Abstract
Candida parapsilosis causes ~35% of all candidemia cases in neonates. High-resolution fingerprinting of C. parapsilosis isolates from neonatal intensive care unit (NICU) patients in Maternity Hospital (MH) was performed to identify epidemiologically related strains. Sixty-eight bloodstream/colonizing strains isolated from 59 NICU patients, two isolates from health care workers (HCWs) from MH and 18 bloodstream isolates from two other hospitals were used. Six microsatellite markers were employed, isolates were assigned a numerical microsatellite genotype (MSG), dendrogram was constructed and similarities between genotypes were visualized by minimum spanning tree. Fifty bloodstream isolates from MH yielded 37 MSGs with 20 isolates clustering in 7 MSGs. Duplicate isolates and colonizing strains yielded same/highly similar MSG as bloodstream isolates. Colonizing strains from two non-candidemia patients yielded unique MSGs while others belonged to a cluster. All isolates from HCWs and from two other hospitals belonged to unique MSGs. Cluster isolates came from patients in NICU-1 or from neonates in NICU-1 and other NICUs. Clonal complexes comprising closely related genotypes indicative of microevolution were also detected. Our data show that some C. parapsilosis strains have persisted in MH environment over several years and these endemic genotypes were transmitted to other patients in NICU-1 and/or other nearby NICUs.
Introduction
Candida spp. colonize human skin, mucosal membranes and the gastrointestinal tract soon after birth1,2. They also cause opportunistic infections in immunocompromised and hospitalized patients3. Candida spp. are also among the four most common cause of late onset septicemia in very-low-birth-weight infants4,5. Although Candida albicans is the most common cause of candidemia/invasive candidiasis, nearly 50% of all Candida infections are now caused by other non-albicans species of Candida6,7. Candida parapsilosis is commonly associated with invasive candidiasis and in some centers/geographical areas it has even surpassed C. albicans in frequency6–9. This species is also commonly isolated from neonatal intensive care units (NICUs), causing ~35% of all candidemia cases in neonates9–12. Candida parapsilosis now comprises three closely-related species and most invasive infections are caused by C. parapsilosis sensu stricto isolates13–15. Although the main reservoir of C. parapsilosis in hospital environment remains unknown, nosocomial infections are facilitated by the spread of the yeast from the hands of health care workers (HCWs) or contaminated patient care equipment to susceptible patients9–12,16,17. This organism also has affinity for glucose-containing parenteral nutrition and forms biofilms on intravascular devices which also promote its spread in hospital settings9–12.
The source of invasive C. parapsilosis infections are investigated by molecular fingerprinting studies9,16–20. Recent studies utilizing multi-locus microsatellite typing have shown that C. parapsilosis infections mostly occur as a result of exogenous transmission of the organism to the patient, particularly from the hands of HCWs and have reported the presence of endemic genotypes in some settings9,16,19–21. However, these studies have mostly been carried out with adult patient populations9,19–21. Although studies involving neonates, including low/very-low-birth-weight neonates, have also been carried out, the number of isolates analyzed from a single hospital in most of these investigations were relatively small (<30)9,11,17,18,22. A total of 152 neonates were treated for culture-confirmed candidemia in various NICUs of Maternity Hospital (MH) in Kuwait during January 2010 to August 2014 and C. parapsilosis was the most common (61 of 152, 40%) Candida spp. isolated from bloodstream specimens of neonates. This study performed high-resolution fingerprinting of 70 bloodstream and colonizing C. parapsilosis isolates from NICU patients and HCWs in MH in Kuwait to identify epidemiologically related strains.
Results
Candidemia cases at Maternity Hospital and phenotypic and molecular characterization of study isolates
The MH in Kuwait is the main government hospital for the care of women during pregnancy and childbirth. Nearly 11,000 deliveries (corresponding to ~70% of all deliveries in Kuwait) are performed every year at MH. The MH has four NICUs (including special care units) which are in close proximity to each other. Premature very-low-birth-weight neonates are treated in NICU-1, low-birth-weight neonates are treated in NICU-2 while other neonates are treated in NICU-3 and NICU-4. The HCWs are assigned to their respective NICUs, however, this is not strictly observed. A total of 152 neonates were treated for culture-confirmed candidemia in various NICUs of MH in Kuwait during the study period of January 2010 to August 2014. Although C. albicans was the most common species during 2010 and 2011, C. parapsilosis surpassed C. albicans as the most common species causing candidemia in MH in Kuwait during January 2012 to August 2014.
