Abstract
Host-defense peptides (HDPs) feature evolution-tested potency against life-threatening pathogens. While piscidin 1 (p1) and piscidin 3 (p3) are homologous and potent fish HDPs, only p1 is strongly membranolytic. Here, we hypothesize that another mechanism imparts p3 strong potency. We demonstrate that the N-termini of both peptides coordinate Cu2+ and p3-Cu cleaves isolated DNA at a rate on par with free Cu2+ but significantly faster than p1-Cu. On planktonic bacteria, p1 is more antimicrobial but only p3 features copper-dependent DNA cleavage. On biofilms and persister cells, p3-Cu is more active than p1-Cu, commensurate with stronger peptide-induced DNA damage. Molecular dynamics and NMR show that more DNA-peptide interactions exist with p3 than p1, and the peptides adopt conformations simultaneously poised for metal- and DNA-binding. These results generate several important conclusions. First, homologous HDPs cannot be assumed to have identical mechanisms since p1 and p3 eradicate bacteria through distinct relative contributions of membrane and DNA-disruptive effects. Second, the nuclease and membrane activities of p1 and p3 show that naturally occurring HDPs can inflict not only physicochemical but also covalent damage. Third, strong nuclease activity is essential for biofilm and persister cell eradication, as shown by p3, the homolog more specific toward bacteria and more expressed in vascularized tissues. Fourth, p3 combines several physicochemical properties (e.g., Amino Terminal Copper and Nickel binding motif; numerous arginines; moderate hydrophobicity) that confer low membranolytic effects, robust copper-scavenging capability, strong interactions with DNA, and fast nuclease activity. This new knowledge could help design novel therapeutics active against hard-to-treat persister cells and biofilms.
Keywords: antimicrobial peptide, copper, DNA damage, host defense, reactive oxygen species
Introduction
Host-defense (antimicrobial) peptides (HDPs), of which more than 2700 have been reported, are innate immune effector molecules that play a critical role as a first line of defense in all kingdoms of life investigated for this function [1–4]. Their relevance to mammalian health, broad-spectrum antimicrobial activity, low prevalence of antibiotic resistance, and immunomodulatory effects have motivated significant efforts to use them as templates to develop novel antimicrobial therapeutics active on drug resistant bacteria [1,2,5,6]. Fighting drug-resistant pathogens is particularly urgent given the failure of conventional drugs to eradicate ‘super bugs’ such as the Klebsiella pneumoniae strain that recently infected an individual and could not be overcome by the 26 antibiotics available in the US [7]. Another difficult challenge associated with pathogenic bacteria is combatting biofilms and persister cells due to their growth properties and resilience to agents otherwise active on planktonic cells (free flowing bacteria in suspension) [8,9]. Here, we focus on the direct antimicrobial effects of two highly potent HDPs from the piscidin family with the goal of explaining their differing efficacy and uncovering mechanistic strategies that go beyond physicochemical disruption of targets to eradicate multiple modes of bacterial growth.
HDPs share physicochemical features (e.g., amphipathicity, cationicity, and conformational plasticity) [2,4,10] that enable them to interact with multiple components in bacterial cells, including anionic cell membranes, nucleic acids, and enzymes [11–13]. Since amphipathic cationic peptides display membrane activity on model membranes and HDPs that directly eradicate bacteria must, at least initially, interact with membranes, mechanistic studies of HDPs generally focus on cell membranes as their primary targets [2,4,10]. However, recent studies have identified a handful of HDPs that act by mechanisms other than membrane disruption. For example, some HDPs exhibit a direct causal relationship between intracellular targeting and cell death [14–16] while human defensin-6 has been demonstrated to perform extracellular attack of bacteria via entrapment in nanonets [17]. Another emerging notion about HDPs is that their damage sometimes goes beyond physicochemical disruption of targets and includes covalent modifications via oxidative stress [14].
It is in the context of this shifting paradigm about how HDPs achieve high antimicrobial potency that we investigated two isoforms from the piscidin family that strikingly vary in therapeutic index (TI) and membrane activity. Piscidins, which were the first HDPs discovered in the mast cells of vertebrates, kill a broad spectrum of microbes, including methicillin-resistant Staphylococcus aureus [18,19]. Initially isolated from the hybrid striped seabass, piscidins have also been found in other teleost fish [18,20]. Their ubiquity is particularly significant knowing that fish are exposed to an onslaught of life-threatening pathogens in their environment.
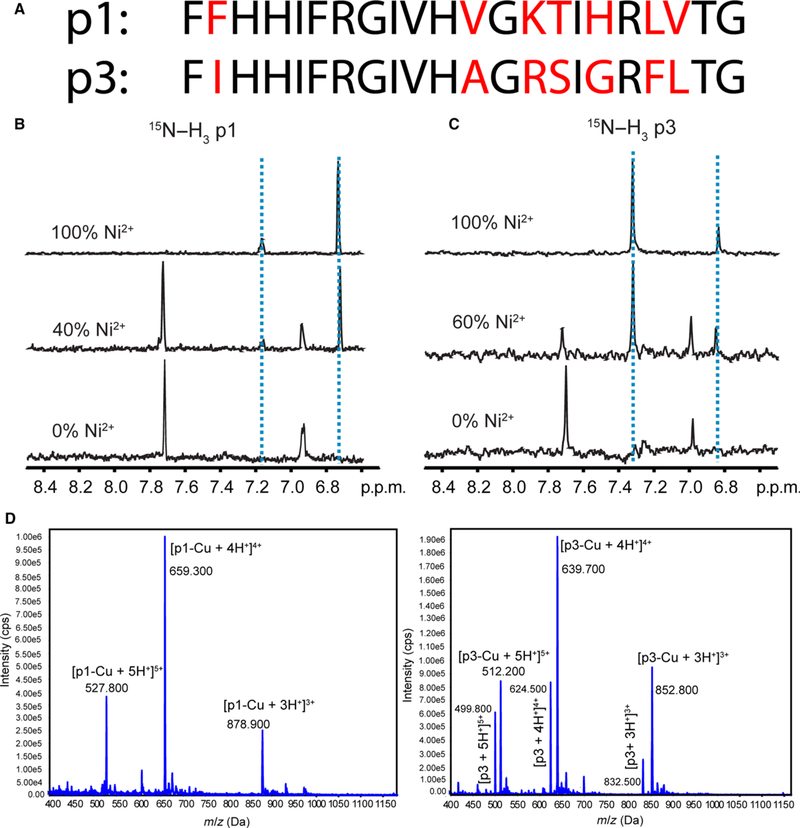
The specific piscidins selected for our studies are piscidin 1 (p1, FFHHIFRGIVHVGKTIHRLVTG-NH2) and piscidin 3 (p3, FIHHIFRGIVHAGRSIGRFLTG-NH2) from hybrid striped seabass (Fig. 1A). They represent an interesting pair of piscidins since they are homologous and highly potent but differentially expressed, and their biological activities differ for unknown reasons [18,19]. P1 is more antimicrobial than p3 on some planktonic bacteria. However, p3 has higher specificity and TI (54 versus 10 for p1 based on activity values given in Ref. [19]), reflecting its particularly low hemolytic activity (EC50 of 300 μg·mL−1 for p3 versus 30 μg·mL−1 for p1 [19]).
Fig. 1.
Structural characterization of copper binding to p1 and p3. (A) Amino acid sequences of p1 and p3 [19]. The nonconserved residues are shown in red. (B) and (C) One dimensional slices from solution NMR heteronuclear multiple quantum correlation (HMQC) spectra of 15N[α, δ, ε]-His3-labeled p1 (B) and 15N[α, δ, ε]-His3-labeled p3 (C) titrated with Ni2+ indicating perturbation of chemical shifts of d-proton (7.0 p.p.m.) and δ-proton (7.7 p.p.m.) of His3. (D) Mass spectrometry of piscidin exposed to Cu2+. Cu2+ and piscidin were mixed in a 1 : 1.2 molar ratio and subsequently injected into an electrospray ionization MS. Peaks corresponding to a 1 : 1 stoichiometry of binding appear in the mass spectra. Peaks at m/z 499.8, 624.5, and 832.5 in the bottom panel correspond to the +5, +4, and +3 charged states of naive p3.
Given the amphipathicity and membrane affinity of p1 [19,21,22], it was initially assumed that piscidins exclusively act by disrupting the structure of bacterial membranes. However, we recently showed that both p1 and p3 localize on bacterial cell membranes as well as intracellularly at sub-inhibitory concentrations [23]. More specifically, we demonstrated that both p1 and p3, which are intrinsically disordered in aqueous solution, fold into similar α-helical conformations when they are bound to model membranes and isolated DNA [21–23]. We also showed that while p1 is more hydrophobic and membrane disruptive, p3 is more neutralizing and condensing to isolated DNA [23]. These results led us to hypothesize in this new study that p3 may be targeting DNA rather than membranes to achieve strong potency.
In exploring how p3 could fatally damage DNA, we noted that known members of the piscidin family contain an Amino Terminal Copper and Nickel (ATCUN) binding motif. The ATCUN motif, with the consensus sequence, XXH- (e.g., FFH for p1 and FIH for p3) binds to Cu2+ and Ni2+ ions with high affinity via the N-terminal amino group, the two subsequent backbone amide nitrogens, and the δ-nitrogen of His-3 [24]. Cowan and colleagues [25] demonstrated that the noncanonical reactive oxygen species (ROS) directly formed by copper-ATCUN complexes retain the oxidizing power of classical ROS and can damage covalent bonds within minutes. When they added the ATCUN motif to a short synthetic peptide that recognizes bacterial Holliday junctions, they observed significant nuclease activity on isolated DNA and improved minimum inhibitory concentrations (MICs) on planktonic cells [26]. The Angeles-Boza group obtained similar results as well as increased DNA damage in bacterial cells by adding the motif to modified DNA-interacting antimicrobial peptides, such as buforin II [27]. While these studies correlated ATCUN-induced DNA damage and antimicrobial effects, they were limited to planktonic cells and did not explore the relevance of this mechanism to naturally occurring HDPs. In particular, it remains unknown whether the ATCUN motif could boost the activity of naturally occurring HDPs in a native setting where other mechanisms, such as membrane lysis, are not well suited due to their cytotoxicity or lack of effectiveness on resilient bacterial forms, such as biofilms and persister cells.
In this investigation, not only did we investigate the relevance of the ATCUN motif for the antimicrobial activity of naturally occurring p1 and p3 but also tested the hypothesis that the ATCUN motif and arginine-rich sequence of p3 afford the peptide a robust mode of DNA targeting that compensates for weak membrane activity. We characterized the antimicrobial activity of p1 and p3 on planktonic, biofilm, and persister cells, and performed the experiments under different conditions of copper availability. We also used isolated DNA to biochemically quantify the nuclease activity of p1 and p3, and biophysically examined their interactions with DNA. The results presented next substantiate our hypothesis, and in turn identify evolutionary and structurally homologous HDPs that act through different relative contributions of physicochemical membrane disruption and covalent DNA damage to induce cell death. They also show that in the presence of copper, p3, which is less active than p1 on planktonic cells, exhibits a nuclease activity phenotype that renders it more active on biofilms and persisters. Since p1 and p3 are differentially expressed in fish tissues, we explore insightful relationships between their mechanisms of action, specificity, and physiological localizations. These combined findings help us better understand the sophisticated strategies by which HDPs contribute to innate immunity.
