Abstract
Natural product evodiamine and its derivatives represent a promising class of multi-target antitumor agents. However, the clinical development of these compounds has been hampered by a poor understanding of their antitumor mechanisms. To tackle this obstacle, herein, novel fluorescent probes were designed to elucidate the antitumor mode of action of 10-hydroxyevodiamine. This compound was proven to be distributed in the mitochondria and lysosomes and to act by autophagy and apoptosis mechanisms.
Abbreviations: Boc, di-tert-butyl dicarbonate; CCK8, cell counting kit-8; DMAP, 4-dimethylaminopyridine; DMSO, dimethylsulfoxide; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride; HBTU, O-benzotriazole-N,N,N,N-tetramethyl-uronium-hexafluorophosphate; 3MA, 3-methyladenine; MMP, mitochondrial membrane potential; NPs, natural products; TEA, trimethylamine; TFA, trifluoroacetic acid
KEY WORDS: Fluorescent probes, 10-Hydroxyevodiamine, Anticancer mechanisms, Autophagy, Apoptosis
Graphical abstract
Novel fluorescent probes were designed to elucidate the antitumor mode of action of 10-hydroxyevodiamine, which was proven to be distributed in the mitochondria and lysosomes and acted by autophagy and apoptosis mechanisms.
1. Introduction
Bioactive molecules derived from natural products (NPs) have been rapidly developed in the past few decades1., 2., 3., 4., 5.. Clarification of the mechanisms of bioactive NPs is of key importance in accelerating the process of drug discovery and development. Nevertheless, the mode of action of most NPs remains unclear6., 7.. Elucidating the mechanisms of bioactive NPs is challenging because of the limitations of analytical approaches8. Recently, fluorescent probes, which often act as a valuable tool to investigate the distribution of compounds in cells9., 10., 11., have been widely used in chemical biology studies of NPs12., 13., 14..
Evodiamine is a natural, multi-targeting antitumor molecule derived from the fruits of Evodiae fructus. However, clinical development of evodiamine has been hampered because of limited antitumor potency and a poor understanding of its molecular targets. Previously, our group designed a number of evodiamine derivatives and clarified the structure—activity relationships (SARs)15., 16.. In particular, a series of novel evodiamine derivatives were prepared that showed excellent in vitro and in vivo antitumor activity and were promising lead compounds for the development of novel antitumor agents12., 13.. Importantly, in silico target prediction in combination with antitumor mechanism studies indicated that evodiamine derivatives were dual topoisomerase 1 (Top1) and topoisomerase 2 (Top2) inhibitors and thio-evodiamine derivatives were triple Top1/Top2/tubulin inhibitors17. Despite these successes, the detailed anticancer mechanism of these highly active evodiamine derivatives is still poorly understood.
Recently, a number of chemical biology approaches have been developed to investigate the mode of action of bioactive small molecules, and among these techniques, fluorescence imaging has been widely applied to monitor changes in cellular morphology and function, and visualize the localization and interactions of bioactive molecules in organelles18., 19., 20., 21.. Herein, novel fluorescent probes based on 10-hydroxyevodiamine (1) were designed and synthesized to explore their subcellular localization and anticancer mechanism. The results provided deeper insights into the anticancer mechanism of evodiamine derivatives and will facilitate further structural optimization and drug development.
2. Results and discussion
2.1. Design and chemical synthesis of probes
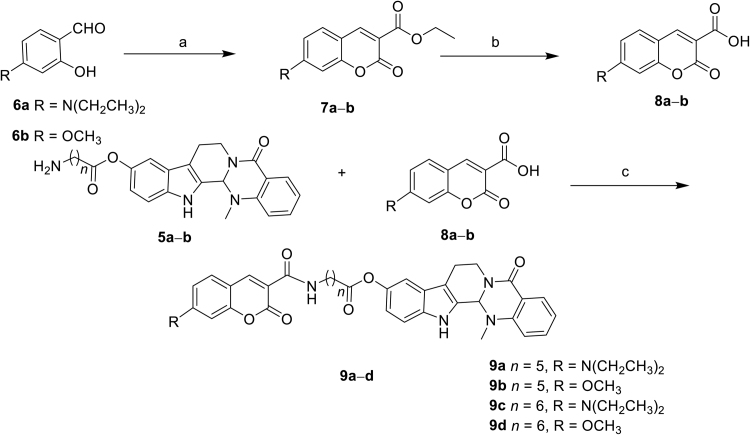
Compound 1, whose 10-hydroxyl group provided the structural basis for probe design, showed potent antitumor activity against a variety of cancer cell lines. Moreover, previous SARs also revealed that the 10-hydroxyl group could be substituted without a significant decrease in the antitumor activity15., 16.. Therefore, novel 10-hydroxyevodiamine-based fluorescent probes 9a—d were designed using substituted coumarins as the fluorophore and an alkylamide as the linker (Fig. 1). The chemical syntheses of the probes are depicted in Scheme 1, Scheme 2. Compound 1 was prepared according to our reported procedure12., 13.. Then, di-tert-butyl dicarbonate was used to protect 6-aminohexanoic acid (2a) to afford compound 3a, which was subsequently reacted with compound 1 in the presence of EDC and DMAP to give compound 4a. Removal of the Boc protecting group in the presence of TFA generated compound 5a. Similarly, 7-aminoheptanoic acid was used as a linker to afford compound 5b. Key intermediate 8a was obtained by treating 4-(diethylamino)-2-hydroxybenzaldehyde (6a) with diethyl propanedioate in methanol using piperidine as the catalyst followed by hydrolysis with 10% NaOH aqueous solution at room temperature. Coupling intermediate 8a with 5a or 5b using HBTU as the coupling reagent gave amides 9a and 9c, respectively. To investigate the effect of different electron-donating groups on the coumarin fragment, probes 9b and 9d were synthesized by a similar procedure (Scheme 2). Amide probe 16 was synthesized by a similar procedure to that used to prepare compound 9a (Scheme 3).
Figure 1.
Probes designed based on evodiamine derivative 1.
Scheme 1.
The synthetic route of intermediates 5a and 5b. Reagents and conditions: (a) (Boc)2O, TEA, MeOH, 60 °C, 10 h, yield 91%—93%; (b) EDC, DMAP, DCM, yield 65%—68%; (c) DCM, TFA, rt, 1.5 h, yield 53%—57%.
Scheme 2.
The synthetic route of target probes 9a—d. Reagents and conditions: (a) ethyl propanedioate, piperidine, MeOH, reflux, 10 h; (b) 10% NaOH, rt, 0.5 h, two steps yield 62%—65%. (c) HBTU, TEA, DMF, rt, 6 h, yield 47%—53%.
Scheme 3.
The synthetic route of probe 16. Reagents and conditions: (a) KNO3, H2SO4; (b) H2, 10% Pd/C, DCM, two steps yield 47%; (c) HBTU, TEA, DMF; (d) LiOH, rt, 4 h, 60%; (e) HBTU, TEA, DMF, rt, 6 h, yield 51%.
2.2. Cytotoxicity and fluorescent properties
The in vitro antiproliferative activities of probes 9a—d against the HCT116 cell line (human colon cancer cell) were evaluated using the cell counting kit-8 (CCK8) assay. As shown in Table 1, all the probes showed modest to good antitumor activities; however, these probes were less active than compound 1. The linker length was important to the antitumor activity of the probe. Generally, probes with six-carbon spacers (probes 9a and 9b) performed better than the probes with seven-carbon spacers (probes 9c and 9d). In contrast, coumarin fluorophores 8a, 8b and 15 were completely inactive (Table 1). Moreover, the maximum excitation and emission wavelengths of the fluorescent probes were detected (Table 1). The stokes shift value of probes 9a—d were 45, 75, 49, and 81 nm, respectively, indicating that they were suitable for fluorescence imaging studies. Considering their in vitro antitumor activities, fluorescent probe 9a was selected for the subcellular localization studies.
