Abstract
MicroRNAs, which regulate mRNAs, operate through a variety of signaling pathways to participate in the development of colorectal cancer (CRC). In this study, we found that microRNA (miR)‐143‐3p expression was significantly lower in both CRC and liver metastatic CRC tissues from liver compared with normal colonic tissues. Functional assays showed that miR‐143‐3p inhibited CRC cell invasion and migration in vitro. Using a bioinformatics approach, we identified miR‐143‐3p target mRNAs. Among the candidate targets, only the expression of integrin alpha 6 (ITGA6) and ArfGAP with the SH3 domain and ankyrin repeat and PH domain 3 (ASAP3) were significantly reduced by miR‐143‐3p mimics as examined by western blot, and the metastasis potential of CRC cells was attenuated by endogenous ITGA6 and ASAP3 knockdown, determined by migration and invasion assays. Both ITGA6 and ASAP3 were upregulated in CRC tissues compared to normal tissues. Analysis of the relationship between clinicopathological features and ITGA6/ASAP3 protein expression in 200 patients with CRC showed a significant difference in positive ITGA6 expression between the early stage (I + II) and the advanced stage (III + IV), and ASAP3 expression levels positively correlated with metastasis in the lymph nodes. These results indicate that miR‐143‐3p acts as an anti‐oncogene by downregulating ITGA6/ASAP3 protein expression and could offer new insight into potential therapeutic targets for CRC.
Keywords: ASAP3, colorectal cancer, ITGA6, metastasis, miR‐143‐3p
1. INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer in both men and women.1 Although early surgery is recommended, this treatment is limited by the fact that 25% of CRC patients have clinically detectable liver metastases at initial diagnosis.2 Even after resection of liver metastases, more than 50% of patients cannot be cured due to recurrence and other distant metastases.3, 4 Many molecular targeted therapies have shown remarkable clinical success in the treatment of many cancer types including CRC; however, common adverse effects include rash, diarrhea, hypertension, hypothyroidism, proteinuria, depigmentation, and hepatotoxicity.5 It is therefore essential to explore now molecular targets for the treatment of CRC.
MicroRNAs (miRNAs) are short endogenous noncoding RNAs that repress gene expression in eukaryotic organisms. Many studies have shown a correlation between dysregulated miRNAs and the aberrant regulation of signaling pathways involved in CRC initiation and progression. The downregulation of miR‐143 and miR‐145 in CRC relative to normal colon epithelial cells was first reported by Michael et al.6 Many other studies have reported the tumor suppressive or oncogenic functions of miRNAs in CRC.7, 8 Therefore, miRNAs could also represent novel therapeutic targets for gene therapy in the treatment of CRC.
Based on previously identified miRNAs that were decreased in CRC compared with normal colonic tissues (see NCBI GBO platform, No. GSE53339), miR‐143‐3p was selected for further study. Here, we show that exogenous miRNA‐143‐3p inhibits CRC invasion and metastasis. Furthermore, we reveal the following target genes of miR‐143‐3p: integrin alpha 6 (ITGA6) and Arf GTPase activating protein (ArfGAP) with SH3 domain, ankyrin repeat, and PH domain 3 (ASAP3), and their reciprocal relationship was confirmed in CRC cells and tissues. Our findings suggest that miR‐143‐3p, ITGA6, and ASAP3 could be valuable targets for the management of CRC progression.
2. MATERIALS AND METHODS
2.1. Patients and tissue specimens
Clinical specimens for the miRNA microarray were obtained from 6 patients who had been diagnosed as having CRC with liver metastasis at the First Affiliated Hospital of Soochow University (Suzhou, China). Liver metastatic CRC tissues were obtained from these patients, and informed written consent was provided. All of the patients underwent surgical resection between March 2014 and December 2014, and none had received preoperative radiotherapy, chemotherapy, or other treatments. Specimen sampling included CRC tissue and corresponding normal colorectal tissue from an area more than 5 cm from the lesion. Specimens were obtained aseptically during surgery and were immediately placed in liquid nitrogen.
2.2. Total RNA extraction and quantitative RT‐PCR
Total RNA was harvested from CRC surgical specimens using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed according to the manufacturer's recommendations. The cDNA acted as the template for the amplification of RNA, and real‐time PCR was used for microRNA (miR)‐143‐3p amplification and TaqMan miRNA assays (Takara Shuzo, Kyoto, Japan) were used for quantification following amplification. The small nuclear RNA U6 was used as a normalization control. The RT reaction comprised of 16°C for 30 minutes, 42°C for 42 minutes, and 85°C for 5 minutes. The following program was used for quantitative PCR: 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and then 60°C for 60 seconds. Relative expression changes were calculated using the method.
2.3. MicroRNA array analysis
Total RNA was extracted from 3 sets of paired liver metastases from CRC tumors, primary CRC tumors, and adjacent normal tissues. RNA was quantified using a NanoDrop 1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA), and the samples were labeled using the miRCURY Hy3/Hy5 Power labeling kit and hybridized on the miRCURY LNA Array (version 6.0) (Exiqon, Vedbaek, Denmark). Following washing, the slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA, USA). Scanned images were then imported into GenePix Pro 6.0 software (Axon Instruments) for grid alignment and data extraction. Expressed data were normalized using median normalization. Finally, hierarchical clustering was carried out to determine distinguishable miRNA expression profiles among the samples.
2.4. Oligonucleotide synthesis and transfection
LNA/DNA hybrid oligonucleotides (miR mimic sequence, UGAGAUGAAGCACUGUAGCUC) and control oligonucleotides (scrambled miR sequence, UUUGAACUACAUAAAAGCACUG) were chemically synthesized (Guangzhou RiboBio, Guangzhou, China). For transfection, 5 μL Lipofectamine 2000 transfection reagent (Invitrogen) in 200 μL serum‐free medium was mixed with 100 pmol oligonucleotides dissolved in 200 μL of the same medium. Target gene‐specific siRNAs were designed using BLOCK‐iT RNAi Designer (Invitrogen). After confirmation by immunoblotting (data not shown), optimal ITGA6 siRNA (GTGGGAAGTTTAATAGAGT), ASAP3 siRNA (CTGTCAAAGTCGCCAACCA), and scrambled siRNA (UUUTGATCAUTGATGAAA) sequences were selected for ITGA6 and ASAP3 RNA interference (synthesized by Guangzhou RiboBio). The siRNA transfection was carried out following the same procedures used for microRNA at a final concentration of 30 nM.
