Abstract
Eyes absent homolog 2 (EYA2), a transcriptional activator, is pivotal for organ development, but aberrant regulation of EYA2 has been reported in multiple human tumors. However, the role of EYA2 in breast cancer is still lack of full understanding. To explore the biological significance of EYA2 in breast cancer, we conducted data analysis on public breast cancer datasets, and performed immunohistochemistry (IHC) analysis, colony-forming unit assays, EdU assay, western blotting, and immunofluorescence (IF). Meta-analysis showed that EYA2 mRNA expression was correlated with tumor grade, the status of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). IHC analysis displayed that EYA2 protein abundance was inversely associated with the status of ER and PR, and enriched in triple-negative breast cancer in comparison with luminal-type tumors. Additionally, correlation analysis reflected that EYA2 mRNA was negatively correlated with luminal markers, and positively associated with markers of basal cells, epithelial-mesenchymal transition and cancer stem cells. Clone-forming assay and EdU experiment showed that EYA2 overexpression enhanced proliferation of breast cancer cells. Results from western blotting and IF displayed that overexpression of EYA2 up-regulated the protein abundance of proliferation markers. Importantly, survival analysis indicated that higher EYA2 mRNA level predicted worse overall survival, relapse-free survival and metastasis-free survival among whole enrolled breast cancer patients. Collectively, EYA2 was closely correlated with clinico-pathological characteristics, and served as a proliferation stimulator for breast cancer cells and an unfavorable prognostic element for breast cancer patients, suggesting that EYA2 is involved in the progression of breast carcinoma.
Keywords: breast cancer, EYA2, tumor grade, molecular subtypes, proliferation, epithelial-mesenchymal transition, cancer stem cells, prognosis
Introduction
Breast cancer is the leading cancer type in women and poses a major threat to public health worldwide (1). During the past decades, many efforts have been exerted to better management of this tumor type (2–7). However, current understanding of molecular mechanisms for this highly heterogenous disease is not fully clear (8). Identification of key molecules that promote and maintain malignant conversion facilitates comprehensive understanding of breast tumor biology, ultimately contributing to novel potential targets for drugs and better management of breast carcinoma (3, 9, 10).
Retinal determination gene network (RDGN), mainly including dachshund (DACH), sine oculis (SIX) and eyes absent (EYA), governs organ development (11). EYA family members, including EYA1, EYA2, EYA3, and EYA4, contain a C-terminal 271 amino-acid region for the interaction with other proteins and a N-terminal domain responsible for the innate immune response with inherent threonine phosphorylation activity (12–14). Recently, the dysregulation of EYA2 has been reported to be involved in several human cancers (15–18). For instance, EYA2 expression was enhanced in multiple tumors in comparison with corresponding normal tissues (16, 17). EYA2 can promote tumor growth in diverse tumor types (16–19). In human astrocytoma, EYA2 promoted cell cycle progression of tumor cells via the up-regulation of cyclin D1 and cyclin E (17). Knockdown of EYA2 by siRNA reduced the proliferation through cell cycle G1 block and enhanced the apoptosis of lung adenocarcinoma cells (20). On the contrary, one previous research demonstrated that EYA2 overexpression was an unfavorable molecule for tumor growth of pancreatic adenocarcinoma in orthotopic models (21). In addition, EYA2 can also contribute to tumor invasion and metastasis for some cancer types, including breast cancer (18), lung adenocarcinoma (22), and astrocytoma (17). Several mechanisms might underlie the role of EYA2 in tumor invasion, including the activation of ERK signaling (17) and the promotion of epithelial-mesenchymal transition (EMT) (18). The association between EYA2 and clinical outcomes is controversial. High EYA2 level has been demonstrated to be a negative element for prognosis in lung cancer (16), while EYA2 predicted better clinical outcomes in colorectal cancer (15) and pancreatic cancer (21).
A recent study has demonstrated that microRNA-338-3p/EYA2 axis led to epidermal growth factor receptor (EGFR)-induced tumor growth and lung metastasis (18). However, the role of EYA2 in breast cancer remains to be further explored. Herein, we performed a meta-analysis of public available breast cancer gene expression omnibus (GEO) datasets to further detect the differential expression of EYA2 in normal vs. breast tumors, and to explore the association between EYA2 and tumor differentiation, the status of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) as well as molecular subtypes at mRNA level. Immunohistochemistry (IHC) analysis on tissue microarray (TMA) was conducted to explore the correlation between EYA2 and the status of ER and PR as well as molecular subtypes at protein level. Furthermore, correlation analysis of GSE25066 was performed to explore the association between EYA2 and markers of luminal, triple-negative breast cancer (TNBC), EMT, cancer stem cells (CSCs) as well as the cell cycle-related gene. Besides, we conducted colony-forming unit assays, EdU experiments, western blotting, and immunofluorescence (IF) to evaluate the role of EYA2 in tumor proliferation and explore EYA2 regulated genes Finally, we employed the Kaplan-Meier Plotter platform to explore the role of EYA2 in the prognosis of breast cancer patients.
Materials and Methods
IHC Staining and Quantification Evaluation
One commercially available TMA slide (HBre-Duc140Sur-01, Shanghai Outdo Biotech Co., Ltd.) was purchased for IHC analysis, which contained histologically confirmed breast cancer tissues with clinico-pathological information, such as tumor grade, clinical stage and the status of ER, PR, and HER2 in IHC (Table 1). Breast tumors with positive status of ER or PR belong to luminal-type, and tumors that do not express ER, PR, and HER2 are TNBC. Due to tissue shedding of 15 cases, the number of actually available tissue points was 125. To evaluate the protein abundance of EYA2 in ER– vs. ER+, PR– vs. PR+, and luminal-type vs. TNBC tissues as well as the prognostic value among breast cancer population, IHC analysis was conducted with a standard protocol described previously (23). The specific primary antibody against EYA2 (ab95875, Abcam) was utilized for IHC at a dilution of 1:100.
Table 1.
Clinico-pathological information of patients in tissue microarray (HBre-Duc140Sur-01).
| Variables | Patient number |
|---|---|
| GRADE | |
| 1 | 9 |
| 1–2 | 21 |
| 2 | 87 |
| 2–3 | 6 |
| 3 | 1 |
| TNM | |
| I | 8 |
| II | 71 |
| III | 44 |
| ER | |
| ER– | 42 |
| ER+ | 76 |
| PR | |
| PR– | 49 |
| PR+ | 68 |
| HER2 | |
| HER2– | 81 |
| HER2+ | 13 |
| MOLECULAR SUBTYPES | |
| Luminal | 81 |
| HER2-enriched | 3 |
| TNBC | 23 |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
Two experienced pathologists performed IHC scoring independently with no prior knowledge of the clinico-pathological information. The multiplication of intensity and proportion of positive-staining tumor cells was exploited to quantify the protein levels of EYA2 according to a standard protocol as described previously (24).
Meta-Analysis
We carried out a meta-analysis of 14 relevant GEO breast cancer databases for the mRNA expression of EYA2 available in ArrayExpress (Table 2) (25–38). Cutoff value for EYA2 was median expression. The STATA software package (version 12.0) (Stata Corp LP, College Station, TX, USA) was employed to perform the meta-analysis. Odds ratio (OR) and 95% confidence intervals (95% CIs) were used to evaluate the association between EYA2 mRNA and clinico-pathological factors. Overall survival (OS), relapse-free survival (RFS) and metastasis-free survival (MFS) were assessed by hazard ratio (HR) and 95% CIs. Heterogeneity of publication was evaluated by means of the inconsistency index I2.
