Abstract
3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase (DAHPS) is a key rate-limiting enzyme in aromatic amino acid anabolism. A new Iβ-type DAHPS gene (aro1A) was identified in a metagenomic library from subtropical marine mangrove sediment. The gene encoded a polypeptide composed of 272 amino acids and had a maximum similarity of 52.4% to a known DAHPS at the amino acid level. Multiple sequence alignment, homologous modeling, and molecular docking showed that Aro1A had the typical (β/α)8 barrel-shaped catalytic structural domain of DAHPS. The motifs and amino acid residues involved in the combination of substrates and metal ligand were highly conservative with the known DAHPS. The putative DAHPS gene was subcloned into a pET-30a(+) vector and was overexpressed in Escherichia coli Rosetta (DE3) cells. The recombinant protein was purified to homogeneity. The maximum activity for the recombinant Aro1A protein occurred at pH 8.0 and 40 °C. Ba2+ and Ca2+ stimulated the activity of Aro1A protein. The enzyme showed high affinity and catalytic efficiency (KPEPm = 19.58 μM, VPEPmax = 29.02 μM min−1, and kPEPcat/KPEPm = 0.88 s−1 μM−1) under optimal reaction conditions. The enzymatic property of Aro1A indicates its potential in aromatic amino acid industrial production.
Electronic supplementary material
The online version of this article (10.1186/s13568-019-0742-4) contains supplementary material, which is available to authorized users.
Keywords: 3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase, Novel gene, Metagenomic library, Subtropical marine mangrove sediment, Biochemical characterization
Introduction
3-Deoxy-d-arabino-heptulosonate-7-phosphate synthase (DAHPS) is a key rate-limiting enzyme in the synthesis of aromatic amino acids, such as phenylalanine, tyrosine, and tryptophan (Herrmann 1995). This enzyme can catalyze phosphoenolpyruvate (PEP), d-erythrose-4-phosphate (E4P), and H2O to form 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) in the first step of shikimic acid approach. Shikimic acid approach mainly exists in bacteria, fungi, and vegetation but not inside higher animals (Herrmann 1995). Therefore, DAHPS from pathogenic microorganisms, including Mycobacterium tuberculosis and Neisseria meningitidis, becomes the antibacterial target candidate (Cross et al. 2013; Webby et al. 2005a). Studies on the engineered strains aim to produce shikimic acid or aromatic amino acid (like L-Phe) to relieve the feedback inhibition of DAHPS and improve the yield (Cui et al. 2014; Liu et al. 2013). Thus, DAHPS received research interest for medical field and industrial production.
DAHPSs can be classified as type I or type II according to their molecular dimension: type I is < 40 kDa, and type II is 50 kDa (Gosset et al. 2001; Jensen et al. 2002). Type I DAHPS is divided into types Iα and Iβ (Jensen et al. 2002). The DAHPSs from Escherichia coli, Saccharomyces cerevisiae, and N. meningitidis represent type Iα. The N terminus of DAHPS has a regulating region that inhibits enzymatic activity by combining with Phe, Tyr, and Trp. Type Iβ DAHPS is divided into two types. The first type includes a feedback regulation domain, whereas the other type does not have a feedback regulation domain. The recently discovered regulation domains include chorismate mutase and ferredoxin-like domains (Table 1). Among these domains, the most common is chorismate mutase located in the N terminus, such as the DAHPSs from Bacillus subtilis and Listeria monocytogenes (Light et al. 2012; Pratap et al. 2017). The feedback inhibition of various type Iβ DAHPSs is more complicated than that of type Iα DAHPS. Type Iβ DAHPS is inhibited by downstream aromatic amino acids, including Phe, Tyr, Trp, chorismate, and prephenate, in independent or cooperative ways. Type Iβ DAHPS without a regulation domain is not generally inhibited by downstream aromatic amino acids (Table 1). Meanwhile, type II DAHPS includes DAHPSs from vegetation and certain microorganisms such as M. tuberculosis, Corynebacterium glutamicum, and Helicobacter pylori. These representatives are inhibited similarly as types Iα and Iβ DAHPS. Types I and II DAHPSs have no apparent sequence similarity (Shumilin et al. 2004). The polymer form also varied among different DAHPSs. Recent research shows that the activity of the DAHPS from Providencia alcalifaciens is affected by its oligomeric state (Sharma et al. 2018). However, all the reported DAHPSs have similar (β/α)8 barrel-shaped catalytic structural domain, and their catalytic activity depends on a divalent metal ion (Wu et al. 2005).
Table 1.
Representative DAHPSs from different types of microorganisms
| Protein | Organism | T | Feedback structure | Feedback inhibitor | PF | References |
|---|---|---|---|---|---|---|
| AroG, AroF, AroH | Escherichia coli | Iα | N-Region | Phe, Tyr, Trp | 2, 4 | Ray and Bauerle (1991); Schoner and Herrmann (1976); Shumilin et al. (1999, 2004) |
| Aro3, Aro4 | Saccharomyces cerevisiae | Iα | N-Region | Phe, Tyr, Trp | 2, 4, 8 | Helmstaedt et al. (2005); König et al. (2004); Künzler et al. (1992); Teshiba et al. (1986) |
| Nme DS | Neisseria meningitidis | Iα | N-Region | Phe, Tyr, Trp | 4 | Cross et al. (2013); Heyes et al. (2014) |
| DAHPPae | Pseudomonas aeruginosa | Iα | – | – | – | Sterritt et al. (2018) |
| Tm DS | Thermotoga maritima | Iβ | N-Ferredoxin-like domain | Phe, Tyr | 2, 4 | Shumilin et al. (2004); Wu et al. (2003) |
| Pfu DS | Pyrococcus furiosus | Iβ | None | None of Phe, Tyr, and Trp | 2, 4 | Schofield et al. (2004, 2005) |
| Gsp DS | Geobacillus sp. | Iβ | N-Chorismate mutase | Prephenate | 4 | Nazmi et al. (2016) |
| Ap DS | Aeropyrum pernix | Iβ | None | None of Phe, Tyr, Trp, chorismate, shikimate, and prephenate, | 4 | Zhou et al. (2012) |
| aroA(Q)168 | Bacillus subtilis | Iβ | N-Chorismate mutase | Chorismate, Prephenate | 4 | Wu et al. (2005) |
| Pg DS | Porphyromonas gingivalis | Iβ | C-Chorismate mutase | Chorismate, prephenate | Wu and Woodard (2006) | |
| Lm DS | Listeria monocytogenes | Iβ | N-Chorismate mutase | Chorismate, prephenate | 4 | Light et al. (2012) |
| Pae DAHPSPA2843 | Pseudomonas aeruginosa | II | N-Region | Trp | 4 | Sterritt et al. (2018) |
| Mt DAHPS | Mycobacterium tuberculosis | II | N-Chorismate mutase | Phe/Tyr, Phe/Trp, Tyr/Trp, Tyr, Trp, and chorismate | 2, 4 | Light et al. (2012); Webby et al. (2010) |
| AroG, AroF, AroH | Corynebacterium glutamicum | II | N-Region | Phe, Tyr, Trp | 2, 4 | Burschowsky et al. (2018); Liu et al. (2008) |
| Hpy DS | Helicobacter pylori | II | Unknown | None of Phe, Tyr, Trp, and chorismate | 2 | Webby et al. (2005b) |
T DAHPS type, PF polymer form
To date, more than 99% microorganisms cannot be cultivated under pure-cultured conditions (Amann et al. 1995). Metagenomic technology that is not cultivation-dependent was developed to overcome limitations in studying genes that come from microorganisms that cannot be cultivated (Amann et al. 1995). Metagenome-derived amylases, cellulases, esterases, polyketide synthases, and alkaline proteases were identified using function-based and sequence-based screening strategies (Leis et al. 2015; Mewis et al. 2013; Niehaus et al. 2011; Seow et al. 1997; Yang et al. 2016; Yun et al. 2004). Most of these enzymes have new physio-biochemical characteristics and provide rich research materials for the improvement of industrial enzymes and for the further investigation of enzyme structures and functions.
Herein, a plasmid metagenomic library was constructed successfully from subtropical marine mangrove wetland sediments by using pUC118 as the cloning vector. A new gene (aro1A) encoding DAHPS was cloned and identified. To our knowledge, this gene is the first metagenome-derived DAHPS from subtropical marine mangrove sediment. The gene provided new materials and theoretical references for the industrial production of aromatic amino acids.
Materials and methods
Strains and plasmids
The host strain of the metagenomic library was E. coli DH5α (Novagen), which was also used to construct and preserve recombinant expression plasmids. E. coli Rosetta (DE3) (Novagen) was used for the expression of recombinant proteins. Plasmid pUC118 HincII/BAP (Takara) was the vector carrying the metagenomic library, and plasmid pET-30a(+) (Novagen) was the expression vector.
Construction of the metagenomic library
A sample of 0–10 cm-deep sediment was collected from a mangrove surrounding the intertidal zone in Beihai City, Guangxi Province, China (N21°26′28″, E109°11′37″). The sediment sample had a temperature of 30 °C and a pH of 5.5. A high-quality metagenomic DNA was extracted from the sample by using a FastDNA SPIN kit (MP Biomedicals, USA) according to the manufacturer’s protocols (Additional file 1: Fig. S1A). The inserted DNA was the 2–6 kb gel-extracted fragments from the mixture of equal amounts of products digested with HincII and SmaI. This inserted DNA was ligated to the pUC118 HincII/BAP (Takara), and 5 μL of ligation products were transformed into 50 μL of electro-competent E. coli DH5α. The transformed cells were recovered using 1 mL of SOC medium at 37 °C and 180 rpm. The same batches of recovery culture were combined. To calculate the size of the DNA fragments in the library, we placed 5 μL of cultured samples in LB agar plates containing 100 μg mL−1 of ampicillin, 40 μg mL−1 of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and 40 μg mL−1 of isopropyl β-d-1-thiogalactopyranoside (IPTG). The combined culture was allowed to grow for 3 h, and then 5 μL of the cultured sample was collected to calculate the proliferation fold. The rest of the cells were stored in 20% glycerinum at − 80 °C after centrifugation. The library was stored at − 80 °C until screening.
Isolation and sequence analysis of aro1A
DNA sequence analysis was performed using a BigDye terminator cycle sequencing kit on an ABI Prism 3700 DNA analyzer (Applied Biosystems, USA). Open reading frame (ORF) analysis of all clone inserts was performed using the ORF Finder (NCBI). ORF annotation was based on the results of blastx and CD-Search from NCBI. The target gene aro1A in this study was derived from a positive clone (pUME11) and was annotated as a DAHPS gene based on sequence analysis.
The molecular weight and the theoretical isoelectric point of the protein were predicted via ProtParam (Gasteiger et al. 2005). Furthermore, sequence identification and conserved domain analysis of the protein were performed using the BLAST and CD-Search programs from NCBI, respectively (Marchler-Bauer et al. 2017). Phylogenetic analysis was performed using the MEGA7 software (Kumar et al. 2016). The evolutionary history was inferred using the neighbor-joining method. Multiple sequence alignment was performed via the Clustal OMEGA program (Sievers et al. 2014), and the alignment result was slightly adjusted to align the conserved sites, according to previous studies. The secondary structure information revealed in the alignment was obtained from the 3D structure data of the protein. The predicted structure of Aro1A was built automatically using the SWISS-MODEL server (Waterhouse et al. 2018).
Combination patterns of Aro1A and ligands
The combination patterns of Aro1A and ligands were predicted via the AutoDock 4.2.6 program (Morris et al. 2009). The receptor was the predicted structure of Aro1A. The ligands PEP and E4P were obtained from the DAHPS structures of Aeropyrum pernix (PDB: 1VS1) and Thermotoga maritima (PDB: 1RZM), respectively. The atoms of the receptor were assigned to “AD4 type.” The “Grid box” was set to maximum, and the “Search Parameter” was the “genetic algorithm.” The “Number of GA Runs” was set to 200, and default values were used for the remaining parameters.
Overexpression and purification of the recombinant DAHPS protein
The plasmid containing aro1A was extracted as the template for the polymerase chain reaction (PCR). The forward primer (5′-CGGAAGCTTGCATGATGGCCCCATTGGTAACACAAA-3′) and the reverse primer (5′-GGACTCGAGCACCAACTCCCTGTCTATAGCTGCC-3′) were designed based on the nucleotide sequence of aro1A, and the restriction enzyme sites for HindIII and XhoI were underlined in the above-mentioned primers, respectively. PCR was performed in a 50 μL reactor consisting of 1× PrimeSTAR buffer (Takara), 1.25 U PrimeSTAR HS DNA polymerase (Takara), 4 μL of dNTP mixture (2.5 mM) (Takara), 0.2 μM forward primer, 0.2 μM reverse primer, 50 ng plasmid, and H2O. The PCR program was as follows: 30 cycles at 98 °C for 10 s and at 68 °C for 60 s. The PCR product (Additional file 1: Fig. S2) was purified after being digested with HindIII and XhoI at 37 °C for 3 h. The purified product was ligated to the HindIII and XhoI double-digested vector pET-30a(+) with T4 ligase (Takara). The recombinant plasmid pET-30a(+)-aro1A was confirmed by double digestion with HindIII and XhoI (Additional file 1: Fig. S3) and was sequenced by Sangon Biotech (Shanghai). The corresponding recombinant plasmid was transformed into competent E. coli Rosetta (DE3) cells. The clone obtained via double-enzyme digestion and sequencing was used for the recombinant protein expression.
A single colony of the protein expression strain E. coli Rosetta (DE3)/pET30a(+)-aro1A was inoculated into 10 mL of LB-kanamycin (50 µg mL−1) and was allowed to grow for 8 h at 37 °C. Then, 3 mL of culture was added to 200 mL of LB-kanamycin (50 µg mL−1) containing 0.5 M sorbitol in a 500 mL flask. The resulting mixture was agitated (180 rpm) at 37 °C. IPTG was added to the final concentration of 0.1 mM when the OD600 was 0.4–0.6, and the culture was agitated (180 rpm) for 8 h at 16 °C. The His-tagged Aro1A protein was purified from the sonicated lysate of harvested cells by using His60 Ni Superflow Resin (Takara) according to the manufacturer’s instructions. The protein concentration was analyzed using the BCA Protein Assay Kit (Solarbio, China). The expression of the protein was detected and analyzed using SDS–PAGE.
Assay of Aro1A activity
The assay method for Aro1A was modified as previously described (Nazmi et al. 2016). The reaction mixture solution (1 mL) was composed of phosphate buffer saline (pH 6.8, 10 mM), PEP (25 μM), E4P (25 μM), and CoCl2 (0.1 mM). The reaction mixture was incubated at 25 °C for 5 min, and the reaction was initiated by adding Aro1A protein (2 μg). The activity of Aro1A was examined by monitoring the PEP consumption at 232 nm. One unit of enzyme was defined as the amount of Aro1A that converts 1 μmol PEP in 1 min at pH 6.8 and 25 °C.
Effect of temperature, pH, and divalent metal ion on enzyme activity
The optimal reaction time was studied at 25 °C and pH 6.8. The enzymatic reaction progress was monitored by the change in OD232 in the reaction system, in which the initial substrate concentration was 0.25 μM. The optimum reaction time of 10 min was observed based on the reaction progress curve (Additional file 1: Fig. S4).
Temperature-dependent assays were performed at 4 °C–55 °C and pH 6.8 for 10 min, and those that are pH dependent were performed at pH 4.0–9.0 and at the optimum reaction temperature for 10 min. To determine thermostability, we incubated the enzyme at 4 °C–55 °C for 2 h. The assays were performed at optimal reaction conditions. To draw the relative enzyme activity curve of the assays above, we measured the highest enzyme activity in each assay at 100%. Furthermore, to determine the activation of Aro1A, we measured the different divalent metal ions in different reactions, each containing 5 mM metal ion, at optimal reaction conditions by using the standard assay (0.1 mM CoCl2) as the control (100%).
Kinetic data
Reaction velocity was measured when 0.1–0.5 μM PEP was used as the substrate under optimal conditions. The KPEPm, VPEPmax, and kPEPcat of Aro1A were calculated from the Lineweaver–Burk plot. All reactions were performed in three independent experiments.
Nucleotide sequence accession number
The aro1A nucleotide sequence was deposited in the GenBank database with the accession number MH757446.
Results
Construction of metagenomic library and isolation of aro1A gene
Two blunt endonucleases of HincII and SmaI for the metagenomic DNA preparation (Additional file 1: Fig. S1B) were used to obtain diverse fragments. The 2–6 kb DNA fragments from the mixture of equal amounts of enzyme-digested products were ligated to linear blunt-end plasmid pUC118 HincII/BAP. Through blue-white screening, 15 white clones were randomly selected to verify the insertion size (Additional file 1: Fig. S1C). The constructed metagenomic library contained approximately 750,000 clones, the average insertion size was roughly 4 kb, and the metagenomes covered approximately 3.0 Gb. The target gene in this study was from a clone named pUME11 and was annotated as a DAHPS gene based on the sequence analysis. The gene was named aro1A, which was 819 bp long.
Phylogenic relationship and primary structure of Aro1A
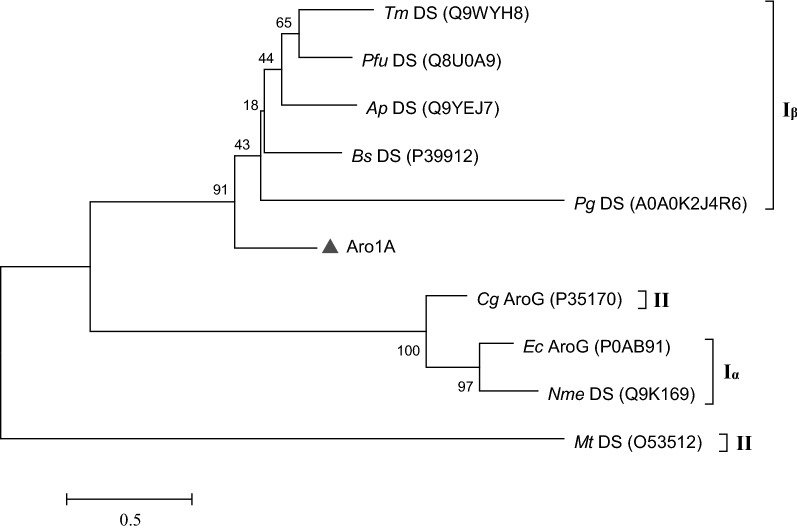
An estimate based on bioinformatics analysis indicated that Aro1A encoded a polypeptide composed of 272 amino acids and had a theoretical isoelectric point of 4.76 and a theoretical molecular weight of 28.82 kDa. The conserved domain analysis tool, CD-Search of NCBI, annotated that Aro1A was a new member of type I DAHPS super family. Aro1A had the highest similarity of 52.4% to the DAHPS from T. maritima MSB8 (Accession number: Q9WYH8). Phylogenetic analysis showed that the evolutionary relationship of Aro1A with type Iβ DAHPS was higher than that with type II or type Iα DAHPS (Fig. 1).
Fig. 1.
Phylogenetic tree of Aro1A and other DAHPSs. These proteins came from T. maritima (Tm DS), P. furiosus (Pfu DS), A. pernix (Ap DS), B. subtilis (Bs DS), P. gingivalis (Pg DS), C. glutamicum (Cg AroG), E. coli (Ec AroG), N. meningitidis (Nme DS), and M. tuberculosis (Mt DS). The percentage of replicate trees in which the associated replicates were clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Protein accession numbers are in the parentheses
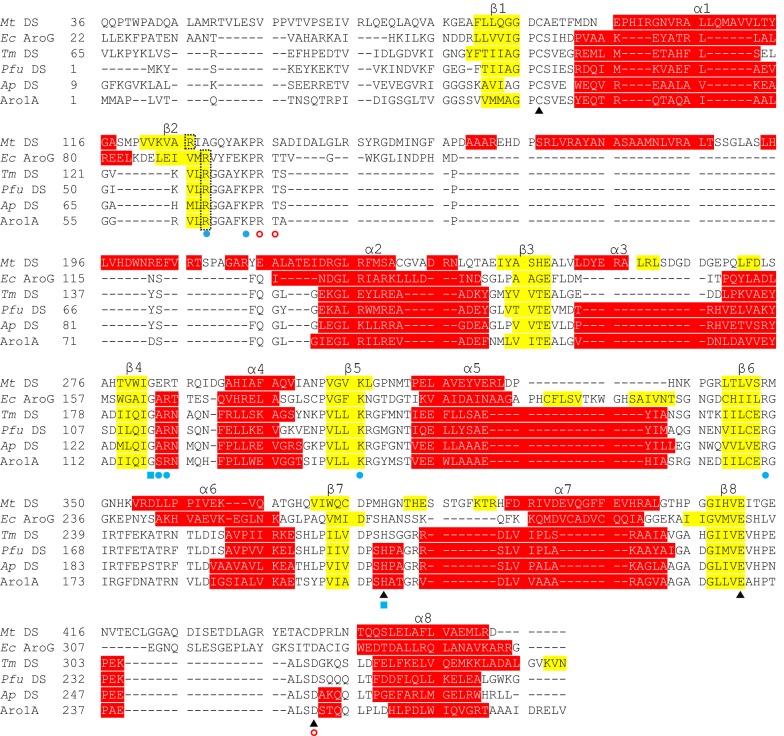
DAHPS enzymes from M. tuberculosis (PDB: 2B7O), E. coli (PDB: 1QR7), T. maritima (PDB: 1RZM), Pyrococcus furiosus (PDB: 4C1K), and A. pernix (PDB: 1VS1) were selected as the representative sequences of types Iα, Iβ, and II, which were multiple-aligned with the Aro1A protein. Multiple sequence alignment results revealed that Aro1A and the other DAHPSs shared similar motif sites (Fig. 2). The divalent metal binding sites of C36, H206, E232, and D243 of Aro1A were consistent with those of the representative DAHPS. The conserved residues of R60, K65, S119, R120, K141, and R171 in Aro1A protein were annotated as the PEP binding sites; G118 and H206 were possibly the conserved amino acid residues in the substrate-binding motif, which had non-bond contact with PEP; R67, T68, and D243 were the possible binding sites of E4P. Figure 2 shows that the properties of secondary structure (e.g. length and amino acid residues) of Aro1A was slightly different from the other DAHPSs, especially Mt DAHPS.