Table 1.
In vitro antitumor activities (IC50, μmol/L) and fluorescence properties of 10-hydroxyevodiamine-based probesa.
| Compd. | HCT116 (IC50, μmol/L) | DMSOb |
||
|---|---|---|---|---|
| λex (nm) | λem (nm) | Stokes shiftd (nm) | ||
| 8a | >50 | 425 | 462 | 37 |
| 8b | >50 | 342 | 413 | 71 |
| 9a | 1.94±0.21 | 432 | 477 | 45 |
| 9b | 4.85±0.62 | 346 | 421 | 75 |
| 9c | 5.48±0.46 | 430 | 479 | 49 |
| 9d | 9.34±0.89 | 343 | 424 | 81 |
| 15 | >50 | 439 | 481 | 42 |
| 16 | 19.11±1.85 | 418 | 465 | 47 |
| 1 | 0.24±0.02 | NTc | NT | NT |
IC50 values are the mean of at least three independent assays and are presented as the mean±SD.
Fluorescence properties were detected in DMSO with the compounds at a concentration of 1 μmol/L, λex: maximum excitation, λem: maximum emission.
NT=not tested.
The difference between the λex and λem wavelengths.
2.3. Subcellular localization studies
To further investigate the location of compound 1 in organelles, various organelle-specific dyes and probe 9a were co-incubated with HCT116 cells. The overlap of red fluorescence from the organelle-specific dyes and the green fluorescence from probe 9a confirmed that 9a accumulates in the mitochondria and lysosomes (Fig. 2 and Supporting Information Fig. S1). Probe 9a began to be absorbed after 4 h of incubation. Meanwhile, when we tested compound 15 as the control, no obvious overlap was detected. However, fluorescent probe 9a contains a phenolic ester linkage that may be prone to cleavage inside live cells. Thus, more stable amide probe 16 was synthesized and assayed (Table 1). Interestingly, probe 16 was also distributed in the mitochondria and lysosomes (Supporting Information Fig. S2), which confirms the reliability of the subcellular localization results. Moreover, the Pearson correlation coefficient (PCC) was calculated by using Image J software based on the pixel intensity of the dye and probe, and the results showed that the PCC value was significantly higher after incubation with the probes than incubation with the control (Fig. 2B and D).
Figure 2.
Subcellular localization of fluorescent probe 9a and compound 15. MitoTracker (mitochondria specific dye) and LysoTracker (lysosomes specific dye) in HCT116 cells. Images were collected by confocal microscopy (TCS-SP5): (A) subcellular localization of fluorescent probe 9a, compound 15 and MitoTracker in HCT116 cells. (a) HCT116 cells were incubated with 0.5 μmol/L of probe 9a for 4 h, left, cellular localization of probe 9a and MitoTracker; Merged, overlay of the two images; Zoom, expanded image of boxed areas in Merged. (b)—(d) HCT116 cells were treated with 0.5 μmol/L of probe 9a for 8 h, 12 h and 24 h, respectively. (e) HCT116 cells were incubated with 20 μmol/L of compound 15 for 24 h. Images were collected with excitation of probe 9a and compound 15 at λex=408 nm and emission at λem=456 nm, MitoTracker and LysoTracker at λex=581 nm and emission at λem=605 nm. (B) PCC analysis of co-localization between MitoTracker and probe 9a. **P<0.01, ***P<0.001 vs the control. Bars denote 10 μm. (C) subcellular localization of fluorescent probe 9a, compound 15 and LysoTracker in HCT116 cells after treatment with 0.5 μmol/L of probe 9a or 20 μmol/L of compound 15 for 4, 8, 12 and 24 h, respectively. (D) PCC analysis of co-localization between LysoTracker and probe 9a. **P<0.01, ***P<0.001 vs the control. Bars denote 10 μm.
2.4. Autophagy induced by compound 1
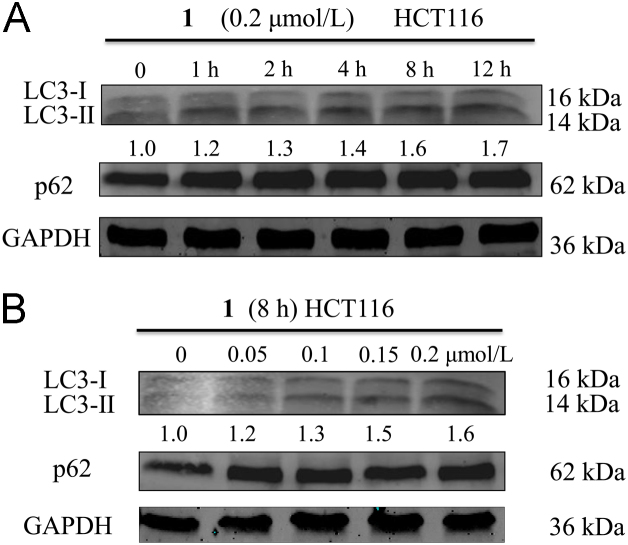
Lysosomes are an essential organelle, as they contain various hydrolytic enzymes22 that can breakdown a variety of biomolecules. However, autophagy is the process by which cells disassemble nonessential or dysfunctional constituents23. Therefore, it is widely accepted that autophagy and lysosomes are inextricably associated24. Thus, the ability of compound 1 to induce autophagy in HCT116 cells was firstly investigated by detecting the level of LC3-II expression using Western blotting analysis. As demonstrated in Fig. 3A, after incubating with compound 1 (0.2 μmol/L) for the indicated time, the expression of LC3-I and LC3-II protein was up-regulated and the ratio between LC3-II and LC3-I increased from 1.0 to 1.7. Based on these results, HCT116 cells were treated with various concentrations of compound 1 for 8 h and then analyzed by Western blotting. The expression of LC3-I and LC3-II was gradually up-regulated as the concentration of compound 1 increased, also the ratio between LC3-II and LC3-I increased from 1.0 to 1.6 (Fig. 3B), indicating that compound 1 induced cell autophagy in HCT116 cells. In addition, the expression level of another autophagy related protein p62 was also detected by Western blotting. The result showed that compound 1 up-regulated the expression of p62 protein. However, it was not clear whether compound 1 could induce autophagic activation or autophagic flux blockade. Thus, the autophagy flux was detected by the transfection of adenovirus harboring mRFP-GFP-LC3. After transfection, autophagosomes were demonstrated as yellow dots (the combination of red and green fluorescence), and autolysosomes were shown as red dots (the extinction of GFP in the acid environment of lysosomes). As shown in Fig. 4, compound 1 increased the number of both yellow dots and red dots, indicating that the number of autophagosomes increased and the autophagy flux was activated. Furthermore, compound 1-induced autophagy was further verified by transmission electron microscopy (TEM). As shown in Fig. 5, the number of autophagosomes and autolysosomes increased dramatically after incubation with compound 1 for 8 h, indicating that compound 1 could activate the autophagy.
Figure 3.
Compound 1-induced autophagy in HCT116 cells. (A) HCT116 cells were treated with 0.2 μmol/L of compound 1 for the indicated time and then analyzed by Western blotting with specific antibodies. (B) HCT116 cells were incubated with various concentrations of compound 1 (0, 0.05, 0.1, 0.15 and 0.2 μmol/L) for 8 h and then analyzed by Western blotting. GAPDH served as an internal control.
Figure 4.
Compound 1 increased the level of autophagy flux in HCT116 cells. HCT116 cells were incubated with or without compound 1 (0.2 μmol/L) and representative images of mRFP-GFP-LC3 vector were collected by confocal microscopy (TCS-SP5). (a) HCT116 cells were incubated with mRFP-GFP-LC3 vector for 6 h; (b) HCT116 cells were incubated with mRFP-GFP-LC3 vector for 6 h, followed by the treatment of compound 1 for 8 h. Bars denote 10 μm.
Figure 5.
