Abstract
Mycobacterium avium subspecies paratuberculosis (MAP), as an obligate intracellular bacterium, causes paratuberculosis (Johne’s disease) in ruminants. Plus, MAP has consistently been isolated from Crohn’s disease (CD) lesions in humans; a notion implying possible direct causative effect for MAP in CD development. Infections caused by MAP are refractory to treatment and in many cases the treatment does not easily resolve the infection. Studying the molecular mechanisms of host-pathogen interaction is helpful in identifying possible drug targets. In this line, it has already been shown that in macrophages infected with various bacteria, including mycobacteria, micro RNA 21 (miR-21) is upregulated, a change that results in diminished macrophages clearance ability and favours pathogens survival within the cells. However, the molecular mechanism(s) by which the intracellular bacteria induce miR-21 expression is not known. In order to verify possible effects from epigenetic changes induced by intracellular bacteria, we studied the cytosine methylation changes at the transcription start regions of miR-21 in THP-1 macrophages infected with MAP. For this purpose, genomic DNA was extracted from infected cells and the methylation status at the region of interest was evaluated by bisulfite conversion method. Our work showed that MAP directs de-methylation of the cystosines at CpG di-nucleotides in this region, while non-CpG cytosines of this region did not show significant changes. Interestingly, the CpG cytosines that were differentially methylated in the infected macrophages occur at the binding sites of the transcription factors already known to regulate miR-21 expression.
Key Words: Crohn’s disease, Johne’s disease, miR-21, Mycobacterium
Introduction
Intracellular microorganisms, including the intracellular bacteria, use various strategies to avoid the host hostile reaction and survive inside the cells. Autophagy, as a sophisticated mechanism that is controlled by many genes and molecules, is one of the main processes by which the cells react against the invading intracellular pathogens (Wileman, 2013).
Micro RNAs (miRNA) are small non-coding RNAs that regulate gene expression, mostly post-transcriptionally by inhibition of translation (Felekkis et al., 2010; Catalanotto et al., 2016). These small RNAs are involved in various biological processes and enable cells to optimize the expression of protein coding genes. Micro RNAs (miRNA) also target long non-coning RNAs (lncRNA) (Jeggari et al., 2012) and the important functions of lncRNAs in normal and pathological conditions are extensively emerging (Schmitz et al., 2016).
Micro RNA 21 (miR-21) has been identified as one of the main and negative controllers of autophagy (Seca et al., 2013; Liu et al., 2015). It has been shown that miR-21 targets several key genes, such as PTEN (He et al., 2015; Peralta-Zaragoza et al., 2016), RAB11a (Liu et al., 2015), Bcl-2 (Wu et al., 2012; Wang et al., 2014; Sims et al., 2017), all involved in the progression of an autophagic reaction. In line with this, several research works have shown that upregulation of miR-21 in macrophage cells results in suppression of autophagy so that the process of clearing the invading agent fails (Wang et al., 2014). In fact, it is now experimentally proven that many bacteria, such as Mycobacteriums (Liu et al., 2012; Wu et al., 2012; Zheng et al., 2015; Das et al., 2016) Salmonella (Schulte et al., 2011) and Helicobacter (Zhang et al., 2008) use this specific strategy and somehow induce the miR-21 expression in the infected macrophages.
Gene expression, including miRNA expression, is controlled at various levels inside the eukaryote cells (Davis and Hata, 2009; Barrett et al., 2012; Bronevetsky and Ansel, 2013) and the epigenetic status of genome is an important determinate in this regard (Liu et al., 2013; Sandoval et al., 2015). Many intracellular pathogens, indeed, affect the normal epigenetic signature of their host to subvert the host killing power (Huang et al., 2012; Marr et al., 2014; Pacis et al., 2015). DNA methylation is one of the epigenetic mechanisms by which cells specifically regulate the expressional behaviour of their genes (Moore et al., 2013). Until recently, it was believed that only cytosine methylation at CpG islands located at the promoter regions of genes are important in gene expression regulation (Bird, 1980). However, there are now indications that cytosines at CpG downstream of transcription start sites (TSS) may be involved in transcription regulation as well (Krinner et al., 2014). Furthermore, Sharma et al. (2015) recently conducted a global methylation analysis on THP-1 macrophages infected with Mycobacterium tuberculosis and showed that this bacterium regulates methylation of cytosines mostly at non-CpG sites that were preferably situated around transcription start sites and in gene bodies; therefore, evidence on the importance of non-CpG methylation is emerging (Patil et al., 2014; Sharma et al., 2015).
Pathogenic mycobacteria, as the cause of several human and animal intractable diseases, are intracellularly residing bacteria (Cosma et al., 2003). These pathogens manage to induce significant debilitating changes in the autophagy processes; a strategy that enables them to survive and propagate for long periods inside the infected host macrophages (Kuehnel et al., 2001; Kathania et al., 2011; Hmama et al., 2015; Weiss and Schaible, 2015). Mycobacterium avium subspecies paratuberculosis (MAP) is an animal pathogen that causes substantial economic losses in livestock worldwide (Lombard., 2011). Though Mycobacterium avium subspecies paratuberculosis has not yet been identified as a direct human pathogen, but this bacterium has reproducibly been isolated from lesions and peripheral blood mononuclear cells in people affected with Crohn’s disease (CD) (Naser et al., 2014; McNees et al., 2015). Since this bacterium survive the usual pasteurization treatments applied to milk (Lund et al., 2002; Paolicchi et al., 2012), scientists believe in a causal link between the milk from the infected animals and the development and/or progression of the CD (Pierce., 2010; Paolicchi et al., 2012; Naser et al., 2014; McNees et al., 2015).
