Abstract
Alterations in central extended amygdala (EAc) function have been linked to anxiety, depression, and anxious temperament (AT), the early-life risk to develop these disorders. The EAc is composed of the central nucleus of the amygdala (Ce), the bed nucleus of the stria terminalis (BST), and the sublenticular extended amygdala (SLEA). Using a non-human primate model of AT and multimodal neuroimaging, the Ce and the BST were identified as key AT-related regions. Both areas are primarily comprised of GABAergic neurons and the lateral Ce (CeL) and lateral BST (BSTL) have among the highest expression of neuropeptides in the brain. Somatostatin (SST) is of particular interest because mouse studies demonstrate that SST neurons, along with corticotropin releasing factor (CRF) neurons, contribute to a threat-relevant EAc microcircuit. Although the distribution of CeL and BSTL SST neurons has been explored in rodents, this system is not well described in non-human primates. In situ hybridization demonstrated an anterior-posterior gradient of SST mRNA in the CeL but not the BSTL of non-human primates. Triple labeling immunofluorescence staining revealed that SST protein expressing cell bodies are a small proportion of the total CeL and BSTL neurons and have considerable co-labeling with CRF. The SLEA exhibited strong SST mRNA and protein expression, suggesting a role for SST in mediating information transfer between the CeL and BSTL. These data provide the foundation for mechanistic non-human primate studies focused on understanding EAc function in neuropsychiatric disorders.
Introduction
The central extended amygdala (EAc) is an integral part of the neural circuit mediating anxiety, depression, and anxious temperament (AT), a childhood risk factor for the development of these disorders [1–4]. As such, understanding the cellular composition of the EAc has far reaching implications for parsing out specific circuits related to the risk for the development and the emergence of anxiety and affective disorders. Using a well-validated rhesus monkey model of AT combined with fluorodeoxyglucose positron emission tomography imaging, our laboratory demonstrated that two major nodes of the EAc, the central nucleus of the amygdala (Ce) and the bed nucleus of the stria terminalis (BST) are key regions in which individual differences in metabolism are associated with individual differences in early life AT [3, 5, 6]. These regions are both structurally [7] and functionally connected [8] and this functional connectivity, as assessed with functional magnetic resonance imaging, is also associated with individual differences in AT [9]. The Ce and BST both contain subdivisions, of which the lateral Ce (CeL) and the laterodorsal BST (BSTL) are critical components of the EAc [10–14]. Numerous connections between these regions course through the sublenticular extended amygdala (SLEA), which lies ventral and medial to the globus pallidus [13–15] and is thought to aid in the transfer of threat-related information between the CeL and the BSTL [16].
The CeL and BSTL are composed of mostly striatal-like medium spiny GABAergic neurons that can be subdivided into multiple types based on their cellular morphology and neuropeptide expression profiles [17–21]. The peptide somatostatin (SST) is of particular interest as studies in mice demonstrate a role for CeL SST neurons in modulating threat-related responses [16, 17, 22–25]. Within the CeL, SST neurons interact with local microcircuits and also modulate the function of more distant regions involved in threat-responding [16, 23, 24]. It is noteworthy that the CeL microcircuit also involves corticotropin-releasing factor (CRF) expressing neurons which are well known to mediate stress, anxiety, and AT [17, 26, 27]. Within the mouse microcircuit, some SST neurons also express CRF [19], and there are also separable populations of SST and CRF expressing neurons [17, 26].
While the EAc is considered a structural and functional unit, evidence suggests that its cellular make-up is comprised of developmentally-distinct neuron populations [28] some of which have well-defined anterior-posterior (A-P) gradients [26, 29]. For example, in the mouse CeL, SST neurons are more concentrated in the posterior CeL than the anterior CeL, suggesting a pattern that is relevant to function and maybe conserved in primates. Although the distribution and function of EAc SST expressing neurons have been explored in rodents, and initial evidence from human studies suggests that the SST system is associated with psychopathology [30, 31], little is known about SST expressing neurons in the non-human primate EAc. Studies in non-human primates are important to further the translational link between rodent studies and an understanding of mechanisms underlying human psychopathology. This is evident because non-human primates and humans have evolutionarily conserved brain structure and function that underlies their similarities in social and emotional behavior.
In this study, we used in situ hybridization and immunofluorescence staining to characterize SST expression in the non-human primate amygdala. We focused on the distribution of SST throughout the EAc, with emphasis on its A-P distribution. Additionally, within the CeL and BSTL, we investigated the extent to which SST and CRF are co-expressed. Understanding the expression and distribution of SST neurons in the non-human primate EAc will aid in translating data from rodent studies and also will provide a basis for future studies using the non-human primate model of AT to investigate SST circuit-based hypotheses relevant to human stress-related psychopathology.
Experimental Procedures
Animals and Tissue Collection
Two highly related macaque species were used for the studies. For the in situ hybridization studies, four rhesus monkeys (macaca mulatta; mean age = 9.5 years, 3 females and 1 male) were euthanized under deep anesthesia with the guidance of veterinary staff using pentobarbital, which is the standard method of euthanasia at these facilities. Fresh frozen tissue was collected and stored at −80°C as previously described [32]. Briefly, upon removal from the skull, each hemisphere of the brain was cut into 14mm slabs and flash frozen in cold isopentane. This method of euthanasia is consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. For the immunofluorescence studies, we use formaldehyde-fixed tissue from two cynomolgus monkeys (macaca fasicularis; ages =3–4 years, 2 males), used in other experiments at the University of Rochester. Animals had been deeply anesthetized and killed by perfusion through the heart with 0.9% saline containing 0.5 ml of heparin sulfate (200 ml/min for 10 minutes), followed by cold 4% paraformaldehyde in a 0.1 M phosphate buffer/30% sucrose solution (100 ml/min for 1 h). The brain was extracted from the skull, placed in a fixative overnight, and then equilibrated in increasing gradients of sucrose (10%, 20%, 30%). Fixed brains were cut on a freezing microtome (40 μm) and all sections were stored in cryoprotectant solution (30% ethylene glycol and 30% sucrose in 0.1 M phosphate buffer) at −20 °C [33]. 1: 24 sections were sent to the University of Wisconsin for further processing.
All experiments were carried out in accordance with National Institute of Health guidelines. Experimental design and techniques were aimed at minimizing animal use and suffering and were reviewed by the University Committees on Use of Animals in Research at the University of Wisconsin-Madison and University of Rochester.