2.5. Cell proliferation assay
A cell suspension (1 × 104 cells/mL) was prepared and seeded in a 96‐well cell culture plate at 100 μL/well. The test wells were then divided into the Mock group, the miR‐143‐3p Mimic group, and the Scrambled Mimic group. Cell proliferation was measured after transfection for 24, 48, 72, and 96 hours, and 5 replicate test wells were set up. Before inspection, 20 μL MTT solution at a concentration of 5 mg/mL was added to each well of the cell culture plate and incubation was continued for 4 hours in a 37°C incubator. Then the medium was carefully aspirated and 150 μL/well DMSO was added. A microplate reader was used to determine the OD value at 560 nm.
2.6. Migration and invasion assay
Transwell cell migration plates and Matrigel invasion chambers (Corning, Corning, NY, USA) were used to investigate the migratory and invasive capabilities of LoVo cells. First, for the invasion assay, 300 μL serum‐free medium was mixed with 60 μL Matrigel glue, and 50 μL of this solution was added to the upper chamber and incubated for 6 hours in a 37°C incubator, until it solidified. Cells (1 × 105 cells/mL) were then seeded in serum‐free medium into the upper chamber, and 20% FBS was added to the lower chamber as a chemoattractant to induce cells to invade the lower chamber. After 24 hours, the cells that had invaded the membrane and adhered to the underside of the membrane were stained by 0.2% crystal violet and the cells in randomized fields were counted. All assays were independently repeated 3 times.
2.7. Wound healing assay
Cells were cultured on 6‐well plates to form a single cell layer, and a straight wound line was made in the middle of the cell layer. After culturing for 24 or 48 hours, cells that migrated into the wound line were observed. Images of the cells along the wound line were then taken by phase‐contrast microscopy.
2.8. Western blot analysis
Cells were collected and lysed in TNES buffer. Equal aliquots of proteins were electrophoresed on SDS‐polyacrylamide gels and electrotransferred onto PVDF membranes. Membranes were blocked with TBS containing 5% nonfat dry milk for at least 1 hour. The blots were then incubated with 1 of the following Abs: anti‐ITGA6, anti‐ASAP3, anti‐musashi homolog 2 (MSI2) (Boster, Wuhan, China), anti‐cysteine‐rich with EGF‐like domains 1 (CRELD1), or anti‐β‐actin (Abcam, Cambridge, UK). Proteins were visualized with peroxidase‐conjugated secondary Abs at 1:2000 for 1 hour, using an enhanced chemiluminescence detection system (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
2.9. Tissue microarray construction
To construct the tissue microarray, CRC paraffin‐embedded tissue samples from 200 cases and normal colorectal tissue samples from 50 cases obtained at the First Affiliated Hospital of Soochow University were used. In the corresponding paraffin block, 1.5‐mm cores were punched out (1 core per case). These cores, each 5‐7 mm high, were then embedded in the donor block using a manually operated tissue microarray device.
2.10. Immunohistochemical analysis
The sections of paraffin‐embedded tissue were incubated with 0.3% hydrogen peroxide for 10 minutes to block endogenous peroxidase activity. The slides were then subjected to antigen retrieval as follows. After boiling in 10 mM citrate buffer for 10 minutes in a microwave oven, the tissue sections were incubated with 10% normal rabbit serum in buffer for 20 minutes to block nonspecific binding sites. Then consecutive sections were incubated with Abs (anti‐ITGA6 and anti‐ASAP3 [Abcam]) at 4°C overnight. The next day, the slides were washed 3 times with PBS and incubated with biotin‐labeled secondary Ab for 40 minutes at room temperature. After washing, DAB color‐substrate solution was added for 30 seconds, and then counterstained with hematoxylin. Expression was scored according to the total immunoreactive score (IRS) as follows: the staining extent was categorized as 0, no positive cells; 1, ≤25% positive cells; 2, >25% and ≤50% positive cells; or 3, >50% positive cells. Staining intensity was categorized as: 0, negative;1, weak; 2, moderate; or 3, strong. An IRS 6–9 and IRS 0–4 were defined as high expression and low expression of proteins, respectively. The immunostained slides were observed by 3 independent pathologists.
2.11. Statistical analysis
All data are presented as the mean ± SD. Statistical analyses were carried out using SPSS, version 20 (SPSS, Chicago, IL, USA). Independent sample t test was used to compare the significance of the differences between 2 groups. P < .05 was considered statistically significant after 2‐tailed t tests and significant differences are denoted by an asterisk in the figures. *P < .05, **P < .01, ***P < .001.
3. RESULTS
3.1. Differential expression of miR‐143‐3p between primary CRC and liver metastatic CRC
In our previous research, we used a miRNA microarray (Exiqon version 18.0) to detect miRNAs that were differentially expressed between 6 CRC tissues and normal colonic tissues and identified miR‐143‐3p as one of the most significantly decreased in CRC tissues (34.8‐fold) compared with normal colonic tissues (Query No. GSE53339). In this study, we used a miRNA microarray to detect the differential expression of miRNAs in 3 primary CRC tissues and liver metastatic CRC tissues (high, moderate, and poorly differentiated adenocarcinomas), and compared these with 3 normal colonic tissues. Hierarchical cluster analysis generated a dendrogram with 2 major branches both in columns (primary CRC vs normal, liver metastatic CRC vs normal) and in rows (upregulated vs downregulated genes). The criteria used to define significantly altered miRNAs were: (i) mean fold change >3 or <0.3333; and (ii) P‐value <0.05. Statistical analysis revealed only 1 miRNA that was significantly decreased in primary CRC tissue compared with normal colonic tissue, but more than 10 miRNAs were decreased in expression in liver metastases compared with normal colonic tissue, of which miR‐143‐3p showed the greatest decrease (59‐fold; Figure 1A and Table 1).
