Abstract
Cholera has affected Haiti with damping waves of outbreaks since October 2010. However, mechanisms behind disease persistence during lull periods remain poorly understood. By mid 2014, cholera transmission seemed to only persist in the northern part of Haiti. Meanwhile, cholera appeared nearly extinct in the capital, Port-au-Prince, where it eventually exploded in September 2014. This study aimed to determine whether this outbreak was caused by local undetected cases or by re-importation of the disease from the north. Applying an integrated approach between November 2013 and November 2014, we assessed the temporal and spatial dynamics of cholera using routine surveillance data and performed population genetics analyses of 178 Vibrio cholerae O1 clinical isolates. The results suggest that the northern part of the country exhibited a persisting metapopulation pattern with roaming oligoclonal outbreaks that could not be effectively controlled. Conversely, undetected and unaddressed autochthonous low-grade transmission persisted in the Port-au-Prince area, which may have been the source of the acute outbreak in late-2014. Cholera genotyping is a simple but powerful tool to adapt control strategies based on epidemic specificities. In Haiti, these data have already yielded significant progress in cholera surveillance, which is a key component of the strategy to eventually eliminate cholera.
Introduction
Since October 2010, Haiti continues to experience one of the most aggressive cholera epidemics recorded worldwide since the beginning of the seventh pandemic in 19611. More than 800,000 suspect cases and 9,000 associated deaths have been notified since the epidemic began2, although casualties may have been much higher than the numbers reported3,4. Following the initial nationwide epidemic explosion5,6, cases have progressively receded2. However, epidemic rebounds have recurrently affected the Port-au-Prince Metropolitan Area, the northern part of the country and, to a lesser degree, the southwestern tip of the country7,8. During epidemic intermissions, Haiti has experienced periods characterized by low levels of cholera incidence periods, as observed in 2014, when an unprecedented lull period in Haiti lasted for over six months2. Low-grade cholera transmission appeared limited to the northern part of the country, while the surveillance system recorded zero cases in the Southern Peninsula and only sporadic cases in the Port-au-Prince Metropolitan Area2. The lull eventually ended with an acute outbreak that erupted in the Port-au-Prince area in September 20142.
The underlying mechanisms of cholera outbreak recurrence after a cholera lull period have remained poorly understood at a local scale, and several hypotheses may thus be proposed to explain this outbreak in the Port-au-Prince Metropolitan Area. The outbreak may have re-emerged from a local persistence of cholera during the lull period. Conversely, the outbreak may have been caused by the re-importation of cholera from the northern part of the country. As each hypothesis implies a distinct strategy to control cholera during lull periods, understanding this prototypical cholera recurrence event in the Port-au-Prince Metropolitan Area may help to improve cholera elimination strategies worldwide.
Molecular epidemiology of Vibrio cholerae strains has contributed to the understanding of cholera dynamics in Asia9–21, Africa22–27, South America28,29 and Haiti. In the latter case, molecular analysis of clinical strains both confirmed the importation of cholera from Nepal in 201014,30 and demonstrated the subsequent clonal differentiation of these Vibrio cholerae O1 atypical El Tor strains31. On a larger scale, other studies have established a phylogenetic relationship between the epidemics of the current seventh pandemic32–37.
We therefore performed an integrated epidemiological and molecular epidemiology study of cholera in Haiti between November 2013 and November 2014. Briefly, routine cholera surveillance data were first analyzed to identify epidemic zones and periods. A subset of V. cholerae O1 clinical isolates collected in the country was then randomly selected for genotyping via MLVA (Multiple Loci VNTR [Variable Number of Tandem Repeats] Analysis). Multilocus genotypes (MLGs) were determined and the spatial and temporal distribution of isolate genotypes was analyzed to identify potential strain movement patterns and epidemiological links between cholera foci, with a particular focus on Port-au-Prince. Finally, V. cholerae populations corresponding to periods and zones were defined, and population genetics analysis, including fixation indices and assignment tests, was performed to test these links.
Methods
Cholera surveillance system in Haiti, and epidemiological data
In 2010, the Haitian Ministry of Public Health and Population (MSPP) established a cholera surveillance system based on the routine daily notification of all suspected cholera cases and associated deaths by up to 416 cholera treatment institutions. According to the WHO standard definition38, suspected cases were defined as a patient with acute watery diarrhea, with or without vomiting, and with or without dehydration. Cases and deaths were separately recorded for patients aged ≥5 and <5 years. The Directorate for Epidemiology Laboratory and Research compiled the number of suspected cholera cases and cholera deaths on a national Excel database at a daily and communal scale.
Routine bacteriological confirmation of suspected cholera cases was performed at the National Laboratory of Public Health (LNSP) using standard culture and phenotyping methods39. A total of 2,454 stool specimens were cultured between November 2013 and November 2014, out of which 30% were sampled via a sentinel surveillance network of four hospitals in three departments40. The remainder was sampled in other cholera treatment centers across the country by the laboratory technicians of each department and medical organizations.
In this study, anonymous MSPP databases of daily reported cases, deaths and stool culture results were used between November 2013 and November 2014. During this period, these data were completed by numerous field investigations performed by epidemiologists from the MSPP, Assistance Publique – Hôpitaux de Marseille (APHM) and UNICEF, in treatment institutions and communities, to confirm or identify cholera outbreaks and determine the underlying risk factors. Their findings were used to help interpret the results of the study.
Country and commune population estimates in 2014 were calculated using 2012 and 2015 population projections of the Haitian Institute of Statistics and Informatics41. Satellite estimates of daily-accumulated rainfall (area-averaged TRMM_3B42_daily v7) and mean daily temperature (mean_GLDAS_CLSM025_D_2_0_Tair_f_tavg) were extracted from NASA websites covering the entire surface of Haiti42.
Selection of V. cholerae O1 clinical isolates, DNA extraction and MLVA-based genotyping
V. cholerae O1 strains isolated at the LNSP were routinely stored at −20 °C in a dedicated biobank. Of the 1,220 isolates collected between November 2013 and November 2014, 275 strains were randomly selected from the laboratory database with commune- and month-specific probabilities calculated beforehand to maximize the spatial and temporal diversity of the sample panel. Among these 275 strains, 191 could be retrieved, successfully subcultured in Haiti and shipped to Marseille, France in Stock Culture Agar vials (Bio-Rad, Hercules, CA, USA) at room temperature. The strains were then recultivated on non-selective Tryptic Soy Agar medium (BD Diagnostic Systems, Heidelberg, Germany) for 24 hours at 37 °C. Suspected V. cholerae colonies were identified via Gram-staining, oxidase reaction and agglutination assessment with V. cholerae O1 polyvalent antisera (Bio-Rad).