In Situ Hybridization, Data Acquisition, and Analysis
Brain slabs containing the amygdala were sectioned at 20μm and stored in a −80°C freezer. Approximately every 25th section from each animal was used for in situ hybridization and adjacent sections were stained with acetylcholinesterase (AChE) to determine the A-P extent of the amygdala. AChE staining was performed as previously described [32]. AChE images were assigned a bregma value that most closely matched that AChE image in the Paxinos Atlas [34]. For each animal, A-P location was calculated as a percentile through the A-P extent of that animal’s amygdala with 0 being the most anterior and 100 being the most posterior slice. AChE stained sections were scanned and used to delineate amygdala nuclei. The rhesus SST probe was amplified from rhesus amygdala cDNA. The sequence was based on the Affymetrix probe set RHESUS:MMUGDNA.7526.1.S1_AT. The PCR products were subcloned into pBluescript II SK(+) (Agilent Technologies, Santa Clara, CA), digested, linearized, transcribed and labeled with [35-S]UTP (Perkin-Elmer, Waltham, MA) and then purified. The tissue was hybridized with either the labeled sense probe or the labeled antisense probe overnight at 55 °C and then exposed to a phosphor screen (Fujifilm, Tokyo, Japan) for 2 weeks. Phosphor screens were scanned using a Typhoon 9410 (GE Healthcare, Chicago, IL). The specificity of the SST probe was demonstrated by the lack of significant signal seen with the sense probe control.
In situ and AChE images were imported into Adobe Illustrator CC 18.0 (Adobe Systems, San Jose, CA) and corresponding AChE sections were overlayed onto the in situ images. Amygdala nuclei regions of interest (ROI) were delineated for each amygdala and EAc nucleus (see Table 1 for nuclei abbreviations) at each A-P level using the AChE stains and these were exported as separate files. The sum of in situ signal intensity for all the pixels within each A-P slice of each nucleus ROI was measured and these values were divided by the area of the ROI. The intensity in each A-P ROI was corrected for both the intensity of the screen as well as non-specific background binding on the tissue. Screen intensity was sampled from the top left corner of the slide which contained no tissue and this was subtracted from the ROI intensity values. These ROI values were further corrected for tissue background intensity by dividing screen corrected ROI values by those from a defined white matter region. All imaging analyses were completed using scripts written in python 3.6 (Python Software Foundation, https://www.python.org/) using IPython [35] and the following packages: scipy [36], scipy-image [37], pandas [38], and matplotlib [39].
Table 1.
Abbreviations of amygdala nuclei
| Amygdala Nuclei | Abbreviation |
|---|---|
| basal | BL |
| basomedial | BM |
| lateral | La |
| medial | Me |
| central, lateral division | CeL |
| central medial division | CeM |
| bed nucleus of the stria terminalis, lateral division | BSTL |
| sublenticular extended amygdala | SLEA |
| central extended amygdala | EAc |
SST mRNA expression was evaluated for all amygdala nuclei. To test whether SST mRNA expression differed across the A-P extent of each region, a linear mixed effects (LME) approach was utilized. LME models account for subject as a random variable while estimating the extent to which SST expression varies across the A-P location of the region. In contrast to the standard ordinary least squares (OLS) approach, this method is not sensitive to variation across subjects. LME models were utilized because initial analyses using OLS regression identified significant variation across subjects. Primary questions focused on the CeL and BSTL. We used an LME model to test the overall distribution of SST across the A-P extent of both regions (CeL and BSTL) as well as the interaction between region and A-P location. In this model, the intercept, slope of region and slope of A-P location was allowed to vary within subjects. Age was not included as a covariate as initial OLS and LME analyses determined that Age did not account for a significant amount of variance. A secondary analysis using a repeated measures ANOVA compared SST mRNA expression in the CeL and BSTL to all other amygdala nuclei. Post-hoc testing was performed on pairwise comparisons to determine amygdala nuclei differences in SST expression and p-values were Bonferroni corrected. Finally, the relationship between SST mRNA expression and A-P location of other amygdala nuclei was also investigated using LME models. One data point from the medial nucleus of one animal was determined to be an outlier based on studentized residuals and was excluded from the analysis. All statistical tests were performed in R Studio (Version 1.1.419) using R [40]. All predictors were mean centered and the statistics reported are from the lmer function in the lme4 package [41] and the ANOVA function from the car package [42]. Graphs were made using the python seaborn (version 0.0.9, https://seaborn.pydata.org) module which uses matplotlib [39].
Immunofluorescence Staining, Image Acquisition and Data Analysis
Two to three tissue sections through the Ce and BST were used to determine the number of SST expressing neurons and their overlap with CRF expressing neurons. Sections were stained using a typical immunofluorescence protocol. Antibodies were previously characterized [43–48]. Prior to triple labelling experiments, all antibodies were first individually optimized in individual single-labeling experiments. Subsequent experiments were carried out to demonstrate a lack of cross reactivity between antibodies.
For triple-labeling experiments, anti-sera to SST, NeuN, and CRF were immunodetected in sequence, rather than pooled. Tissue was removed from cryoprotectant solution 24 hours before staining and rinsed at least 3 times in 1x PBS. An avidin/biotin blocking kit (Vectorlabs, Burlingame, CA) was used to remove the possibility of endogenous biotin binding. Tissue was first incubated in 5% donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour, washed in 1x PBS, and then incubated in a somatostatin primary antibody, raised in goat (Santa Cruz, Dallas, TX, cat# SC7819, 1:4000), solution for 24 hours at 4°C. Sections were then washed three times for 5 minutes each time and incubated in an AlexaFluor 568 secondary antibody (donkey anti-goat, ThermoFisher, Waltham, MA) for 1 hour at room temperature. Following this, sections were washed thoroughly three times for 5 minutes each time and incubated for 24 hours in mouse monoclonal NeuN (mouse, Millipore, Burlington, MA, cat# MAB377, 1:1000) primary antibody. The following day, sections were washed in 1x PBS and incubated in an AlexaFluor 647 secondary antibody (donkey anti-mouse, ThermoFisher). Sections were thoroughly rinsed three times for 5 minutes each time. Finally, tissue was incubated in 5% goat serum (Vectorlabs) for 1 hour before being stained with a CRF primary antibody, raised in guinea pig (Bachem, Bubendorf, Switzerland, cat#T-5007, 1:4000), overnight. Sections were washed three times for five minutes each time and then incubated in AlexaFluor 488 secondary antibody (goat anti-guinea pig, ThermoFisher) for 1 hour. CRF signal was amplified using a biotinylated anti-streptavidin antibody (Vectorlabs). To decrease endogenously fluorescing lipofuscin, sections were further incubated in an autofluorescence eliminator reagent (EMD Millipore, cat# 2160) at the end of the protocol and then rinsed in 70% ethanol for 1–3 minutes followed by one 1x PBS wash. Sections were mounted and cover slipped using ProLong Gold (ThermoFisher).