Results
Both p1 and p3 bind Cu2+ in vitro
Both p1 and p3 are ATCUN-containing peptides that have the potential to coordinate Cu2+. To demonstrate Cu2+-binding and obtain the stoichiometry of each piscidin-Cu complex, we incubated an equimolar solution of each peptide and Cu2+, and subjected the mixture to electrospray ionization-mass spectrometry (ESI-MS) analysis. We observed m/z values of 878.9, 659.3, and 527.8 for p1-Cu, and m/z 852.8, 639.7, and 512.2 for p3-Cu. These values are consistent with the [M + 3H]3+, [M + 4H]4+, and [M + 5H]5+ multiply charged species of a 1 : 1 complex for each isoform (Fig. 1D). We also note that the molecular weight of each peptide changes by exactly the amounts expected when the ATCUN motif is involved in binding the metal. Indeed, metal coordination by the ATCUN motif, which occurs at the amino end of the peptide, involves the deprotonated form of the amino end and the deprotonation of the amide sites at positions 2 and 3 [24]. Compared to the monoisotopic molecular weight of the neutral peptide, we thus expect m/z values of the metal-bound peptides to reflect the loss of two protons due to amide deprotonation (−2) and the gain of a copper ion (+63). The net change of 61 is exactly what is observed. For instance, for the [M + 3H]+3 species associated with p3, the change in m/z upon metallation is exactly 20.3, indicating that the ATCUN motif rather than only side chains are coordinating the metal. We then employed solution NMR to demonstrate that the metal ion specifically binds to the ATCUN motif. Because Cu2+ is paramagnetic, any proton in its vicinity will exhibit NMR signal loss. To circumvent this problem, we used Ni2+ as a spectroscopic probe (both Cu2+ and Ni2+ bind to the ATCUN motif in a similar fashion and with comparable dissociation constants [24]). Upon titrating each peptide with Ni2+, we observed an upfield shift of the aromatic proton signals in the histidine at position 3, His3 (Fig. 1A–C), consistent with metal binding to the ATCUN motif. Each titration was characterized by a 1 : 1 ratio between peptide and nickel ions and a complete disappearance of the downfield imidazole protons resonances from His3.
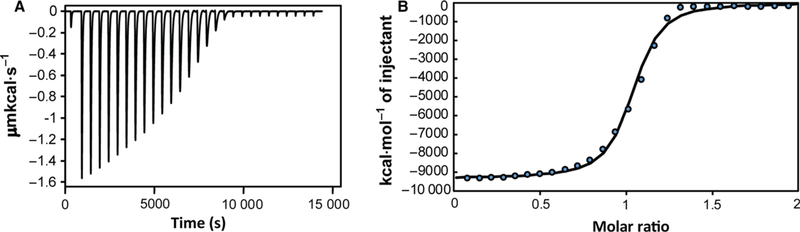
Piscidin was then titrated with Cu2+ at pH 7.4 using isothermal titration calorimetry (ITC), as shown in Fig. 2. Due to the high binding affinity of Cu2+ to ATCUN sequences, a displacement experiment with a competitor that binds copper was necessary [28–32]. To avoid issues associated with precipitation of copper hydroxide at basic pH and account for secondary effects that could affect the thermodynamics measured by ITC (e.g., copper binding to other molecules than the peptide; proton exchange with the buffer), we followed the comprehensive approach used by Trapaidze et al. [28] and obtained not only the apparent binding constant but also the corrected (‘conditional’) binding constant between copper and the ATCUN motif of piscidin at pH 7.4. Similarly to prior investigations of copper-binding proteins [29,30,32], this protocol uses glycine as a competitor and HEPES as the buffer. But the authors went a step further and derived a complete set of equations to take into account that a Cu-Gly2 complex is formed, Cu2+ interacts with HEPES, and there may be buffer protonation/deprotonation effects in the reaction mixture. Our ITC data obtained at pH 7.4 (Fig. 2) yielded an apparent dissociation constant, of 5.63 (± 0.09) × 10−7 M and stoichiometry of 1.0. Using this value in Eqn (1) together with 5 mM for the total glycine concentration, 10 mM for the HEPES concentration, , , , , and ([28] and references therein), we extracted a dissociation constant, of 3.29 (± 0.05) × 10−13, corresponding to a binding association constant, of 3.04 (± 0.05) × 1012. This value for p1 agrees well with that obtained for DAHK [28] that was studied by the same approach and various ATCUN peptides investigated by visible spectroscopy [29]. Since p1 behaves similarly to other ATCUN peptides and the residues are highly conserved at the amino end of p1 and p3, we expect a comparable binding affinity of p3 for copper. Overall, the combined MS, NMR, and ITC results indicate that in vitro p1 and p3 bind to Cu2+ in a 1 : 1 fashion using their ATCUN motif, and that no other potential ligands in the peptide backbone compete for the Cu2+ ions.
Fig. 2.
Titration of piscidin by Cu2+ in the presence of glycine as a competitor. (A) ITC data showing twenty-eight 10 μL-injections of 900 μM Cu2+ into 90 μM p1 performed in the presence of 5 mM glycine and 10 mM HEPES buffer (pH 7.4) at 25 °C. (B) Integrated titration data at increasing molar ratio of copper:piscidin showing the binding isotherm (with the individual experimental data points showing as blue circles) and the least-square fit of the data assuming a one binding-site model (solid line). This isotherm yielded an apparent binding constant between piscidin and Cu2+ that was used to obtain the corrected binding constant between p1 and Cu2+ at pH 7.4, as described in the text.
p3-Cu cleaves DNA faster than p1-Cu in vitro
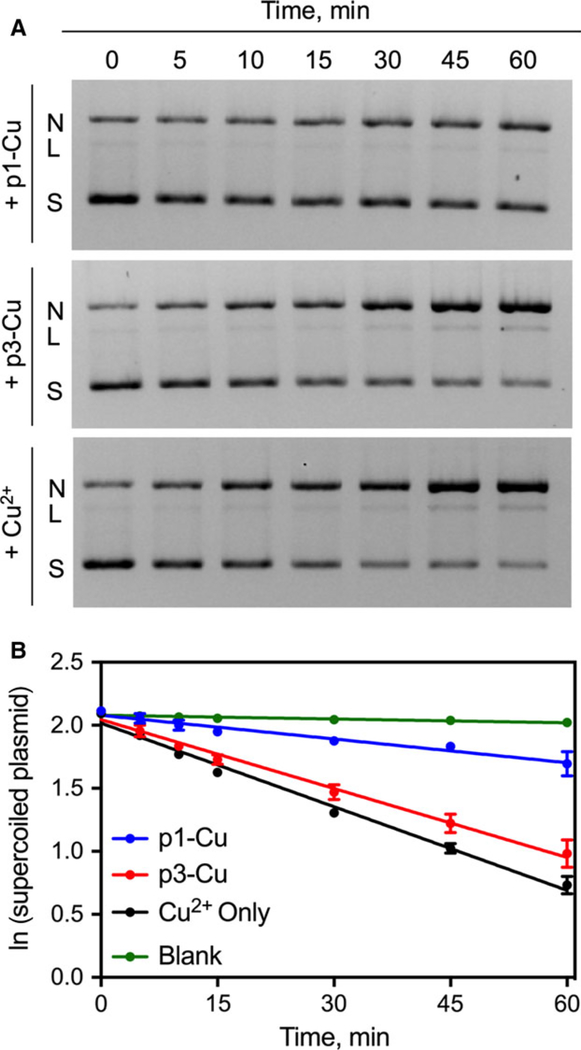
Nuclease activity is a phenotype of p1 and p3 that is expected only if the peptides are metallated. To quantify the rate at which the metallated p1 and p3 nick DNA, we monitored the time-dependent cleavage of the plasmid pUC19 when incubated with the metallated peptides in the presence of H2O2 and ascorbic acid. We observed a progressive loss of the supercoiled (S) form with a concomitant increase in the nicked (N) and linearized (L) forms of the plasmid (Fig. 3A). When the data were fitted into a pseudo-first order rate equation (, Fig. 3B), p3-Cu was found to cleave DNA about three-fold faster than p1-Cu and in par with free Cu2+ (Table 1). Interestingly, our voltammetric studies provided redox potentials (Table 1) indicating that thermodynamically, p1-Cu is slightly superior for the conversion of Cu3+ to Cu2+. To further analyze the DNA cleavage data, we applied the Freifelder-Trumbo (F–T) statistical analysis. The F–T relationship suggests that if the cleavage occurs via random mechanisms, n1/n2 ≥ 100, where n1 = number of single-strand breaks and n2 = number of double-strand breaks [33]. We calculated n1 and n2 values using Eqns (2) and (3) given in the Materials and methods. The lower n1/n2 ratio for p3-Cu relative to p1-Cu (Table 1) is consistent with a more concerted mechanism of DNA cleavage by p3-Cu. Overall, these experiments demonstrate that p3-Cu has faster nuclease activity than p1-Cu and appears to operate via a more concerted mechanism.
Fig. 3.
Kinetic characterization of DNA Cleavage by p1-Cu and p3-Cu. (A) Electrophoretic monitoring of piscidin-induced cleavage of pUC19 showing loss of supercoiled (S) plasmid with corresponding increase in the nicked (N) and linearized (L) form. (B) Pseudo first-order integrated rate law plots of in vitro DNA cleavage promoted by the piscidins. Blank runs represent experiments with H2O2 and ascorbic acid but without any metallopeptide. Data represent mean ± SEM (n = 3).
Table 1.
Summary of values obtained from electrochemal, kinetic, and statistical analysis of metallopeptides and their in vitro DNA cleavage activity.
| Parameter | p1-Cu | p3-Cu | Cu2+ |
|---|---|---|---|
| E° vs NHEa (V) | 0.990 | 0.972 | Nd |
| kcleavage (× 10−3 min−1) | 6.3 ± 0.5 | 18.2 ± 0.7 | 22.1 ± 0.6 |
| n1/n2 | 34 | 27 | 27 |
The standard potential was referenced to H+/H2 couple, the normal hydrogen electrode (NHE).
The bactericidal activity of p3 strongly depends on Cu2+ and involves DNA damage in E. coli
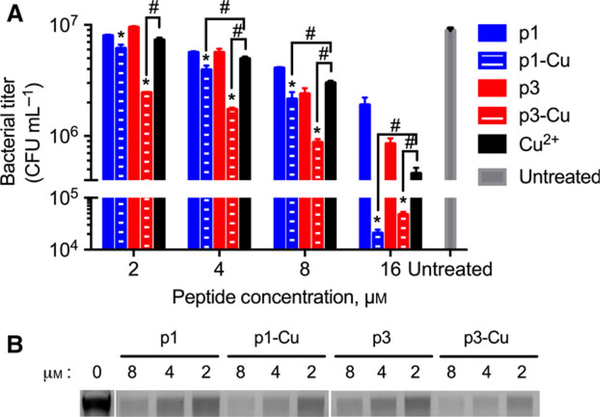
Since both p1-Cu and p3-Cu nick DNA, we measured their MICs on planktonic cells with or without preincubation with an equimolar amount of Cu2+. Consistent with previous studies [19,34], we found that for all Gram-negative bacteria tested, p1 was more active than p3, and that against the Gram-positive S. aureus, both peptides exhibited identical MICs (Table 2). Interestingly, no change in MIC was observed upon preincubation of the peptides with Cu2+. The Mueller-Hinton Broth (MHB) contains ~78 μM of copper [35], a concentration in excess of the highest peptide concentration tested (16 μM). Thus, both p1 and p3 are very likely copper-saturated in the growth medium, explaining the identical MICs. When tested against the isogenic mutant strain Escherichia coli TD172 (a strain harboring a deletion of the recA gene, essential for DNA damage repair), a four-fold increase in p3 activity was observed (relative to wild-type E. coli) whereas the activity of p1 was unaffected. Thus, a strong correlation exists between DNA damage and the antimicrobial efficacy of p3.
Table 2.