For DNA isolation, an aliquot of approximately 50 cultured colonies was suspended in 500 μL NucliSENS easyMAG lysis buffer (bioMérieux, Marcy l’Etoile, France). Total nucleic acid was extracted from the V. cholerae culture samples using a NucliSENS easyMAG platform (bioMérieux) according to the manufacturer’s instructions. The supernatants (100 μL) were then stored at −20 °C for downstream applications43.
MLVA-based genotyping of the V. cholerae isolates was performed using six VNTRs summarized in S1 Appendix. Each VNTR locus was amplified separately. DNA amplification was carried out by preparing a PCR mix containing the following components: 0.375 μL of each primer (20 μM), 1 X LightCycler 480 Probes Master (Roche Diagnostics, Meylan, France) and approximately 100 ng of template DNA. The PCR mix was then brought to a total volume of 30 μL with sterile water. PCRs were performed using a LightCycler 480 System (Roche Diagnostics), with thermal cycling conditions as follows: 95 °C for 5 min; 30 cycles of 95 °C for 30 sec, 58 °C for 30 sec and 72 °C for 45 sec; and 72 °C for 5 min. Aliquots of the PCR products were diluted 1:30 in sterile water. Next, 1 μL of the diluted PCR reaction was aliquoted into a solution containing 25 μL Hi-Di Formamide 3500 Dx Series (Applied Biosystems, Foster City, CA, USA) and 0.5 μL GeneScan 500 LIZ Size Standard (Applied Biosystems). The fluorescent end-labeled PCR amplicons were then separated via capillary electrophoresis using an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) with POP-7 Polymer (Applied Biosystems). Finally, amplicon size was determined using GeneMapper v.3.0 software (Applied Biosystems), and the results were exported to Microsoft Excel for further analysis.
Temporal and spatial analysis of epidemiological data
Identification of the dry season between November 2013 and November 2014 was computed using temporal scan statistics in SaTScan v9.4.244. A purely temporal retrospective analysis of the daily Haiti-averaged rainfall time series was run with a discrete Poisson model in search for low-rate clusters. Similarly, subdivision of the study window into epidemic periods was computed using SaTScan by searching for low-rate clusters in the time series of daily numbers of suspected cholera cases recorded in Haiti.
As many of the 140 administrative communes lacked cholera treatment institutions and some patients may rather seek treatment outside their living commune, data from several neighboring communes were aggregated after interviewing local health actors and analyzing local reports. The weighted mean coordinates of the 117 merged communes were computed using QGIS v3.0.345, a shapefile of communal administrative boundaries46, and a high resolution raster of the population from the WorldPop project47. Mapping of data was performed using QGIS v3.0.3 and shapefiles of administrative boundaries and roads provided by CNIGS46. Spatial cluster analysis of suspected cholera case distribution for each epidemic period was performed using SaTScan44 with a discrete Poisson model. The Port-au-Prince Metropolitan Area hosts over one-third of the Haitian population, and it is the single major communication passage between the northern part of the country and the southern Peninsula. As the main study objective aimed to decipher the origin of the cholera outbreak that hit the capital in September 2014, the 117 merged communes were therefore grouped into three zones before the analyses were carried out: the PaP zone (corresponding to the communes of the Port-au-Prince Metropolitan Area); the North zone (grouping the northern and eastern communes of Ouest Department as well as the communes of the Artibonite, Centre, Nord-Est, Nord and Nord-Ouest Departments); and the South zone (grouping the western communes of Ouest Department as well as the communes of the Sud-Est, Nippes, Sud and Grand’Anse Departments).
MLVA result analysis
Using VNTR allele sizes, MLGs were identified using excel. A network of MLGs with single-locus variants was assembled using R version 3.2.1 for Mac48, and the igraph package. The temporal and spatial distribution of MLGs were illustrated using Excel and QGIS, respectively.
The genetic diversity of the V. cholerae O1 populations defined from the identified epidemic periods and the three geographic zones was estimated using the Nei’s index (He)49, calculated from allelic frequencies with R and the poppr package. The genotypic diversity (Gd), which corresponds to the probability of drawing two different MLGs in a population, was calculated from genotypic frequencies. Genetic differentiation between populations was assessed using the Wright’s FST fixation index50. This was estimated by the Weir & Cockerham θ51, which is based on within- and among-population variance components and was computed in FSTAT version 2.9.452. Genetic assignment probability of isolates to previous populations was calculated using the Rannala & Mountain Bayesian algorithm53 on GENECLASS2 version 2.054.
The MLG distribution and population genetics analysis findings were confirmed by additional analyses provided as supporting information: comparison between FST and other genetic differentiation indices (S5 Appendix); sensitivity analysis of FST using a different spatial and temporal aggregation of isolates (S6 Appendix); multiple component analysis and hierarchical classification of MLVA results (S7 Appendix); multiple linear regression analysis of genetic, spatial and temporal distances between isolates (S8 Appendix); and Bayesian clustering for spatial population genetics (S9 Appendix).
Ethical statement
All materials used in the study, including clinical V. cholerae strains, were collected by the MSPP during routine surveillance activities to control and prevent cholera. Stool specimens had been obtained from cholera patients with patient and/or legal guardian informed consent. The data were analyzed anonymously with the authorization of the MSPP and in close collaboration with MSPP epidemiologists and biologists. The protocol received authorization from the MSPP National Bioethics Comity (authorization #1415-52). All research was performed in accordance with relevant regulations.
Results
Cholera dynamics during the study period
The study period covered 395 days from November 1, 2013 to November 30, 2014. Based on a temporal cluster analysis of daily accumulated rainfall over Haiti, a significant dry season lasted from November 6, 2013 to March 29, 2014 (Fig. 1), which was the longest dry spell since the beginning of the epidemic (see S2 Appendix for details). Overall, 33,428 suspected cases were recorded by the cholera surveillance system, which corresponded to an average of 2.83 cases/1000 person-years. Forty-nine percent (95%-confidence interval, 47–51) of the 2,415 collected stool samples yielded V. cholerae O1 growth upon culture. Specific characteristics of the three spatial zones of the country, North, PaP and South, are summarized in S3 Appendix.
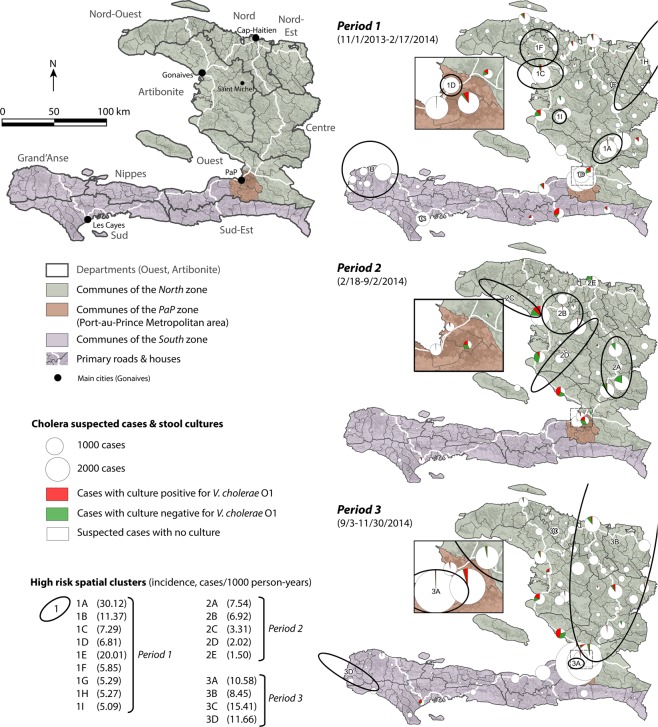
Figure 1.
Daily evolution of cholera in Haiti between November 2013 and November 2014: (Panel A) averaged daily accumulated rainfall in Haiti and derived dry season (see S2 Appendix for details); (Panel B) daily number of suspected cholera cases in the North, PaP (Port-au-Prince) and South zones as well as derived epidemic periods identified by the temporal cluster analysis; and (Panel C) daily number of stool samples for culture confirmation of cholera and monthly culture positivity ratio. Period P1 covers the late 2013 incidence peak and the abrupt decrease in cases during the 2013–2014 dry season; Period P2 covers a lull phase that extended through the end of the dry season (P2D) and the first months of the rainy season (P2R); Period P3 represents the intense epidemic recurrence that started in September 2014.
Temporal cluster analysis of daily suspected cases revealed three distinct epidemic periods (Fig. 1, S4 Appendix). Period P1 (11/1/2013-2/17/2014) included the end of the late-2013 incidence peak and the abrupt decrease in cases during the 2013–2014 dry season. An average of 3.81 cases/1000 person-years was recorded, and the culture positivity ratio was 58% (54–62). Cases were clustered in the North zone (in the Artibonite, Centre and Nord-Est Departments), the PaP zone (Port-au-Prince Metropolitan Area) and the western tip of the South zone (Grand’Anse Department) (Fig. 2), although cases rapidly receded from most foci.
Figure 2.
Situation map and communal distribution of cholera in Haiti between November 2013 and November 2014: number of suspected cholera cases; proportion of stool sampling and culture results; and high-risk spatial clusters. The situation map shows the three zones of the study in green, brown and purple, the 10 departments, primary roads and main cities and towns cited in the text. For each of the three periods identified by the temporal cluster analysis (Fig. 1), pie charts represent the number of suspected cholera cases notified by each commune grouped into culture-positive cases, culture-negative cases and non-sampled cases. Black ellipses exhibit significant high-risk spatial clusters for each period.
Period P2 (2/18-9/2/2014) was a lull phase, which extended over the last 40 days of the dry season (P2D) and the first five months of the 2014 rainy season (P2R). Only 1.08 cases/1000 person-years were recorded, with no significant difference between the dry and rainy sub-periods (Fig. 1 and S4 Table). The culture positivity ratio during period P2 was only 34% (31–37), although it represented a significant increase from a markedly low 19% (11–27) during the dry sub-period P2D to 35% (32–39) after the return of the rainy season. Cases primarily retracted in residual foci in Artibonite Department (notably Gonaives, the most affected commune, and Saint Michel) and Centre Department (e.g., Hinche, Mirebalais and Lascahobas) (Fig. 1 and Fig. 2). Cholera was apparently extinct in the South zone. The PaP zone recorded only 572 suspected cases (0.39 cases/1000 person-years) during period P2, and 41% of the 87 cultures tested positive for V. cholerae O1 during period P2 (Figs 1, 2 and S3 Table).
Period P3 (9/3-11/30/2014) corresponded to an intense epidemic recurrence with 5.70 cases/1000 person-years, which started in early September during the rainy season and peaked at over 300 cases per day on November 5, 2014. The positive culture ratio increased to 59% (55–63) during this period. Many cases clustered in a vast area of the North zone and the western end of the South zone (Fig. 2). Most importantly, nearly half of the 14,923 suspected cases recorded significantly clustered in the PaP zone (Fig. 1), with 10.11 cases/1000 person-years. In the PaP zone, the epidemic exploded in the Martissant region after case numbers had increased in Corail and Cité Soleil for nearly two months with no epidemiological surveillance. However, the origin of this epidemic recurrence remained unknown.
Stool sampling and MLVA results
During the 13-month study period, stool cultures for cholera confirmation were performed on 7% of suspected cases throughout the country (S4 Appendix). Significantly more effort was focused on culture confirmation during the lull period P2 than during periods P1 and P3. However, this disproportion was corrected in our genotype analysis, as the number of complete MLGs was not significantly different between the three periods (S4 Appendix). Of the 275 randomly selected V. cholerae clinical isolates, 177 isolates were successfully genotyped via MLVA, including three isolates with mixed infections by more than one V. cholerae O1 clone. Overall, 178 complete MLGs were reconstructed.
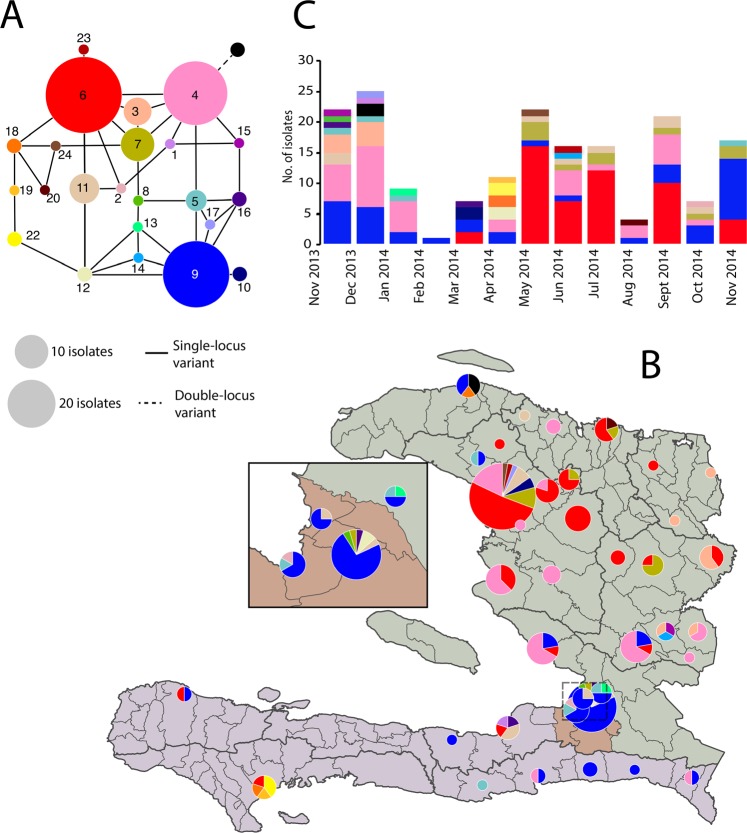
According to the results, the VC9 microsatellite locus was monoallelic, while the VC1, VC4, VC5, LAV6 and VCMS12 assays revealed two, five, two, six and three allelic variants, respectively. The observed genetic diversity (Nei’s index) of the entire population was 0.273. MLVA identified 24 closely related MLGs: all but one formed a network of single-locus variants (Fig. 3 Panel A). Three frequent MLGs – #4 (pink), #6 (red) and #9 (blue) – were identified in 126 (71%) of 178 isolates.
Figure 3.
MLVA-based multilocus genotypes (MLGs) of V. cholerae O1 clinical isolates in Haiti between November 2013 and November 2014: (Panel A) clonal complex, (Panel B) spatial distribution and (Panel C) temporal distribution. Each color represents a unique MLG. The size of the nodes in the network (Panel A), the size of pie-charts on the map (Panel B) and the height of stacked histograms on the graph (Panel C) are proportional to the number of isolates. Solid edges in the network (Panel A) represent single-locus variants between two MLGs (i.e., allele difference at one of the six loci), and dashed edges double-locus variants (i.e., allele difference at two of the six loci).
Spatial and temporal distribution of multilocus genotypes
MLGs #9 and #4 were frequently isolated throughout the study period, while other major MLGs (such as MLG #3) were only detected during the initial months. MLG #6 and MLG #7, two single-locus variants of MLG #4, appeared later during the study period (Fig. 3, Panel C). MLG #5 seemed to temporally disappear during period P2. Several other MLGs (e.g., #1, #2, #8, #13, #14 and #15) that were closely related to the main MLGs were only isolated once during the study (Fig. 3, Panels B and C). MLG distribution also exhibited a strong spatial pattern: 34 of the 36 MLG #4 isolates, 41/51 of MLG #6, 7/7 of MLG #3 and 9/10 of MLG #7 were located in the North zone, while 23 of the 39 MLG #9 were isolated in the PaP zone (Fig. 3, Panel B). MLGs #19 and #22 were only isolated in the South zone.
Consequently, the most frequently detected genotypes in the North zone were MLGs #4, #6, #3 and #7. MLG #6 likely emerged from MLG #4 in Gonaives during the dry season. It successfully multiplied locally during the second part of period P2, then expanded in Artibonite Department (notably Saint Michel) to the rest of the North zone and the South zone (Table 1 and Fig. 3). Similarly, MLG #7 likely emerged in Gonaives from MLG #6 or MLG #4 during period P2 before spreading to the nearby regions.
Table 1.
Genetic diversity (Nei’s He index) and differentiation (fixation index FST) of V. cholerae O1 populations in Haiti between November 2013 and November 2014.
| Period-based populations | No. isolates | Diversity | FST | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gd | He | P2 | P3 | ||||||||
| P1 | 57 | 0.763 | 0.242 | 0.213* | 0.061* | ||||||
| P2 | 79 | 0.731 | 0.270 | 0.084* | |||||||
| P3 | 42 | 0.735 | 0.238 | ||||||||
| Zone-based populations | No. isolates | Diversity | FST | ||||||||
| Gd | He | PaP | South | ||||||||
| North | 125 | 0.763 | 0.208 | 0.499* | 0.220* | ||||||
| PaP | 32 | 0.471 | 0.148 | 0.150* | |||||||
| South | 21 | 0.830 | 0.326 | ||||||||
| Period and zone-based populations | No. isolates | Diversity | FST | ||||||||
| Gd | He | North2 | North3 | PaP1 | PaP2 | PaP3 | South1 | South2 | South3 | ||
| North1 | 38 | 0.691 | 0.208 | 0.304* | 0.149* | 0.421* | 0.489* | 0.336 | 0.202* | 0.522* | 0.095 |
| North2 | 64 | 0.623 | 0.152 | 0.028 | 0.671* | 0.689* | 0.577* | 0.505* | 0.505* | 0.002 | |
| North3 | 23 | 0.691 | 0.212 | 0.507* | 0.552* | 0.394 | 0.296 | 0.399 | −0.169 | ||
| PaP1 | 6 | 0.278 | 0.111 | −0.042 | −0.027 | 0.049 | 0.604 | 0.431 | |||
| PaP2 | 10 | 0.460 | 0.093 | 0.009 | 0.110 | 0.649* | 0.516 | ||||
| PaP3 | 16 | 0.500 | 0.194 | −0.028 | 0.510 | 0.236 | |||||
| South1 | 13 | 0.722 | 0.233 | 0.464* | 0.108 | ||||||
| South2 | 5 | 0.720 | 0.267 | 0.174 | |||||||
| South3 | 3 | 0.444 | 0.333 | ||||||||
P1, P2 and P3 correspond to the V. cholerae O1 populations of the three periods. North1 represents the population from the North zone during P1, etc.
