Abstract
Childhood psychotic experiences (PEs), such as seeing or hearing things that others do not, or extreme paranoia, are relatively common with around 1 in 20 children reporting them at age 12. Childhood PEs are often distressing and can be predictive of schizophrenia, other psychiatric disorders, and suicide attempts in adulthood, particularly if they persist during adolescence. Previous research has demonstrated that methylomic signatures in blood could be potential biomarkers of psychotic phenomena. This study explores the association between DNA methylation (DNAm) and the emergence, persistence, and remission of PEs in childhood and adolescence. DNAm profiles were obtained from the ALSPAC cohort at birth, age 7, and age 15/17 (n = 901). PEs were assessed through interviews with participants at ages 12 and 18. We identified PE-associated probes (p < 5 × 10−5) and regions (corrected p < 0.05) at ages 12 and 18. Several of the differentially methylated probes were also associated with the continuity of PEs across adolescence. One probe (cg16459265), detected consistently at multiple timepoints in the study sample, was replicated in an independent sample of twins (n = 1658). Six regions, including those spanning the HLA-DBP2 and GDF7 genes, were consistently differentially methylated at ages 7 and 15–17. Findings from this large, population-based study suggest that DNAm at multiple stages of development may be associated with PEs in late childhood and adolescence, though further replication is required. Research uncovering biomarkers associated with pre-clinical PEs is important as it has the potential to facilitate early identification of individuals at increased risk who could benefit from preventive interventions.
Introduction
Psychotic experiences (PEs), such as hearing voices, seeing visions, or being extremely paranoid, are experienced by around 1 in 20 children1, usually without a diagnosable illness. In most children these experiences remit, but nonetheless they are associated with later psychiatric problems such as psychotic disorders2, including schizophrenia3,4, and post-traumatic stress disorder3, as well as suicidality and self-harm in adolescence1,5, and suicide attempts in adulthood3. The likelihood of developing a psychotic disorder6 or post-traumatic stress disorder7 is especially increased if PEs persist during adolescence. Therefore, it is important to identify as early as possible which children are at risk of developing these experiences. Uncovering biomarkers associated with the onset and persistence of pre-clinical PEs in childhood can increase our understanding of the aetiological factors accompanying the development and progression of PEs, and may in the future facilitate early identification of individuals at increased psychiatric risk, and improve targeting of preventive interventions.
Biomarkers for the emergence and persistence of PEs might be evident at the level of epigenetics, that is by any of the many cellular mechanisms capable of regulating gene expression without changes in the underlying genetic sequence. DNA methylation (DNAm) patterns have, for example, been associated with a broad range of psychiatric phenotypes8–14, including schizophrenia and other psychotic disorders. Research in monozygotic twin pairs discordant for a clinical diagnosis of psychosis15 and in case–control samples16–22 has identified DNAm differences associated with psychosis. Importantly, these findings were identified not only in post-mortem brain tissue from diagnosed individuals19,21,22, but also in peripheral samples from living patients15–18,20, and studies have replicated findings from peripheral samples in post-mortem tissue15.
To date, the majority of studies have relied on cross-sectional analyses of DNAm and psychotic disorder status. Cross-sectional studies with affected individuals are likely to be highly confounded by factors associated with the disorder, such as medication. Crucially, few epigenetic studies have utilised prospective designs to focus on pre-clinical psychotic phenomena in the general population, which are important to optimise early detection efforts. To this end, research from our group recently demonstrated specific DNAm differences at age 10 between monozygotic twin pairs discordant for psychotic symptoms at age 12 (n = 48;23). Of note, hypomethylation of the top-ranked CpG site was replicated in post-mortem brain tissue in an independent sample of schizophrenia patients compared to controls, and several of the findings of interest were located in, or near, genes previously implicated in neurodevelopment and psychiatric disorders. Furthermore, a recent study found that conversion to psychosis in young help-seeking individuals (an ultra-high risk group, n = 39) was associated with differential changes in DNAm, with top results occurring in pathways relevant for psychosis such as oxidative stress regulation, axon guidance and inflammatory pathways24. These studies indicate that the PEs in pre-diagnosis populations may be associated with differential DNAm, and that the pattern of PEs may correspond with changes in DNAm in relevant pathways. However, it is difficult to draw firm conclusions based on such small samples that may not be representative of the broader population.
The aim of this study was to explore associations between genome-wide DNAm patterns and the emergence and persistence of PEs in childhood and adolescence utilising a longitudinal design covering key points in development in a large, population-based sample. First, we investigated the association between DNAm at specific timepoints during childhood and adolescence (birth, age 7, and age 15–17) and reports of PEs at ages 12 and 18. Second, we examined the role of DNAm in the continuity of PEs by comparing individuals whose experiences persisted between 12 and 18 years with those whose experiences remitted, emerged for the first time during adolescence, and those with no history of such experiences. Third, we explored the association between longitudinal trajectories of DNAm and PEs across early development. Fourth, we investigated differentially methylated regions (DMRs) associated with PEs. Finally, we tested the robustness of these findings in an independent sample.
Methods
Sample
The Avon Longitudinal Study of Parents and Children (ALSPAC) is a large, prospective cohort study that recruited 14,541 pregnant women resident in Avon, UK with expected dates of delivery between 1 April 1991 and 31 December 199225,26. Further details of the study and available data are provided on the study website through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/). The current study focusses on the Accessible Resource for Integrated Epigenomic Studies (ARIES) sub-study, which consists of 1018 mother–offspring pairs who provided DNA samples at multiple timepoints27. All data are available by request from the Avon Longitudinal Study of Parents and Children Executive Committee (http://www.bristol.ac.uk/alspac/researchers/access/) for researchers who meet the criteria for access to confidential data.
Written informed consent was provided by parents and assent by children during the childhood phases of the study, and then by the children themselves at age 18. Ethical approval for the study was obtained from the ALSPAC Law and Ethics Committee and the Local Research Ethics Committees. Ethical approvals are in place for all sources of biological samples and data in ARIES in accordance with the Declaration of Helsinki.
Measures
Psychotic experiences
PEs at ages 12 and 18 were assessed via semi-structured interviews (Psychosis-like symptoms semi-structured interview—PLIKSi) that have been described previously2,28. At 12 years participants were asked about PEs over the previous 6 months; at 18 years participants were asked about experiences since the age of 12. Further details regarding determination of PEs in this sample are provided in the Supplementary Information. For longitudinal analyses, individuals were included if they had PE data available at both timepoints, and the nature of PEs from ages 12 to 18 was determined as persistent (present at both timepoints), remitted (present at 12 but not 18), emergent (present at 18 but not 12), and none (no experiences at either timepoint).
DNA methylation
DNA samples were extracted from cord blood on delivery, and from peripheral blood samples in childhood (age 7) and in adolescence (age 15–17) according to established procedures27. DNA was bisulfite-converted using the Zymo EZ DNA MethylationTM kit (Zymo, Irvine, CA) and then DNA methylation of over 485,000 CpG sites was quantified using the Illumina Infinium HumanMethylation450K BeadChip assay (HM450; Illumina Inc., CA). Arrays were scanned using the Illumina iScan, and GenomeStudio (version 2011.1; Illumina Inc.) was used to extract signal intensities and assess initial quality review.
