Abstract
Cyanobacteria are among only a few organisms that naturally synthesize long-chain alkane and alkene hydrocarbons. Cyanobacteria use one of two pathways to synthesize alka/enes, either acyl-ACP reductase (Aar) and aldehyde deformylating oxygenase (Ado) or olefin synthase (Ols). The genomes of cyanobacteria encode one of these pathways but never both, suggesting a mutual exclusivity. We studied hydrocarbon pathway compatibility using the model cyanobacterium Synechococcus sp. PCC 7002 (S7002) by co-expressing Ado/Aar and Ols and by entirely replacing Ols with three other types of hydrocarbon biosynthetic pathways. We find that Ado/Aar and Ols can co-exist and that slower growth occurs only when Ado/Aar are overexpressed at 38 °C. Furthermore, Ado/Aar and the non-cyanobacterial enzymes UndA and fatty acid photodecarboxylase are able to substitute for Ols in a knockout strain and conditionally rescue slow growth. Production of hydrocarbons by UndA in S7002 required a rational mutation to increase substrate range. Expression of the non-native enzymes in S7002 afforded unique hydrocarbon profiles and alka/enes not naturally produced by cyanobacteria. This suggests that the biosynthetic enzyme and the resulting types of hydrocarbons are not critical to supporting growth. Exchanging or mixing hydrocarbon pathways could enable production of novel types of CO2-derived hydrocarbons in cyanobacteria.
Introduction
Cyanobacteria are an ancient and highly diverse phylum of photosynthetic prokaryotes with the capability to reproduce using (sun)light and carbon dioxide (CO2) as their primary energy and carbon sources, respectively. Extant cyanobacteria are key players in the global carbon and nitrogen cycles and as primary producers in the open ocean1,2. It has been known since the late 1960s that cyanobacteria naturally synthesize branched and linear C15 to C19 alkanes and alkenes3,4. The total amount of hydrocarbon produced per cell is around 0.02–0.35 percent of dry cell weight (% DCW)5,6. Importantly, the carbon chain lengths of most cyanobacterial hydrocarbons are within the range of fuels used currently in jet (C8–C16) and diesel engines (C9–C36)7,8, and there is, accordingly, interest in developing these organisms as bio-production platforms for CO2-derived biofuels9–13. In cyanobacteria, alka/ene hydrocarbons most likely localize to the thylakoid and plasma membranes14 where they serve numerous important roles. Knock-out strains that are unable to produce alka/enes exhibit a variety of phenotypes including greatly decreased growth rates, increased cyclic electron flow in the photosystems, changes in cell morphology, and salt sensitivity14–16. Typically, the slower growth of the knockout strains is more pronounced at lower growth temperatures15,17. Although many of the impacts of hydrocarbons on cells are known, the exact mechanism by which they produce these effects in vivo remains enigmatic.
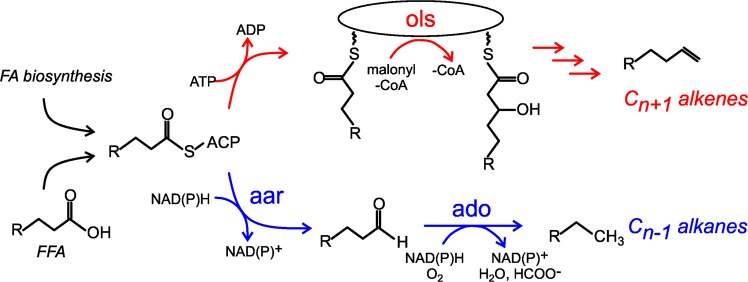
Two distinct biosynthetic pathways for alka/enes have been characterized in cyanobacteria18,19 (Fig. 1). The substrates for these native pathways are fatty acids (FAs) covalently linked to acyl carrier protein (fatty acyl-ACPs) produced by the FA synthase pathway20. One type of hydrocarbon biosynthesis pathway relies on two enzymes to convert acyl-ACP to alkane (Fig. 1, blue path). The first enzyme, acyl-ACP reductase (Aar), reduces substrate to the corresponding free fatty aldehyde using NAD(P)H as a cofactor18,21. The second enzyme, aldehyde deformylating oxygenase (Ado), uses molecular dioxygen and NAD(P)H to deformylate the Aar-produced fatty aldehyde to the corresponding Cn-1 alkane18,22,23. Depending on the substrate captured by Aar, this pathway can result in the production of methyl-branched alkanes or alkenes with an internal desaturation5.
Figure 1.
The two major alka/ene hydrocarbon biosynthetic pathways in cyanobacteria are the Ols (red) and Ado/Aar (blue) pathways. Fatty acid (FA) biosynthesis or activation from free fatty acids (FFA) provides acyl-ACP substrates for both pathways.
The second type utilizes a large, modular multi-domain enzyme called the olefin synthase (Ols) that is related to polyketide synthases19 (Fig. 1, red path). Ols captures fatty acyl-ACP substrates and extends the carbon chain via a decarboxylative condensation reaction with malonyl-CoA to generate a covalently bound Cn+2 acyl chain. This intermediate is decarboxylated to generate the Cn+1 alkene product with a terminal double bond, an α-olefin19,24. Enzymatic desaturation of acyl-ACP prior to capture by Ols results in production of doubly-desaturated 1,14-nonadecadiene17. The ratio of 1,14-nonadecadiene to 1-nonadecene is dramatically higher at lower growth temperatures due to increased production of singly desaturated acyl-ACP precursor17,19.
Genomic comparisons of cyanobacteria have shown that Ols or Ado/Aar genes are present in nearly all characterized members, though the Ado/Aar pathway is much more prevalent than Ols5. Interestingly, it was found during these studies that no cyanobacterium encodes genes for both of the pathways5. The reason for this apparent mutual exclusivity remains unknown but it could be due to a negative selection mechanism that precludes the co-existence of Ado/Aar and Ols in a cell. In order to gain insights into the compatibility between the different hydrocarbon pathways, we first studied the effects of co-expressing the Ols and Ado/Aar pathways in the model unicellular cyanobacterium Synechococcus sp. PCC 7002 (S7002). We then tested whether non-native hydrocarbon biosynthetic pathways could functionally substitute for Ols in a knockout strain of S7002. Our data show that Ado/Aar from other model cyanobacteria as well as disparate hydrocarbon biosynthetic enzymes from non-cyanobacteria including Pseudomonas and eukaryotic green algae are able to replace Ols and rescue the slow growth of alkene-deficient S7002. This suggests that these diverse pathways are interchangeable to some extent. Our results also show that the biosynthetic source of alka/enes is not critical to serving their role and various types of alka/ene hydrocarbons are able to support growth of S7002. This work implies malleability in bacterial hydrocarbon synthesis and could enable strategies for expanding the biosynthetic range of accessible alka/ene biofuels from CO2 using cyanobacteria.
Results
Introduction of the Ado/Aar genes into S7002
To express the Ado/Aar pathway in S7002, we constructed a series of autonomously replicating plasmids derived from the broad host-range vector pVZ32125. The Materials and Methods section provides details on plasmid assembly and Supplementary Figure S1 diagrams the construction workflow. Briefly, we first generated a derivative vector pSL3067 from the broad-host range plasmid pVZ321 that contains a kanamycin resistance cassette and the mobilization/replication genes needed for passage through Escherichia coli and for conjugation into S7002. Alkane expression vectors were then generated from pSL3067 using Gibson Assembly26. Genotypes for strains characterized in this study are presented in Supplementary Table S1 and oligos in Supplementary Table S2. To construct the initial set of vectors, we cloned the alkane biosynthetic operons from the two model cyanobacteria Synechococcus elongatus PCC 7942 (S7942) and Synechocystis sp. PCC 6803 (S6803) for introduction into S7002 via tri-parental mating. The empty plasmid pSL3067 was introduced into S7002 to be used as a control (strain 7002::control). The S6803 ado and aar genes were introduced into S7002 on plasmid pSL3071 to make strain 7002::6803alk. The alkane operon from S7942 containing genes ado, aar, and accA was initially introduced in vector pSL3070 under control of the native S7942 promoter. However, this strain did not produce detectable amounts of non-native hydrocarbons under any growth conditions. We determined that this was due to the S7942 alkane gene promoters being inactive in S7002 (Supplementary Fig. S2) and re-designed the expression vector by placing the genes under control of the synthetic promoter Ptrc1O27. This vector, pSL3072, was introduced into S7002 to make strain 7002::trc7942alk.
Effects of ado/aar and ols co-expression on S7002 growth and hydrocarbon content
We compared the growth of the control, 7002::6803alk and 7002::trc7942alk strains in a bioreactor at 38 °C and 27 °C while bubbling with air supplemented with 5% CO2 under 300 μmol LED photons s−1 m−2 illumination. We determined the maximum exponential and linear growth rates for the cultures and a wild-type (WT) S7002 as a reference (Table 1). At 38 °C, growth of 7002::control and 7002::6803alk were essentially the same but 7002::trc7942alk grew more slowly, exhibiting a longer exponential doubling time and slower linear growth rate (Fig. 2A). In contrast, growth rates of all three strains were comparable 27 °C (Fig. 2B). This was initially surprising since alka/enes had previously been shown to have a more important effect on strain growth as temperature is lowered15,17. Given the apparent temperature-dependent effect, we tested whether a large decrease in growth temperature and light intensity during culturing differentially affected the three strains. Each recovered from the two-day cold period at a similar rate (Fig. 2C), suggesting that Ado/Aar, even when overexpressed, do not impact S7002 temperature adaptation.
Table 1.
Exponential and linear growth rates of S7002 strains.
| Strain | Exponential doubling time in hours at 38 °C [27 °C] | Linear phase growth rate at 38 °C [27 °C]* |
|---|---|---|
| WT S7002 | 4.7 ± 0.1 [7.1 ± 0.3]† | 0.045(2) [0.028(1)] |
| 7002::control | 6.1 ± 1 [11 ± 1.4] | 0.037(2) [0.023(1)] |
| 7002::6803alk | 6.8 ± 1 [12 ± 0.7] | 0.038(1) [0.023(1)] |
| 7002::trc7942alk | 7.2 ± 1 [11 ± 0.7] | 0.024(4) [0.020(2)] |
| 7002ΔOls | 9.7 ± 2 [ND]‡ | 0.0190(6) [ND] |
| ΔOls::control | 21 ± 2 [ND] | 0.011(1) [ND] |
| ΔOls::6803alk | 8.0 ± 1 [27 ± 10] | 0.022(2) [0.003] |
| ΔOls::7942alk | 7.1 ± 1 [21 ± 2] | 0.029(2) [0.005] |
| ΔOls::UndA | 7.7 ± 1 [28 ± 10] | 0.023(1) [0.003] |
| ΔOls::FAP | 7.5 ± 0.8 [16 ± 2] | 0.029(2) [0.008] |
*Measured in O.D. 720 nm/ hour.
†Errors are 1 SD based on three biological replicates.
‡‘ND’, not determined.
Figure 2.
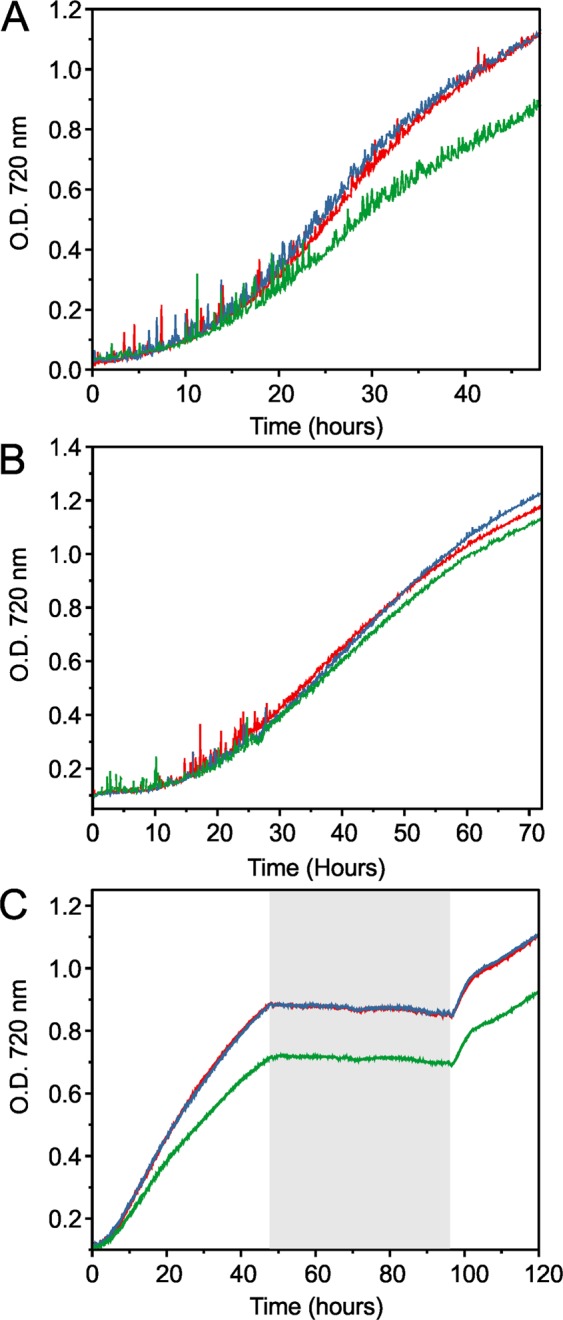
Representative growth curves of S7002 strains co-expressing Ols and the Ado/Aar pathways. In all panels, 7002::control is shown in red, 7002::6803alk in blue, and 7002::trc7942alk in green. (A) Strains grown at 38 °C. (B) Strains grown at 27 °C. (C) Strains exposed to a temperature and light change during growth. Strains were grown in a bioreactor under 300 μmol LED photons s−1m−2 and bubbled with air supplement with 1% CO2 (panels A and B) or only air (panel C). In panel C, the gray box indicates the two-day 15 °C dark period. Cells were grown in a bioreactor under 300 μmol LED photons s−1m−2 and bubbled with air supplement with 5% CO2.