Gd, genotypic diversity; He, Nei’s diversity index; FST, Weir & Cockerham fixation index.
* significant FST.
By contrast, the most frequent genotype in the PaP zone remained MLG #9: 5 of 18 isolates during period P1, 7 of 18 isolates during P2, and 11 of 18 isolates sampled during the epidemic of period P3 (Table 1 and Fig. 3). While MLG #9 was present in the three zones during period P1, it appeared to retract in the PaP zone during the lull period P2, before expanding again to the North and South zones during P3 (Table 1 and Fig. 3).
Genetic diversity and structure of V. cholerae populations
Population genetics statistical analyses based on allele frequencies showed that the diversity of V. cholerae O1 isolates remained stable between November 2013 and November 2014 (Table 1), even during the lull period P2, which interestingly did not exhibit any decrease in genetic diversity (Table 1). However, the differentiation index FST showed a marked temporal structure between P1 and P2 (Table 1). Spatially, the genetic diversity of V. cholerae clinical isolates appeared greater in the North and South zones than in the PaP zone (Table 1). Additionally, PaP isolate populations exhibited a strong genetic differentiation with the North population (FST index = 0.499, Table 1).
Combining the three periods and the three zones, high genetic differentiation was observed between the North1 (isolates from period P1 in the North zone) and North2 populations, while FST was low between North2 and North3. FST indexes remained very high between the North and PaP zones throughout the study period. Conversely, no significant genetic differentiation was observed between PaP1, PaP2 and PaP3. Although too few isolates were collected in the South zone to perform significant population genetics comparisons, South2 appeared differentiated from all other populations.
FST appeared highly correlated with three other genetic differentiation indices: D (Host), GST (Nei) and GST (Hedrick) (S5 Appendix).
Assignment of V. cholerae isolates during epidemic recurrence in Port-au-Prince
The origin of the outbreak that started in September 2014 was assessed using assignment analysis of isolates collected during period P3. Most of the 16 isolates sampled in the PaP zone during the period P3 (PaP3 isolates) were preferentially assigned to the previous local populations PaP1 and PaP2 (Fig. 4, Panel B). Conversely, probability of assignment to the previous populations North1 and North2 was very low (Fig. 4, Panel B). Additionally, no major difference was observed in the assignment probabilities of PaP3 isolates between the populations PaP1 and PaP2. Assignment of PaP3 isolates to the South1 population was strong, while it was negligible to the South2 population.
Figure 4.
Assignment probabilities of North3 (Panel A) and PaP3 (Panel B) isolates to previous populations. Dots represent the assignment probability of each isolate of the analyzed population to each of the other populations. Black bars represent the average assignment probability of the analyzed population to the other populations, with standard errors.
Assignment of North3 isolates was higher with the North2 population, although some isolates probably originated from the PaP2 population. The South3 population comprised only three isolates that were likely imported from the North2 or PaP2 populations.
Discussion
Cholera incidence in Haiti exhibited a dramatic reduction during the very dry period of late 2013. During several months of lull period, the disease appeared nearly extinct, especially in the South zone and in the Port-au-Prince area where only a few sporadic confirmed cases occurred. Following the return of rainfall in April 2014, the national positivity ratio of stool culture for V. cholerae O1 gradually increased, before the epidemic resurged in September mainly in the Port-au-Prince Metropolitan Area.
Our genotyping results confirm the intense diversification of the V. cholerae O1 clone that was introduced in Haiti in 201031,55. The temporospatial distribution of genotypes (MLG) suggests two different mechanisms of cholera persistence in the northern part of the country and the Port-au-Prince Metropolitan Area. The North zone appeared affected by oligoclonal outbreaks of various genotypes, which either rapidly vanished or successfully persisted over time, exhibiting a metapopulation pattern (i.e., a group of connected local populations). Despite the low incidence in this area, genotypic diversity was significant and represented a source of cholera spread across the country, as previously described56. Conversely, cholera in the PaP zone showed little genetic diversification. A few autochthonous genotypes persisted during the marked lull period, before causing the subsequent epidemic peak in September 2014. This acute outbreak in PaP was thus likely not re-imported from persisting foci in the North zone. Instead, it rather likely re-emerged locally, by a mechanism that our study was not designed to determine more precisely. Some authors have stated the existence of established environmental reservoirs of cholera in Haiti55,57–59. However, isolation of toxigenic V. cholerae O1 in Haitian surface waters has remained sporadic55,57–62, and usually concomitant to local cholera cases, persisting rainfall and widespread open defecation and laundry in the rivers63. Such conditions render it difficult to determine whether V. cholerae O1–positive surface waters constitute perennial reservoirs at the source of cholera outbreaks or only transient vectors of cholera following contamination by nearby infected patients57. The outbreak in PaP could also have come out from a low-grade persistent interhuman transmission of the disease. Indeed, considering the number of suspected cholera cases recorded by the MSPP during the lull period P2 in the PaP zone, the likelihood of additional suspected cases undetected by the surveillance system, and the concomitant positivity ratio of stool cultures for V. cholera O1, the number of “true” symptomatic cholera patients may theoretically have been sufficient to sustain continuous chains of interhuman and peridomestic transmission (S3 Appendix).