Staining was visualized on a Leica macrofocal microscope with a 10x objective for a large overview image, and then with a 20x objective for quantification. Edges of CeL and BSTL were identified using the high density of SST neuropil that clearly demarcate their borders. 200x images of the whole Ce and BSTL were acquired as stacks with a 0.5μm step and tiled together using the Leica software. Additional images were acquired with a 60x objective using a Nikon A1 confocal microscope. Images were acquired using the same settings for each animal. Each channel for each image stack went through a processing pipeline consisting of a maximum intensity projection followed by a 3μm gaussian blurr, watershed segmentation, and thresholding in Fiji [49]. Identification of neurons was first performed by defining NeuN positive staining that was at least 5μm in diameter and had an intensity that was greater than non-neuronal background staining. Non-neuronal background staining was defined as visible groups of pixels that were not dense enough to form the circular shape of the nucleus or did not meet the criteria of being at least 5μm in diameter. This was most obvious after a gaussian blur and watershed segmentation was applied to this channel as these groups of pixels did not stay merged. The neuronal size criterion was based on previous stereological volumetric estimates of neurons in non-human primate Ce [50]. Neurons were then evaluated for co-labelling with SST or CRF immunoreactivity [50]. Neurons were considered to be SST or CRF expressing if half of the cytoplasmic compartment of the neuron (identified with NeuN staining) expressed the peptide in the maximum intensity projection image. When examining the immunofluorescence signal for SST and CRF staining there was also signal in the surrounding neuropil. This signal is strikingly pronounced in the EAc compared to non-specific background binding in regions that do not contain or have low levels of SST or CRF. The Fiji ROI Selection Tool was used to identify stained neurons and an experimenter confirmed whether cells were double or triple labeled with NeuN and the peptide markers. The number of SST, CRF, and SST/CRF expressing neurons was calculated as a proportion of the number of neurons counted for each section (n=2) of each animal (n=2). Data was averaged across the A-P extent and across animals.
Results
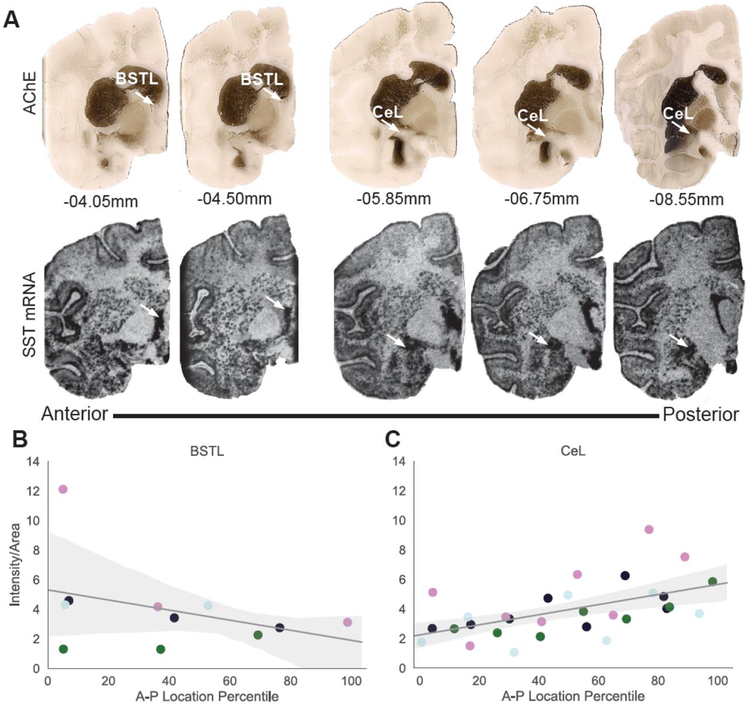
We first characterized SST mRNA expression in the two major nodes of the non-human primate EAc, the CeL and the BSTL. Previous research in rodents described robust SST expression in both the BST and the Ce. More specifically, in the mouse CeL, SST expression follows an A-P gradient with significantly more SST expression in the posterior Ce [29, 51]. Data from our rhesus monkey in situ hybridization experiment revealed robust and similar expression of SST mRNA in the BSTL and CeL (Figure 1A; F1,18=0.03, p=1.0, Table 2). A linear mixed effects model was used to determine potential differences in AP distribution of SST expression across and between the BSTL and CeL. This analysis confirmed the lack of difference in overall SST expression between these structures (F1,3=0.024, p=0.89) and further revealed a significant main effect of A-P extent such that higher concentrations of SST mRNA were found in the more posterior sections of the EAc (F1,5.2=11.3, p=0.02). Importantly, a significant region by A-P location interaction was found (F1, 31.5=13.5, p=0.0009; Figure 1B–C) and post-hoc analyses demonstrated that within the CeL, SST mRNA expression showed a significant increase in the posterior compared to anterior sections (Figure 1C, F1,3=10.9, p=0.046). This finding is consistent with the demonstration of greater numbers of SST expressing neurons in the mouse posterior CeL [29]. The A-P gradient that we found in the rhesus CeL was not observed in the BSTL (Figure 1B, F1,2.6= 0.956, p=0.41).
Figure 1.
Anterior-Posterior Distribution of SST mRNA in the non-human primate EAc. A. Top: AChE through the EAc of a representative animal. Arrows point to the laterodorsal bed nucleus of the stria terminalis (BSTL) or the lateral division of the central nucleus of the amygdala (CeL). Bottom: adjacent sections displaying SST mRNA expression through the extended amygdala visualized by in situ hybridization. B. BSTL SST mRNA intensity from in situ images. C. CeL SST mRNA intensity from in situ images. Each color represents an individual animal. For each animal, the location was calculated as a percentile through the A-P extent of that animal’s amygdala with 0 being the most anterior and 100 being the most posterior slice.
Table 2.
Mean differences (MD) of intensity values for SST between amygdala nuclei (columns – rows) with standard errors (SE) in parentheses. Bonferroni corrected p-values of regional differences are noted and significant differences are bolded. See Table 1 for abbreviations.