Minimum Inhibitory Concentration (MIC) of piscidin isoforms against selected bacteria obtained from a standard broth microdilution method.
| Bacteria | MIC, μM |
|||
|---|---|---|---|---|
| p1 | p1-Cu | p3 | p3-Cu | |
| E. coli (WT) | 4 | 4 | 8 | 8 |
| E. coli (ΔrecA) | 4 | 4 | 2 | 2 |
| S. aureus | 2 | 2 | 2 | 2 |
| P. aeruginosa | 16 | 16 | 32 | 32 |
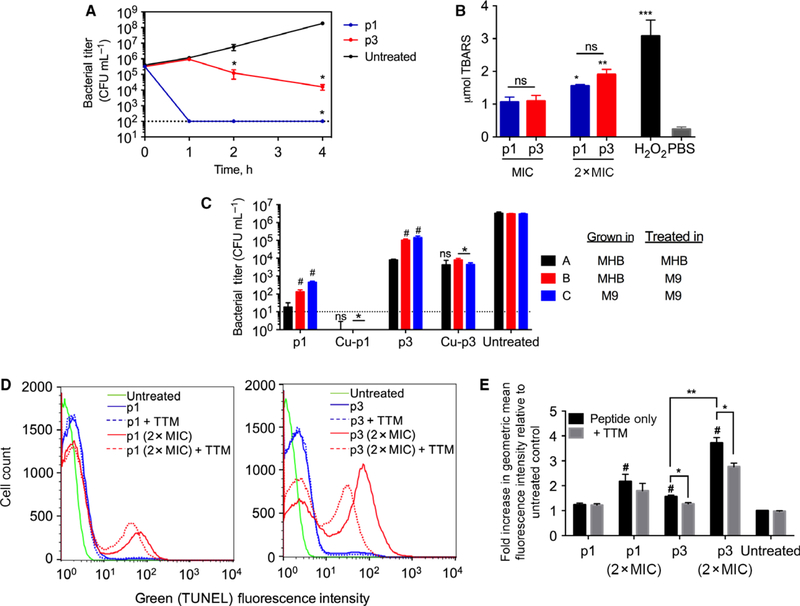
We subsequently obtained the time-kill curves of p1 and p3 at their MIC. Within the first hour of exposure to p1, bacterial viability decreased by 4 orders of magnitude relative to the control (Fig. 4A), leading to rapid sterilization of the culture. After 4 h, p1 reduced bacterial viability by 6 orders of magnitude. The behavior of p3 was remarkably different from that of p1 as it did not induce bacterial death during the first hour after addition. Only after a one-hour lag phase during which bacteria grew by 0.5 log did p3 strongly reduce bacterial survival (bacterial viability decreased by ~ 2 logs over the 3 h after the lag phase). These experiments show that the mechanism of cell death is slower with p3 than p1.
Fig. 4.
Activity of p1 and p3 against planktonic bacteria. (A) Time-kill kinetics curves of piscidin-treated E. coli. The dotted line represents the limit of detection of CFU enumeration (mean ± SEM, n = 3 duplicates). (B) E. coli cells were treated with p1 or p3 at the indicated concentrations and subsequently harvested for peroxidized lipid quantification using a standard assay measuring thiobarbituric acid reactive substances (TBARS). *P < 0.05; **P < 0.01; ***P < 0.005 compared to the PBS control. ns = not significant. Bars represent mean ± SEM (n = 3 duplicates). (C) E. coli cells were grown in either complex (MHB) or minimal media (M9 + glucose) and treated with peptides at their MIC for 30 min (p1 and p1-Cu) or 4 h (p3 and p3-Cu), based on the time-kill traces in (A). The limit of detection for the assay is shown by the dotted line; bars for p1-Cu fall below this limit. Data represent mean ± SEM (n = 3 duplicates). ns = not significant; *P < 0.05 compared to corresponding nonmetallated peptide; #P < 0.05 compared to condition A. (D) Representative histograms obtained from flow cytometric analysis of piscidin-treated E. coli. The fluorescence intensity is directly proportional to the amount of DNA strand breaks. (E) The geometric mean fluorescence intensity from (D) was normalized against that of the untreated cells (assigned a value of 1). Bars represent mean ± SEM (n = 3 duplicates). *P < 0.05, **P < 0.01, and #P < 0.05 compared to untreated control.
Knowing the MICs of p1 and p3 in copper-rich media and their time-kill profiles, we performed experiments to investigate the importance of copper depletion on peptide efficacy. Bacteria were grown and exposed to piscidin under three sets of conditions: (a) grown and treated with HDP in MHB media; (b) grown in MHB and treated with HDP in minimal media, M9; (c) grown and treated with HDP in M9. Each peptide was tested at its MIC (4 and 8 μM for p1 and p3, respectively) and a time when peptide-induced cell death was clearly occurring based on the time-kill curves (30 min for p1 and 4 h for p3). In all of these experiments, Cu2+ was present at nonantimicrobial concentrations since its MIC on E. coli is > 2 mM [26]. As indicated in Fig. 4C, naïve peptides in MHB and premetallated peptides were similarly effective within experimental error, in agreement with peptide metallation readily occurring in the MHB media. For both p1 and p3, the drop in efficacy from highest (condition A) to lowest (condition C) copper availability was on the order of 2 logs at the investigated time points. Since the two piscidin-Cu2+ combinations were more antimicrobial than either Cu2+ or piscidin, both peptides act synergistically with Cu2+ to kill planktonic cells. Synergy may occur because the peptide portion of the Cu-peptide complex hauls the oxidizing agent to vital molecules (e.g., DNA, lipids). Interestingly, p1-Cu and p3-Cu were as effective in MHB as M9 media, thus they must tightly bind copper and not release it even when copper is scarce in the surrounding. Notably, after 4 h at its MIC, p3-Cu was far from having completely inhibited growth while p1-Cu at its MIC had eradicated all bacteria after 30 min. This further underscores the slow killing process of p3-Cu and its ability to progressively catch up and inhibit growth even after a lag phase. Overall, these assays reveal synergy between copper and the two piscidin homologs, and confirm the different timescales of their killing effects on planktonic cells (Fig. 4A).
p1 and p3 produce similar levels of phospholipid peroxidation
Using a standard thiobarbituric acid (TBA) assay on planktonic E. coli, we tested the ability of p1 and p3 to covalently damage phospholipids via peroxidation. We found a three-fold increase in peroxidized lipids in E. coli treated with each peptide at its MIC (Fig. 4B), indicating that both peptides sensitize bacterial cell membranes via lipid peroxidation. However, no statistical difference between p1- and p3-treated cells was observed. Thus, lipid peroxidation occurs in the presence of p1 and p3 but does not explain why their MICs and time-kill curve behaviors differ on planktonic cells.
p3 cleaves DNA in live E. coli in a Cu2+-dependent manner
We utilized the Terminal deoxynucleotidyl transferase dUTP Nick End Labeling (TUNEL) assay to assess covalent damage to DNA in planktonic wild-type E. coli exposed to p1 and p3 for 2 h, i.e., once the peptides have initiated cell death. Since preincubating the peptides did not alter their MICs, we mixed the naïve peptides (i.e., without preincubation with Cu2+) with bacteria in the presence or absence of 100 μM tetrathiomolybdate (TTM), a copper-selective, cell-permeable chelator [36]. At 100 μM, TTM does not affect the viability of cells (our assays determined the MIC to be 10 mM), thus it is an appropriate concentration to generate nonlethal metal depletion and test if this reduction affects peptide-induced DNA damage. At 1× MIC, p1 had no notable effect on the quantity of DNA with nicked ends relative to untreated cells. When it was used at 2× MIC, a two-fold increase in nicked DNA was observed, which was unchanged within the margin of error when TTM was present (Fig. 4D,E). In sharp contrast, p3 used at its MIC induced a statistically significant 1.5-fold increase in TUNEL fluorescence, which decreased slightly in the presence of TTM (P < 0.05). When used at 2× MIC, p3 generated a ~four-fold greater DNA damage, which dropped considerably in the presence of TTM. Notably, TTM treatment alone did not alter the TUNEL fluorescence of the control cells, indicating that the impact of TTM on p3 is strictly due to decreased copper availability. Overall, these TUNEL data show that p1, which cleaves DNA much more slowly than p3, generates relatively small amounts of nicked DNA and the effects are not Cu2+-dependent while they are for p3 tested under the same conditions. This may be due to the ability of p1 to indirectly induce DNA damage (e.g., induction of ROS-generating pathways) and/or directly damage DNA using a source of copper unaffected by 100 μM TTM. In the case of p3, the results presented thus far reveal a robust in vitro nuclease activity, large magnitude of DNA damage at the time of cell death, and strong copper-dependence of DNA damage and cell death. There is thus a clear relationship between the metallation of p3, its killing efficacy, and the extent of p3-induced DNA damage in planktonic bacteria.
Synergy between p1 and p3 requires an attenuated DNA repair system
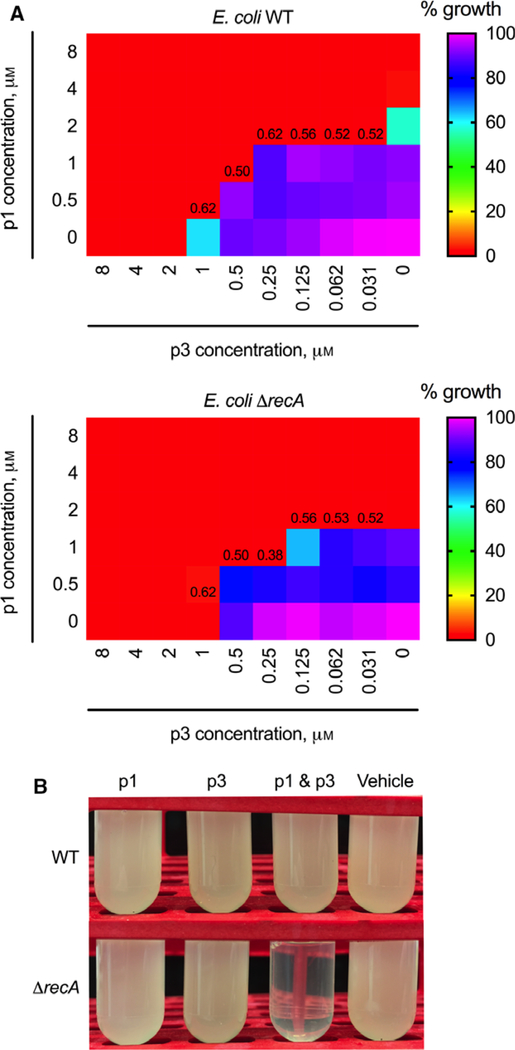
Given the contrasted mechanistic behaviors of p1 and p3, we tested whether they are synergistic on planktonic cells. We utilized the checkerboard assay to obtain their fractional inhibitory concentrations (FIC) and corresponding FIC indices. We observed additive interactions between p1 and p3 when tested against WT E. coli. However, when the ΔrecA mutant was used, we observed a synergistic interaction with a minimum FIC index of 0.38 (Fig. 5A), confirming the importance of targeting DNA to exacerbate cell death. The synergy was also obvious in growth inhibition assays showing that sterilization occurred at sub-MIC concentrations of combined p1 and p3 but not when either peptide was present alone (Fig. 5B).
Fig. 5.
Antimicrobial action of combined p1 and p3. (A) The checkerboard assay was used to assess synergy between p1 and p3 following an overnight incubation with varying ratios of both peptides to either WT E. coli (top) or ΔrecA mutant (bottom). Numbers at the boundary of the zone of inhibition and zone of growth represent FIC indices calculated from the corresponding wells. Values correspond to the mean collected on three duplicates (n = 3). (B) p1 and p3 at sub-inhibitory concentrations were used either alone or in combination to treat E. coli. Growth was inhibited when the ΔrecA mutant was treated with both p1 and p3. Data are representative of three duplicates (n = 3).
Both p1 and p3 act synergistically with Cu2+ to eradicate biofilms with p3-Cu being more effective than p1-Cu
Biofilms are implicated in recurring and persistent infections due to their recalcitrance to antibiotics [8]. To determine if p1 and p3 can inhibit Pseudomonas aeruginosa PA01 biofilms, we treated preformed biofilms (grown in rich medium) with peptides and their corresponding copper complexes. We rinsed the biofilms with copper-free MOPS buffer before exposing them to the peptides dissolved in the same buffer. We used this buffer so that the only source of copper for the peptides is the exogenously added Cu2+ or the bacterial bioavailable pool. We observed a modest inhibitory activity for both nonmetallated piscidins, effecting one order of magnitude decrease in bacterial viability at 16 μM HDP (Fig. 6A). When the peptides were preincubated with Cu2+ to form peptide-Cu complexes, a stronger biofilm clearance effect was observed and became more pronounced at higher concentrations. While P. aeruginosa biofilms are susceptible to the toxic effects of free Cu2+, we obtained a statistically significant (P < 0.01) difference between the inhibitory activities of the metallated piscidins and free Cu2+. Thus, the two antimicrobial agents act synergistically. Importantly, these assays also show that p3, which is less active than p1 on planktonic bacteria, is the stronger antibiofilm agent, especially in the presence of copper.