Pre-processing and QC procedures
HM450 detects the proportion of molecules methylated at each CpG site on the array. For each sample, the estimated methylation level at each CpG site is expressed as a beta value (β), which is the ratio of the methylated probe intensity to the overall intensity and ranges from 0 (no cytosine methylation) to 1 (complete cytosine methylation). Background correction and functional normalisation were performed using meffil29. Samples with >10% of sites with a detection p value >0.01 or a bead count <3 in >10% of probes were removed from further analysis. Non-specific probes and probes on sex chromosomes were removed30,31. Following QC procedures, data were available for 381,871 probes. Probes were annotated using information provided by Illumina (genome build: hg19).
Batch effects
Samples from all timepoints in ARIES were distributed across slides using a semi-random approach, with all samples from each individual presented on the same array to minimise confounding by batch. Further batch variables were recorded using a purpose-built laboratory information management system, which also recorded QC metrics from the standard control probes on the HM450 array. Samples failing QC (>20% probes with p ≥ 0.01) were repeated and, if unsuccessful, excluded from further analysis.
Cell heterogeneity
To account for potential differences in methylation arising from cell composition in whole-blood samples, cell counts were estimated using the Houseman algorithm32 and included as covariates. However, for longitudinal analyses including data from cord blood and whole-blood samples, analyses instead included the first 20 independent surrogate variable components to account for both heterogeneity between cord blood and peripheral blood samples as well as batch effects. Previous research has indicated that surrogate variables derived in this way account for cell count heterogeneity as well as estimated cell counts33,34.
Potential confounders
Pregnancy and birth-related variables were considered as potential confounders, including parity (number of pregnancies resulting in live birth), maternal age at child’s birth (derived from date of birth), maternal smoking during pregnancy (never, stopped first trimester, continued throughout pregnancy), infections during pregnancy (at any time), and delivery method (caesarean; yes/no). Factors relating to the child included gender, smoking, and use of psychiatric medications. Data concerning child smoking behaviour during adolescence were not available for all samples; therefore, a proxy smoking variable was derived using DNAm signals from probes previously associated with smoking35,36. This method has been shown to distinguish between smokers and non-smokers (e.g. see 20). One child reported use of psychiatric medications across the study period and was removed from the analysis sample.
Statistical analyses
We investigated the association between DNAm and PEs in four stages. All analyses were conducted using R (version 3.3.1) unless otherwise stated.
Epigenome-wide Association Study
The Epigenome-wide Association Study (EWAS) function in meffil29 was used to conduct six epigenome-wide association studies of PEs at ages 12 and 18 with cord blood, age-7 peripheral blood, and age-15/17 peripheral blood DNAm. Parity, maternal age, maternal smoking during pregnancy, infections during pregnancy, delivery methods, and child’s gender were included as covariates, as well as estimated cell counts. Analyses using DNAm data from age 7 or age 15–17 also included child age as a covariate. Probes with values at the extremes of the distribution (5%) were winsorised. Statistical significance was determined using a Bonferroni correction, giving a threshold of p < 1.3 × 10−7. Tests with p < 5 × 10−5 were defined as reaching suggestive significance.
Continuity of PEs across adolescence
The CpG sites most strongly associated with PEs at either timepoint (12 or 18, p < 5 × 10−5) were assessed for association with the continuity of PEs between 12 and 18 (persistent, remitted, emergent, none) using ANOVA. Post hoc tests were used to determine the relationships between groups. Significant results (p < 0.00045; p = 0.05/110 CpG sites) were re-run with linear models to include all previously described covariates.
Longitudinal DNAm trajectories and continuity of PEs
Multilevel models were constructed to test the association between the methylomic trajectories of the top CpG sites and the continuity of PEs across adolescence, where DNAm was the dependent variable. A linear spline term with a knot at 7 was included to allow for different linear changes from 0 to 7 and from 7 to 15–17. Models were also adjusted for all previously described pregnancy and birth-related covariates, and the first 20 independent surrogate variable components to account for heterogeneity between cord blood and peripheral blood samples (e.g. cell composition and batch effects). Models were repeated including derived smoking scores as previously described.
Differentially methylated regions
The comb-p module in Python was used to identify DMRs by combining spatially correlated p values from each EWAS. We used a seed p value of <0.001, a maximum distance of 500 bp, and a minimum of three probes. Statistical significance was determined using a Šidák correction for multiple testing, where p < 0.05 is significant.
Gene Ontology (GO) enrichment analysis
Enrichment analyses were conducted to assess the over-representation of GO37 biological processes and functions in CpG sites of each EWAS where p < 0.001. A lenient significance threshold was used to maximise our probes of interest. We used a methodology that controls for the number of probes that are annotated to each gene, as well as taking into account the hierarchical structure of ontological categories by grouping terms where the significant enrichment was explained by the overlap with a more significant term (as described in ref. 20).
Replication
CpG sites of interest (detailed below) were tested for association with PEs in the Environmental Risk (E-Risk) Longitudinal Twin Study, a nationally representative cohort of 2232 British-born twins, which has been previously described in detail38. In this sample, DNAm from whole-blood samples obtained at age 18 was also quantified using the Illumina Infinium HumanMethylation450K BeadChip assay (HM450; Illumina Inc., CA)39, and PEs between 12 and 18 years were obtained from an interview that is similar to the PLIKSi40. Following stringent quality control procedures using the wateRmelon package41, data were available for 1658 individuals at age 18. The associations between DNAm at our CpG sites of interest and concurrent PEs at age 18 were tested using linear regression, controlling for gender, zygosity, batch, and cell counts (determined using the Houseman algorithm as previously described). Clustered standard errors were used to calculate p values to account for the effect of family, and were implemented using the plm package42. For the closest consistency between timepoints in the two studies, CpG sites associated with PEs at age 18 (p < 5 × 10−5) in the age 15–17 DNAm data from ALSPAC were tested in the E-Risk sample. Consistently detected CpG sites, that is, those detected in the top 100 most significant results of more than one EWAS in ALSPAC, were also tested in the E-Risk sample.
Results
Sample characteristics
DNAm data were available on 901 samples from cord blood, 966 at age 7, and 966 at age 15–17 (n = 845 with all three). In those with at least one DNA sample (n = 999), 978 had PE data available for at least one timepoint (age 12: n = 929; age 18: n = 816). At age 12, 12.5% (n = 116) reported PEs. At age 18, 8.8% (n = 72) reported PEs. In total, 767 individuals had DNAm and PE data available for all timepoints. Of those, 81.4% had no PEs at either timepoint; 10.2% had PEs that remitted between 12 and 18; 6.2% had PEs that emerged between 12 and 18; and 2.2% had PEs that were persistent between 12 and 18. Sample characteristics are displayed in Table 1.
Table 1.
Sample descriptives
| DNA methylation from cord blood | DNA methylation at age 7 | DNA methylation at age 17 | ||
|---|---|---|---|---|
| PEs at age 12 (n) | 833 | 900 | 899 | |
| PEs at age 18 (n) | 739 | 791 | 787 | |
| Characteristic | n | % | (missing n ) | |
| PEs at age 12 | 116 | 12.5 | 69 | |
| PEs at age 18 | 72 | 8.8 | 182 | |
| Parity | 35 | |||
| 0 | 451 | 46.8 | ||
| 1 | 358 | 37.2 | ||
| 2 | 117 | 12.1 | ||
| 3 | 37 | 3.8 | ||
| Maternal age | Mean = 29.5 | sd = 4.4 | 6 | |
| Maternal smoking | 18 | |||
| Never | 847 | 86.4 | ||
| Stopped during pregnancy | 36 | 3.7 | ||
| Continued throughout pregnancy | 97 | 9.9 | ||
| Infections during pregnancy | 445 | 44.6 | ||
| Caesarean section delivery | 92 | 9.6 | 40 | |
| Sex (male) | 485 | 48.6 |
PEs psychotic experiences
DNAm and PEs—overview
Differences in DNAm at birth, age 7, and ages 15–17 between those with and without PEs at ages 12 and 18 did not survive correction for multiple testing (p < 1.3 × 10−7; reflecting 381,871 sites in each analysis). However, a number of methylation probes were nominally significant in each analysis (p < 5 × 10−5, detailed in the Supplementary Information). The top 20 differentially methylated positions (DMPs) for each analysis are detailed in Table 2a–f, with a more extensive list provided in Supplementary Tables (top 100 DMPs: Tables S1–S6). Comparison of the top 100 DMPs across analyses showed three CpG sites (cg16459265, cg24940155, cg25184754) detected in more than one analysis and with the same direction of effect, and nine genes that were detected in more than one analysis but with different CpG sites (Supplementary Table S7).
Table 2a.
Cord blood methylation and age-12 psychotic experiences: top 20 probes
| Probe | p value | t statistic | Chromosome | Annotated gene |
|---|---|---|---|---|
| cg20862283 | 8.66E-06 | 4.48 | chr2 | |
| cg10490202 | 1.21E-05 | −4.4 | chr17 | RPTOR |
| cg21040096 | 2.15E-05 | 4.27 | chr17 | RPH3AL |
| cg18752363 | 2.21E-05 | −4.27 | chr4 | C4orf10;NOP14 |
| cg11271415 | 3.15E-05 | 4.19 | chr8 | FBXO32 |
| cg20083186 | 4.06E-05 | 4.13 | chr3 | |
| cg00712792 | 4.28E-05 | −4.11 | chr16 | SPIRE2 |
| cg11936556 | 5.14E-05 | 4.07 | chr5 | |
| cg14892222 | 5.15E-05 | 4.07 | chr15 | GOLGA8A |
| cg26672652 | 5.25E-05 | 4.07 | chr8 | PXDNL |
| cg13751188 | 5.26E-05 | −4.07 | chr11 | TECTA |
| cg14829814 | 5.39E-05 | 4.06 | chr12 | |
| cg26302009 | 6.39E-05 | 4.02 | chr6 | PHACTR1 |
| cg01840115 | 6.94E-05 | −4 | chr13 | |
| cg06893379 | 7.29E-05 | 3.99 | chr5 | |
| cg04409048 | 7.41E-05 | 3.98 | chr6 | |
| cg08105378 | 7.57E-05 | −3.98 | chr19 | HOMER3 |
| cg07438586 | 7.92E-05 | −3.97 | chr6 | GNL1 |
| cg04114269 | 8.76E-05 | −3.94 | chr3 | C3orf26;FILIP1L;MIR548G |
Italic rows indicate p < 5 × 10−5
Table 2f.
Age 15–17 methylation and age-18 psychotic experiences: top 20 probes
| Probe | p value | t statistic | Chromosome | Annotated gene |
|---|---|---|---|---|
| cg25975712 | 1.13E-05 | 4.42 | chr22 | FAM19A5 |
| cg24177611 | 1.15E-05 | −4.42 | chr10 | KNDC1 |
| cg25324164 | 1.57E-05 | −4.35 | chr11 | FADS2 |
| cg15986671 | 4.00E-05 | −4.13 | chr7 | |
| cg16073378 | 4.74E-05 | 4.09 | chr2 | |
| cg04942547 | 4.82E-05 | −4.09 | chr20 | ZFP64 |
| cg11333576 | 5.26E-05 | −4.07 | chr19 | SHC2 |
| cg16459265 | 5.51E-05 | −4.06 | chr7 | C7orf40;SNORA9 |
| cg09222367 | 7.68E-05 | −3.98 | chr15 | IQGAP1 |
| cg23415756 | 1.02E-04 | 3.91 | chr17 | NTN1 |
| cg18656829 | 1.14E-04 | 3.88 | chr13 | |
| cg06046431 | 1.15E-04 | 3.88 | chr11 | BDNF |
| cg16791444 | 1.17E-04 | −3.87 | chr17 | |
| cg11604728 | 1.17E-04 | −3.87 | chr7 | POU6F2;POU6F2 |
| cg22012759 | 1.19E-04 | −3.87 | chr1 | |
| cg19052355 | 1.25E-04 | 3.86 | chr2 | GBX2 |
| cg05580655 | 1.30E-04 | 3.85 | chr3 | SCHIP1 |
| cg16740157 | 1.43E-04 | −3.82 | chr6 | |
| cg26530497 | 0.000156178 | −3.8 | chr13 | SOHLH2 |
| cg19769301 | 0.000156717 | 3.8 | chr1 |
Italic rows indicate p < 5 × 10−5
Cord blood DNAm and PEs
Analysis of cord blood DNAm identified seven nominally significant CpG sites associated with age-12 PEs (Table 2a), and five with age-18 PEs (Table 2b). The top CpG site at age 12 was cg20862283 (p = 8.66 × 10-6), which was not mapped to a RefSeq gene. GO enrichment analysis of the CpG sites where p < 0.001 from analysis of PEs at age 12 identified 104 groups of related GO categories (p < 0.05, Table S8). At age 18, the top CpG site was cg00407329 (p = 1.84 × 10−5), which was annotated with the gene SIM1. Fifty-one groups of GO enrichment terms were identified (Table S9). Ranked highly among the grouped terms were “Wnt-activated receptor activity” and “central nervous system neuron development”. The top-ranked terms did not show any similarities between timepoints.
Table 2b.