Figure 1.
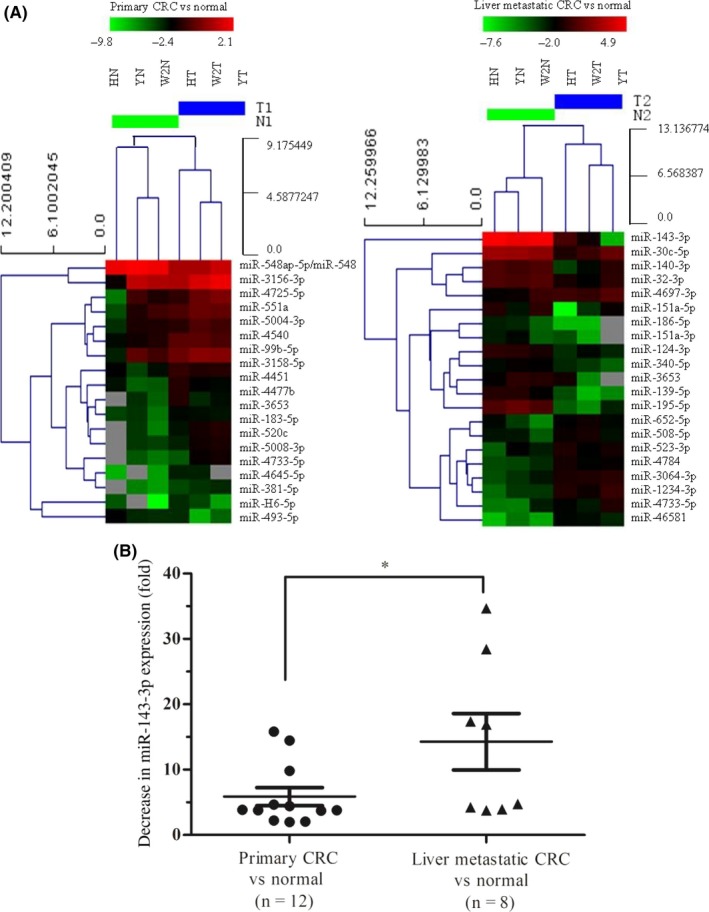
Differentially expressed microRNAs (miRNAs) in primary colorectal cancer (CRC) tissues and liver metastatic CRC tissues compared with normal colonic tissues. A, Clustering of differentially expressed miRNAs (pass volcano plot) in primary CRC tissues (HN, YN, and W2N) vs normal colonic tissues (HT, W2T, and YT) and liver metastatic CRC tissues (HN, YN, and W2N) vs normal colonic tissues (HT, W2T, and YT). B, Downregulated expression of miR‐143‐3p in the tissue samples of 12 cases of primary CRC and 8 cases of liver metastatic CRC compared with normal colonic tissue through RT‐PCR analysis. Results were analyzed with Student's t test (*P < .05)
Table 1.
MicroRNA (miRNA) microarray data showing miRNAs that were decreased more than 3‐fold between primary colorectal cancer (CRC) and liver metastatic CRC tissue and normal colonic tissue
| MiRNA | Fold change | |
|---|---|---|
| Primary CRC vs normal | miR‐493‐5p | 3 |
| Liver metastatic CRC vs normal | miR‐143‐3p | 59 |
| miR‐195‐5p | 16 | |
| miR‐139‐5p | 10 | |
| miR‐186‐5p | 8 | |
| miR‐30c‐5p | 4 | |
| miR‐140‐3p | 3 | |
| miR‐151a‐3p | 3 | |
| miR‐124‐3p | 3 |
Reverse transcription‐PCR was used to detect the expression level of miR‐143‐3p in 12 primary CRC tissues and 8 liver metastatic CRC tissues, compared with normal colonic tissue samples. The results showed that the proportion of primary CRC cases in which miR‐143‐3p was decreased by more than 5‐fold was 25% (3/12), whereas this proportion in liver metastatic CRC cases was 50% (4/8) (Figure 1B). This significant reduction in miR‐143‐3p expression in liver metastatic CRC suggests some underlying mechanism involving this miRNA.
3.2. MicroRNA‐143‐3p inhibits the invasion and migration of CRC cells in vitro
We transiently transfected synthesized miR‐143‐3p mimics into LoVo cells (which have low endogenous miR‐143‐3p expression) and verified that miR‐143‐3p expression was significantly increased in the transfected cells (Figure S1). To explore the effect of miR‐143‐3p on the biological behavior of CRC, we used an MTT assay to determine the proliferation ability of LoVo cells 24, 48, 72, and 96 hours after transfection with miR‐143‐3p‐mimics, but no significant differences between the miR‐143‐3p mimic‐treated group and the control group were detected (Figure 2A). We then used a Transwell assay to detect the migration and invasion ability of LoVo cells. As shown in Figure 2B, both the migration and invasion ability of LoVo cells significantly decreased 24 hours after transfection with miR‐143‐3p mimics compared with the control groups. These results indicated that miR‐143‐3p has no effect on the proliferation of LoVo cells, but has a regulatory role in the metastasis of CRC.
Figure 2.
