Abstract
Staphylococcus pseudintermedius is an opportunistic and emerging zoonotic pathogen that primarily colonises the skin of dogs. Many common variants are methicillin resistant (MRSP) or multidrug resistant (MDR), and drug resistance is increasingly reported across the globe. In New Zealand, MRSP isolation remains rare in clinics. To pre-emptively inform diagnostic and antimicrobial stewardship practices, we examine isolates of S. pseudintermedius, MRSP and MDR-MRSP from New Zealand dogs using a combination of methodologies. Genetic and genomic data combined with antimicrobial susceptibility screening identify common drug-resistance profiles and their genetic determinants. We demonstrate that sensitive and specific species-level identification of S. pseudintermedius can be achieved using Bruker MALDI-TOF MS and, further, that this technique can be used to identify some common subtype variants, providing a level of categorical precision that falls somewhere between single-locus and multi-locus sequence typing. Comparative genomics analysis of global S. pseudintermedius data shows that MRSP moves frequently across the globe, but that horizontal gene transfer events resulting in the acquisition of the SCCmec cassette (responsible for beta-lactam antibiotic resistance) are infrequent. This suggests that biosecurity and surveillance in addition to antibiotic stewardship should play important roles in mitigating the risk of MRSP, especially in countries such as New Zealand where MRSP is still rare.
Introduction
Staphylococci are ubiquitous gram-positive bacteria, of which many are opportunistic pathogens that commonly constitute part of the natural skin flora of diverse animal species1,2. In humans, the main infecting species is Staphylococcus aureus, while in dogs, and to a lesser extent cats, Staphylococcus pseudintermedius has been shown to be the most common skin-associated coagulase-positive staphylococcal species2. Taxonomically, S. pseudintermedius belongs to the Staphylococcus Intermedius Group (SIG), which includes two other coagulase-positive species; Staphylococcus delphini and Staphylococcus intermedius3. These species share many phenotypic characteristics, and S. pseudintermedius was only recognised as a novel species in 2005, previous to which it was referred to as Staphylococcus intermedius4. S. pseudintermedius is also an emerging zoonotic pathogen and cases of S. intermedius in humans existed before the reclassification of S. pseudintermedius5–8.
As in many bacteria, antibiotic resistance is an increasing problem in Staphylococcus infections leading to major challenges to combat disease in both human and veterinary medicine9. Methicillin-resistant S. pseudintermedius (MRSP) and multidrug-resistant S. pseudintermedius including methicillin-susceptible and resistant variants (MDR-MSSP and MDR-MRSP) are found worldwide10. MRSP is of particular concern because beta-lactam antibiotics are often the first choice of treatment for Staphylococcus-associated infections11. While there are no known differences in virulence or invasiveness between MSSP and MRSP strains12, favourable clinical outcomes from MRSP infection largely rely on the timely identification of antimicrobial susceptibilities followed by suitable antibiotic treatment13. However, many of the predominant lineages of MRSP, such as members of clonal complexes (CCs) 71 and 45, have resistance to as many as 7 different classes of antibiotic and their prevalence is reported to be increasing in multiple geographic settings10. In veterinary medicine, where the use of important human antimicrobials such as vancomycin, daptomycin and linezolid is increasingly discouraged (eg.14), this effectively renders some of these lineages extensively, or pan-drug resistant (XDR, PDR) to their recommended and available antibiotics. These variants are commonly carried on the skin and oral mucus membranes of healthy animals, and no effective strategy exists for the removal of commensal association13 leaving surveillance and monitoring as the only effective approaches to reduce the burden of antimicrobial resistance15.
MRSP was first identified in New Zealand in 201416,17 and it remains an uncommon laboratory isolate. Routine diagnostic methods currently used for identification of S. pseudintermedius and MRSP are relatively slow. The current diagnostic procedure for S. pseudintermedius differs between veterinary microbiology laboratories, and involves either the use of hyaluronidase and DNase biochemical tests or an initial test of polymyxin B susceptibility. Members of the SIG are most often hyaluronidase negative, DNAse positive and susceptible to polymyxin B, whereas S. aureus is intrinsically resistant to polymyxin B18 and demonstrates hyaluronidase activity19. Presumed S. aureus are tested for cefoxitin susceptibility and reported as either S. aureus or methicillin-resistant S. aureus (MRSA). Presumed members of the SIG are tested for oxacillin susceptibility and are reported as either SIG or methicillin-resistant SIG, however this classification may include other similar coagulase positive staphylococci such as Staphylococcus schleiferi. The use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for pathogen identification is increasing, but there are varying reports of its specificity for members of the SIG20–22. Antimicrobial susceptibility testing of isolates by standard methods still takes several hours of incubation (commonly overnight).
The objectives of this study were to validate a rapid diagnostic test for S. pseudintermedius for application in New Zealand veterinary laboratories and to provide additional information surrounding antimicrobial resistance in New Zealand MRSP in order to inform antibiotic stewardship practices. We tested the discriminatory power of MALDI-TOF MS and its utility in differentiating variants of S. pseudintermedius found in New Zealand. Additionally, we used whole genome sequencing (WGS) and comparative genomics to characterise the resistance profiles, diversity and origins of New Zealand S. pseudintermedius strains. We show that MALDI-TOF MS accurately identifies S. pseudintermedius and some of its more common variants. Additionally, we find that MRSP is likely to have been imported into New Zealand on numerous occasions and provide genetic evidence supporting autochthonous transmission of some MRSP lineages. Using WGS-derived phylogenies we demonstrate that acquisition of the SCCmec cassette by horizontal gene transfer has occurred less frequently than has previously been estimated and that this difference in estimation is due to limitations in the phylogenetic resolution provided by seven gene MLST data.
Materials and Methods
Sample collection and isolation
In response to earlier observations of MRSP infection in dogs in New Zealand16,17, laboratory cultures of coagulase positive Staphylococcus spp. opportunistically collected from routine practice cases were supplied to mEpiLab, Massey University by Gribbles and NZVP-IDEXX veterinary laboratories. Isolates came from across New Zealand between December 2015 and May 2017, and included the earliest isolate originating from the index study in 201416; 117 samples in total, of which 106 were from dogs and 11 from cats. Where possible, two independent colonies were isolated from each sample on Tryptic Soy Agar (TSA) plates at 37 °C to ensure clonality, resulting in 229 separate isolates.
MALDI-TOF
Isolate identification was carried out using full protein extraction (FPE) in the Bruker Biotyper MALDI-TOF. Ethanolic suspensions of overnight TSA culture were generated by homogenisation of approximately 2 mm3 of bacterial material in 300 µL of sterile high grade water, followed by addition of 900 µL of high grade 100% ethanol. Targets were prepared following the manufacturer’s recommendations. MALDI-TOF spectra, predictions of species, and log-score estimates of certainty were obtained in triplicate for each isolate using Bruker’s software and version 7 of Bruker’s isolate database (7311 RUO).
Antimicrobial resistance screening
All isolates were assessed for their susceptibility to a panel of antimicrobials. The panel included oxacillin, amoxycillin-clavulanate (Australian approved name), trimethoprim, doxycycline, enrofloxacin, clindamycin, ciprofloxacin, chloramphenicol, cefpodoxime, cephalexin, cefovecin, amoxycillin (Australian approved name), novobiocin, oxytetracycline, gentamicin and ampicillin using the standard CLSI disk diffusion protocol, and validation control strain ATCC 25923 (ESR 917).