A total of 61 neonates with culture confirmed candidemia due to C. parapsilosis were treated in the four NICUs during the study period. All patients were low/very-low-birth-weight (0.8 to 1.5 kg) preterm neonates on parenteral nutrition. Fifty C. parapsilosis bloodstream isolates from 50 neonates (representing 82% of all bloodstream isolates at MH during the study period) were available for fingerprinting studies. Of the 50 neonates, 34, five, five and six were treated in NICU-1, NICU-2, NICU-3 and NICU-4, respectively. Two duplicate bloodstream isolates from two neonates (collected within twelve days of isolation of the first isolate due to persistence of candidemia), six colonizing strains from other body sites of five neonates with candidemia, 10 colonizing strains from body sites of nine non-candidemia NICU patients and two surveillance cultures from hands of two HCWs attending neonates in NICUs were also used. Duplicate isolates were used to confirm whether the persistence of candidemia was due to the same or a different C. parapsilosis strain. Colonizing strains from candidemia patients, non-candidemia NICU patients and isolates from hands of two HCWs attending neonates in NICUs were used to see their relationship with strains causing candidemia. Additionally, nine randomly selected bloodstream isolates from eight adult and one pediatric candidemia patient (HA1 to HA9 cultured in Hospital A located within 1 km from MH) and representing 43% of all bloodstream C. parapsilosis from Hospital A during the study period were also used. Similarly, nine randomly selected bloodstream isolates from eight adult and one pediatric candidemia patient (HB1 to HB9 cultured in Hospital B located 17 km from MH) and representing 35% of all bloodstream C. parapsilosis from Hospital B during the study period were also used. The isolates from Hospital A and Hospital B were used for comparison purpose and to ascertain the discriminatory power of the fingerprinting technique employed in this study (Supplementary Table 1).
All 88 isolates were initially identified at the respective hospital microbiology laboratory as C. parapsilosis sensu lato by Vitek 2 (Biomerieux, Marcy l’Etoile, France). The multiplex PCR assay identified all 88 isolates as C. parapsilosis sensu stricto. The identification of selected isolates was further confirmed by species-specific PCR amplification of rDNA and/or by PCR-sequencing of ITS region of rDNA.
Antifungal Susceptibility Testing
All 88 C. parapsilosis isolates were susceptible to amphotericin B, fluconazole, and caspofungin according to the CLSI breakpoints for C. parapsilosis23. The range of minimum inhibitory concentration (MIC) values of 88 C. parapsilosis isolates are summarized in Supplementary Table 2. The MIC50 and MIC90 values are also provided in Supplementary Table 2.
Fingerprinting of C. parapsilosis isolates
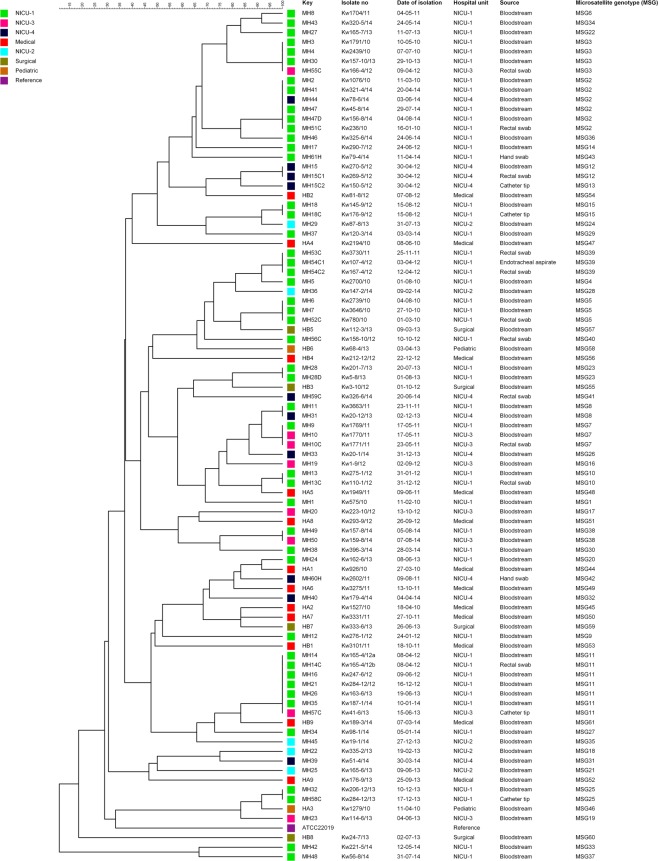
All 18 C. parapsilosis strains isolated from 18 candidemia patients at Hospital A and Hospital B analyzed in this study exhibited 18 unique microsatellite genotypes (MSGs) (Fig. 1, Supplementary Table 1). These studies confirmed that the six microsatellite loci-based fingerprinting method used in this study yields highly discriminatory patterns as all 18 isolates belonged to 18 unique genotypes. On the other hand, fingerprinting of 50 bloodstream C. parapsilosis isolates from 50 neonates from MH yielded only 37 MSGs. Interestingly, 30 isolates yielded 30 unique MSGs while 20 isolates were clustered in 7 MSGs (Supplementary Table 1). The three largest clusters included MSG11 with five isolates, MSG2 with four isolates and MSG3 with three isolates. The remaining four clusters (MSG5, MSG7, MSG8 and MSG38) contained two isolates each (Supplementary Table 1).
Figure 1.
An UPGMA-derived dendrogram based on microsatellite fragments from 88 C. parapsilosis isolates. Similarity is presented in percentages using the scale bar in the upper left corner. The columns after the patient number refer to isolate number, date of isolation, hospital unit, source of the isolates and microsatellite-based genotype (MSG).
Duplicate isolates yielded the same MSG as the first isolate from the same patient. Similarly, the five colonizing strains from five neonates also yielded the same MSG as the bloodstream isolates from these patients. Interestingly, a second colonizing strain from one patient (MH15C2) yielded a unique MSG (MSG13). However, MSG13 differed from MSG12 in only one marker. Of the 10 colonizing strains from nine non-candidemia neonates, two isolates belonged to two unique MSGs (MSG40 and MSG41), five isolates from five neonates exhibited the same genotype as the unique or cluster MSG exhibited by the bloodstream isolates while the three colonizing strains from two neonates belonged to a new cluster (MSG39). The two C. parapsilosis strains from the hands of two HCWs belonged to unique genotypes (Supplementary Table 1).
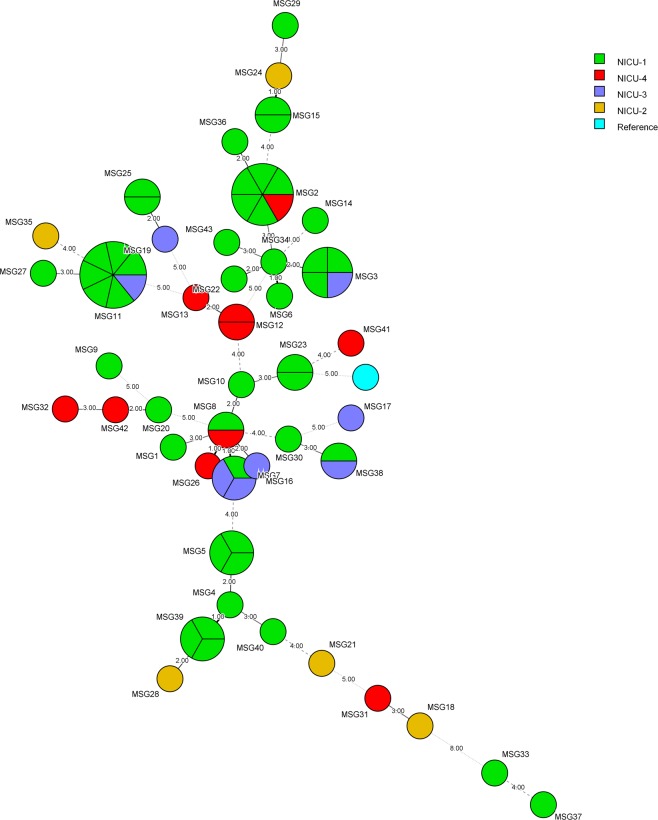
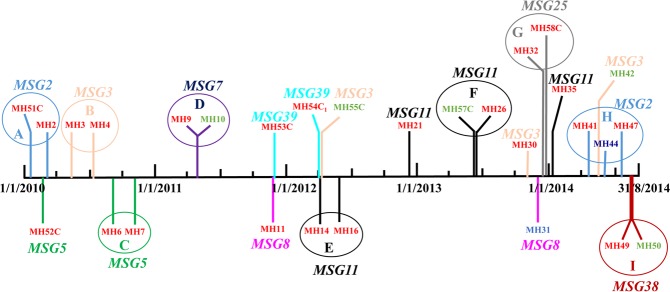
Overall, fingerprinting of 88 C. parapsilosis isolates from Kuwait identified 61 MSGs (MSG1 to MSG61; Fig. 1, Supplementary Table 1). Interestingly, 29 isolates from 29 candidemia and non-candidemia patients from MH clustered into 9 MSGs (excluding duplicate bloodstream isolates and colonizing isolates from the same candidemia patients which exhibited identical MSGs) (Fig. 1). The minimum spanning tree showed that all cluster isolates were obtained either exclusively from NICU-1 patients (isolates in MSG5, MSG25 and MSG39 clusters) or included isolates mostly collected from NICU-1 in addition to few isolates from other NICUs (isolates in MSG2, MSG3, MSG7, MSG8, MSG11, and MSG38 clusters) (Fig. 2). The data also showed that four clonal complexes (designated A to D) existed among C. parapsilosis isolates from MH which included closely related genotypes that differed in only one of the six microsatellite markers (Table 1). The color-coded time-line of isolation of cluster isolates obtained from different patients is presented in Fig. 3. An arbitrary window period of ≤62 days was used to define epidemiologically related isolates with identical MSG collected from two or more patients from the same or nearby NICU within MH. Based on this criteria, all isolates from neonates in NICU-1 for MSG2 (isolates from MH2 and MH51 patients in circle A), MSG3 (isolates from MH3 and MH4 patients in circle B), MSG5 (isolates from MH6 and MH7 patients in circle C), MSG11 (isolates from MH14 and MH16 patients in circle E) and MSG25 (isolates from MH32 and MH58 patients in circle G) were considered as epidemiologically related strains. Since HCWs are not strictly confined to their respective NICU, cluster isolates from neonates in different NICUs for MSG7 (isolates from MH9 and MH10 patients in circle D), MSG11 (isolates from MH26 and MH57 patients in circle F), MSG2 (isolates from MH41, MH44 and MH47 patients in circle H) and MSG38 (isolates from MH49 and MH50 patients in circle I) may also be considered as epidemiologically related strains. Furthermore, some genotypes (such as MSG3 and, to some extent, MSG8) were detected sporadically during a long period while others (MSG5, MSG7, MSG39, MSG25 and MSG38) were detected during a short period only. Interestingly, MSG11 persisted in the NICUs of MH for nearly two years (April 2012 to January 2014). On the other hand, MSG2, detected in early 2010, was not detected for four years (April 2010 to March 2014) until it re-appeared in April 2014 (Fig. 3).