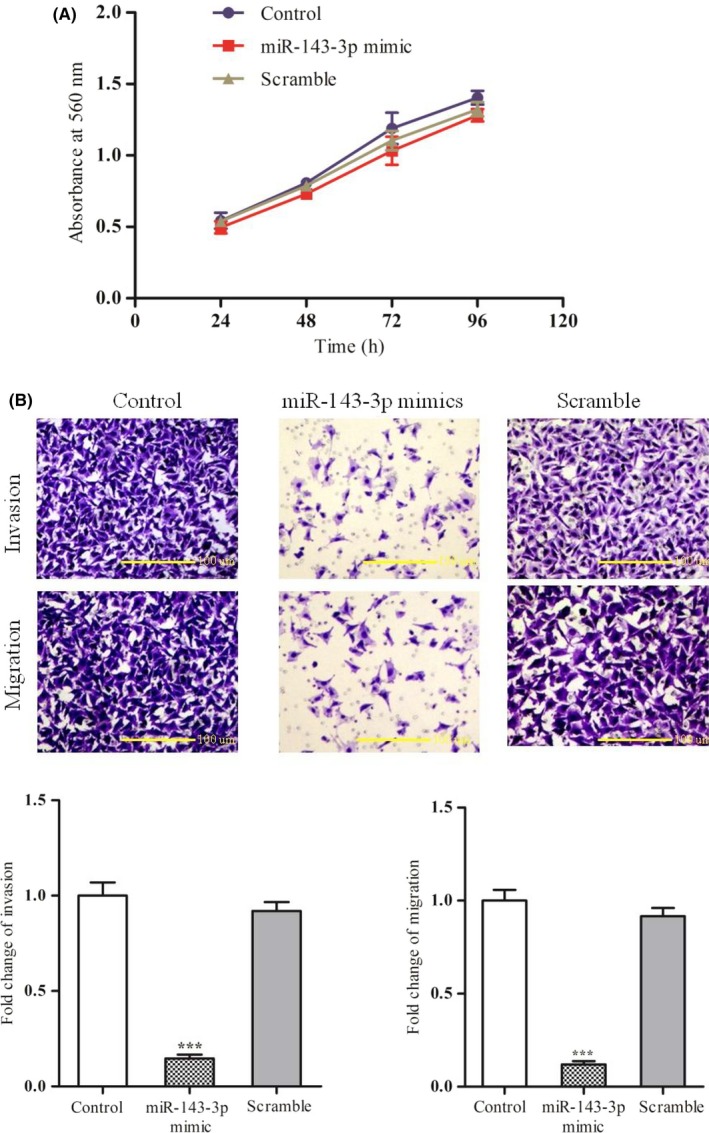
Effects of exogenous microRNA (miR)‐143‐3p on the behavior of colorectal cancer (CRC) cell lines. A, Transfection of miR‐143‐3p mimics verified the effect of overexpression of miR‐143‐3p on the proliferation of LoVo cells. B, Effect of miR‐143‐3p on the migration and invasion of LoVo cells was determined by invasion and migration assays in LoVo cells infected with miR‐143‐3p or scrambled mimics or the control group. Upper panels, images of each group. Lower panels, statistical calculations. Each assay was undertaken at least 3 times independently. Error bars indicate SD. ***P < .005 vs corresponding Scramble
3.3. ITGA6 and ASAP3 are targets of miR‐143‐3p
Bioinformatic searches using the online resources TargetScan, MIRDB, DIANA‐MICROT, and MICRORNA.ORG, identified 20 predicted targets of miR‐143‐3p that overlapped between these 4 bioinformatics websites. Then, based on the analysis of function using data from the miR‐Ontology database, we identified 4 target genes of miR‐143‐3p: ASAP3, MSI2, CRELD1, and ITGA6 (Figure 3A). The correlation between these target genes and CRC has not previously been reported.
Figure 3.
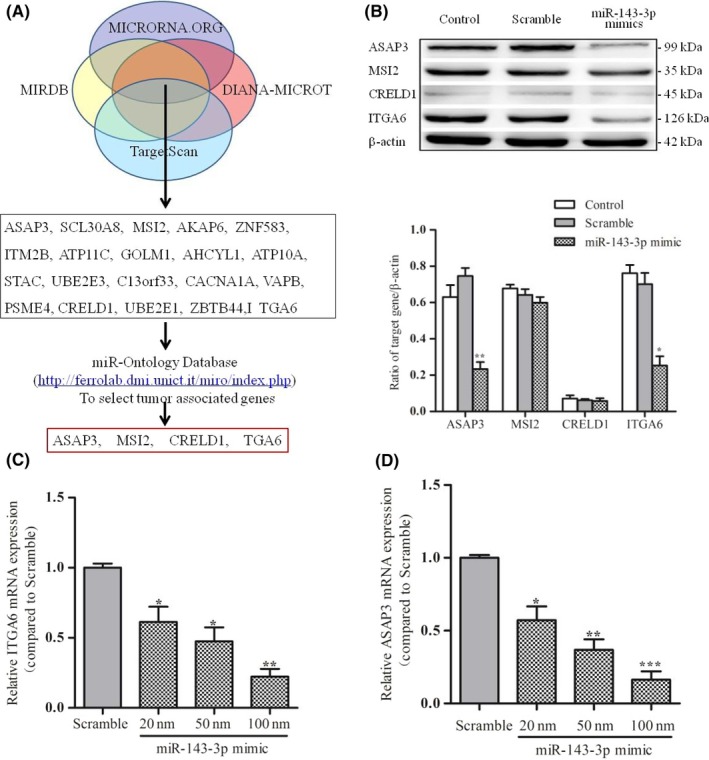
ITGA6 and ASAP3 are direct targets of microRNA (miR)‐143‐3p. A, Target genes of miR‐143‐3p were predicted using 4 online bioinformatics resources, TargetScan, MIRDB, DIANA‐MICROT, and MICRORNA.ORG. B, Protein expression levels of each candidate target gene were detected by western blotting in LoVo cells transfected for 48 hours with miR‐143‐3p mimics, scrambled mimics, or the control (empty) group. Upper panel, electrophorograms of each group. Lower panels calculations. C,D, Real‐time PCR was used to detect the expression of ITGA6 and ASAP3 in LoVo cells transfected with different concentrations of miR‐143‐3p mimics (scrambled mimic group and miR‐143‐3p mimic group). Each assay was undertaken at least 3 times independently. Error bars indicate SD. *P < .05, **P < .01, ***P < .005 vs corresponding Scramble
To determine whether miR‐143‐3p was negatively correlated with the expression of candidate target genes, we used western blot analysis to detect the expression of each candidate target gene in LoVo cells transfected with miR‐143‐3p mimics. The results showed that only ITGA6 and ASAP3 were significantly reduced in expression among these predicted target genes (Figure 3B). Real‐time PCR was then carried out and revealed that the increase in miR‐143‐3p concentration following the transfection of miR‐143‐3p mimics resulted in a gradual decrease in the mRNA expression of the ITGA6 and ASAP3 genes. When the concentration of transfected miR‐143‐3p mimics was 100 nmol, the expression level of ITGA6 mRNA was decreased by approximately 4.5‐fold, and the expression level of ASAP3 mRNA was decreased by more than 5 times that in the control groups (P < .01) (Figure 3C,D). These results suggested that ITGA6 and ASAP3 are likely target genes of miR‐143‐3p.