Since mycobacterial infections are relatively refractory to treatment and considering the consistent emergence of antibiotic resistance, molecular delineation of pathobiology of the diseases caused by these bacteria would be helpful in defining new therapeutic targets. Considering the documented role of miR-21 in autophagy (Seca et al., 2013; He et al., 2015; Liu et al., 2015; Song et al., 2016) and the notion that this miRNA is differentially regulated in many bacterial infections including in that of Mycobacterium (Liu et al., 2012; Wu et al., 2012; Vegh et al., 2015) we studied the cytosine methylation changes at the TSS area of miR-21 in THP-1 macrophages infected with MAP. Our specific aim here was to verify whether MAP may be inducing epigenetic changes at miR-21 locus as a negative regulator of autophagy.
Materials and Methods
MAP culture
For this study, a MAP sample previously isolated at the Microbiology Department of the School of Veterinary Medicine, Shiraz University, Iran was used. Herrold’s egg yolk (HEY) solid media agar (Herrold, 1931) supplemented with mycobactin J (MJ) (2 mg/L) was used to culture MAP cells. Four weeks after seeding, MAP cultures were verified by acid-fast staining using Ziehl-Neelsen method and polymerase chain reaction (PCR) using primers amplifying IS900 insertion element (Pillai and Jayarao, 2002; Bhide et al., 2006). For liquid cultures, a loop full sample from the bacteria grown on the solid medium was dissolved in 1 ml of Middle Brook 7H9 and 250 µL of this preparation was used to seed 2.5 ml Middle Brook 7H9 medium (Cat. No. M198, Himedia, India) supplemented with 2 mg/L of MJ and OADC [OADC fractions in final medium preparation: 10% oleic acid, (Cas No. 112-80-1, Sigma, Germany); 0.4% glycerol, (Cat. No. 1040921000, Merck, Germany); 2.5 g/500 ml of albumin fraction V, (Cat. No. 12657, Merck, Germany); 1 g/500 ml of dextrose, (Cat. No. 50-99-7, Sigma, Germany); 0.0015 g/500 ml of catalase, (Cas No. 9001-05-2, Sigma, Germany); 0.05% tween 80, (Cat. No. 1.46376.0006, Merck, Germany)]. The liquid cultures were grown in round bottom screw cap glass tubes in a microbiology shaker incubator at 37°C while being shaken at 7 rpm. The presence of MAP in fully grown liquid cultures was confirmed by acid fast staining and PCR as explained earlier.
MAP growth pattern
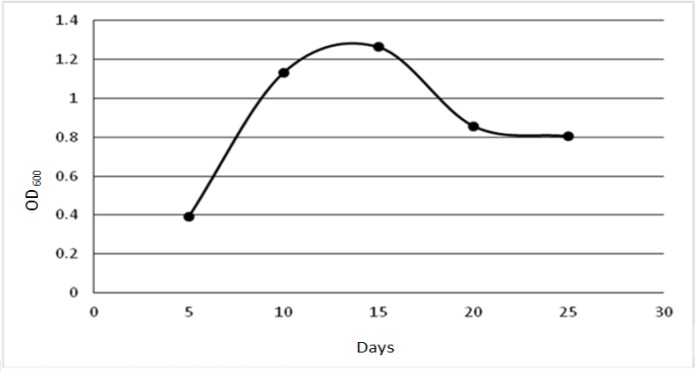
For determining the growth pattern of the MAP used in this study, 10 liquid cultures were seeded as explained earlier, and at five time points, i.e. on days 5, 10, 15, 20 and 25 of seeding two cultures were used to estimate the MAP cells number. For this purpose, at each time point the cultures were homogenized by vortexing for several minutes and the content of each tube was then centrifuged in two 1.5 ml Eppendorf microtubes at 8000 × g for 10 min. The pellets were washed twice in PBS by vortexing and pelleting as explained earlier and after the final wash the pellets in each microtube were resuspended in 1.25 ml of PBS. The MAP cells were dispersed by sequentially passing the content of each mircotube through syringes gauge 24 (5 times), 26 (5 times) and 30 (15 times). The final preparations were used to read the OD600 using an Eppendorf spectrophotometer (Biophotometer Uv/Vis, Eppendorf, USA). The MAP growth curve was prepared using the average ODs for each time point (Fig. 1).
Fig. 1.
This figure depicts the growth pattern of the MAP that was used in this study
Preparing MAP for THP-1 infection
Five hundred µL of the homogenized (by votrexing) MAP liquid cultures at their late logarithmic phase was used to pellet MAP cells as described earlier under Growth Pattern section. The MAP cells were washed twice in plain RPMI1640 (Cat. No. L0500-500, Biowest, France) and resuspended in 1 ml of plain RPMI1640 by vortexing. The preparations were then sequentially passed through syringes gauge 24 (5 times), 26 (5 times) and 30 (15 times), and were left to sit at room temperature for 5 min. The top one third of the preparations was then used to estimate the cell numbers using a spectrophotometer (OD600 of 0.6 is estimated to have 1 × 108 cells/ml (Kumar et al., 2010) and to infect the cells at the multiplicity of infection (MOI) of 10.
Cell culture and infection
THP-1 cells were obtained from the National Cell Bank of Iran (NCBI), Pasteur Institute, Iran. The cells were maintained in their exponential growth phase and under five passages in complete RPMI1640 (Cat. No. L0500-500, Biowest, France). Complete RPMI1640 contained: 10% fetal bovine serum (FBS) (Cat. No. 10439-024, Gibco, Germany), 100 IU/ml penicillin (Cat. No. L0022-100, Biowest, France), 100 µg/ml streptomycin (Cat. No. L0022-100, Biowest, France), 2 mm L-glutamine (Cat. No. X0550-100, Biowest, France), and 50 µM 2-mercaptoethanol (2-ME) (Cas. No. 60-24-2, Sigma, USA).