Figure 2.
Minimum spanning tree of 70 C. parapsilosis isolates, from Maternity Hospital only, derived from microsatellite-based genotyping data. Each circle corresponds to a unique genotype, and lines between circles represent relative distance between isolates. The sizes of the circles correspond to the number of isolates in the same MSG. Connecting lines correspond to the number of differences between genotypes, with a solid thick line connecting genotypes that differ in one locus, a solid thin line connecting genotypes that differ in two or three loci, a dashed line connecting genotypes that differ in four loci, and a dotted line connecting genotypes that differ in more than four loci.
Table 1.
Candida parapsilosis genotypes identified from the Maternity Hospital and forming clonal complex.
| Microsatellite genotype (MSG) | Patient no.a | Microsatellite fragment length-based allelic profileb | Clonal complex | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CanpaSTR3A/a | CanpaSTR3A/b | CanpaSTR3B/a | CanpaSTR3B/b | CanpaSTR3C/a | CanpaSTR3C/b | CanpaSTR6A/a | CanpaSTR6A/b | CanpaSTR6B/a | CanpaSTR6B/b | CanpaSTR6C/a | CanpaSTR6C/b | |||
| MSG2 | MH2, MH41, MH44, MH47, MH47D, MH51C | 26 | 26 | 37 | 46 | 39 | 52 | 7 | 7 | 6 | 6 | 7 | 7 | A |
| MSG36 | MH46 | 26 | 26 | 37 | 46 | 39 | 52 | 7 | 7 | 9 | 14 | 7 | 7 | |
| MSG3 | MH3, MH4, MH30, MH55C | 26 | 26 | 47 | 47 | 40 | 40 | 7 | 7 | 6 | 6 | 7 | 7 | B |
| MSG34 | MH43 | 26 | 26 | 47 | 47 | 39 | 39 | 7 | 7 | 6 | 6 | 7 | 7 | |
| MSG7 | MH9, MH10, MH10C | 25 | 27 | 32 | 32 | 38 | 42 | 11 | 11 | 6 | 6 | 7 | 7 | C |
| MSG8 | MH11, MH31 | 25 | 27 | 32 | 32 | 36 | 42 | 11 | 11 | 6 | 6 | 7 | 7 | |
| MSG1 | MH1 | 25 | 25 | 32 | 32 | 27 | 27 | 11 | 11 | 6 | 6 | 7 | 7 | |
| MSG12 | MH15, MH15C1 | 26 | 32 | 32 | 32 | 44 | 44 | 7 | 7 | 6 | 6 | 7 | 7 | D |
| MSG13 | MH15C2 | 26 | 32 | 32 | 32 | 44 | 44 | 12 | 12 | 6 | 6 | 7 | 7 | |
aMH: patients from Maternity Hospital; MH51C: colonizing isolate from patient 51 at Maternity Hospital; MH47D: duplicate bloodstream isolate from patient 47 at Maternity Hospital. bBold numbers indicate marker differences within the clonal complex.
Figure 3.
Color-coded timeline showing the distribution of 29 C. parapsilosis isolates exhibiting cluster fingerprinting patterns obtained from 29 patients in NICUs of the Maternity Hospital in Kuwait. The patient number (MH1, MH2, MH3 etc.) yielding C. parapsilosis at the indicated time points (vertical colored lines) are shown along with the corresponding cluster microsatellite genotype (MSG) in bold and italicized letters of same color. The location of patients in the four NICUs is indicated by patient number color: red font, patients in NICU-1; green font, patients in NICU-3 and blue font; patients in NICU-4. Colonizing strains are indicated by letter “C” after patient number. The patients yielding isolates belonging to same MSG that were recovered within 62 days of each other are shown in circles (color coded with each MSG) marked ‘A’ to ‘I’.
Discussion
The origin of nosocomial Candida infections in adult patients could be colonizing endogenous strains or exogenous strains transmitted to the patients from biomedical devises, contaminated infusates, hospital personnel and environments24–26. Among preterm (low/very-low-birth-weight) neonates, early onset bloodstream infections are usually caused by organisms transmitted vertically from mother to infants during or shortly after delivery, however, late onset bloodstream infections may also be caused by organisms acquired during the course of hospital care5,27. C. parapsilosis sensu stricto isolates have a remarkable capability to easily colonize human skin (such as the hands of HCWs) and contaminate hospital surroundings10,12. Genotyping methods based on length variations associated with microsatellites have been developed for C. parapsilosis and are now considered as the most popular and versatile typing schemes due to their rapid evolution and ability to detect microevolutionary variations28,29. These methods, however, differ in the number of polymorphic microsatellite markers (varying from 4 to 6 loci) and consequently the discriminatory power of the typing scheme11,17,28–30. In this study, we have used the six-marker microsatellite panel which yields 12 alleles per strain, due to diploid nature of the organism, with maximum discriminatory power described so far30. Application of six-marker microsatellite panel yielded high-resolution fingerprinting of C. parapsilosis isolates in Kuwait. This is evident from the fact that all 18 randomly selected isolates collected from candidemia patients from two major hospitals belonged to different genotypes (MSG44 to MSG61).
