1111/cas.12698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wing SS. Deubiquitinases in skeletal muscle atrophy. The International Journal of Biochemistry & Cell Biology. 2013;45(10):2130–2135. 10.1016/j.biocel.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu Y, Adegoke OaJ, Nepveu A, Nakayama KI, Bedard N, Cheng D, et al. USP19 deubiquitinating enzyme supports cell proliferation by stabilizing KPC1, a ubiquitin ligase for p27Kip1. Molecular and cellular biology. 2009;29(2):547–58. 10.1128/MCB.00329-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mei Y, Hahn AA, Hu S, Yang X. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. The Journal of biological chemistry. 2011;286(41):35380–7. 10.1074/jbc.M111.282020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake S, Damato B, Taktak A, Lloyd B, Olohan L, Liu X, et al. Combining SNP microarray analysis and whole exome sequencing to identify regulators of metastasis in uveal melanoma. NCRI Cancer Conferences. 2012;.
- 20. Hiriart E, Gruffat H, Buisson M, Mikaelian I, Keppler S, Meresse P, et al. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. The Journal of biological chemistry. 2005;280(44):36935–45. 10.1074/jbc.M501725200 [DOI] [PubMed] [Google Scholar]
- 21. Uranishi H, Zolotukhin AS, Lindtner S, Warming S, Zhang GM, Bear J, et al. The RNA-binding Motif Protein 15B (RBM15B/OTT3) Acts as Cofactor of the Nuclear Export Receptor NXF1. Journal of Biological Chemistry. 2009;284(38):26106–26116. 10.1074/jbc.M109.040113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Loyer P, Busson A, Trembley JH, Hyle J, Grenet J, Zhao W, et al. The RNA binding motif protein 15B (RBM15B/OTT3) is a functional competitor of serine-arginine (SR) proteins and antagonizes the positive effect of the CDK11p110-cyclin L2α complex on splicing. The Journal of biological chemistry. 2011;286(1):147–59. 10.1074/jbc.M110.192518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiang J, Lahti JM, Grenet J, Easton J, Kidd VJ. Molecular cloning and expression of alternatively spliced PITSLRE protein kinase isoforms. The Journal of biological chemistry. 1994;269(22):15786–94. [PubMed] [Google Scholar]
- 24. Abdel-Rahman MH, Cebulla CM, Verma V, Christopher BN, Carson WE, Olencki T, et al. Monosomy 3 status of uveal melanoma metastases is associated with rapidly progressive tumors and short survival. Experimental eye research. 2012;100:26–31. 10.1016/j.exer.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsai MH, Fang WH, Lin SH, Tzeng ST, Huang CS, Yen SJ, et al. Mapping of Genetic Deletions on Chromosome 3 in Colorectal Cancer: Loss of 3p25-pter is Associated with Distant Metastasis and Poor Survival. Annals of Surgical Oncology. 2011;18(9):2662–2670. 10.1245/s10434-011-1603-9 [DOI] [PubMed] [Google Scholar]
- 26.Langholtz J. MDS Patients With Chromosome 3 Abnormalities May Have High Risk Of AML Progression And Shorter Survival; 2011.
- 27. Angeloni D. Molecular analysis of deletions in human chromosome 3p21 and the role of resident cancer genes in disease. Briefings in functional genomics & proteomics. 2007;6(1):19–39. 10.1093/bfgp/elm007 [DOI] [PubMed] [Google Scholar]
- 28. Shahriyari L. Effect of normalization methods on the performance of supervised learning algorithms applied to HTSeq-FPKM-UQ data sets: 7SK RNA expression as a predictor of survival in patients with colon adenocarcinoma. Briefings in Bioinformatics. 2017; p. 1–10. [DOI] [PubMed] [Google Scholar]
- 29. Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. The Annals of Mathematical Statistics. 1947;18(1):50–60. 10.1214/aoms/1177730491 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying the results presented in the study are available from the TCGA-UVM, TCGA-BRCA, and TCGA-COAD projects at the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov).