Abstract
There has been a limited number of systematic reviews conducted to summarize the overview of the relationship between DNA methylation and depression, and to critically appraise the roles of major study characteristics in the accuracy of study findings. This systematic review aims to critically appraise the impact of study characteristics on the association between DNA methylation and depression, and summarize the overview of this association. Electronic databases and gray literatures until December 2017 were searched for English-language studies with standard diagnostic criteria of depression. A total of 67 studies were included in this review along with a summary of their study characteristics. We grouped the findings into etiological and treatment studies. Majority of these selected studies were recently published and from developed countries. Whole blood samples were the most studied common tissues. Bisulfite conversion, along with pyrosequencing, was widely used to test the DNA methylation level across all the studies. High heterogeneity existed among the studies in terms of experimental and statistical methodologies and study designs. As recommended by the Cochrane guideline, a systematic review without meta-analysis should be undertaken. This review has, in general, found that DNA methylation modifications were associated with depression. Subgroup analyses showed that most studies found BDNF and SLC6A4 hypermethylations to be associated with MDD or depression in general. In contrast, studies on NR3C1, OXTR, and other genes, which were tested by only few studies, reported mixed findings. More longitudinal studies using standardized experimental and laboratory methodologies are needed in future studies to enable more systematical comparisons and quantitative synthesis.
Introduction
A number of systematic reviews on susceptible genes and gene–environment interplay provide a comprehensive list of putative genetic and environmental risk factors for depression1–6. In contrast, there has been little compilation of our knowledge of DNA methylation modifications and depression.
To our knowledge, there are five reviews, including only one systematic review so far on the relationship between DNA methylation and depression7–11. Generally, they suggested that altered DNA methylations may be associated with the etiology of depression. Lockwood et al. in their narrative review of epigenetic findings in both animal and human studies concluded that epigenetics could play an important role in depression and suicide in humans7. Again, Uddin et al8., using a similar approach, studied the role of sex in DNA methylation and post-traumatic stress disorder and major depressive disorder (MDD), and suggested that sex differences in DNA methylation among those genes known to influence brain development may explain the sexually dimorphic risk for developing post-traumatic stress disorder and MDD. Another narrative review found the inverse association between adverse environmental factors, i.e., early-life stress, and the epigenetic modification of gene expression9. A review examined the association between DNA methylation of seven candidate genes and depression, and found that brain-derived neurotropic factor (BDNF) and nuclear receptor subfamily 3 group C member 1 (NR3C1) gene methylation levels may be related to depression, whereas the relationship between serotonin transporter gene (SLC6A4; synonyms: 5-HTT and SERT) and depression was inconsistent11. One recent systematic review assessed both animal and human studies and identified the correlation between burnout/depression and global and candidate-gene DNA methylation10. However, this review did not examine the influence of experimental and statistical methodologies and analyses on findings.
Although a few reviews are published to explore the relationships between DNA methylation modifications and depression, there has been no review critically examining experimental methodologies and verification of laboratory testing in humans. The experimental methodologies and laboratory testing are closely linked with the accuracy of results. In addition, these reviews only focused on some aspects; for example, exploring the roles of sex and stress in this relationship. In this review, we aimed to (1) systematically synthesize the major findings on DNA methylation and depression, (2) compare the similarities and differences across different studies, including experimental and laboratory factors and statistical analyses used, which might partially explain some inconsistencies in the results, and (3) discuss the challenges and opportunities for future studies.
Materials and methods
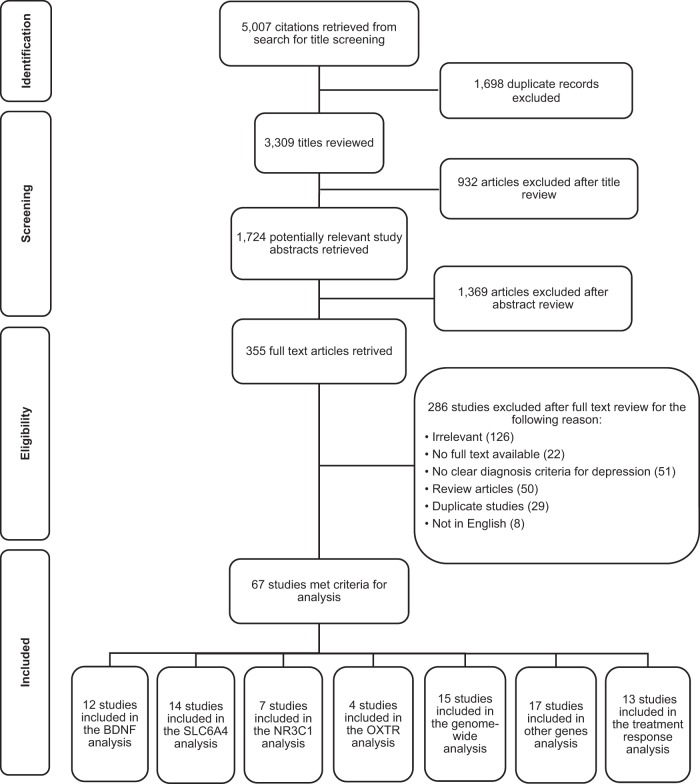
The processing and reporting of the results of this systematic review were guided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines 2009 revision 12. To ensure a thorough and systematic review of the literature, two methods were used to retrieve all the studies on relevant topics. We conducted a search of the computerized bibliographic databases PubMed, Web of Science, EMBASE, Medline, and Cochrane Library. The search strategy is detailed in Supplementary Appendix 1. The literature search comprised articles published until December 2017. A snowball technique was then applied to identify further studies. In addition, we manually searched other resources for other relevant studies. The reference lists of selected articles, review articles on relevant topics, and the gray literatures were screened. Figure 1 presents the process of study selection.
Fig. 1.
PRISMA flow diagram: DNA methylation and depression. Some selected studies had more than one study topic (i.e., BDNF); therefore, the total of these subgroups were bigger than the final number eligible for the review
All suitable articles were evaluated with regard to their internal validity based on the four selection criteria as follows: (1) if they used a clear diagnosis criteria for depression (e.g., depression in general, major depressive disorder, depressive symptoms, or other types of depression), specifically the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) and its updates13, and the International Statistical Classification of Diseases and Related Health Problems, 10th revision (ICD-10)14 or other generally accepted diagnostic criteria; (2) if they examined the association between DNA methylation and depression; and (3) if they provided a statistical indicator (i.e., coefficient) or original data to estimate the relationship between DNA methylation and depression. Articles were excluded if (1) they did not specify a clear diagnosis for major depression, major depressive disorder, unipolar depression, or other types of depression or (2) they were not written in English.
Two authors (M. Li and X. Li) independently screened all the retrieved articles. Inconsistencies in interpretation were resolved through group discussions (X. Li, M. Li, and X. Meng). Endnote and RefWorks were the bibliographic softwares used. Data on author(s), year of publication, sample size, study design, study cohort, experimental methods, type of tissues, candidate genes or genome, DNA purification method, DNA methylation method, DNA methylation validation, genotyping, gene expression, experimental factors, statistical methods, and major findings were extracted independently. For those studies with multiple reports, a single record denoted one study with the information extracted from multiple reports. Group discussions dealt with all the inconsistent interpretations. The reviewers endeavored to contact the original authors of the studies for any missing information in order to gather complete and consistent study information. Open-ended questions were used to prevent misleading answers.
Because of the divergence of candidate genes and genomes, for example, some studies used the candidate-gene approach and others examined the whole genome, we grouped the summarized findings according to the number of studies available, including etiological studies and treatment studies. The etiological studies were then further divided into the following subgroups, including (1) BDNF, (2) SLC6A4, (3) NR3C1, (4) oxytocin receptor (OXTR), (5) other genes, and (6) genome-wide. Some articles were involved in multiple separate analyses as their data permitted.
Results
A total of 67 articles met our eligibility criteria. Figure 1 shows the detailed information of the process of study selection. Table 1 presents a summary of study characteristics of these selected studies. Supplementary Appendix 2 provides a list of the references for all the selected articles corresponding to their order in Table 1. Most of the reviewed articles were published between 2014 and 2017, especially in the past 4 years. The selected studies mainly focused on adults and seniors (58/67), covering a total number of 11,935 subjects worldwide (North America: 18/67, Asia: 21/67, Europe: 24/67, and Australia: 6/67). We also evaluated study quality, including design (study design, sample size, and subject characteristics), implementation (biological sample, DNA methylation method, purification of DNA extraction, and validation of methylation), analysis (analytical method, batch effect, genotyping, and gene expression), and interpretation of results. Most studies in this review were case–control with hospital- or general population-based cohorts. There was a wide variety in terms of sample size, ranging from 11 to 1024. Whole blood was the most commonly used biological sample analyzed by generally accepted DNA methylation methods, such as bisulfite conversion with pyrosequencing. Both parametric and non-parametric statistics were used. Importantly, most of these studies did not analyze the influence of batch effect on their results (64/67), except the three studies targeted on genome-wide variations.
