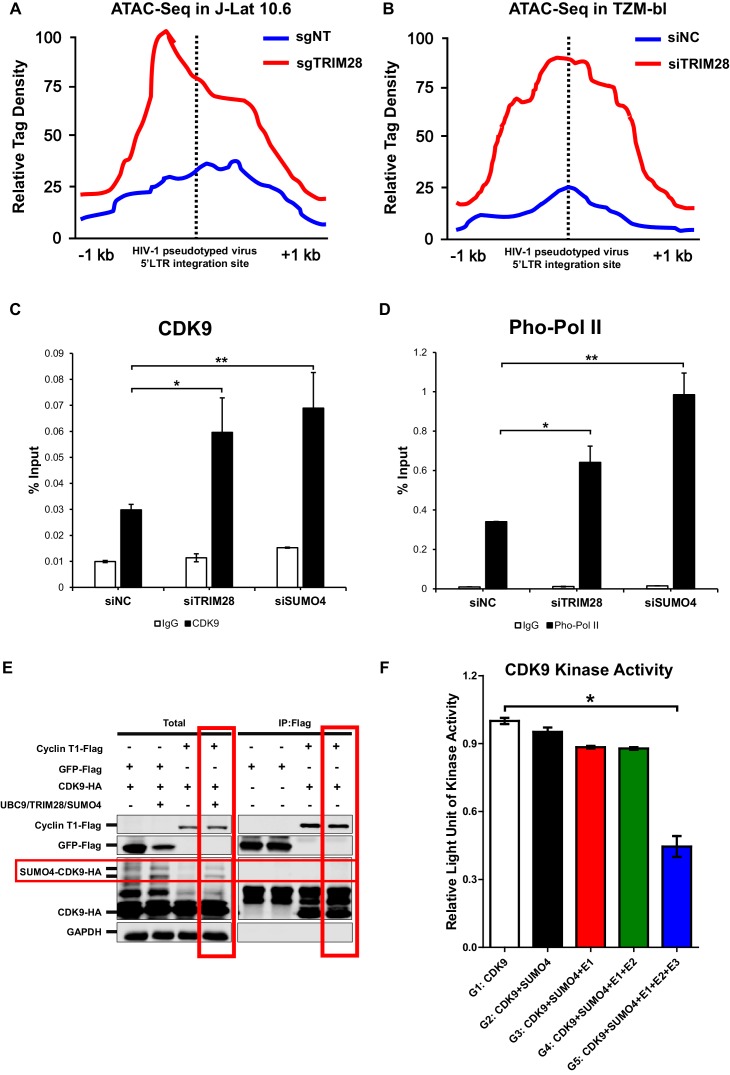
Figure 8. CDK9 function is reduced when SUMOylated by TRIM28.
(A–B) TRIM28-defective (sgTRIM28) J-Lat 10.6 cell line was generated by CRISPR-CAS9 technique. ATAC-Seq was conducted with sgNT and sgTRIM28 J-Lat 10.6 cell lines, as well as siNC and siTRIM28 TZM-bl cell lines. The tag reads of the HIV-1 pseudotyped virus/minigenome 5’LTR integration sites were counted and normalized to the total mapped reads, and represented as relative tag density. The highest tag density was set as 100. Figures showed 2 kb range centered the 5’LTR integration sites. (C–D) ChIP assays with antibodies against CDK9 and Ser2 Pho-Pol II were performed in TZM-bl cell lines which were treated with siNC, siSUMO4 and siTRIM28, respectively. (E) Cyclin T1 or GFP was co-overexpressed with CDK9 in the absence or presence of SUMO4, UBC9 and TRIM28. Cyclin T1 and GFP were IP followed by IB. (F) Fold change of kinase activity when CDK9 was SUMOylated. Data represents mean ±SEM in triplicates. p-Values were calculated by Student’s t-test. *p<0.05, **p<0.01.