Table 2.
A list of enrolled GEO datasets in meta-analysis.
| GEO datasets | First author | Year | Number of patients | References |
|---|---|---|---|---|
| GSE10885 | Hennessy BT | 2009 | 89 | (25) |
| GSE17907 | Sircoulomb F | 2010 | 51 | (26) |
| GSE20711 | Dedeurwaerder S | 2011 | 88 | (27) |
| GSE39004 | Terunuma A | 2014 | 61 | (28) |
| GSE25066 | Hatzis C | 2011 | 508 | (29) |
| GSE45255 | Nagalla S | 2013 | 139 | (30) |
| GSE6532 | Loi S | 2010 | 327 | (31) |
| GSE7390 | Desmedt C | 2007 | 198 | (32) |
| GSE58644 | Tofigh A | 2014 | 321 | (33) |
| GSE16446 | Desmedt C | 2011 | 120 | (34) |
| GSE2603 | Minn AJ | 2005 | 99 | (35) |
| GSE2034 | Wang Y | 2005 | 286 | (36) |
| GSE1456 | Pawitan Y | 2005 | 159 | (37) |
| GSE20685 | Kao KJ | 2011 | 327 | (38) |
GEO, gene expression omnibus.
Correlation Analysis of Gene Expression Data
GEO dataset GSE25066, containing 508 breast carcinoma patients, was analyzed to evaluate the correlation between EYA2 mRNA level and the mRNA expression of ESR1, PGR, forkhead box A1 (FOXA1), keratin 5 (KRT5), KRT6B, EGFR, SNAI2, Y-box binding protein 1 (YBX1), kruppel like factor 5 (KLF5), sex determining region Y-box 10 (SOX10), CCNE1, and DACH1.
Cell Culture and Establishment of EYA2 Stable Cell Lines
The breast cancer cell lines (MCF-7 and MDA-MB-231) were cultured in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Life Technologies, Inc.) under the condition of 37°C and 5% CO2 in a humidified incubator. Retrovirus expression vector for EYA2 (MSCV-EYA2, #49265) was purchased from Addgene. Human embryonic kidney 293T cells were transfected with the combination of expression vector or control vector with human package plasmids by Lipofectamine™ 2000 (Invitrogen, Carlsbad CA, USA). After transfection, the viral supernatant was harvested and filtered to infect MCF-7 and MDA-MB-231 cells for three times as described previously (39). EYA2 stable cells were selected by green fluorescent protein.
Western Blotting
Protein extraction from MCF-7 and MDA-MB-231 cells and western blotting were conducted according to a standard protocol previously described (24). The primary antibodies used were as following: EYA2 (ab95875, Abcam), YBX1 (sc-101198, Santa Cruz), EGFR (sc-03, Santa Cruz), cyclin E (sc-25303, Santa Cruz), PCNA (sc-7907, Santa Cruz), and β-actin (sc-47778, Santa Cruz).
Cellular IF
IF staining was performed based on published methods (39, 40). Primary antibodies were used at 1:150 dilution as following: EYA2 (ab95875, Abcam), YBX1 (sc-101198, Santa Cruz), EGFR (sc-03, Santa Cruz) and PCNA (sc-7907, Santa Cruz). The goat anti-mouse and the goat anti-rabbit secondary antibodies (Alexa Fluor-568) were both used at 1:300. Cell nuclei were stained with Hoechst 33342 at the dilution of 1:1,000.
Colony-Forming Unit Assays
The colony-forming assay was performed as previously described (41). Breast cancer cell lines (MCF-7 and MDA-MB-231) with EYA2 overexpression or vector control were seeded in 3.5 cm dishes (3 × 103 cells). Culture medium was changed every three days. At day 12, cells were fixed with 4% paraformaldehyde for 20 min and stained with 0.5% crystal violet for visualization.
EdU Assay
Cell-LightTM EdU Apollo567 In Vitro Kit (C10310-1, RIBOBIO, Guangzhou) was purchased for EdU experiment. Firstly, cells were seeded in 24-well plates at a density of 3 × 103 per well and incubated for 2 days. Secondly, 250 μl of EdU medium was added to each well and incubated for 2.5 h. Next, cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 20 min and subsequently added glycine solution (2 mg/ml) to neutralize aldehyde group. After increasing cell membrane permeability with 0.5% TritonX-100 in PBS, Apollo dyeing reaction solution was added and incubated in dark place at room temperature for 30 min. Then, dye nucleus with Hoechst 33342 in dark place at room temperature for 30 min. Finally, fluorescence microscope was employed for observation and counting.
Statistical Analysis
The Student's t-test was applied to evaluate the differences in groups. Correlation analysis was performed using Statistical Product and Service Solutions (SPSS) 20 statistical software (SPSS Inc., Chicago, IL, USA). A two-tailed p-value < 0.05 was considered statistically significant.
Results
EYA2 mRNA Expression in Normal Breast and Breast Tumors
In order to evaluate EYA2 mRNA level in normal breast tissues vs. malignant tissues, grade 3 vs. grade 1–2 tumors, and HER2– vs. HER2+ tumors, we conducted a comprehensive meta-analysis of 14 GSE datasets. The results showed that EYA2 mRNA expression was remarkably lower in cancerous tissues than in non-cancerous tissues [OR: 0.21 (0.10–0.43), I2 = 0.0%; Figure 1A]. EYA2 mRNA level was significantly higher in high-grade cancer tissues [OR: 1.48 (1.22–1.80), I2 = 38.0%; Figure 1B) and HER2+ tumors [OR: 1.76 (1.28–2.42), I2 = 11.0%; Figure 1C] in comparison with low-grade tumor tissues and HER2– tumor tissues, respectively.
Figure 1.
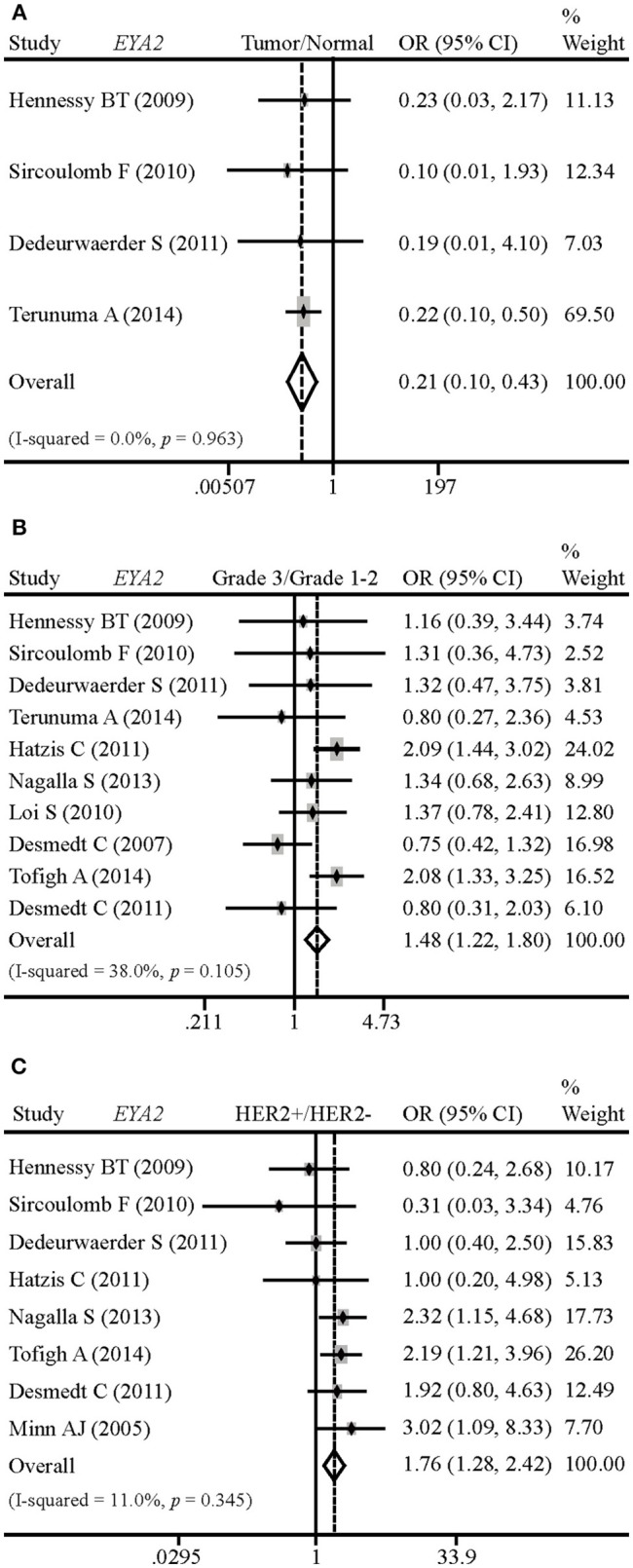
Differences of EYA2 mRNA in breast tumors vs. normal breast, and the correlation between EYA2 mRNA and tumor grade and HER2 status. EYA2 mRNA level was remarkably lower in cancerous tissues than non-cancerous tissues (A). EYA2 mRNA expression was significantly higher in high-grade cancer tissues (B) and HER2+ tumors (C) in comparison with low-grade and HER2− tumors, respectively.
Correlation Between EYA2 Expression and the Status of ER and PR
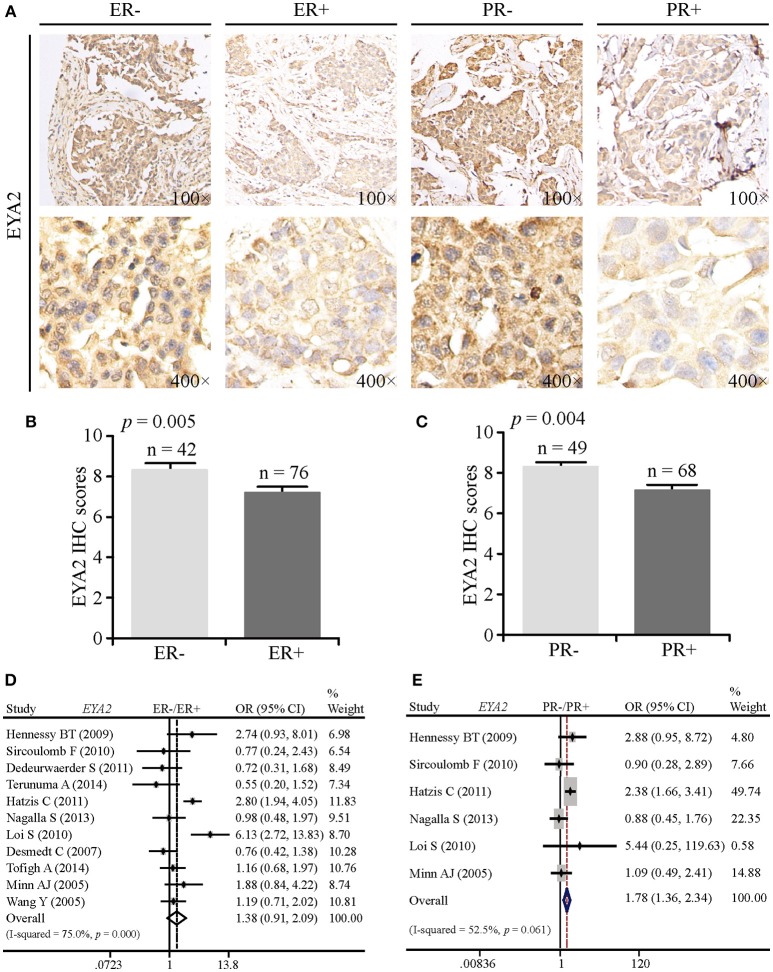
To compare EYA2 protein abundance in ER– vs. ER+ and PR– vs. PR+, we analyzed a TMA containing 125 informative cancer tissue points by IHC. EYA2 was majorly detected in the cytoplasm of breast cancer cells. Representative images of IHC staining were shown in Figure 2A. Next, IHC scores by using semi-quantitative criteria were also examined. The results indicated that protein abundance of EYA2 was significantly higher in ER– (p = 0.005) (Figure 2B) or PR– (p = 0.004) (Figure 2C) in comparison with ER+ or PR+ cancer tissues, respectively.
Figure 2.
The correlation between EYA2 expression and the status of ER and PR. (A) Representative images of IHC staining for ER− vs. ER+ and PR− vs. PR+ were shown. Statistical analysis of IHC scores indicated that protein abundance of EYA2 was significantly higher in ER− (B) or PR− (C) in comparison with ER+ or PR+ cancer tissues, respectively. The results were shown as mean + SEM. (D) There was no significant difference in the EYA2 mRNA level between ER− and ER+ cancer tissues. (E) EYA2 mRNA expression was significantly enhanced in PR− tumors in comparison with PR+ breast cancer.
To assess whether the mRNA expression of EYA2 is consistent with the protein expression, meta-analysis was performed. There was an increasing tendency of EYA2 mRNA expression in ER– tumors in comparison with ER+ tumors. However, the difference was not statistically significant [OR: 1.38 (0.91–2.09), I2 = 75.0%; Figure 2D]. The mRNA expression of EYA2 was higher in PR– tumors than PR+ tumors [OR: 1.78 (1.36–2.34), I2 = 52.5%; Figure 2E].
EYA2 Is Associated With Molecular Subtypes of Breast Cancer
To elucidate whether there was any association between EYA2 protein abundance and molecular subtypes, we conducted IHC analysis on the TMA. Representative images of IHC staining for luminal-type and TNBC tissues were, respectively, showed in Figure 3A. Statistical analysis on IHC scores revealed that EYA2 protein level was significantly enhanced in TNBC tissues in comparison with luminal-type tissues (p = 0.033) (Figure 3B).
Figure 3.
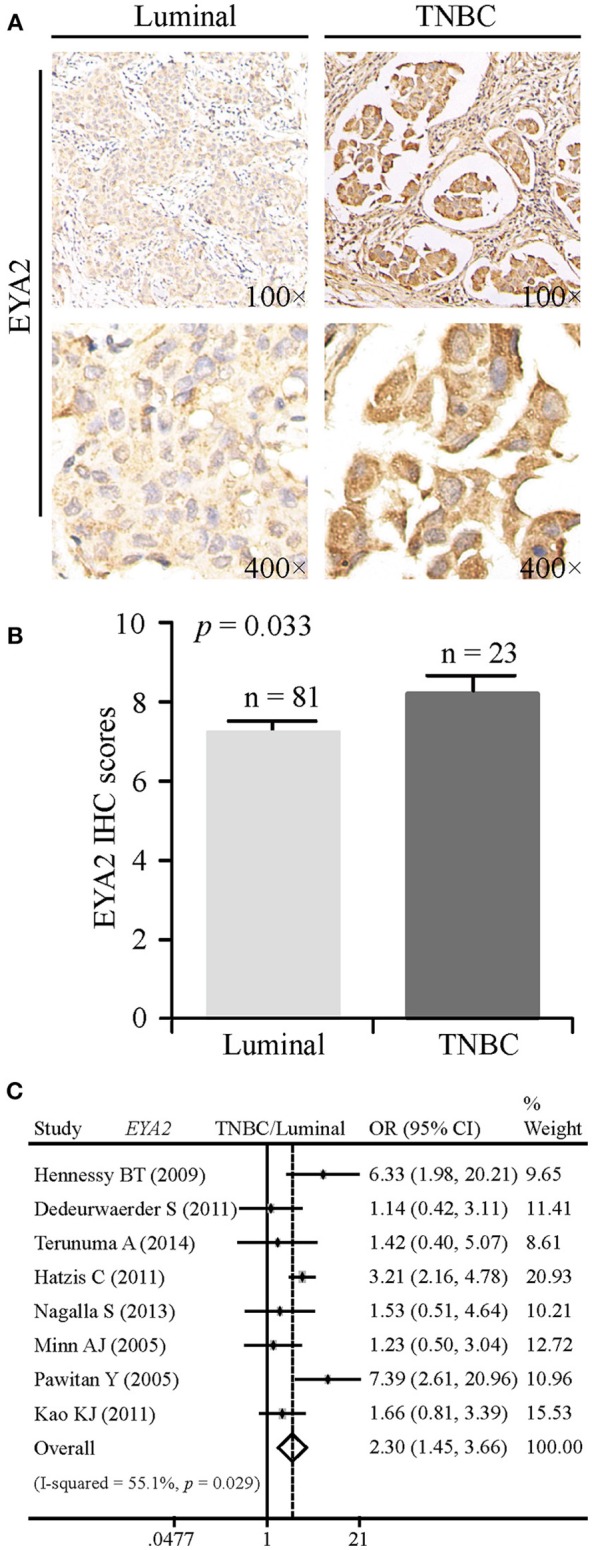
EYA2 is associated with molecular subtypes of breast cancer. (A) Representative images of IHC staining for luminal-type and TNBC breast cancer tissues were showed. (B) IHC scores revealed that higher level of EYA2 protein was significantly enhanced in TNBC tissues in comparison with luminal-type tissues, which was displayed as mean + SEM. (C) The mRNA level of EYA2 in TNBC was much higher than that in luminal-type.
In order to explore whether the mRNA level of EYA2 is consistent with the protein expression in distinct molecular subtypes, meta-analysis was performed. The mRNA level of EYA2 in TNBC was much higher than in luminal-type [OR: 2.30 (1.45–3.66), I2 = 55.1%; Figure 3C]. We further evaluated the EYA2 mRNA expression in the Cancer Genome Atlas breast cancer dataset downloaded from UCSC Xena. We found that EYA2 expression was significantly higher in basal-like tumors than luminal-type cancer tissues (p < 0.0001) (Supplementary Figure 1A), which was consistent with the results from our meta-analysis. Collectively, we drew a conclusion that EYA2 was enriched in TNBC tissues in comparison with luminal-type tissues at both protein and mRNA levels.
The Correlations Between EYA2 mRNA and the Markers of Luminal, TNBC, Mesenchymal, CSCs as Well as Cancer-Related Genes
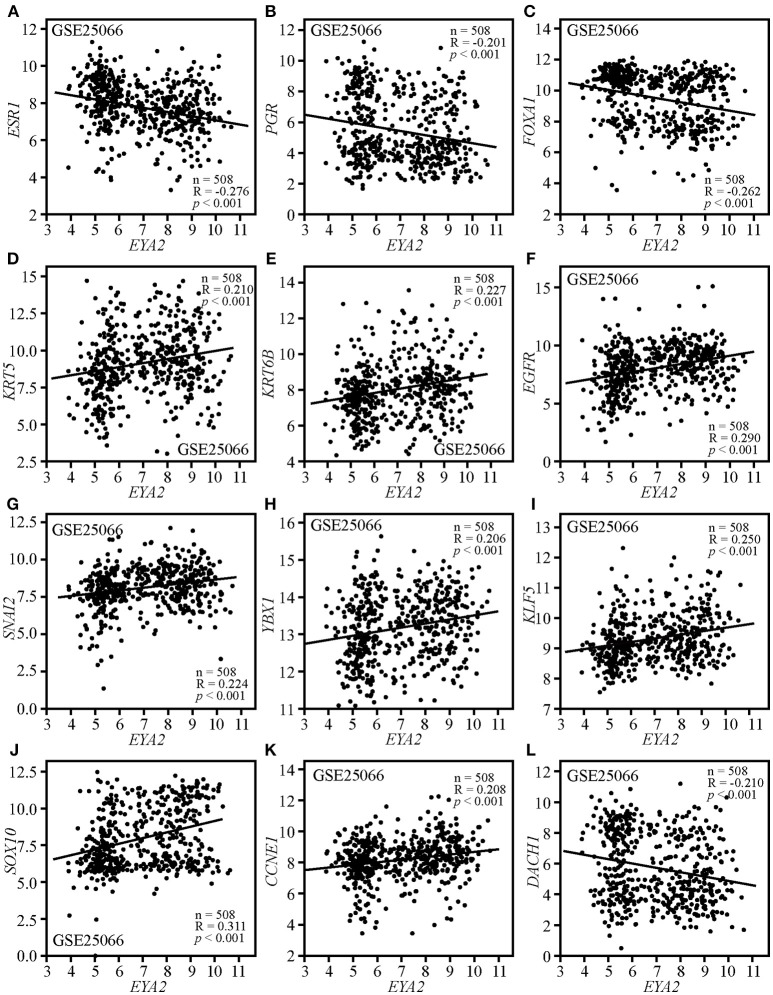
Correlation analysis was conducted on GSE25066, containing 508 breast cancer patients with distinct molecular subtypes. The results showed that EYA2 mRNA level was negatively associated with the mRNA expression of ESR1 (R = −0.276, p < 0.001; Figure 4A), PGR (R = −0.201, p < 0.001; Figure 4B) and FOXA1 (R = −0.262, p < 0.001; Figure 4C), while it was positively correlated with KRT5 (R = 0.210, p < 0.001; Figure 4D), KRT6B (R = 0.227, p < 0.001; Figure 4E), and EGFR (R = 0.290, p < 0.001; Figure 4F). In addition, the mRNA expression of EYA2 was positively associated with SNAI2 (R = 0.224, p < 0.001; Figure 4G), YBX1 (R = 0.206, p < 0.001; Figure 4H), KLF5 (R = 0.250, p < 0.001; Figure 4I), SOX10 (R = 0.311, p < 0.001; Figure 4J), and CCNE1 (R = 0.208, p < 0.001; Figure 4K), while EYA2 mRNA was inversely correlated with RDGN gene DACH1 (R = −0.210, p < 0.001; Figure 4L).
Figure 4.
The correlations between EYA2 with the markers of luminal, TNBC, mesenchymal, CSCs, proliferation as well as DACH1. EYA2 mRNA level was negatively associated with the mRNA expression of ESR1 (A), PGR (B), FOXA1 (C), and DACH1 (L), while it was positively correlated with KRT5 (D), KRT6B (E), EGFR (F), SNAI2 (G), YBX1 (H), KLF5 (I), SOX10 (J), and CCNE1 (K).
EYA2 Promotes the Proliferation of Breast Cancer Cells With the Regulation of PCNA, EGFR, and YBX1
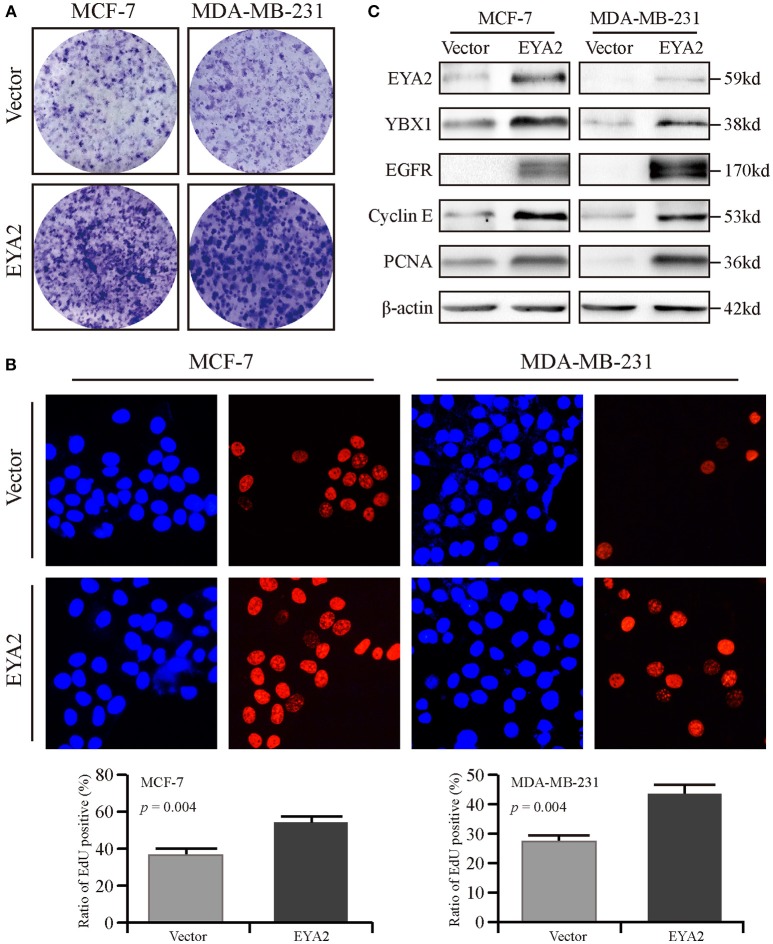
We also assessed the mRNA level of EYA2 in distinct breast cancer cell types (42), showing that there was no remarkable difference of EYA2 mRNA between luminal-type and basal-like cell lines (p = 0.666) (Supplementary Figure 1B). Therefore, two representative breast cancer cell lines (MCF-7 and MDA-MB-231) were selected to be transfected with EYA2 or empty vectors. Colony-forming unit assays showed that both MCF-7 and MDA-MB-231 with EYA2 overexpression formed more clones with the same number of initiating cells in comparison with the controls (Figure 5A). EdU proliferation assay displayed that the ratio of proliferative cells was much higher among EYA2-overexpressing cancer cells than empty vector controls for both MCF-7 (p = 0.004) and MDA-MB-231 (p = 0.004) (Figure 5B). Western blotting showed that EYA2 overexpression induced the up-regulation of YBX1, EGFR, cyclin E, and PCNA in both these two breast cancer cell lines (Figure 5C). Cellular IF assay also showed that EYA2 overexpression (Figure 6A) enhanced the protein abundance of YBX1 (Figure 6B), EGFR (Figure 6C), and PCNA (Figure 6D) in both breast cancer cell lines.
Figure 5.
EYA2 promotes the proliferation of breast cancer cells. (A) Colony-forming unit assays showed that both MCF-7 and MDA-MB-231 with EYA2 overexpression formed more clones with the same number of initiating tumor cells than the controls. (B) EdU cell proliferation assay displayed that the ratio of proliferative cells is much higher among EYA2-overexpressing cancer cells than empty vector controls for both MCF-7 and MDA-MB-231. (C) Western blotting showed that EYA2 overexpression contributed to up-regulation of the cancer stem cell marker YBX1, proliferative markers of EGFR, cyclin E, and PCNA in both MCF-7 and MDA-MB-231 at protein level.
Figure 6.