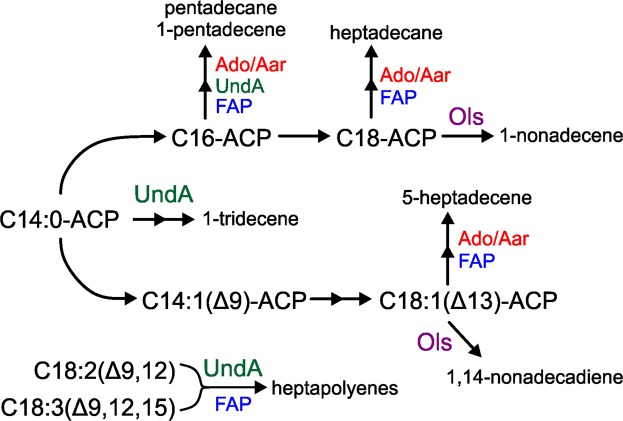
In order to determine the extent to which Ado/Aar affect hydrocarbon pools in S7002, we quantified hydrocarbons in methanolic extracts of cells grown in the bioreactor. Methanolic extracts of dried cells were analyzed using gas chromatography (GC) and hydrocarbon content was normalized as percent dry cell weight (% DCW, Fig. 3). Hydrocarbon content of WT S7942 and WT S6803 were also determined for comparison (Fig. 3A and B). S7002 natively produces two types of α-olefins: 1-nonadecene (C19:1) and 1,14-nonadecadiene (C19:2)5,19. At 38 °C, the 7002::control strain produced 0.35% DCW hydrocarbons (Fig. 3C). In transitioning from 38 °C to 27 °C, the ratio of C19:2 to C19:1 increased from 0.25 to 1.2 while the overall alkene content decreased to 0.26% DCW. Co-expression of S6803 Ado/Aar in 7002::6803alk afforded production of C17 hydrocarbons in addition to the native α-olefins (Fig. 3D). Heptadecane was the major product but we also observed another metabolite at 38 °C that eluted shortly before heptadecane (starred in Supplementary Fig. S3). Due to the elution time and the available acyl-ACP substrates in S7002, we hypothesized that this metabolite was likely 5-heptadecene produced by action of Ado/Aar on the internally desaturated C18:1(Δ13) acyl-ACP precursor of 1,14-nonadecadiene17,19. The mass spectrum of this molecules was consistent with a singly desaturated C17 hydrocarbon (Supplementary Fig. S4), either 8- or 5-heptadecene. Since S7942 naturally produces traces of 8-heptadecene11, we compared extracts of WT S7942 to 7002::6803alk which showed that the metabolite observed was not 8-heptadecene (Supplementary Fig. S5). At 38 °C in 7002::6803alk, cells contained predominantly native C19 alkenes and Ado/Aar-derived hydrocarbons amounted to only 10% of the total in the strain (Fig. 3D). The effects of Ado/Aar were further reduced at 27 °C where C17 hydrocarbons were barely detectable, constituting only 1.5% of alka/enes in cells (Fig. 3G). At 38 °C, 7002::trc7942alk produced a high level of non-native alka/enes, and a total hydrocarbon content of 0.75% DCW (Fig. 3E). Three hydrocarbons were detected in addition to the native alkenes: pentadecane, 5-heptadecene, and heptadecane (Supplementary Fig. S3). The amounts of the non-native alka/enes in 7002::trc7942alk exceeded the levels of native alkenes by more than two-fold and the total alkane content was also two- to three-fold that of the control strain. However, growth at 27 °C dramatically reduced production of the non-native hydrocarbons in 7002::trc7942alk to only 4% of the total (Fig. 3H). The hydrocarbon profile was close to that of the control strain, providing a plausible explanation for the reduced growth rate of 7002::trc7942alk at 38 °C but not 27 °C. We considered that the relatively high light intensity used to grow the strains at 27 °C could have affected hydrocarbon production. To test this, we cultured the three strains in shaker flasks at 25 °C in air under fluorescent lights at 50 μmol photons s−1 m−2 and quantified alka/ene content (Fig. 3I to K). The strains again grew at similar rates (Supplementary Fig. S6) but 7002::trc7942alk contained higher quantities of Ado/Aar-derived C17 hydrocarbons. Compared to at 27 °C in the bioreactor, heptadecane levels were increased 10-fold to 0.054% DCW and 5-heptadecene 8-fold to 0.039% DCW. Notably, these quantities were still only 18% of Ado/Aar-derived hydrocarbons produced at 38 °C. 7002::6803alk did not produce appreciable quantities of non-native hydrocarbons when grown at these conditions.
Figure 3.
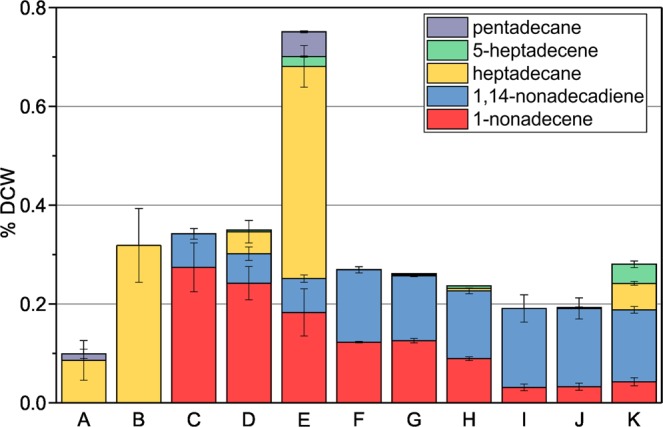
Alka/ene hydrocarbon content of the S7002 strains expressing Ols and Ado/Aar pathways and WT references. Hydrocarbon content was normalized as percent of dry cell weight (% DCW). (A) S7942 at 38 °C. (B) S6803 at 30 °C. (C) 7002::control at 38 °C. (D) 7002::6803alk at 38 °C. (E) 7002::trc7942alk at 38 °C. (F) 7002::control at 27 °C. (G) 7002::6803alk at 27 °C. (H) 7002::trc7942alk at 27 °C. (I) 7002::control at 25 °C. (J) 7002::6803alk at 25 °C. (K) 7002::trc7942alk at 25 °C. A to H were cultured under 300 μmol photons s−1 m−2 and bubbled with 5% CO2-supplemented air and I to K at 50 μmol photons s−1 m−2 in air. S6803 was cultured at 30 °C since the strains is not tolerant of 38 °C. Error bars are 1 SD based on at least three biological replicates.
We hypothesized that the marginal effects of S6803 Ado/Aar might be due to S7002 up-regulating ols to maintain a near-normal level of alkenes in response to consumption of acyl-ACPs by Ado/Aar. Using quantitative RT-PCR, we determined that ols was expressed at similar levels in all strains and is not up-regulated even when Ado/Aar were overexpressed (Supplementary Fig. S7). Furthermore, transcript levels for the key fatty acid biosynthetic genes accB and fab20 were also mostly unaffected by the presence of Ado/Aar. Ols is apparently able to continue competing effectively for acyl-ACP substrates without up-regulation. We conclude from these observations that Ado/Aar and Ols can coexist in S7002 and that even overexpression of the non-native pathways is not obviously harmful to S7002.
Substitution of S7002 Ols with Ado/Aar enzymes and effects on growth and hydrocarbon content
Since S7002 was able to tolerate Ado/Aar, we were curious to determine the limits of hydrocarbon pathway inter-compatibility and thus asked the question of whether Ado/Aar could functionally replace Ols in a knockout strain of S7002. We knocked out olefin production in S7002 by replacing the start codon and promoter region of the Ols gene with a spectinomycin resistance cassette using natural transformation and double homologous recombination with a linear PCR fragment. This 7002ΔOls strain was fully segregated after two weeks of re-patching on selective media and no longer produced either type of C19 alkene (Supplementary Fig. S8). In a bioreactor, growth of 7002ΔOls at 38 °C was markedly reduced relative to WT 7002 and at 27 °C 7002ΔOls was unable to grow (Fig. 4A and Table 1). We tested whether Ado/Aar could rescue this phenotype by introducing the S6803 and S7942 Ado/Aar pathways into 7002ΔOls on plasmids. S6803 genes were expressed from pSL3071 (strain ΔOls::6803alk). The S7942 ado, aar, and accA genes were expressed from plasmid pSL3258 that uses the endogenous S7002 A2813 promoter described by Ruffing et al.28 (strain ΔOls::7942alk). We assumed that A2813 would afford more modest expression levels than trc and transcriptomic data indicated that this promoter did not significantly respond to temperature change29. Finally, the empty plasmid pSL3067 was introduced into 7002ΔOls and used as a control strain for comparison (ΔOls::control).
Figure 4.
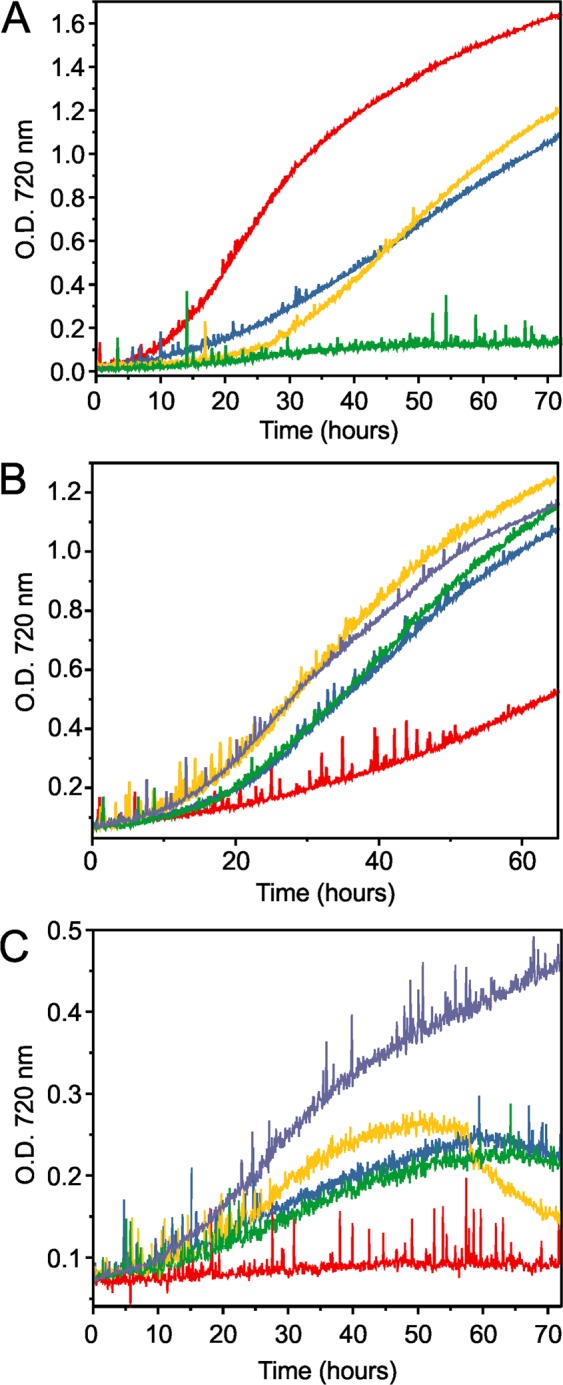
Representative growth curves of WT S7002, 7002ΔOls and Ols-replacement strains at 38 °C and 27 °C. (A) WT S7002 and 7002ΔOls at 38 °C (red and blue traces, respectively) and at 27 °C (yellow and green traces). (B) ΔOls strains grown at 38 °C. ΔOls::control (red trace), ΔOls::6803alk (blue), ΔOls::7942alk (yellow), ΔOls::UndA (green), and ΔOls::FAP (grey). (C) ΔOls strains grown at 27 °C, indicated using the same colors as in panel B. Strains were cultured as in Fig. 3.
Growth of ΔOls::6803alk, ΔOls::7942alk and ΔOls::control was assessed in a multicultivator at 38 °C and 27 °C. Like 7002ΔOls, ΔOls::control grew slowly at 38 °C and no growth occurred at 27 °C (Fig. 4B). Growth rates of ΔOls::control at 38 °C were significantly reduced relative to 7002ΔOls (21 hr vs. 10 hr exponential doubling time, Table 1). We are uncertain of the exact cause of this but it could be due to the additional metabolic burden imposed by plasmid replication and antibiotic resistance. 7002::control also exhibited reduction in growth rate relative to WT S7002 but this was not as pronounced as ΔOls (Table 1). Since ΔOls was already compromised, perhaps the effect was enhanced in this background. Interestingly, the expression of Ado/Aar in trans rescued this phenotype and both strains grew at both temperatures (Fig. 4B). At 38 °C, exponential doubling times were greatly improved to 8 and 7.1 hrs for ΔOls::6803alk and ΔOls::7942alk, respectively, compared to 21 hours for the knockout control strain. Linear phase growth rates were similarly improved (Table 1). However, expressing Ado/Aar had only marginal positive effect on growth at 27 °C. While the strains were able to be cultured better than ΔOls::control at this temperature, they only survived for a few days before growth halted and cell density began to decrease (Fig. 4C). Growth rates at this temperature were also markedly reduced relative to the strains expressing Ols at 27 °C (compare to Fig. 2C).
We quantified alkane production in these Ado/Aar-replacement strains. ΔOls::6803alk produced exclusively C17 hydrocarbons, mostly heptadecane and some 5-heptadecene (Fig. 5A and Supplementary Fig. S9). At 38 °C, these hydrocarbons amounted to 0.075% DCW. Similarly, ΔOls::7942alk produced 0.17% DCW alka/enes at 38 °C consisting of heptadecane, 5-heptadecene, and pentadecane, in decreasing amounts (Fig. 5B and Supplementary Fig. S9). In contrast, when grown at 27 °C, these strains produced very few alka/enes. In ΔOls::6803alk, total hydrocarbons were reduced to 0.012% DCW and in ΔOls::7942alk to only 0.005% DCW (Fig. 5E and F). This corresponds to an 84% reduction in alka/enes content for ΔOls::6803alk and >97% in ΔOls::7942alk. We considered that decreased expression of the ado and aar genes at 27 °C might be responsible and performed semi-quantitative RT-PCR on ΔOls::6803alk and ΔOls::7942alk. However, the results indicated that the expression levels were not significantly impacted by the temperature change in either strain (Fig. S10).
Figure 5.
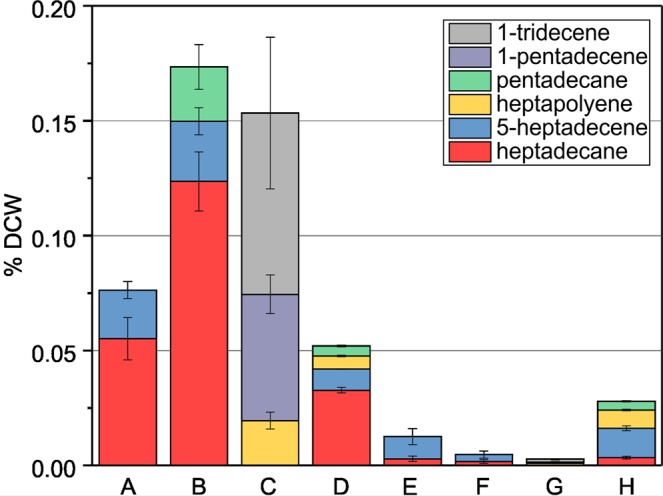
Alka/ene content of 7002ΔOls substituted with non-native hydrocarbon biosynthetic genes presented as % DCW. (A) ΔOls::6803alk at 38 °C. (B) ΔOls::7942alk at 38 °C. (C) ΔOls::UndA at 38 °C. (D) ΔOls::FAP at 38 °C. (E) ΔOls::6803alk at 27 °C. (F) ΔOls::7942alk at 27 °C. (G) ΔOls::UndA at 27 °C. (H) ΔOls::FAP at 27 °C. Strains were cultured as in Fig. 3. Error bars are 1 SD based on at least three biological replicates.
Replacement of Ols with non-cyanobacterial alka/ene-producing enzymes UndA and fatty acid photodecarboxylase
Given the success of replacing Ols with Ado/Aar at 38 °C, we asked whether non-cyanobacterial alka/ene-producing pathways would be able to similarly substitute for Ols and recover the slow-growth phenotype of ΔOls. This experimental approach also afforded us the opportunity to find enzymes that might not exhibit the temperature-dependent effects displayed by Ado/Aar. To test this, we chose two other alka/ene-synthesizing enzymes, UndA from Pseudomonas aeruginosa30 and the fatty acid photodecarboxylase (FAP) from green algae31 to test in the 7002ΔOls background. FAP catalyzes the blue light-driven synthesis of alkanes from fatty acids via decarboxylation31. The non-heme iron enzyme UndA and its homologues catalyze the oxidative decarboxylation of fatty acids to produce terminal alkenes in Pseudomonas species30. The UndA gene from Pseudomonas aeruginosa PA14 (PA14_53120) was synthesized and placed under control of the A2813 promoter on a vector (pSL3208). The FAP gene from Chlorella variabilis NC64A (GenBank accession KY511411) was synthesized and cloned into an expression vector under control of the A2813 promoter (pSL3244).
We initially tested UndA for activity in WT S7002. Expression of UndA afforded minimal production of non-native hydrocarbons and only traces of 1-tridecene were detected (Fig. 6A, red trace). We hypothesized that the low activity of UndA was due to the limited substrate range of the native enzyme and its preference for mid-chain fatty acids (C10–C14) which were present in too-low amounts in S700230. To increase the substrate range of UndA, we generated a single mutation based on the crystal structure of the Pseudomonas fluorescens Pf-5 homologue (PDB ID 4WWZ, Fig. 6B). We predicted that a mutation of phenylalanine 239 to alanine would enlarge the substrate pocket (Fig. 6C) and allow reactions with longer substrates such as hexadecanoic acid, which is the major fatty acid in S700232. This rational enzyme engineering approach was successful and UndA-F239A produced 1-pentadecene and increased quantities of 1-tridecene in WT S7002 (Fig. 6A, blue trace). Detection of 1-pentadecene indicates a reaction of the mutant enzyme with hexadecanoic acid substrate. The increased production of 1-tridecene also suggests that the activity towards tetradecane was improved by the mutation. UndA-F239A was introduced in trans into 7002ΔOls on plasmid pSL3209 to make strain ΔOls::UndA.
Figure 6.
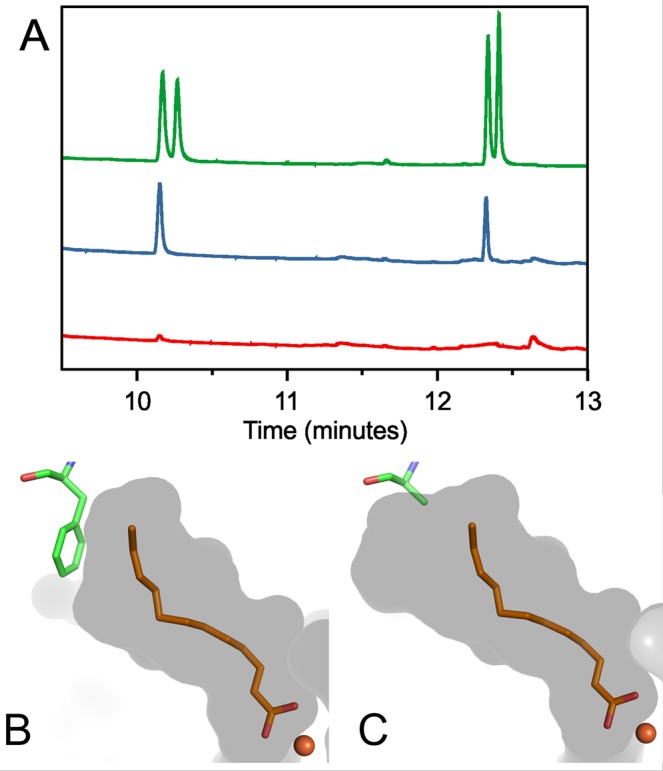
Mutation of Pseudomonas aeruginosa UndA to increase substrate range. (A) GC traces of S7002 cell extracts expressing native UndA (red trace) and the UndA-F239A mutant (blue). The green trace shows hydrocarbon standards for C13:1, C13, C15:1, and C15 in order of elution. (B) Substrate pocket in WT UndA. (C) Predicted expanded substrate pocket in UndA F239A. In B and C, the substrate pocket is shown as a grey surface, 2,3-dodecenoic acid substrate in orange and the iron ion as a brown sphere. UndA structure images were made using Pymol version 2.0.7, Schrödinger, LLC. Sample inputs for GC in panel A were normalized to dry cell biomass.
Growth rates and hydrocarbon content of UndA- and FAP-substituted 7002Δols
Both UndA and FAP were able to support growth of 7002ΔOls at 38 °C resulting in similarly improved doubling times as the Ado/Aar-substituted strains (Fig. 4B and Table 1). However, at 27 °C, we observed a differential response of the two strains. ΔOls::UndA performed poorly and growth ceased after about two days, similar to ΔOls::6803alk and ΔOls::7942alk (Fig. 4C). In contrast, ΔOls::FAP growth was markedly improved compared to the other strains (Fig. 4C), doubling in 16 hours compared to >21 hours for the others. The strain also did not cease growth after a few days but did reach a lower culture density than strains with Ols.
We quantified alka/ene content in ΔOls::UndA and ΔOls::FAP under the two conditions in order to determine why FAP afforded improved growth at 27 °C relative to the other strains. ΔOls::UndA produced mainly the shorter alkenes 1-tridecene and 1-pentadecene (Fig. 5C and Supplementary Fig. S9). ΔOls::FAP produced primarily heptadecane, 5-heptadecene and pentadecane resulting in a hydrocarbon profile similar to that of ΔOls::7942alk but with a lower overall content (Fig. 5D, and Supplementary Fig. S9). In both strains, we observed metabolites in the GC traces that were not seen in any of the other strains (starred in Supplementary Fig. S9). The elution time suggested that these were C17 compounds and we hypothesized that they were alkenes derived from reaction of UndA F239A with linoleic (C18:2(Δ9,12)) and α-linolenic (C18:3(Δ9,12,15)) fatty acids present in S700232. Feeding of these fatty acids to ΔOls::UndA cultures resulted in increased production of the two metabolites (Supplementary Fig. S11), confirming our hypothesis and identifying these compounds as heptapolyenes containing two to four desaturation sites. ΔOls::UndA also contained traces of heptadecane, but these were too low to accurately quantify. The production of C17 alkenes by UndA F239A suggests that the mutation successfully extended the substrate range from C14 up to C18 fatty acids, which has not been previously shown. At 38 °C, ΔOls::UndA contained 0.15% DCW hydrocarbons and ΔOls::FAP contained 0.05% DCW (Fig. 5C and D). Although ΔOls::FAP produced the lowest total alka/ene content of the replacement strains at 38 °C (less than one third that of ΔOls::7942alk), the strain was still among the best-performing. At 27 °C, ΔOls::UndA hydrocarbons were reduced >98% whereas ΔOls::FAP maintained 0.028% DCW alka/enes corresponding to only a 54% reduction relative to 38 °C (Fig. 5G and H). In ΔOls::FAP, the decrease in hydrocarbons was mainly due to a major reduction of heptadecane production. Content of the other hydrocarbons 5-heptadecene and heptapolyenes was slightly increased. Of the four strains tested, ΔOls::FAP maintained the highest level of hydrocarbons upon shifting to 27 °C, providing a likely explanation for its superior growth relative to the other Ols-substituted strains. The various products generated by the non-native pathways in S7002 and their substrates are summarized in Fig. 7.
Figure 7.