Fig. 6.
P. aeruginosa biofilm growth inhibition by p1, p3, and their respective copper complexes. (A) 24-Hour old PA01 biofilms were treated with p1 and p3 at the indicated concentrations for 3 h followed by CFU enumeration. Bars represent mean ± SEM (n = 3 quadruplicates). *P < 0.05, #P < 0.01 compared to the corresponding nonmetallated peptide. (B) Electrophoretic characterization of cleavage of biofilm extracellular DNA (eDNA) following exposure to the indicated piscidin species at the indicated concentrations.
Cleavage of biofilm extracellular DNA occurs during piscidin treatment and p3-Cu is more effective than p1-Cu at cleaving extracellular DNA
The extracellular DNA (eDNA) in biofilms strengthens the matrix of biofilms [8], and thus could be an important point of attack for antibiofilm agents, such as p1 and p3. We treated 24-h old P. aeruginosa PA01 biofilms with p1 and p3 with and without preincubation with Cu2+, extracted the biofilm eDNA, and characterized it using electrophoresis. Untreated biofilms contained eDNA of ~ 11 kbp in length (Fig. 6B), far from the size of the PA01 genomic DNA (6.3 Mbp [37]) confirming intracellular DNA did not contaminate the assay. Piscidin treatment of the biofilms resulted in eDNA cleavage as evidenced by the loss of intensity in the treated lanes. Treatment of biofilms with naïve p1 and p3 cleaved eDNA in an equivalent dose-dependent manner. Thus, p1 and p3 were able to sequester some copper from the biofilm’s bioavailable pool, most likely because the exopolysaccharide matrix of P. aeruginosa biofilms is known to retain Cu2+ under similar experimental conditions [38]. The comparable efficiency of p1 and p3 at cleaving eDNA arises probably because DNA is more accessible when it is extracellular rather than intracellular and the cleavage experiments were run overnight, allowing p1 to have noticeable effects even though its nuclease activity is slower than that of p3. When Cu2+ was added, eDNA cleavage was more effective with p3-Cu cleaving more efficiently than p1-Cu. Since this trend parallels that of the nuclease and biofilm inhibition assays (Fig. 6A), the relative efficacy of p1-Cu and p3-Cu against biofilms can be rationalized based on how effective each complex is at cleaving eDNA.
Only p3 synergizes with copper against P. aeruginosa persisters
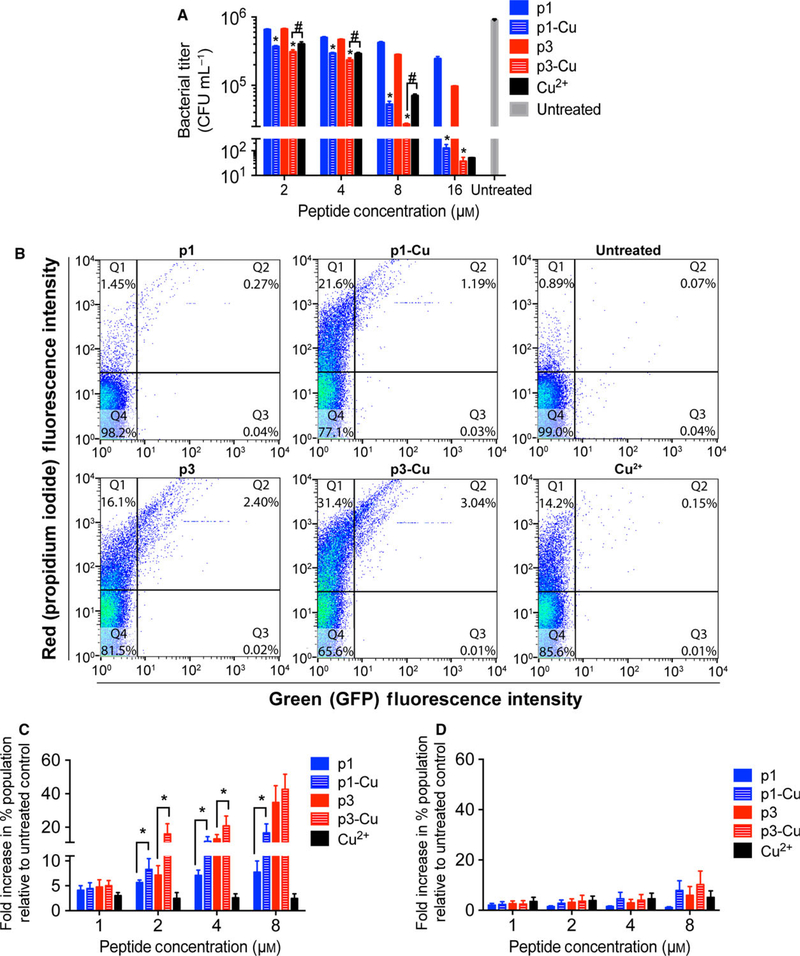
Persister cells are a subpopulation of transiently antibiotic-tolerant bacteria that are often the cause of recurring infections. Due to their metabolic idleness, persisters are less susceptible to antibiotics than normal cells [9]. Recent studies showed efficient persister cell eradication via ROS production [39]. It has also been suggested that multihit agents that damage DNA can be cytotoxic to persisters [9]. For this reason, we tested the activity of p1, p3, and their corresponding copper complexes against P. aeruginosa PA01 persisters. The cells were pelleted and washed with copper-free MOPS buffer before treatment with the peptides. Naïve p1 and p3 had slight bactericidal activity against persisters, reaching a maximum of 1 log of reduction in viability at 16 μM (Fig. 7A). While several studies have reported different MICs of Cu2+ against P. aeruginosa persisters ranging from low micromolar to millimolar values [38], the PA01 persisters used here succumbed to concentrations as low as 4 μM Cu2+. Across all concentrations, we observed a similar activity between free Cu2+ and p1-Cu, indicating that p1-Cu toxicity has the same bactericidal effect on PA01 as cupric ion. In sharp contrast, p3-Cu exhibited a stronger bactericidal effect than Cu2+ or p3 alone, and thus acted synergistically with the metal in eradicating persisters.
Fig. 7.
Activity of p1 and p3 against persister cells. (A) P. aeruginosa PA01 persisters were treated with the indicated concentrations of p1 and p3 for 3 h prior to CFU enumeration. Bars represent mean ± SEM (n = 3 triplicates). *P < 0.01, #P < 0.05 compared to the corresponding nonmetallated peptide. (B) Representative scatter plots from flow cytometric analysis of E. coli AT15 persisters treated with the indicated piscidin species and stained with PI. (C) Quantitative analysis of the increase in doubly fluorescent – GFP(+), PI(+) – cells (dead cells bearing DNA damage) following E. coli AT15 persister cell treatment with the indicated peptide concentrations. Bars represent mean ± SEM (n = 3). *P < 0.05. (D) Analysis of DNA damage in E. coli AT15 persister cells shows that number of GFP(−), propidium iodide (+) cells (dead cells without a damaged DNA; Q1 in scatter plots in (B)) is unchanged upon addition of copper.
p1-Cu and p3-Cu trigger the SOS response in E. coli persister cells and the response is stronger with p3-Cu
Knowing the strong activity of p3-Cu on persister cells and the significant magnitude of p3-induced DNA damage in planktonic bacteria, we postulated that DNA damage plays a role in piscidin-induced persister cell death. Exposure to DNA-damaging agents leads to the induction of the SOS response, which signals DNA damage and induction of the DNA repair machinery. To determine whether DNA damage is part of the bactericidal mechanism of p1 and p3, we used the GFP fluorescence of the E. coli reporter strain AT15 [40] (Δattλ::sulApΩgfp-mut2 TetR) as a direct readout of the SOS response. We exposed E. coli AT15 persisters to varying concentrations of p1, p3 and their corresponding Cu2+ complex for 2 h and then counterstained the cells with propidium iodide (PI). This setup allowed us to monitor the number of dead cells bearing damaged DNA by comparing the levels of doubly fluorescent cells, that is, both GFP and PI positive cells. Our treatments resulted in a significant increase in GFP(+), PI (+) cells (Q2 in the scatter plots of Fig. 7B). At peptide concentrations greater than 1 μM, we observed an increase in GFP(+), PI(+) cells with metallated p1 and p3 (Fig. 7C). Interestingly, treatment with naïve p3 also elicited a strong SOS response, suggesting that some p3 was effective at scavenging intrinsic Cu2+ from the persisters and caused direct DNA cleavage; or, that p3 in the nonmetallated form was strongly bound to genomic DNA and elicited the SOS response as observed for some non-DNA cleaving fluoroquinolones [41]. We also note that treatment with metallated or nonmetallated peptides produce similar counts of dead cells with no SOS response (Q1) (Fig. 7D), possibly because membrane activity is implicated in killing these cells. These combined results show that the SOS response following p1-Cu and p3-Cu treatment is commensurate with their efficacy at killing E. coli persister cells and cleaving isolated DNA.
Solid-state NMR experiments show that the ATCUN motif of piscidin adopts a conformation simultaneously poised for DNA and metal-ion binding
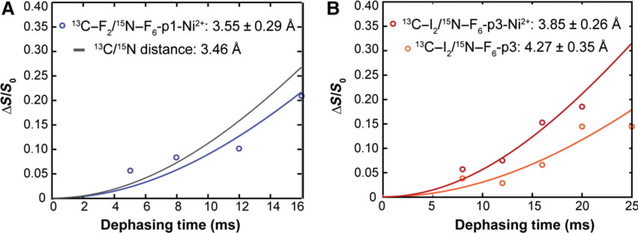
To understand the structural features that predispose piscidin for nuclease activity, we employed solid-state NMR on metallated piscidin/DNA samples. We previously used rotational-echo double-resonance (REDOR) experiments to show that the central domain of p1 and p3 (position 8 through 12) is a-helical when bound to the DNA duplex AAATACACTTTTGGT [23]. The α-helical structure of p1 and p3 is amphipathic, and therefore well suited to binding the anionic surface of DNA. The carbonyl 13C and amide 15N isotopic labels were placed in i and i + 4 positions, respectively, to span the length of hydrogen-bonding partners in a canonical α-helix (~ 4.2 Å). We also observed that the N-terminal domain of p1 bound to DNA features a 13C-F2/15N-F6 distance of 3.46 ± 0.51 A, indicating a well folded conformation, unlike the fraying observed when the peptide is bound to lipid bilayers [21]. Here, we also used REDOR and the same DNA duplex to determine if the folding of the amino end is affected by metal binding. p1-Ni exhibited a 13C-F2 – 15N-F6 distance of 3.55 ± 0.29 A (Fig. 8A), comparable to the aforementioned distance in nonmetallated p1. In the case of p3 (Fig. 8B), the 13C-I2 – 15N-F6 distances in the presence and absence of metal were 3.85 ± 0.26 and 4.27 ± 0.35 A, respectively. Based on these measurements, the carbonyl to amide distance between residues 2 and 6 is not significantly affected by metal binding. Overall, these structural studies show that remarkably, the N-termini of p1 and p3 adopt conformations simultaneously poised for metal- and DNA-binding.
Fig. 8.
Spectroscopic characterization of DNA-piscidin interactions via solid-state NMR. Distance measurements obtained by solid-state NMR for the hydrogen-bond between 13C-carbonyl of Phe-2 and 15N-amide of Phe-6 in p1 (A) and between the 13C-carbonyl of Ile-2 and 15N-amide of Phe-6 in p3 (B) in the presence of Ni2+. For p1, the simulated line for 3.46 Å corresponds to the fit to the previously measured distance in the absence of Ni2+ [23]. For p3, the Ni2+-free data are plotted in lighter red. Error bars were calculated from the signal-to-noise ratio in the NMR spectra.