| BL | BM | BSTL | CeL | CeM | La | Me | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pvalue | pvalue | pvalue | pvalue | pvalue | pvalue | pvalue | |||||||
| BL | - | 1.0 | 0.0004 | 0.00061 | 1.0 | 1.0 | 1.0 | ||||||
| BM | 1.0 | - | 0.001 | 0.002 | 1.0 | 1.0 | 1.0 | ||||||
| BSTL | 0.0004 | 0.001 | - | 1.0 | 0.008 | 0.001 | 0.001 | ||||||
| CeL | 0.0006 | 0.002 | 1.0 | - | 0.01 | 0.002 | 0.002 | ||||||
| CeM | 1.0 | 1.0 | 0.008 | 0.01 | - | 1.0 | 1.0 | ||||||
| La | 1.0 | 1.0 | 0.001 | 0.002 | 1.0 | - | 1.0 | ||||||
| Me | 1.0 | 1.0 | 0.001 | 0.002* | 1.0 | 1.0 | - | ||||||
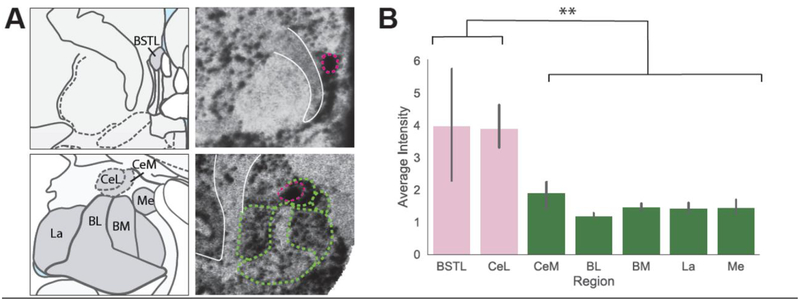
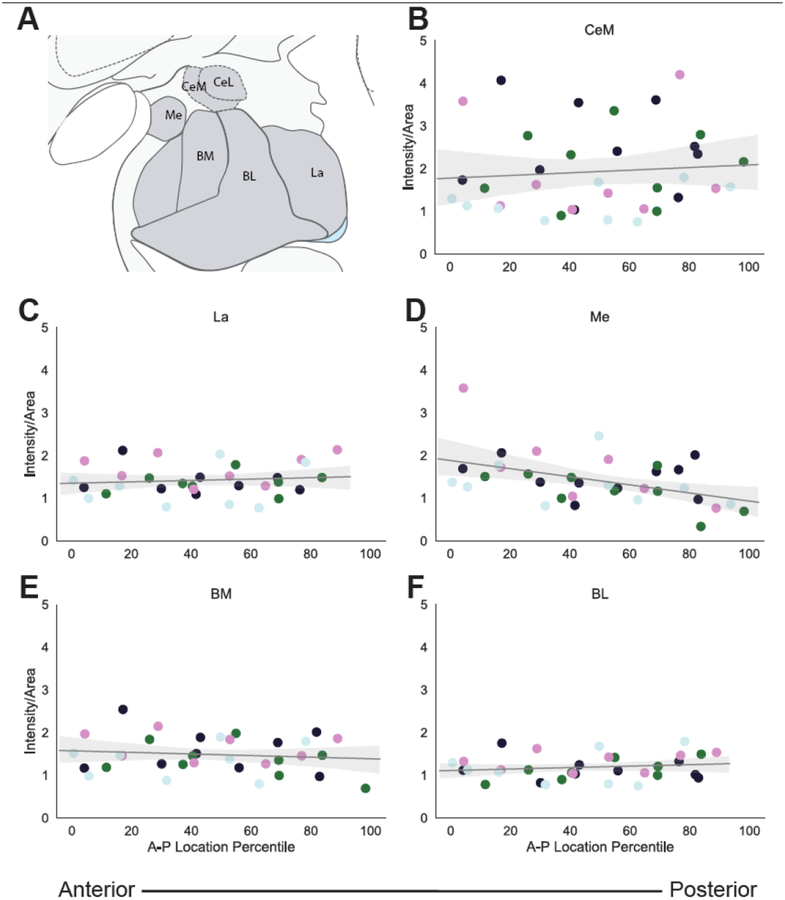
While our primary analysis focused on the CeL and BSTL, SST mRNA is expressed throughout the brain, including in other amygdala nuclei (Figure 2A). Since SST expressing neurons have been implicated in modulating fear learning in other amygdala regions [52], we investigated the level of expression in these nuclei compared to the CeL and BSTL and characterized SST A-P distribution in these nuclei. A repeated measures ANOVA (regions-CeL, BSTL, BL, BM, Me, La, CeM) demonstrated that SST mRNA expression levels differed between amygdala nuclei (Figure 2B, F6,18=10.21, p<0.001). Post-hoc analyses revealed that SST mRNA expression was significantly higher in the CeL and BSTL compared to each of the other amygdala nuclei, but that the other nuclei did not differ from each other (Figure 2B; Table 2). Like in the CeL and BSTL, SST mRNA was expressed throughout the A-P extent of the other amygdala nuclei (Figure 3). Separate analyses for each nucleus revealed no effects of A-P extent on SST mRNA: CeM (F1,2.9=0.1038, p=0.769), Me (F1,2.9=3.65, p=0.153), BL (F1,2.8=0.7072, p=0.47), La (F1,2.8=0.2432, p=0.658) and BM (F1,2.9=0.4922, p=0.53) nuclei (Figure 3B–C).
Figure 2.
Average SST mRNA Expression Across the Non-human Primate Amygdala and EAc. A. Atlas slices and SST in situ images of the primate BST (top) and amygdala (bottom) with nuclei outlined in dotted lines. B. Average intensity of SST expression in EAc regions and amygdala nuclei. See Table 2 for mean differences and statistics. ** p<0.001.
Figure 3.
Anterior-Posterior Distribution of SST mRNA in the Non-human Primate Amygdala. A. Atlas figure adapted from [34] depicting the different amygdala nuclei investigated. SST mRNA expression in the B. medial division of the central nucleus of the amygdala (CeM) C. lateral nucleus (La) D. medial nucleus (Me) E. basomedial nucleus (BM) F. basolateral nucleus (BL). On the x-axis, 0 is the most anterior slice and 100 is the most posterior slice. Each color represents an individual animal.
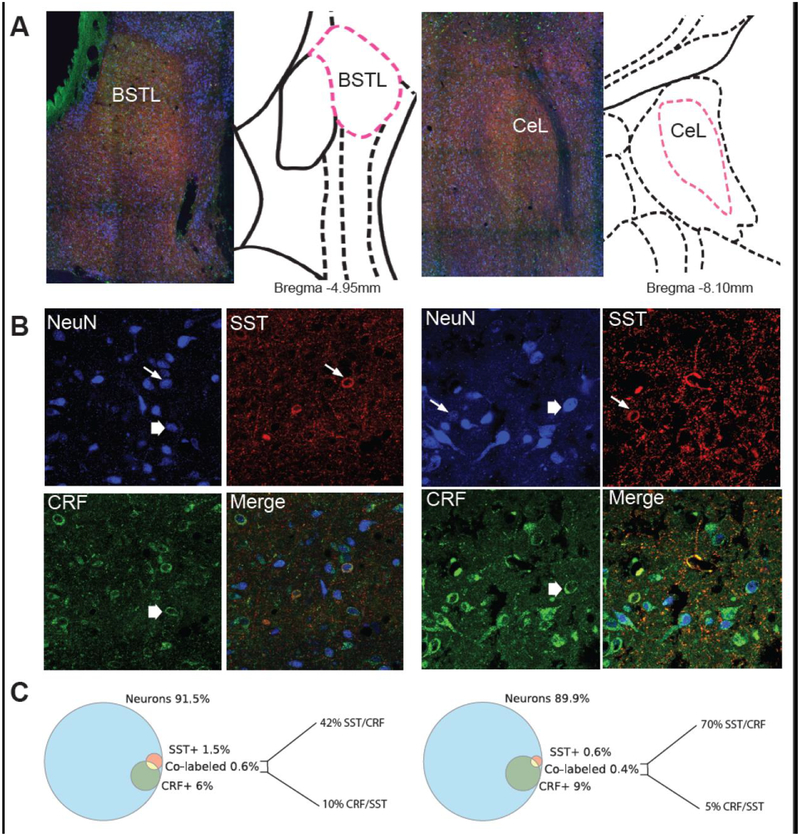
To build on our characterization of SST mRNA distribution, we quantified the proportion of neurons within the EAc that express SST protein. Using confocal microscopy, we attempted to not only understand the degree to which SST immunoreactivity was localized in cell bodies but also the extent to which SST was co-expressed with CRF. The CeL and BSTL were identified on 100x stitched images (Figure 4A). Within the CeL and BSTL there were relatively few neuronal cell bodies expressing SST protein, however, these were embedded in a dense SST neuropil. Less than 2% of the neurons counted in each of these structures expressed SST (Figure 4B–C). In both regions, CRF expressing neurons made up a larger population: 6% of the neurons counted in the BSTL and 9% of the neurons counted in the CeL. CRF neuropil was also observed in both regions but was less dense than the SST neuropil. The number of co-labeled SST/CRF neurons accounted for a small percentage of the neurons within the CeL (0.4 %) or BSTL (0.6%). In the CeL, 70% of SST neurons also expressed CRF while 5% of CRF neurons also expressed SST. In the BSTL, 42% of SST neurons expressed CRF and 10% of CRF neurons expressed SST (Figure 4C). In both regions however, the majority of neurons were not accounted for by either SST or CRF staining: 91.5% of neurons in the BSTL and 89.9% of neurons in the CeL did not express either SST or CRF.