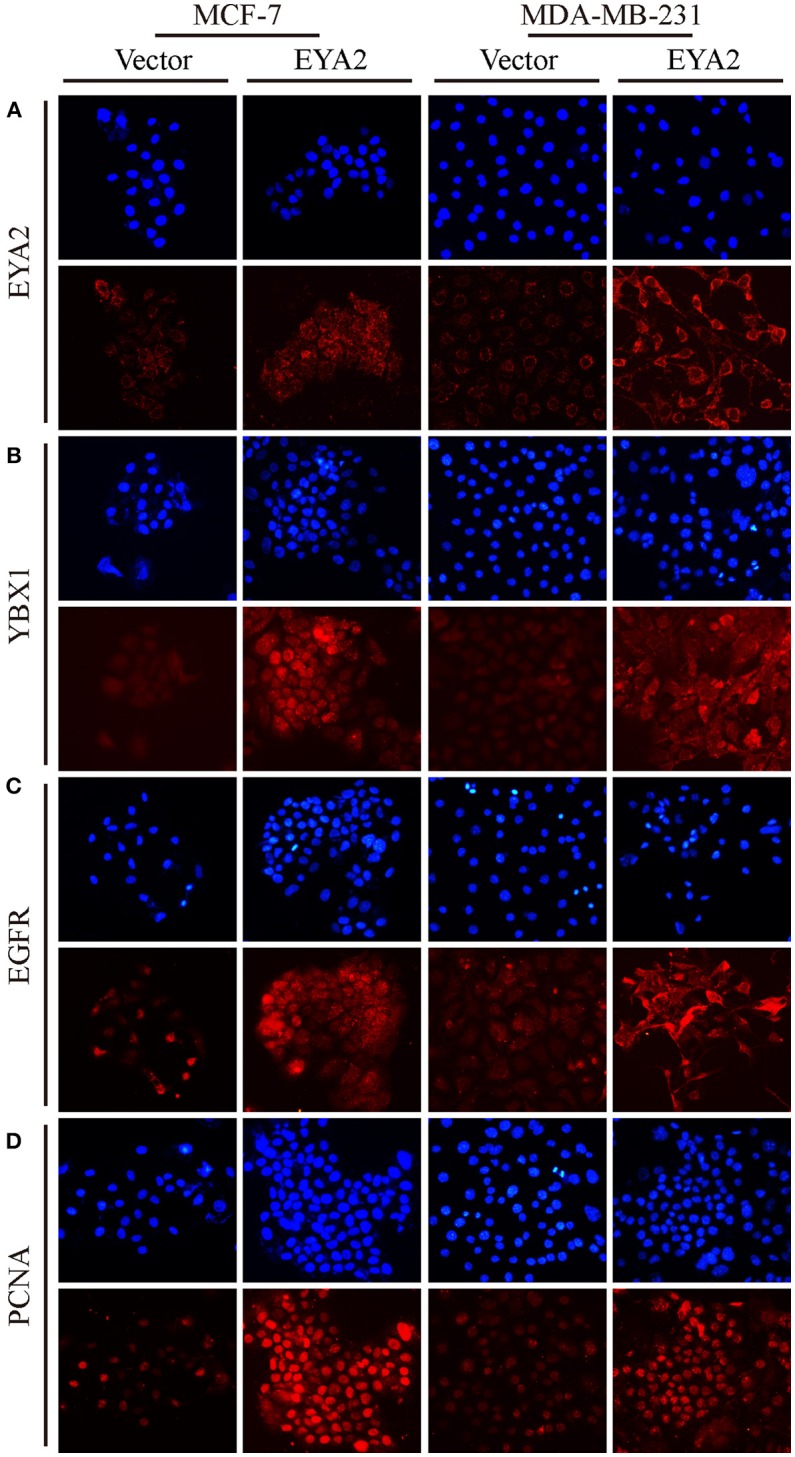
EYA2 over-expression up-regulates YBX1, EGFR, and PCNA. Cellular IF assay showed that EYA2 over-expression (A) enhanced the protein abundance of YBX1 (B), EGFR (C) and PCNA (D) in both MCF-7 and MDA-MB-231 cells.
High EYA2 mRNA Predicted Poor Prognosis of Breast Cancer
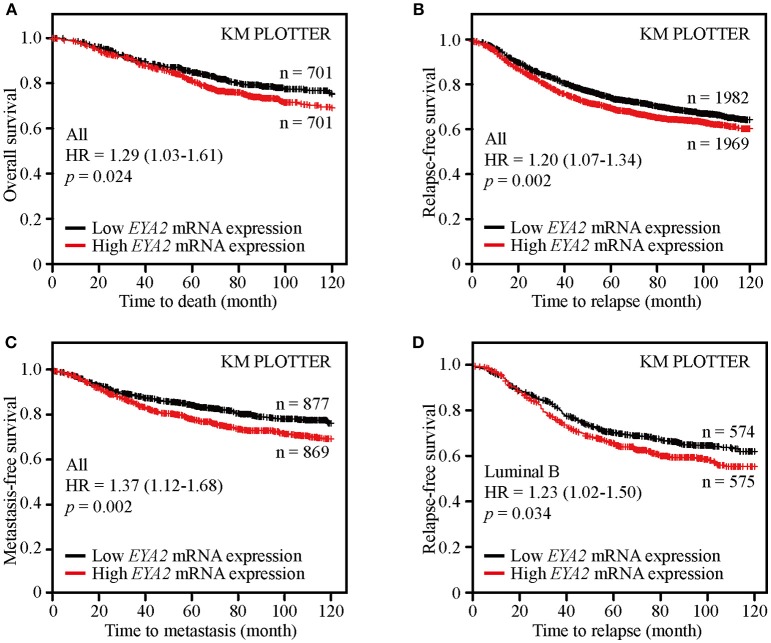
Survival analysis of public available breast cancer datasets was conducted using the Kaplan-Meier Plotter platform to examine the clinical significance of EYA2 in breast cancer. The results showed that patients with higher EYA2 mRNA expression had worse OS [HR = 1.29 (1.03–1.61), p = 0.024; Figure 7A], RFS [HR = 1.20 (1.07–1.34), p = 0.002; Figure 7B] and MFS [HR = 1.37 (1.12–1.68), p = 0.002] (Figure 7C) among whole breast cancer population. Further subgroup analysis showed that higher EYA2 mRNA level was correlated with worse RFS among luminal B subgroup [HR = 1.23 (1.02–1.50), p = 0.034; Figure 7D]. However, there was no statistically significant association between EYA2 mRNA and clinical outcomes in basal-like breast cancer patients, including OS [HR = 1.49 (0.91–2.45), p = 0.11], RFS [HR = 1.04 (0.81–1.34), p = 0.75], and MFS [HR = 1.52 (0.91–2.55), p = 0.11].
Figure 7.
EYA2 was an unfavorable prognostic element for breast cancer patients. KM plotter analysis showed that higher EYA2 mRNA expression was correlated with worse OS (A), RFS (B) and MFS (C) at whole level, and patients with lower EYA2 mRNA level enjoyed longer time free from tumor relapse among luminal B subgroup (D).
Discussion
EYA family was firstly identified as a key regulator for proper eye development in Drosophila (11). EYA2 has been implied in tumorigenesis and progression of some cancer types (14). Our results indicated that EYA2 was closely associated with tumor grade and molecular subtypes of breast cancer. Cellular experiments revealed that EYA2 promoted proliferation of breast cancer cell lines. Survival analysis based on the public database indicated that EYA2 was an unfavorable prognostic element. Surprisingly, our analysis of GEO datasets showed that EYA2 mRNA was dramatically lower in breast cancer tissues than normal breast. However, previous study indicated that EYA2 protein abundance increased in breast cancer tissues (43). This inverse tendency of EYA2 at mRNA and protein levels may arise from the complex regulatory processes from mRNA to protein.
Our results showed that EYA2 mRNA was higher in grade 3 breast tumors than grade 1–2 cancer, indicating that EYA2 was correlated with poor-differentiation in breast carcinoma. FOXA1 is a marker for luminal epithelium and plays pivotal roles in mammary duct formation (44). Our correlation analysis displayed that EYA2 was inversely associated with FOXA1, which further supported the correlation between high EYA2 expression and poor tumor differentiation.ER, PR and HER2 are critical pathological markers in breast cancer. According to the status of ER, PR, and HER2, this highly heterogeneous disease can be roughly classified into three major molecular subtypes, including luminal-type, HER2-enriched, and TNBC. Among these three major subtypes, luminal type accounts for the most part of breast cancer population with relatively better prognosis, while TNBC group show more progressively malignant manifestation with worse clinical outcomes. Our results displayed that EYA2 expression was higher in hormone receptor (HR)-negativebreast cancer tissues in comparison with HR-positive cancerous tissues, while EYA2 mRNA was positively associated with HER2 expression. In addition, EYA2 expression was remarkably enhanced in TNBC in comparison with luminal-type. In accordance, correlation analysis showed that EYA2 mRNA was inversely correlated with the mRNA levels of luminal markers ESR1, PGR and FOXA1, and positively associated with TNBC markers KRT5, KRT6B, and EGFR. In consistence, it was reported that EGFR positively regulated EYA2 through the regulation of microRNA-338-3P to promote breast tumor growth and metastasis (18).