Compound 1-induced autophagosomes (green arrow) and autolysosomes (red arrow) accumulation in HCT116 cells. HCT116 cells were incubated without (A) or with (B and C) compound 1 for 8 h and then were fixed. (D) Expanded image of boxed areas and the double membrane was observed in autophagosomes. The changes of ultrastructure were observed under TEM. Magnification, 5000×.
To further verify the role of autophagy in compound 1-induced cell death, HCT116 cells were incubated with the autophagy inhibitors25 wortmannin (0.25 μmol/L), LY294002 (5 μmol/L), chloroquine (CQ, 5 μmol/L) and 3MA (5 μmol/L) for 2 h before treatment with compound 1. Compared with cells incubated with compound 1 alone, the apoptosis was considerably higher in cells treated with both an autophagy inhibitor and compound 1. However, the autophagy inhibitors alone had no significant effect on cell apoptosis (Fig. 6B). The in vitro antiproliferative activity assay also demonstrated that the cell viability decreased after co-incubation with compound 1 and an autophagy inhibitor (Table 2). Taken together, these results indicate that compound 1 induced cytoprotective autophagy in HCT116 cells. Notably, combination drug therapy with autophagy inhibitors may provide a new strategy to improve the therapeutic effects of compound 1.
Figure 6.
Effects of autophagy inhibitors on compound 1-induced cell apoptosis. HCT116 cells were pretreated with autophagy inhibitors (wortmannin, LY294002, 3MA and CQ) for 2 h and then incubated with 0.2 μmol/L of compound 1 for 48 h. Apoptosis was detected by flow cytometry with annexin V/PI (propidium iodide) double-staining. (A) Representative data shown in Figure A were derived from three independent experiments, and the statistical results are presented in Fig. 6B. Percent of cell apoptotic equals the percentage of apoptotic cells when incubated with compound 1 in combination with the autophagy inhibitor minus the percentage of apoptotic cells when treated with the autophagy inhibitor alone. “1-Control” is the percentage of apoptotic cells after treatment with compound 1 minus the percentage in untreated cells. Student׳s t-test was used to compare the difference between the experimental groups and control groups. *P<0.05, **P<0.01 vs the control.
Table 2.
In vitro antiproliferative activities of compound 1 in combination with different autophagy inhibitors (IC50, μmol/L)a.
| Compd. | HCT116 (IC50, μmol/L) |
|---|---|
| 1 | 0.24±0.02 |
| 1+LY294002 | 0.11±0.01 |
| 1+3MA | 0.09±0.01 |
| 1+wortmannin | 0.12±0.01 |
| 1+CQ | 0.08±0.01 |
IC50 values are the mean of at least three independent assays and are presented as the mean±SD.
2.5. Apoptosis and mitochondrial membrane potential assay
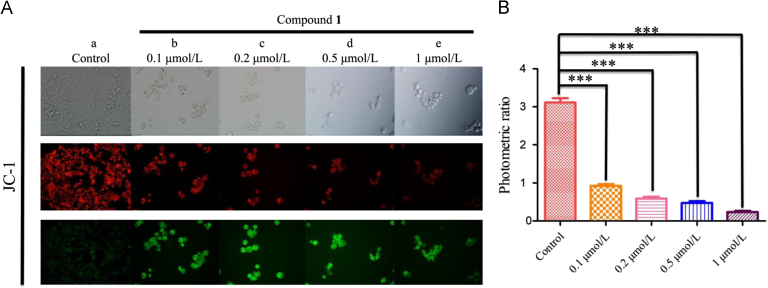
Apoptosis is the process of programmed cell death26 and usually plays a crucial role in the death of tumor cells. Moreover, the mitochondria are important organelles that are closely associated with energy production and whose dysfunction may cause cell apoptosis27., 28.. Given that compound 1 was distributed in the mitochondrion of HCT116 cells, it was determined that if the apoptosis was induced by dysfunction of the mitochondria. Firstly, the apoptosis induced by compound 1 in HCT116 cells was detected by flow cytometry with annexin V/PI staining. As shown in Fig. 7, the percentage of apoptotic cells had a remarkable increase in a concentration-dependent manner. Furthermore, JC-1, a commercially available fluorescent probe, was used to measure the mitochondrial membrane potential (MMP) of HCT116 cells. In healthy cells, JC-1 accumulates in the mitochondria and forms complexes that emit red fluorescence. In contrast, JC-1 exists as a monomer in the cytosol and fluoresces green in apoptotic cells. Thus, the HCT116 cells were treated with the indicated concentration of compound 1 for 24 h and then analyzed by fluorescence microscopy. As shown in Fig. 8A, the red fluorescence decreased, and the green fluorescence increased, indicating that compound 1 induced the dissipation of MMP in HCT116 cells. As shown in Fig. 8B, the ratio of red to green fluorescent intensity was calculated, and incubation with 1 μmol/L of compound 1 decreased the ratio from 3.11 (control) to 0.23. Collectively, these results confirmed the mitochondrial apoptosis mechanism of compound 1.
Figure 7.
Compound 1 induces apoptosis in HCT116 cells. HCT116 cells were treated with the indicated concentration of compound 1 for 24 h and then analyzed by flow cytometry with annexin V/PI staining. (A) show the results of three independent experiments, and the statistical results are presented in (B). Student׳s t-test was used to determine whether the results of the experimental groups significantly differed from those of the control. **P < 0.01, ***P < 0.001 vs the control.
Figure 8.
Compound 1-induced loss of MMP in HCT116 cells. (A) HCT116 cells were incubated with the indicated concentration of compound 1 for 48 h, and then images of the cells were collected using fluorescence microscopy. (a) HCT116 cells were incubated without compound 1 for 48 h, upper panel: cell vision selected, the middle panel: the red fluorescence of cells, bottom panel: the green fluorescence of cells; (b)—(e) HCT116 cells were incubated with 0.1, 0.2, 0.5 and 1 μmol/L of compound 1 for 48 h, respectively. As the concentration of compound 1 increased, the fluorescence intensity noticeably changed from red to green. The results were analyzed using a JC-1 MMP Assay Kit. (B) The bar chart shows the ratios of red to green fluorescent intensity. ***P < 0.01 vs the control.
2.6. Effects of compound 1 on the expression of apoptosis-related proteins
The above results revealed that compound 1-induced apoptosis was closely related to the mitochondria. It was reported that the BCL-2 family of proteins, which includes various proteins (e.g., BAX and BCL-2)29, played an important role in controlling mitochondrial apoptosis30. BCL-2 is a type of membrane protein that can directly suppress apoptosis31. BAX protein is usually located in the cytoplasm; however, when the apoptotic signal appears, BAX can shift from the cytoplasm to the mitochondrial membrane to increase mitochondrial permeability, releasing several cellular components (e.g., Cytochrome C)32., 33., 34.. To investigate whether such a mechanism is related to the apoptosis induced by compound 1, the expression levels of BAX, BCL-2, and Cytochrome C were determined by Western blotting. As demonstrated in Fig. 9, the expression level of BAX was up-regulated in a concentration-dependent manner after incubation with compound 1 for 8 h, while BCL-2 expression was down-regulated, and the ratio between BAX and BCL-2 increased from 1.0 to 10.6 as the incubation concentration increased. Furthermore, the released Cytochrome C can combine with other biomolecules to form a caspase-activating complex, which is of great significance for inducing caspase-dependent apoptosis35. In particular, caspase-3 is believed to play a key role in the apoptosis of tumor cells36. Therefore, Western blotting analysis was conducted to determine if caspase-3 is involved in compound 1-induced apoptosis. As shown in Fig. 9, compound 1 increased the expression of cleaved caspase-3 in a concentration-dependent manner, which was consistent with the mitochondrial depolarization results described above. In addition, increased expression of caspase-9 was also observed, and caspase-9 can interact with caspase-3 and induce apoptosis. Subsequently, we determined the expression level of apoptosis-related protein P53. Compound 1 could up-regulate the expression of P53 (Fig. 9), which contributed to cell apoptosis.
