Abstract
Multiple genome-wide association studies have shown that the single-nucleotide polymorphism (SNP) rs2281808 TT variant, present within the signal regulatory protein gamma (SIRPG) gene, is associated with autoimmune diseases, such as type 1 diabetes. SIRPγ is the only SIRP expressed on T cells. The role of SIRPγ in human T-cells or the effect of the TT variant are poorly understood. In this short report, we demonstrate the rather unusual finding that this intronic SNP is associated with a reduction of SIRPγ expression on T cells, both in healthy subjects as well as patients with type 1 diabetes. Using this information, we propose that a simple flow cytometric detection of SIRPγ could be a potential diagnostic testing approach for the presence of SNP in the appropriate clinical context.
Keywords: SNP rs2281808, SIRPγ, autoimmunity, diagnostic, flow cytometry
1. Introduction
Signal Regulatory Protein γ (SIRPγ) is the only SIRP expressed by human T-cells and is absent in rodents [1, 2]. While its function is poorly understood, SIRPγ is one of the ligands for CD47, expressed on antigen-presenting cells and a potential target for cancer immunotherapy [3]. SIRPγ may also be involved in trans-endothelial migration of human T-cells [4].
rs2281808 is an intronic SNP present between exons 5 and 6 of the SIRPG gene and causes a C/T variant. The TT variant is associated with a greater risk for autoimmunity, particularly type 1 diabetes [5–7]. To gain a functional understanding of this SNP, we recently demonstrated that the rs2281808 TT SNP was associated with heightened effector state in CD8+ T-cells [8].
Interestingly, while conducting phenotypic and functional studies in the context of the CC, CT and TT genotypes, we discovered the rather unusual finding that this intronic SNP resulted in changed expression of the protein on the surface of both CD4+ and CD8+ T-cells. In this short report, we evaluated whether the evaluation of protein expression by this simple flow cytometric approach could be “diagnostic” of the CC, CT or TT genotypes.
2. Materials and Methods
2.1. Human Subjects
Ethics statement:
All studies were approved by the University of Iowa IRB according to Declaration of Helsinki principles.
Patients and control subjects:
After obtaining informed consent, 19 type 1 diabetes (T1D) patients were recruited at the pediatric endocrinology clinics, University of Iowa. De-identified leukoreduction buffy coat samples from 145 healthy donors (HD) were obtained from the University of Iowa DeGowin Blood Center, Department of Pathology. Age range of HD was 25-76 y (52±15) and T1D subjects was 18–29 y (20±3).
Cell preparation and genotyping:
PBMC were isolated from all the samples using Ficoll Hypaque (GE Healthcare Biosciences, Pittsburg, PA) density gradient. The cells were stored in liquid nitrogen until further use. An aliquot of PBMC were used to extract genomic DNA using Qiagen mini DNA prep kit. For rs2281808 genotyping, allelic discrimination PCR was done using TaqMan assay and probes.
Flow cytometry staining for SIRPγ:
PBMC were stained with anti-human CD3-Alexa700, CD4-APC, CD8-BV786 and SIRPγ-PE. All the antibodies were obtained from either BD Biosciences (San Jose, CA), or Biolegend (San Diego, CA). Following staining, cells were washed and data was acquired on 4-Laser LSRII using FACSDiva software (Becton Dickinson). Data were analyzed using Flow Jo (TreeStar, Ashland, OR).
Statistical analysis:
Data between the groups was analyzed with 2-way ANOVA with Tukey’s posthoc test and p<0.05 was considered significant. Chi-Square test was used to compare the HD vs. T1D genotyping data and p<0.05 was considered significant.
3. Results
Low SIRPγ expression is diagnostic of TT genotype in healthy subjects
PBMC samples from 145 healthy donors were subjected to rs2281808 genotyping and flow cytometry staining to study the surface expression of SIRPγ expression on CD4 and CD8 T-cells. As reported earlier and now confirmed in greater number of samples, this intronic SNP resulted in changed expression of the protein on the surface of both CD4+ and CD8+ T-cells (Fig 1A-D). Thus, individuals with the CC genotype showed the highest expression of SIRPγ, the CT genotype corresponded to medium-level expression, whereas the TT genotype showed the lowest expression.
Figure 1. Low SIRPγ expression is diagnostic of TT genotype in healthy subjects.