Figure 4.
SST and CRF Protein Expression in the CeL and BSTL. A. 10x stitched images of the BSTL (left) and the CeL (right) along with their approximate rhesus monkey atlas slice [34]. B. NeuN, SST, CRF and merged images in the BSTL (left) and the CeL (right). Thin arrows point to SST neurons and thick arrows point to CRF neurons. Images were taken at 60x magnification. C. Venn diagrams of the percentages of SST (red), CRF (green), and SST/CRF co-labeled neurons (yellow) out of the total number of counted neurons (blue), along with the percentage breakdown of the co-labeling between SST and CRF in BSTL (left) and CeL (right).
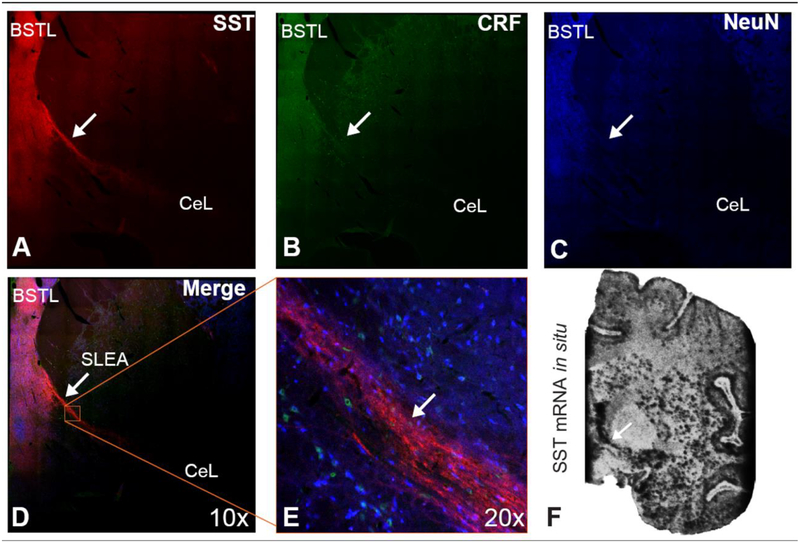
A striking feature of the SST immunoreactivity comprising the EAc neuropil was the observation of densely labeled varicose fibers (Figure 4A–B). Importantly, in tissue sections that included the anterior portion of the CeL, the SLEA, and the posterior portion of the BSTL, we observed SST expressing fibers that extended between the CeL and BSTL (Figure 5A–E). We also observed SST mRNA expression in this SLEA region (Figure 5F). CRF immunoreactivity was also within the CeL and BSTL but not in the fibers extending between these regions (Figure 5B, D–E).
Figure 5.
SST expression in the fibers that run through SLEA. A. SST B. CRF C. NeuN D. Merged stitched 10x image of the BST and SLEA. E. 20x image of SLEA fibers expressing SST but not CRF. F. SST mRNA expression is robust throughout the SLEA.
Discussion
Because the CeL and BSTL have important roles in threat-processing and EAc function has been linked to psychiatric illness, we characterized SST expression and distribution in these structures. While brain SST systems have been well characterized in rodents [17, 22, 53–55], less has been done in non-human primates. SST has a putative role in neuropsychiatric disorders [31] and in modulating function in various brain regions including the CeL, thalamus, cortex, and striatum [17, 22–24, 56–60]. Studies have also implicated other neuropeptides in modulating EAc function, for example via CRF and dynorphin signaling [27, 61]. The current data are focused on the SST system demonstrating that in the non-human primate EAc, SST is highly expressed, is predominantly found in neuronal extensions and not in cell bodies and has an A-P gradient within the CeL. The demonstration that SST mRNA in the non-human primate CeL has an A-P gradient is consistent with previous reports in rodents using different methods of quantification [29]. Results from the analyses of other amygdala nuclei are also consistent with earlier data, demonstrating that SST immunoreactivity is lower in the rest of the amygdala when compared to the CeL and BSTL [10, 62]. This finding suggests that SST, although not specific to the EAc, is heavily utilized by EAc neurons and manipulations to this system would result in alterations to threat-related behaviors.
The quantitative approach used in this study, similar to that used in rodent studies [17, 19, 26, 51], revealed that less than 2% of cell bodies in the CeL, or in the BSTL, express SST protein. While other studies in non-human primates and humans have examined SST expression in these regions, to our knowledge, our study is the first to use quantitative methods establishing the proportion of SST expressing cell bodies out of total counted neurons. Our finding is in contrast to those from studies of the mouse CeL, where across various studies SST containing cell bodies are reported to range from between 20–50% of total neurons [19, 51]. In our study, we also find that the overlap between SST and CRF neurons accounts for a small population of overall EAc neurons, a pattern that is consistent with some studies in mice [17]. With regard to co-labeling, a relatively high percentage of SST neurons also expressed CRF, while only a small percentage of CRF neurons expressed SST, similar to what has been reported in previous work in the mouse [19, 51]. We emphasize that further studies should be performed in non-human primates and mice to confirm the species differences we observe here.
Alterations in EAc function have been implicated in neuropsychiatric disorders and mouse studies investigating its microcircuitry have shed light on the contribution of specific EAc neuronal populations to the expression of threat-related behaviors [16, 17, 22–25, 30, 31, 63, 64]. It has been hypothesized that SST expressing cell bodies comprise an intrinsic circuit within the EAc [10, 65–67]. Although we observed few SST cell bodies in the CeL and BSTL, we and others [10, 65–67] found heavy SST fiber labeling within these regions, as well as in the SLEA. Previous studies proposed the existence of a somatostatinergic CeL to BST projection [10, 55, 67], which has been verified with double labeling studies in rodents [16, 68]. While it has been proposed that a complimentary somatostatinergic BST to CeL projection may exist [65], we are unaware of any double labeling tract-tracing studies delineating this pathway. It is noteworthy that the primate EAc receives significant input from other regions of the brain that have not been thoroughly explored as possible sources of EAc SST neuropil [69–71]. In mice, SST neurons in the thalamus project to the CeL, pointing to the possibility of at least one source of SST input [24]. In contrast to SST, we did not detect CRF expressing fibers in the SLEA. While this could be interpreted as a failure to demonstrate CRF fibers communicating between the CeL and BSTL, we note that previous studies reported SLEA CRF staining [72–74]. Differences in the CRF antibody used here, as well as the level of SLEA that we examined, could account for this discrepancy.