When microsatellite genotyping was applied to 50 bloodstream isolates from neonates from MH, 30 isolates were identified as unique strains while 20 isolates were clustered in 7 MSGs. Duplicate isolates from two patients and colonizing strains from five neonates yielded the same genotype as the bloodstream isolate. However, a colonizing strain (MH15C2 from the catheter tip) from one patient yielded a different genotype than bloodstream and gastrointestinal tract isolates (MH15 and MH15C1) but differed in a single microsatellite marker only. Only few studies have analyzed multiple isolates from bloodstream and/or other body sites from the same patient. Multiple isolates from nine of 18 patients in one study yielded the same MSG, two patients yielded closely-related MSGs differing in only one marker while seven patients yielded unrelated MSGs from multiple sites30. Another study reported identical MSGs in serial isolates from the same or multiple anatomic sites of four of six patients while in the remaining two cases, the isolates differed in one or two markers20. Multiple isolates from two patients also yielded closely-related MSGs differing in only one marker in another study11. Taken together, these findings are consistent with spontaneous variation in microsatellite markers leading to microevolution within the same patient or in different patients infected with the same strain9,30.
Further analysis of 10 colonizing strains from nine non-candidemia patients admitted in the same NICUs showed that only two isolates belonged to a unique MSG (MSG40 and MSG41), three isolates from two neonates belonged to a new cluster (MSG39) while the remaining five isolates belonged to other genotypes prevalent in the same or adjoining NICUs. Thus a total of nine clusters comprising MSG2, MSG3, MSG5, MSG7, MSG8, MSG11, MSG25, MSG38 and MSG39 were identified. Interestingly all isolates in three clusters (comprising MSG5, MSG25 and MSG39) and nearly all isolates in the remaining six clusters (comprising MSG2, MSG3, MSG7, MSG8, MSG11 and MSG38) were obtained from neonates in NICU-1 which houses very-low-birth-weight neonates. Furthermore, some genotypes were observed during a short window period (MSG7, MSG25, MSG38 and to some extent MSG5 and MSG39) while others (MSG2, MSG3, MSG8 and MSG11) showed a more extended distribution spread over several years. In many cases (denoted by circles “A” to ‘I’ in Fig. 3), cluster isolates were obtained from patients admitted to NICUs within 62 days of each other, typically in the same NICU, suggesting persistence of some genotypes in MH in Kuwait. A window period of 60–90 days has also been used previously by other investigators for defining epidemiologically-linked cluster isolates of Candida albicans causing bloodstream infections24,31. Endemic genotypes have also been reported to persist within the hospital environment for several years causing nosocomial outbreaks of invasive Candida infections in several studies18,25,32. The isolation of clonally-related genotypes in both time and space supports nosocomial transmission of C. parapsilosis among some neonates in MH in Kuwait. Similar observations were reported from NICU patients in Italy and among neonates in one (Hospital B) of three hospitals from USA11,18. Although microbiological testing of HCWs or hospital environment was not carried out in these studies, it is probable that patient care and manipulation of catheters by HCWs was the source of C. parapsilosis in very-low-birth-weight neonates in NICU as most infants are not colonized by yeast species at birth1,2,10,12. The involvement of HCWs in cross-transmission of C. parapsilosis infection has been demonstrated in few studies9,16,17. One recent study has shown that C. parapsilosis isolates from the hands of most of the HCWs belonged to a unique genotype. However, genotypes of some C. parapsilosis isolates obtained from the hospital environment and the hands of HCWs were also common with bloodstream isolates obtained from NICU patients implicating the environment and the hands of HCWs as a cause of nosocomial infections9. However, the source of infection among our cluster isolates remained unknown as the two C. parapsilosis isolates recovered from the hands of two HCWs belonged to unique genotypes which were different from the cluster genotypes.
The presence of clonal complexes of closely related genotypes as a result of microevolution due to intrinsic instability of microsatellite loci has been observed in few studies9,18,21,30. However, many of these studies were based on analysis of only four microsatellite loci and the close association may have resulted from fortuitous grouping of the isolates due to lower discriminatory power of these typing schemes9,18,21. Our data based on analysis of six microsatellite loci also identified some genotypes in which minor changes had occurred in a single locus compared to the genotype of the cluster isolates. These closely related genotypes were obtained either from the same patient (MH15) or were found mainly among isolates from NICU-1 patients strongly supporting the possibility of microevolution from the parental strain30. The results of antifungal drug susceptibility testing showed that all 88 C. parapsilosis isolates were susceptible to amphotericin B, fluconazole and caspofungin and therefore, no association of antifungal resistance with specific genotypes was found. On the contrary, resistance to fluconazole was found in 77 of 143 (54%) C. parapsilosis isolates from NICU patients in South Africa and vast majority of isolates in four clusters comprised fluconazole-resistant strains33.
Our study has few limitations. i) Surveillance cultures from the mothers of neonates were not available. ii) The hands of the HCWs from the four NICUs in MH were not screened systematically and without prior notice which may have resulted in the recovery of C. parapsilosis from the hands of only two HCWs. iii) Environmental samples from the NICUs were not screened to trace an epidemiological link. iv) Clinical characteristics, major risk factors (other than very-low-birth-weight of the neonates) and the outcome of candidemia were not evaluated.