Figure 9.
Effects of compound 1 on the expression of apoptosis-related proteins in HCT116 cells. HCT116 cells were incubated with different concentrations of compound 1 for 8 h, and then the expression levels of cleaved caspase-3, caspase-9, BCL-2, BAX, P53 and cytochrome C were analyzed by Western blotting with the appropriate specific antibodies. GAPDH served as an internal control.
2.7. Summary of mitochondrial pathway of compound 1-induced apoptosis
On the basis of the above results, the hypothetical mitochondrial pathway for compound 1-induced apoptosis in HCT116 cells is summarized in Fig. 10. We hypothesize that compound 1 could up-regulate the P53 protein and down-regulate the BCL-2 protein, leading to the shift of BAX protein from cytoplasm to the membranes of mitochondria. Then, Cytochrome C is released from mitochondria and activates caspase 9 and 3, finally triggering cell apoptosis.
Figure 10.
The hypothetical mitochondrial pathway for compound 1-induced apoptosis in HCT116 cells.
3. Conclusions
In summary, novel fluorescent probes were designed to elucidate the antitumor mechanism of compound 1. The subcellular localization study confirmed that compound 1 was distributed in the mitochondria and lysosomes. Inspired by these findings, the autophagy and apoptosis mechanisms were clarified, which provided important information for the structural optimization and drug development of evodiamine derivatives. Compound 1 acted as an autophagy inducer with cytoprotective response, which had negative effects on the apoptosis. When the cytoprotective autophagy was inhibited by combinational use of autophagy inhibitors, both apoptosis and antitumor activity of compound 1 could be significantly improved37., 38., 39.. Thus, the combination therapy with autophagy inhibitors was an effective strategy for improving the antitumor activities of evodiamine derivatives. The results also highlighted the multi-targeting profile of evodiamine derivatives. The identification of new molecular targeting is currently in progress in our lab.
4. Experimental
4.1. Chemistry
4.1.1. Materials and instruments
1H NMR and 13C NMR spectra were recorded on Bruker AVANCE300 and AVANCE600 spectrometer (Bruker Company, Germany), using TMS as an internal standard and CDCl3 or DMSO-d6 as solvents. Chemical shifts are given in ppm (δ). The mass spectra were recorded on an Esquire 3000 LC—MS mass spectrometer. TLC analysis was carried out on silica gel plates GF254 (Qingdao Haiyang Chemical, China). Silica gel column chromatography was performed with Silica gel 60G (Qingdao Haiyang Chemical, China). Unless otherwise noted, all materials were obtained from commercial suppliers and used without further purification.
4.1.2. Synthesis procedures
4.1.2.1. Synthesis procedure of 6-((tert-butoxycarbonyl)amino)hexanoic acid (3a)
Di-tert-butyl dicarbonate (10 g, 46 mmol) was added to a solution of 6-aminohexanoic acid (5.0 g, 38 mmol) in dry MeOH (90 mL), and then TEA (8.0 mL) was added in one portion under stirring. The reaction mixture was refluxing at 60 °C for 10 h. Subsequently 2 mol/L HCl (20 mL) was added to the reaction mixture and the pH was adjusted to 4 and extracted with EtOAc (80 mL × 3). The combined organic layers were washed with saturated NaCl aqueous solution (80 mL × 3) followed by extracted with ethyl acetate. The aqueous layer was washed with 1 mol/L HCl (80 mL × 3) followed by the extraction with EtOAc (50 mL × 3). The organic phase was washed with water (80 mL×3), brine (80 mL), dried over anhydrous Na2SO4 and the solvent was removed under the reduced pressure to afford 3a as a colorless oil (8.2 g, 93%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.99 (s, 1 H), 6.78 (s, 1 H), 2.87 (q, J=6.64 Hz, 2 H), 2.17 (t, J=7.43 Hz, 2 H), 1.42—1.51 (m, 2 H), 1.30—1.39 (m, 11 H), 1.17—1.26 (m, 2 H). (ESI): m/z [M+Na]+=254.40.
4.1.2.2. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-10-yl-6-((tert-butoxycarbonyl)amino)hexanoate (4a)
EDC (0.86 g, 4.5 mmol) and DMAP (0.55 g, 4.5 mmol) were added to a solution of 3a (1.1 g, 4.5 mmol) in dry DCM. The reaction mixture was stirred for 30 min and compound 1 (0.96 g, 3.0 mmol) was subsequently added. The resulting solution was stirred at room temperature for 2 h. The solvent was removed under the reduced pressure and the residue was purified by silica column chromatography (DCM: MeOH=100:2, v/v) to afford compound 4a as a white solid (1.1 g, 68%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.18 (s, 1 H), 7.79 (dd, J=1.47 Hz, 7.71 Hz, 1 H), 7.45—7.50 (m, 1 H), 7.34 (d, J=8.73 Hz, 1 H), 7.18 (d, J=2.15 Hz, 1 H), 7.05 (d, J=8.05 Hz, 1 H), 6.96 (t, J=7.71 Hz, 1 H), 6.81—6.84 (m, 2 H), 6.14 (s, 1 H), 4.59—4.64 (m, 1 H), 3.15—3.24 (m, 1 H), 2.92 (q, J=6.46 Hz, 2 H), 2.89 (s, 3 H), 2.83—2.86 (m, 1 H), 2.72—2.78 (m, 1 H), 2.54 (t, J=7.37 Hz, 2 H), 1.59—1.68 (m, 2 H), 1.33—1.41 (m, 13 H). (ESI): m/z [M—H]—= 531.43.
4.1.2.3. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-10-yl-6-aminohexanoate (5a)
TFA (1.0 mL) was added to a suspension of 4a (0.30 g, 0.56 mmol) in DCM (3.0 mL) and the reaction mixture was stirring for 1.5 h at room temperature. DCM and TFA were removed under the reduced pressure. The crude product was purified by silica column chromatography (DCM: MeOH=100: 2, v/v) to afford compound 5a as a pale yellow solid (0.14 g, 57%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.22 (s, 1 H), 7.80 (dd, J=1.47 Hz, 7.80 Hz, 1 H), 7.72 (s, 2 H), 7.48—7.53 (m, 1 H), 7.36 (d, J=8.97 Hz, 1 H), 7.20 (d, J=2.03 Hz, 1 H), 7.07 (d, J=8.06 Hz, 1 H), 6.98 (t, J=7.65 Hz, 1 H), 6.86 (dd, J=2.18 Hz, 8.82 Hz, 1 H), 6.17 (s, 1 H), 4.61—4.67 (m, 1 H), 3.18—3.25 (m, 1 H), 2.92 (s, 3 H), 2.75—2.86 (m, 4 H), 2.60 (t, J=7.20 Hz, 2 H), 1.54—1.73 (m, 4 H), 1.40—1.78 (m, 2 H). (ESI): m/z [M+H]+=433.51.
The synthetic method for compounds 4b and 5b were similar to that of compound 4a and 5a.
4.1.2.4. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo[2′,3′:3,4] pyrido[2,1-b]quinazolin-10-yl-7-((tert-butoxycarbonyl)amino)heptanoate (4b)
White solid, (1.2 g, 65%). 1H NMR (DMSO-d6, 600 MHz) δ: 1H NMR (DMSO-d6, 600 MHz) δ: 11.18 (s, 1 H), 7.79 (d, J=8.09 Hz, 1 H), 7.34 (t, J=8.15 Hz, 1 H), 7.18 (d, J=8.96 Hz, 1 H), 7.18 (s, 1 H), 7.05 (d, J=8.38 Hz, 1 H), 6.96 (t, J=7.81 Hz, 1 H), 6.81—6.84 (m, 2 H), 6.14 (s, 1 H), 4.59—4.64 (m, 1 H), 3.15—3.24 (m, 1 H), 2.91—2.94 (m, 2 H), 2.89 (s, 4 H), 2.72—2.78 (m, 1 H), 2.55—2.57 (m, 2 H), 1.58—1.65 (m, 2 H), 1.23—1.41 (m, 15 H). (ESI): m/z [M—H]—=545.44.