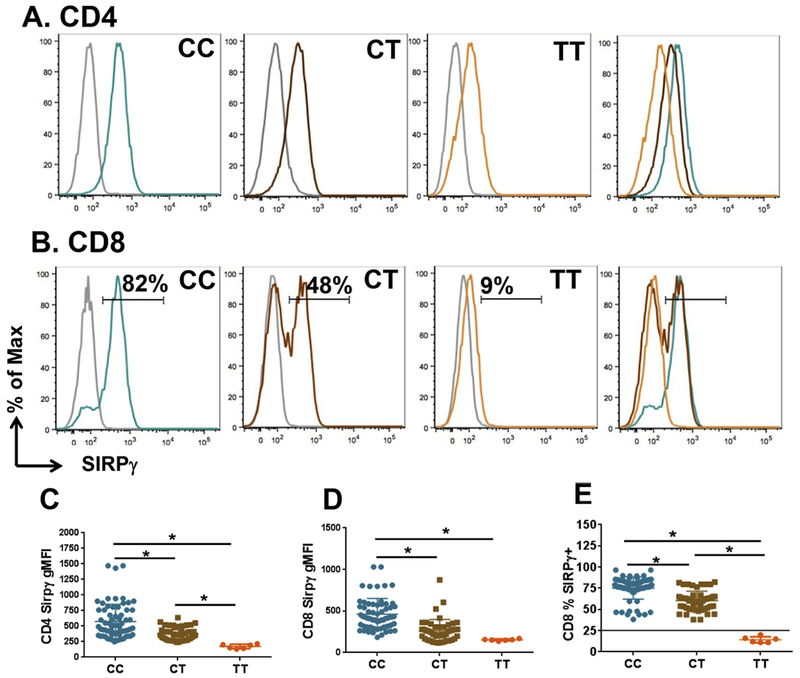
PBMC derived from de-identified healthy blood donors were stained for CD3, CD4, CD8 and SIRPγ. Panel A shows representative examples of SIRPγ expression on gated CD4+ T-cells from subjects with a CC, CT or TT genotype, as indicated. The grey histogram depicts isotypic control staining and the rightmost panel shows overlapping histograms from CC, CT and TT, maintaining the same colors, showing decreasing expression of SIRPγ from CC to TT. Panel B shows similar representative examples of SIRPγ expression on gated CD8+ T-cells. The bimodal expression pattern SIRPγ expression on CD8+ T-cells allowed quantification of % SIRPγ+ cells within CD8+ T-cells, indicated by the numbers. Panels C-E show cumulative SIRPγ expression data from 79 CC, 60 CT and 6 TT subjects. Panels C and D represent mean fluorescence intensity (MFI) on CD4 and CD8 T-cells, whereas Panel E shows %SIRPγ+CD8+ T-cells. The horizontal black line in Panel E represents a cutoff at 26% expression, based on 3SD interval of the CT expression data, as described in the text.
While the mean fluorescence intensity (MFI) was statistically different amongst the three groups, both on CD4 and CD8 T-cells (Figs 1C-D), this readout was not amenable to a clean cutoff due to overlap at the lower ends, particularly on CD8+ T-cells (Figs 1C-D). However, we observed that the bimodal expression of this molecule on CD8+ T-cells allowed us to quantify the percentage of cells that were positive/high vs. negative/low for this marker (Fig 1B). Specifically, looking at % SIRPγ-positive CD8+ T-cells, we observed that CC individuals showed a mean of 74% with a 2SD interval of 45-102 and a 3SD interval of 31-117 (Fig. 1E). The CT genotype showed a mean of 61% SIRPγ+CD8+ T-cells with a 2SD interval of 38-84 and a 3SD interval of 26–96. Again, while this difference between CC and CT was statistically significant, it showed overlap between the two groups, with no clear cutoff.
Interestingly, the TT genotype, which is associated with autoimmunity, was prominent in having a significantly lower proportion of % SIRPγ+CD8+ T-cells, with a mean of 14%, a 2SD interval of 7–22 and 3SD interval of 4–25. This allowed for a clean cutoff of 26%, based on the lower end of 3SD interval of the CT group, where 100% of the non-TT cohorts were above the cutoff and vice versa, as shown by the horizontal black line in Fig 1E.
Low SIRPγ expression is diagnostic of TT genotype in T1D
The rs2281808 SNP is known to be over-represented in autoimmune diseases, particularly in type 1 diabetes (T1D). Therefore, we recruited a cohort of patients with T1D (n=19) to test the whether this “diagnostic cutoff’ would also work in the context of autoimmune disease, where there could potentially be a larger spread of SIRPγ expression.