As suggested by a previous study, MLVA results may have been marginally influenced by the sub-culturing process64. In addition, our population genetics analysis was limited by the partial sampling of cholera isolates. Indeed, many communes never sent stool specimens for cholera culture confirmation, mainly for logistic reasons, as was the case in Grand’Anse Department. Furthermore, strain availability in the LNSP biobank also reflected the cholera dynamics: samples were very scarce in the South zone during Period P2 because suspected cases were sporadic. This may explain why our results did not exhibit any significant bottleneck during the lull period55. We tried to optimize the representativeness of our strain panel by randomly selecting isolates in a way that maximized the spatial and temporal range, so that only 9% of our isolates originated from the four sentinel sites. Additionally, FST and assignment probabilities were also partly biased by the fact that the large outbreak in Port-au-Prince was oligoclonal. However, we did not expect that the PaP3 population would be almost exclusively composed of genotypes already present in PaP2 but not in the North2 population. Although we cannot be sure that these genotypes did not circulate outside PaP during Period P2 before returning to cause the outbreak during Period P3, our analyses found this to be a less probable scenario than local persistence of low-grade transmission cases. Finally, our results may have been influenced by the partitioning of our isolate population. The spatial division in PaP, North and South was determined by the theoretical objective of the study (which was to assess the origin of the late 2014 outbreak in the Port-au-Prince Metropolitan Area) and by the fact that the capital is a required crossroad between the northern part of the country and the Southern Peninsula. The temporal division was obtained by cluster analysis. We thus conducted a sensitivity FST analysis using an alternative time and space aggregation of isolates according to their sampling trimester and department, which exhibited the same structure pattern (S6 Appendix). We also performed a multiple component analysis and hierarchical classification of MLVA results; this grouped the isolates into three genetic clusters, for which distribution confirmed the marked genetic differentiation between the North and PaP zones (S7 Appendix). A similar pattern was obtained when analyzing the relationship between genetic, spatial and temporal distances between isolates using multiple linear regression (S8 Appendix). Finally, a Bayesian clustering algorithm for spatial population genetics without assuming predefined populations exhibited a remarkable match with our chosen populations (S9 Appendix).
Although such biases have been a concern in most of the retrospective molecular epidemiology studies dealing with cholera in resource-constrained countries, such challenges did not prevent researchers to draw definite evidence concerning the origin of other cholera epidemics as well as the overall dynamics of the seventh cholera pandemic33,36,37. Whole genome sequencing (WGS)-based phylogenic analysis is a powerful tool that could help minimize such sampling biases. However, MLVA has been shown to reflect the same genetic relatedness of isolates as WGS in several cholera studies65,66 and at a much lower cost. Additionally, due to the slow mutation rate of the core genome of V. cholerae33, WGS may not have provided a sufficient resolution over the short period and narrow space of our study.
Despite these limitations, we believe that the persistence of cholera in the northern part of Haiti and the Port-au-Prince Metropolitan Area during the 2014 lull period may be attributed to two distinct weaknesses in the cholera elimination strategy67 and the nationwide cholera rapid-response strategy that was implemented in mid-201368. In the North zone, outbreaks were well detected but not effectively controlled. Response teams in charge of targeting cholera-affected communities with awareness campaigns, soap distribution and water chlorination had only been implemented from July 2013 onward. Response interventions remained slow and few in number during this period68. By contrast, epidemiological surveillance in the PaP zone appeared deficient, as gradually increasing local transmission remained undetected and unaddressed, especially in unsafe neighborhoods, such as Cité-Soleil, or in rapidly expanding and disorganized slums in the north of the Port-au-Prince area, such as Corail (authors personal data from field investigations). Following the epidemic recurrence of September 2014, cholera spread towards the Southern Peninsula and the northern departments.
Remarkably, cholera incidence remained low during five months after the onset of the 2014 rainy season. Although our study was not designed to analyze mechanisms underlying this extended lull, we believe this may be due to the combination of several factors: (1) a low cholera incidence at the end of the 2013–2014 dry season, (2) a relatively dryer rainy season in 2014 compared to previous years (S2 Appendix); and (3) the implementation of the nationwide cholera rapid-response strategy68. Although this response strategy was not able to eliminate cholera during the 2014 lull period, we believe it may have mitigated the effect of rainfall on transmission during a few months, until the outbreak finally exploded in the Port-au-Prince.
Our results remain relevant today and highlight the necessity to improve both the quality of epidemiological cholera surveillance69 and the effectiveness of the struggle to limit cholera transmission in Haiti. Four years later, suspected cholera cases records have become more exhaustive in Haitian treatment institutions, even though cholera surveillance has still not been extended to the community level, as planned by the 2016–2018 mid-term development of the national plan for cholera elimination by the Haitian government70. Thanks to important logistical efforts, stool sampling between January and August 2018 has been performed for 81% of the 2,961 reported suspected cholera cases, and culture results are becoming more often considered to better target active transmission foci and prevent cholera dissemination. In the current context of a new remarkable lull in cholera transmission in Haiti2, this progress in surveillance is becoming critical in order to promptly identify and target every single and last case, to eventually reach the goal of cholera elimination in Haiti by the planned deadline of 202267.
Supplementary information
Acknowledgements
The authors would like to thank all staff who took part in routine patient care; data reporting and compilation; as well as sample collection, transport, culture and analysis. We are especially grateful to the PRESEPI team of the National Public Health Laboratory in Haiti for collecting cholera clinical samples.
Author Contributions
E.R., M.J.L., P.A. and J.B. provided cholera isolates and epidemiological data. S.M. and A.C.N. performed the genotyping of V. cholerae isolates. S.R., J.G. and H.B. performed the statistical and population genetics analyses. S.R. and R.P. wrote the main manuscript text. S.R. prepared all figures and tables. All authors reviewed the manuscript.
Data Availability
All data generated or analyzed during this study are included in this published article (and in Supplementary Dataset file).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37706-0.
References
- 1.World Health Organization. Cholera [every year since1968]. Wkly Epidemiol Rec.
- 2.Republic of Haiti. Ministry of Public Health and Population. Centre de Documentation. Bulletins choléra 2010-2018 (French). http://mspp.gouv.ht/newsite/documentation.php (2018).