4.1.2.5. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo[2′,3′:3,4]pyrido[2,1-b]quinazolin-10-yl-7-aminoheptanoate (5b)
Pale yellow solid, (0.22 g, 53%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.18 (s, 1 H), 7.77 (dd, J=1.53 Hz, 7.71 Hz, 1 H), 7.72 (s, 2 H), 7.44—7.50 (m, 1 H), 7.33 (d, J=8.68 Hz, 1 H), 7.16 (d, J=2.17 Hz, 1 H), 7.04 (d, J=8.04 Hz, 1 H), 6.95 (t, J=7.28 Hz, 1 H), 6.81 (dd, J=2.43 Hz, 8.81 Hz, 1 H), 6.14 (s, 1 H), 4.58—4.64 (m, 1 H), 3.14—3.24 (m, 1 H), 2.89 (s, 3 H), 2.71—2.84 (m, 3 H), 2.56 (t, J=6.55 Hz, 2 H), 1.61—1.66 (m, 2 H), 1.52—1.56 (m, 2 H), 1.32—1.41 (m, 4 H). (ESI): m/z [M+H]+=447.47.
4.1.2.6. Synthesis procedure of 7-(diethylamino)-2-oxo-2H-chromene-3-carboxylic acid (8a)
4-(Diethylamino)-2-hydroxybenzaldehyde (3.0 g, 16 mmol) was added to a mixture of ethyl propanedioate (6.2 g, 39 mmol) and MeOH (50 mL), followed by the addition of piperidine (3.1 mL, 31 mmol) in one portion under stirring, and then the suspension was reflux at 70 °C for 10 h. The MeOH was removed under the reduced pressure, and then the residue was dissolved in water, followed by pouring 10% NaOH aqueous solution (200 mL) into the reaction mixture, and stirring for another 30 min. Subsequently, the concentrated HCl was added into the reaction mixture and then the precipitate was collected by filtration, washed with water (2 × 30 mL) and dried to give compound 8a as a yellow solid (2.5 g, 62%, two steps). 1H NMR (CDCl3, 600 MHz) δ: 12.33 (s, 1H), 8.65 (s, 1H), 7.45 (d, J=9.08 Hz, 1H), 6.70 (dd, J=2.59 Hz, 9.19 Hz, 1H), 6.53 (d, J=2.49 Hz, 1H), 3.49 (q, J=7.13 Hz, 4H), 1.27 (t, J=7.35 Hz, 6H), (ESI): m/z [M+H]+=262.14.
The synthetic method for compound 8b was similar to the synthesis of compound 8a.
4.1.2.7. Synthesis procedure of 7-methoxy-2-oxo-2H-chromene-3-carboxylic acid (8b)
Pale yellow solid, (2.5 g, 65%). 1H NMR (CDCl3, 600 MHz) δ: 12.23 (s, 1H), 8.88 (s, 1H), 7.66 (d, J=8.91 Hz, 1H), 7.04 (dd, J=2.55 Hz, 8.91 Hz, 1 H), 6.94 (d, J=2.51 Hz, 1H), 3.97 (s, 3H), (ESI): m/z [M+H]+=221.26.
4.1.2.8. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo[2׳,3׳:3,4]pyrido[2,1-b] quinazolin-10-yl-6-(7-(diethylamino) -2-oxo-2H-chromene-3-carboxamido)hexanoate (9a)
To a solution of intermediate 8a (0.070 g, 0.28 mmol) in DMF, HBTU (0.13 g, 0.35 mmol) was added. Then TEA (0.050 mL, 0.35 mmol) was added under stirring and the reaction mixture was stirred for 0.5 h at room temperature. Subsequently, intermediate 5a (0.10 g, 0.23 mmol) was added. The reaction was stirred at room temperature for 6 h, followed by pouring the reaction mixture into ice/water (150 mL). Then, the precipitate was collected by filtration, washed with water (2 × 30 mL) and dried to give the crude product, which was purified by silica column chromatography (DCM: MeOH=100:5v/v) to afford compound 9a as a yellow solid (0.079 g, 50%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.15 (s, 1H), 8.64–8.68 (m, 2H), 7.79 (dd, J=4.50 Hz, 7.72 Hz, 1H), 7.67 (d, J=9.13 Hz, 1H), 7.46—7.49 (m, 1H), 7.33 (d, J=8.88 Hz, 1H), 7.17 (d, J=2.22 Hz, 1H), 7.05 (d, J=8.69 Hz, 1H), 6.96 (t, J=8.31 Hz, 1H), 6.83 (dd, J=2.32 Hz, 8.30 Hz, 1H), 6.79 (dd, J=2.70 Hz, 9.07 Hz, 1H), 6.61 (d, J=2.51 Hz, 1H), 6.12 (s, 1H), 4.59—4.62 (m, 1H), 3.47 (dd, J=6.95 Hz, 13.90 Hz, 4H), 3.16—3.21 (m, 1H), 2.87 (s, 4H), 2.73—2.76 (m, 1H), 2.57 (t, J=7.36 Hz, 2H), 1.66—1.71 (m, 2H), 1.55—1.60 (m, 2H), 1.40—1.45 (m, 2H), 1.21—1.26 (m, 2H), 1.13 (t, J=7.23 Hz, 6H). 13C NMR (DMSO-d6, 150 MHz) δ: 172.88, 164.65, 162.57, 162.27, 157.67, 152.88, 149.22, 148.10, 144.14, 134.66, 133.93, 132.58, 132.01, 128.47, 126.49, 120.82, 119.75, 118.00, 116.73, 112.45, 112.20, 110.96, 110.60, 110.00, 108.14, 96.33, 70.15, 44.79, 41.28, 39.17, 36.98, 33.96, 29.32, 26.40, 24.69, 19.89, 12.78. HR-MS (ESI) m/z: Calcd. for C39H41N5O6 [M+H]+ 676.3135, Found 676.3141.
The synthetic method for compounds 9b—d were similar to that of compound 9a.
4.1.2.9. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo [2′,3′:3,4] pyrido[2,1-b] quinazolin-10-yl-7-(7-(diethylamino)-2-oxo-2H-chromene-3-carboxamido)heptanoate (9b)
Pale yellow solid, (0.071 g, 52%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.16 (s, 1 H), 8.83 (s, 1 H), 8.68 (t, J=5.15 Hz, 1 H), 7.90 (d, J=8.89 Hz, 1 H), 7.80 (d, J=8.42 Hz, 1 H), 7.48 (t, J=7.49 Hz, 1 H), 7.33 (d, J=8.42 Hz, 1 H), 7.18 (d, J=1.76 Hz, 1 H), 7.12 (d, J=1.95 Hz, 1 H), 7.05 (t, J=7.61 Hz, 2 H), 6.97 (t, J=7.41 Hz, 1 H), 6.83 (dd, J=2.34 Hz, 8.78 Hz, 1 H), 6.13 (s, 1 H), 5.76 (s, 1 H), 4.60—4.63 (m, 1 H), 3.91 (s, 3 H), 3.17—3.22 (m, 1 H), 2.89 (s, 4 H), 2.74—2.77 (m, 1 H), 2.58 (t, J=8.01 Hz, 2 H), 1.69—1.73 (m, 2 H), 1.58—1.62 (m, 2 H), 1.43—1.47 (m, 2 H), 1.23—1.26 (m, 1 H). 13C NMR (DMSO-d6, 150 MHz) δ: 172.87, 164.84, 161.82, 156.60, 149.20, 148.14, 144.13, 134.65, 133.93, 132.61, 131.97, 128.47, 126.49, 120.81, 119.72, 117.97, 116.71, 115.48, 114.10, 112.62, 112.44, 112.19, 110.94, 100.74, 70.17, 56.72, 55.37, 41.29, 40.56, 39.32, 36.99, 33.96, 29.16, 26.34, 24.67, 19.89. HR-MS (ESI) m/z: Calcd. for C36H34N4O7 [M+H]+ 635.2505, Found 635.2503.