For infecting the THP-1 cells, 35 mm culture dishes were used to differentiate 6 × 105 THP-1 cells at their exponential phase of growth using Phorbol 12-myristate 13-acetate (PMA) (Cat. No. P 1585, Sigma) at a final concentration of 25 ng/ml. 48 h after PMA treatment the cells were gently washed with antibiotic free complete medium and 1 ml of antibiotic free complete medium was placed over the cells. Mycobacterium avium subspecies paratuberculosis cells were added to the cells at MOI of 10 and the dishes were then centrifuged at 100 × g for 5 min before being incubated in a cell culture incubator at 37°C in the presence of 5% CO2. Four h after infection cells were gently washed twice with complete RPMI and incubated in RPMI containing 10 μg/ml gentamicin (Sina Darou, Iran) for 2 h to remove any remaining extra-cellular bacteria. Subsequently, the media were replaced with complete RPMI and 72 h after infection cells were used for DNA extraction.
Cell culture staining
Thirty-five mm dishes containing the infected THP-1 cells were used to stain using Kinyoun modified Ziehl-Neelsen method (Kinyoun, 1915). Briefly, the cell culture medium was removed and the cells were left in open air to dry. The cells were then covered with carbol fuchsin (Cat. No. 108512, Merck Millipore, Germany) for 5 min while being briefly touch-heated followed by decolourization in acid/alcohol (1 ml HCl in 99 ml ethanol) for 3 min. Finally, the cells were stained in methylene blue (Cat. No. 115943, Merck Millipore, Germany) preparation for 3 min.
DNA extraction and bisulfite conversion
For DNA extraction from MAP cells, a boiling method was used. Briefly, 1 ml culture was centrifuged at 13000 rpm for 10 min in an Eppendorf microtube then the pellet was resuspended in 400 µL of 50 mM EDTA aqueous solution by vortexing. The preparation was left at room temperature for 30 min followed by 1 h of incubation at 37°C and then placed in boiling water for 8 min. 5 µL from the top of the preparation was used for each PCR reaction of 20 µL final volume.
For DNA extraction from THP-1 cells, QIAamp DNA Mini Kit (Cat. No. 51304, Qiagen, Germany) was used and the DNA was extracted as per the manufacturer’s instruction. The extracted DNA was determined to be of good quality by measurements obtained using a NanoDrop 2000 instrument (Thermo Scientific, USA). For bisulfite conversion, the EpiTect Bisulfite kit (Cat. No. 59104, Qiagen, Germany) was used and the instructions given by the manufacturer were followed. For each conversion reaction 300 ng of DNA in 140 µL final reaction volume was used.
Primers and PCR amplification of converted DNA
Primers used for amplification of IS900 insertion element of MAP are shown in (Table 1). The PCR condition for IS900 amplification was as follows: 94°C/5 min, 30 × (94°C/1 min, 59°C/1 min, 72ºC/9 min). The MethPrimer (Li and Dahiya, 2002) tool was used to design primers for amplification of the converted DNA and after addition of M13 virus sequences to the 5´ ends of the forward and reverse primers (Table 1) the primers were ordered to be synthesized by Macrogen (http://www.macrogen.com, Korea). 1.5 µL of the converted DNA was used for PCR amplification at the following conditions: 95°C/10 min, 5 × (95°C/20 s, 52°C/2 min, 72°C/3 min), 40 × (95°C/20 s, 52°C/60 s, 72°C/3 min), and 60°C/60 min.
Table 1.
The sequences of the primers used for PCR. Bold sequences show the M13 sequences
| IS900 Primers | 5´ to 3´ Sequences |
|---|---|
| P90 | GTTCGGGGCCGTCGCTTAGG |
| P91 | GAGGTCGATCGCCCACGTGA |
| miR-21 promoter primers | |
| Forward _ large segment | TGTAAAACGACGGCCAGTTATGTTTTATTGGGAAATTTGT |
| Reverse _ large segment | CAGGAAACAGCTATGACCCCAATATTAAAAACTATTTTTAAAACA |
| Forward _ small segment | TGTAAAACGACGGCCAGTTTTTTTAAATGTGTTTTTTTT |
| Reverse _ small segment | CAGGAAACAGCTATGACCAAATAAAAACTATAAATATAATTTCAACC |
Cloning of the amplified converted DNA and sequencing
The PCR products were run on 1% agarose gel and the amplified product with the expected size was excised out. The DNA in the excised bands was purified using MinElute Gel Extraction kit (Cat. No. 28604, Qiagen, Germany) and used to clone into the T-Vector PMD20 (Cat. No. 3270, Takara, Japan) according to the manufacturer’s instructions. 2 µL of the cloning reaction was used to transform 50 µL of DH5α competent cells by heat shock. After growing the transformed bacteria in 1 ml S.O.C. broth (Hanahan., 1983) for 3 h at 37°C, the culture was used to seed selective Luria-Bertani (LB) agar plates containing 100 µg/ml of ampicillin (Jabber Ebne Hayyan, Iran), 100 µM of IPTG (Cat. No. PR911706, Sinaclon, Iran) and 40 µg/ml of X-Gal (Cat. No. PR911716, Sinaclon, Iran). Following 24 h incubation, the plates were inspected for the presence of white colonies and several white colonies were used to seed 5 ml selective LB broth containing 100 µg/ml ampicillin for plasmid DNA extraction. The following day the cultures were used to extract plasmid using EZ-10 Spin Column PCR Products Purification kit (Cat. No. BS363, Bio Basic, Canada) and the extracted DNA were sent to Macrogen (Korea) for sequencing, after being qualitatively and quantitatively evaluated by Nanodrop. The M13 forward and reverse primers were used for sequencing.
The sequence data were qualitatively controlled and further checked for the status of cytosine conversion, manually for non-CpG sites or by using BISMA Analyzer (Rohde et al., 2010) for CpG sites. The methylated sites repeated in more than one clone (i.e. detected in two or more clones) were considered for the statistical analysis.