The density of SST positive fibers in the EAc suggests a role for SST in broadly modulating GABAergic neurons which predominate in the EAc. Additionally, some of these GABAergic neurons express SST, and SST receptors are expressed throughout the EAc [75]. While not directly studied in the EAc, evidence demonstrates that SST can modulate the activity of other types of neurons [59, 76]. Similarly, EAc SST could be important in modulating the mutual inhibitory GABAergic circuits within and between the CeL and BSTL that mediate threat-responses. It is noteworthy that activation of SST neurons in the mouse CeL results in inhibition of other CeL GABAergic neurons and in freezing [22]. Other neuropeptides, including CRF and dynorphin, likely play a modulatory role in EAc function. For example, in rodents, dynorphin signaling via kappa opioid receptors appears to modulate GABAergic and glutamatergic neurotransmission in the BST [61, 77] and activation of CRF neurons in the CeL leads to flight behavior [17]. While we focused on SST and its co-expression with CRF in this study, future research examining dynorphin projections from the Ce to BST in non-human primates will be informative especially in relation to possible co-expression with SST and CRF. Along with SST and CRF systems, this suggests the potential of dynorphin receptors as serving as novel drug targets.
The present study replicates and extends previous research in non-human primates on the distribution of SST in the EAc. In the context of rodent research implicating SST and other neuropeptides, our study suggests that EAc SST neurons may be a good candidate for future anatomical and mechanistic studies in non-human primates. As the SST distribution in non-human primates is quite similar to that in humans, using non-human primates to investigate EAc SST function is an important translational step to understand the role of SST in human psychopathology. The data from our current study has the potential to guide mechanistic non-human primate studies focused on altering EAc SST function with the ultimate goal of developing new treatments to reduce the suffering associated with anxiety and other stress-related disorders.
Somatostatin mRNA and protein is abundant in the primate extended amygdala
Somatostatin mRNA is expressed more densely in the posterior lateral central nucleus
Somatostatin neurons make up a small proportion of primate extended amygdala neurons
A large percentage of somatostatin neurons express corticotropin-releasing factor
Acknowledgements and funding
This work was supported by funding from the NIMH (R01MH081884, P50MH100031) awarded to NHK and an NIMH T32 training grant (5T32MH018931) awarded to RK. We thank the personnel of the Harlow Center for Biological Psychology, the HealthEmotions Research Institute, the Waisman Laboratory for Brain Imaging and Behavior, the Wisconsin National Primate Research Center, the Wisconsin Institutes for Medical Research, M. Reidel and E. Fekete, D. McFarlin, C. Cleveland, A. Meier, L. Gerow, The Waisman Cellular Imaging Core at UW-Madison and The Neuroscience Training Program at UW-Madison.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Shackman AJ, Fox AS, Contributions of the Central Extended Amygdala to Fear and Anxiety, J Neurosci, 36 (2016) 8050–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fox AS, Kalin NH, A translational neuroscience approach to understanding the development of social anxiety disorder and its pathophysiology, Am J Psychiatry, 171 (2014) 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fox AS, Oler JA, Tromp do PM, Fudge JL, Kalin NH, Extending the amygdala in theories of threat processing, Trends Neurosci, 38 (2015) 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Davis M, The role of the amygdala in fear and anxiety, Annu Rev Neurosci, 15 (1992) 353–375. [DOI] [PubMed] [Google Scholar]
- [5].Fox AS, Oler JA, Shackman AJ, Shelton SE, Raveendran M, McKay DR, Converse AK, Alexander A, Davidson RJ, Blangero J, Rogers J, Kalin NH, Intergenerational neural mediators of early-life anxious temperament, Proc Natl Acad Sci U S A, 112 (2015) 9118–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fox AS, Shelton SE, Oakes TR, Davidson RJ, Kalin NH, Trait-like brain activity during adolescence predicts anxious temperament in primates, PLoS ONE, 3 (2008) e2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Oler JA, Tromp DP, Fox AS, Kovner R, Davidson RJ, Alexander AL, McFarlin DR, Birn RM, B EB, deCampo DM, Kalin NH, Fudge JL, Connectivity between the central nucleus of the amygdala and the bed nucleus of the stria terminalis in the non-human primate: neuronal tract tracing and developmental neuroimaging studies, Brain Struct Funct, 222 (2017) 21–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Oler JA, Birn RM, Patriat R, Fox AS, Shelton SE, Burghy CA, Stodola DE, Essex MJ, Davidson RJ, Kalin NH, Evidence for coordinated functional activity within the extended amygdala of non-human and human primates, Neuroimage, 61 (2012) 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fox AS, Oler JA, Birn RM, Shackman AJ, Alexander AL, Kalin NH, Functional connectivity within the primate extended amygdala is heritable and associated with early-life anxious temperament, J Neurosci, DOI 10.1523/JNEUROSCI.0102-18.2018(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Amaral DG, Avendano C, Benoit R, Distribution of somatostatin-like immunoreactivity in the monkey amygdala, J Comp Neurol, 284 (1989) 294–313. [DOI] [PubMed] [Google Scholar]