Table 1.
A summary of selected articles in this systematic review
| ID | First author | Publication year | Country | Sample size | Sample characteristics | Study design | Diagnoses of depression | Biological samples | DNA methylation methods/kits | Targeted genetic locations | Markers found in genome-wide studies/ CpG sites for candidate-gene studies | Major findings |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Bostrom et al. | 2017 | Sweden | 223 | Population-based adolescent cohort | Case–control | Depression in general | Whole blood | Illumina 450k | Genome-wide | The promoter region of miRNA4646 and TSS of ZSWIM5 | Two CpG sites (cg13227623 and cg04102384) predicted depression in adolescents. cg04102384 was hypomethylated |
| 2 | Roy et al. | 2017 | USA | 34 | Hospital-based cohort | Case–control | MDD | Peripheral blood mononuclear cells | Immunoprecipitate the 5-methyl cytosine-enriched and qPCR | BDNF, FKBP5, CRHBP, CRHR1, NR3C1 | Promoters, CpG islands | BDNF, FKBP5, CRHBP, CRHR1, NR3C1 gene promoters were significantly hypermethylated in MDD |
| 3 | Meng et al. | 2017 | China | 162 | Hospital-based cohort | Case–control | MDD | White blood cells | Bisulfite conversion, pyrosequencing | NET or SLC6A2 | Promoters, other CpG sites | There were no significant differences in DNA methylation of the NET gene promoter between healthy controls and patients with MDD |
| 4 | Kaut et al. | 2017 | Germany | 12 | Senior cases and controls | Case–control | MDD | Brain tissue | Bisulfite conversion, pyrosequencing | PSD-95 and GLA-1 | Promoters, CpG islands | There were no significant differences in DNA methylation of PSD-95 and GJA-1 between controls and cases |
| 5 | Ryan et al. | 2017 | Australia | 380 | Late-life MDD and controls | Case–control | MDD | Buccal cells | Bisulfite conversion, pyrosequencing | IL-6 and treatment responses | Promoters, CpG islands | Individuals with depression (current MDD or high depressive symptoms) had lower IL6 methylation levels at one of the four sites investigated. Antidepressant use was independently associated with higher IL-6 methylation at the same site |
| 6 | Shi et al. | 2017 | China | 161 | Hospital-based cohort | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing | 5-HTT or SLC6A4 | Promoters, CpG islands | Methylation (hypo- and hyper-) at positions 4 and 5 was significantly associated with MDD |
| 7 | Han et al. | 2017 | South Korea | 145 | Hospital-based cohort | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing | TESC | Gene body, other CpG sites | MDD had significantly higher methylation on CpG2 position of TESC gene-regulating genetic variant (rs7294919) than controls |
| 8 | Takeuchi et al. | 2017 | Japan | 20 | Cases with best and worst treatment responses | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing | Genome-wide | PPF1A4, HS3ST1 | Patients’ DNA-methylation profile at specific genes such as PPF1A4 and HS3ST1 was associated with individual variations in therapeutic responses |
| 9 | Crueanu et al. | 2016 | Canada | 32 | White Caucasians, cases and controls | Case–control | MDD | PFC brain tissue | Bisulfite conversion, quantified with EpiTYPER | SYN2 | Promoters and gene body, CpG islands | Hypomethylation of synapsins (SYN2) was linked to depression |
| 10 | Won et al. | 2016 | South Korea | 74 | Antidepressant-free cases and controls | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing | SLC6A4 | Promoter region, other CpG sites | Significant inverse correlations were observed between SLC6A4 DNA methylation and fractional anisotropy. SLC6A4 DNA methylation was significantly higher at CpG2 in MDD |
| 11 | Walker et al. | 2016 | Scotland | 29 | Members of a large family multiply affected by BD and MDD | Case–control | MDD | Whole blood | Sodium bisulphite using the EZ-96 DNA Methylation Kit, bead array using the Infinium HumanMethylation450 BeadChip | Genome-wide | Three DMR regions (promoter region of HOXA5 for hypomethylation, 5’ end of RNF39 for hypermethylation, and promoter and first exon of AGPAT1 and RNF5 for hypo-methylation) | Nominally significant differences in DNA methylation were observed; altered DNA methylation was a potential mechanism for mood disorders |
| 12 | Osborne et al. | 2016 | USA | 291 | Derived from two prospective cohorts designed to study PPD and two cohorts from which DNA was taken long after pregnancy | Case–control | PPD | Whole blood | Illumina Human Methylation 450 (HM450) bead array for 51 women with mood disorders (existing data); bisulfite conversion pyrosequencing using PyroMark MD system for the rest of the samples | Genome-wide | No site identified | Epigenetic variation at PPD biomarker loci was likely to be associated with expression |
| 13 | Bustamante et al. | 2016 | USA | 147 | Lifetime MDD and controls | Case–control | MDD | Whole blood | Bisulfite conversion using EpiTect Bisulfite Kit, pyrosequencing using PyroMark Q24 Assay Design Software | NR3C1 | Promoters, CpG islands | DNA methylation was significantly lower over CpG sites 5–13 in those with vs without MDD |
| 14 | Na et al. | 2016 | South Korea | 117 | Recurrent MDD and controls | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing, using PyroMark ID system with the Pyro Gold reagents kit (Qiagen, Valencia, CA, USA) | BDNF and treatment response | Promoters, CpG islands | Patients with MDD had significantly higher rates of methylation at CpG2 and CpG4 than healthy controls. No difference was found in naive or on-medication patients |
| 15 | Kimmel et al. | 2016 | USA | 352 | Caucasian women | Cohort | PPD | Whole blood | Bisulfite conversion by EZ DNA Methylation-Gold Kit and pyrosequencing using PyroMark MD system | OXTR | 5’-UTR, CpG islands | CpG (cg12695586) positioned in the middle of SP1 transcription factor binding site. Its methylation had a negative correlation with PPD |
| 16 | Kahl et al. | 2016 | Germany | 70 | Treated MDD in patients and university announcements for controls | Case–control | MDD | Whole blood | Bisulfite conversion, PCR and sequencing. Sodium-bisulfite conversion using the EpiTect® Bisulfite Kit | GLU1, GLU4 | Promoters, CpG islands | Increased methylation of GLUT1 in MDD |
| 17 | Iga et al. | 2016 | Japan | 57 | Unmediated cases and controls | Case–control | MDD | Leukocytes | Bisulfite conversion, pyrosequencing, EpiTect Plus DNA Bisulfite Kit (Qiagen) | SLC6A4 | Promoters, CpG islands | Mean methylation level was significantly increased in patients compared with controls, p = 0.04. No significant difference was found in single CpG site |
| 18 | Oh et al. | 2015 | Peripheral blood samples from Australia, The Netherlands, and UK; prefrontal cortex and sperm samples from Canada | 260 | Cases and matched controls | Case–control | MDD | Peripheral blood, prefrontal cortex, and sperm | Bisulfite conversion, pyrosequencing using Gold Q96 reagents, and Pyromark Q24 | Genome-wide | No site identified | Hypermethylated loci were found in the white blood cells of MDD twins. The brain and the sperm showed higher proportions of hypomethylated regions in MDD patients compared with the controls |
| 19 | Nagy et al. | 2015 | Canada | 121 | Cases with MDD and died from suicide, and controls, not died from suicide and with no MDD | Case–control | MDD | Brain tissue | Bisulfite conversion using EpiTect Bisulfite kit from Qiagen, PCR, and sequencing | Genome-wide | 115 DMRs | Significant differences (decrease) in the methylation patterns specific to astrocytic dysfunction associated with depressive psychopathology |
| 20 | van der Knapp et al. | 2015 | The Netherlands | 954 | Adolescents cohort | Case–control | Depression in general | Whole blood | Methylation levels analyzed using EpiTYPER method; bisulfite conversion using EZ-96 DNA Methylation Kit, followed by PCR | NR3C1 and SLC6A4 | Promoters, CpG islands | NR3C1 methylation levels at NR3C1_1 were positively associated with the risk of a depressive disorder and were positively associated with depressive symptom scores at follow-up, but became non-significant when accounted for depressive symptom scores at the baseline |