Statistical analysis
The methylation experiment data were first evaluated using the Kolmogorov-Smirnov test (Wilcox, 2005) and since the distribution followed Poisson distribution, the Poisson regression (Zou, 2004) was used for the statistical analysis.
Results
MAP growth pattern
Ten MAP cultures in Middle brook 7H9 broth media were prepared as detailed in M&M section. On days 5, 10, 15, 20, 25 of seeding two cultures were used to prepare samples for spectrophotometry as explained in M&M section, Fig. 1 depicts the growth kinetic of the MAP cells used in this work.
MAP staining in cultured cells
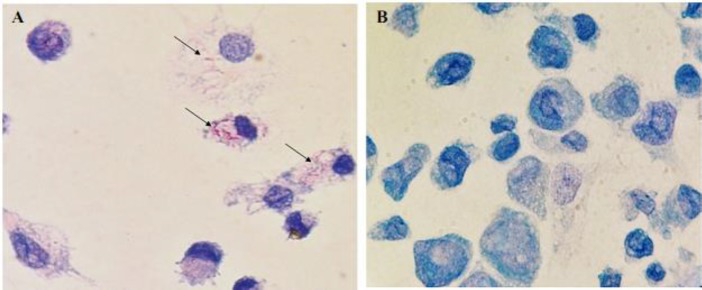
We used a modified method of Ziehl-Neelsen staining (See M&M) to visualize MAP inside the THP-1 macrophages in culture dishes. Figures 2A-B show MAP cells stained red within infected THP-1 macrophages.
Fig. 2.
Ziehl-Neelsen staining of THP-1 macrophages infected with MAP. (A) THP-1 cell infected with MAP (arrows), and (B) Control cell culture (magnification ×100)
Cytosine composition of miR-21 TSS region
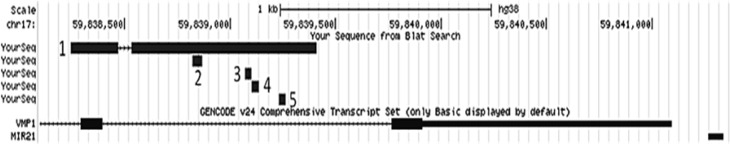
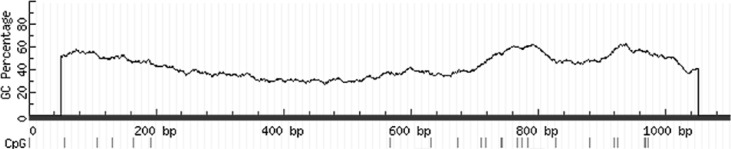
In a global differential methylation analysis on THP-1 macrophages infected with M. tuberculosis, Sharma et al. (2015) report a consensus motif (GCCTCC) over-represented in the regions hypo-methylated for non-CpG cytosines in the infected cells. Figure 3 depicts the miR-21 TSS region, showing the area analysed for differential methylation in this work and the locations where three of Sharma et al.’s (2015) consensus motif occurs. Moreover, regarding the recent reports on the importance of CpG di-nucleotides downstream of TSS sites in gene expression regulation (Krinner et al., 2014), MethPrimer software (Li and Dahiya, 2002) was used to analyse the CpG composition of the sequence corresponding to the miR-21 TSS region (Team, 2002; Cai et al., 2004; Ribas and Lupold, 2010) as well. This latter analysis detected enrichment of CpG di-nucleotides in the region (Fig. 4).
Fig. 3.
Genome browser overview of the miR-21 TSS region elements. This UCSC genome browser (Kent et al., 2002) view of the miR-21 TSS depicts the following genomic elements. 1: The genomic region evaluated for methylation status in this work, 2: the genomic segment in which the miR-21 TSS is located as reported by three independent research groups (Team, 2002; Cai et al., 2004; Ribas and Lupold, 2010); 3, 4, and 5: the position of Sharma et al.’s (2015) motif indicated for preferential non-CpG de-methylation upon M. tuberculosis infection
Fig. 4.
CpG di-nucleotides composition of the miR-21 TSS region. MethPrimer software was used to evaluate the CpG composition of the region evaluated for differential methylation in this study. This analysis detected enrichment of CpG di-nucleotides (short perpendicular bars below the graph) in the region
MAP induces de-methylation of the cytosine at CpG sites of miR-21 TSS region
Thirty-five mm cell culture dishes were seeded with 6 × 105 THP-1 cells at their exponential phase of growth (see M&M for details). These cells were differentiated and infected with 6 × 106 of MAP cells per dish and 70 h after infection, the cells were used to extract the genomic DNA. The miR-21 TSS region was amplified using two primer sets resulting in two amplicons (short, 222 bps and long, 878 bps; see Supplementary Table 1 (ST1) for the sequence), that were used to verify the cytosine methylation status in control uninfected and MAP infected THP-1 macrophages as detailed in M&M section. Our work showed that in THP-1 macrophages, MAP induces significant (P=0.007) de-methylation of the CpG sites in the miR-21 TSS region corresponding to the longer amplicon. However, neither the shorter amplicon, for cytosines at the CpG sites, nor the whole region, for the cytosines at the non-CpG sites, showed any significant difference between the control and MAP infected cells.