3.4. Invasive and migration of CRC cells were attenuated by endogenous ITGA6 and ASAP3 knockdown
To investigate whether miR‐143‐3p acts as a tumor suppressor through inhibiting the expression of ITGA6 and ASAP3 to weaken their proto‐oncogene effect, we used RNA interference technology to knockdown the expression of ITGA6 and ASAP3 in LoVo cells and explored the resulting invasion and migration of CRC cells. Quantitative PCR confirmed that the expression of ITGA6 and ASAP3 was decreased by more than 10‐fold in siRNA‐treated LoVo cells compared with control groups at the mRNA level (Figure S2). Similarly, both ITGA6 siRNA and ASAP3 siRNA were significantly reduced in expression in LoVo cells at the protein level (Figure 4A).
Figure 4.
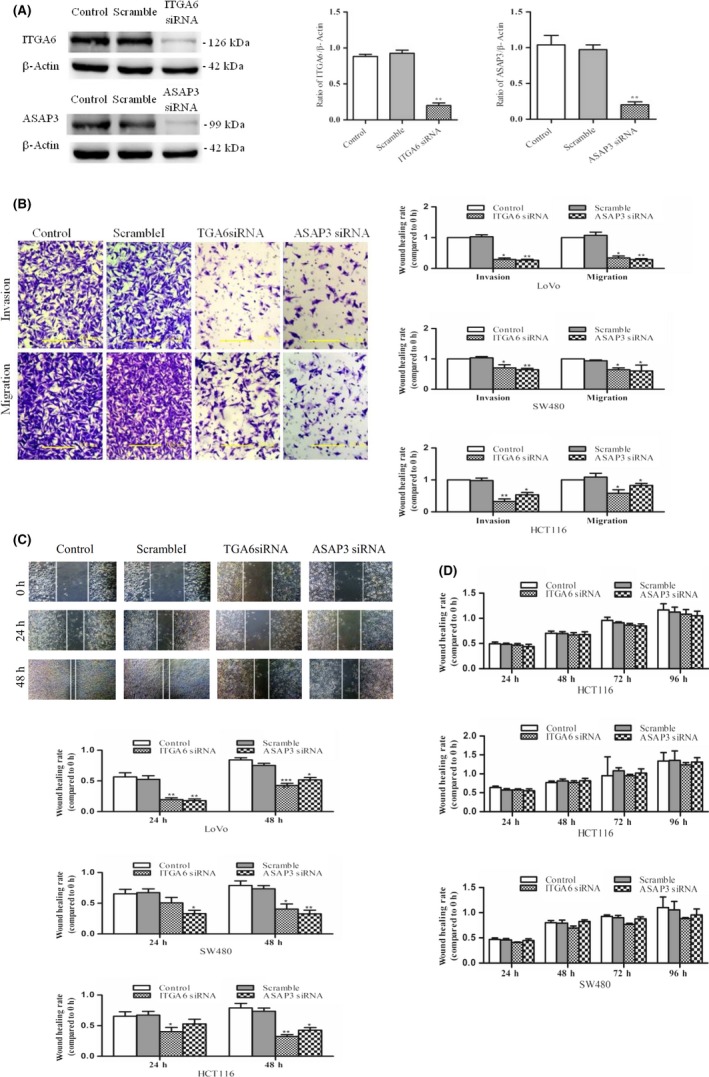
Invasion and metastasis of colorectal cancer (CRC) cells were attenuated by endogenous ITGA6 and ASAP3 knockdown. A, ITGA6 and ASAP3 were detected by western blotting in LoVo cells transfected with ITGA6 or ASAP3 siRNA, scrambled siRNA (Scramble), and the empty group (Control). B, Transwell assay was used to detect the invasive and migratory potential of CRC cell lines (LoVo, SW480, and HCT116) transfected with ITGA6 or ASAP3 siRNA. Left panels, representative photographs of each group. Right panels, statistical calculations. C, Effects of ITGA6 or ASAP3 siRNA on the migratory ability of CRC cell lines were examined by wound healing assays. Upper panels, representative photographs of each group. Lower panels, statistical calculations. D, MTT assay was used to detect the effect of ITGA6 or ASAP3 siRNA on the proliferation of CRC cell lines. Each assay was undertaken at least 3 times independently. Error bars indicate SD. *P < .05, **P < .01, ***P < .005 vs corresponding Scramble
The Transwell chamber assay was used to determine the effect of ITGA6 and ASAP3 siRNA transfection on the invasion and migration capacities of CRC cell lines (LoVo, SW480, and HCT116). As shown in Figure 4B, the number of transmembrane cells was significantly reduced in groups transfected with ITGA6 and ASAP3 siRNA compared with control groups as determined by migration and invasion assays. Statistical analysis showed that application of ITGA6 and ASAP3 siRNA had inhibitory effects on CRC cell lines (LoVo, SW480, and HCT116), influencing migration or invasion ability to various extents.