Summary of substrates and hydrocarbon products generated by Ols and non-native pathways in S7002. The substrates for Ado/Aar and Ols are acyl-ACPs and those for UndA and FAP are the equivalent fatty acids likely derived from turnover of membrane lipids.
Discussion
The ability of disparate hydrocarbon biosynthetic pathways from bacteria and eukaryotes to conditionally substitute for Ols in S7002 has several important implications. The effect of the non-native pathways on S7002 growth rates and hydrocarbon pools and growth rate is critically dependent on growth temperature. With the exception of FAP, growth at 27 °C dramatically reduced the production of alka/enes by the non-native pathways in S7002 by 84–98%. It is likely that there is a threshold quantity of hydrocarbons needed per cell in order to facilitate growth and the low quantities produced by most of the ΔOls strains at 27 °C are insufficient to support growth. There are several plausible explanations for the reduction in alka/ene production. The first is that Ado/Aar preferentially react with saturated acyl-ACP substrates that are in increased supply at 38 °C. This preference is evidenced, in part, by the ratio of 5-heptadecene to heptadecane (0.07, 0.05) vs. the ratio of 1,14-nonadecadiene to 1-nonadecene (0.24, 0.37) produced in 7002::6803alk and 7002::trc7942alk at 38 °C. Due to this preference, the increased desaturation of acyl-ACPs in S7002 at lower temperature reduced the impact of Ado/Aar due to a shift to less favorable desaturated substrates. This essentially has the effect sequestering a pool of desaturated acyl-ACPs for reaction with Ols and not Ado/Aar. The production of fatty acids for UndA could be similarly affected by the increased acyl-ACP desaturation. We observed mainly production of 1-tridecene and 1-pentadecene in ΔOls::UndA, suggesting that the enzyme is reactive towards tetradecanoic acid and hexadecanoic acid and not their internally desaturated analogues which would have resulted in production of alkadienes. Since myristic acid is the major substrate for UndA at 38 °C, increased desaturation of C14-ACP to C14:1(Δ9)-ACP at 27 °C would reduce production of the corresponding saturated fatty acids and thereby limit production of UndA-derived alkenes. In contrast, FAP is able to produce predominantly 5-heptadecene at 27 °C by reaction with C18:1(Δ9) fatty acid, giving it an advantage over UndA since it is more compatible with the internally desaturated substrates.
The second reason for the reduction may be tied to a differential availability of reducing equivalents for the enzymes at 27 °C vs. 38 °C. Previous reports show that S7002 still contains quantities of palmitic and stearic acid at 22 °C33 which would be viable substrates for FAP and, if activated to acyl-ACPs, for Ado/Aar. The delivery of reducing equivalents to Ado and/or Aar could be compromised at the lower temperature due to photoinhibition and/or redox imbalance at 27 °C. We see some evidence for this. At 27 °C, production of 1-nonadecene in 7002::trc7942alk is reduced relative to the control, possibly due to capture by Aar. However, there is little production of Ado/Aar -derived alka/enes (Fig. 3H). If electron delivery to Ado is compromised at 27 °C, the coupling between Aar acyl-ACP reduction and Ado decarbonylation would result in incomplete turnover of acyl-ACPs into alka/enes. Furthermore, when 7002::trc7942alk was cultured under low light conditions in shake flasks at 25 °C, it contained eight- to ten-fold more non-native hydrocarbons. The decreased light intensity and overall slower growth rate of the strains could have resulted in less photoinhibition and higher Ado/Aar activity. While the electron-donating co-substrate for the UndA reaction is still in question30, a similar inefficient delivery of reducing equivalents to the enzyme at 27 °C could contribute to its low activity at this temperature. Interestingly, the use of light as a substrate in the decarboxylation reaction by FAP obviates the need for electron delivery by ferredoxin or other redox partners31. This could provide another explanation for this enzyme’s better relative performance at 27 °C.
The story is quite different at the higher growth temperature. The improved growth exhibited by the Ols-replacement strains compared to the knockout indicates that these disparate biosynthetic pathways are able to support healthy growth of S7002 at 38 °C. This observation has several implications. First, the biological function of hydrocarbons is not necessarily tied into the specific structure of the alka/ene molecules. Expression of UndA and FAP in S7002 afforded unique hydrocarbon profiles not observed in any other wild cyanobacteria5. Hydrocarbons with various chain lengths from C13 to C17 containing different desaturation profiles are able to serve the same role as the native C19 α-olefins in S7002. Secondly, it means the producing enzyme and the source of the substrates are not critical. The biosynthetic mechanisms of these enzymes are distinct, as are their preferred substrates. The Ols and Ado/Aar enzymes use acyl-ACP substrates whereas UndA and FAP react with fatty acids. Since free fatty acids are activated to acyl-ACPs in cyanobacteria, the source for the UndA and FAP substrates is likely to derive from turnover of membrane lipids34. Capture and consumption of these molecules apparently does not greatly impact growth of S7002 compared to Ado/Aar and both types of enzymes are able to support growth of S7002 at 38 °C.
One of prevailing hypotheses for the role of alka/enes is that they partition into the interior of membrane bilayers and thereby afford cells a means to modulate membrane fluidity and structure in response to a variety of stimuli including light and temperature changes, during cell division, and fluctuating thylakoid lumen pressure resulting from proton pumping14,15,35–37. The ability of different types of hydrocarbons to support growth of S7002 in consistent with the hypothesis that these molecules primarily serve basal structural roles in cells. Although different kinds of hydrocarbons can serve this role, regulated and steady production of alka/enes in response to changing growth conditions is critical. In a less controlled growth environment, the native Ols pathway is likely to afford an advantage over Ado/Aar, UndA, and FAP for S7002 since it is able to continue producing hydrocarbons in response to changing temperature. This provides a plausible explanation for why Ols is maintained over Ado/Aar in S7002 and that enzyme compatibility with endogenous acyl-ACP and fatty acid profiles may be an important determinant of which pathway is preferred in a cyanobacterial cell.
In conclusion, we found that Ado/Aar and Ols pathways are not mutually exclusive and S7002 is able to support the co-existence of both pathways. Far from being incompatible, alka/ene biosynthetic pathways from disparate organisms and domains of life are able to support growth of S7002. While the native Ols pathway in S7002 does display an advantage over the exogenous pathways, we hypothesize that this has more to do with a compatibility of Ols with the native acyl-ACP substrates over a variety of growth conditions rather than a strict requirement of α-olefin production in cells. These data suggest that the specific biosynthetic mechanism and enzyme pathway for hydrocarbon production is not critical as long as sufficient hydrocarbon production is maintained in response to changing environmental conditions. The unique hydrocarbon profiles of ΔOls::UndA and ΔOls::FAP evidence an opportunity to expand the hydrocarbon production range of cyanobacteria to chain lengths not accessed by WT strains. Our experiments only tested a subset of the hydrocarbon biosynthetic pathways found in nature38,39, and it will be interesting to determine to what extent these diverse pathways are fundamentally interchangeable in vivo.