- [11].Pitkanen A, Amaral DG, The distribution of GABAergic cells, fibers, and terminals in the monkey amygdaloid complex: an immunohistochemical and in situ hybridization study, J Neurosci, 14 (1994) 2200–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Price JL, Russchen FT, Amaral DG, The limbic region. II. The amygdaloid complex, in: Hokfelt BT, Swanson LW (Eds.) Handbook of Chemical Neuroanatomy, Elsevier, Amsterdam, 1987, pp. 279–381. [Google Scholar]
- [13].Alheid GF, Extended amygdala and basal forebrain, Ann N Y Acad Sci, 985 (2003) 185–205. [DOI] [PubMed] [Google Scholar]
- [14].Alheid GF, Heimer L, New perspectives in basal forebrain organization of special relevance for neuropsychiatric disorders: the striatopallidal, amygdaloid, and corticopetal components of substantia innominata, Neuroscience, 27 (1988) 1–39. [DOI] [PubMed] [Google Scholar]
- [15].deCampo DM, Fudge JL, Amygdala projections to the lateral bed nucleus of the stria terminalis in the macaque: comparison with ventral striatal afferents, J Comp Neurol, 521 (2013) 3191–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ahrens S, Wu MV, Furlan A, Hwang GR, Paik R, Li H, Penzo MA, Tollkuhn J, Li B, A Central Extended Amygdala Circuit That Modulates Anxiety, J Neurosci, 38 (2018) 5567–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fadok JP, Krabbe S, Markovic M, Courtin J, Xu C, Massi L, Botta P, Bylund K, Muller C, Kovacevic A, Tovote P, Luthi A, A competitive inhibitory circuit for selection of active and passive fear responses, Nature, 542 (2017) 96–100. [DOI] [PubMed] [Google Scholar]
- [18].Kim J, Pignatelli M, Xu S, Itohara S, Tonegawa S, Antagonistic negative and positive neurons of the basolateral amygdala, Nat Neurosci, 19 (2016) 1636–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kim J, Zhang X, Muralidhar S, LeBlanc SA, Tonegawa S, Basolateral to Central Amygdala Neural Circuits for Appetitive Behaviors, Neuron, 93 (2017) 1464–1479. e1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Price DL, Martin LJ, Powers RE, Dellovade TL, The bed nucleus-amygdala continuum in human and monkey, The Journal of Comparative Neurology, 309 (1991) 445–485. [DOI] [PubMed] [Google Scholar]
- [21].Cassel MD, Gray TS, Morphology of Peptide Immunoreactive neurons in the rat central nucleus of the amygdala, Journal of Comparative Neurology, DOI (1989) 320–333. [DOI] [PubMed] [Google Scholar]
- [22].Li H, Penzo MA, Taniguchi H, Kopec CD, Huang ZJ, Li B, Experience-dependent modification of a central amygdala fear circuit, Nat Neurosci, 16 (2013) 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Penzo MA, Robert V, Li B, Fear conditioning potentiates synaptic transmission onto long-range projection neurons in the lateral subdivision of central amygdala, J Neurosci, 34 (2014) 2432–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Penzo MA, Robert V, Tucciarone J, De Bundel D, Wang M, Van Aelst L, Darvas M, Parada LF, Palmiter RD, He M, Huang ZJ, Li B, The paraventricular thalamus controls a central amygdala fear circuit, Nature, 519 (2015) 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yu K, Garcia da Silva P, Albeanu DF, Li B, Central Amygdala Somatostatin Neurons Gate Passive and Active Defensive Behaviors, J Neurosci, 36 (2016) 6488–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sanford CA, Soden ME, Baird MA, Miller SM, Schulkin J, Palmiter RD, Clark M, Zweifel LS, A Central Amygdala CRF Circuit Facilitates Learning about Weak Threats, Neuron, 93 (2017) 164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kalin NH, Fox AS, Kovner R, Riedel MK, Fekete EM, Roseboom PH, Tromp do PM, Grabow BP, Olsen ME, Brodsky EK, McFarlin DR, Alexander AL, Emborg ME, Block WF, Fudge JL, Oler JA, Overexpressing Corticotropin-Releasing Factor in the Primate Amygdala Increases Anxious Temperament and Alters Its Neural Circuit, Biol Psychiatry, 80 (2016) 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bupesh M, Abellán A, Medina L, Genetic and experimental evidence supports the continuum of the central extended amygdala and a mutiple embryonic origin of its principal neurons, Journal of Comparative Neurology, 519 (2011) 3507–3531. [DOI] [PubMed] [Google Scholar]
- [29].Han S, Soleiman MT, Soden ME, Zweifel LS, Palmiter RD, Elucidating an Affective Pain Circuit that Creates a Threat Memory, Cell, 162 (2015) 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Morris HM, Hashimoto T, Lewis DA, Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder, Cereb Cortex, 18 (2008) 1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sibille E, Reduced Somatostatin Expression or Somatostatin-Positive Gamma-Aminobutyric Acid Neurons: A Shared Pathology Across Brain Disorders, Biol Psychiatry, 81 (2017) 467–469. [DOI] [PubMed] [Google Scholar]
- [32].Fox AS, Oler JA, Shelton SE, Nanda SA, Davidson RJ, Roseboom PH, Kalin NH, Central amygdala nucleus (Ce) gene expression linked to increased trait-like Ce metabolism and anxious temperament in young primates, Proc Natl Acad Sci U S A, 109 (2012) 18108–18113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rosene DL, Roy NJ, Davis BJ, A cryoprotection method that facilitates cutting frozen sections of whole monkey brains for histological and histochemical processing without freezing artifact, J. Histochem. Cytochem, Vol. 34, No. 10 (1986) 1301–1315. [DOI] [PubMed] [Google Scholar]
- [34].Paxinos G, Huang X, Petrides M, Toga AW, The Rhesus Monkey Brain: Stereotaxic Coordinates, Elsevier; 2009. [Google Scholar]
- [35].Granger F.P.a.B.E., IPython: A System for Interactive Scientific Computing, Computing in Science & Engineering, 9 (2007) 21–29. [Google Scholar]
- [36].O T. Jones Eric, Pearu Peterson, others, SciPy: Open source scientific tools for Python, 2001.