A wound healing assay showed that the migration distance of LoVo cells was significantly shorter in the two siRNA interference groups compared with the control groups (Figure 4C). The same results were confirmed in other cell lines (SW480 and HCT116). Then we applied an MTT assay to determine the proliferative ability of CRC cells following transfection with ITGA6 or ASAP3 siRNA. However, it was found that both ITGA6 and ASAP3 siRNA had no effect on the proliferation of CRC cell lines (LoVo, SW480, and HCT116; Figure 4D). Taken together, these results suggest that the overexpression of both ITGA6 and ASAP3 in CRC cells could play a key role in the progression of CRC and especially in the metastasis of this cancer.
3.5. ITGA6 and ASAP3 are highly expressed in CRC tissues
To investigate the expression of ITGA6 and ASAP3 in CRC, we prepared a tissue microarray with 200 CRC surgical specimens and 50 normal tissues (Figures 5 and S3) and the ITGA6 and ASAP3 protein expression levels are shown in Table 2. Compared with normal tissues, both ITGA6 and ASAP3 were upregulated in CRC tissues. Immunohistochemistry showed positive expression of ITGA6 protein mainly in the membrane and cytoplasm of cancer cells, compared with low expression of ITGA6 protein in the cell membrane of normal intestinal epithelial cells (Figure 5A). Similar to ITGA6 expression, ASAP3 was also positively expressed in the cytoplasm of cancer cells, but rarely in normal tissues (Figure 5B). Statistical analyses showed that the positive detection rates of ITGA6 in tumor tissues and normal tissues were 36% (72/200) and 20% (10/50), respectively, revealing a significant difference (P < .05, Table 2). Furthermore, the positive detection rates of ASAP3 in tumor tissues and normal tissues were 39.5% (79/200) and 16% (8/50), respectively; again these differences were statistically significant (P < .05, Table 2).
Figure 5.
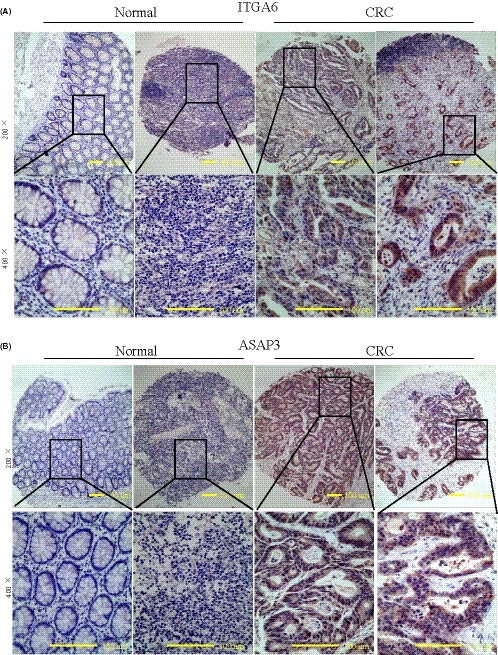
ITGA6 and ASAP3 are highly expressed in 200 colorectal cancer (CRC) tissue samples. A, Representative immunohistochemical staining of ITGA6 in CRC samples and normal intestine epithelium samples. B, Representative immunohistochemical staining of ASAP3 in CRC samples and normal intestine epithelium samples. Original magnification, ×100 on upper; ×400 on lower
Table 2.
Expression of ITGA6 and ASAP3 protein in colorectal cancer tissues and normal tissues
| ITGA6 expression | ASAP3 expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Low, n (%) | High, n (%) | χ2 | P | Low, n (%) | High, n (%) | χ2 | P | |
| Tumor (n = 200) | 128 (64) | 72 (36) | 4.646 | .031* | 121 (60.5) | 79 (39.5) | 10.253 | .008** |
| Normal (n = 50) | 40 (80) | 10 (20) | 42 (84) | 8 (16) | ||||
P < .05 indicates significance. *P < .05, **P < .01.
3.6. Relationship between clinicopathological features and ITGA6/ASAP3 protein expression
The results of immunohistochemistry of the 200 CRC tissue specimens on the tissue microarray showed that high ITGA6 expression positively correlated with the American Joint Committee on Cancer (AJCC) stage. The positive rate of ITGA6 expression was 43.14% in stage III + IV cancer specimens and only 28.57% in stage I + II cancer specimens, thereby showing a significant difference between the 2 staging groups (P = .032), whereas no significant differences were observed with patients’ gender, age, tumor size, differentiation, depth of invasion, or metastasis of lymph nodes (Table 3). High ASAP3 expression levels positively correlated with the metastasis of lymph nodes and the AJCC stage of CRC. The positive rate of ASAP3 expression was 29.70% in the nonlymph node metastasis group, and 49.49% in the lymph node metastasis group (P = .006). The positive rate of ASAP3 expression was 47.06% in the AJCC stage III + IV stage group of specimens compared with 31.63% in the group I + II stage specimens (P = .030), whereas no significant differences were observed with patients’ gender, age, tumor size, depth of invasion, or differentiation (Table 3).
Table 3.