Materials and Methods
Assembly of expression vectors
A diagram illustrating vector construction is shown in Supplementary Figure S1. Vector pSL3067 was constructed by sequential digestion of the broad host-range vector pVZ32125 with the restriction enzymes XbaI and PstI. The doubly digested fragment was ligated with a linker molecule composed of the annealed oligos linker1 and linker2 using T4 DNA Ligase (Thermo Scientific). pSL3067 contains the aphA gene conferring kanamycin resistance and RSF1010-derived replication, mobilization genes and ori sites25. pSL3067 was cut with XbaI, purified and used in Gibson Assembly to generate the expression vectors. Alkane genes were PCR-amplified from S7942 and S6803 genomic DNA (gDNA). To construct pSL3070, the ado-aar-accA genes were amplified from S7942 gDNA using primers alk1/alk2 and combined with XbaI-linearized pSL3067. The fragments were joined using Gibson assembly and the product transformed into XL1-Blue Escherichia coli. Transformants were selected on LB media with 50 μg/ml kanamycin. Resulting colonies were PCR screened for the correctly assembled plasmid and positive colonies cultured overnight in liquid LB with 50 μg/ml kanamycin. Plasmids were purified using a GeneJet Plasmid Miniprep Kit (Thermo Fisher Scientific) and sequences verified using Sanger Sequencing (Genewiz, www.genewiz.com). pSL3071 was constructed by amplifying ado-aar and their native promoters from S6803 gDNA using primers alk3/alk4 and combining the PCR fragment with XbaI-digested pSL3067 for Gibson assembly and transformation. For pSL3072, the Ptrc10 sequence was amplified from vector template pSL280140 using primers alk5/alk6 and ado-aar-accA genes from S7942 gDNA using primers alk7/alk2. The fragments were Gibson assembled, transformed and isolated as described for pSL3070 and pSL3071 above. The UndA and FAP gene coding sequences were synthesized by Genewiz. pSL3208 containing the UndA coding sequence was generated using primers alk8/alk9 to amplify the A2813 promoter from S7002 gDNA and alk10/alk11 to amplify UndA from the synthesized DNA fragment. The UndA F239A point mutation was generated by amplifying two fragments from pSL3208 using the error-introducing primer pairs alk8/alk12 and alk13/alk11 followed by Gibson assembly and isolation as described for pSL3070. pSL3244 containing the FAP coding sequence was made using primer pairs alk8/alk14 to amplify the A2813 promoter sequence and alk15/alk16 to amplify FAP from the synthesized DNA fragment and with terminal overlaps for Gibson Assembly. The fragments were mixed with XbaI-cut pSL3067 and assembled and isolated as described above. Vector pSL3258 was generated by first double-digesting vector pSL3070 with KpnI and BamHI. The A2813 promoter sequence was amplified from S7002 gDNA using primers alk17/alk18 and mixed with linearized pSL3070 for assembly. To make the ols knockout PCR fragment, the homology regions were amplified from S7002 gDNA using primers olsUS-F/olsUS-R and olsDS-F/olsDS-R and the spectinomycin cassette from vector pAM1303 (Addgene) using primers spec-F/spec-R. These three fragments were joined using overlap extension PCR and transformed into S7002 as described28. To generate vector pSL3068, we digested pSL3067 with XbaI enzyme. The eYFP gene was amplified from vector pSL280140 using primers eYFP-F/eYFP-R and used in Gibson assembly with the XbaI-linearized pSL3067. The forward primer introduces an AarI restriction site upstream of eYFP. Promoter sequences were amplified using the following primer pairs and inserted into AarI-digested pSL3068 using Gibson assembly: Pado, Pado-F/Pado-R; Palk, Palk-F/Palk-R; Ptrc1O, Ptrc1O-F/Ptrc1O-R. Cells were grown under 500 μmol LED photons s−1m−2 and 1% CO2-enriched air at 38 °C overnight.
Cyanobacterial genetic manipulation and culturing conditions
S7002 was cultured in medium A supplemented with 4 μg/L Vitamin B12. Antibiotic concentrations in the media were 60–100 μg/ml for kanamycin and/or 40–80 μg/ml for spectinomycin. Vectors were transferred into S7002 by conjugation using tri-parental mating as described previously41. Optical density of cells at 730 nm was determined in 96-well culture trays using 150 μl cells and measured using a BioTek μQuant plate reader. For bioreactor growth, S7002 was cultured in a Multi-Cultivator MC1000 instrument (Photon Systems Instruments). Cells were grown in 50 mL volume in 200 × 30 mm Pyrex test tubes. Strains were acclimated to growth in the multicultivator for a few days at the requisite temperature and then re-diluted to start the experiment. The ols knockout was generated by natural transformation of S7002 using previously reported methods28. Exponential doubling times were determined from fitting the exponential growth phase using equations y = Aekt and t1/2 = ln(2)/k where t1/2 is the doubling time. Linear growth rates were calculated directly from the linear phase after exponential growth.
Metabolite extraction and GC/MS analysis
S7002 cells were grown to O.D. 730 nm of 0.8–1.2 and harvested by centrifugation. The pelleted cells were washed with water twice, re-pelleted and frozen in liquid N2. Frozen cells were lyophilized overnight and weighed to determine dry biomass. Alka/enes were extracted in 100% methanol spiked with an internal hexadecane standard (at 77 μg/ml). The cell debris was pelleted and the supernatant transferred to a glass vial for analysis. Alka/enes were quantified using a HP/Agilent 5890 N gas chromatography instrument equipped with a 12 m DB5-MS column and run using the following method: 80 °C for 5 mins, ramp to 200 °C at 15 °C/min, hold for 4 mins, then ramp to 300 °C at 33 °C/min and hold for 3 mins. We injected 6–8 μl of extract for each run. Helium was used as the carrier gas. Alka/enes were identified by comparison to authentic standards (Sigma-Aldrich) when available and/or using GC/MS analysis. Metabolites were quantified from a standard curve of authentic alka/enes and normalized to an internal hexadecane standard. Concentrations of 1,14-nonadecadience and 5-heptadecene were calculated using the standard curves for 1-nonadecene and heptadecane, respectively. GC/MS analysis was performed at the Washington University Biomedical Mass Spectrometry Resource on an Agilent 7890 GC instrument running in positive electrospray ionization mode. The LC/MS data was analyzed using Agilent Mass Hunter Software.
Supplementary information
Acknowledgements
We would like to thank all members of the Pakrasi group for helpful discussions. This research was supported by funding from the Department of Energy, Office of Science, Biological and Environmental Research (DE-SC0012722) to HBP. CJK was partially supported by a postdoctoral fellowship from the Arnold and Mabel Beckman Foundation (Irvine, CA). The Washington University Biomedical Mass Spectrometry Resource was supported by a grant from the National Institute of General Medical Sciences (8 P41 GM103422) from the National Institutes of Health. pAM1303 was a gift from Susan Golden (Addgene plasmid # 40243).
Author Contributions
C.J.K. and H.B.P designed the experiments. C.J.K. performed the experiments and analyzed data. C.J.K. and H.B.P. wrote the paper. All authors approve the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-38124-y.
References
- 1.Flombaum P, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA. 2013;110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitton, B. A. & Potts, M. The ecology of cyanobacteria: their diversity in time and space. (Springer Science & Business Media, 2007).
- 3.Han, J., McCarthy, E. D., Calvin, M. & Benn, M. H. Hydrocarbon constituents of the blue-green algae Nostoc muscorum, Anacystis nidulans, Phormidium luridium and Chlorogloea fritschii. Journal of the Chemical Society C: Organic, 2785–2791, 10.1039/J39680002785 (1968).