Resistant, intermediate and susceptible classes of resistance were assigned for each antibiotic referring to CLSI standards from CLSI VET08, Edition 4 or CLSI M100, Edition 27 where appropriate. Inducible clindamycin resistance was assessed by disk diffusion using the D-zone test as described by23.
In addition, we assessed the ability of the disk diffusion assay to predict the resistance genotype of each isolate from our data set where gene presence/absence data were available (either from genomic or PCR data, see below). Zone diameter upper and lower bounds were calculated that maximised the positive predictive value of phenotype (diameter) for genotype (presence/absence).
Molecular Biology
Crude DNA extracts were prepared by boiling approximately 2 mm3 of bacterial material for 10 minutes in 1 mL of 2% Chelex™ (BioRad) suspension. The species of each isolate was verified using PCR primers and protocols for Staphylococcus aureus, Staphylococcus intermedius and Staphylococcus pseudintermedius described by Sasaki et al.24. PCR conditions were optimised using positive control strains ATCC 25923 (S. aureus), ATCC 29663 (S. intermedius Hajek) and in-house S. pseudintermedius controls that had been characterised by whole genome sequencing as well as negative control strains PNAL 93/21525 (S. felis), 87/1493 (S. schleiferi) and ATCC l2228 (S. epidermidis). Presence/absence of the mecA gene was screened in each isolate using the protocol of Lee et al.25, which was validated using in-house positive and negative control strains for which both phenotypic and genomic data were available.
Purified genomic DNA was extracted using a QIAamp Mini kit (Qiagen) following the manufacturer’s instructions with minor modifications. Genomic DNA was prepared for sequencing using the Nextera XT library kit (Illumina, San Diego, USA). Libraries were sequenced on an Illumina HiSeq using 2 × 250 bp paired end sequencing by the Massey Genome Service (New Zealand).
Bioinformatics
Illumina reads were trimmed using a combination of in-house software and Trimmomatic26 and assembled using SPAdes version 3.9.027 as part of the Nullarbor28 pipeline (https://github.com/tseemann/nullarbor). Nullarbor and seqkit29 were also used to extract the assembly statistics (Table S1). Assembled draft genomes were deposited to NCBI under BioProject number PRJNA473042.
Seven-gene MLST alignments and isolate data were obtained for all isolates recorded in PubMLST30 using the mlst software package (https://github.com/tseemann/mlst). Phylogenetic analysis of concatenated MLST sequences was performed in BEAST231 with an MCMC chain length of 500,000,000 using an HKY substitution model accounting for invariant sites, a strict molecular clock and a Yule population size model. Convergence of the Bayesian phylogenetic inference was verified in Tracer v1.6 (http://beast.bio.ed.ac.uk/Tracer), with all estimated parameters returning estimated sample sizes values greater than 200 after a 10% burn-in.
For comparative genomic analyses, an extensive literature survey was performed at the start of March 2018 to identify potential sources of Staphylococcus pseudintermedius genomes, which were subsequently downloaded from ENA, NCBI and SRA databases or supplied directly from the authors of these published works. A total of 248 genomes were obtained for comparative analyses, including the 29 genomes generated in this study. SRA datasets were assembled using the methodology presented above. Clusters of orthologous groups were defined using the R package ‘pewit’ (https://github.com/iferres/pewit)32. Preliminary analyses showed that many published assembled genomes were highly fragmented, leading to errors in gene prediction and significant increase in per-genome gene number and a reduction in the number of identifiable core genes. Therefore, genomes were excluded from our comparative analysis based on the N50 statistic until all 7-gene MLST loci were identified as core genes in our clustering pipeline. The final threshold was an N50 value higher than 20,000. This resulted in 223 genomes being included in the final analysis. Gubbins33 was used to trim predicted mutation hotspot regions from the concatenated core gene alignment, which was then used for whole genome sequence (WGS) phylogeny. WGS phylogeny was performed using BEAST231, with an MCMC chain length of 2 * 109 using an HKY substitution model accounting for invariant sites, a strict molecular clock and a Yule population size model. As above, all estimated parameters returned estimated sample size values greater than 200 after a 50% burn-in. Time to most recent common ancestor estimates were performed in a similar way, integrating tip dates into the analysis and optimising the parameter posteriors for both strict and relaxed clock models over 108 MCMC chains. Ancestral state reconstructions were performed from the output trees of the BEAST analysis in Mesquite34, defining either geographical origin or SCCmec variant as a categorical variable associated with each tip. Ancestral states were inferred using a parsimony model and the average number of estimated state changes was calculated over all trees. Network analysis was performed in the ‘igraph’ R package35, community analysis of geographic exchanges was performed using fast greedy clustering after summing directional measures to generate an undirected graph.
Presence/absence of antibiotic resistance genes was assessed using abricate (https://github.com/tseemann/abricate) and the ResFinder database36. Resistance associated point mutations at gyrA and glrA loci were identified by examining alignments generated using the Geneious R10 software package37.
The SCCmec type of each MRSP isolate was assigned based on its ccrA and ccrB gene composition using abricate (as above) and relevant reference sequences as specified by http://www.staphylococcus.net/Pages/SCCmecTypingEN.html and http://www.sccmec.org. Unambiguous matches of >95% identity were considered sufficient to attribute the presence of a ccr gene type. Ambiguous or inconclusive matches were curated manually using the Geneious R10 software package37. Data manipulation and visualisation was performed using the R packages ‘ggtree’38 and ‘ggmap’39.
MALDI-TOF spectral data analysis was performed using the ‘MALDIquant’ R package40. Peaks were aligned using the Lowess warping function at a tolerance of 0.01, and binned at a tolerance of 0.002. Linear discriminant analysis and peptide profiling were performed using the ‘sda’ R package41 and discriminant performance of peak combinations was assessed using the ‘crossval’ R package42.
Results
MALDI-TOF accurately identifies New Zealand S. pseudintermedius isolates
This study included 229 coagulase positive Staphylococcus spp. isolates obtained from veterinary pathology laboratories around New Zealand. MALDI-TOF MS was used for species-level identification of all isolates and the accuracy of these results was evaluated using PCR with primers specific for S. aureus, S. intermedius and S. pseudintermedius24. Isolates were also tested for novobiocin, polymyxin B and acriflavine sensitivities which have been shown to discriminate between coagulase positive/negative staphylococci or between S. aureus and members of the SIG43.
A total of 174 isolates were confirmed as S. pseudintermedius by PCR. MALDI-TOF MS identified 176 isolates as S. pseudintermedius with a median MALDI-TOF log-score of 2.33 (IQR: 2.23–2.42), corresponding to a sensitivity of 0.99 and a specificity of 0.93 when setting the PCR result as the gold standard. Five S. pseudintermedius isolates confirmed by MALDI-TOF MS and PCR were also positive with S. aureus PCR, suggesting a specificity of the S. aureus PCR of less than 100%. None of the isolates collected were identified as S. intermedius by either MALDI-TOF MS or PCR. We found that polymyxin B, acriflavine and novobiocin sensitivity cut-offs were all imperfect methods for the identification of S. pseudintermedius (Fig. S1).
Beta-lactam antibiotic susceptibility and its determinants in New Zealand S. pseudintermedius
Beta-lactam resistance is conferred by the mecA or, less commonly, mecC genes that encode a penicillin-binding-protein (PBP2a) carried on a mobile chromosomal cassette called SCCmec44. MecA confers broad-spectrum beta-lactam resistance to many commonly used antibiotics. We assigned methicillin resistance or susceptibility based on the result of the CLSI disk diffusion protocol to oxacillin, and identified the presence or absence of the mecA gene by PCR for each isolate. Sixty-seven of the 176 presumptive S. pseudintermedius isolates (38%) were mecA positive. All oxacillin resistant isolates but one were identified as carrying the mecA gene.