MAP induces cytosine de-methylation at the transcription factor binding sites (TFBS) controlling miR-21 expression
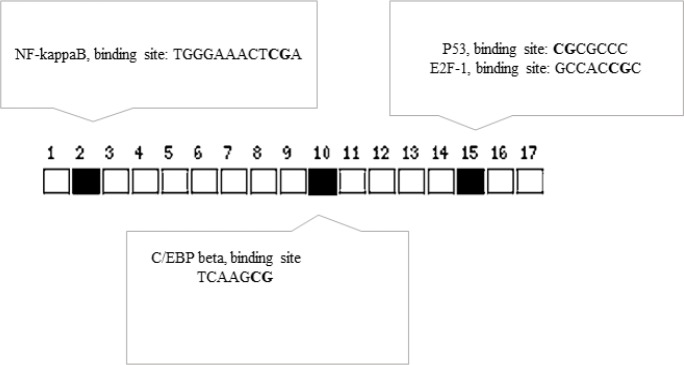
We used Promo (Messeguer et al., 2002) to detect possible TFBS at the miR-21 TSS region that showed significant differential cytosine methylation upon MAP infection (Supplementary Table 2 (ST2)). The predicted TFBS at this region were then filtered based on the presence of binding sites for known 83 core autophagy related transcription factors (Füllgrabe et al., 2016) (Supplementary Table 3 (ST3)). Interestingly, as shown in Table 2, this region contains predicted sites for transcription factors that are known to upregulate miR-21 (Papagiannakopoulos et al., 2008; Bhat-Nakshatri et al., 2009; Kida et al., 2011; Liu et al., 2014; Yang et al., 2015; McClure et al., 2017). Of note, these sites, indeed, contained the cytosines differentially methylated in MAP infected THP-1 macrophages (Fig. 5).
Table 2.
List of the predicted binding sites for the transcription factors known to control miR-21 expression occurring at miR-21 TSS. The genomic region corresponding to the large segment analysed for differential methylation in this work was used for the analysis using Promo software (Messeguer et al., 2002; Farré et al., 2003)
| Factor name | Start position | End position | String | RE equally | RE query | mir-21 up regulation |
|---|---|---|---|---|---|---|
| NF-kappaB [T00590] | 401 | 412 | TGGGAAACTCGA | 0.009 | 0.00704 | (Yang et al., 2015) |
| HIF-1 [T01609] | 605 | 613 | ACGTGCCAC | 0.0201 | 0.01428 | (Liu et al., 2014) |
| HIF-1 [T01609] | 600 | 608 | CAGGCACGT | 0.04019 | 0.0242 | (Liu et al., 2014) |
| p53 [T00671] | 745 | 751 | CGCGCCC | 0.10718 | 0.03485 | (Papagiannakopoulos et al., 2008) |
| PPAR-alpha:RXR-alpha [T05221] | 497 | 507 | TTGCCCAAGTT | 0.06782 | 0.05126 | (Kida et al., 2011) |
| E2F-1 [T01542] | 693 | 700 | TGATCCGC | 0.14737 | 0.06899 | (Bhat-Nakshatri et al., 2009) |
| E2F-1 [T01542] | 540 | 547 | ACCTCCGC | 0.20096 | 0.07366 | (Bhat-Nakshatri et al., 2009) |
| E2F-1 [T01542] | 697 | 704 | CCGCCCGC | 0.20096 | 0.07989 | (Bhat-Nakshatri et al., 2009) |
| p53 [T00671] | 696 | 702 | TCCGCCC | 0.21436 | 0.09784 | (Papagiannakopoulos et al., 2008) |
| p53 [T00671] | 496 | 502 | GTTGCCC | 0.21436 | 0.10711 | (Papagiannakopoulos et al., 2008) |
| p53 [T00671] | 803 | 809 | GCTGCCC | 0.21436 | 0.10711 | (Papagiannakopoulos et al., 2008) |
| E2F-1 [T01542] | 740 | 747 | GCCACCGC | 0.32153 | 0.14301 | (Bhat-Nakshatri et al., 2009) |
| C/EBPbeta [T00581] | 557 | 560 | TCAA | 6.85938 | 7.77501 | (McClure et al., 2017) |
Fig. 5.
Predicted binding sites for TFs known to be involved in miR-21 regulation are differentially methylated at the miR-21 TSS region in MAP infected THP-1 macrophages. Squares show the position of the CpG di-nucleotides in the amplified sequences analysed for differential methylation. Solid squares show differentially methylated cytosines (bolded in the TF binding sequences indicated in the boxes)
Discussion
Macrophages are immune cells specialized for detection and killing of pathogens including the disease causing bacteria. Though these cells are mostly successful in clearing the invading pathogens, but many intracellular bacteria have developed strategies to manipulate the intracellular environment of macrophages in their favor (Weiss and Schaible, 2015; Mitchell et al., 2016). It has been shown that intracellular bacteria counteract the hostile attitude of the macrophages, at least partly, by inducing epigenetic alterations in their host genome (Bierne et al., 2012; Yaseen et al., 2015). The new epigenetic state dictated by the intracellular bacteria makes the macrophages permissible to infection by changing the gene expression pattern in the infected cells (Bierne et al., 2012).
miR-21 targets and down-regulates several genes involved in autophagy and bacterial clearance, such as PTEN (He et al., 2015; Peralta-Zaragoza et al., 2016), BCL2 (Wu et al., 2012; Wang et al., 2014; Sims et al., 2017), Rab11a (Liu et al., 2015). Interestingly, in vivo and in vitro studies have reproducibly indicated this miRNA as a target used by various bacterial pathogens (Zhang et al., 2008; Schulte et al., 2011), including Mycobacteria (Liu et al., 2012; Wu et al., 2012; Zheng et al., 2015; Das et al., 2016) for upregulation to suppress the host macrophage defense capability. miR-21 is also reported to be upregulated in the blood from people infected with active tuberculosis (Wang et al., 2011) and epigenetic changes at the miR-21 promoter region has also been identified in blood cells obtained from patient suffering from CD (Adams et al., 2014). It is interesting to note here that MAP has long been considered as a possible cause in the development and/or progress of CD (Naser et al., 2014).