In conclusion, all 18 C. parapsilosis isolates from candidemia patients from two major hospitals belonged to different genotypes showing high discriminatory power of the six microsatellite marker-based fingerprinting method used in the present study. Our fingerprinting data from MH showed that 19 of 68 C. parapsilosis isolates from 19 preterm neonates in NICUs with/without candidemia (excluding duplicate bloodstream isolates and colonizing isolates from the same candidemia patients which exhibited identical MSGs) belonged to a cluster genotype. Furthermore, these cluster isolates were obtained from different patients within a short (62 days) time period. These findings suggest that some strains have persisted in the MH environment, particularly in NICU-1, over several years and these endemic genotypes were transmitted to other patients in NICU-1 and other nearby NICUs in MH in Kuwait. However, the source of infection among neonates remained unidentified as only two isolates were obtained from the hands of HCWs which belonged to different genotypes. This is the first report describing high-resolution fingerprinting of C. parapsilosis sensu stricto isolates from NICU patients from the Middle East.
Materials and Methods
Study setting and clinical isolates
This retrospective study was carried out on bloodstream C. parapsilosis strains isolated at Maternity Hospital (MH), Kuwait. A total of 70 C. parapsilosis strains isolated during January 2010 to August 2014 were analyzed. These included 50 C. parapsilosis bloodstream isolates from 50 neonates, two duplicate bloodstream isolates from two neonates (collected within twelve days of isolation of the first isolate), six colonizing strains from other (rectal swab, n = 4; catheter tip, n = 2) body sites of five neonates with candidemia, 10 colonizing strains from body sites (rectal swab, n = 7; catheter tip, n = 2 and endotracheal aspirate, n = 1) of nine non-candidemia NICU patients and two surveillance cultures from hands of two HCWs attending neonates in NICUs. Additionally, 18 bloodstream isolates from 16 adults and two pediatric candidemia patients from two other hospitals were also used for comparison purpose. The clinical specimens yielding C. parapsilosis were collected from neonates/patients after obtaining informed verbal consent from a parent and/or legal guardian (for patients under the age of 18 years old) as part of routine diagnostic work-up for the isolation of bacterial/fungal pathogens. The isolates were sent to the Mycology Reference Laboratory (MRL), Department of Microbiology, Faculty of Medicine, Kuwait University for identification and antifungal susceptibility testing and data on deidentified samples are reported in this study. C. parapsilosis (ATCC22019 = CBS604) and C. albicans (ATCC90028 = CBS8837) were used as reference strains. All isolates were subjected to species-specific identification by phenotypic and molecular methods, antifungal drug susceptibility testing and high-resolution fingerprinting by six loci-based multilocus microsatellite typing.
Phenotypic and molecular identification
The isolates were initially identified as C. parapsilosis sensu lato at the original hospital by Vitek 2 yeast identification system (bioMérieux, Marcy-l’Etoile, France). All isolates were grown at MRL for 48 h on Sabouraud dextrose agar medium at 37 °C. The genomic DNA from the isolates was extracted by using the Gentra Puregene Yeast DNA extraction kit (Qiagen, Hilden, Germany) according to the instructions supplied by the manufacturer. Molecular identification was performed by a multiplex PCR assay that discriminates C. parapsilosis sensu stricto from Candida orthopsilosis and Candida metapsilosis among C. parapsilosis sensu lato strains, as described previously32. The identity of randomly selected isolates was further confirmed by C. parapsilosis sensu stricto-specific PCR assay described previously14 and DNA sequencing of internal transcribed spacer (ITS) region of rDNA by using panfungal ITS1 and ITS4 primers, as described previously34,35. The study was approved by the Joint Committee for the Protection of Human Subjects in Research, Health Sciences Center, Kuwait University and Ministry of Health, Kuwait and all the methods and investigations reported in this study were carried out according to their guidelines. The clinical samples yielding the isolates used in this study were taken after obtaining informed verbal consent of the patients as part of routine diagnostic work-up and this procedure was approved by the relevant ethical committees.
Antifungal drug susceptibility testing
The susceptibility of C. parapsilosis isolates against antifungal drugs, amphotericin B, fluconazole and caspofungin was determined by Etest (bioMérieux, Marcy-L´Etoile, France) as described previously36. The revised interpretive susceptibility breakpoints as recommended by Clinical Laboratory Standards Institute (CLSI) were used for fluconazole (≤2 µg/ml, susceptible; 4 µg/ml, susceptible dose-dependent and >8 µg/ml, resistant) and caspofungin (≤2 µg/ml, susceptible; 4 µg/ml, intermediate and >8 µg/ml, resistant)23. Due to lack of defined breakpoints for amphotericin B, an isolate showing an MIC ≤ 1 µg/ml was considered as susceptible. Quality control was ensured by testing C. parapsilosis ATCC22019 (=CBS604) and C. albicans ATCC90028 (=CBS8837), as recommended by CLSI23.