| 21 | Melas et al. | 2015 | Sweden | 44 | Female cases and controls | Case–control | Depression in general | Saliva | Bisulfite conversion using EZ-96 DNA Methylation-Gold Kit, PCR, and sequencing | MAOA | Gene body, other CpG sites | Subjects with a history of depression were hypomethylated, compared to controls. Female individuals were hypermethylated at the MAOA region compared to males |
| 22 | Hohne et al. | 2015 | Germany | 116 | Remitted MDD and healthy controls | Case–control | MDD | Peripheral blood cells | Bisulfite conversion, PCR, and sequencing using EpiTYPER assay | FKBP5 | Gene body, other CpG sites | Subjects with TT genotype and a lifetime history of MD had a 10% higher DNA methylation rate than healthy controls with the same FKBP5 genotype |
| 23 | Choi et al. | 2015 | South Korea | 113 | MDD with a mixed history of treatment | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing was performed on a PyroMark ID system using the Pyro Gold reagent kit (Qiagen) | BDNF | Promoters, other CpG sites | There were no significant differences in the BDNF DNA methylation status at CpG1, CpG2, CpG3, and CpG4 between patients with MDD and healthy controls |
| 24 | Domschke et al. | 2015 | Germany | 94 | Caucasian case cohort with antidepressants | Cohort | MDD | Whole blood | Sodium bisulfite converted using EZ-96 DNA methylation kit, PCR, and sequencing using BigDye Terminator | MAOA and treatment response | Promoters and gene body, not mentioned for CpG sites | The study did not find a major influence of MAOA DNA methylation on antidepressant treatment response. However, the presently observed trend towards CpG-specific MAOA gene hypomethylation might potentially drive impaired antidepressant treatment response in females—larger pharmacoepigenetic studies are needed |
| 25 | Córdova-Palomera et al. | 2015 | Spain | 34 | Caucasian MZ twins | Twin study | Depression in general | Whole blood | Bisulfite conversion, bead array using The Illumina Infinium HumanMethylation450 (450K) BeadChip | DEPDC7 | Gene body, other CpG sites | A hypomethylation of cg09090376 in a co-twin would be associated with an increase in his/her depressive symptom score |
| 26 | Reiner et al. | 2015 | Germany | 85 | Female inpatients and controls | Case–control | Depression and/or dysthymia | Leukocytes | Bisulfite conversion using EpiTect Bisulfite Kit, PCR, and sequencing using BigDye Terminator v3.1 Cycle Sequencing Kit | OXTR | Gene body, other CpG sites | Depressed female patients had decreased OXTR exon 1 DNA methylation compared to non-depressed women. Exon 1 methylation appears to be associated with depressive phenotypes, whereas exon 2 methylation was influenced by genotype rs53576 |
| 27 | Haghighi et al. | 2015 | USA | 120 | Age- and sex-matched cases and controls | Case–control | MDD | Buffy coat of blood | Bisulfite conversion by EpiTect Bisulfite Kit, pyrosequencing using PyroMark Q96 MD | FADS1, FADS2, and ELOVL5 | 5’-UTR, CpG islands, and shores | MDD patients had a lower methylation in FADS2, but higher in ELOVL5 |
| 28 | Chagnon et al. | 2015 | Canada | 43 | Women aged 65 years and plus | Case–control | Depression (major and minor) and/or anxiety | Saliva | Bisulfite conversion, pyrosequencing using Pyromark 96, except for APOE analyzed on Illumina Beadchip | BDNF, OXTR, SLC6A4, and APOE | Gene body, other CpG sites | A higher BDNF and OXTR DNA methylation was observed in subjects with anxiety/depression compared to controls |
| 29 | Córdova-Palomera et al. | 2015 | Spain | 34 | Twin pairs with MDD and healthy controls | Case–control twin study | MDD | Whole blood | Bisulfite conversion using Illumina Infinium HumanMethylation450 Beadchip | Genome-wide | cg01122889 (WDR26) | Hypomethylation in WDR26 gene was associated with a lifetime diagnosis of depression |
| 30 | Bell et al. | 2015 | USA | 545 | Nested case–control study in a longitudinal cohort | Nested case–control | PPD | Whole blood | Bisulfite conversion, pyrosequencing using PyroMark Gold Q24 | OXTR | Gene body, other CpG sites | Methylation was not significantly associated with postpartum depression |