The converging controlling effects of various bacteria from different classes on miR-21 locus and the importance of miR-21 in autophagy processes, suggest this gene as a common target for bacteria to manipulate host immune system. Therefore, delineation of the method(s) by which bacteria control the expression of this gene can be helpful in the process of establishing broad spectrum therapeutic approaches. Considering the similarities between MAP and some other members of Mycobaterium family such as M. bovis and M. tuberculosis, known to induce miR-21 expression (Vegh et al., 2015; Zheng et al., 2015), we decided to evaluate possible epigenetic effects by MAP infection on cytosine methylation status at the miR-21 locus.
Recently, in a global genome methylation analysis Sharma et al. (2015) showed that, in addition to changes at the CpG sites, M. tuberculosis alters cytosine methylation status at the non-CpG sites that were mostly located in gene bodies and close to the TSS (Team, 2002; Cai et al., 2004; Ribas and Lupold, 2010). Sharma et al. (2015) reported a consensus sequence (GCCTCC) to be over-represented in genomic regions containing the non-CpG cytosines hypo-methylated in THP-1 macrophages infected with M. tuberculosis. We noticed that Sharma et al.’s (2015) motif happens three times at miR-21 TSS region (Fig. 3), that suggested for a possible regulatory domain based on Sharma’s findings. Therefore, we evaluated the methylation status of the cytosine at this region in THP-1 macrophages infected with MAP. Our analyses did not detect significant differential methylation of non-CPG cytosine in this region, however, a significant de-methylation (P=0.007) of cytosines at the CpG site downstream TSS was detected. This is an interesting finding, considering a recent work showing the importance of methylation of the cytosine at the CpG sites downstream TSS in gene regulation (Krinner et al., 2014).
Several transcription factors active in autophagy processes have predicted binding sites at the miR-21 TSS region (ST2). Interestingly, the CpG cytosines that showed significant (P=0.007) de-methylation in our work happen in the predicted binding sites for transcription factors known to be involved in miR-21 upregulation (Fig. 5). Therefore, our work shows that MAP induces epigenetic changes at the genomic regions capable of controlling miR-21 expression.
Acknowledgment
This work was financially supported by a grant from Shiraz University, Shiraz, Iran.
Conflict of interest
Authors declare no conflict of interest.
Supporting Online Material
Refer to web version on PubMed Central® (PMC) for Supplementary Material.
ST2: List of the predicted transcription factor binding sites occurring at miR-21 TSS
ST3: List of the predicted binding sites for the transcription factors known to control autophagy
References
- 1.Adams, AT , Kennedy, NA , Hansen, R , Ventham, NT , O’Leary, KR , Drummond, HE , Noble, CL , El-Omar, E , Russell, RK , Wilson, DC Two-stage genome-wide methylation profiling in childhood-onset Crohn’s disease implicates epigenetic alterations at the VMP1/MIR21 and HLA loci. Inflamm Bowel Dis. 2014;20:1784–1793. doi: 10.1097/MIB.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, LW , Fletcher, S , Wilton, SD Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol. Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat-Nakshatri, P , Wang, G , Collins, NR , Thomson, MJ , Geistlinger, TR , Carroll, JS , Brown, M , Hammond, S , Srour, EF , Liu, Y Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhide, M , Chakurkar, E , Tkacikova, L , Barbuddhe, S , Novak, M , Mikula, I IS900-PCR-based detection and characterization of Mycobacterium avium subsp paratuberculosis from buffy coat of cattle and sheep. Vet. Microbiol. 2006;112:33–41. doi: 10.1016/j.vetmic.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Bierne, H , Hamon, M , Cossart, P Epigenetics and bacterial infections. Cold Spring Harb Perspect Med. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird, AP DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bronevetsky, Y , Ansel, KM Regulation of miRNA biogenesis and turnover in the immune system. Immunol. Rev. 2013;253:304–316. doi: 10.1111/imr.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, X , Hagedorn, CH , Cullen, BR Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. 2004;RNA. 10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catalanotto, C , Cogoni, C , Zardo, G MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci. 2016;17:1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosma, CL , Sherman, DR , Ramakrishnan, L The secret lives of the pathogenic mycobacteria. Annu. Rev. Microbiol. 2003;57:641–676. doi: 10.1146/annurev.micro.57.030502.091033. [DOI] [PubMed] [Google Scholar]
- 11.Das, K , Garnica, O , Dhandayuthapani, S Modulation of host miRNAs by intracellular bacterial pathogens. Front Cell Infect. Microbiol. 2016;6:Article 79. doi: 10.3389/fcimb.2016.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, BN , Hata, A Regulation of MicroRNA biogenesis: a miRiad of mechanisms. J. Cell Commun. Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farré, D , Roset, R , Huerta, M , Adsuara, JE , Roselló, L , Albà, MM , Messeguer, X Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felekkis, K , Touvana, E , Stefanou, C , Deltas, C MicroRNAs: a newly described class of encoded molecules that play a role in health and disease. 2010;Hippokratia. 14:236–240. [PMC free article] [PubMed] [Google Scholar]
- 15.Füllgrabe, J , Ghislat, G , Cho, DH , Rubinsztein, DC Transcriptional regulation of mammalian autophagy at a glance. J. Cell Sci. 2016;129:3059–3066. doi: 10.1242/jcs.188920. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.He, C , Dong, X , Zhai, B , Jiang, X , Dong, D , Li, B , Jiang, H , Xu, S , Sun, X MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrold, RD Egg yolk agar medium for the growth of tubercle bacilli. J. Infect. 1931;48:236–241. [Google Scholar]