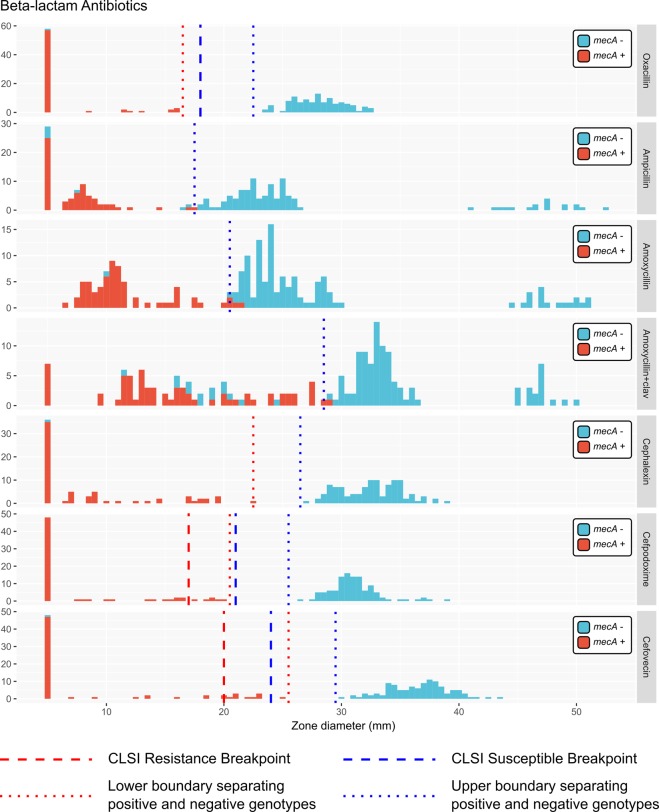
We correlated the mecA presence/absence genotype with the results of the disk diffusion assay for other beta-lactam antibiotics, assigning isolates as susceptible, intermediate or resistant where CLSI breakpoints were available. Within our dataset, cephalexin, cefpodoxime and cefovecin zone diameters were all reliable predictors of the presence of the mecA gene (Table 1) with clear, well-separated upper and lower diameter boundaries that separated mecA+ and mecA− isolates. Numerous isolates with intermediate resistance to cefpodoxime and cefovecin and a single isolate classified as susceptible to cefovecin possessed the mecA gene.
Table 1.
Optimal upper and lower bounds of zone diameter for predicting antibiotic resistance genotype from disk diffusion phenotype in our dataset.
| Antibiotic | Lower Bound (mm) | Upper Bound (mm) | Positive Predictive Value for genotype* | Resistance genotype |
|---|---|---|---|---|
| Oxacillin | 16.5 | 22.5 | 0.98 | mecA+ |
| Ampicillin | 17.5 | 17.5 | 0.9 | mecA+ |
| Amoxycillin | 20.5 | 20.5 | 0.97 | mecA+ |
| Amoxycillin + clavulanate | 28.5 | 28.5 | 0.82 | mecA+ |
| Cephalexin | 22.5 | 26.5 | 0.97 | mecA+ |
| Cefpodoxime | 20.5 | 25.5 | 0.98 | mecA+ |
| Cefovecin | 25.5 | 29.5 | 0.97 | mecA+ |
| Oxytetracycline | 11.5 | 27 | 1 (S = 1, n = 12) | tetM+ |
| Doxycycline | 17.5 | 28 | 1 (S = 1, n = 12) | tetM+ |
| Ciprofloxacin | 11.5 | 28.5 | 1 (S = 1, n = 16) | GyrA 84 L and Glr 80I |
| Enrofloxacin | 12.5 | 25 | 1 (S = 1, n = 16) | GyrA 84 L and Glr 80I |
| Trimethoprim | 5.5 | 19.5 | 1 (S = 1, n = 19) | dfrG+ |
| Clindamycin | 19 | 25 | 1 (S = 0.94, n = 18) | ermB+ |
| Chloramphenicol | 10.5 | 23.5 | 1 (S = 1, n = 15) | cat(pC221)+ |
| Gentamicin | 18.5 | 23 | 1 (S = 1, n = 18) | aac(6)/aph(2) + ** |
*For non-beta-lactam class antibiotics, positive predictive values are calculated for the 29 isolates which had genomes sequenced as part of this study. The sensitivity (S) of the test and the number (n) of resistance gene-containing genomes is shown in parentheses.
**Gentamicin resistance is also associated with genes aphIII and antIV in the sequenced NZ isolates.
Many isolates showed no sensitivity to the ampicillin disk but lacked the mecA gene. Ampicillin resistance may be conferred by a variety of other beta-lactamase encoding (bla) genes in staphylococci45. A subset of isolates lacking the mecA gene showed increased resistance to amoxycillin/clavulanate. To our knowledge there is no known specific mechanism for amoxycillin/clavulanate resistance, and interestingly all mecA− isolates with increased amoxycillin/clavulanate resistance showed similar susceptibilities to amoxicillin as all other mecA− isolates. A second subset of oxacillin sensitive isolates were also hypersensitive (showed approximately 100% increased zone diameters) to ampicillin, amoxycillin and amoxycillin/clavulanate (Fig. 1).
Figure 1.
Beta-lactam antibiotic zone diameter measurements for all S. pseudintermedius isolates in this study. Histograms are presented for each antibiotic. A zone diameter of 5 mm is used to indicate a “no zone” result. Available genetic data are shown by the colour of each histogram, where antibiotic resistance gene positive genotypes are coloured light red, and negative genotypes are coloured light blue. Individual legends specify the relevant antibiotic resistance gene for each plot. Dotted lines specify the calculated optimal bounds for separating positive and negative genotypes in our data. Dashed lines show the R/I/S CLSI boundaries for antibiotics where breakpoint data were available at the time of publication.
Whole genome sequencing identifies New Zealand MRSP diversity and resistance genotypes
MDR variants of S. pseudintermedius, including MDR-MRSP, have been associated with specific genetic lineages or clonal complexes10. In order to examine the diversity of drug resistant variants obtained in New Zealand, we conducted sensitivity measurements for eight additional antibiotics from six antimicrobial classes commonly used to describe S. aureus resistance profiles46, and assigned susceptible, intermediate and resistant profiles where CLSI breakpoints were available.