4.1.2.10. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo[2′,3′:3,4] pyrido[2,1-b]quinazolin-10-yl-6-(7-methoxy-2-oxo-2H-chromene-3-carboxamido)hexanoate (9c)
Yellow solid, (0.080 g, 53%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.18 (s, 1 H), 8.67 (s, 1 H), 7.82 (dd, J=1.51 Hz, 7.55 Hz, 1 H), 7.69 (d, J=9.05 Hz, 1 H), 7.48—7.51 (m, 1 H), 7.36 (d, J=8.69 Hz, 1 H), 7.20 (d, J=2.24 Hz, 1 H), 7.07 (d, J=8.29 Hz, 1 H), 6.97—7.00 (m, 1 H), 6.85 (dd, J=2.27 Hz, 8.69 Hz, 1 H), 6.81 (dd, J=2.38 Hz, 9.01 Hz, 1 H), 6.62 (d, J=2.27 Hz, 1 H), 6.15 (s, 1 H), 4.62—4.65 (m, 1 H), 3.49 (dd, J=7.03 Hz, 14.13 Hz, 4 H), 3.20—3.23 (m, 1 H), 2.91 (s, 4 H), 2.76—2.79 (m, 2 H), 2.71 (s, 2 H), 2.58 (t, J=7.03 Hz, 2 H), 1.67—1.69 (m, 2 H), 1.54—1.59 (m, 2 H), 1.37—1.44 (m, 4 H), 1.15 (t, J=6.95 Hz, 6 H). 13C NMR (DMSO-d6, 150 MHz) δ: 170.67, 164.33, 162.20, 161.93, 157.32, 152.53, 148.83, 147.75, 133.58, 133.23, 131.67, 131.46, 128.11, 125.88, 120.30, 119.26, 117.43, 115.83, 111.55, 110.25, 109.66, 108.95, 107.80, 96.00, 69.95, 44.45, 41.04, 40.21, 38.92, 36.57, 36.40, 29.13, 28.82, 26.33, 25.17, 22.21, 19.58, 12.44. HR-MS (ESI) m/z: Calcd. for C40H43N5O6 [M + H]+ 690.3291, Found 690.3284.
4.1.2.11. Synthesis procedure of 14-methyl-5-oxo-5,7,8,13,13b,14-hexahydroindolo [2′,3′:3,4] pyrido[2,1-b]quinazolin-10-yl-7-(7-methoxy-2-oxo-2H-chromene-3-carboxamido)heptanoate (9d)
Pale yellow solid, (0.067 g, 47%). 1H NMR (DMSO-d6, 600 MHz) δ: 11.16 (s, 1 H), 8.82 (s, 1 H), 8.66 (t, J=5.90 Hz, 1 H), 7.90 (d, J=8.66 Hz, 1 H), 7.79 (d, J=7.84 Hz, 1 H), 7.48 (t, J=7.56 Hz, 1 H), 7.34 (d, J=9.03 Hz, 1 H), 7.18 (d, J=1.84 Hz, 1 H), 7.11 (d, J=2.40 Hz, 1 H), 7.03—7.07 (m, 2 H), 6.97 (t, J=7.56 Hz, 1 H), 6.83 (dd, J=2.58 Hz, 8.85 Hz, 1 H), 6.14 (s, 1 H), 4.61—4.63 (m, 1 H), 3.90 (s, 3 H), 3.17—3.22 (m, 1 H), 2.89 (s, 4 H), 2.75—2.78 (m, 1 H), 2.57 (t, J=7.74 Hz, 2 H), 1.66—1.68 (m, 2 H), 1.56—1.58 (m, 2 H), 1.38—1.42 (m, 4 H), 1.23—1.25 (m, 2 H). 13C NMR (DMSO-d6, 150 MHz) δ: 172.90, 164.82, 161.80, 161.38, 156.59, 149.20, 148.11, 144.15, 134.65, 133.93, 132.62, 131.95, 128.47, 126.50, 120.80, 119.72, 117.97, 116.72, 115.51, 114.08, 112.62, 112.45, 112.19, 110.94, 100.75, 70.18, 56.72, 41.30, 39.41, 36.99, 33.96, 29.45, 29.31, 28.57, 26.55, 24.87, 19.89. HR-MS (ESI) m/z: Calcd. for C37H36N4O7 [M+H]+ 649.2662, Found 649.2657.
4.1.2.12. Synthesis procedure of 6-(7-(diethylamino)-2-oxo-2H-chromene-3-carboxamido)hexanoic acid (15)
To a solution of intermediate 8a (0.21 g, 0.84 mmol) in DMF, HBTU (0.39 g, 1.1 mmol) was added. Then TEA (0.15 mL, 1.1 mmol) was added under stirring and the reaction mixture was stirred for 0.5 h at room temperature. Subsequently, methyl 6-aminohexanoate (0.15 g, 1.0 mmol) was added. The reaction was stirred at room temperature for 6 h, followed by pouring the reaction mixture into ice/water (150 mL). Then, the precipitate was collected by filtration, washed with water (2 × 30 mL) and dried to give the crude product (compound 14). Then LiOH (0.051 g, 2.1 mmol) was added to a suspension of 14 (0.20 g, 0.52 mmol) in THF (6 mL), MeOH (4 mL), H2O (2 mL) and the reaction mixture was stirred for 4 h at room temperature. THF and MeOH were removed under the reduced pressure, followed by adjusting pH to 4—5 using 1 mol/L HCl. Then, the precipitate was collected by filtration and dried to give the crude product which was purified by silica column chromatography (DCM: MeOH=100:3, v/v) to afford compound 15 as a yellow solid (0.11 g, 60%).1H NMR (DMSO-d6, 600 MHz) δ: 8.64 (s, 2 H), 7.67 (d, J=8.99 Hz, 1 H), 6.79 (d, J=7.93 Hz, 1 H), 6.61 (s, 1 H), 3.47 (dd, J=6.87, 14.81 Hz, 4 H), 3.27 (dd, J=6.87, 12.68 Hz, 2 H), 2.30 (t, J=6.87 Hz, 2 H), 1.45—1.59 (m, 4 H), 1.24—1.34 (m, 2 H), 1.13 (t, J=6.35 Hz, 6 H). 13C NMR (DMSO-d6, 150 MHz) δ: 173.78, 162.51, 162.24, 157.64, 152.83, 148.10, 132.01, 110.56, 109.90, 108.10, 96.29, 51.64, 44.77, 33.62, 29.27, 26.35, 24.59, 12.77.