Cord blood methylation and age-18 psychotic experiences: top 20 probes
| Probe | p value | t statistic | Chromosome | Annotated gene |
|---|---|---|---|---|
| cg00407329 | 1.84E-05 | 4.31 | chr6 | SIM1 |
| cg17972930 | 2.76E-05 | 4.22 | chr15 | USP50 |
| cg07571954 | 3.22E-05 | −4.18 | chr10 | C10orf131 |
| cg05138918 | 4.56E-05 | −4.1 | chr3 | |
| cg17843418 | 4.64E-05 | −4.1 | chr17 | TBC1D16 |
| cg21167905 | 8.29E-05 | 3.96 | chr1 | C1orf9 |
| cg09825080 | 8.67E-05 | −3.95 | chr16 | |
| cg02424103 | 9.58E-05 | 3.92 | chr3 | |
| cg16426764 | 0.0001 | −3.9 | chr10 | LHPP |
| cg00680277 | 0.00012 | 3.87 | chr2 | TTC15 |
| cg23463608 | 0.00012 | −3.86 | chr19 | GNG7 |
| cg25930186 | 0.00012 | −3.86 | chr20 | MOCS3;DPM1 |
| cg01395281 | 0.00013 | −3.86 | chr1 | ZNF593 |
| cg25275750 | 0.00015 | 3.82 | chr13 | TBC1D4 |
| cg16327647 | 0.00016 | −3.8 | chr1 | CENPF |
| cg01496136 | 0.00017 | −3.78 | chr16 | |
| cg09568647 | 0.00017 | −3.77 | chr5 | |
| cg22694818 | 0.00018 | 3.77 | chr1 | LHX8 |
| cg06475223 | 0.00018 | −3.76 | chr18 | LAMA3 |
| cg12859112 | 0.00019 | 3.76 | chr6 | TMEM14B |
Italic rows indicate p < 5 × 10−5
Age-7 DNAm and PEs
Analysis of age-7 DNAm identified 63 nominally significant CpG sites associated with age-12 PEs (Table 2c), and 19 with age-18 PEs (Table 2d). At age 12, the top CpG site was cg22499215 (p = 2.97 × 10−7), which was not annotated with a RefSeq gene. In the top CpG sites, 92 groups of GO terms were identified (Table S10), including a number of terms related to brain development and “Wnt signalling pathway”. At age 18, the top CpG site was cg00995854 (p = 2.74 × 10−6, CD5L). In GO enrichment analysis, 137 groups of terms were identified (Table S11). The top-ranked term was “proteoglycan binding”, including a number of terms related to immune processes, as well as “neurogenesis” and terms representing neuron development and projection. The top-ranked terms did not show any similarities between timepoints.
Table 2c.
Age-7 methylation and age-12 psychotic experiences: top 20 probes
| Probe | p value | t statistic | Chromosome | Annotated gene |
|---|---|---|---|---|
| cg22499215 | 2.97E-07 | −5.16 | chr22 | |
| cg14263553 | 1.12E-06 | −4.9 | chr8 | |
| cg07366553 | 2.35E-06 | −4.75 | chr22 | SELO |
| cg09553452 | 2.75E-06 | −4.72 | chr16 | CDH5 |
| cg02284814 | 4.82E-06 | −4.6 | chr1 | PPP1R12B |
| cg17160666 | 4.95E-06 | −4.6 | chr10 | |
| cg17139666 | 5.04E-06 | 4.59 | chr4 | |
| cg08032135 | 5.08E-06 | 4.59 | chr8 | NRG1 |
| cg04584761 | 1.00E-05 | −4.56 | chr2 | SUPT7L |
| cg10756647 | 1.00E-05 | 4.55 | chr7 | PSPH |
| cg16451872 | 1.00E-05 | 4.54 | chr15 | UBE3A |
| cg10725301 | 1.00E-05 | −4.52 | chr6 | ZKSCAN3 |
| cg18477816 | 1.00E-05 | −4.52 | chr11 | IRF7 |
| cg10143301 | 1.00E-05 | 4.51 | chr5 | TXNDC15 |
| cg02572956 | 1.00E-05 | −4.48 | chr7 | CHPF2;MIR671 |
| cg01040786 | 1.00E-05 | −4.41 | chr5 | |
| cg02975060 | 1.00E-05 | −4.4 | chr14 | MIR1185-2 |
| cg23658045 | 1.00E-05 | 4.4 | chr12 | DCTN2 |
| cg10151454 | 1.00E-05 | −4.36 | chr10 | EBF3 |
| cg10303842 | 2.00E-05 | 4.35 | chr5 | CDH12 |
Italic rows indicate p < 5 × 10−5
Table 2d.
Age-7 methylation and age-18 psychotic experiences: top 20 probes
| Probe | p value | t statistic | Chromosome | Annotated gene |
|---|---|---|---|---|
| cg00995854 | 2.74E-06 | −4.72 | chr1 | CD5L |
| cg13845105 | 3.08E-06 | −4.7 | chr12 | RPL6 |
| cg27190398 | 8.76E-06 | −4.48 | chr16 | STUB1;JMJD8 |
| cg07143863 | 9.18E-06 | −4.47 | chr12 | |
| cg10468951 | 1.33E-05 | −4.38 | chr3 | C3orf45 |
| cg05927274 | 1.79E-05 | −4.32 | chr1 | |
| cg09936919 | 1.90E-05 | −4.3 | chr2 | |
| cg15129144 | 1.95E-05 | −4.3 | chr2 | EPAS1 |
| cg03332469 | 2.00E-05 | 4.28 | chr7 | |
| cg02743632 | 2.00E-05 | −4.28 | chr1 | HLX |
| cg20477259 | 2.00E-05 | −4.25 | chr6 | TNF |
| cg24110396 | 3.00E-05 | −4.22 | chr10 | CCNY |
| cg27185423 | 3.00E-05 | −4.21 | chr15 | |
| cg08522143 | 3.00E-05 | −4.19 | chr19 | ZNF14 |
| cg04936274 | 3.00E-05 | −4.18 | chr10 | |
| cg14468692 | 3.00E-05 | −4.18 | chr11 | RNH1 |
| cg15031103 | 4.00E-05 | −4.15 | chr10 | LRRC27 |
| cg05072413 | 4.00E-05 | −4.11 | chr22 | FAM19A5 |
| cg16459265 | 4.00E-05 | −4.11 | chr7 | C7orf40;SNORA9 |
| cg02108135 | 5.00E-05 | −4.06 | chr17 |
Italic rows indicate p < 5 × 10−5
Age-15–17 DNAm and PEs
Analysis of age-15–17 DNAm identified ten nominally significant CpG sites associated with age-12 PEs (Table 2e), and six with age-18 PEs (Table 2f). At age 12, the top CpG site was cg14284469 (p = 1.87 × 10−6, STMN2), and 114 grouped GO terms were identified (Table S12). At age 18, the top CpG site was cg25975712 (p = 1.13 × 10−5, FAM19A5), and 123 grouped GO terms were identified (Table S13). The top-ranked term was “rostrocaudal neural tube patterning”, which largely included terms related to neuronal and physical development and morphogenesis. Also within the top grouped terms were “behavioural fear response” and “behavioural defense response”. The top-ranked terms did not show any similarities between timepoints.