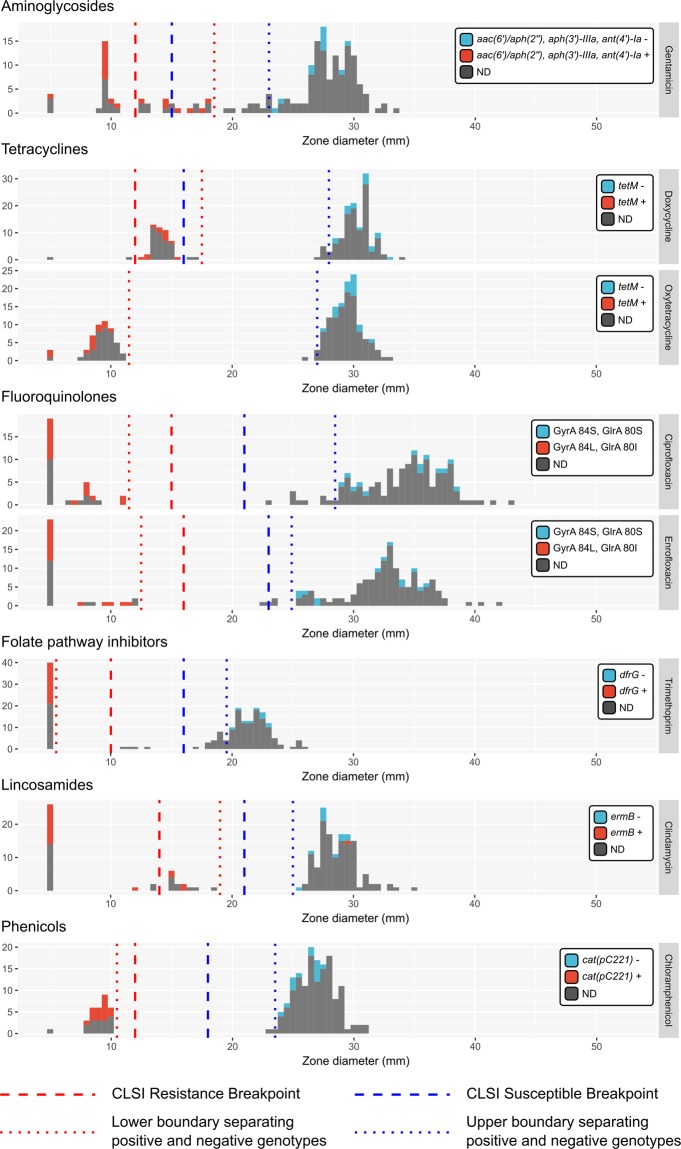
Based on the obtained distribution of antibiotic resistance measurements, we randomly selected 29 MRSP and MDR-MRSP isolates for typing by whole genome sequencing (WGS). Illumina sequencing resulted in high-quality draft genome assemblies from all 29 isolates (Table S1). As was done for beta-lactam antibiotics, we examined the susceptibility profiles of each isolate and correlated zone diameter measurements with genotypes from sequenced genomes (Fig. 2 and Table 1).
Figure 2.
Non-beta-lactam antibiotic zone diameter measurements for all S. pseudintermedius isolates in this study. Histograms are presented for each antibiotic. A zone diameter of 5 mm is used to indicate a “no zone” result. Available genetic data are shown by the colour of each histogram, where antibiotic resistance gene positive genotypes are coloured light red, and negative genotypes are coloured light blue. Individual legends specify the relevant antibiotic resistance gene for each plot. Dotted lines specify the calculated optimal bounds for separating positive and negative genotypes in our data. Dashed lines show the R/I/S CLSI boundaries for antibiotics where breakpoint data were available at the time of publication.
Oxytetracycline and doxycycline were used to test for tetracycline-class resistance. Zone diameters for both these antibiotics showed a strong positive correlation (data not shown), and two distinct groups of zone diameter were obtained (Fig. 2). Tetracycline resistance is known to be conferred by the presence of the tetM, tetL and tetK genes in S. pseudintermedius47,48. Within isolates from our study, only tetM was identifiable in sequenced genomes, and we obtained a perfect correlation between genotype and phenotype between susceptible and intermediate classes. All tetM+ isolates fell in the intermediate category of resistance for doxycycline based on CLSI breakpoints.
Enrofloxacin and ciprofloxacin were used to test for fluoroquinolone-class resistance. Zone diameters for both enrofloxacin and ciprofloxacin showed a strong positive correlation (data not shown) and two distinct groups of zone diameter were obtained (Fig. 2). In S. pseudintermedius, fluoroquinolone resistance is thought to be associated with mutations at codon positions 84 and 80 in the gyrA and glrA genes, respectively49. Isolates from our study were either GyrA-84S:GlrA-80S or GyrA-84L:GlrA-80I. We obtained a perfect correlation between genotype and phenotype between susceptible and resistant classes, with GyrA-84L:GlrA-80I being the resistant genotype.
Similarly, trimethoprim and chloramphenicol showed a perfect correlation with the dfrG and cat(pC221) genotypes, respectively (Fig. 2). Intermediate levels of trimethoprim resistance have previously been seen in S. aureus with point mutations in dfrA and dfrB genes50,51. However, genomic data were not available to resolve the resistance phenotype of four isolates that showed intermediate levels of trimethoprim resistance.
Clindamycin resistance in S. pseudintermedius is conferred by the ermB gene and its expression can be constitutive or inducible52. Here, the relationship between the ermB genotype and constitutive resistance was imperfect. All isolates that were ermB+ and fell into either susceptible or intermediate categories for constitutive resistance also gave evidence of D-zone formation when tested for inducible resistance (Fig. 2 and Table 2); however, we noted that S. pseudintermedius generates relatively diffuse disk borders by this method, complicating interpretation of the assay.
Table 2.
Resistance profiles of all isolates based on CLSI disk diffusion break points (VET08 4th Edition and M100 27th Edition) for Staphylococcus pseudintermedius (oxacillin and doxycycline) and Staphylococcus spp. (all other antibiotics). “ind = #” denotes the number of isolates in each category that gave evidence of an inducible clindamycin phenotype via D-formation in the erythromycin/clindamycin test.
| Oxacillin | Doxycycline | Ciprofloxacin | Clindamycin | Trimethoprim | Chloramphenicol | Gentamicin | Resistance Profile | Count | Category | ST by WGS |
|---|---|---|---|---|---|---|---|---|---|---|
| R | I | R | I (ind = 2) | R | R | I | RIRIRRI | 2 | MDR-MRSP | 496 (1) |
| R | I | R | I (ind = 2) | R | R | S | RIRIRRS | 2 | MDR-MRSP | 496 (1) |
| R | I | R | R | R | R | S | RIRRRRS | 6 | MDR-MRSP | 496 (3) |
| R | S | R | R | R | R | R | RSRRRRR | 18 | MDR-MRSP | 71 (9) |
| R | S | R | S | R | R | R | RSRSRRR | 2 | MDR-MRSP | 71 (1) |
| R | S | R | S | R | S | R | RSRSRSR | 2 | MDR-MRSP | 71 (1) |
| R | R | S | S | R | S | S | RRSSRSS | 1 | MDR-MRSP | |
| R | I | S | I (ind = 3) | S | S | S | RISISSS | 3 | MRSP | 84 (1) |
| R | I | S | R (ind = 3) | S | S | S | RISRSSS | 3 | MRSP | 84 (1) |
| R | I | S | S | R | S | S | RISSRSS | 3 | MRSP | 258 (1), 1173 (1) |
| R | I | S | S | S | S | S | RISSSSS | 6 | MRSP | 1172 (1), 84 (1) |
| R | I | S | I (ind = 2) | R | S | S | RISIRSS | 2 | MRSP | 277 (1) |
| R | S | S | I | S | S | R | RSSISSR | 1 | MRSP | |
| R | S | S | I (ind = 1) | S | S | S | RSSISSS | 1 | MRSP | |
| R | S | S | S | S | R | I | RSSSSRI | 1 | MRSP | |
| R | S | S | S | S | S | I | RSSSSSI | 5 | MRSP | 64 (2) |
| R | S | S | S (ind = 1) | S | S | S | RSSSSSS | 10 | MRSP | 1171 (1), 498 (1), 749 (2) |
| S | I | S | I | S | S | S | SISISSS | 2 | MSSP | |
| S | I | S | S | S | S | S | SISSSSS | 18 | MSSP | |
| S | R | S | S | S | S | S | SRSSSSS | 1 | MSSP | |
| S | S | S | R | S | S | S | SSSRSSS | 2 | MSSP | |
| S | S | S | S | I | S | S | SSSSISS | 4 | MSSP | |
| S | S | S | S | R | S | S | SSSSRSS | 2 | MSSP | |
| S | S | S | S | S | S | R | SSSSSSR | 2 | MSSP | |
| S | S | S | S | S | S | S | SSSSSSS | 81 | PSSP |
Sequence Type (ST), derived from alleles of seven MLST genes is given for isolates belonging to each resistance profile for which genomic data were available, the number of isolates sequenced is shown in parentheses.