4.1.2.13. Synthesis procedure of 7-(diethylamino)-N-(6-((14-methyl -5-oxo- 5,7,8,13,13b,14- hexahydroindolo[2׳,3׳:3,4] pyrido[2,1-b]quinazolin-10-yl)amino)-6-oxohexyl)-2-oxo-2H- chromene-3-carboxamide (16)
To a solution of intermediate 15 (0.10 g, 0.27 mmol) in DMF, HBTU (0.11 g, 0.32 mmol) was added. Then TEA (0.043 mL, 0.32 mmol) was added under stirring and the reaction mixture was stirred for 0.5 h at room temperature. Subsequently, compound 12 (0.10 g, 0.32 mmol) was added. The reaction was stirred at room temperature for 3 h, followed by pouring the reaction mixture into ice/water (150 mL). Then, the precipitate was collected by filtration, washed with water (2×30 mL) and dried to give the crude product. The crude product was purified by silica column chromatography (DCM: MeOH=100:5, v/v) to afford compound 16 as a yellow solid (0.092 g, 51%). 1H NMR (DMSO-d6, 600 MHz) δ: 10.96 (s, 1 H), 9.70 (s, 1 H), 8.66 (s, 2 H), 7.81 (s, 2 H), 7.69 (d, J=9.38 Hz, 1 H), 7.49 (t, J=7.29 Hz, 1 H), 7.21—7.28 (m, 2 H), 7.05 (d, J=8.34 Hz, 1 H), 6.97 (t, J=7.29 Hz, 1 H), 6.82 (d, J=8.86 Hz, 1 H), 6.63 (s, 1 H), 6.12 (s, 1 H), 4.62—4.65 (m, 1 H), 3.49 (m, 4 H), 3.19—3.22 (m, 1 H), 2.90 (s, 4 H), 2.72—2.74 (m, 1 H), 2.31 (t, J=7.59 Hz, 2 H), 1.63—1.77 (m, 2 H), 1.56—1.59 (m, 2 H), 1.35—1.37 (m, 2 H), 1.25 (m, 6 H), 1.13—1.16 (m, 2 H). 13C NMR (DMSO-d6, 150 MHz) δ: 171.01, 164.67, 162.53, 162.27, 157.67, 152.87, 149.17, 148.29, 133.92, 133.57, 132.01, 131.80, 128.45, 126.22, 120.64, 119.60, 117.78, 116.16, 111.89, 110.59, 110.00, 109.29, 108.14, 96.34, 70.29, 44.79, 41.38, 39.26, 36.91, 31.75, 29.47, 26.67, 25.51, 22.56, 19.92, 14.42, 12.79. HR-MS (ESI) m/z: Calcd. for C39H42N6O5 [M+H]+ 675.3295, Found 675.3292.
4.2. Biological studies
4.2.1. In vitro cytotoxicity assay
The effect of compound 1 and its cytotoxicity towards cancer cells were determined by the CCK-8 method. HCT116 cells in the log phase of their growth cycle were plated in 96-well microtiter plates at a density of 8×103 cells/well and incubated in a humidified atmosphere with 5% CO2 at 37 °C for 24 h. Test compounds were double gradient diluted then added into triplicate wells, and 0.1% DMSO was used as the control. After incubating for 48 h, 10 μL of the CCK-8 solution was added to each well, and the plate was incubated for another 1—4 h. Subsequently, the absorbance (OD) of each well was measured at 450 nm with a WellscanMK-2 Microplate reader (Labsystems). The concentration causing 50% inhibition of cell growth (IC50) was determined by the logit method. All experiments were performed in triplicate.
4.2.2. Subcellular iocalization studies
HCT116 cells were seeded in a glass-bottom cell culture dish (NEST), pretreated with 0.5 μmol/L 9a for 4, 8, 12, and 24 h, and then washed twice with phosphate-buffered saline (PBS). The cells were then incubated with MitoTracker (YEASEN, MitoTracker Red CM-H2XRos, 50 nmol/L) and LysoTracker, (YEASEN, LysoTracker Red DND-99, 100 nmol/L) for another 0.5 h. After washing three times with cold PBS, the cells were fixed with paraformaldehyde and washed before they were imaged with a confocal laser scanning microscope (TCS-SP5). Fluorescence excitation and emission wavelengths for MitoTracker Red CM-H2XRos are 581 and 605 nm, and those for probe 9a are 408 and 456 nm, respectively.
4.2.3. JC-1 assay to determine MMP
The MMP was determined with the MMP Assay Kit JC-1(YEASEN). HCT116 cells were plated in 6-well cell culture plates (Corning) at a density of 2 × 105 cells/well and incubated in a humidified atmosphere with 5% CO2 at 37 °C for 24 h, and then, the cells were treated with various concentrations of compound 1 for 48 h. After that, the cells were washed twice with PBS, incubated with 10 µg/mL JC-1 dye for 30 min at 37 °C, and then washed three times with PBS. After incubation, samples were immediately analyzed using a confocal microscope. All the experiments were performed in triplicate.
4.2.4. Cell apoptosis detection assay
HCT116 cells (2 × 105 cells/well) were seeded in six-well plates and treated with compounds at the indicated concentration for 48 h. The cells were then harvested by trypsinization and washed twice with cold PBS. After centrifugation and removal of the supernatants, the cells were re-suspended in 400 μL of 1×binding buffer, combined with 5 μL of annexin V-FITC, and then incubated at room temperature for 15 min. After adding 10 μL of PI, the cells were incubated at room temperature for another 15 min in the dark. The stained cells were analyzed by flow cytometry (BD Accuri C6).
4.2.5. Characterization of autophagy
HCT116 cells (2 × 105 cells/well) were seeded in six-well plates and treated with the autophagy inhibitors Wortmannin (0.25 μmol/L), LY294002 (5 μmol/L), CQ (5 μmol/L) and 3MA (5 mmol/L) for 2 h before compound 1 was added. Then, the cells were incubated with compound 1 at a concentration of 0.2 μmol/L for 48 h. After incubation, the cells were harvested to detect the cell apoptosis according to the above method.
4.2.6. Western blotting analysis
HCT116 cells were incubated with various concentration of compound 1 and collected after 8 h. The cells were washed twice with ice cold PBS and incubated with lysis buffer. After the cells were lysed on ice for 20 min, the lysates were centrifuged at 12,000×g at 4 °C for 15 min. The protein concentration in the supernatant was determined using the BCA protein assay reagents. Equal amounts of protein (30 μg) were resolved using SDS-PAGE (8%−12% acrylamide gels) and transferred to polyvinylidene difluoride (PVDF) Hybond-P membrane. Membranes were blocked with BSA for 2 h at room temperature. Membranes were then incubated with primary antibodies against LC3-II, cleaved caspase-3, caspase-9, BAX, BCL-2, P53, Cytochrome C, and GAPDH with gentle rotation overnight at 4 °C. Membranes were next incubated with fluorescent secondary antibodies for 1 h and then analyzed using an Odyssey Infrared Imaging System.
4.2.7. Autophagy flux assessment
HCT116 cells were seeded in a glass-bottom cell culture dish (NEST) and transfected with adenovirus harboring mRFP-GFP-LC3 (HanBio, Shanghai, China) when the confluence reached to 50%–70%. After incubating with adenovirus for 6 h, cells were then incubated with fresh medium supplemented with 10% (v/v) FBS for another 48 h to ensure the high efficiency of transfection. After transfection, cells were cultured with 0.2 μmol/L of compound 1 for 8 h, and then were imaged with a confocal laser scanning microscope (TCS-SP5).
4.2.8. Transmission electron microscopy analysis
HCT116 cells (2×105 cells/well) were seeded in six-well plates and treated with or without compound 1 for 8 h. And then cells were harvested and fixed overnight at 4 °C in 2.5% glutaraldehyde in 0.1 mol/L PBS, and then post-fxed in 1% buffered osmium tetroxide for 2 h. Subsequently, specimens were examined under a transmission electron microscope (H-700; Hitachi, Tokyo, Japan).