EMT is a pivotal process for tumor invasion and metastasis. Our results indicated that EYA2 was positively correlated with the mesenchymal marker SNAI2 at mRNA level. Liang et al.'s work demonstrated that EYA2 promoted EMT in lung cancer (18). In pancreatic adenocarcinoma, stable overexpression of EYA2 up-regulated transforming growth factor-β (TGF-β) signaling which is an important inducer of EMT (21). In agreement with our finding, knockdown of EYA2 antagonized the SIX1-induced TGF-β signaling, and partially restored epithelial properties with a decrease of the mesenchymal marker fibronectin in breast cancer MCF-7 cells (45).
CSCs are inherently endowed with potent self-renewal capacity, and contribute to tumor initiation and progression. In our study, we showed that EYA2 was associated with CSCs markers YBX1, KLF5, and SOX10 in breast tumor tissues, and EYA2 overexpression up-regulated YBX1 in breast cancer cell lines. In pancreatic cancer, EYA2 overexpression enhanced the level of stem cell marker CD133 (21). Silencing of EYA2 led to a decrease of cells with CD44+ and CD24– among MCF-7 cells with exogenous overexpression of SIX1, indicating that EYA2 was required for SIX1 in the enhancement of CSCs features (45).
Cancer cells are characterized with unlimited proliferation. Results of EdU cell proliferation assay indicated that EYA2 promoted proliferation of tumor cells. EYA2 mRNA was positively associated with the proliferative marker CCNE1, and EYA2 overexpression enhanced the expression of EGFR and PCNA. Previous studies elaborated that EYA2 drove the proliferation in multiple cancer types, including breast cancer (43), lung cancer (16, 20), and astrocytoma (17). In lung cancer, EYA2 promoted tumor cell proliferation through microRNA-93-mediated inhibition of phosphatase and tension homolog (16). As a downstream target of microRNA-30a, EYA2 could boost the proliferation of breast cancer cells through driving G1/S cell cycle progression with up-regulation of cell cycle-related proteins cyclin A, cyclin D1, and cyclin E (43).
DACH and EYA are the key members of RDGN. There are feedback regulations between DACH and EYA in both physiological and pathological situations (11). DACH1 was reported to be an anti-tumor protein in breast cancer (46), while EYA2 served as a tumor-driving molecule in breast carcinoma (18). Our results indicated that EYA2 was inversely associated with DACH1 in breast cancer. The imbalance of DACH1 and EYA2 may contribute to tumor initiation and development.
In some cancers, EYA2 acted as a prognostic predictor (15, 19, 21, 45). Although EYA2 cannot serve as an independent prognostic biomarker, high EYA2 expression was correlated with poor prognosis for pancreatic cancer patients (21). Among patients with advanced ovarian cancer, higher EYA2 level was correlated with worse OS (19). In agreement with our analysis, Farabaugh et al. found that higher expression of EYA2 was associated with worse RFS, MFS, and disease-specific survival (DSS) among 295 patients with invasive breast cancer (45), and high expression of both SIX1 and EYA2 represent the type with the worst RFS, MFS, and DSS in comparison with another three types including SIX1low/EYA2low (with the best prognosis), SIX1high/EYA2low and SIX1low/EYA2high (45).
In conclusion, EYA2 was significantly correlated with clinico-pathological features of breast cancer, including tumor differentiation and the status of ER, PR, and HER2, and it was enriched in TNBC tumors. Furthermore, EYA2 mRNA was positively associated with markers of TNBC, EMT, and CSCs. Besides, ectopic expression of EYA2 promoted proliferation of breast cancer cells accompanied with the up-regulation of EGFR, cyclin E, and PCNA. Importantly, Kaplan-Meier analysis of public datasets showed that higher EYA2 mRNA level predicted worse prognosis among breast cancer population. Taken together, EYA2 promoted malignant behavior of breast cancer.
Author Contributions
HX performed experiments and data analysis, drafted the manuscript, and prepared the figures. MY, YJ, and WZ carried out immunohistochemistry analysis. KW designed experiments and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- RDGN
retinal determination gene network
- DACH
dachshund
- SIX
sine oculis
- EYA
eyes absent
- EMT
epithelial-mesenchymal transition
- EGFR
epidermal growth factor receptor
- GEO
gene expression omnibus
- ER
estrogen receptor
- PR
progesterone receptor
- HER2
human epidermal growth factor receptor 2
- IHC
immunohistochemistry
- TMA
tissue microarray
- TNBC
triple-negative breast cancer
- CSCs
cancer stem cells
- IF
immunofluorescence
- OR
odds ratio
- 95% CIs
95% confidence intervals
- OS
overall survival
- RFS
relapse-free survival
- MFS
metastasis-free survival
- HR
hazard ratio
- FOXA1
forkhead box A1
- KRT5
keratin 5
- YBX1
Y-box binding protein 1
- KLF5
kruppel like factor 5
- SOX10
sex determining region Y-box 10
- PCNA
proliferating cell nuclear antigen
- PBS
phosphate buffered saline
- SPSS
Statistical Product and Service Solutions
- TGF-β
transforming growth factor-β
- DSS
disease-specific survival.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China No. 81572608 and 81874120 KW, and supported by Wuhan Science and Technology Bureau (No. 2017060201010170) KW.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.00026/full#supplementary-material
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Jiang W, Zhu F, Mao X, Agrawal S. Modulation of the tumor microenvironment by intratumoral administration of IMO-2125, a novel TLR9 agonist, for cancer immunotherapy. Int J Oncol. (2018) 53:1193–203. 10.3892/ijo.2018.4456 [DOI] [PubMed] [Google Scholar]
- 3.Xu H, Yu S, Liu Q, Yuan X, Mani S, Pestell RG, et al. Recent advances of highly selective CDK4/6 inhibitors in breast cancer. J Hematol Oncol. (2017) 10:97. 10.1186/s13045-017-0467-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu S, Li A, Liu Q, Yuan X, Xu H, Jiao D, et al. Recent advances of bispecific antibodies in solid tumors. J Hematol Oncol. (2017) 10:155. 10.1186/s13045-017-0522-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballinger TJ, Meier JB, Jansen VM. Current landscape of targeted therapies for hormone-receptor positive, HER2 negative metastatic breast cancer. Front Oncol. (2018) 8:308. 10.3389/fonc.2018.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tong CWS, Wu M, Cho WCS, To KKW. Recent advances in the treatment of breast cancer. Front Oncol. (2018) 8:227. 10.3389/fonc.2018.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franchino F, Ruda R, Soffietti R. Mechanisms and therapy for cancer metastasis to the brain. Front Oncol. (2018) 8:161. 10.3389/fonc.2018.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. (2016) 66:271–89. 10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Goldstein D, Crowe PJ, Yang JL. Antitumour effects and mechanisms of action of the panHER inhibitor, dacomitinib, alone and in combination with the STAT3 inhibitor, S3I-201, in human sarcoma cell lines. Int J Oncol. (2018) 52:2143–54. 10.3892/ijo.2018.4337 [DOI] [PubMed] [Google Scholar]