Gentamicin zone diameters were relatively evenly distributed and did not fall into obvious groups. Gentamicin resistance is associated with the aac(6′)/aph(2″):aph(3′)-IIIa:ant(4′)-Ia genotype in S. pseudintermedius. These genes are most frequently acquired together as part of a resistance cassette in staphylococci53. Several isolates that were defined as susceptible according to CLSI guidelines were observed to carry the aac(6′)/aph(2″):aph(3′)-IIIa:ant(4′)-Ia genes, although the zone diameter measurements were capable of separating positive and negative genotypes at values slightly higher than those of the CLSI standards (Fig. 2).
Full resistance profiles were defined for seven classes of antibiotic using available CLSI breakpoints (Table 2). Pan-susceptibility was the most commonly identified resistance profile. MRSP isolates possessed between 0 and 6 classes of resistance in addition to mecA-mediated broad-spectrum beta lactam resistance (including intermediate resistance). All MDR isolates (possessing three or more classes of antibiotic resistance) were MDR-MRSP; MDR-MSSP has been reported elsewhere10 but was not identified in this study.
Seven-gene MLST locus data were extracted from each sequenced genome in order to assign a sequence-type (ST) to each isolate. A total of 11 different STs were identified among the 29 sequenced isolates; STs 64, 71, 84, 258, 277, 496, 498 and 749 as well as three previously undefined STs 1171, 1172 and 1173 (Table 2). The most highly drug resistant variants that we identified all belonged to either ST 71 or ST 496.
MALDI-TOF spectra distinguish between some common sequence types of S. pseudintermedius
MALDI-TOF spectroscopy has revolutionised veterinary and medical diagnostics, making rapid species identification robust, simple and affordable54. Having demonstrated that S. pseudintermedius species detection by MALDI-TOF is both sensitive and specific for isolates in our study, we tested its ability to distinguish identified subtypes of S. pseudintermedius.
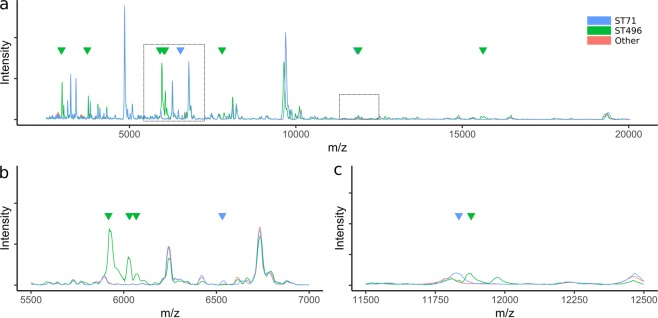
MRSP was indistinguishable from MSSP by either variance-based methods (Fig. S2) or peak-specific discriminant analysis (data not shown). However, 98% and 99% accuracy was achieved when discriminating ST 71 isolates and ST 496 isolates, respectively. Major variations at multiple peak positions were visible for all ST 496 isolates, which were specific markers of this group (Figs 3 and S3). ST 71 isolates showed much more subtle variation in their spectra; characteristic low-level peaks were observed at m/z values of 6543 and 11799. The latter of these appeared to be the result of a slight peak-shift, with all other isolates demonstrating a similar intensity peak at m/z 11770 (Fig. 3c).
Figure 3.
MALDI-TOF spectra of S. pseudintermedius belonging to ST 71 and ST 496 compared to other S. pseudintermedius isolates. Discriminatory peaks are indicated with arrows; two distinct peaks were present in ST 71 spectra and multiple distinct peaks in ST 496 spectra. Zoomed-in views of the highlighted areas are presented in panels b and c.
WGS predicts infrequent, ancestral SCCmec acquisition, followed by subsequent global dispersal
Understanding the dynamics of antibiotic resistance acquisition is important when developing antibiotic stewardship or biosecurity strategies. The availability of genomic data for S. pseudintermedius is rapidly increasing (eg.55–58) but, to date, population structure estimates and genealogies for this species have principally been constructed using seven-gene MLST data10,59. To test how genomic data influenced estimates of resistance gene acquisition frequency, we constructed phylogenies from a comprehensive collection of global MLST profiles and WGS data for S. pseudintermedius. At the time of analysis, a total of 949 different seven-gene MLST definitions were identified by combining PubMLST and genomic data. The 223 published genome sequences came from a subset of 50 unique MLST-types. With the exception of some datasets from the UK, the vast majority of published genomic data come from MRSP variants.
Our S. pseudintermedius genealogy reconstructions using seven-gene MLST loci produced a star-like phylogeny with poor support (Fig. S4). This has been observed previously, and the lack of phylogenetic resolution could be explained by either insufficient variant sites or high rates of recombination at these loci60.
Contrastingly, whole genome phylogenetic analysis generated a well-supported bifurcating topology for the subset of available strain types (Fig. 4A). Members of MLST-based clonal complexes (CCs) 71, 112 and 45 were identifiable as monophyletic clades in this analysis, whereas members of clonal complexes 68 and 258 formed paraphyletic groups that clustered loosely by resistance gene profile. Resistance gene profiles showed that multidrug resistance genotypes to numerous classes of antibiotics were most strongly associated with CC 45, CC 112, CC 71, ST 316 and ST 496. TetK-based tetracycline resistance was common but not conserved in members of ST71, whereas TetM-based tetracycline resistance was more common in other lineages. Ciprofloxacin resistance-associated mutations GyrA(84 L) and GlrA(80I) were most frequently present together, with the exception of ST 316 and ST 497 which possessed only the GyrA(84 L) mutation.
Figure 4.
Comparative genomics analysis of global MRSP. (A) Predicted phylogenetic relationship between 223 genomes. Dendrogram nodes with posterior probabilities of 1 are indicated by circles. Stars denote isolates generated as part of this study. Resistome profiles are displayed as a heatmap indicating presence (red) or absence (white) of the genotype indicated at the head of each column. (B) Summary of global exchange predicted from the presented phylogeny. Network arrow thickness denotes an estimate of the frequency of exchange between each country in the analysis, and is corrected for reported sequence-type diversity from each location. Node size indicates the number of different MLST types of S. pseudintermedius that are present in published genomes from each location. Coloured boundaries represent discriminatory groupings defined by community analysis of the undirected network. Country abbreviations used are; UK: United Kingdom, SWE: Sweden, DEN: Denmark, GER: Germany, NL: Netherlands, AUS: Australia, NZ: New Zealand, SL: Sri Lanka, BOT: Botswana, USA: United States of America.
MRSP variants formed mainly monophyletic groups which clustered by SCCmec type. Despite the over-representation of MRSP strains in the global genomic data set, parsimony-based ancestral reconstructions of each lineage’s SCCmec state identified a methicillin susceptible state at the root of the phylogeny (Fig. S5). Furthermore, MRSP diversity associated with all S. pseudintermedius strains was estimated to have derived from as few as 14 independent acquisitions of different SCCmec cassettes (Table S2). These analyses also predicted infrequent loss of the SCCmec cassette, however, loss events were associated with nodes that had lower posterior support in the phylogeny. Most monophyletic SCCmec clades consisted of isolates derived from multiple countries across the world, showing that stable methicillin-resistant lineages are likely to have circulated the world on multiple occasions.