Table 2e.
Age 15–17 methylation and age-12 psychotic experiences: top 20 probes
| Probe | p value | t statistic | Chromosome | Annotated gene |
|---|---|---|---|---|
| cg14284469 | 1.87E-06 | 4.8 | chr8 | STMN2 |
| cg00956759 | 2.94E-06 | −4.71 | chr17 | |
| cg12009697 | 4.13E-06 | −4.63 | chr2 | |
| cg21602768 | 8.15E-06 | −4.49 | chr20 | ABHD12 |
| cg17027353 | 1.25E-05 | 4.39 | chr6 | |
| cg14386808 | 1.34E-05 | −4.38 | chr8 | |
| cg22035229 | 1.77E-05 | −4.32 | chr1 | MSH4 |
| cg16817992 | 2.11E-05 | 4.28 | chr7 | KCNH2 |
| cg25344401 | 3.26E-05 | 4.18 | chr7 | FOXK1 |
| cg05942128 | 4.71E-05 | 4.09 | chr2 | HOXD11 |
| cg26370729 | 5.19E-05 | 4.07 | chr5 | F2RL1 |
| cg25024734 | 5.84E-05 | 4.04 | chr10 | |
| cg03945301 | 6.32E-05 | −4.02 | chr6 | EHMT2 |
| cg17103467 | 7.84E-05 | −3.97 | chr1 | WDR8 |
| cg24695977 | 7.92E-05 | 3.97 | chr1 | |
| cg04554240 | 8.12E-05 | −3.96 | chr3 | ESYT3 |
| cg08621773 | 1.02E-04 | −3.9 | chr2 | |
| cg27280332 | 1.04E-04 | −3.9 | chr12 | |
| cg25827710 | 0.00011 | −3.88 | chr13 | LCP1 |
| cg15318176 | 0.00012 | −3.86 | chr7 | PTPRN2 |
Italic rows indicate p < 5 × 10−5
DNAm from cord blood, at age 7, and at ages 15–17 and continuity of PEs between ages 12 and 18
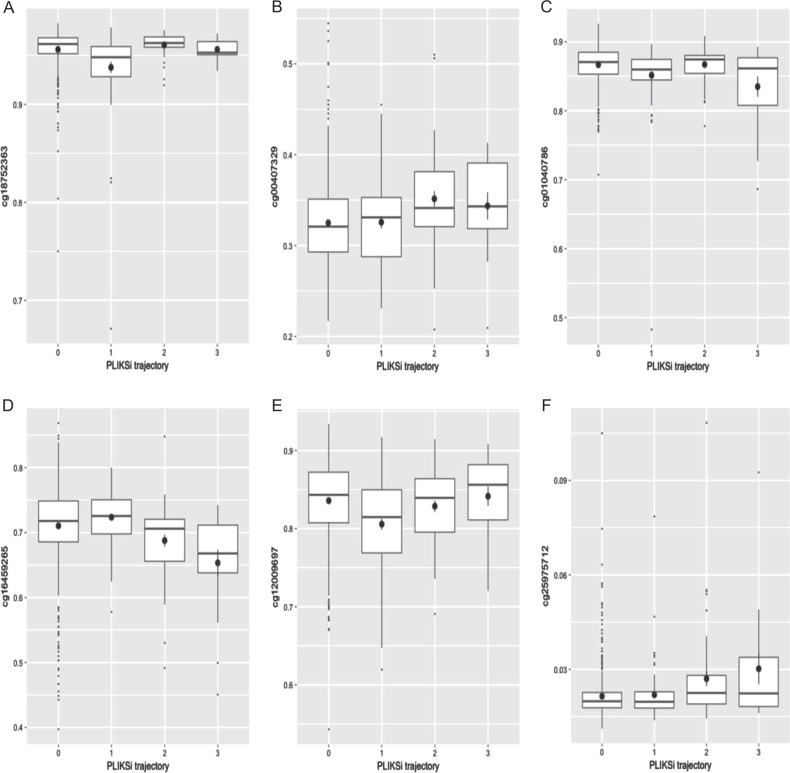
Next, we tested the association between timepoint-specific DNAm and continuity of PEs across adolescence for each of the top CpG sites in each EWAS (p < 5 × 10−5). Results for these analyses are provided in the Supplementary Information, Table S14. Different patterns of DNAm at each timepoint were observed in relation to PE continuity. At some CpG sites, statistical differences between the groups were limited to one group (e.g. cg18752363, Fig. 1a). At other CpG sites, individuals whose PEs had remitted by age 18 had DNAm profiles more similar to those who never had any PEs, whereas those with persistent PEs tended to show the greatest differences in DNAm profiles, while those whose PEs emerged between 12 and 18 often had intermediary differences between the none/remitted groups and the persistent group (e.g. cg16459265, Fig. 1d; full statistical results reported in Table S14, Fig. 1a–f).
Fig. 1.
a–f DNA methylation and the continuity of psychotic experiences (PEs) between ages 12 and 18. Boxplots represent the most strongly associated CpG site (tested using ANOVA) at each timepoint. a DNA methylation from cord blood, CpG site detected at age 12; b DNA methylation from cord blood, CpG site detected at age 18; c DNA methylation at age 7, CpG site detected at age 12; d DNA methylation at age 7, CpG site detected at age 18; e DNA methylation at age 15–17, CpG site detected at age 12; f DNA methylation at age 15–17, CpG site detected at age 18. For each boxplot, 0 = never had PEs, 1 = PEs remitted between 12 and 18, 2 = PEs emerged between 12 and 18, and 3 = PEs persisted between 12 and 18 years. Circles represent group means. PLIKSi Psychosis-like symptoms semi-structured interview
Longitudinal DNAm changes and continuity of PEs
We tested the association between longitudinal DNAm and continuity of PEs between 12 and 18 for the previously identified top CpG sites. Results are provided in Supplementary Table 15 for the full longitudinal model. Longitudinal models were unaffected by the inclusion of a DNAm-derived smoking score. For several CpG sites, the methylation trajectory was nominally different between groups (remitted, emergent, or persistent PEs—examples are given in Fig. 2).
Fig. 2.