- 19.Hmama, Z , Peña-Díaz, S , Joseph, S , Av-Gay, Y Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol. Rev. 2015;264:220–232. doi: 10.1111/imr.12268. [DOI] [PubMed] [Google Scholar]
- 20.Huang, FY , Chan, AO , Rashid, A , Wong, DK , Cho, CH , Yuen, MF Helicobacter pylori induces promoter methylation of E-cadherin via interleukin-1β activation of nitric oxide production in gastric cancer cells. Cancer. 2012;118:4969–4980. doi: 10.1002/cncr.27519. [DOI] [PubMed] [Google Scholar]
- 21.Jeggari, A , Marks, DS , Larsson, E miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kathania, M , Raje, CI , Raje, M , Dutta, RK , Majumdar, S Bfl-1/A1 acts as a negative regulator of autophagy in mycobacteria infected macrophages. Int. J. Biochem. Cell Biol. 2011;43:573–585. doi: 10.1016/j.biocel.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 23.Kent, WJ , Sugnet, CW , Furey, TS , Roskin, KM , Pringle, TH , Zahler, AM , Haussler, D The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kida, K , Nakajima, M , Mohri, T , Oda, Y , Takagi, S , Fukami, T , Yokoi, T PPARα is regulated by miR-21 and miR-27b in human liver. J. Lipid. Res. 2011;28:2467–2476. doi: 10.1007/s11095-011-0473-y. [DOI] [PubMed] [Google Scholar]
- 25.Kinyoun, J A note on Uhlenhuths method for sputum examination, for tubercle bacilli. Am. J. Public Health (NY) 1915;5:867–870. doi: 10.2105/ajph.5.9.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krinner, S , Heitzer, AP , Diermeier, SD , Obermeier, I , Längst, G , Wagner, R CpG domains downstream of TSSs promote high levels of gene expression. Nucleic Acids Res. 2014;42:3551–3564. doi: 10.1093/nar/gkt1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuehnel, MP , Goethe, R , Habermann, A , Mueller, E , Rohde, M , Griffiths, G , Valentin-Weigand, P Characterization of the intracellular survival of Mycobacterium avium ssp paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell Microbiol. 2001;3:551–566. doi: 10.1046/j.1462-5822.2001.00139.x. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, D , Nath, L , Kamal, MA , Varshney, A , Jain, A , Singh, S , Rao, KV Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140:731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Li, LC , Dahiya, R MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 30.Liu, X , Chen, X , Yu, X , Tao, Y , Bode, AM , Dong, Z , Cao, Y Regulation of microRNAs by epigenetics and their interplay involved in cancer. J. Exp. Clin. Cancer Res. 2013;32:96. doi: 10.1186/1756-9966-32-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, X , Hong, Q , Wang, Z , Yu, Y , Zou, X , Xu, L MiR-21 inhibits autophagy by targeting Rab11a in renal ischemia/reperfusion. Exp. Cell Res. 2015;338:64–69. doi: 10.1016/j.yexcr.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y , Nie, H , Zhang, K , Ma, D , Yang, G , Zheng, Z , Liu, K , Yu, B , Zhai, C , Yang, S A feedback regulatory loop between HIF-1α and miR-21 in response to hypoxia in cardiomyocytes. FEBS Lett. 2014;588:3137–3146. doi: 10.1016/j.febslet.2014.05.067. [DOI] [PubMed] [Google Scholar]
- 33.Liu, PT , Wheelwright, M , Teles, R , Komisopoulou, E , Edfeldt, K , Ferguson, B , Mehta, MD , Vazirnia, A , Rea, TH , Sarno, EN MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012;18:267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombard, JE Epidemiology and economics of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 2011;27:525–535. doi: 10.1016/j.cvfa.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 35.Lund, BM , Gould, GW , Rampling, AM Pasteurization of milk and the heat resistance of Mycobacterium avium subsp paratuberculosis: a critical review of the data. Int. J. Food Microbiol. 2002;77:135–145. doi: 10.1016/s0168-1605(02)00057-0. [DOI] [PubMed] [Google Scholar]
- 36.Marr, AK , MacIsaac, JL , Jiang, R , Airo, AM , Kobor, MS , McMaster, WR Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PLoS Pathog. 2014;10:e1004419. doi: 10.1371/journal.ppat.1004419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClure, C , McPeak, MB , Youssef, D , Yao, ZQ , McCall, CE , El Gazzar, M Stat3 and C/EBPβ synergize to induce miR-21 and miR-181b expression during sepsis. Immunol. Cell Biol. 2017;95:42–55. doi: 10.1038/icb.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNees, AL , Markesich, D , Zayyani, NR , Graham, DY Mycobacterium paratuberculosis as a cause of Crohn’s disease. Expert Rev. Gastroenterol. Hepatol. 2015;9:1523–1534. doi: 10.1586/17474124.2015.1093931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messeguer, X , Escudero, R , Farré, D , Núñez, O , Martínez, J , Albà, MM PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell, G , Chen, C , Portnoy, DA Strategies used by bacteria to grow in macrophages. Microbiol. Specter. 2016;4:1–2. doi: 10.1128/microbiolspec.MCHD-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore, LD , Le, T , Fan, G DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naser, SA , Sagramsingh, SR , Naser, AS , Thanigachalam, S Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients. World J. Gastroenterol. 2014;20:7403–7415. doi: 10.3748/wjg.v20.i23.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pacis, A , Tailleux, L , Morin, AM , Lambourne, J , MacIsaac, JL , Yotova, V , Dumaine, A , Danckaert, A , Luca, F , Grenier, JC Bacterial infection remodels the DNA methylation landscape of human dendritic cells. Genome Res. 2015;25:1801–1811. doi: 10.1101/gr.192005.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paolicchi, F , Cirone, K , Morsella, C , Gioffré, A First isolation of Mycobacterium avium subsp paratuberculosis from commercial pasteurized milk in Argentina. Braz. J. Microbiol. 2012;43:1034–1037. doi: 10.1590/S1517-838220120003000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papagiannakopoulos, T , Shapiro, A , Kosik, KS MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 46.Patil, V , Ward, RL , Hesson, LB The evidence for functional non-CpG methylation in mammalian cells. Epigenetics. 