4.2.9. Statistical analysis
All data are presented as mean ± standard deviation (SD). Student's t-test was used to compare the difference between the experimental groups and control groups. Statistical significance was defined as a P value: *P < 0.05, **P < 0.01, ***P < 0.001.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81725020), Shanghai Education Development Foundation and Shanghai Municipal Education Commission (Chenguang Program, Grant No. 15CG42).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.apsb.2018.08.003.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Schenone M., Dančik V., Wagner B.K., Clemons P.A. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuatrecasas P., Wilchek M., Anfinsen C.B. Selective enzyme purification by affinity chromatography. Proc Natl Acad Sci U S A. 1968;61:636–643. doi: 10.1073/pnas.61.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Titov D.V., Liu J.O. Identification and validation of protein targets of bioactive small molecules. Biorg Med Chem. 2012;20:1902–1909. doi: 10.1016/j.bmc.2011.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Z. The modification of natural products for medical use. Acta Pharm Sin B. 2017;7:119–136. doi: 10.1016/j.apsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Q., Liu S., Gao L., Sun S., Huan Y., Li C. Anti-diabetic effects and mechanisms of action of a Chinese herbal medicine preparation JQ-R in vitro and in diabetic KKAy mice. Acta Pharm Sin B. 2017;7:461–469. doi: 10.1016/j.apsb.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eder J., Sedrani R., Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat Rev Drug Discov. 2014;13:577–587. doi: 10.1038/nrd4336. [DOI] [PubMed] [Google Scholar]
- 7.Moffat J.G., Rudolph J., Bailey D. Phenotypic screening in cancer drug discovery—past, present and future. Nat Rev Drug Discov. 2014;13:588–602. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- 8.Robles O., Romo D. Chemo- and site-selective derivatizations of natural products enabling biological studies. Nat Prod Rep. 2014;31:318–334. doi: 10.1039/c3np70087a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu S., Hu H.Y. Fluorogen-activating proteins: beyond classical fluorescent proteins. Acta Pharm Sin B. 2018;8:339–348. doi: 10.1016/j.apsb.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deguire S.M., Earl D.C., Du Y., Crews B.A., Jacobs A.T., Ustione A. Fluorescent probes of the apoptolidins and their utility in cellular localization studies. Angew Chem. 2015;54:961–964. doi: 10.1002/anie.201408906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J., Chen J., Ma S., Liu Q., Huang L., Chen X. Recent developments in multimodality fluorescence imaging probes. Acta Pharm Sin B. 2018;8:320–338. doi: 10.1016/j.apsb.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegler S., Pries V., Hedberg C., Waldmann H. Target identification for small bioactive molecules: finding the needle in the haystack. Angew Chem Int Ed. 2013;52:2744–2792. doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]
- 13.DeGuire S.M., Earl D.C., Du Y., Crews B.A., Jacobs A.T., Ustione A. Fluorescent probes of the apoptolidins and their utility in cellular localization studies. Angew Chem. 2015;127:975–978. doi: 10.1002/anie.201408906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Böttcher T., Pitscheider M., Sieber S.A. Natural products and their biological targets: proteomic and metabolomic labeling strategies. Angew Chem Int Ed. 2010;49:2680–2698. doi: 10.1002/anie.200905352. [DOI] [PubMed] [Google Scholar]
- 15.Dong G., Sheng C., Wang S., Miao Z., Yao J., Zhang W. Selection of evodiamine as a novel topoisomerase I inhibitor by structure-based virtual screening and hit optimization of evodiamine derivatives as antitumor agents. J Med Chem. 2010;53:7521–7531. doi: 10.1021/jm100387d. [DOI] [PubMed] [Google Scholar]
- 16.Dong G., Wang S., Miao Z., Yao J., Zhang Y., Guo Z. New tricks for an old natural product: discovery of highly potent evodiamine derivatives as novel antitumor agents by systemic structure–activity relationship analysis and biological evaluations. J Med Chem. 2012;55:7593–7613. doi: 10.1021/jm300605m. [DOI] [PubMed] [Google Scholar]
- 17.Wang S., Fang K., Dong G., Chen S., Liu N., Miao Z. Scaffold diversity inspired by the natural product evodiamine: discovery of highly potent and multitargeting antitumor agents. J Med Chem. 2015;58:6678–6696. doi: 10.1021/acs.jmedchem.5b00910. [DOI] [PubMed] [Google Scholar]
- 18.Yun S.W., Kang N.Y., Park S.J., Ha H.H., Kim Y.K., Lee J.S. Diversity oriented fluorescence library approach (DOFLA) for live cell imaging probe development. Acc Chem Res. 2014;47:1277–1286. doi: 10.1021/ar400285f. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Lok C.N., Ko B.C., Shum T.Y., Wong M.K., Che C.M. Subcellular localization of a fluorescent artemisinin derivative to endoplasmic reticulum. Org Lett. 2010;12:1420–1423. doi: 10.1021/ol902890j. [DOI] [PubMed] [Google Scholar]
- 20.Wu J., Shen Q., Wang Y., Zhao D., Peng C., Li J.X. Fluorescent probes for subcellular localization during osteclast formation. ACS Med Chem Lett. 2014;5:911–914. doi: 10.1021/ml500181e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu S., Luo S., Yao H., Cai H., Miao X., Wu F. Probing the anticancer action of oridonin with fluorescent analogues: visualizing subcellular localization to mitochondria. J Med Chem. 2016;59:5022–5034. doi: 10.1021/acs.jmedchem.6b00408. [DOI] [PubMed] [Google Scholar]
- 22.Xu H., Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klionsky D.J. Autophagy revisited: a conversation with christian de Duve. Autophagy. 2008;4:740–743. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 24.Deter R.L., Baudhuin P., de Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nahimana A., Attinger A., Aubry D., Greaney P., Ireson C., Thougaard A.V. The NAD biosynthesis inhibitor APO866 has potent antitumor activity against hematologic malignancies. Blood. 2009;113:3276–3286. doi: 10.1182/blood-2008-08-173369. [DOI] [PubMed] [Google Scholar]
- 26.Green D.R. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2011. Means to an end: apoptosis and other cell death mechanisms. [Google Scholar]
- 27.Huang Z. The chemical biology of apoptosis: exploring protein—protein interactions and the life and death of cells with small molecules. Chem Biol. 2002;9:1059–1072. doi: 10.1016/s1074-5521(02)00247-8. [DOI] [PubMed] [Google Scholar]
- 28.Hikim A.P., Lue Y., Yamamoto C.M., Vera Y., Rodriguez S., Yen P.H. Key apoptotic pathways for heat-induced programmed germ cell death in the testis. Endocrinology. 2003;144:3167–3175. doi: 10.1210/en.2003-0175. [DOI] [PubMed] [Google Scholar]
- 29.Oltval Z.N., Milliman C.L., Korsmeyer S.J. BCL-2 heterodimerizes in vivo with a conserved homolog, BAX, that accelerates programed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 30.Moldoveanu T., Follis A.V., Kriwacki R.W., Green D.R. Many players in BCL-2 family affairs. Trends Biochem Sci. 2014;39:101–111. doi: 10.1016/j.tibs.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaux D.L., Cory S., Adams J.M. BCL-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 32.Debatin K.M. Chronic lymphocytic leukemia: keeping cell death at bay. Cell. 2007;129:853–855. doi: 10.1016/j.cell.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 33.Wong C.K., Chen Z., So K.L., Li D., Li P. Polycomb group protein RING1B is a direct substrate of caspases-3 and -9. Biochim Biophys Acta. 2007;1773:844–852. doi: 10.1016/j.bbamcr.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Bao Q., Lu W., Rabinowitz J.D., Shi Y. Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol Cell. 2007;25:181–192. doi: 10.1016/j.molcel.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Monian P., Jiang X. Clearing the final hurdles to mitochondrial apoptosis: regulation post cytochrome C release. Exp Oncol. 2012;34:185–191. [PubMed] [Google Scholar]
- 36.Tian H.Y., Li Z.X., Li H.Y., Wang H.J., Zhu X.W., Dou Z.H. Effects of 14 single herbs on the induction of caspase-3 in tumor cells: a brief review. Chin J Integr Med. 2013;19:636–640. doi: 10.1007/s11655-013-1539-y. [DOI] [PubMed] [Google Scholar]
- 37.Levy J.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claerhout S., Verschooten L., Van Kelst S., De Vos R., Proby C., Agostinis P. Concomitant inhibition of AKT and autophagy is required for efficient cisplatin-induced apoptosis of metastatic skin carcinoma. Int J Cancer. 2010;127:2790–2803. doi: 10.1002/ijc.25300. [DOI] [PubMed] [Google Scholar]
- 39.Wang J., Wu G. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289:17163–17173. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.