- 10.Matsuhashi N, Takahashi T, Matsui S, Tanahashi T, Imai H, Tanaka Y, et al. A novel therapeutic strategy of personalized medicine based on anti-epidermal growth factor receptor monoclonal antibodies in patients with metastatic colorectal cancer. Int J Oncol. (2018). [Epub ahead of print]. 10.3892/ijo.2018.4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Han N, Zhou S, Zhou R, Yuan X, Xu H, et al. The DACH/EYA/SIX gene network and its role in tumor initiation and progression. Int J Cancer (2016) 138:1067–75. 10.1002/ijc.29560 [DOI] [PubMed] [Google Scholar]
- 12.Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, et al. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature (2003) 426:299–302. 10.1038/nature02097 [DOI] [PubMed] [Google Scholar]
- 13.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature (2003) 426:247–54. 10.1038/nature02083 [DOI] [PubMed] [Google Scholar]
- 14.Kong D, Liu Y, Liu Q, Han N, Zhang C, Pestell RG, et al. The retinal determination gene network: from developmental regulator to cancer therapeutic target. Oncotarget (2016) 7:50755–65. 10.18632/oncotarget.9394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J, Cao F, Huang X, Ramen K, Xu X, Zhu Y, et al. Eyes absent homologue 2 predicts a favorable prognosis in colorectal cancer. Onco Targets Ther. (2018) 11:4661–71. 10.2147/ott.s164149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Qiu R, Qiu X, Tian T. EYA2 promotes lung cancer cell proliferation by downregulating the expression of PTEN. Oncotarget (2017) 8:110837–48. 10.18632/oncotarget.22860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen Z, Liang C, Pan Q, Wang Y. Eya2 overexpression promotes the invasion of human astrocytoma through the regulation of ERK/MMP9 signaling. Int J Mol Med. (2017) 40:1315–22. 10.3892/ijmm.2017.3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Y, Xu X, Wang T, Li Y, You W, Fu J, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. (2017) 8:e2928. 10.1038/cddis.2017.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Yang N, Huang J, Buckanovich RJ, Liang S, Barchetti A, et al. Transcriptional coactivator Drosophila eyes absent homologue 2 is up-regulated in epithelial ovarian cancer and promotes tumor growth. Cancer Res. (2005) 65:925–32. 10.1158/0008-5472.CAN-04-4368 [DOI] [PubMed] [Google Scholar]
- 20.Gao T, Zheng S, Li Q, Ran P, Sun L, Yuan Y, et al. Aberrant hypomethylation and overexpression of the eyes absent homologue 2 suppresses tumor cell growth of human lung adenocarcinoma cells. Oncol Rep. (2015) 34:2333–42. 10.3892/or.2015.4245 [DOI] [PubMed] [Google Scholar]
- 21.Vincent A, Hong SM, Hu C, Omura N, Young A, Kim H, et al. Epigenetic silencing of EYA2 in pancreatic adenocarcinomas promotes tumor growth. Oncotarget (2014) 5:2575–87. 10.18632/oncotarget.1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Y, Zheng S, Li Q, Xiang X, Gao T, Ran P, et al. Overexpression of miR-30a in lung adenocarcinoma A549 cell line inhibits migration and invasion via targeting EYA2. Acta Biochim Biophys Sin (2016) 48:220–8. 10.1093/abbs/gmv139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Wu K, Tian Y, Liu Q, Han N, Yuan X, et al. CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int J Oncol. (2016) 49:1343–50. 10.3892/ijo.2016.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Yu S, Yuan X, Xiong J, Kuang D, Pestell RG, et al. DACH1 suppresses breast cancer as a negative regulator of CD44. Sci Rep. (2017) 7:4361. 10.1038/s41598-017-04709-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. (2009) 69:4116–24. 10.1158/0008-5472.can-08-3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sircoulomb F, Bekhouche I, Finetti P, Adelaide J, Ben Hamida A, Bonansea J, et al. Genome profiling of ERBB2-amplified breast cancers. BMC Cancer (2010) 10:539. 10.1186/1471-2407-10-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dedeurwaerder S, Desmedt C, Calonne E, Singhal SK, Haibe-Kains B, Defrance M, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. (2011) 3:726–41. 10.1002/emmm.201100801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. (2014) 124:398–412. 10.1172/jci71180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA (2011) 305:1873–81. 10.1001/jama.2011.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagalla S, Chou JW, Willingham MC, Ruiz J, Vaughn JP, Dubey P, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. (2013) 14:R34. 10.1186/gb-2013-14-4-r34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci USA. (2010) 107:10208–13. 10.1073/pnas.0907011107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. (2007) 13:3207–14. 10.1158/1078-0432.ccr-06-2765 [DOI] [PubMed] [Google Scholar]
- 33.Tofigh A, Suderman M, Paquet ER, Livingstone J, Bertos N, Saleh SM, et al. The prognostic ease and difficulty of invasive breast carcinoma. Cell Rep. (2014) 9:129–42. 10.1016/j.celrep.2014.08.073 [DOI] [PubMed] [Google Scholar]
- 34.Desmedt C, Di Leo A, de Azambuja E, Larsimont D, Haibe-Kains B, Selleslags J, et al. Multifactorial approach to predicting resistance to anthracyclines. J Clin Oncol. (2011) 29:1578–86. 10.1200/jco.2010.31.2231 [DOI] [PubMed] [Google Scholar]
- 35.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature (2005) 436:518–24. 10.1038/nature03799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet (2005) 365:671–9. 10.1016/s0140-6736(05)17947-1 [DOI] [PubMed] [Google Scholar]
- 37.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. (2005) 7:R953–64. 10.1186/bcr1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer (2011) 11:143. 10.1186/1471-2407-11-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Bai X, Yu S, Liu Q, Pestell RG, Wu K. MAT1 correlates with molecular subtypes and predicts poor survival in breast cancer. Chin J Cancer Res. (2018) 30:351–63. 10.21147/j.issn.1000-9604.2018.03.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Li A, Yu S, Qin S, Han N, Pestell RG, et al. DACH1 antagonizes CXCL8 to repress tumorigenesis of lung adenocarcinoma and improve prognosis. J Hematol Oncol. (2018) 11:53. 10.1186/s13045-018-0597-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Zhou R, Yuan X, Han N, Zhou S, Xu H, et al. DACH1 is a novel predictive and prognostic biomarker in hepatocellular carcinoma as a negative regulator of Wnt/beta-catenin signaling. Oncotarget (2015) 6:8621–34. 10.18632/oncotarget.3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell (2006) 10:515–27. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J, et al. miR-30a suppresses breast cancer cell proliferation and migration by targeting Eya2. Biochem Biophys Res Commun. (2014) 445:314–9. 10.1016/j.bbrc.2014.01.174 [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Zhao Y, Skerry B, Wang X, Colin-Cassin C, Radisky DC, et al. Foxa1 is essential for mammary duct formation. Genesis (2016) 54:277–85. 10.1002/dvg.22929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-beta signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene (2012) 31:552–62. 10.1038/onc.2011.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu K, Katiyar S, Li A, Liu M, Ju X, Popov VM, et al. Dachshund inhibits oncogene-induced breast cancer cellular migration and invasion through suppression of interleukin-8. Proc Natl Acad Sci USA. (2008) 105:6924–9. 10.1073/pnas.0802085105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.