Fingerprinting by multilocus microsatellite typing
Fingerprinting of C. parapsilosis isolates was performed by using a panel of six short tandem repeat (STR) markers as described by Diab-Elschahawi et al.30. The alleles were designated by their sizes which were recorded by using GeneScan 500 LIZ size standard (Applied Biosystems) in the 35–500 nucleotide range. The number of repeats in each marker was determined by comparing the relative size of each allele to the reference C. parapsilosis strain CDC317 that has been subjected to whole genome sequencing (GenBank accession no. HE605206) and the combination of allele numbers for all six diploid loci was assigned an arbitrary microsatellite genotype (MSG). Based on the allele number for the six diploid loci for each isolate, a dendrogram was constructed by using BioNumerics v7.6 software (Applied Maths, Saint-Martens-Latem, Belgium) and standard unweighted pair group method with arithmetic mean (UPGMA) settings, as described previously24. The genetic relationship between the genotypes was studied by constructing a minimum spanning tree. The isolates from the same or different patients were considered belonging to the same MSG (cluster) when they possessed the same alleles for all six diploid loci. Closely related MSGs which differed in only one of six diploid microsatellite marker were grouped into clonal complexes. The reproducibility of the fingerprinting method was ensured by including the reference C. parapsilosis strain (ATCC22019 = CBS604) in each run.
Supplementary information
Acknowledgements
This study was supported by College of Graduate Studies, Kuwait University, internal departmental funding, Department of Microbiology and Research Core Facility Grant No. SRUL 02/13. We thank Dr. Seema Khan for her help in collection of data from the neonates.
Author Contributions
Conceived and designed the experiments: M.A., S.A., N.A. and Z.K. Performed the experiments: M.A. and F.H. Analyzed the data: M.A., S.A., N.A., F.H., J.F.M. and Z.K. Contributed reagents/materials/analysis tools: J.F.M. and Z.K. Wrote the paper: M.A., S.A., N.A., F.H., J.F.M. and Z.K.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37855-2.
References
- 1.Kaufman DA, Manzoni P. Strategies to prevent invasive candidal infection in extremely preterm infants. Clin. Perinatol. 2010;37:611–628. doi: 10.1016/j.clp.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Ward TL, et al. Development of the human mycobiome over the first month of life and across body sites. mSystems. 2018;3:e00140–17. doi: 10.1128/mSystems.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown GD, et al. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4:165rv13–165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Hornik CP, et al. Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Hum. Dev. 2012;88:S69–S74. doi: 10.1016/S0378-3782(12)70019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenberg RG, et al. Late-onset sepsis in extremely premature infants: 2000-2011. Pediatr. Infect. Dis. J. 2017;36:774–779. doi: 10.1097/INF.0000000000001570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart SR, et al. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J. Clin. Microbiol. 2012;50:3435–3442. doi: 10.1128/JCM.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaller MA, et al. Epidemiology and outcomes of invasive candidiasis due to non-albicans species of Candida in 2,496 patients: data from the Prospective Antifungal Therapy (PATH) registry 2004-2008. PLoS One. 2014;9:e101510. doi: 10.1371/journal.pone.0101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nucci M, et al. Epidemiology of candidemia in Latin America: a laboratory-based survey. PLoS One. 2013;8:e59373. doi: 10.1371/journal.pone.0059373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delfino D, et al. Potential association of specific Candida parapsilosis genotypes, bloodstream infections and colonization of health workers’ hands. Clin. Microbiol. Infect. 2014;20:O946–O951. doi: 10.1111/1469-0691.12685. [DOI] [PubMed] [Google Scholar]
- 10.van Asbeck EC, Clemons KV, Stevens DA. Candida parapsilosis: a review of its epidemiology, pathogenesis, clinical aspects, typing and antimicrobial susceptibility. Crit. Rev. Microbiol. 2009;35:283–309. doi: 10.3109/10408410903213393. [DOI] [PubMed] [Google Scholar]
- 11.Reiss E, et al. Genotyping of Candida parapsilosis from three neonatal intensive care units (NICUs) using a panel of five multilocus microsatellite markers: broad genetic diversity and a cluster of related strains in one NICU. Infect. Genet. Evol. 2012;12:1654–1660. doi: 10.1016/j.meegid.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Pammi M, Holland L, Butler G, Gacser A. Candida parapsilosis is a significant neonatal pathogen: a systematic review and meta-analysis. Pediatr. Infect. Dis. J. 2014;32:1–23. doi: 10.1097/INF.0b013e3182863a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tavanti A, Davidson A, Gow N, Maiden MC, Odds FC. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 2005;43:284–292. doi: 10.1128/JCM.43.1.284-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asadzadeh M, Ahmad S, Al-Sweih N, Khan ZU. Rapid molecular differentiation and genotypic heterogeneity among Candida parapsilosis and Candida orthopsilosis strains isolated from clinical specimens in Kuwait. J. Med. Microbiol. 2009;58:745–752. doi: 10.1099/jmm.0.008235-0. [DOI] [PubMed] [Google Scholar]