- 3.Allan M, et al. High-resolution spatial analysis of cholera patients reported in Artibonite department, Haiti in 2010–2011. Epidemics. 2016;14:1–10. doi: 10.1016/j.epidem.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Luquero FJ, et al. Mortality Rates during Cholera Epidemic, Haiti, 2010-2011. Emerging Infect. Dis. 2016;22:410–416. doi: 10.3201/eid2203.141970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piarroux R, et al. Understanding the cholera epidemic, Haiti. Emerg Infect Dis. 2011;17:1161–1168. doi: 10.3201/eid1707.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudart J, et al. Spatio-Temporal Dynamics of Cholera during the First Year of the Epidemic in Haiti. PLoS Negl Trop Dis. 2013;7:e2145. doi: 10.1371/journal.pntd.0002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barzilay EJ, et al. Cholera Surveillance during the Haiti Epidemic - The First 2 Years. N. Engl. J. Med. 2013;368:599–609. doi: 10.1056/NEJMoa1204927. [DOI] [PubMed] [Google Scholar]
- 8.Rebaudet, S. et al. The dry season in Haiti: a window of opportunity to eliminate cholera. PLoS Curr Outbreaks5 (2013). [DOI] [PMC free article] [PubMed]
- 9.Stine OC, et al. Seasonal cholera from multiple small outbreaks, rural Bangladesh. Emerg Infect Dis. 2008;14:831–833. doi: 10.3201/eid1405.071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashed SM, et al. Genetic Variation of Vibrio cholerae during Outbreaks, Bangladesh, 2010-2011. Emerging Infect. Dis. 2014;20:54–60. doi: 10.3201/eid2001.130796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh R, et al. Epidemiological study of Vibrio cholerae using variable number of tandem repeats. FEMS Microbiol. Lett. 2008;288:196–201. doi: 10.1111/j.1574-6968.2008.01352.x. [DOI] [PubMed] [Google Scholar]
- 12.Abd El Ghany M, et al. The Population Structure of Vibrio cholerae from the Chandigarh Region of Northern India. PLoS Negl Trop Dis. 2014;8:e2981. doi: 10.1371/journal.pntd.0002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh R, et al. Phenotypic and Genetic Heterogeneity in Vibrio cholerae O139 Isolated from Cholera Cases in Delhi, India during 2001-2006. Front Microbiol. 2016;7:1250. doi: 10.3389/fmicb.2016.01250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendriksen RS, et al. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. MBio. 2011;2:e00157–00111. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit SM, et al. Cholera outbreaks (2012) in three districts of Nepal reveal clonal transmission of multi-drug resistant Vibrio cholerae O1. BMC Infect. Dis. 2014;14:392. doi: 10.1186/1471-2334-14-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah MA, et al. Genomic Epidemiology of Vibrio cholerae O1 Associated with Floods, Pakistan, 2010. Emerging Infect. Dis. 2014;20:13–20. doi: 10.3201/eid2001.130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, et al. A molecular surveillance reveals the prevalence of Vibrio cholerae O139 isolates in China from 1993 to 2012. J. Clin. Microbiol. 2014;52:1146–1152. doi: 10.1128/JCM.03354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou H, et al. Population structural analysis of O1 El Tor Vibrio cholerae isolated in China among the seventh cholera pandemic on the basis of multilocus sequence typing and virulence gene profiles. Infect. Genet. Evol. 2014;22:72–80. doi: 10.1016/j.meegid.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Okada K, Roobthaisong A, Nakagawa I, Hamada S, Chantaroj S. Genotypic and PFGE/MLVA analyses of Vibrio cholerae O1: geographical spread and temporal changes during the 2007-2010 cholera outbreaks in Thailand. PLoS ONE. 2012;7:e30863. doi: 10.1371/journal.pone.0030863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teh CSJ, et al. Outbreak-associated Vibrio cholerae Genotypes with Identical Pulsotypes, Malaysia, 2009. Emerg Infect Dis. 2012;18:1177–1179. doi: 10.3201/eid1807.111656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwood PF, et al. Clonal Origins of Vibrio cholerae O1 El Tor Strains, Papua New Guinea, 2009-2011. Emerg Infect Dis. 2011;17:2063–2065. doi: 10.3201/eid1711.110782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohamed AA, et al. Molecular Epidemiology of Geographically Dispersed Vibrio cholerae, Kenya, January 2009-May 2010. Emerg Infect Dis. 2012;18:925–931. doi: 10.3201/eid1806.111774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiiru J, et al. A Study on the Geophylogeny of Clinical and Environmental Vibrio cholerae in Kenya. PLoS ONE. 2013;8:e74829. doi: 10.1371/journal.pone.0074829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valia R, et al. Vibrio cholerae O1 epidemic variants in Angola: a retrospective study between 1992 and 2006. Front Microbiol. 2013;4:354. doi: 10.3389/fmicb.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngwa, M. C. et al. Genetic Studies of Vibrio cholerae in South West Cameroon-A Phylogenetic Analysis of Isolates from the 2010–2011 Epidemic. PLoS Curr8 (2016). [DOI] [PMC free article] [PubMed]
- 26.Kaas RS, et al. The Lake Chad Basin, an Isolated and Persistent Reservoir of Vibrio cholerae O1: A Genomic Insight into the Outbreak in Cameroon, 2010. PLOS ONE. 2016;11:e0155691. doi: 10.1371/journal.pone.0155691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebaudet S, et al. Deciphering the Origin of the 2012 Cholera Epidemic in Guinea by Integrating Epidemiological and Molecular Analyses. PLoS Negl Trop Dis. 2014;8:e2898. doi: 10.1371/journal.pntd.0002898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beltrán P, et al. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999;37:581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam C, Octavia S, Reeves P, Wang L, Lan R. Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America. Emerg Infect Dis. 2010;16:1130–1132. doi: 10.3201/eid1607.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orata FD, Keim PS, Boucher Y. The 2010 Cholera Outbreak in Haiti: How Science Solved a Controversy. PLoS Pathog. 2014;10:e1003967. doi: 10.1371/journal.ppat.1003967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz LS, et al. Evolutionary Dynamics of Vibrio cholerae O1 following a Single-Source Introduction to Haiti. mBio. 2013;4:e00398–13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan R, Reeves PR. Pandemic spread of cholera: genetic diversity and relationships within the seventh pandemic clone of Vibrio cholerae determined by amplified fragment length polymorphism. J. Clin. Microbiol. 2002;40:172–181. doi: 10.1128/JCM.40.1.172-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam C, Octavia S, Reeves PR, Lan R. Multi-locus variable number tandem repeat analysis of 7th pandemic Vibrio cholerae. BMC Microbiol. 2012;12:82. doi: 10.1186/1471-2180-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robins, W. P. & Mekalanos, J. J. Genomic Science in Understanding Cholera Outbreaks and Evolution of Vibrio cholerae as a Human Pathogen. Curr. Top. Microbiol. Immunol. 10.1007/82_2014_366 (2014). [DOI] [PMC free article] [PubMed]
- 36.Weill F-X, et al. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017;358:785–789. doi: 10.1126/science.aad5901. [DOI] [PubMed] [Google Scholar]
- 37.Domman D, et al. Integrated view of Vibrio cholerae in the Americas. Science. 2017;358:789–793. doi: 10.1126/science.aao2136. [DOI] [PubMed] [Google Scholar]
- 38.Global Task Force on Cholera Control. Cholera outbreak: assessing the outbreak response and improving preparedness. 87 (World Health Organization (WHO), 2010).