Relationship between clinicopathological features and ITGA6/ASAP3 protein expression in 200 colorectal cancer patients
| Pathological features | ITGA6 expression | Positive rate (%) | P‐value | ASAP3 expression | Positive rate (%) | P‐value | ||
|---|---|---|---|---|---|---|---|---|
| Low (n = 128) | High (n = 72) | Low (n = 121) | High (n = 79) | |||||
| Gender and age | ||||||||
| Female | 60 | 34 | 36.17 | .962 | 52 | 42 | 44.68 | .158 |
| Male | 68 | 38 | 35.85 | 69 | 37 | 34.91 | ||
| <65 y | 60 | 37 | 38.14 | .540 | 58 | 39 | 40.21 | .843 |
| ≥65 y | 68 | 35 | 34.10 | 63 | 40 | 38.83 | ||
| Tumor size and depth | ||||||||
| <5 cm | 83 | 44 | 34.65 | .599 | 78 | 49 | 38.58 | .726 |
| ≥5 cm | 45 | 28 | 38.36 | 43 | 30 | 41.10 | ||
| Mucosa involved | 2 | 1 | 33.33 | .563 | 3 | 0 | 0.00 | .128 |
| Muscle involved | 15 | 6 | 28.57 | 13 | 8 | 38.10 | ||
| Serosa involved | 46 | 21 | 31.34 | 46 | 21 | 31.34 | ||
| Differentiation | ||||||||
| Well | 17 | 11 | 39.29 | .369 | 23 | 5 | 17.86 | .050 |
| Moderate | 102 | 52 | 33.77 | 91 | 63 | 40.91 | ||
| Poor | 9 | 9 | 50.00 | 7 | 11 | 61.11 | ||
| Lymph node involved | ||||||||
| No metastasis | 69 | 32 | 31.68 | .239 | 71 | 30 | 29.70 | .006* |
| Metastasis | 59 | 40 | 40.40 | 50 | 49 | 49.49 | ||
| AJCC stage | ||||||||
| I + II | 70 | 28 | 28.57 | .032* | 67 | 31 | 31.63 | .030* |
| III + IV | 58 | 44 | 43.14 | 54 | 48 | 47.06 | ||
P < .05 indicates significance.
AJCC, American Joint Committee on Cancer.
4. DISCUSSION
The downregulation of miR‐143‐3p has been reported in endometrial cancer,9 non‐small cell lung cancer,10 bladder cancer,11 and in liver metastases compared with their primary CRC,12 but the mechanisms are not well established. Here, we revealed that the expression of miR‐143‐3p was most significantly decreased in liver metastases CRC tissues compared with primary CRC tissues and miR‐143‐3p overexpression showed the inhibitory effect of miR‐143‐3p on the migration and invasion potential of cells in vivo. Therefore, we hypothesized that miR‐143‐3p is a tumor suppressor for CRC liver metastases.
MicroRNAs are known to carry out their biological functions through regulating the protein expression of downstream target genes, thus affecting the occurrence and development of tumors. To date, a number of possible target genes have been identified for miR‐143‐3p including quaking I‐5 protein (QKI‐5),13 AKT serine/threonine kinase 2 (AKT2),14 mitogen‐activated protein kinase 7 (MAPK7),15 and DNA methyltransferase 3A (DNMT3A).16 In this study, we used bioinformatics software to predict 108 possible target genes of miR‐143‐3p and by real‐time quantitative PCR confirmed that the expression levels of both ITGA6 and ASAP3 negatively correlated with miR‐143‐3p in LoVo cells transfected with miR‐143‐3p mimics. Subsequent western blot analysis further confirmed the negative correlation between ITGA6, ASAP3, and miR‐143‐3p. We then showed that knockdown of endogenous ITGA6 and ASAP3 reduced CRC cell invasion and migration in vitro, similar to miR‐143‐3p restoration. If the formation of CRC is a consequence of multiple gene changes in the cells of the colorectal epithelium, as generally believed, then expression of the proto‐oncogenes ITGA6 and ASAP3 might contribute to the progression of CRC, including the later stages involving metastasis.
As a transmembrane glycoprotein‐adhering receptor, ITGA6 mediates cell‐matrix and cell‐cell adhesion.17, 18 Several studies have shown that ITGA6 is involved in the regulation of biological behavior including tumor cell adhesion, migration, invasion, and survival.19, 20 The expression of ITGA6 in breast cancer is more effective than estrogen receptor levels at predicting decreased survival rates.21 ITGA6 is overexpressed in 70% of cases of advanced prostate cancer and prostate cancer with metastases.22 As a known tumor suppressor in many cancer types, including CRC, p53 was shown to inhibit CRC invasion and metastasis by targeting ITGA6 and ITGB1.23 Also known as UPLCl, ACAP4, or DDEFLl, ASAP3 is a newly discovered member of the ArfGAP family, has multiple structural domains, and could play an important role in cytoskeletal assembly, cell growth, and migration. A few studies have confirmed that the upregulated expression of ASAP3 is associated with the development and clinical outcomes of mammary carcinoma,24 non‐small cell lung cancer cells, and ovarian serous carcinoma.25, 26 A recent report showed that upregulation of ASAP3 contributes to colorectal carcinogenesis and indicates poor survival outcomes.27
We used a tissue microarray constructed from 200 cases of CRC and 50 cases of colorectal adenoma tissues to analyze the clinicopathological correlation with ITGA6 and ASAP3 expression. The frequency and extent of ITGA6 and ASAP3 expression were enhanced in the transition from adenoma to CRC, whereas increased expression was not observed in colorectal adenomas. Analysis of this clinicopathological correlation revealed that higher expression of ITGA6 was closely related to the advanced clinical stage, and higher expression of ASAP3 was closely related to lymph node metastasis and an advanced clinical stage in CRC patients. These results indicated that ITGA6 and ASAP3 could be valuable prognostic biomarkers for CRC patients. However, no significant difference was observed in different degrees of differentiation, with classification criteria based only on morphological features.