- 4.Winters K, Parker PL, Baalen CV. Hydrocarbons of Blue-Green Algae: Geochemical Significance. Science. 1969;163:467–468. doi: 10.1126/science.163.3866.467. [DOI] [PubMed] [Google Scholar]
- 5.Coates, R. C. et al. Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PLoS One9, 10.1371/journal.pone.0085140 (2014). [DOI] [PMC free article] [PubMed]
- 6.Lea-Smith DJ, et al. Contribution of cyanobacterial alkane production to the ocean hydrocarbon cycle. Proc. Natl. Acad. Sci. USA. 2015;2015:07274. doi: 10.1073/pnas.1507274112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu WJ, et al. Hydrocarbons, the advanced biofuels produced by different organisms, the evidence that alkanes in petroleum can be renewable. Appl. Microbiol. Biotechnol. 2015;99:7481–7494. doi: 10.1007/s00253-015-6840-6. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez-Díaz L, Caballero A, Perez-Hernandez N, Segura A. Microbial alkane production for jet fuel industry: Motivation, state of the art and perspectives. Microb. Biotechnol. 2016;10:103–124. doi: 10.1111/1751-7915.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Liu X, Lu X. Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol. Biofuels. 2013;6:69. doi: 10.1186/1754-6834-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshino T, et al. Alkane production by the marine cyanobacterium Synechococcus sp. NKBG15041c possessing the α-olefin biosynthesis pathway. Appl. Microbiol. Biotechnol. 2015;99:1521–1529. doi: 10.1007/s00253-014-6286-2. [DOI] [PubMed] [Google Scholar]
- 11.Tan X, et al. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 2011;13:169–176. doi: 10.1016/j.ymben.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Kageyama H, et al. Improved Alkane Production in Nitrogen-Fixing and Halotolerant Cyanobacteria via Abiotic Stresses and Genetic Manipulation of Alkane Synthetic Genes. Curr. Microbiol. 2015;71:115–120. doi: 10.1007/s00284-015-0833-7. [DOI] [PubMed] [Google Scholar]
- 13.Knoot, C. J., Ungerer, J. L., Wangikar, P. P. & Pakrasi, H. B. Cyanobacteria: promising biocatalysts for sustainable chemical production. J. Biol. Chem., 10.1074/jbc.R117.815886 (2017). [DOI] [PMC free article] [PubMed]
- 14.Lea-Smith DJ, et al. Hydrocarbons are essential for optimal cell size, division and growth of cyanobacteria. Plant Physiol. 2016;172:01205.02016. doi: 10.1104/pp.16.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berla BM, Saha R, Maranas CD, Pakrasi HB. Cyanobacterial Alkanes Modulate Photosynthetic Cyclic Electron Flow to Assist Growth under Cold Stress. Sci. Rep. 2015;5:14894. doi: 10.1038/srep14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamori T, Kageyama H, Tanaka Y, Takabe T. Requirement of alkanes for salt tolerance of Cyanobacteria: characterization of alkane synthesis genes from salt-sensitive Synechococcus elongatus PCC7942 and salt-tolerant Aphanothece halophytica. Lett. Appl. Microbiol. 2018;67:299–305. doi: 10.1111/lam.13038. [DOI] [PubMed] [Google Scholar]
- 17.Mendez-Perez D, Herman NA, Pfleger BF. A desaturase gene involved in the formation of 1,14-nonadecadiene in Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 2014;80:6073–6079. doi: 10.1128/AEM.01615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schirmer, A., Rude, M. A., Li, X., Popova, E. & del Cardayre, S. B. Microbial Biosynthesis of Alkanes. Science, 559–562 (2010). [DOI] [PubMed]
- 19.Mendez-Perez D, Begemann MB, Pfleger BF. Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl. Environ. Microbiol. 2011;77:4264–4267. doi: 10.1128/AEM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rock CO, Jackowski S. Forty Years of Bacterial Fatty Acid Synthesis. Biochem. Biophys. Res. Commun. 2002;292:1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser, B. K. et al. Fatty Aldehydes in Cyanobacteria Are a Metabolically Flexible Precursor for a Diversity of Biofuel Products. PLoS One8, 10.1371/journal.pone.0058307 (2013). [DOI] [PMC free article] [PubMed]
- 22.Aukema KG, et al. Cyanobacterial aldehyde deformylase oxygenation of aldehydes yields n - 1 aldehydes and alcohols in addition to alkanes. ACS Catalysis. 2013;3:2228–2238. doi: 10.1021/cs400484m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N, et al. Conversion of fatty aldehydes to alka(e)nes and formate by a cyanobacterial aldehyde decarbonylase: cryptic redox by an unusual dimetal oxygenase. J. Am. Chem. Soc. 2011;133:6158–6161. doi: 10.1021/ja2013517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu L, et al. Polyketide decarboxylative chain termination preceded by O-sulfonation in curacin A biosynthesis. J. Am. Chem. Soc. 2009;131:16033–16035. doi: 10.1021/ja9071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taton A, et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 2014;42:e136. doi: 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 27.Brosius J, Erfle M, Storella J. Spacing of the −10 and −35 regions in the tac promoter. Effect on its in vivo activity. J. Biol. Chem. 1985;260:3539–3541. [PubMed] [Google Scholar]
- 28.Ruffing AM, Jensen TJ, Strickland LM. Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis. Microb. Cell Fact. 2016;15:190. doi: 10.1186/s12934-016-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig M, Bryant D. Synechococcus sp. Strain PCC 7002 Transcriptome: Acclimation to Temperature, Salinity, Oxidative Stress, and Mixotrophic Growth Conditions. Front. Microbiol. 2012;3:354. doi: 10.3389/fmicb.2012.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rui Z, et al. Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc. Natl. Acad. Sci. USA. 2014;111:18237. doi: 10.1073/pnas.1419701112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sorigué D, et al. An algal photoenzyme converts fatty acids to hydrocarbons. Science. 2017;357:903. doi: 10.1126/science.aan6349. [DOI] [PubMed] [Google Scholar]
- 32.Zhu, T., Scalvenzi, T., Sassoon, N., Lu, X. & Gugger, M. Terminal Olefin Profiles and Phylogenetic Analyses of Olefin Synthases of Diverse Cyanobacterial Species. Appl. Environ. Microbiol. 84 (2018). [DOI] [PMC free article] [PubMed]
- 33.Sakamoto T, Higashi S, Wada H, Murata N, Bryant DA. Low-temperature-induced desaturation of fatty acids and expression of desaturase genes in the cyanobacterium Synechococcus sp. PCC 7002. FEMS Microbiol. Lett. 1997;152:313–320. doi: 10.1111/j.1574-6968.1997.tb10445.x. [DOI] [PubMed] [Google Scholar]
- 34.Kaczmarzyk D, Fulda M. Fatty Acid Activation in Cyanobacteria Mediated by Acyl-Acyl Carrier Protein Synthetase Enables Fatty Acid Recycling. Plant Physiol. 2010;152:1598–1610. doi: 10.1104/pp.109.148007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klähn S, et al. Alkane Biosynthesis Genes in Cyanobacteria and Their Transcriptional Organization. Front. Bioeng. Biotechnol. 2014;2:24. doi: 10.3389/fbioe.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberton M, et al. Organization and flexibility of cyanobacterial thylakoid membranes examined by neutron scattering. J. Biol. Chem. 2013;288:3632–3640. doi: 10.1074/jbc.M112.416933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stingaciu L-R, et al. Revealing the Dynamics of Thylakoid Membranes in Living Cyanobacterial Cells. Sci. Rep. 2016;6:19627. doi: 10.1038/srep19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christenson, J. K. et al. Active Multienzyme Assemblies for Long-Chain Olefinic Hydrocarbon Biosynthesis. J. Bacteriol. 199 (2017). [DOI] [PMC free article] [PubMed]
- 39.Grant JL, Hsieh CH, Makris TM. Decarboxylation of Fatty Acids to Terminal Alkenes by Cytochrome P450 Compound I. J. Am. Chem. Soc. 2015;137:4940–4943. doi: 10.1021/jacs.5b01965. [DOI] [PubMed] [Google Scholar]
- 40.Ungerer J, Pakrasi HB. Cpf1 Is A Versatile Tool for CRISPR Genome Editing Across Diverse Species of Cyanobacteria. Sci. Rep. 2016;6:39681. doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wendt KE, Ungerer J, Cobb RE, Zhao H, Pakrasi HB. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb. Cell Fact. 2016;15:115. doi: 10.1186/s12934-016-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.