Piscidin-DNA interactions reveal a more extensive network of intermolecular interactions with p3 than p1
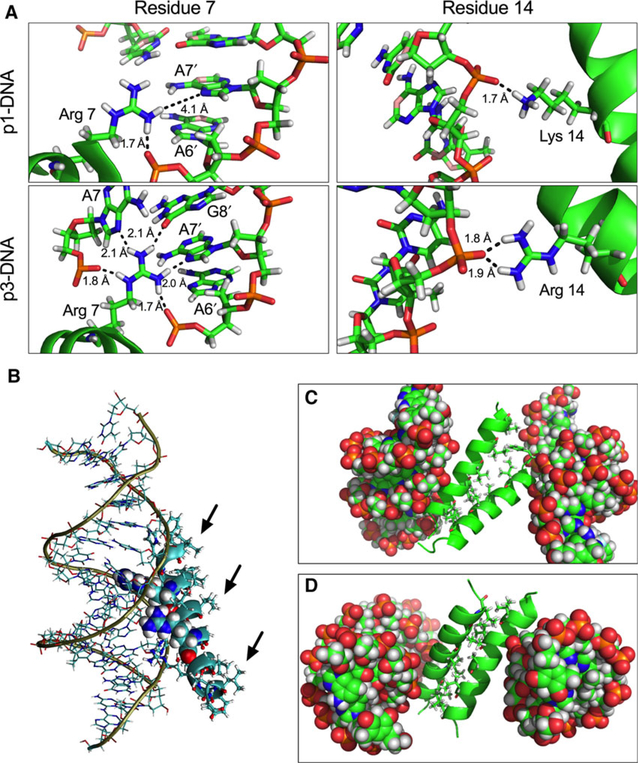
We used molecular dynamics (MD) and the AAATACACTTTTGGT duplex to scrutinize piscidin-DNA interactions and better understand how p3 achieves stronger nuclease effects than p1. First, we focused on the binding of monomeric (nonaggregated) piscidin to DNA. Under these conditions, the most stable conformation of DNA-p3 was found to have a binding energy to DNA that was more favorable than with p1 (Table 3). Furthermore, the Arg7 in p3 engages in five H-bonds with DNA while Arg7 in p1 only has two such bonds (Fig. 9A, left panels). Moreover, the Arg14 residue in p3 experiences more interactions with DNA than the Lys14 in p1 (Fig. 9A, right panels). The more extensive network of DNA-peptide interactions observed with p3 relative to p1 is consistent with its faster nuclease activity and lower n1/n2 values. We also note that these simulations yield distances at the N-termini of p1 and p3 (Table 4) that are consistent within their standard deviations with those obtained by solid-state NMR. Second, we considered the aggregation experimentally observed in samples containing DNA-piscidin complexes [23]. The MD simulations clearly illustrate that DNA-bound peptides unfavorably expose their hydrophobic surfaces to the solvent (black arrows in Fig. 9B), leading to aggregation. They also yield highly favorable interaction energies for aggregated (piscidin-DNA)2 complexes (Table 3), especially with p3. Interestingly, the most stable conformation of the dimers (Fig. 9C,D) shows the hydrophobic residues interacting in a manner analogous to leucine zippers. Altogether, these simulations of p1 and p3 indicate that more extensive intermolecular interactions with DNA correlate well with faster nuclease activity and stronger efficacy at damaging DNA. They also illustrate how aggregation between cationic peptides and DNA takes place on a molecular level.
Table 3.
Binding energy for the most stable conformation based on molecular dynamics of piscidin bound to DNA in the monomeric (nonaggregated) and dimeric (aggregated) states.
| Binding energy (kcal·mol−1) ± SD | |
|---|---|
| p1-DNA | 81.4 ± 14.1 |
| p3-DNA | −1.4 ± 10.1 |
| (p1-DNA)2 | −69.6 ± 27.6 |
| (p3-DNA)2 | −189.3 ± 18.7 |
Fig. 9.
Molecular dynamics simulations of DNA-piscidin interactions. (A) Extensive hydrogen-bonding network formed by the 7th and 14th residue in p1 (top panels) and p3 (bottom panels) with both the phosphate backbone and the major groove surface of the DNA bases. Hydrogen-bonding distances are shown in Angstroms. (B) Depiction of the most stable configuration of the p3-DNA complex. The bases in the duplex are shown as lines, Arg7 and Arg14 are shown as spheres. The other p3 side chains are displayed as sticks while the backbone appears as a ribbon. The amino end of the peptide is closer to the DNA backbone than the carboxylic end. (C and D) Exposure of the hydrophobic residues of p3 (black arrows) contributes to aggregation. The most stable configurations of (p1-DNA)2 (C) and (p3-DNA)2 (D) show that the hydrophobic face of the peptides interacts via hydrophobic zippering during oligomerization. DNA is shown as spheres while the peptide backbones appear as ribbons. Hydrophobic residues of the peptides are displayed as sticks.
Table 4.
Distances obtained from molecular dynamics for p1 and p3 bound to DNA.a
| Distance (Å) ± SD | |
|---|---|
| p1-DNA: C1-F2/N-F6 | 4.6 ± 0.8 |
| p1-DNA: C1-G8/N-V12 | 4.2 ± 0.4 |
| p3-DNA: C1-I2/N-F6 | 4.1 ± 0.2 |
| p3-DNA: C1-G8/N-A12 | 4.1 ± 0.2 |
Simulations were done over 100 ns.
Discussion
HDPs are powerful innate immune effector molecules that feature evolution-tested efficacy against a broad range of pathogens, including drug-resistant bacteria. HDPs that directly kill bacteria are traditionally evaluated for their membrane activity. However, the inability to explain the high potency of nonmembranolytic HDPs is leading to a paradigm shift in mechanistic studies of HDPs. Considering the homologous p1/p3 pair, both HDPs are highly potent and adopt similar structures bound to lipid bilayers. Surprisingly, only p1 is strongly membranolytic on microbial and model membranes [23], and hemolytic [19]. The investigations presented here validate the hypothesis that p3 uses another effective strategy to achieve high potency and specificity, as summarized next.
First, in vitro studies clearly identify p3-Cu as a complex that cleaves isolated DNA much faster than p1-Cu and at a pace comparable to that of free Cu2+. Second, on planktonic cells, p3 is the only member of the pair that cleaves large amounts of DNA in a Cu2+-dependent fashion (as measured by a TUNEL assay) in spite of the lag phase before initiating cell death. Third, on biofilms, the metallated form of p3 is more active than that of p1 and features stronger synergy with Cu2+. It is commensurately more effective than p1-Cu at cleaving the eDNA of biofilms. Fourth, on persister cells, only p3 is synergistic with Cu2+, rendering it more efficacious than p1. Furthermore, p3-Cu induces significant DNA damage (as measured by the SOS response). Fifth, MD simulations demonstrate that the network of DNA-peptide interactions is more extensive with p3 than p1, due to an additional arginine in p3. Additionally, solid-state NMR distance measurements show that the N-terminus of piscidin folds in a conformation that is poised for not only DNA but also metal binding. Overall, these new results demonstrate that stronger nuclease activity and interactions with DNA in vitro translate into more pronounced DNA damage in planktonic cells, biofilms, and persister cells when copper is available. Importantly, the stronger nuclease activity phenotype of p3 correlates with its slower initiation of cell death, and enhanced eradication of biofilms and persisters relative to p1, which is more membranolytic. Overall, the combination of low membranolytic effects and strong nuclease activity allows p3 to achieve high efficacy and specificity.
To our knowledge, there is no precedent of homologous HDPs that kill bacteria through different relative contributions of membrane and DNA-disruptive effects, and directly inflict not only physicochemical but also covalent damage. Equally significant is the discovery that while p1 is largely celebrated for its strong potency against planktonic bacteria, p3 stands out when we consider other bacterial modes of growth, such as persisters and biofilms. An important corollary to this finding is that HDP homologs and mutants should be tested on various modes of bacterial growth before concluding which version is more efficacious and advantageous in a natural setting and for therapeutic applications. Similarly, homologous HDPs should not be automatically considered to have identical mechanisms of action – an assumption largely practiced in structure-activity studies of HDPs.
Whereas p1 does not induce as much DNA damage as p3, it still demonstrates some metal coordination since it is synergistic with Cu2+ on planktonic cells and biofilms. Thus, the copper-dependent pathways that generate lipid peroxidation and a modest amount of DNA damage in the presence of p1 and copper must lead to a more versatile mechanism of action for p1-Cu relative to naïve p1. Since p1 is strongly membranolytic, its lipid disruption and peroxidation effects could synergize to affect cell survival. In the case of p3, low copper does not completely abolish its activity after 4 h of exposure at the MIC. Since naïve p3 is known to be more condensing to DNA than p1 [23], this may be how it inhibits the growth of planktonic cells under conditions of low copper availability, and eradicates some persisters without inducing the SOS response. Overall, neither peptide acts through a single mode of action. Rather, both p1 and p3 are multihit HDPs with differing killing timescales and contrasted dominance of their membrane- and DNA-disruptive effects. In vivo, their multihit mechanisms and complementarity could help curtail bacterial resistance since targeted cells would face the difficult task of having to modify multiple pathways to circumvent the multipronged effects of the peptides.
While the membrane- and DNA-disruptive effects are complementary, p1 and p3 are not synergistic on wild-type E. coli. The different timescale of their action may affect their ability to act synergistically unless DNA repair is impaired as it is the case for the ΔrecA mutant. Since access to the membrane precedes that to intracellular DNA, it is not surprising that p1 acts more quickly than p3. In the case of p3, acting on intracellular DNA requires not only translocating across membranes but also circumventing the redundant DNA damage repair pathways on wild-type cells, giving rise to slower killing kinetics. Nevertheless, p3 does have a sustained and efficacious mechanism of cell killing that catches up over time. Its properties are reminiscent of the benefits put forward for catalytic metallodrugs compared to traditional agents, including (a) lower doses due to irreversible modification of the target; (b) multiple turnovers due to enzyme-like properties; (c) improved selectivity due to the cumulative effects of being attracted to the targets and chemically modifying them; (d) enhanced chemical and structural stability due to metal coordination [25].
Noga and colleagues have demonstrated the essential role of piscidin for fish health [18,20]. Here, we uncover their nuclease activity and realize that it is physiologically relevant to fish on multiple accounts. First, the potency of metallated piscidin on fish pathogens has been demonstrated. Indeed, antimicrobial assays previously done on the fish parasites Saprolegnia sp. and Tetrahymena pyriformis [42] showed synergistic effects between piscidin 2 (FFHHIFRGIVHVGKTIHKLVTG) and copper sulfate, a chemical commonly used in fish farms to eradicate molds and parasites. While the molecular underpinning of this synergy on fish pathogens was enigmatic at the time, our studies indicate that it can be rationalized through the binding of copper to the ATCUN motif of piscidin.
Second, in terms of the likelihood that piscidin undergoes metallation in vivo, there is evidence supporting that p1 and p3 can be secreted in their metallated form following expression. Indeed, genomic information indicates that piscidins are expressed as prepropeptides that need to be proteolytically cleaved to liberate their active form [18]. Moreover, piscidins were originally isolated in mast cells [19] and were later found to be stored in granules [43]. Both proteolysis and trafficking to cytoplasmic vesicles involve the trans-Golgi network, an organelle that houses the copper-transporters ATP7A and 7B, which metallate newly synthesized cuproenzymes [44]. Therefore, the fundamental processes that act to ensure proper piscidin expression and localization place these peptides in copper-rich sites of cells. For this reason, it is very likely that both p1 and p3 exist posttranslationally as metallopeptides.