2014;9:823–828. doi: 10.4161/epi.28741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peralta-Zaragoza, O , Deas, J , Meneses-Acosta, A , De la O-Gómez, F , Fernández-Tilapa, G , Gómez-Cerón, C , Benítez-Boijseauneau, O , Burguete-García, A , Torres-Poveda, K , Bermúdez-Morales, VH Relevance of miR-21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells. BMC Cancer. 2016;16 doi: 10.1186/s12885-016-2231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce, ES Ulcerative colitis and Crohn’s disease: is Mycobacterium avium subspecies paratuberculosis the common villain? Gut Pathogens. 2010;2:21. doi: 10.1186/1757-4749-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pillai, S , Jayarao, B Application of IS900 PCR for detection of Mycobacterium avium subsp paratuberculosis directly from raw milk. J. Dairy Sci. 2002;85:1052–1057. doi: 10.3168/jds.S0022-0302(02)74165-9. [DOI] [PubMed] [Google Scholar]
- 50.Ribas, J , Lupold, SE The transcriptional regulation of miR-21, its multiple transcripts and their implication in prostate cancer. Cell Cycle. 2010;9:923–929. doi: 10.4161/cc.9.5.10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohde, C , Zhang, Y , Reinhardt, R , Jeltsch, A BISMA-Fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandoval, J , Díaz-Lagares, A , Salgado, R , Servitje, O , Climent, F , Ortiz-Romero, PL , Pérez-Ferriols, A , Garcia-Muret, MP , Estrach, T , Garcia, M MicroRNA expression profiling and DNA methylation signature for deregulated microRNA in cutaneous T-cell lymphoma. J. Invest. Dermatol. 2015;135:1128–1137. doi: 10.1038/jid.2014.487. [DOI] [PubMed] [Google Scholar]
- 53.Schmitz, SU , Grote, P , Herrmann, BG Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulte, LN , Eulalio, A , Mollenkopf, HJ , Reinhardt, R , Vogel, J Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seca, H , Lima, RT , Lopes-Rodrigues, V , Guimaraes, JE , Almeida, GM , Vasconcelos, MH Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr. Drug Targets. 2013;14:1135–1143. doi: 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]
- 56.Sharma, G , Upadhyay, S , Srilalitha, M , Nandicoori, VK , Khosla, S The interaction of mycobacterial protein Rv2966c with host chromatin is mediated through non-CpG methylation and histone H3/H4 binding. Nucleic Acids Res. 2015;43:3922–3937. doi: 10.1093/nar/gkv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sims, EK , Lakhter, AJ , Anderson-Baucum, E , Kono, T , Tong, X , Evans-Molina, C MicroRNA 21 targets BCL2 mRNA to increase apoptosis in rat and human beta cells. Diabetologia. 2017;60:1057–1065. doi: 10.1007/s00125-017-4237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song, L , Liu, S , Zhang, L , Yao, H , Gao, F , Xu, D , Li, Q MiR-21 modulates radiosensitivity of cervicalcancer through inhibiting autophagy via the PTEN/Akt/HIF-1α feedback loop and the Akt-mTOR signaling pathway. Tumor Biol. 2016;37:12161–12168. doi: 10.1007/s13277-016-5073-3. [DOI] [PubMed] [Google Scholar]
- 59.Team, MGCP Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vegh, P , Magee, DA , Nalpas, NC , Bryan, K , McCabe, MS , Browne, JA , Conlon, KM , Gordon, SV , Bradley, DG , MacHugh, DE MicroRNA profiling of the bovine alveolar macrophage response to Mycobacterium bovis infection suggests pathogen survival is enhanced by microRNA regulation of endocytosis and lysosome trafficking. Tuberculosis (Edinb) 2015;95:60–67. doi: 10.1016/j.tube.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 61.Wang, Q , Liu, S , Tang, Y , Liu, Q , Yao, Y MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS One. 2014;9:e100949. doi: 10.1371/journal.pone.0100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, C , Yang, S , Sun, G , Tang, X , Lu, S , Neyrolles, O , Gao, Q Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PloS One. 2011;6:e25832. doi: 10.1371/journal.pone.0025832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss, G , Schaible, UE Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015;264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilcox, R Kolmogorov-smirnov test. Encyclopedia of Biostatistics. 2005 10.1002/0470011815. [Google Scholar]
- 65.Wileman, T Autophagy as a defence against intracellular pathogens. Essays Biochem. 2013;55:153–163. doi: 10.1042/bse0550153. [DOI] [PubMed] [Google Scholar]
- 66.Wu, Z , Lu, H , Sheng, J , Li, L Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012;586:2459–2467. doi: 10.1016/j.febslet.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Yang, Z , Fang, S , Di, Y , Ying, W , Tan, Y , Gu, W Modulation of NF-κB/miR-21/PTEN pathway sensitizes non-small cell lung cancer to cisplatin. PloS One. 2015;10:e0121547. doi: 10.1371/journal.pone.0121547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yaseen, I , Kaur, P , Nandicoori, VK , Khosla, S Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat. Commun. 2015;6:8922. doi: 10.1038/ncomms9922. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Z , Li, Z , Gao, C , Chen, P , Chen, J , Liu, W , Xiao, S , Lu, H miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab. Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 70.Zheng, L , Leung, E , Lee, N , Lui, G , To, KF , Chan, RC , Ip, M Differential microRNA expression in human macrophages with Mycobacterium tuberculosis infection of Beijing/W and non-Beijing/W strain types. PloS One. 2015;10:e0126018. doi: 10.1371/journal.pone.0126018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zou, G A modified Poisson regression approach to prospective studies with binary data. Am. J. Epidemiol. 2004;159:702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ST2: List of the predicted transcription factor binding sites occurring at miR-21 TSS
ST3: List of the predicted binding sites for the transcription factors known to control autophagy