Fig. 2.
Sequence alignment based on the structure of Aro1A and the five representative DAHPSs. These proteins came from M. tuberculosis (Mt DS), E. coli (Ec AroG), T. maritima (Tm DS), P. furiosus (Pfu DS), and A. pernix (Ap DS). The α-helices are highlighted in red, and the β-strands are highlighted in yellow. Conserved divalent metal binding sites are indicated with “black triangle”; conserved PEP binding sites are indicated with “blue circle”; conserved PEP non-bonded contact sites are indicated with “blue square”; and conserved E4P binding sites are indicated with “empty circle”
Molecular model and substrate docking analysis of Aro1A
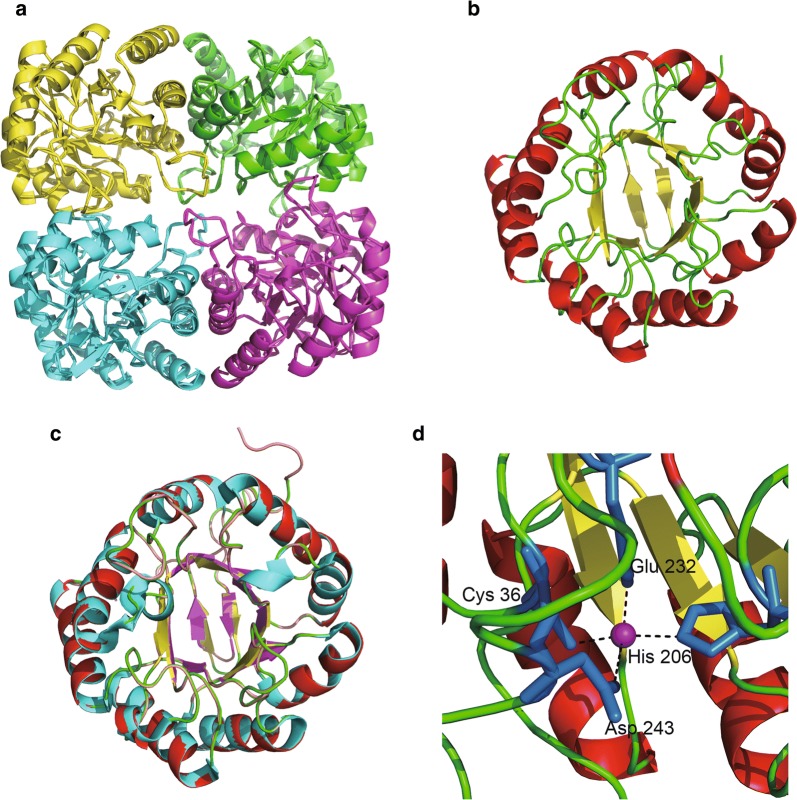
The optimal complexus crystal template of a DAHPS from A. pernix (Accession number: 1VS1.1.A) was selected for the homologous modeling of Aro1A on the basis of the SWISS-MODEL analysis. This template has the best Global Model Quality Estimate (0.77) and Quaternary Structure Quality Estimate (0.81) (Waterhouse et al. 2018). Figure 3a shows the tetramer of Aro1A that resulted from homology modeling. The monomeric structure of Aro1A is a (β/α)8 barrel structure (Fig. 3b), which was highly similar to that of 1VS1 (Fig. 3c). The results of homologous modeling showed a divalent metal ion (Mn2+) among four conserved metal binding residues (C36, H206, E232, and D243) (Fig. 3d).
Fig. 3.
Predicting structure and ligand interaction sites of Aro1A. a Tetrameric form of Aro1A. b Monomeric form of Aro1A. c Superposition of Aro1A monomer on the Ap DS (PDB: 1VS1). d Binding interaction model of divalent metal and Aro1A. The purple ball represents Mn2+; the blue stick represents the four binding residues; the black dotted lines represent inferred coordinating interactions
The results of molecular docking analysis showed that PEP combined with five residues (R60, Q116, S119, K141, and R171) through eight hydrogen bonds (Fig. 4a). Furthermore, E4P combined with six residues (R60, K65, Q116, S119, K141, and R171) through nine hydrogen bonds (Fig. 4b).
Fig. 4.
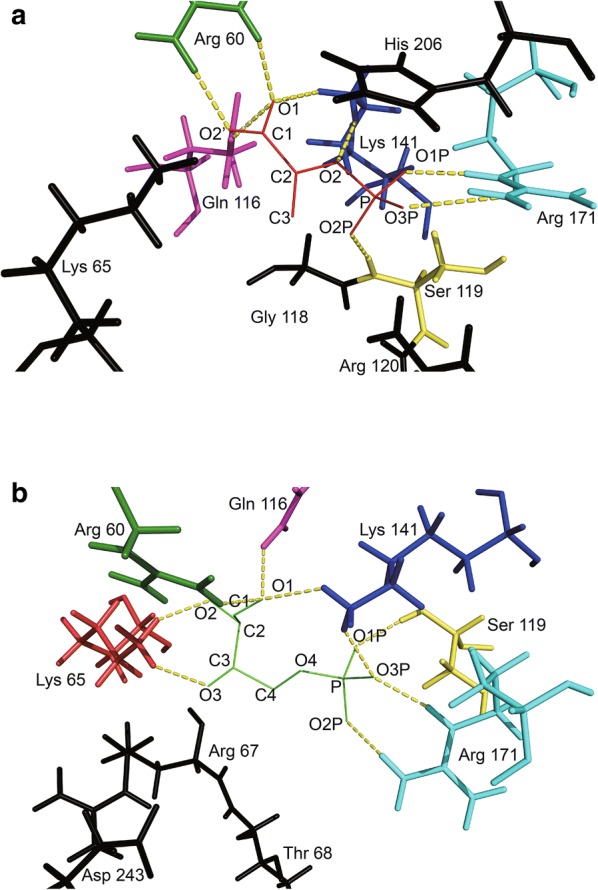
Docking model of Aro1A with substrates. a Docking model of PEP and Aro1A. b Docking model of E4P and Aro1A. PEP is shown as the red line, E4P is shown as the green line. The atomic names of these two substrates are labeled. The yellow dotted lines represent the hydrogen-bonding interactions. Residues are drawn as stick: Arg 60 (green), Lys 65 (red), Gln 116 (magenta), Ser 119 (yellow), Lys 141 (blue), and Arg 171 (cyan). Black sticks are the residues predicted to be associated with substrate binding in the alignment but not in docking
Expression and purification of Aro1A in E. coli
Plasmid pET30a(+) with aro1A was transformed into competent cells of E. coli Rosetta (DE3). The transformed cells were cultivated by introducing IPTG. Cell extracts expressing Aro1A were subjected to SDS–PAGE. The results of SDS–PAGE indicated that cell lysate contained the target protein with a size of approximately 37 kDa (Fig. 5a). The protein was consistent with the predicted molecular weight. Furthermore, Fig. 5a shows that the quantity of soluble protein was more than 80%. The recombinant Aro1A protein was purified with Ni-IDA and analyzed via magnetic agarose chromatography (Fig. 5b).
Fig. 5.
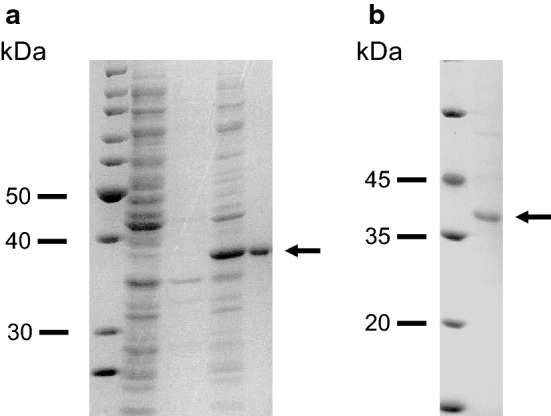
Analysis of expression and purification of Aro1A through SDS–PAGE. a Lane 1, molecular mass standards; lane 2, lysate supernatant of empty vector-carried expression strain; lane 3, lysate precipitate of empty vector-carried expression strain; lane 4, lysate supernatant of Aro1A-expressed strain; and Lane 5, lysate precipitate of Aro1A-expressed clone. b Lane 1, molecular mass standards; and Lane 2, purified Aro1A
Effects of temperature, pH, and divalent metal ion on Aro1A
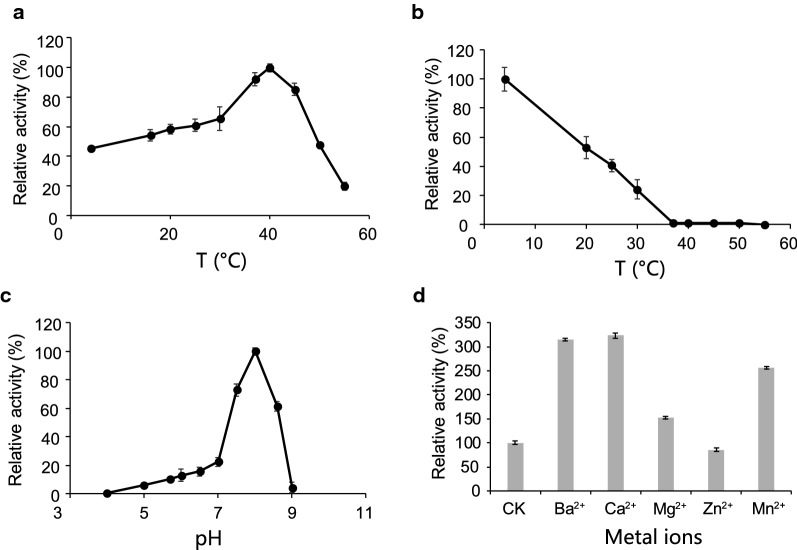
Figure 6 shows the influence of temperature, pH, and divalent metal ions on the activity of Aro1A. The enzymatic activity of Aro1A was examined at different temperatures (4 °C, 16 °C, 20 °C, 25 °C, 30 °C, 37 °C, 40 °C, 45 °C, 50 °C, and 55 °C) and pH 6.8. Results showed that the optimal temperature was 40 °C. The enzymatic activity was more than 60% when the temperature was within 30 °C–47 °C (Fig. 6a). The thermostability of Aro1A was also tested. Furthermore, the enzymatic activity of Aro1A was analyzed under optimal reaction conditions after incubation at 4 °C, 20 °C, 25 °C, 30 °C, 37 °C, 40 °C, 45 °C, 50 °C, and 55 °C for 2 h, and the relative enzymatic activity at 4 °C was marked as 100%. Figure 6b shows that Aro1A had approximately 50% enzymatic activity at 20 °C. This activity greatly decreased to less than 5% when maintained at 37 °C–50 °C for 2 h. Furthermore, Aro1A lost its enzymatic activity when the temperature was increased to 55 °C.
Fig. 6.
Effects of temperature, pH, and divalent metal ion on Aro1A. a Optimum reaction temperature of Aro1A, b thermostability of Aro1A, c optimum reaction pH of Aro1A, and d activation of Aro1A by divalent metal ions
The enzymatic activity of Aro1A at different pH levels (4.0, 5.0, 5.6, 6.0, 6.8, 7.0, 7.5, 8.0, 8.5 and 9.0) and 40 °C was also evaluated. Figure 6c shows that the optimal pH for Aro1A was 8.0, and the enzymatic activity was more than 60% when pH was within 7.3–8.5.
Ba2+, Ca2+, Mg2+, Zn2+, and Mn2+ were used in the enzymatic reaction system to determine the activation of Aro1A (Fig. 6d). Ba2+ and Ca2+ stimulated the activity of Aro1A to more than 300%, and Mn2+ stimulated such activity to more than 250%. Meanwhile, Mg2+ and Zn2+ had no substantial effect on the activity.
Kinetic analysis
The enzymatic reaction rate was analyzed when the substrate concentration was 0.1–0.5 μM at optimal reaction conditions. The molecular kinetic parameters of Aro1A were examined via the Lineweaver–Burk double-reciprocal graphing method (Additional file 1: Fig. S4). The measured parameters were as follows: KPEPm = 19.58 μM, VPEPmax = 29.02 μM·min−1, kPEPcat value = 17.31 s−1, and kPEPcat/KPEPm = 0.88 s−1 μM−1 (Table 2).
Table 2.
Enzymatic property of representative DAHPSs that were expressed in the E. coli system
| Organism | Protein | T | OpH | OT (°C) | KPEPm (µM) | kPEPcat (s−1) | kPEPcat/KPEPm (s−1 µM−1) | References |
|---|---|---|---|---|---|---|---|---|
| N. meningitidis | Nme DS | Iα | – | 40 | 11 | 25 | 2.3 | Cross et al. (2013) |
| Unculture Microorganisms | Aro1A | Iβ | 8.0 | 40 | 19.58 | 17.31 | 0.88 | This study |
| Geobacillus sp. | Gsp DS | Iβ | – | – | 87 | 45 | 0.52 | Nazmi et al. (2016) |
| T. maritima | Tm DS | Iβ | 6.3 | 90 | 9.5 | 7.6 | 0.8 | Wu et al. (2003) |
| P. furiosus | Pfu DS | Iβ | – | – | 120 | 1.5 | 0.01 | Schofield et al. (2004) |
| B. subtilis | Bs DS | Iβ | 9.0 | – | 139 | 4.6 | 0.03 | Wu et al. (2005) |
| P. gingivalis | Pg DS | Iβ | 9.0 | – | 421 | 337 | 0.80 | Wu and Woodard (2006) |
| A. pernix | Ap DS | Iβ | 5.7 | 95 | 891 | 1.0 | 0.001 | Zhou et al. (2012) |
| M. tuberculosis | Mt DS | II | – | – | 37 | 3.1 | 0.08 | Webby et al. (2005a) |
| C. glutamicum | Cg AroF | II | – | – | 160 | 0.35 | 0.002 | Liu et al. (2008) |
| Cg AroG | 8520 | 1.65 | 0.0002 |
T DAHPS type, OpH optimal pH, OT optimal temperature
Discussion
Construction of the metagenomic library
The metagenomic DNA was directly extracted from the subtropical mangrove coastal wetland sediments. The constructed library contained a genome pool of the microorganisms in the wetland sediments, including that of uncultured microorganisms. Further analysis of randomly selected recombinant plasmids revealed that the foreign DNA fragments in pUC118 vector were highly diverse. This result also confirmed that the metagenomic library contained DNA molecules from uncharacterized genomes and that the metagenome of naturally occurring microbacteria contained an immense pool of genes; most of these genes could not be represented by pure and enrichment cultures established under certain selective conditions (Westmann et al. 2018). A new type Iβ DAHPS gene (aro1A) was identified in a metagenomic library by using a sequence-based screening strategy from the subtropical mangrove sediment.
Bioinformatics analysis of Aro1A protein
Relatively low consistence of sequence existed among DAHPSs; in particular, the sequence consistence between type I and type II is only 10% (Webby et al. 2005a). However, different DAHPSs have highly similar catalytic structural domain of the (β/α)8 barrel structure (König et al. 2004; Light et al. 2012; Nazmi et al. 2016; Shumilin et al. 1999, 2004; Sterritt et al. 2018; Webby et al. 2005a). The results of multiple sequence alignment reflected a similar situation. Only metal ion binding sites were totally conserved, and most of the DAHPSs had low sequence consistence. α-Helix and β chains of the catalytic structural domain shared a similar motif (Fig. 2).
The results of multiple sequence alignment revealed that the highly conserved residues in Aro1A involved in the combination of substrate binding sites and divalent metal ligands in other DAHPS enzymes (König et al. 2004; Wu and Woodard 2006) were completely conserved (Fig. 2). Four conserved binding residues (C36, H206, E232, and D243) were found with Mn2+ in the Aro1A protein (Fig. 3d). The motif 58VLRGGAFKPRT68 in Aro1A was highly conserved in type Iβ DAHPS. R60 and K65 in this motif combined with PEP in Mt DS, Tm DS, Pfu DS, and Ap DS and had non-bonded contact with PEP in Ec AroG. In addition, R67 and T68 in the abovementioned motif were the binding sites of E4P in Tm DS and were predicted to be the binding sites of E4P in Mt DS, Ec AroG, and Pfu DS. This finding implied that 58VLRGGAFKPRT68 was the motif that participated in the PEP and E4P binding for Aro1A protein. The G118 in motif 118GSR120 was a highly conserved non-bond-contacting residue of PEP. The corresponding residue of S119 was Ala in Ec AroG, Tm DS, Pfu DS, and Ap DS and Glu in Mt DS. These residues were the binding sites of PEP, and R120 was the completely conserved binding site of PEP. Therefore, 118GSR120 also participated in the binding of PEP in Aro1A. H206 was another highly conserved residue that had non-bonded contact with PEP. This residue was the binding site of metal ligand and was annotated as a catalytic site in the analysis of other DAHPSs. The metal ligand binding site D243 bound with E4P in Tm DS and Pfu DS. All the above conserved residues, including K141 and R171, covered most of the ligand binding sites for proteins in multiple sequence alignments (Nazmi et al. 2014; Schofield et al. 2005; Shumilin et al. 1999, 2004; Webby et al. 2005a; Zhou et al. 2012). A previous research also indicated that the catalytic capacity of DAHPS was mainly based on the same (β/α)8 structure.
Six binding sites (R60, S119, K141, R171, K65, and R120) of PEP were found in the multiple sequence alignment based on the conservative property (Fig. 2). Based on the results of molecular docking analysis, R60, S119, K141, and R171 bound with PEP through a hydrogen bond. K65 bound with PEP in Pfu DS, Ec AroG, and Ap DS. R120 bound with PEP in all five reference DAHPSs (Nazmi et al. 2014; Schofield et al. 2005; Shumilin et al. 1999, 2004; Webby et al. 2005a; Zhou et al. 2012). Furthermore, Q116 was predicted to bind with PEP in molecular docking. This residue was conserved among type Iβ DAHPS in the multiple sequence alignment and bound with PEP in Pfu DS and Ap DS (Nazmi et al. 2014; Schofield et al. 2005; Zhou et al. 2012). The results of molecular docking analysis revealed that G118 and H206, which were conserved residues having non-bonded contact with PEP, were near PEP in Aro1A (Fig. 4a). This finding implied the importance of the two residues in PEP binding.
The predicted binding sites of E4P in the multiple sequence alignment were only R67, T68, and D243 residues, which were adjacent, but not bound, to E4P in molecular docking. In the molecular docking, the six residues bonded with E4P were R60, K65, Q116, S119, K141, and R171, which were nearly identical to the residues bonded with PEP. This result may be attributed to the similarity in the molecular structures of the two substrates. The two substrates were spatially closed in all reference protein structures. Furthermore, the binding mode of the five reference proteins with E4P in multiple sequence alignment is rarely researched. Among these proteins, only Tm DS with E4P was studied with crystal analysis of complexus (Shumilin et al. 2004). Mt DS and Ec DS were the binding sites of E4P based on the similarity of sulfate and phosphate groups (Shumilin et al. 1999, 2004; Webby et al. 2005a). The binding of E4P was not analyzed for Pfu DS and Ap DS (Nazmi et al. 2014; Schofield et al. 2005; Zhou et al. 2012). Although the binding mode of E4P and DAHPS is unclear, the possible binding sites were analyzed via molecular docking.
Based on the combined results of molecular docking and multiple sequence alignment, Aro1A was similar with other DAHPSs because they all had a “conserved” ligand-binding space to accommodate a divalent metal ion (PEP and E4P). The space included, but not limited to, totally conserved residues and a motif. We speculated that Aro1A was similar with Pfu DS and ApDS based on the following: (1) only a catalytic part composed of (β/α)8 barrel existed, and (2) no part for the regulation on the N terminus or C terminus, indicating that Aro1A was not inhibited by the feedback of downstream aromatic amino acid.
Enzymatic property of Aro1A
The optimal temperature (40 °C) of Aro1A was close to that of the DAHPS from N. meningitis (Table 2). The temperature activity was similar to that of N. meningitis (Cross et al. 2013). The sequence length, the amino acid composition of key motif, and the secondary structural arrangement of Aro1A (Fig. 2) were almost consistent with those of DAHPS from A. pernix and P. furiosus. The similarity of Aro1A and these two DAHPS was around 50%. In addition, Aro1A and the DAHPS from T. maritima (ACCESSION: Q9WYH8.1) had the highest similarity. However, the optimal temperatures of the DAHPS from A. pernix and T. maritima were 95 and 90 °C, respectively (Table 2). These enzymes have good thermostability (at least 60 °C) (Schofield et al. 2004; Wu et al. 2003; Zhou et al. 2012). Hence, although Aro1A and these DAHPS from thermophiles had highly similar sequence and structure, they apparently had different optimal temperature and thermostability. The difference among these DAHPS in temperature response requires further evaluation.
The optimal pH of Aro1A was 8.0. This pH was higher than that of the acidic DAHPs from Tm DS and Ap DS and was similar with that of DAHPS from Bacillus subtilis and Porphyromonas gingivalis (Table 2).
To date, all the reported DAHPSs are metalloenzymes (Wu et al. 2005) that can be activated by a series of divalent metal ions. However, the activation mechanism of different metal ions considerably varies for different DAHPSs. Similar with the Aro1A protein, DAHPSs from C. glutamicum, P. furiosus, T. maritima, Actinosynnema, M. tuberculosis, H. pylori, Pseudomonas aeruginosa, and N. meningitidis (Cross et al. 2013; Liu et al. 2008; Ma et al. 2012; Schofield et al. 2004; Sterritt et al. 2018; Webby et al. 2005a, b; Wu et al. 2003) can be stimulated with Mn2+ ion. Ba2+ and Ca2+ had no effect for the DAHPS from N. meningitis (Cross et al. 2013). However, Ba2+ and Ca2+ can stimulate the activity of Aro1A protein. Furthermore, Mg2+ had relatively weak activation action on Aro1A but had better effect on DAHPS from C. glutamicum and Actinosynnema (Liu et al. 2008; Ma et al. 2012). Similar results can be found when the activation capacities of metal ions are compared.
The catalytic capacity of Aro1A was higher than that of the other type Iβ DAHPSs (Table 1), and Aro1A had relatively moderate optimal temperature (Table 2). Furthermore, the enzyme was an inherent DAHPS without feedback inhibition structure. Hence, Aro1A can be potentially used in the industrial production of aromatic amino acids, provided that the thermostability was solved by molecular modification.
Additional file
Authors’ contributions
HZ, NL, and CJ set up and designed the study, and BY and MZ collected the sediment sample. HZ and HG constructed the metagenomic library. HG, SM, and QO performed the protein expression and purification experiments, while QL, KJ, and HZ performed the bioinformatics analysis. HZ and HG wrote the manuscript, and finally, BW, NL, and CJ made the revisions. All authors discussed and commented the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data analyzed in this study has been included in the main article. And the aro1A nucleotide sequence was deposited in the GenBank database with the Accession number MH757446.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 31760437), the Science and Technology Basic Resources Investigation Program of China (Grant No. 2017FY100704), the Basic Scientific Fund for National Public Research Institutes of China (Grant No. 2016Q07), the Natural Science Foundation of Guangxi Zhuang Autonomous Region of China (Grant No. 2017JJB130020), the Open Research Fund Program of Guangxi Key Lab of Mangrove Conservation and Utilization (Grant No. GKLMC-201702), and the Distinguished Employment Offered Unit of Guangxi for Conservation and Ecological Monitoring of Mangroves and Sea-grasses.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DAHP
3-deoxy-d-arabino-heptulosonate-7-phosphate
- DAHPS
3-deoxy-d-arabino-heptulosonate-7-phosphate synthase
- PEP
phosphoenolpyruvate
- E4P
d-erythrose 4-phosphate
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- PCR
polymerase chain reaction
Contributor Information
Huaxian Zhao, Email: wstdklr@163.com.
Hua Gao, Email: 1726469989@qq.com.
Kai Ji, Email: 409650242@qq.com.
Bing Yan, Email: gxybing@tom.com.
Quanwen Li, Email: 344560867@qq.com.
Shuming Mo, Email: 772083034@qq.com.
Minggang Zheng, Email: zmg@fio.org.cn.
Qian Ou, Email: ouqian510@126.com.
Bo Wu, Email: 657575168@qq.com.
Nan Li, Phone: +86-771-3239403, Email: nli@yic.ac.cn.
Chengjian Jiang, Phone: +86-771-3239403, Email: jiangcj0520@gmail.com.
References
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burschowsky D, Thorbjørnsrud HV, Heim JB, Fahrig-Kamarauskaitė J, Würth-Roderer K, Kast P, Krengel U. Inter-enzyme allosteric regulation of chorismate mutase in corynebacterium glutamicum: structural basis of feedback activation by Trp. Biochemistry. 2018;57:557–573. doi: 10.1021/acs.biochem.7b01018. [DOI] [PubMed] [Google Scholar]
- Cross PJ, Pietersma AL, Allison TM, Wilson-Coutts SM, Cochrane FC, Parker EJ. Neisseria meningitidis expresses a single 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase that is inhibited primarily by phenylalanine. Protein Sci. 2013;22:1087–1099. doi: 10.1002/pro.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Ling C, Zhang Y, Huang J, Liu J. Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering. Microb Cell Fact. 2014;13:21. doi: 10.1186/1475-2859-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Totowa: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gosset G, Bonner CA, Jensen RA. Microbial origin of plant-type 2-keto-3-deoxy-d-arabino-heptulosonate 7-phosphate synthases, exemplified by the chorismate- and tryptophan-regulated enzyme from Xanthomonas campestris. J Bacteriol. 2001;183:4061–4070. doi: 10.1128/JB.183.13.4061-4070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedt K, Strittmatter A, Lipscomb WN, Braus GH. Evolution of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase-encoding genes in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci. 2005;102:9784–9789. doi: 10.1073/pnas.0504238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM. The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 1995;107:7–12. doi: 10.1104/pp.107.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyes LC, Reichau S, Cross PJ, Jameson GB, Parker EJ. Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase—the importance of accommodating the active site water. Bioorg Chem. 2014;57:242–250. doi: 10.1016/j.bioorg.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Jensen RA, Xie G, Calhoun DH, Bonner CA. The correct phylogenetic relationship of KdsA (3-deoxy-d-manno-octulosonate 8-phosphate synthase) with one of two independently evolved classes of AroA (3-deoxy-d-arabino-heptulosonate 7-phosphate synthase) J Mol Evol. 2002;54:416–423. doi: 10.1007/s00239-001-0031-z. [DOI] [PubMed] [Google Scholar]
- König V, Pfeil A, Braus GH, Schneider TR. Substrate and metal complexes of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Saccharomyces cerevisiae provide new insights into the catalytic mechanism. J Mol Biol. 2004;337:675–690. doi: 10.1016/j.jmb.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzler M, Paravicini G, Egli CM, Irniger S, Braus GH. Cloning, primary structure and regulation of the ARO4 gene, encoding the tyrosine-inhibited 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Saccharomyces cerevisiae. Gene. 1992;113:67–74. doi: 10.1016/0378-1119(92)90670-K. [DOI] [PubMed] [Google Scholar]
- Leis B, Angelov A, Mientus M, Li H, Pham VTT, Lauinger B, Bongen P, Pietruszka J, Gonalves LG, Santos H, Liebl W. Identification of novel esterase-active enzymes from hot environments by use of the host bacterium Thermus thermophilus. Front Microbiol. 2015;6:275. doi: 10.3389/fmicb.2015.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light SH, Halavaty AS, Minasov G, Shuvalova L, Anderson WF. Structural analysis of a 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase with an N-terminal chorismate mutase-like regulatory domain. Protein Sci. 2012;21:887–895. doi: 10.1002/pro.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Li PP, Zhao KX, Wang BJ, Jiang CY, Drake HL, Liu SJ. Corynebacterium glutamicum contains 3-deoxy-d-arabino-heptulosonate 7-phosphate synthases that display novel biochemical features. Appl Environ Microbiol. 2008;74:5497–5503. doi: 10.1128/AEM.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Xiao M, Zhang L, Xu J, Ding Z, Gu Z, Shi G. Production of l-phenylalanine from glucose by metabolic engineering of wild type Escherichia coli W3110. Process Biochem. 2013;48:413–419. doi: 10.1016/j.procbio.2013.02.016. [DOI] [Google Scholar]
- Ma N, Wei L, Fan Y, Hua Q. Heterologous expression and characterization of soluble recombinant 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Actinosynnema pretiosum ssp. auranticum ATCC31565 through co-expression with Chaperones in Escherichia coli. Protein Expr Purif. 2012;82:263–269. doi: 10.1016/j.pep.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis K, Armstrong Z, Song YC, Baldwin SA, Withers SG, Hallam SJ. Biomining active cellulases from a mining bioremediation system. J Biotechnol. 2013;167:462–471. doi: 10.1016/j.jbiotec.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazmi AR, Schofield LR, Dobson RCJ, Jameson GB, Parker EJ. Destabilization of the homotetrameric assembly of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from the hyperthermophile pyrococcus furiosus enhances enzymatic activity. J Mol Biol. 2014;426:656–673. doi: 10.1016/j.jmb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Nazmi AR, Lang EJM, Bai Y, Allison TM, Othman MH, Panjikar S, Arcus VL, Parker EJ. Interdomain conformational changes provide allosteric regulation en route to chorismate. J Biol Chem. 2016;291:21836–21847. doi: 10.1074/jbc.M116.741637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus F, Gabor E, Wieland S, Siegert P, Maurer KH, Eck J. Enzymes for the laundry industries: tapping the vast metagenomic pool of alkaline proteases. Microb Biotechnol. 2011;4:767–776. doi: 10.1111/j.1751-7915.2011.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap S, Dev A, Kumar V, Yadav R, Narwal M, Tomar S, Kumar P. Structure of chorismate mutase-like domain of DAHPS from Bacillus subtilis complexed with novel inhibitor reveals conformational plasticity of active site. Sci Rep. 2017;7:6364. doi: 10.1038/s41598-017-06578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray JM, Bauerle R. Purification and properties of tryptophan-sensitive 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli. J Bacteriol. 1991;173:1894–1901. doi: 10.1128/jb.173.6.1894-1901.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield LR, Patchett ML, Parker EJ. Expression, purification, and characterization of 3-deoxy-d-arabino- heptulosonate 7-phosphate synthase from Pyrococcus furiosus. Protein Expr Purif. 2004;34:17–27. doi: 10.1016/j.pep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Schofield LR, Anderson BF, Patchett ML, Norris GE, Jameson GB, Parker EJ. Substrate ambiguity and crystal structure of Pyrococcus furiosus 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase: an ancestral 3-deoxyald-2-ulosonate-phosphate synthase? Biochemistry. 2005;44:11950–11962. doi: 10.1021/bi050577z. [DOI] [PubMed] [Google Scholar]
- Schoner R, Herrmann KM. 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase. Purification, properties, and kinetics of the tyrosine-sensitive isoenzyme from Escherichia coli. J Biol Chem. 1976;251:5440–5447. [PubMed] [Google Scholar]
- Seow KT, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson CR, Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Kumar V, Chatrath A, Dev A, Prasad R, Sharma AK, Tomar S, Kumar P. In vitro metal catalyzed oxidative stress in DAH7PS: methionine modification leads to structure destabilization and induce amorphous aggregation. Int J Biol Macromol. 2018;106:1089–1106. doi: 10.1016/j.ijbiomac.2017.08.105. [DOI] [PubMed] [Google Scholar]
- Shumilin IA, Kretsinger RH, Bauerle RH. Crystal structure of phenylalanine-regulated 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli. Structure. 1999;7:865–875. doi: 10.1016/S0969-2126(99)80109-9. [DOI] [PubMed] [Google Scholar]
- Shumilin IA, Bauerle R, Wu J, Woodard RW, Kretsinger RH. Crystal structure of the reaction complex of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Thermotoga maritima refines the catalytic mechanism and indicates a new mechanism of allosteric regulation. J Mol Biol. 2004;341:455–466. doi: 10.1016/j.jmb.2004.05.077. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2014;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterritt OW, Kessans SA, Jameson GB, Parker EJ. A pseudoisostructural type II DAH7PS enzyme from Pseudomonas aeruginosa: alternative evolutionary strategies to control shikimate pathway flux. Biochemistry. 2018;57:2667–2678. doi: 10.1021/acs.biochem.8b00082. [DOI] [PubMed] [Google Scholar]
- Teshiba S, Furter R, Niederberger P, Braus G, Paravicini G, Hütter R. Cloning of the ARO3 gene of Saccharomyces cerevisiae and its regulation. Mol Gen Genet. 1986;205:353–357. doi: 10.1007/BF00430450. [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby CJ, Baker HM, Lott JS, Baker EN, Parker EJ. The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis reveals a common catalytic scaffold and ancestry for type i and type ii enzymes. J Mol Biol. 2005;354:927–939. doi: 10.1016/j.jmb.2005.09.093. [DOI] [PubMed] [Google Scholar]
- Webby CJ, Patchett ML, Parker EJ. Characterization of a recombinant type II 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Helicobacter pylori. Biochem J. 2005;390:223–230. doi: 10.1042/BJ20050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby CJ, Jiao W, Hutton RD, Blackmore NJ, Baker HM, Baker EN, Jameson GB, Parker EJ. Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2010;285:30567–30576. doi: 10.1074/jbc.M110.111856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmann CA, de Alves L, Silva-Rocha R, Guazzaroni M. Mining novel constitutive promoter elements in soil metagenomic libraries in Escherichia coli. Front Microbiol. 2018;9:1344. doi: 10.3389/fmicb.2018.01344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Woodard RW. New Insights into the evolutionary links relating to the 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subfamilies. J Biol Chem. 2006;281:4042–4048. doi: 10.1074/jbc.M512223200. [DOI] [PubMed] [Google Scholar]
- Wu J, Howe DL, Woodard RW. Thermotoga maritima 3-deoxy-d-arabino-heptulosonate 7-phosphate (DAHP) synthase. J Biol Chem. 2003;278:27525–27531. doi: 10.1074/jbc.M304631200. [DOI] [PubMed] [Google Scholar]
- Wu J, Sheflyan GY, Woodard RW. Bacillus subtilis 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase revisited: resolution of two long-standing enigmas. Biochem J. 2005;390:583–590. doi: 10.1042/BJ20050294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Xia Y, Qu H, Li A, Liu R, Wang Y, Zhang T. Discovery of new cellulases from the metagenome by a metagenomics-guided strategy. Biotechnol Biofuels. 2016;9:138. doi: 10.1186/s13068-016-0557-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Kang S, Park S, Yoon H, Kim M, Heu S, Ryu S. Characterization of a novel amylolytic enzyme encoded by a gene from a soil-derived metagenomic library. Appl Environ Microbiol. 2004;70:7229–7235. doi: 10.1128/AEM.70.12.7229-7235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wu J, Janakiraman V, Shumilin IA, Bauerle R, Kretsinger RH, Woodard RW. Structure and characterization of the 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from Aeropyrum pernix. Bioorg Chem. 2012;40:79–86. doi: 10.1016/j.bioorg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study has been included in the main article. And the aro1A nucleotide sequence was deposited in the GenBank database with the Accession number MH757446.