In summary, we detected a more significant decrease in miR‐143‐3p expression levels in clinical specimens of CRC than in normal tissues, and revealed that miR‐143‐3p mimics inhibit the invasion and migration of CRC cells in vitro. Using a bioinformatics approach and functional assays, we identified ITGA6 and ASAP3 as the target genes of miR‐143‐3p, and the metastatic potential of the CRC cells was attenuated by endogenous ITGA6 and ASAP3 knockdown. Hence, our data provide a new molecular mechanism for CRC progression and miR‐143‐3p could prove to have therapeutic or prognostic value for the management of CRC in the future.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Guo L, Fu J, Sun S, et al. MicroRNA‐143‐3p inhibits colorectal cancer metastases by targeting ITGA6 and ASAP3. Cancer Sci. 2019;110:805–816. 10.1111/cas.13910
Funding information
This study was supported by the National Natural Science Foundation of China (81472297) and the Priority Academic Program Development of Jiangsu Higher Education Institutions
Guo, Fu and Sun contributed equally to this study.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. De Greef K, Rolfo C, Russo A, et al. Multisciplinary management of patients with liver metastasis from colorectal cancer. World J Gastroenterol. 2016;22:7215‐7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adam R, De Gramont A, Figueras J, et al. The oncosurgery approach to managing liver metastases from colorectal cancer: a multidisciplinary international consensus. Oncologist. 2012;17:1225‐1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adam R, Haller DG, Poston G, et al. Toward optimized front‐line therapeutic strategies in patients with metastatic colorectal cancer–an expert review from the International Congress on Anti‐Cancer Treatment (ICACT) 2009. Ann Oncol. 2010;21:1579‐1584. [DOI] [PubMed] [Google Scholar]
- 5. Liu S, Kurzrock R. Toxicity of targeted therapy: implications for response and impact of genetic polymorphisms. Cancer Treat Rev. 2014;40:883‐891. [DOI] [PubMed] [Google Scholar]
- 6. Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882‐891. [PubMed] [Google Scholar]
- 7. Dassow H, Aigner A. MicroRNAs (miRNAs) in colorectal cancer: from aberrant expression towards therapy. Curr Pharm Des. 2013;19:1242‐1252. [DOI] [PubMed] [Google Scholar]
- 8. To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: from bench to bedside. World J Gastroenterol. 2018;24:2949‐2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X, Dong Y, Ti H, et al. Down‐regulation of miR‐145 and miR‐143 might be associated with DNA methyltransferase 3B overexpression and worse prognosis in endometrioid carcinomas. Hum Pathol. 2013;44:2571‐2580. [DOI] [PubMed] [Google Scholar]
- 10. Zhang HB, Sun LC, Ling L, Cong LH, Lian R. miR‐143 suppresses the proliferation of NSCLC cells by inhibiting the epidermal growth factor receptor. Exp Ther Med. 2016;12:1795‐1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang H, Li Q, Niu X, et al. miR‐143 inhibits bladder cancer cell proliferation and enhances their sensitivity to gemcitabine by repressing IGF‐1R signaling. Oncol Lett. 2017;13:435‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vychytilova‐Faltejskova P, Pesta M, Radova L, et al. Genome‐wide microRNA expression profiling in primary tumors and matched liver metastasis of patients with colorectal cancer. Cancer Genomics Proteomics. 2016;13:311‐316. [PubMed] [Google Scholar]
- 13. He Z, Yi J, Liu X, et al. MiR‐143‐3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial‐mesenchymal transition by targeting QKI‐5 in esophageal squamous cell carcinoma. Mol Cancer. 2016;15:51. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Wang F, Liu J, Zou Y, et al. MicroRNA‐143‐3p, up‐regulated in H. pylori‐positive gastric cancer, suppresses tumor growth, migration and invasion by directly targeting AKT2. Oncotarget. 2017;8:28711‐28724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia C, Yang Y, Kong F, Kong Q, Shan C. MiR‐143‐3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of MAPK7. Biochimie. 2018;147:98‐104. [DOI] [PubMed] [Google Scholar]
- 16. Ng EK, Li R, Shin VY, Siu JM, Ma ES, Kwong A. MicroRNA‐143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumour Biol. 2014;35:2591‐2598. [DOI] [PubMed] [Google Scholar]
- 17. Hehlgans S, Haase M, Cordes N. Signalling via integrins: implications for cell survival and anticancer strategies. Biochim Biophys Acta. 2007;1775:163‐180. [DOI] [PubMed] [Google Scholar]
- 18. Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028‐1032. [DOI] [PubMed] [Google Scholar]
- 19. Mukhopadhyay R, Theriault RL, Price JE. Increased levels of alpha6 integrins are associated with the metastatic phenotype of human breast cancer cells. Clin Exp Metastasis. 1999;17:325‐332. [DOI] [PubMed] [Google Scholar]
- 20. Kim HI, Huang H, Cheepala S, Huang S, Chung J. Curcumin inhibition of integrin (alpha6beta4)‐dependent breast cancer cell motility and invasion. Cancer Prev Res (Phila). 2008;1:385‐391. [DOI] [PubMed] [Google Scholar]
- 21. Hu T, Zhou R, Zhao Y, Wu G. Integrin alpha6/Akt/Erk signaling is essential for human breast cancer resistance to radiotherapy. Sci Rep. 2016;6:33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Putz E, Witter K, Offner S, et al. Phenotypic characteristics of cell lines derived from disseminated cancer cells in bone marrow of patients with solid epithelial tumors: establishment of working models for human micrometastases. Cancer Res. 1999;59:241‐248. [PubMed] [Google Scholar]
- 23. Laudato S, Patil N, Abba ML, et al. P53‐induced miR‐30e‐5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int J Cancer. 2017;141:1879‐1890. [DOI] [PubMed] [Google Scholar]
- 24. Ha VL, Bharti S, Inoue H, et al. ASAP3 is a focal adhesion‐associated Arf GAP that functions in cell migration and invasion. J Biol Chem. 2008;283:14915‐14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fan C, Tian Y, Miao Y, et al. ASAP3 expression in non‐small cell lung cancer: association with cancer development and patients’ clinical outcome. Tumour Biol. 2014;35:1489‐1494. [DOI] [PubMed] [Google Scholar]
- 26. Willis S, Villalobos VM, Gevaert O, et al. Single gene prognostic biomarkers in ovarian cancer: a meta‐analysis. PLoS One. 2016;11:e0149183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tian H, Qian J, Ai L, et al. Upregulation of ASAP3 contributes to colorectal carcinogenesis and indicates poor survival outcome. Cancer Sci. 2017;108:1544‐1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