As anticipated, we found a significantly higher proportion of T1D patients expressing the TT genotype (4/19, 21%), compared to healthy subjects (6/145, 4%, p<0.05) (Fig 2A). Once again, similar to healthy subjects, CD4 and CD8 T-cells from T1D patients also showed statistically different levels of SIRPγ expression between CC, CT and TT individuals (Fig 2B-E). While expression in CC and CT subjects was statistically different in terms of MFI expression as well as % SIRPγ+CD8+ T-cells, the data were not amenable to a diagnostic cutoff between these two groups due to significant overlap. However, once again, we were able to use the cutoff for % SIRPγ+CD8+ T-cells determined from 3SD intervals of healthy subjects (26%, Fig 1E) to “diagnose” the TT genotype with 100% sensitivity and specificity, as shown in Fig 2F.
Figure 2. Low SIRPγ expression is diagnostic of TT genotype in T1D.
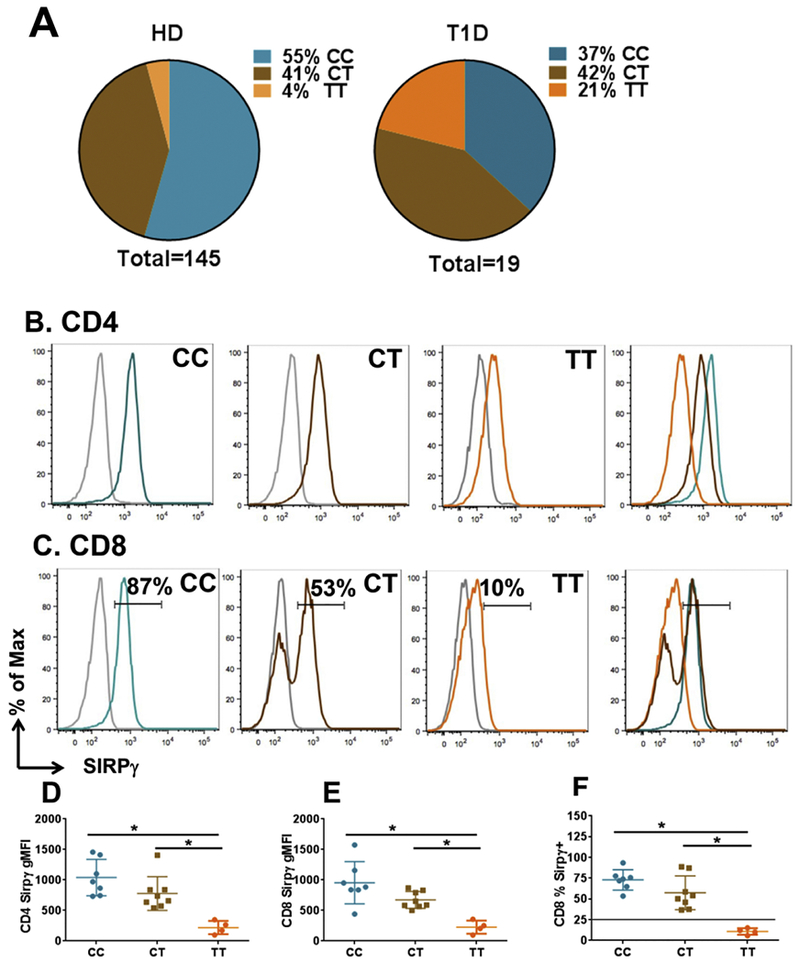
Panel A shows the distribution of CC, CT and TT genotypes amongst 145 healthy donors and 19 patients with type 1 diabetes (T1D). Panels B-F are displayed in a pattern similar to Panels A-E of Fig 1, showing SIRPγ expression on CD4 and CD8 T-cells from T1D subjects. Fig 2F uses the same cutoff (black line) of 26% from Fig 1E, showing that all T1D subjects with the TT genotype had %SIRPγ+CD8+ T-cells lower than the cutoff.
4. Discussion
In this Brief Communication, we present this interesting example of a simple phenotypic assay of protein expression tightly correlating with an intronic SNP. While we are aware of examples of certain point mutations in coding sequences being detected by changed proteins (e.g., sickle hemoglobin) and also SNPs resulting in different isoforms, to our knowledge, this is the first example of a SNP (especially an intronic SNP) resulting in a dramatic change in protein expression, to the extent of potentially being used as a robust “diagnostic/prognostic test,” in the appropriate clinical context.
Despite significant age differences between the HD and T1D groups, SIRPγ expression on T-cells from T1D subjects with the TT genotype was significantly lower than those with CC genotype, demonstrating that neither the younger age range nor the autoimmune disease status of these subjects alter this association. In corroboration of this finding, we also did not observe a correlation between age and SIRPγ expression within HD. In the same vein, we have also observed the same tight correlation between the TT genotype and low SIRPγ expression in the setting on multiple sclerosis, a different immune-mediated disease in a patient population with age range similar to our HD cohort (data not shown).