Third, regarding the relevance of piscidin metallation to host defense in live fish, piscidins are ubiquitously available throughout fish tissues and can kill both extracellular (via mast cell degranulation) and intracellular (following phagocytosis) microbes since the peptides are present in mast cells and acidophilic granulocytes that circulate systemically in fish [43]. Compartmentalization in the phagosome of immune cells and secretion through degranulation in infected areas where bacteria outnumber host cells very likely contribute to the protection of host cells from the cytotoxicity of the peptides. Previous immunolocalization studies showed high p1, p2, and p3 concentrations near the luminal surface of several species of teleost fish, and that p1 was highly concentrated in epithelial cells of the Atlantic cod [45]. These surfaces are lined with a protective layer of mucous which can support bacterial growth [46]. Thus, the combination of multiple metallated piscidins at high concentrations in regions prone to biofilm formation might lead to advantageous synergistic effects with copper and other HDPs.
Fourth, differential expression of p1 and p3 provides the fish with an opportunity to produce p3, the homolog with more pronounced nuclease activity, in tissues not well suited to p1 due to its strong hemolytic effects and/or lower efficacy on persisters and biofilms. Indeed, higher levels of p3 RNA relative to p1 RNA have been found in highly vascularized organs (e.g., gills; intestines; spleen) of hybrid striped bass [18]. Moreover, in the context of an infection, the bacteria that infiltrate connective tissues can circulate through the bloodstream, causing recurring infections and persisting within the fish. We find here that p3-Cu is particularly active against bacterial persisters, making the expression of p3 in highly vascularized organs all the more profitable. Clearly, the differential expression and localization of p1 and p3 track well with their mechanistic and functional properties, indicating that fish are equipped with a clever host-defense mechanism to eradicate invading pathogens without harming host cells.
In conclusion, we have demonstrated that p1 and p3 boast contrasted mechanisms of action in spite of being homologous. It is often assumed that peptides that share similar residues exert identical bactericidal mechanisms but we establish here that this assumption is dogmatic. We uncover that p3 features remarkable physicochemical properties (e.g., ATCUN motif; large number of arginines; moderate hydrophobicity) that bestow low membranolytic effects, high specificity, robust copper-scavenging capability, strong interactions with DNA, and fast nuclease activity in the context of living cells. Its nuclease activity phenotype simultaneously remedies its low membrane activity and bestows strong efficacy against persisters and biofilms without preventing high specificity. In the context of fish, these studies reveal a salient relationship between the differential expression, tissue localization, and mechanistic behavior of p1 and p3. They also underscore that the co-expression of complementary multihit HDPs that inflict both physicochemical and covalent damage to their targets may be a powerful way for fish to reduce bacterial drug resistance. Overall, these findings identify new avenues of research to investigate the sophisticated defense strategies employed by HDPs to knock out pathogens. They also provide principles that could be useful to design novel anti-infective drugs that are highly potent on life-threatening pathogens and nontoxic to host cells.
Materials and methods
Materials
Unless otherwise indicated, the reagents and chemicals were ordered from Fisher Scientific (Waltham, MA, USA) or Sigma Aldrich (Saint Louis, MO, USA).
Peptide synthesis and purification
The peptides p1 and p3 were synthesized by Fmoc solid-phase peptide synthesis at the University of Texas Southwestern Medical Center (Houston, TX, USA) and purified on a C18 column by reverse phase HPLC, as previously described [21,22]. In the case of the site-specifically labeled peptides, the labeled amino acids were purchased from Cambridge Isotopes Laboratories (Tewksbury, MA, USA) or Sigma Aldrich.
Strains and culture conditions
E. coli MG1655, TD172 (DrecA) and AT15 (Δattλ::sulApΩgfpmut2 TetR) were a generous gift from Kim Lewis (Northeastern University, Boston, USA) while P. aeruginosa PA01 was kindly provided by Michael Lynes (University of Connecticut, Storrs, USA). S. aureus (ATCC 25923) was obtained from American Type Culture Collection. Unless otherwise specifies, all cell lines were initially streaked in Luria-Bertani (LB) agar plates prior to inoculation of single colonies into Mueller-Hinton Broth (MHB) for peptide testing. Bacteria were grown until mid-logarithmic phase (OD600 ~ 0.6) at 37 °C and 225 rpm.
Mass spectrometry and cyclic voltammetry
To determine the stoichiometry of binding, mass spectrometry was employed. One hundred micromolar of p1 or p3 was mixed with an equal volume of 100 μM Cu2+ and incubated at room temperature for 30 min. The mixture was diluted 1 : 500 with 1% formic acid in acetonitrile (ACN) and injected into an electrospray ionization-mass spectrometer. Resulting spectra were reported without further manipulation. To determine the standard reduction potential of the respective copper complexes, we used cyclic voltammetry. A 250 μM solution of p1 or p3 was prepared in 20 mM MOPS, 100 mM KCl, pH 7.40. An equal volume of 225 μM Cu2+ was then added (slight excess of peptide is present to ensure that no free Cu2+ exists) and the mixture was incubated for 30 min. The solution was transferred into a microcell for voltammetry using Ag/AgCl as reference electrode, glassy carbon as working electrode and a platinum mesh as counter electrode; a 100 mVs−1 scan rate over a 1.1 V range was used.
Isothermal titration calorimetry
The ITC experiments were completed using a VP-ITC (MicroCal, Northhampton, MA, USA) containing a cell volume of 1.4 mL. Measurements were carried out at constant atmospheric pressure and constant temperature (298 K). The Cu2+ and peptide solutions were dissolved in HEPES buffer (10 mM, pH 7.4). Solutions were degassed to avoid bubbles and their pH was checked before and after the titrations. Reference power was set at 19.5 μcal·s−1, stir speed was maintained constant at 307 rpm, and a 300-s pretitration delay was utilized. For each titration, up to 30 injections containing 10 μL of titrant were done. The peptide piscidin was titrated with Cu2+ in the absence of glycine the confirm high-affinity binding, and thus a displacement experiment was performed in the presence of 5 mM glycine. The large excess of glycine compared to copper allowed us to assume that throughout the titration the glycine concentration was constant. Likewise, the HEPES buffer was in large excess of the metal that it interacts with, thus the total buffer concentration could be assumed to be constant during the titration. Raw data were processed in ORIGIN 7.0 (Northampton, MA, USA) and AFFINIMETER software (Campus Vida, Santiago de Compostela, Spain). We obtained a good response using 0.9 mM Cu2+ in the syringe and 90 μM piscidin in the cell (Fig. 2). The thermodynamics of binding, including the binding association constant (Kb), stoichiometry (n, the number of Cu2+ bound per peptide molecule), enthalpy change (ΔH), and corresponding error bars (standard deviations) were calculated using the least-squares regression model for the one-binding site model. This fitting procedure yielded the apparent association constant, Kapp. Next, this value was used in the equation derived by Trapaidze et al. [28] to derive corrected for the contributions of glycine and HEPES to the thermodynamic parameters measured by ITC. This equation, adapted to the piscidin case, is as follows:
| (1) |
where [glycine]0 is the total glycine concentration, [HEPES]0 is the total HEPES buffer concentration, is the binding dissociation constant of glycine-Cu2+, is the binding dissociation constant of glycine2-Cu2+, pH is the pH of the solution, is the pKa of glycine, is the pKa of the HEPES buffer, and is the binding dissociation constant of the HEPES-Cu2+ complex.
In vitro DNA cleavage assay
The cleavage reactions consisted of 10 μM base pair pUC19, 100 nM Cu-peptide (or Cu2+), 1 mM sodium ascorbate, and 1 mM H2O2 in 20 mM HEPES, 100 mM NaCl, pH 7.40. Reactions were run at room temperature for the indicated time points when 10 μL was withdrawn. The reactions were stopped using 3× loading dye containing 1 mM EDTA. The samples were loaded onto a 1% agarose gel containing ethidium bromide and ran for 60 min at 100 V. The gels were imaged using a Bio-Rad GelDoc XR+ imager and the bands were quantified using the accompanying IMAGE LAB 5.0 software (Bio-Rad, Hercules, CA, USA). Due to the diminished capacity of supercoiled DNA to intercalate ethidium bromide, the intensity of the bands corresponding to the supercoiled form was multiplied by 1.47 [25]. Data shown represent Mean ± SEM obtained from three independent trials. When single-strand breaks occur at sites that are close to each other (as a result of localized ROS formation), it results in linearization of pUC19. The n1/n2 values were calculated using (Eqn 2) and (Eqn 3) from the Poisson distribution and Freifelder–Trumbo equation where f(I) and f(III) are the respective fractions of supercoiled and linearized plasmid remaining per time point.
| (2) |
| (3) |
Antimicrobial assays and time-kill kinetics
Antimicrobial susceptibility testing was done using the broth microdilution method as suggested by Hancock [47]. Gram-positive S. aureus (ATCC 25923); Gram-negative E. coli (MG1655, TD172 – ΔrecA) and P. aeruginosa (PAO1) were grown in Mueller–Hinton Broth (MHB; Difco, Detroit, MI, USA) until mid-log phase for 3–5 h. Peptide stock solutions were serially diluted in PBS (Gibco, Gaithersburg, MD, USA) pH 7.40. Assays were done in 96-well poplypropylene plate (Greiner, Mount Joy, PA, USA). Each well received 50 μL of the peptide solution needed to reach the desired final concentration. Next, 50 μL of a bacterial suspension was added resulting to a final inoculum of 5 × 105 CFUmL−1 per well. Plates were incubated at 37 °C for 18–20 h. MIC was defined as the concentration that prevented visual growth of bacteria. The reported MICs are the average from three independent trials, each done in triplicates. For time-kill experiments, 350 μL of each peptide at 2× the MIC was mixed with bacteria to yield a 1× MIC-peptide concentration and the same inoculum density as in the antimicrobial assays. Every time point, 10 μL was withdrawn and diluted 1000-fold. Then, 100 μL of either the 1 : 1000 or the 1 : 100 dilution was plated on LB agar plates. Colonies were counted manually after overnight incubation. Plotted curves display bacterial titer data obtained from two independent trials done in duplicates. Titer values represent quantities in undiluted solutions.
To characterize the effect of copper at specific time points after adding p1 and p3, the following protocol was followed. Single colonies of E. coli MG1655 were taken from a LB agar plate and inoculated into MHB or M9 (i.e., minimal media supplemented with glucose). To encourage growth of bacteria in M9, cells were passaged thrice prior to being used in the experiment. This also ensured infinite dilution of the copper that may have originated from the LB agar plate. Cells were grown in liquid medium to mid-log phase after which time cells were harvested and washed in the same growth medium as the medium used to expose them to piscidin. A 350 μL, 5 × 105 CFUmL−1 suspension was mixed with an equal volume of peptides (nonmetallated or preincubated with equimolar concentration of Cu2+) for 30 min (for p1 and P1-Cu), 4 h (for p3 and P3-Cu) or 2 h (for the untreated controls). The cells were incubated at 37 °C and diluted up to 1 : 1000. Then, a 100 μL-volume of each dilution was plated into a LB agar plate and incubated overnight for colony enumeration. Bars represent data obtained from three trials done in duplicates.
Lipid peroxidation assay
The extent of oxidative damage in the surface of the cell was measured by quantifying the peroxidation products of unsaturated phospholipids. E. coli strain MWF1, fabR::kan recD:: Tn10 (containing a higher level of unsaturated fatty acid in its membrane) were grown, washed and resuspended in 20 mM HEPES, 100 mM NaCl pH 7.40 and was incubated with peptides at their MIC without addition of exogenous Cu2+ for 2 h. Next, the suspension was subjected to a standard thiobarbituric acid (TBA) assay to determine amount of malonyldialdehyde (MDA) product. We used tetramethoxypropane as external standard for MDA. Briefly, 50 μL of the sample/standard was added to 150 μL of 0.1 M perchloric acid and 150 μL of 40 mM TBA. The mixture was heated at 97 °C for 60 min followed by a 20 min incubation at 20 °C. Then, 300 μL of methanol was added followed by 100 μL of 20% trichloroacetic acid. The tubes were spun down and the 100 μL of the mixture was injected in an analytical HPLC using a 72 : 17 : 11 50 mM KH2PO4:MeOH:ACN as mobile phase with a flow rate of 1 mL·min−1. Bars represent mean ± SEM obtained from three independent trials.