- 15.Barbedo LS, et al. Different scenarios for Candida parapsilosis fungaemia reveal high numbers of mixed C. parapsilosis and Candida orthopsilosis infections. J. Med. Microbiol. 2015;64:7–17. doi: 10.1099/jmm.0.080655-0. [DOI] [PubMed] [Google Scholar]
- 16.Van Asbeck EC, Huang YC, Markham AN, Clemons KV, Stevens DA. Candida parapsilosis fungemia in neonates: genotyping results suggest healthcare workers hands as source, and review of published studies. Mycopathologia. 2007;164:287–293. doi: 10.1007/s11046-007-9054-3. [DOI] [PubMed] [Google Scholar]
- 17.Vaz C, et al. Microsatellite multilocus genotyping clarifies the relationship of Candida parapsilosis strains involved in a neonatal intensive care unit outbreak. Diagn. Microbiol. Infect. Dis. 2011;71:159–62. doi: 10.1016/j.diagmicrobio.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Romeo O, et al. Microsatellite-based genotyping of Candida parapsilosis sensu stricto isolates reveals dominance and persistence of a particular epidemiological clone among neonatal intensive care unit patients. Infect. Genet. Evol. 2013;13:105–108. doi: 10.1016/j.meegid.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Marcos-Zambrano LJ, et al. Clusters of patients with candidaemia due to genotypes of Candida albicans and Candida parapsilosis: differences in frequency between hospitals. Clin. Microbiol. Infect. 2015;21:677–683. doi: 10.1016/j.cmi.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Sabino R, et al. Analysis of clinical and environmental Candida parapsilosis isolates by microsatellite genotyping-a tool for hospital infection surveillance. Clin. Microbiol. Infect. 2015;21:954.e1–954.e8. doi: 10.1016/j.cmi.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, et al. Investigation of an unrecognized large-scale outbreak of Candida parapsilosis sensu stricto fungaemia in a tertiary-care hospital in China. Sci. Rep. 2016;6:1–11. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pulcrano G, et al. MALDI-TOF mass spectrometry and microsatellite markers to evaluate Candida parapsilosis transmission in neonatal intensive care units. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:2919–2928. doi: 10.1007/s10096-012-1642-6. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts; fourth informational supplement, M27-S4. Wayne, PA, USA. (2012).
- 24.Asadzadeh M, Ahmad S, Al-Sweih N, Khan Z. Molecular fingerprinting studies do not support intrahospital transmission of Candida albicans among candidemia patients in Kuwait. Front. Microbiol. 2017;8:247. doi: 10.3389/fmicb.2017.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ásmundsdóttir LR, et al. Molecular epidemiology of candidemia: evidence of clusters of smoldering nosocomial infections. Clin. Infect. Dis. 2008;47:e17–e24. doi: 10.1086/589298. [DOI] [PubMed] [Google Scholar]
- 26.Suleyman G, Alangaden GJ. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect. Dis. Clin. North Am. 2016;30:1023–1052. doi: 10.1016/j.idc.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Stoll BJ, et al. Changes in pathogens causing early-onset sepsis in very-low-birth-weight infants. N. Engl. J. Med. 2002;347:240–247. doi: 10.1056/NEJMoa012657. [DOI] [PubMed] [Google Scholar]
- 28.Lasker, B. A., Butler, G. & Lott, T. J. Molecular genotyping of Candida parapsilosis group I clinical isolates by analysis of polymorphic microsatellite markers. 44, 750–759 (2006). [DOI] [PMC free article] [PubMed]
- 29.Sabino R, et al. New polymorphic microsatellite markers able to distinguish among Candida parapsilosis sensu stricto isolates. J. Clin. Microbiol. 2010;48:1677–1682. doi: 10.1128/JCM.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diab-Elschahawi M, et al. Microsatellite genotyping clarified conspicuous accumulation of Candida parapsilosis at a cardiothoracic surgery intensive care unit. J. Clin. Microbiol. 2012;50:3422–3426. doi: 10.1128/JCM.01179-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai MH, et al. Clinical and molecular characteristics of bloodstream infections caused by Candida albicans in children from 2003 to 2011. Clin. Microbiol. Infect. 2015;21:e1–e8. doi: 10.1016/j.cmi.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 32.Asadzadeh M, et al. Simple, low-cost detection of Candida parapsilosis complex isolates and molecular fingerprinting of Candida orthopsilosis strains in Kuwait by ITS region sequencing and amplified fragment length polymorphism analysis. PLoS One. 2015;10:e0142880. doi: 10.1371/journal.pone.0142880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magobo RE, et al. Detection of neonatal unit clusters of Candida parapsilosis fungaemia by microsatellite genotyping: results from laboratory-based sentinel surveillance, South Africa, 2009-2010. Mycoses. 2017;60:320–327. doi: 10.1111/myc.12596. [DOI] [PubMed] [Google Scholar]
- 34.Khan ZU, et al. Actinomucor elegans var. kuwaitiensis isolated from the wound of a diabetic patient. Antonie Van Leeuwenhoek. 2008;94:343–352. doi: 10.1007/s10482-008-9251-1. [DOI] [PubMed] [Google Scholar]
- 35.Khan ZU, et al. Cryptococcus randhawai sp. nov., a novel anamorphic basidiomycetous yeast isolated from tree trunk hollow of Ficus religiosa (peepal tree) from New Delhi, India. Antonie Van Leeuwenhoek. 2010;97:253–259. doi: 10.1007/s10482-009-9406-8. [DOI] [PubMed] [Google Scholar]
- 36.Asadzadeh M, Al-Sweih NA, Ahmad S, Khan ZU. Antifungal susceptibility of clinical Candida parapsilosis isolates in Kuwait. Mycoses. 2008;51:318–23. doi: 10.1111/j.1439-0507.2008.01492.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.