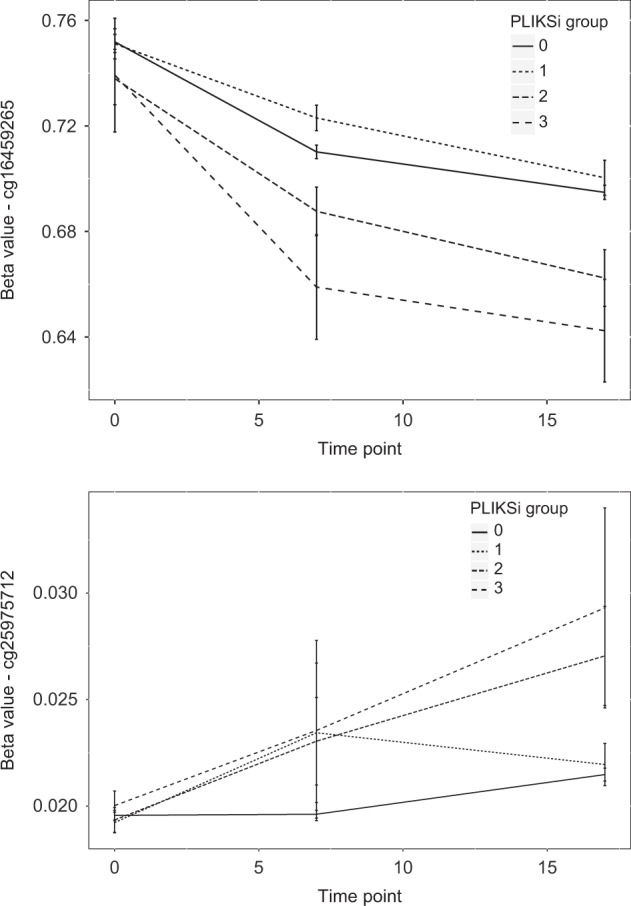
a, b DNA methylation trajectories by PLIKSi psychotic experiences (PEs) group: 0 = no PEs at either timepoint; 1 = PEs at age 12 that remitted by age 18; 2 = no PEs at age 12 but emerged by age 18; 3 = PEs that persisted from age 12 to age 18. a cg16459265 methylation and PEs. b cg25975712 methylation and PEs. Lines represent mean DNA methylation per sample timepoint; bars represent standard errors. PLIKSi Psychosis-like symptoms semi-structured interview
Differentially methylated regions
We tested for DMRs using the comb-p algorithm. Significant DMRs (Šidák corrected p < 0.05) were detected at all timepoints, and are detailed in the Supplementary Information (Table S16). Nine DMRs were identified in cord blood associated with age-12 PEs; four with age-18 PEs. Thirty-five DMRs were identified in age-7 samples associated with age-12 PEs, 11 with age-18 PEs. Fourteen DMRs were identified in age 15–17 samples associated with age-12 PEs, 11 with age-18 PEs. Of note, six regions were differentially methylation in association with either age-12 or age-18 PEs in both age-7 and age 15–17 samples, spanning regions including HLA-DPB2 and HIVEP3 (associated with age-12 PEs), and GDF7 (age-18 PEs). A full list of significant DMRs at each timepoint is available in the Supplementary Information, Table S16.
Replication
No associations between DNAm at age 15–17 and PEs at age 18 in the ALSPAC sample were replicated in the E-Risk sample (all p’s > 0.05). However, DNAm at the CpG site cg16459265 (consistently detected among the top 100 CpG sites in the ALSPAC sample across the different ages) was found to be associated with age-18 PEs in the E-Risk sample. At this probe, individuals with any PEs at age 18 had marginally lower DNAm than those without PEs (difference = −0.76%, p = 0.0029) in keeping with the hypomethylation found in the ALSPAC sample.
Discussion
Summary
This study utilised longitudinal data in a large, population-based sample to investigate the association between epigenome-wide DNAm patterns and the emergence and persistence of PEs in childhood and adolescence. While no CpG sites reached Bonferroni-corrected significance in this study, a number of CpG sites and genes in biological pathways relevant to psychosis appeared highly ranked in these analyses. Genes detected in the top 100 CpG sites of more than one analysis (though not with the same CpG site) included BAIAP2, FGFR3 and MAD1L1, and genetic mutations in these genes have previously been implicated in susceptibility to psychotic disorders43–45. Also consistently detected were LFNG and LRP5, part of the Notch and Wnt signalling pathways (respectively), which have been reported in previous studies of psychiatric phenotypes including schizophrenia46,47. None of the individual CpG sites associated with PEs at age 18 in the ALSPAC sample were replicated in the independent E-Risk sample. Nonetheless, attempts to replicate these results should be undertaken in other large population-based longitudinal samples. Using a DMR approach, six regions associated with PEs were detected consistently in both age-7 and age 15–17 samples. The top DMR associated with age-12 PEs at age 7 (and also significant at age 15–17) spanned 18 probes in the HLA-DPB2, part of the major histocompatibility complex on chromosome 6, which has been widely implicated in schizophrenia48. The top DMR associated with age-18 PEs in age-7 samples overlapped substantially with the top DMR in age 15–17 samples, spanning 7–9 probes on chromosome 2 at exon 2 of the GDF7 gene. DNAm at this gene has previously been associated with early age of onset in anorexia nervosa49 and is thought to play a role in nervous system development.
The top CpG sites identified showed interesting patterns of change in DNAm across development according to the continuity of PEs across adolescence. Some CpG sites distinguished individuals whose PEs remitted or never had PEs from those whose experiences persisted across or emerged during adolescence. An example is CpG site cg16459265 (annotated to the RefSeq genes C7orf40;SNORA9), a top CpG site associated with age-18 PEs in DNA samples from both childhood (age 7) and adolescence (ages 15–17). Lower DNAm at this CpG site was also associated with PEs at age 18 in the E-Risk sample. Similarly, at the CpG site cg25975712 (FAM19A5), DNAm patterns diverged during childhood according to PE status, but by adolescence, those whose PEs remitted had DNAm profiles more similar to those with no PEs (though it should be noted that DNAm is generally low at this CpG site).
Comparison to the literature
Two studies have previously investigated the association between DNAm and subclinical psychotic phenomena. However, there are some differences in the study design between the current study and this previous research. We utilised DNAm from birth to adolescence in a large, population-based, unrelated sample, whereas earlier studies were conducted in smaller samples of monozygotic twin pairs discordant for childhood psychotic symptoms23 or examined conversion to psychosis in ultra-high risk, help-seeking individuals24. Study participants also differed in age at sample collection (ages 5 and 10 in ref. 23; age 16–30 in ref. 24) and sample type (buccal swabs in ref. 23). The differences in time periods are an important consideration when comparing these studies, as there is little concurrence. The current study considers a broad span of ages, compared to shorter intervals or more specific timepoints reported in the previous reports. While the ages included in this study cover important timepoints developmentally, it is not possible to accurately infer what is happening in the intervening years, or identify changes in DNAm accompanying the specific period that the PEs were measured. As such, it is perhaps unsurprising that none of the CpG site associations detected here are seen in either previous study. Furthermore, we do not have information regarding psychotic phenomena in the years following those included in this study. Nevertheless, associations linked to a small number of genes are observed across the studies. One example is the gene PTPRN2, which is thought to be necessary for normal neurotransmitter activity in the brain. Of note, the DMR located on chromosome 6 (and annotated to the gene HLA-DPB2) which was associated with age-12 PEs in our study substantially overlaps with a similar DMR detected in prefrontal cortex samples associated with polygenic risk for schizophrenia50. Taken together, these studies provide tentative early evidence for potential methylomic changes accompanying the development of early psychosis-related phenomena, albeit with some inconsistency regarding the location of these changes.