- [37].van der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T, scikit-image: image processing in Python, PeerJ, 2 (2014) e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].McKinney W, Data Structures for Statistical Computing in Python, Proceedings of the 9th Python in Science Conference, 2010, pp. 51–56. [Google Scholar]
- [39].Hunter JD, Matplotlib: A 2D Graphics Environment, Computing in Science & Engineering, 9 (2007) 90–95. [Google Scholar]
- [40].R.C. Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2015. [Google Scholar]
- [41].Bates D, Mächler M, Bolker B, Walker S, Fitting Linear Mixed-Effects Models Using lme4, 2015, 67 (2015) 48. [Google Scholar]
- [42].Fox J, Weisberg S, An R Companion to Applied Regression, Second ed., Sage; 2011. [Google Scholar]
- [43].Weyer A, Schilling K, Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum, J Neurosci Res, 73 (2003) 400–409. [DOI] [PubMed] [Google Scholar]
- [44].Chee MJ, Pissios P, Maratos-Flier E, Neurochemical characterization of neurons expressing melanin-concentrating hormone receptor 1 in the mouse hypothalamus, J Comp Neurol, 521 (2013) 2208–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Treweek JB, Jaferi A, Colago EE, Zhou P, Pickel VM, Electron microscopic localization of corticotropin-releasing factor (CRF) and CRF receptor in rat and mouse central nucleus of the amygdala, J Comp Neurol, 512 (2009) 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Das M, Vihlen CS, Legradi G, Hypothalamic and brainstem sources of pituitary adenylate cyclase-activating polypeptide nerve fibers innervating the hypothalamic paraventricular nucleus in the rat, J Comp Neurol, 500 (2007) 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Garcia-Lopez M, Abellan A, Legaz I, Rubenstein JL, Puelles L, Medina L, Histogenetic compartments of the mouse centromedial and extended amygdala based on gene expression patterns during development, J Comp Neurol, 506 (2008) 46–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lepousez G, Csaba Z, Bernard V, Loudes C, Videau C, Lacombe J, Epelbaum J, Viollet C, Somatostatin interneurons delineate the inner part of the external plexiform layer in the mouse main olfactory bulb, J Comp Neurol, 518 (2010) 1976–1994. [DOI] [PubMed] [Google Scholar]
- [49].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nature Methods, 9 (2012) 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chareyron LJ, Banta Lavenex P, Amaral DG, Lavenex P, Stereological analysis of the rat and monkey amygdala, J Comp Neurol, 519 (2011) 3218–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].McCullough KM, Morrison FG, Hartmann J, Carlezon WA Jr., Ressler KJ, Quantified Coexpression Analysis of Central Amygdala Subpopulations, eNeuro, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wolff SB, Grundemann J, Tovote P, Krabbe S, Jacobson GA, Muller C, Herry C, Ehrlich I, Friedrich RW, Letzkus JJ, Luthi A, Amygdala interneuron subtypes control fear learning through disinhibition, Nature, 509 (2014) 453–458. [DOI] [PubMed] [Google Scholar]
- [53].Cassell MD, Gray TS, Morphology of peptide-immunoreactive neurons in the rat central nucleus of the amygdala, J Comp Neurol, 281 (1989) 320–333. [DOI] [PubMed] [Google Scholar]
- [54].Lin LC, Sibille E, Somatostatin, neuronal vulnerability and behavioral emotionality, Molecular psychiatry, 20 (2015) 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Palkovits M, Tapia-Arancibia L, Kordon C, Epelbaum J, Somatostatin connections between the hypothalamus and the limbic system of the rat brain, Brain Res, 250 (1982) 223–228. [DOI] [PubMed] [Google Scholar]
- [56].Momiyama T, Zaborszky L, Somatostatin presynaptically inhibits both GABA and glutamate release onto rat basal forebrain cholinergic neurons, J Neurophysiol, 96 (2006) 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sun QQ, Huguenard JR, Prince DA, Somatostatin inhibits thalamic network oscillations in vitro: actions on the GABAergic neurons of the reticular nucleus, J Neurosci, 22 (2002) 5374–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lopez-Huerta VG, Tecuapetla F, Guzman JN, Bargas J, Galarraga E, Presynaptic modulation by somatostatin in the neostriatum, Neurochem Res, 33 (2008) 1452–1458. [DOI] [PubMed] [Google Scholar]
- [59].Urban-Ciecko J, Barth AL, Somatostatin-expressing neurons in cortical networks, Nat Rev Neurosci, 17 (2016) 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Urban-Ciecko J, Fanselow EE, Barth AL, Neocortical somatostatin neurons reversibly silence excitatory transmission via GABAb receptors, Curr Biol, 25 (2015) 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li C, Pleil KE, Stamatakis AM, Busan S, Vong L, Lowell BB, Stuber GD, Kash TL, Presynaptic inhibition of gamma-aminobutyric acid release in the bed nucleus of the stria terminalis by kappa opioid receptor signaling, Biol Psychiatry, 71 (2012) 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].McDonald AJ, Mascagni F, Augustine JR, Neuropeptide Y and somatostatin-like immunoreactivity in neurons of the monkey amygdala, Neuroscience, 66 (1995) 959–982. [DOI] [PubMed] [Google Scholar]
- [63].Seney ML, Tripp A, McCune S, Lewis DA, Sibille E, Laminar and cellular analyses of reduced somatostatin gene expression in the subgenual anterior cingulate cortex in major depression, Neurobiology of disease, 73 (2015) 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tripp A, Kota RS, Lewis DA, Sibille E, Reduced somatostatin in subgenual anterior cingulate cortex in major depression, Neurobiology of disease, 42 (2011) 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Martin LJ, Powers RE, Dellovade TL, Price DL, The bed nucleus-amygdala continuum in human and monkey, J Comp Neurol, 309 (1991) 445–485. [DOI] [PubMed] [Google Scholar]
- [66].Candy JM, Perry RH, Thompson JE, Johnson M, Oakley AE, Neuropeptide localisation in the substantia innominata and adjacent regions of the human brain, Journal of Anatomy, 140 (1985) 309–327. [PMC free article] [PubMed] [Google Scholar]
- [67].Mufson EJ, Benoit R, Mesulam MM, Immunohistochemical evidence for a possible somatostatin-containing amygdalostriatal pathway in normal and Alzheimer’s disease brain, Brain Res, 453 (1988) 117–128. [DOI] [PubMed] [Google Scholar]
- [68].McDonald AJ, Somatostatinergic projections from the amygdala to the bed nucleus of the stria terminalis and medial preoptic-hypothalamic region, Neurosci Lett, 75 (1987) 271–277. [DOI] [PubMed] [Google Scholar]
- [69].Amaral DG, Insausti R, Retrograde transport of D-[3H]-aspartate injected into the monkey amygdaloid complex, Exp Brain Res, 88 (1992) 375–388. [DOI] [PubMed] [Google Scholar]
- [70].Canteras NS, Swanson LW, Projections of the ventral subiculum to the amygdala, septum, and hypothalamus: a PHAL anterograde tract-tracing study in the rat, J Comp Neurol, 324 (1992) 180–194. [DOI] [PubMed] [Google Scholar]
- [71].Fudge JL, Tucker T, Amygdala projections to central amygdaloid nucleus subdivisions and transition zones in the primate, Neuroscience, 159 (2009) 819–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Moga MM, Gray TS, Evidence for corticotropin-releasing factor, neurotensin, and somatostatin in the neural pathway from the central nucleus of the amygdala to the parabrachial nucleus, J Comp Neurol, 241 (1985) 275–284. [DOI] [PubMed] [Google Scholar]
- [73].Bassett JL, Foote SL, Distribution of corticotropin-releasing factor-like immunoreactivity in squirrel monkey (Saimiri sciureus) amygdala, J Comp Neurol, 323 (1992) 91–102. [DOI] [PubMed] [Google Scholar]
- [74].Gray TS, Magnuson DJ, Peptide immunoreactive neurons in the amygdala and the bed nucleus of the stria terminalis project to the midbrain central gray in the rat, Peptides, 13 (1992) 451–460. [DOI] [PubMed] [Google Scholar]
- [75].Breder CD, Yamada Y, Yasuda K, Seino S, Saper CB, Bell GI, Differential expression of somatostatin receptor subtypes in brain, J Neurosci, 12 (1992) 3920–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Moore S, Madamba S, Joels M, Siggins G, Somatostatin augments the M-current in hippocampal neurons, Science, 239 (1988) 278–280. [DOI] [PubMed] [Google Scholar]
- [77].Crowley NA, Bloodgood DW, Hardaway JA, Kendra AM, McCall JG, Al-Hasani R, McCall NM, Yu W, Schools ZL, Krashes MJ, Lowell BB, Whistler JL, Bruchas MR, Kash TL, Dynorphin Controls the Gain of an Amygdalar Anxiety Circuit, Cell reports, 14 (2016) 2774–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]