Community analysis of geographic exchange estimates based on similar ancestral state reconstructions, and corrected for reported ST diversity per country, allowed us to estimate the frequency of exchange of S. pseudintermedius lineages between countries. Significant inter-connectivity was observed between S. pseudintermedius lineages in European countries, as well as frequent bidirectional exchanges between New Zealand and Australia, and Australia and the Netherlands (Fig. 4B). However, it should be noted that sample diversity was heavily biased towards isolates of European and Australasian origin.
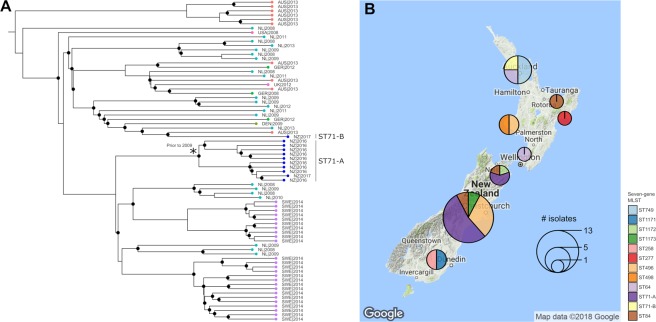
The frequent global exchange of S. pseudintermedius lineages was most evident in highly sampled lineage ST 71. We identified two independent New Zealand lineages of ST 71 in these data: ST 71-A (n = 10) and ST 71-B (n = 1) (Fig. 5A). ST 71-A formed a monophyletic, New Zealand-specific group indicative of endemic circulation following a single introduction of a common ancestral strain. Divergence estimates for lineage ST 71-A suggested that the latest possible date for this common ancestor was around 2009, five years prior to the first reported observation of MRSP in New Zealand16. However, date parameter estimates showed very broad posterior distributions (median = 1977, 95% HPD = 1944–2004) thus precise estimates of the dates of introduction of the different lineages were not possible. Examination of the geographical distribution of identified MRSP lineages from across New Zealand showed that ST 71-A had only been reported in the South Island while ST 84 and ST 496 had been reported on multiple occasions in distinct geographical settings in both the North and South islands, suggesting multiple lineages may be endemic in the country (Fig. 5B). ST 71-B appeared to be recently introduced from Australia and only reported in the North Island.
Figure 5.
Comparative genomics analysis of ST 71. (A) Predicted phylogenetic relationship between genomes belonging to MLST type 71. Dendrogram nodes with posterior probabilities greater than 0.9 are indicated by circles. Leaf tips are coloured by geographical origin. Distinct monophyletic lineages of New Zealand isolates “ST 71-A” and “ST 71-B” are highlighted. The star highlights the latest predicted date of emergence of ST 71-A, corresponding to the upper range of the posterior distribution of predicted date. All other dating information is left off due to lack of support. (B) The geographical distribution of distinct lineages of MRSP types that has been reported from New Zealand to date.
Discussion
MRSP is becoming a major pathogen in veterinary hospitals across the world and is difficult to treat as many MRSP isolates are also MDR61. Global studies of MRSP carriage in healthy animals report highly varied observations, ranging from approximately 2% to 50% prevalence although surveillance data is limited62,63.
In this study, we performed a multifaceted analysis of New Zealand S. pseudintermedius isolates to examine how technology may be used to improve our understanding and surveillance practices surrounding MRSP. MALDI-TOF identification is a valuable tool for diagnostic laboratories, however there are mixed reports in the literature about the specificity of MALDI-TOF typing for the identification of S. pseudintermedius20–22. Our work on New Zealand S. pseudintermedius isolates from dogs demonstrates both sensitive and specific species-level identification of S. pseudintermedius by Bruker MALDI-TOF MS, most likely corresponding to improvements in the Bruker reference database. Additionally, we used PCR and genomics to correlate resistance genotypes to antibiotic susceptibilities as measured by disk diffusion. For isolates in our dataset, we found that oxacillin, cephalexin, cefpodoxime and cefovecin could be used as reliable indicators of the mecA genotype while ampicillin, amoxycillin and amoxycillin/clavulanate were poor indicators due to the existence of non-mecA-mediated resistance mechanisms. Many of these mechanisms have been previously described for ampicillin resistance45; however, we identified a cluster of S. pseudintermedius isolates that demonstrated an unknown mechanism resulting in increased resistance to amoxycillin-clavulanate with no increased resistance to similar concentrations of amoxycillin. We found that existing CLSI breakpoints classified many mecA+ isolates as having intermediate levels of resistance to both cefpodoxime and cefovecin, and noted a single observation of a likely misclassification where an isolate possessing mecA was identified as susceptible to cefovecin. For gentamicin, we also observed many genotype positive isolates of S. pseudintermedius that were classified as susceptible when using CLSI breakpoints established for “Staphylococcus spp.”. Together, these observations suggest that S. pseudintermedius may require further establishment of species-specific break points for disk diffusion experiments to many antibiotics. This is perhaps unsurprising, as reference standards are still evolving for S. pseudintermedius due to its relatively recent reclassification4.
We performed in-depth analysis of the MALDI-TOF spectral data from more than 100 S. pseudintermedius isolates and coupled this with WGS analysis where possible. Within our sample set, MALDI-TOF MS could be used to discriminate between sequence types ST 71 and ST 496. This level of categorical precision is comparable to that of single-locus or multi-locus sequence typing, but can be achieved more rapidly and at much lower cost. Although our analysis was constrained to strain variants that we could isolate in New Zealand at the time of this study (and therefore the number of isolates tested was low), our observations highlight a valuable use of MALDI-TOF MS in the surveillance of S. pseudintermedius at species and genetic variant level. However, it would be necessary to validate such discriminatory analyses in local settings against up-to-date panels of S. pseudintermedius isolates belonging to all regionally circulating lineages.
One of the prerequisites to successful and actionable surveillance is precise typing. At the individual level, standardised strain identification aids our ability to treat infection by drawing on prior stain-specific examples. At the population level, understanding the incidence of circulating strains and their epidemiology allows biosecurity measures to be adapted in the face of changing infection landscapes. While MLST-based schemes have been used to establish useful classifications for S. pseudintermedius, our analyses suggest that increasingly recognised species diversity and the lack of phylogenetic resolution provided by such loci does not allow for accurate definitions of population structure. This ultimately limits our understanding of the origins of different lineages and the evolution of antibiotic resistance in S. pseudintermedius. WGS offers an improved method for addressing such questions, and here we used available global data to examine the frequency of historical SCCmec acquisition, demonstrating that the main circulating lineages acquired resistance ancestrally on as few as 14 occasions. This estimate is greatly reduced compared to that derived from 7-gene MLST data60 and is more comparable with similar estimates in S. aureus64, challenging the assumption that antibiotic resistance acquisition by horizontal gene transfer is commonplace for this species (at least for SCCmec). However, the observed reduction is due to both resolved phylogenetic relationships between lineages and the availability of data from only a subset of STs. Global reports suggest that these resistant strain variants are becoming increasingly prevalent worldwide60. However, there is geographic bias and a dearth of MSSP in available WGS data. Coupled with the demonstrated imprecisions of MLST phylogeny, this currently makes it unclear which lineages are susceptible precursors to many of the dominant MRSP lineages and whether or not they are still in circulation. This, in turn, complicates the evaluation of selection pressures that are likely driving the worldwide increase in MRSP and MDR-MRSP observations.