Methodological considerations
This study has a number of strengths. Analyses were conducted in the relatively large ARIES sample drawn from a population-based cohort study, with DNAm profiled at multiple timepoints across early development and PEs assessed in childhood and adolescence. Prospective collection of high-quality phenotypic data in this cohort allowed for investigation of the temporal relationship between DNAm and PEs and repeated assessments enabled investigation of trajectories of methylation and continuity of PEs. Extensive data regarding potential confounding factors were also available and included in the analyses. Furthermore, this is the largest study to date to examine the association between DNAm and pre-clinical PEs. Research investigating DNAm in psychotic disorders is typically performed in adult samples, where the confounding effects of medication use are difficult to avoid. As ARIES is a young, population-based sample, only one participant was using psychiatric medication (and was excluded from these analyses). Additionally, the longitudinal investigation of early pre-clinical psychotic experiences is particularly important to better understand biological factors associated with the aetiology of psychotic and other psychiatric disorders and facilitate early identification of those at risk.
However, there are methodological factors that should be taken into consideration when interpreting these results. Firstly, DNAm data were generated from peripheral samples (cord blood and whole blood). This approach is necessary when conducting research in large longitudinal samples (i.e. with live participants), where the most relevant tissue (brain) is not available. Previous research has demonstrated tissue specificity in DNAm patterns51, limiting the conclusions that can be drawn from studies performed in blood. However, comparison of DNAm from blood and brain samples is possible in reference datasets. For example, for the CpG site cg16459265, although DNAm in blood samples is lower and shows more variation than in brain tissue, DNAm in blood is strongly correlated with DNAm in matched samples of prefrontal cortex, entorhinal cortex, superior temporal gyrus, and cerebellum52. Nevertheless, for an epigenetic factor to have utility as a biomarker or to facilitate early intervention, it must be detectable in peripheral samples. Secondly, despite the size and high longitudinal retention of the ARIES subsample (n~1000), numbers in the current analyses were reduced by the availability of both biological and phenotypic data. Previous research in ALSPAC has demonstrated non-random attrition, whereby participants with higher polygenic risk scores for schizophrenia were less likely to complete questionnaires and attend data collection sessions53, which may have reduced the power to detect factors associated with psychosis-related phenotypes in this study. Thirdly, it is difficult to identify the biological plausibility of these results, especially given the small differences detected which may represent technical artefacts rather than variation attributable to differences in PEs. However, given the focus here on pre-clinical symptoms of a complex psychiatric phenotype, it is to be expected that relatively small differences in DNAm would be observed, likely reflecting many genes of small effect. In addition, the scale of genome-wide epigenetic profiling techniques means that stringent multiple testing correction is required when interpreting the results. In this study, no CpG site in the EWAS analyses reached Bonferroni-corrected levels of significance. However, we were able to replicate one association (cg16459265) in an independent sample and observed effects in related gene sets such as PTPRN2 and HLA-DPB2 in other studies. DNAm profiles across the array are not independent, and therefore adherence to strict multiple-testing thresholds, particularly in complex phenotypes, may lead to disregarding potentially interesting findings. Finally, only one of the CpG sites associated with PEs in the ALSPAC sample was replicated in the independent E-Risk sample. However, this may be due to methodological differences in the samples, such as the differences in timings between DNA sample collection and the data collection for PEs (DNA sample collection preceded the measurement of PEs in ALSPAC; in E-Risk they were collected concurrently).
Conclusions
In conclusion, we found that DNAm across childhood and adolescence may be associated with the emergence and continuity of PEs between the ages of 12 and 18. Previous research has identified epigenetic patterns related to clinical diagnoses of psychotic disorders15–22, although to date very few have focussed on pre-clinical symptoms23,24 and thus the current findings substantially extend this literature. Research uncovering early biomarkers associated with PEs is important as it may, in the future, have the potential to facilitate early identification of individuals at increased risk of a range of mental health problems, and facilitate targeting of preventive interventions. Considered together with previous research, our findings provide tentative evidence for potential methylomic changes across timepoints spanning the development of early psychotic phenomena.
Supplementary information
Acknowledgements
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We also greatly appreciate the participation of all the study mothers and fathers, and the twins in the E-Risk study. Our thanks to members of the E-Risk team for their dedication, hard work and insights, and especially to Professors Terrie Moffitt and Avshalom Caspi who founded the study. This study was supported by an MQ Fellows Award to H.L.F. (MQ14F40). Core programme support for ALSPAC is provided by the Medical Research Council (MRC) and the Wellcome Trust (Grant ref: 102215/2/13/1) and the University of Bristol; ARIES was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) (Grant refs: BBI025751/1 and BB/I025263/1). The study was also supported by the NIHR Bristol Biomedical Research Centre. Supplementary funding to generate DNAm data which are (or will be) included in ARIES has been obtained from the MRC, ESRC, NIH, and other sources. ARIES is maintained under the auspices of the MRC Integrative Epidemiology Unit at the University of Bristol (grant numbers MC_UU_12013/2, MC_UU_12013/8, and MC_UU_12013/9). The E-Risk Study is funded by the Medical Research Council (G1002190). Additional support was provided by National Institute of Child Health and Human Development (HD077482) and the Jacobs Foundation. L.A. is the Mental Health Leadership Fellow for the UK ESRC.
Code availability
All packages, programmes and pipelines used are detailed above and freely available. Any additional code is available from the authors on request.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-019-0407-8).