At this point, we are not sure how this intronic SNP causes reduction in protein expression. It might create an alternative splice variant of SIRPG resulting in unstable RNA or loss of protein. It has been suggested that in the thymus, this SNP is located within the predicted enhancer [9], raising the possibility that its presence could potentially alter the transcription of SIRPG. This can be a focus of future investigation.
In addition, the bimodal expression of this protein on CD8 T-cells (but not CD4 T-cells), particularly in CT individuals, also suggests distinct regulatory mechanisms. While the mechanism of this bimodal expression is not understood, it does provide the ability to create an expression cutoff that can be used to predict the TT genotype. As shown recently [8], in CC individuals, the vast majority (>90%) of SIRPγlow CD8 T-cells were present in memory/terminally-differentiated fraction and not in the naïve T-cell fraction, indicating an acquired SIRPγlow phenotype post-antigenic exposure. However, in CT as well as TT subjects, a significant proportion of naïve CD8 T-cells show low SIRPγ expression, indicating the genotypic influence even as these cells emerge from the thymus. It will be important to understand the intrinsic regulation of expression in naïve T-cells versus one that happens post-antigenic response.
Our finding could also spur investigation of correlation between such SNPs and protein expression to see if there is a broader applicability of this principle, both in the biology of genotype-phenotype conversion and for potential diagnostic testing.
Highlights.
This is an interesting example of a simple phenotypic assay of protein expression tightly correlating with an intronic SNP.
A single-nucleotide polymorphism (SNP) in the SIRPG gene (rs2281808) is known to be associated with autoimmune disease.
this intronic SNP (rs2281808) can be detected by a simple phenotypic flow cytometric assay detecting the expression of the corresponding protein on T-cells.
The expression pattern may allow such detection to be utilized as a clinical diagnostic/prognostic test for this polymorphism in the appropriate clinical context.
Acknowledgments
Funding
These studies were supported, in part, by grants to NJK from the National Institutes of Health (R01AI092106) and the National Multiple Sclerosis Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability statement
All relevant datasets generated for this study are included in the manuscript.
References
- [1].Barclay AN, Brown MH, The SIRP family of receptors and immune regulation, Nature reviews. Immunology, 6 (2006) 457–464. [DOI] [PubMed] [Google Scholar]
- [2].Piccio L, Vermi W, Boles KS, Fuchs A, Strader CA, Facchetti F, Cella M, Colonna M, Adhesion of human T cells to antigen-presenting cells through SIRPbeta2-CD47 interaction costimulates T-cell proliferation, Blood, 105 (2005) 2421–2427. [DOI] [PubMed] [Google Scholar]
- [3].Huang Y, Ma Y, Gao P, Yao Z, Targeting CD47: the achievements and concerns of current studies on cancer immunotherapy, Journal of thoracic disease, 9 (2017) E168–E174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stefanidakis M, Newton G, Lee WY, Parkos CA, Luscinskas FW, Endothelial CD47 interaction with SIRPgamma is required for human T-cell transendothelial migration under shear flow conditions in vitro, Blood, 112 (2008) 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS, Type C 1 Diabetes Genetics, Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes, Nature genetics, 41 (2009) 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kiani AK, John P, Bhatti A, Zia A, Shahid G, Akhtar P, Wang X, Demirci FY, Kamboh MI, Association of 32 type 1 diabetes risk loci in Pakistani patients, Diabetes research and clinical practice, 108 (2015) 137–142. [DOI] [PubMed] [Google Scholar]
- [7].Reddy MV, Wang H, Liu S, Bode B, Reed JC, Steed RD, Anderson SW, Steed L, Hopkins D, She JX, Association between type 1 diabetes and GWAS SNPs in the southeast US Caucasian population, Genes and immunity, 12 (2011) 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sinha S, Borcherding N, Renavikar PS, Crawford MP, Tsalikian E, Tansey M, Shivapour ET, Bittner F, Kamholz J, Olalde H, Gibson E, Karandikar NJ, An autoimmune disease risk SNP, rs2281808, in SIRPG is associated with reduced expression of SIRPgamma and heightened effector state in human CD8 T-cells, Scientific reports, 8 (2018) 15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gabrielsen IS, Amundsen SS, Helgeland H, Flam ST, Hatinoor N, Holm K, Viken MK, Lie BA, Genetic risk variants for autoimmune diseases that influence gene expression in thymus, Human molecular genetics, 25 (2016) 3117–3124. [DOI] [PubMed] [Google Scholar]