DNA damage assays in live bacterial cells
The Terminal deoxyribonucleotidyl transferase (TdT)-mediated dUTP Nick End Labeling (TUNEL) assay was used to assess extent of DNA damage in live E. coli. A 350 μL aliquot of a 108 CFUmL−1 E. coli culture was mixed with an equal volume of the peptides at 2× its MIC (to reach 1× and 2× concentrations in the final samples) and incubated for 2 h. The cells were pelleted and washed twice with ice-cold PBS. Next, the cells were fixed with 4% paraformaldehyde for 30 min, washed, and permeabilized with 0.1% triton X-100 in 0.1% sodium citrate for 2 min. The cells were then repelleted and resuspended in 1× TUNEL reaction mixture (one part TdT enzyme, nine parts label solution; Roche Molecular Biochemicals, Indianapolis, IN, USA) and incubated in the dark for 1 h at 37 °C. After pelleting, the cells were washed twice with ice-cold PBS and resuspended in PBS containing PI for DNA staining. The green and red fluorescence of 100 000 cells were obtained using a BD FACSCalibur flow cytometer and data were analyzed using FLOWJO v. 10 software (Ashland, OR, USA). Histograms are representative of three independent trials, and bars represent mean ± SEM.
Checkerboard assay and synergy testing
A method analogous to the MIC determinations was used to measure FIC indices. Two-fold serial dilutions of p3 (in MHB broth) were added to the columns of a sterile 96-well polypropylene plate (except the last column in order to generate the appropriate controls). Then, 50 μL of two-fold serial dilutions of p1 (in MHB) was added to the rows of the plate (except the last row in order to generate the appropriate controls). Next, the plate was inoculated with a suspension of 106 CFU·mL−1 of either E. coli MG1655 (WT) or E. coli TD172 (ΔrecA) and incubated overnight at 37 °C. The growth was determined by OD600 measurements using a plate reader. To obtain % growth, absorbance values were divided by the value from the appropriate control (i.e., well receiving no p1 and/or p3). The heat map represents mean ± SEM obtained from three trials done in duplicates. For visual assessment of synergy, a 3.0 mL culture of 5 × 105 CFU·mL−1 of bacteria was treated with 0.5 MIC of p1 or p3 either alone or in combination. The culture tubes were incubated in a shaker at 37 °C overnight and photographed.
Biofilm inhibition and biofilm eDNA cleavage assay
An exponentially growing P. aeruginosa PAO1 culture was used to inoculate 25 mL of fresh LB broth to an OD600 of 0.05. Biofilms were grown in glass cover slips for 24 h at 37 °C. The cover slips were rinsed by dipping carefully in 20 mM MOPS buffer, pH 7.40, before transferring to a 10 mm sterile petri plate containing piscidin at varying concentrations. Following 3 h of incubation at 37 °C, the coverslips were transferred into conical tubes containing 10 mL of MOPS buffer, sonicated for 4 min (at 15 s intervals), and vortexed. The cells were concentrated by centrifugation and resuspended in 100 μL of MOPS buffer. Four 10 μL portions (of each treatment) were then plated into separated LB agar plates, incubated overnight after which the colonies were counted manually. Bars represent data obtained from three independent trials done in triplicates. For the extracellular DNA (eDNA) cleavage experiments, the same treatments were done, following an overnight incubation with the peptides. Coverslips were transferred to conical tubes containing 20 mM Tris-HCl, 5 mM CaCl2 buffer, pH 8.0, and vortexed for 1 min. The coverslips were removed and 5 μg·mL−1 Proteinase K was added and incubated at 37 °C for 1 h. After which, the suspension was centrifuged at maximum speed for 10 min (to remove planktonic cells and other debris) and the supernatant was collected. Qiagen spin columns were used to purify eDNA, using the Qiagen wash buffer provided. The eDNA was eluted using 50 μL of sterile nanopure water. The DNA was quantified by nanodrop spectroscopy (A260/A280 were all > 1.80) and a total of 150 μg of DNA was loaded into a 1% agarose gel containing 1× Sybr Green, ran at 150 V for 2 h. Data are representative of three trials performed independently of each other and done in triplicates.
Persister cell inhibition assay
An overnight P. aeruginosa PAO1 culture was treated with 200 μg·mL−1 ciprofloxacin for 3.5 h with shaking at 37 °C to kill off nonpersister cells. The culture was spun down and washed with MOPS buffer three times before resuspending in MOPS buffer at 4 °C. A total of 500 μL total volume containing the indicated peptide concentrations was used, and treatment was done for 3 h at 37 °C with shaking. Aliquots were diluted in room temperature MOPS buffer and dilutions were plated in LB agar plates. After overnight incubation, colonies were counted manually. Bars represent data from three independent experiments done in triplicates.
Measurement of SOS response via flow cytometry
An exponentially growing culture of E. coli AT15 (a kind gift from Prof Kim Lewis, Northeastern University, Boston, USA) was treated with 50 μg·mL−1 of Ampicillin for 4 h at 37 °C to kill nonpersister cells. The culture was spun down and cells were washed three times with MOPS buffer before resuspending to an OD600 of 0.1. A total of 700 μL total volume was used for piscidin treatment at the indicated concentrations. After 2 h of exposure to the peptides, the cells were spun down, washed twice, and resuspended in 15 μL of MOPS buffer containing 0.5 mg·mL−1 of PI. A 10 μL aliquot was used for flow cytometry analysis in a BD FACSCalibur cytometer, and fluorescence of 100 000 cells was measured. The gates were established using the untreated control, and % populations were calculated (by FLOWJO v 10) after applying the same gating parameters through all treatments. Bars represent data from three independent experiments done in triplicates.
Nuclear magnetic resonance – solution NMR
NMR-monitored metal titrations of p1 and p3 were achieved using a buffered solution of 15N3-His3-containing peptide and a standardized nickel chloride aqueous solution (Fluka, Morris Plains, NJ, USA). Each peptide was dissolved at 0.50 mM in Bis-Tris (10 mM, pH 6.8) and aliquots of nickel chloride were added to achieve specific peptide to nickel molar ratios. Progress of the titrations was monitored using the well-established Heteronuclear Multiple Quantum Coherence experiments [48,49]. The experiments were performed at the Rensselaer Polytechnic Institute (Troy, NY) on 800 and 600 MHz Bruker instruments fitted with cryoprobes.
Nuclear magnetic resonance – solid-state NMR
Solid-state samples were made as previously described [23], with the exception that each peptide (about 2–3 mg) was titrated with an equimolar amount of Ni2+ prior to binding to DNA. The sequence of the duplex DNA was AAATACACTTTTGGT (IDT, Coralville, IA, USA). Upon addition of the peptide to the DNA, aggregation occurred, allowing us to collect via centrifugation at 960 g hydrated peptide-DNA complexes for solid-state NMR. The NMR experiments were performed on a midbore 800 MHz magnet equipped with a Bruker Avance III console and fitted with a 3.2 mm low-E triple-resonance biosolids MAS probe at the National High Magnetic Field Laboratory (NHMFL). Experimental conditions for the NMR experiments were as previously published [23]. Briefly, for each dephasing time (i.e., a multiple of rotor periods), two sets of data were recorded: one without the train of 15N 180° pulses and one with the train of pulses, corresponding to the 13C signals without (S0) and with (S) 15N dephasing, respectively. We collected 10 240 to 30 720 transients depending on the dephasing time. Since the difference ΔS (S–S0) over S0 depends exclusively on the 13C–15N dipolar coupling, we used a MATLAB program to fit the ΔS/S0 ratio as a function of the dephasing time, and obtained the internuclear 13C/15N distances. Additional details about fitting the data and obtaining error bars were previously published [23].
Molecular dynamics simulations – simulations setup
The initial p1 and p3 three-dimensional structures were taken from NMR structures (PDB code 2ojm (p1) and 2mcx (p3)). Two piscidin charged states were simulated, with neutral and protonated histidine residues, respectively. The simulation for the isolated piscidins was done over 100 ns at 310 K. For the peptide-DNA complexes, the simulations were 100 ns for neutral histidine simulation and 30 ns for protonated histidines in the complexes. MD simulations of the solvated variant models were performed in NPT ensemble using the NAMD program [50] with the CHARMM36 force-field. The models were explicitly solvated with TIP3P water molecules, [51,52] with a minimum distance of 10 Å from any edge of the box to any protein atom. Sodium chloride ions were added at random locations to neutralize the peptide charge and to maintain the ionic strength at 150 mM salt concentrations. The Langevin piston method [50,53,54] with a decay period of 100 fs, and a damping time of 50 fs was used to maintain a constant pressure of 1 atm. The temperature (300 K) was controlled by Langevin thermostat with a damping coefficient of 10 ps−1 [50]. The short-range van der Waals (VDW) interactions were calculated using the switching function, with a twin range cutoff of 10.0 and 12.0 Å. Long-range electrostatic interactions were calculated using the particle mesh Ewald method with a cut-off of 12.0 A for all simulations [55,56]. The equations of motion were integrated using the leapfrog integrator with a step of 2 fs. The hydrogen atoms were constrained to the equilibrium bond using the SHAKE algorithm [57]. To obtain the relative structural stability of the variant models, the trajectories were first extracted from the explicit MD simulations excluding water molecules. The solvation energies of all systems were calculated using the Generalized Born Method with Molecular Volume (GBMV) [58,59]. In the GBMV calculations, the dielectric constant of water was set to 80.0. The hydrophobic solvent-accessible surface area (SASA) term factor was set to 0.00592 kcalmol−1Å−2. Each variant is minimized 500 cycles and the conformation energy was evaluated by grid-based GBMV.
Molecular dynamics simulations – characterization of piscidin-DNA conformations
The DNA structure was initially modeled using DNA response element E-box structure as template and mutating sequence to AAATACACTTTTGGT used in the NMR experiments. With constraints of hydrogen bonds between base pairs, the DNA duplex structure was refined using MD simulations. The piscidin-DNA complex structures were searched using Haddock docking and additional manual adjustment to accommodate/optimize the charge interactions between piscidin and DNA. Six starting conformations with best initial energy were used to run MD simulations for p3-DNA complex, and three of them were replaced by p1 to compare their DNA interactions. The most stable conformation was selected from the MD simulations using protonated histidines based on free energies. Starting from this selected conformation, a round of MD simulations was performed to test the structural stability of the peptide-DNA complex at 300 K. The simulations were stopped after confirming the convergence of the conformation in the MD simulations. For the unstable docking pose, the simulations were terminated when peptide moved out of the docked binding pocket. Images were prepared in VMD and Pymol.
Statistical analysis
Data from assays were analyzed for statistical differences using GRAPH-PAD PRISM® v6.0 software (La Jolla, CA, USA). One-way or two-way ANOVA was used to determine statistical significance and was set at P < 0.05.
Acknowledgements
MLC thanks the National Science Foundation (CHE-0832571) and Camille and Henry Dreyfus Foundation (Teacher-Scholar Award) for funding, and the Hamilton College Departments of Biology and Chemistry for access to laboratory resources. AMA-B thanks the Lyme Disease Association for funding. The related research of DR is partially supported by the National Science Foundation (CAREER-1455644) and National Institutes of Health (1R21EY025750–01A1). The solid-state NMR experiments were carried out at the National High Magnetic Field Lab (NHMFL) supported by the NSF Cooperative agreement DMR1157490 and the State of Florida. The molecular dynamics was funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the NIH, Bethesda, MD (http://biowulf.nih.gov). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Abbreviations
- AMPs
antimicrobial peptides
- ATCUN
Amino Terminal Copper and Nickel
- CD
circular dichroism
- eDNA
extracellular DNA
- EPS
exopolysaccharide
- MD
molecular dynamics
- MIC
minimum inhibitory concentration
- p1
piscidin 1
- p3
piscidin 3
- PI
propidium iodide
- REDOR
rotational-echo double-resonance
- ROS
reactive oxygen species
- SASA
solvent-accessible surface area
- TI
therapeutic index
- TUNEL
terminal deoxynucleotidyl transferase (TdT) dUTP nicked end labeling
Footnotes
Conflict of Interest
The authors declare no conflicts of interest with the contents of this article.