- 39.Centers for Disease Control and Prevention (CDC). Laboratory Methods for the Diagnosis of Epidemic Dysentery and Cholera. 108 (Centers for Disease Control and Prevention (CDC), 1999).
- 40.Steenland MW, et al. Laboratory-Confirmed Cholera and Rotavirus among Patients with Acute Diarrhea in Four Hospitals in Haiti, 2012–2013. Am J Trop Med Hyg. 2013;89:641–646. doi: 10.4269/ajtmh.13-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Institut Haitien de Statistique et d’Informatique (IHSI). Statistique Démographiques et Sociales. http://www.ihsi.ht/produit_demo_soc.htm (2018).
- 42.NASA GES DISC. Giovanni online data system. https://giovanni.gsfc.nasa.gov/giovanni/ (2018).
- 43.Moore S, et al. Relationship between Distinct African Cholera Epidemics Revealed via MLVA Haplotyping of 337 Vibrio cholerae Isolates. PLoS neglected tropical diseases. 2015;9:e0003817. doi: 10.1371/journal.pntd.0003817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulldorff, M. & Information Management Services Inc. SaTScan - Software for the spatial, temporal, and space-time scan statistics. https://www.satscan.org/ (2018).
- 45.Open Source Geospatial Foundation Project. Système d’Information Géographique QGIS. http://www.qgis.org/fr/site/ (2018).
- 46.Centre National de l’Information Géospatiale (CNIGS). HaitiData. http://haitidata.org/layers/ (2018)
- 47.Worldpop. Haiti 100m Population. http://www.worldpop.org.uk/ (2018).
- 48.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/ (2018).
- 49.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution. 1965;19:395–420. doi: 10.1111/j.1558-5646.1965.tb01731.x. [DOI] [Google Scholar]
- 51.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 52.Goudet, J. FSTAT. https://www2.unil.ch/popgen/softwares/fstat.htm (2018).
- 53.Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piry, S. et al. GeneClass2: A Software for Genetic Assignment and First-Generation Migrant Detection. http://www1.montpellier.inra.fr/CBGP/software/GeneClass/ (2018) [DOI] [PubMed]
- 55.Azarian, T. et al. Phylodynamic Analysis of Clinical and Environmental Vibrio cholerae Isolates from Haiti Reveals Diversification Driven by Positive Selection. MBio5 (2014). [DOI] [PMC free article] [PubMed]
- 56.Stine, O. C. & Morris, J. G. Circulation and Transmission of Clones of Vibrio Cholerae During Cholera Outbreaks. Curr. Top. Microbiol. Immunol. 10.1007/82_2013_360 (2014). [DOI] [PMC free article] [PubMed]
- 57.Alam MT, et al. Increased Isolation Frequency of Toxigenic Vibrio cholerae O1 from Environmental Monitoring Sites in Haiti. PLoS ONE. 2015;10:e0124098. doi: 10.1371/journal.pone.0124098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kahler AM, et al. Environmental surveillance for toxigenic Vibrio cholerae in surface waters of Haiti. Am. J. Trop. Med. Hyg. 2015;92:118–125. doi: 10.4269/ajtmh.13-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alam MT, et al. Major Shift of Toxigenic V. cholerae O1 from Ogawa to Inaba Serotype Isolated from Clinical and Environmental Samples in Haiti. PLoS Negl Trop Dis. 2016;10:e0005045. doi: 10.1371/journal.pntd.0005045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baron, S. et al. No evidence of significant levels of toxigenic V. cholerae O1 in the Haitian aquatic environment during the 2012 rainy season. PLoS Curr Outbreaks5 (2013). [DOI] [PMC free article] [PubMed]
- 61.Alam, M. T. et al. Monitoring Water Sources for Environmental Reservoirs of Toxigenic Vibrio cholerae O1, Haiti. Emerging Infect. Dis. 20 (2014). [DOI] [PMC free article] [PubMed]
- 62.Briquaire R, et al. Application of a paper based device containing a new culture medium to detect Vibrio cholerae in water samples collected in Haiti. J. Microbiol. Methods. 2016;133:23–31. doi: 10.1016/j.mimet.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 63.Rebaudet S, Piarroux R. Monitoring water sources for environmental reservoirs of toxigenic Vibrio cholerae O1, Haiti. Emerging Infect. Dis. 2015;21:169–170. doi: 10.3201/eid2101.140627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Debes AK, et al. Evaluation in Cameroon of a Novel, Simplified Methodology to Assist Molecular Microbiological Analysis of V. cholerae in Resource-Limited Settings. PLoS Negl Trop Dis. 2016;10:e0004307. doi: 10.1371/journal.pntd.0004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rashid, M. et al. Comparison of Inferred Relatedness Based on Multilocus Variable-number Tandem-repeat Analysis (MLVA) and Whole Genome Sequencing (WGS) of Vibrio cholerae O1. FEMS Microbiology Letters fnw116 10.1093/femsle/fnw116 (2016). [DOI] [PMC free article] [PubMed]
- 66.Garrine M, et al. Minimal genetic change in Vibrio cholerae in Mozambique over time: Multilocus variable number tandem repeat analysis and whole genome sequencing. PLOS Negl Trop Dis. 2017;11:e0005671. doi: 10.1371/journal.pntd.0005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Republic of Haiti, Ministry of Public Health and Population & National Directorate for Water Supply and Sanitation. National Plan for the Elimination of Cholera in Haiti 2013–2022. 114 (2013).
- 68.Rebaudet, S. et al. The national alert-response strategy against cholera in Haiti: a four-year assessment of its implementation. bioRxiv259366, 10.1101/259366 (2018).
- 69.Koski-Karell V, et al. Haiti’s progress in achieving its 10-year plan to eliminate cholera: hidden sickness cannot be cured. Risk Manag Healthc Policy. 2016;9:87–100. doi: 10.2147/RMHP.S75919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.République d’Haïti, Ministère de la Santé Publique et de la Population & Direction Nationale de l’Eau Potable et de l’Assainissement. Plan national d’élimination du choléra. Développement du moyen terme. Juillet 2016–Décembre2018 (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and in Supplementary Dataset file).