References
- 1.Polanczyk G, et al. Etiological and clinical features of childhood psychotic symptoms: results from a birth cohort. Arch. Gen. Psychiatry. 2010;67:328–338. doi: 10.1001/archgenpsychiatry.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horwood J, et al. IQ and non-clinical psychotic symptoms in 12-year-olds: results from the ALSPAC birth cohort. Br. J. Psychiatry. 2008;193:185–191. doi: 10.1192/bjp.bp.108.051904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher HL, et al. Specificity of childhood psychotic symptoms for predicting schizophrenia by 38 years of age: a birth cohort study. Psychol. Med. 2013;43:2077–2086. doi: 10.1017/S0033291712003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poulton R, et al. Children’s self-reported psychotic symptoms and adult schizophreniform disorder: a 15-year longitudinal study. Arch. Gen. Psychiatry. 2000;57:1053–1058. doi: 10.1001/archpsyc.57.11.1053. [DOI] [PubMed] [Google Scholar]
- 5.Kelleher I, et al. Psychotic symptoms and population risk for suicide attempt: a prospective cohort study. JAMA Psychiatry. 2013;70:940–948. doi: 10.1001/jamapsychiatry.2013.140. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez MDG, Wichers M, Lieb R, Wittchen HU, van Os J. Evidence that onset of clinical psychosis is an outcome of progressively more persistent subclinical psychotic experiences: an 8-year cohort study. Schizophr. Bull. 2011;37:84–93. doi: 10.1093/schbul/sbp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartels-Velthuis AA, Wigman JTW, Jenner JA, Bruggeman R, van Os J. Course of auditory vocal hallucinations in childhood: 11-year follow-up study. Acta Psychiatr. Scand. 2016;134:6–15. doi: 10.1111/acps.12571. [DOI] [PubMed] [Google Scholar]
- 8.Menke A, Binder EB. Epigenetic alterations in depression and antidepressant treatment. Dialogues Clin. Neurosci. 2014;16:395–404. doi: 10.31887/DCNS.2014.16.3/amenke. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakusic J, Schaufeli W, Claes S, Godderis L. Stress, burnout and depression: a systematic review on DNA methylation mechanisms. J. Psychosom. Res. 2017;92:34–44. doi: 10.1016/j.jpsychores.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 10.El-Sayed AM, Haloossim MR, Galea S, Koenen KC. Epigenetic modifications associated with suicide and common mood and anxiety disorders: a systematic review of the literature. Biol. Mood Anxiety Disord. 2012;2:10. doi: 10.1186/2045-5380-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uddin M, Sipahi L, Li J, Koenen KC. Sex differences in DNA methylation may contribute to risk of PTSD and depression: a review of existing evidence. Depress Anxiety. 2013;30:1151–1160. doi: 10.1002/da.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loke YJ, Hannan AJ, Craig JM. The role of epigenetic change in autism spectrum disorders. Front. Neurol. 2015;6:107. doi: 10.3389/fneur.2015.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teroganova N, Girshkin L, Suter CM, Green MJ. DNA methylation in peripheral tissue of schizophrenia and bipolar disorder: a systematic review. BMC Genet. 2016;17:27. doi: 10.1186/s12863-016-0332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pidsley R, Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol. Psychiatry. 2011;69:146–156. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Dempster EL, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum. Mol. Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aberg KA, et al. Methylome-wide association study of schizophrenia: identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishioka M, et al. Comprehensive DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia. J. Hum. Genet. 2013;58:91–97. doi: 10.1038/jhg.2012.140. [DOI] [PubMed] [Google Scholar]
- 18.van Eijk KR, et al. Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood. Eur. J. Hum. Genet. 2015;23:1106–1110. doi: 10.1038/ejhg.2014.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pidsley R, et al. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannon E, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17:176. doi: 10.1186/s13059-016-1041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mill J, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wockner LF, et al. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl. Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisher HL, et al. Methylomic analysis of monozygotic twins discordant for childhood psychotic symptoms. Epigenetics. 2015;10:1014–1023. doi: 10.1080/15592294.2015.1099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kebir O, et al. Methylomic changes during conversion to psychosis. Mol. Psychiatry. 2017;22:512–518. doi: 10.1038/mp.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd A, et al. Cohort profile: the ‘Children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2012;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser A, et al. Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int. J. Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relton CL, et al. Data Resource Profile: Accessible Resource for Integrated Epigenomic Studies (ARIES) Int. J. Epidemiol. 2015;44:1181–1190. doi: 10.1093/ije/dyv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zammit S, et al. Psychotic experiences and psychotic disorders at age 18 in relation to psychotic experiences at age 12 in a longitudinal population-based cohort study. Am. J. Psychiatry. 2013;170:742–750. doi: 10.1176/appi.ajp.2013.12060768. [DOI] [PubMed] [Google Scholar]
- 29.Min JL, Hemani G, Davey Smith G, Relton C, Suderman M. Meffil: efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics. 2018;34:3983–3989. doi: 10.1093/bioinformatics/bty476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Ya, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naeem H, et al. Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics. 2014;15:51. doi: 10.1186/1471-2164-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaushal A, et al. Comparison of different cell type correction methods for genome-scale epigenetics studies. BMC Bioinformatics. 2017;18:216. doi: 10.1186/s12859-017-1611-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGregor K, et al. An evaluation of methods correcting for cell-type heterogeneity in DNA methylation studies. Genome Biol. 2016;17:84. doi: 10.1186/s13059-016-0935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elliott HR, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin. Epigenet. 2014;6:4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeilinger S, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gene Ontology Consortium. Gene ontology consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffitt TE, the E. Risk Study Team. Teen-aged mothers in contemporary Britain. J. Child Psychol. Psychiatry. 2002;43:727–742. doi: 10.1111/1469-7610.00082. [DOI] [PubMed] [Google Scholar]
- 39.Marzi SJ, et al. Analysis of DNA methylation in young people reveals limited evidence for an association between victimization stress and epigenetic variation in blood. Am. J. Psychiatry. 2018;175:517–529. doi: 10.1176/appi.ajp.2017.17060693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newbury J, et al. Cumulative effects of neighborhood social adversity and personal crime victimization on adolescent psychotic experiences. Schizophr. Bull. 2018;44:348–358. doi: 10.1093/schbul/sbx060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pidsley R, et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croissant Y, Millo G. Panel data econometrics in R: the plm package. J. Stat. Softw. 2008;27:1–43. [Google Scholar]
- 43.Ikeda M, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry. 2018;23:639–647. doi: 10.1038/mp.2016.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang J, Park H, Kim E. IRSp53/BAIAP2 in dendritic spine development, NMDA receptor regulation, and psychiatric disorders. Neuropharmacology. 2016;100:27–39. doi: 10.1016/j.neuropharm.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 45.van Scheltinga AFT, Bakker SC, Kahn RS. Fibroblast growth factors in schizophrenia. Schizophr. Bull. 2010;36:1157–1166. doi: 10.1093/schbul/sbp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shayevitz C, Cohen OS, Faraone SV, Glatt SJ. A re-review of the association between the NOTCH4 locus and schizophrenia. Am. J. Med Genet Part B, Neuropsychiatr. Genet. 2012;159:477–483. doi: 10.1002/ajmg.b.32050. [DOI] [PubMed] [Google Scholar]
- 47.Singh KK. An emerging role for Wnt and GSK3 signaling pathways in schizophrenia. Clin. Genet. 2013;83:511–517. doi: 10.1111/cge.12111. [DOI] [PubMed] [Google Scholar]
- 48.Debnath M, Berk M, Leboyer M, Tamouza R. The MHC/HLA gene complex in major psychiatric disorders: emerging roles and implications. Curr. Behav. Neurosci. Rep. 2018;5:179–188. [Google Scholar]
- 49.Booij L, et al. DNA methylation in individuals with anorexia nervosa and in matched normal-eater controls: a genome-wide study. Int J. Eat. Disord. 2015;48:874–882. doi: 10.1002/eat.22374. [DOI] [PubMed] [Google Scholar]
- 50.Viana J, et al. Schizophrenia-associated methylomic variation: molecular signatures of disease and polygenic risk burden across multiple brain regions. Hum. Mol. Genet. 2016;26:210–225. doi: 10.1093/hmg/ddw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies MN, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–1032. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin J, et al. Association of genetic risk for schizophrenia with nonparticipation over time in a population-based cohort study. Am. J. Epidemiol. 2016;183:1149–1158. doi: 10.1093/aje/kww009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.