Our genomic analyses of MRSP isolates suggest that multiple types (sequence types 64, 71, 84, 258, 277, 496, 498, 749 1171, 1172 and 1173) have been imported into New Zealand and that some infections are likely to be the result of autochthonous transmission. In this and other settings where MRSP is an established endemic pathogen, unbiased population-level evaluations of prevalence will be required to assess the scale of the burden to public (and veterinary) health. Antibiotic treatments that precede microbiological diagnostics commonly increase the relative abundance of on-animal MRSP, which in turn may lead to increased population-level MRSP prevalence59. This problem is amplified when strains with multiple resistance phenotypes become prevalent as the use of single antibiotics or antimicrobials co-selects for multiple classes of resistance65. Faced with such a challenge to public health, an important step to combat MRSP and similar healthcare-associated MDR pathogens is real-time surveillance and the use of precise diagnostic techniques such as MALDI-TOF MS and WGS in a context that can be linked to clinical and epidemiological data15. Standardised surveillance and reporting can effectively inform antibiotic stewardship programmes, infection control strategies, interventions and help to stabilise or reduce antibiotic resistance prevalence66.
Supplementary information
Acknowledgements
We are very grateful to Dr Kate Worthing for supplying genomic data from her study in Australia. DW and NF are supported by the New Zealand Food Safety Science and Research Centre.
Author Contributions
D.A.W. designed the study. D.A.W., S.N., A.C.M., C.B., I.B., C.F.G., P.V., A.B., N.P.F., J.B. and K.M.B. conducted the study. D.A.W. and S.N. performed the analyses. D.A.W. and S.N. wrote the manuscript.
Data Availability
Assembled draft genomes were deposited to NCBI under BioProject number PRJNA473042.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37503-9.
References
- 1.Foster, T. Staphylococcus. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston Chapter 12 (1996). [PubMed]
- 2.Moodley A, Damborg P, Nielsen SS. Antimicrobial resistance in methicillin susceptible and methicillin resistant Staphylococcus pseudintermedius of canine origin: Literature review from 1980 to 2013. Veterinary Microbiology. 2014;171:337–341. doi: 10.1016/j.vetmic.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Lee J, et al. Improved Detection of Staphylococcus intermedius Group in a Routine Diagnostic Laboratory. Journal of Clinical Microbiology. 2015;53:961–963. doi: 10.1128/jcm.02474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devriese LA, et al. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. International Journal of Systematic and Evolutionary Microbiology. 2005;55:1569–1573. doi: 10.1099/ijs.0.63413-0. [DOI] [PubMed] [Google Scholar]
- 5.Somayaji R, Priyantha MAR, Rubin JE, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagnostic Microbiology and Infectious Disease. 2016;85:471–476. doi: 10.1016/j.diagmicrobio.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Robb AR, Wright ED, Foster AME, Walker R, Malone C. Skin infection caused by a novel strain of Staphylococcus pseudintermedius in a Siberian husky dog owner. JMM Case Reports. 2017;4:jmmcr005087. doi: 10.1099/jmmcr.0.005087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talan DA, Goldstein EJ, Staatz D, Overturf GD. Staphylococcus intermedius: clinical presentation of a new human dog bite pathogen. Ann Emerg Med. 1989;18:410–413. doi: 10.1016/S0196-0644(89)80582-7. [DOI] [PubMed] [Google Scholar]
- 8.Tanner MA, Everett CL, Youvan DC. Molecular phylogenetic evidence for noninvasive zoonotic transmission of Staphylococcus intermedius from a canine pet to a human. J Clin Microbiol. 2000;38:1628–1631. doi: 10.1128/jcm.38.4.1628-1631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neu HC. The Crisis in Antibiotic Resistance. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 10.Pires dos Santos, T., Damborg, P., Moodley, A. & Guardabassi, L. Systematic Review on Global Epidemiology of Methicillin-Resistant Staphylococcus pseudintermedius: Inference of Population Structure from Multilocus Sequence TypingData. Frontiers in Microbiology 7, 1599, doi:10.3389/fmicb.2016.01599 (2016). [DOI] [PMC free article] [PubMed]
- 11.Chatterjee SS, Otto M. Improved understanding of factors driving methicillin-resistant Staphylococcus aureus epidemic waves. Clinical Epidemiology. 2013;5:205–217. doi: 10.2147/CLEP.S37071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryan J, et al. Treatment outcome of dogs with meticillin-resistant and meticillin-susceptible Staphylococcus pseudintermedius pyoderma. Vet Dermatol. 2012;23:361–368, e365. doi: 10.1111/j.1365-3164.2012.01034.x. [DOI] [PubMed] [Google Scholar]
- 13.Morris DO, Loeffler A, Davis MF, Guardabassi L, Weese JS. Recommendations for approaches to meticillin-resistant staphylococcal infections of small animals: diagnosis, therapeutic considerations and preventative measures.: Clinical Consensus Guidelines of the World Association for Veterinary Dermatology. Vet Dermatol. 2017;28:304–e369. doi: 10.1111/vde.12444. [DOI] [PubMed] [Google Scholar]
- 14.Aidara-Kane A, et al. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob Resist Infect Control. 2018;7:7. doi: 10.1186/s13756-017-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tacconelli E, et al. Surveillance for control of antimicrobial resistance. The Lancet Infectious Diseases. 2018;18:e99–e106. doi: 10.1016/S1473-3099(17)30485-1. [DOI] [PubMed] [Google Scholar]
- 16.Bailey, K. Methicillin-resistant Staphylococcus pseudointermedius in New Zealand. Vol. 28 (New Zealand Veterinary Association, 2015).
- 17.Bell AG, Coombs GW, Cater B, Douglass C. First report of a mecA-positive multidrug-resistant Staphylococcus pseudintermedius isolated from a dog in New Zealand. N Z Vet J. 2016;64:253–256. doi: 10.1080/00480169.2016.1146171. [DOI] [PubMed] [Google Scholar]
- 18.Vestergaard, M. et al. Inhibition of the ATP Synthase Eliminates the Intrinsic Resistance of Staphylococcus aureus towards Polymyxins. mBio8, 10.1128/mBio.01114-17 (2017). [DOI] [PMC free article] [PubMed]
- 19.Skalka B. [Hyaluronidase test in the diagnosis of staphylococci] Vet Med (Praha) 1985;30:373–378. [PubMed] [Google Scholar]
- 20.Decristophoris P, Fasola A, Benagli C, Tonolla M, Petrini O. Identification of Staphylococcus intermedius Group by MALDI-TOF MS. Systematic and Applied Microbiology. 2011;34:45–51. doi: 10.1016/j.syapm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Murugaiyan J, et al. Species differentiation within the Staphylococcus intermedius group using a refined MALDI-TOF MS database. Clinical Microbiology and Infection. 2014;20:1007–1014. doi: 10.1111/1469-0691.12662. [DOI] [PubMed] [Google Scholar]
- 22.Silva MB, et al. An evaluation of matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the identification of Staphylococcus pseudintermedius isolates from canine infections. Journal of Veterinary Diagnostic Investigation. 2015;27:231–235. doi: 10.1177/1040638715573297. [DOI] [PubMed] [Google Scholar]
- 23.Prabhu K, Rao S, Rao V. Inducible Clindamycin Resistance in Staphylococcus aureus Isolated from Clinical Samples. J Lab Physicians. 2011;3:25–27. doi: 10.4103/0974-2727.78558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki T, et al. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. 2010;48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH. Methicillin (Oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 2003;69:6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seemann, T. et al. Nullarbor, https://github.com/tseemann/nullarbor.