References
- 1.Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. [DOI] [PubMed] [Google Scholar]
- 2.Fjell CD, Hiss JA, Hancock REW & Schneider G (2012) Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov 11, 37–51. [DOI] [PubMed] [Google Scholar]
- 3.Boman HG (2003) Antibacterial peptides: basic facts and emerging concepts. J Intern Med 254, 197–215. [DOI] [PubMed] [Google Scholar]
- 4.Shai Y (2002) Mode of action of membrane active antimicrobial peptides. Biopolymers 66, 236–248. [DOI] [PubMed] [Google Scholar]
- 5.Wang G (2013) Database-guided discovery of potent peptides to combat HIV-1 or superbugs. Pharmaceuticals (Basel) 6, 728–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox JL (2013) Antimicrobial peptides stage a comeback. Nat Biotechnol 31, 379–382. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Todd R, Kiehlbauch J, Walters M & Kallen A (2017) Notes from the Field: Pan-resistant new delhi metallo-beta-lactamase-producing Klebsiella pneumoniae. MMWR Morb Mortal Wkly Rep 66, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitchurch CB, Tolker-Nielsen T, Ragas PC & Mattick JS (2002) Extracellular DNA required for bacterial biofilm formation. Science 295, 1487. [DOI] [PubMed] [Google Scholar]
- 9.Lewis K (2010) Persister cells. Annu Rev Microbiol 64, 357–372. [DOI] [PubMed] [Google Scholar]
- 10.Wimley WC (2010) Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol 5, 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahar AA & Ren D (2013) Antimicrobial peptides. Pharmaceuticals (Basel) 6, 1543–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yount N & Yeaman M (2005) Immunocontinuum: perspectives in antimicrobial peptide mechanisms of action and resistance. Protein Pept Lett 12, 49–67. [DOI] [PubMed] [Google Scholar]
- 13.Hilchie AL, Wuerth K & Hancock RE (2013) Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol 9, 761–768. [DOI] [PubMed] [Google Scholar]
- 14.Nicolas P (2009) Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J 276, 6483–6496. [DOI] [PubMed] [Google Scholar]
- 15.Mardirossian M, Grzela R, Giglione C, Meinnel T, Gennaro R, Mergaert P & Scocchi M (2014) The host antimicrobial peptide Bac71–35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem Biol 21, 1639–1647. [DOI] [PubMed] [Google Scholar]
- 16.Sim S, Wang P, Beyer BN, Cutrona KJ, Radhakrishnan ML & Elmore DE (2017) Investigating the nucleic acid interactions of histone-derived antimicrobial peptides. FEBS Lett 591, 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu H, Pazgier M, Jung G, Nuccio S-P, Castillo PA, de Jong MF, Winter MG, Winter SE, Wehkamp J, Shen B et al. (2012) Human a-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salger SA, Cassady KR, Reading BJ & Noga EJ (2016) A diverse family of host-defense peptides (piscidins) exhibit specialized anti-bacterial and anti-protozoal activities in fishes. PLoS One 11, e0159423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silphaduang U & Noga EJ (2001) Peptide antibiotics in mast cells of fish. Nature 414, 268–269. [DOI] [PubMed] [Google Scholar]
- 20.Silphaduang U, Colorni A & Noga EJ (2006) Evidence for widespread distribution of piscidin antimicrobial peptides in teleost fish. Dis Aquat Organ 72, 241–252. [DOI] [PubMed] [Google Scholar]
- 21.Perrin BS Jr, Tian Y, Fu R, Grant CV, Chekmenev EY, Wieczorek WE, Dao AE, Hayden RM, Burzynski CM, Venable RM et al. (2014) High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion. J Am Chem Soc 136, 3491–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chekmenev EY, Jones SM, Nikolayeva YN, Vollmar BS, Wagner TJ, Gor’kov PL, Brey WW, Manion MN, Daugherty KC & Cotten M (2006) High-field NMR studies of molecular recognition and structure-function relationships in antimicrobial piscidins at the waterlipid bilayer interface. J Am Chem Soc 128, 5308–5309. [DOI] [PubMed] [Google Scholar]
- 23.Hayden RM, Goldberg GK, Ferguson BM, Schoeneck MW, Libardo MD, Mayeux SE, Shrestha A, Bogardus KA, Hammer J, Pryshchep S et al. (2015) Complementary effects of host defense peptides Piscidin 1 and Piscidin 3 on DNA and lipid membranes: biophysical insights into contrasting biological activities. J Phys Chem B 119, 15235–15246. [DOI] [PubMed] [Google Scholar]
- 24.Harford CSB (1997) Amino terminal Cu(II)- and Ni (II)-binding (ATCUN) motif of proteins and peptides: metal binding, DNA cleavage, and other properties. Acc Chem Res, 30, 123–130. [Google Scholar]
- 25.Joyner JC, Reichfield J & Cowan JA (2011) Factors influencing the DNA nuclease activity of iron, cobalt, nickel, and copper chelates. J Am Chem Soc 133, 15613–15626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joyner JC, Hodnick WF, Cowan AS, Tamuly D, Boyd R & Cowan JA (2013) Antimicrobial metallopeptides with broad nuclease and ribonuclease activity. Chem Commun (Camb) 49, 2118–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libardo MD, Nagella S, Lugo A, Pierce S & AngelesBoza AM (2015) Copper-binding tripeptide motif increases potency of the antimicrobial peptide anoplin via reactive oxygen species generation. Biochem Biophys Res Commun 456, 446–451. [DOI] [PubMed] [Google Scholar]
- 28.Trapaidze A, Hureau C, Bal W, Winterhalter M & Faller P (2012) Thermodynamic study of Cu2 + binding to the DAHK and GHK peptides by isothermal titration calorimetry (ITC) with the weaker competitor glycine. J Biol Inorg Chem 17, 37–47. [DOI] [PubMed] [Google Scholar]
- 29.Haas KL, Putterman AB, White DR, Thiele DJ & Franz KJ (2011) Model peptides provide new insights into the role of histidine residues as potential ligands in human cellular copper acquisition via Ctr1. J Am Chem Soc 133, 4427–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatcher LQ, Hong L, Bush WD, Carducci T & Simon JD (2008) Quantification of the binding constant of copper(II) to the amyloid-beta peptide. J Phys Chem B 112, 8160–8164. [DOI] [PubMed] [Google Scholar]
- 31.Jackson GS, Murray I, Hosszu LL, Gibbs N, Waltho JP, Clarke AR & Collinge J (2001) Location and properties of metal-binding sites on the human prion protein. Proc Natl Acad Sci USA 98, 8531–8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompsett AR, Abdelraheim SR, Daniels M & Brown DR (2005) High affinity binding between copper and full-length prion protein identified by two different techniques. J Biol Chem 280, 42750–42758. [DOI] [PubMed] [Google Scholar]
- 33.Freifelder D & Trumbo B (1969) Matching of singlestrand breaks to form double-strand breaks in DNA. Biopolymers 7, 681–693. [Google Scholar]
- 34.Chekmenev EY, Vollmar BS, Forseth KT, Manion MN, Jones SM, Wagner TJ, Endicott RM, Kyriss BP, Homem LM, Pate M et al. (2006) Investigating molecular recognition and biological function at interfaces using piscidins, antimicrobial peptides from fish. Biochim Biophys Acta 1758, 1359–1372. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez-Mazarrasa C, Mazarrasa O, Calvo J, del Arco A & Martinez-Martinez L (2009) High concentrations of manganese in Mueller-Hinton agar increase MICs of tigecycline determined by Etest. J Clin Microbiol 47, 827–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brewer GJ (2009) Zinc and tetrathiomolybdate for the treatment of Wilson’s disease and the potential efficacy of anticopper therapy in a wide variety of diseases. Metallomics 1, 199–206. [DOI] [PubMed] [Google Scholar]
- 37.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. [DOI] [PubMed] [Google Scholar]
- 38.Harrison JJ, Turner RJ & Ceri H (2005) Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Environ Microbiol 7, 981–994. [DOI] [PubMed] [Google Scholar]
- 39.Grant SS, Kaufmann BB, Chand NS, Haseley N & Hung DT (2012) Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci USA 109, 12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theodore A, Lewis K & Vulic M (2013) Tolerance of Escherichia coli to fluoroquinolone antibiotics depends on specific components of the SOS response pathway. Genetics 195, 1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin TT, Kang HQ, Ma P, Li PP, Huang LY & Gu B (2015) SOS response and its regulation on the fluoroquinolone resistance. Ann Transl Med 3, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zahran E & Noga EJ (2010) Evidence for synergism of the antimicrobial peptide piscidin 2 with antiparasitic and antioomycete drugs. J Fish Dis 33, 995–1003. [DOI] [PubMed] [Google Scholar]
- 43.Mulero I, Noga EJ, Meseguer J, Garcia-Ayala A & Mulero V (2008) The antimicrobial peptides piscidins are stored in the granules of professional phagocytic granulocytes of fish and are delivered to the bacteriacontaining phagosome upon phagocytosis. Dev Comp Immunol 32, 1531–1538. [DOI] [PubMed] [Google Scholar]
- 44.Polishchuk R & Lutsenko S (2013) Golgi in copper homeostasis: a view from the membrane trafficking field. Histochem Cell Biol 140, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruangsri J, Fernandes JM, Rombout JH, Brinchmann MF & Kiron V (2012) Ubiquitous presence of piscidin1 in Atlantic cod as evidenced by immunolocalisation. BMC Vet Res 8, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rakers S, Niklasson L, Steinhagen D, Kruse C, Schauber J, Sundell K & Paus R (2013) Antimicrobial peptides (AMPs) from fish epidermis: perspectives for investigative dermatology. J Invest Dermatol 133, 1140–1149. [DOI] [PubMed] [Google Scholar]
- 47.Wiegand I, Hilpert K & Hancock REW (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3, 163–175. [DOI] [PubMed] [Google Scholar]
- 48.Dawson JE, Seckute J, De S, Schueler SA, Oswald AB & Nicholson LK (2009) Elucidation of a pH-folding switch in the Pseudomonas syringae effector protein AvrPto. Proc Natl Acad Sci USA 106, 8543–8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelton JG, Torchia DA, Meadow ND & Roseman S (1993) Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signaltransducing protein from Escherichia coli, using twodimensional heteronuclear NMR techniques. Protein Sci 2, 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K & Schulten K (1999) NAMD2: greater scalability for parallel molecular dynamics. J Comput Phys 151, 283–312. [Google Scholar]
- 51.Mahoney MW & Jorgensen WL (2000) A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J Chem Phys 112, 8910–8922. [Google Scholar]
- 52.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79, 926–935. [Google Scholar]
- 53.Martyna GJ, Tobias DJ & Klein ML (1994) Constantpressure molecular-dynamics algorithms. J Chem Phys 101, 4177–4189. [Google Scholar]
- 54.Feller SE, Zhang YH, Pastor RW & Brooks BR (1995) Constant-pressure molecular-dynamics simulation – the Langevin Piston method. J Chem Phys 103, 4613–4621. [Google Scholar]
- 55.Darden T, York D & Pedersen L (1993) Particle Mesh Ewald – an N.Log(N) method for ewald sums in large systems. J Chem Phys 98, 10089–10092. [Google Scholar]
- 56.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H & Pedersen LG (1995) A smooth particle mesh Ewald Method. J Chem Phys 103, 8577–8593. [Google Scholar]
- 57.Ryckaert JP, Ciccotti G & Berendsen HJC (1977) Numerical-integration of Cartesian equations of motion of a system with constraints – molecular-dynamics of N-alkanes. J Comput Phys 23, 327–341. [Google Scholar]
- 58.Lee MS, Salsbury FR & Brooks CL (2002) Novel generalized Born methods. J Chem Phys 116, 10606–10614. [Google Scholar]
- 59.Lee MS, Feig M, Salsbury FR & Brooks CL (2003) New analytic approximation to the standard molecular volume definition and its application to generalized born calculations. J Comput Chem 24, 1348–1356. [DOI] [PubMed] [Google Scholar]