- 29.Shen W, Le S, Li Y, Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS One. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolley KA. & Maiden, M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouckaert R, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thibeaux, R. et al. Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb Genom4, 10.1099/mgen.0.000144 (2018). [DOI] [PMC free article] [PubMed]
- 33.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddison, W. P. & Maddison, D. R. Mesquite: a modular system for evolutionary analysis. Version 3.40, http://mesquiteproject.org (2018).
- 35.Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal Complex Systems, 1695 (2006).
- 36.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu G, et al. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 39.Kahle DW. H. ggmap: Spatial Visualization withggplot2. The R Journal. 2013;5:144–161. [Google Scholar]
- 40.Gibb S. & Strimmer, K. MALDIquant: a versatile R package for the analysis of mass spectrometry data. Bioinformatics. 2012;28:2270–2271. doi: 10.1093/bioinformatics/bts447. [DOI] [PubMed] [Google Scholar]
- 41.Ahdesmaki M. & Strimmer, K. Feature selection in omics prediction problems using cat scores and false nondiscovery rate control. Ann. Appl. Stat. 2010;4:503–519. doi: 10.1214/09-AOAS277. [DOI] [Google Scholar]
- 42.crossval: Generic Functios for Cross Validation v. R package version 1. 0.3, https://CRAN.R-project.org/package=crossval (2015).
- 43.Hébert GA, Crowder CG, Hancock GA, Jarvis WR, Thornsberry C. Characteristics of coagulase-negative staphylococci that help differentiate these species and other members of the family Micrococcaceae. Journal of Clinical Microbiology. 1988;26:1939–1949. doi: 10.1128/jcm.26.10.1939-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, et al. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microbial Pathogenesis. 2016;101:56–67. doi: 10.1016/j.micpath.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 45.Munita, J. M. & Arias, C. A. Mechanisms of Antibiotic Resistance. Microbiology spectrum4, 10.1128/microbiolspec.VMBF-0016-2015 (2016). [DOI] [PMC free article] [PubMed]
- 46.Magiorakos, A. P. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection18, 268–281, 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed]
- 47.Cain CL. Antimicrobial resistance in staphylococci in small animals. Vet Clin North Am Small Anim Pract. 2013;43:19–40. doi: 10.1016/j.cvsm.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Weese JS, Sweetman K, Edson H, Rousseau J. Evaluation of minocycline susceptibility of methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol. 2013;162:968–971. doi: 10.1016/j.vetmic.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Loiacono M, et al. High-resolution melting analysis of gyrA codon 84 and grlA codon 80 mutations conferring resistance to fluoroquinolones in Staphylococcus pseudintermedius isolates from canine clinical samples. J Vet Diagn Invest. 2017;29:711–715. doi: 10.1177/1040638717712330. [DOI] [PubMed] [Google Scholar]
- 50.Nurjadi D, et al. Emergence of trimethoprim resistance gene dfrG in Staphylococcus aureus causing human infection and colonization in sub-Saharan Africa and its import to Europe. Journal of Antimicrobial Chemotherapy. 2014;69:2361–2368. doi: 10.1093/jac/dku174. [DOI] [PubMed] [Google Scholar]
- 51.Vickers AA, Potter NJ, Fishwick CWG, Chopra I, O’Neill AJ. Analysis of mutational resistance to trimethoprim in Staphylococcus aureus by genetic and structural modelling techniques. Journal of Antimicrobial Chemotherapy. 2009;63:1112–1117. doi: 10.1093/jac/dkp090. [DOI] [PubMed] [Google Scholar]
- 52.Gold RM, Lawhon SD. Incidence of Inducible Clindamycin Resistance in Staphylococcus pseudintermedius from Dogs. Journal of Clinical Microbiology. 2013;51:4196–4199. doi: 10.1128/jcm.02251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khosravi AD, Jenabi A, Montazeri EA. Distribution of genes encoding resistance to aminoglycoside modifying enzymes in methicillin-resistant Staphylococcus aureus (MRSA) strains. The Kaohsiung Journal of Medical Sciences. 2017;33:587–593. doi: 10.1016/j.kjms.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 54.van Belkum A, Welker M, Pincus D, Charrier J-P, Girard V. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Clinical Microbiology: What Are the Current Issues? Annals of Laboratory Medicine. 2017;37:475–483. doi: 10.3343/alm.2017.37.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abouelkhair, M. A., Thompson, R., Riley, M. C., Bemis, D. A. & Kania, S. A. Complete Genome Sequences of Three Staphylococcus pseudintermedius Strains Isolated from Botswana. Genome Announc6, 10.1128/genomeA.01599-17 (2018). [DOI] [PMC free article] [PubMed]
- 56.Worthing KA, et al. Clonal diversity and geographic distribution of methicillin-resistant Staphylococcus pseudintermedius from Australian animals: Discovery of novel sequence types. Vet Microbiol. 2018;213:58–65. doi: 10.1016/j.vetmic.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 57.Abouelkhair, M. A., Riley, M. C., Bemis, D. A. & Kania, S. A. Complete Genome Sequence of Staphylococcus pseudintermedius Type Strain LMG 22219. Genome Announc5, 10.1128/genomeA.01651-16 (2017). [DOI] [PMC free article] [PubMed]
- 58.Windahl U, Gren J, Holst BS, Borjesson S. Colonization with methicillin-resistant Staphylococcus pseudintermedius in multi-dog households: A longitudinal study using whole genome sequencing. Vet Microbiol. 2016;189:8–14. doi: 10.1016/j.vetmic.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 59.McCarthy AJ, et al. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. Journal of Antimicrobial Chemotherapy. 2015;70:997–1007. doi: 10.1093/jac/dku496. [DOI] [PubMed] [Google Scholar]
- 60.Pires Dos Santos T, Damborg P, Moodley A, Guardabassi L. Systematic Review on Global Epidemiology of Methicillin-Resistant Staphylococcus pseudintermedius: Inference of Population Structure from Multilocus Sequence Typing Data. Front Microbiol. 2016;7:1599. doi: 10.3389/fmicb.2016.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grönthal T, et al. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. Journal of Antimicrobial Chemotherapy. 2017;72:1021–1030. doi: 10.1093/jac/dkw559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kjellman EE, Slettemeas JS, Small H, Sunde M. Methicillin-resistant Staphylococcus pseudintermedius (MRSP) from healthy dogs in Norway - occurrence, genotypes and comparison to clinical MRSP. Microbiologyopen. 2015;4:857–866. doi: 10.1002/mbo3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gronthal T, et al. Epidemiology of methicillin resistant Staphylococcus pseudintermedius in guide dogs in Finland. Acta Vet Scand. 2015;57:37. doi: 10.1186/s13028-015-0129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:3926–3934. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carter, G. P. et al. Topical Antibiotic Use Coselects for the Carriage of Mobile Genetic Elements Conferring Resistance to Unrelated Antimicrobials in Staphylococcus aureus. Antimicrob Agents Chemother62, 10.1128/AAC.02000-17 (2018). [DOI] [PMC free article] [PubMed]
- 66.Lee CR, Cho IH, Jeong BC, Lee SH. Strategies to minimize antibiotic resistance. Int J Environ Res Public Health. 2013;10:4274–4305. doi: 10.3390/ijerph10094274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Assembled draft genomes were deposited to NCBI under BioProject number PRJNA473042.