| 31 | Zhang et al. | 2015 | China | 125 | MDD only, with or without suicide attempts | Case–control | MDD | Whole blood | Bisulfite conversion, methylation-specific PCR | TPH2 | Promoters, other CpG sites | The TPH2 promoter was methylated in 36.0% (18/50) of MDD + suicide patients, as compared with that in 13.0% (10/75) of MDD patients |
| 32 | Nantharat et al. | 2015 | Thailand | 62 | Untreated MDD and controls | Case–control | MDD | Whole blood | Bisulfite pyrosequencing. PyroMark LINE-1 kit (Biotage-Qiagen, Uppsala, Sweden) | NR3C1 | Promoters, CpG islands | Hypermethylation levels at CpG7 were found in MDD in females but not in males |
| 33 | Kleimann et al. | 2015 | Germany | 11 | Treatment-resistant cases | Perspective cohort | MDD | Whole blood | Bisulfite conversion using EpiTect Bisulfite Kit, PCR, and sequencing using BigDye Terminator Cycle Sequencing Kit | Treatment responses on BDNF | Promoters, CpG islands | Remitters had a significantly lower mean promoter methylation rate than non-remitters, especially exon I |
| 34 | Kim et al. | 2015 | South Korea | 969 | Patients with recent acute coronary syndrome | Longitudinal | Mix of major and minor depression | Leukocytes | Bisulfite conversion using EpiTech Bisulfite Kit, pyrosequencing using PSQ 96M System | BDNF | Promoters, CpG islands | At baseline, a higher methylation percentage in MDD compared with no depression. Higher BDNF methylation independently associated with prevalent depressive disorder at baseline and follow-up |
| 35 | Kaut et al. | 2015 | The Netherlands | 12 | Recurrent MDD and controls | Pilot–replication | MDD | Postmortem brain, HIP, PFC tissue | Bisulfite conversion with a ZymoResearch bisulfite kit and Ininium Human Methylation 450K bead arrays | Genome-wide, selected genes for replication | three CpG sites on GRIN2A | 11 genes in the hypocampus and 20 genes in the prefrontal cortex revealed differential methyaltion. In replication, GRIN2A was found hypermethylated in both tissues and single CpG level |
| 36 | Kang et al. | 2015 | South Korea | 631 | Aged 65 years and plus for cases and controls | Longitudinal | Depression in general | Leukocytes | Bisulfite conversion using EpiTech Bisulfite Kit, pyrosequencing using the PSQ 96M System | BDNF | Promoters, CpG islands | Higher BDNF methylation was independently associated with depression and severe depressive symptoms |
| 37 | Kang et al. | 2015 | South Korea | 309 | Hospital-based, all women with breast cancer undergoing breast surgery | Longitudinal | Mix of major and minor depression | Leukocytes | Bisulfite conversion using EpiTech Bisulfite Kit, pyrosequencing using the PSQ 96M System | BDNF | Promoters, CpG islands | A higher methylation percentage at CpG9 with depression, both 1 week and 1 year after breast cancer |
| 38 | Januar et al. | 2015 | France | 1024 | Aged 65 years and plus for cases and controls | Case–control | MDD | Buccal cells | Bisulfite conversion, PCR, and sequencing. Sodium-bisulfite conversion using the EpiTect® Bisulfite Kit); sequencing was performed using a BigDye Terminator v3.1 Cycle Sequencing Kit | BDNF | Promoters, CpG islands | Depression at baseline and chronic late life was associated with higher BDNF methylation |
| 39 | Frodl et al. | 2015 | Ireland | 60 | Cases had experienced acute depressive episodes, matched on age and sex with controls | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing; PyroMark Q24 | SLC6A4 | Promoters, CpG islands | MDD was not significantly associated with methylation |
| 40 | Booij et al. | 2015 | Canada | 69 | Adults, matched on sex and gender between cases and controls, cases not taking antipsychotics or mood stabilizers | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing; PyroMark Q24 Software (Qiagen) for methylation percentage at each site. | SLC6A4, treatment response | Gene body, CpG islands | MDD diagnosis was not significantly associated with DNA methylation. Patients with SSRIs had greater methylation |
| 41 | Numata et al. | 2015 | Japan | 63 | Hospital-based cases and matched controls | Case–control | MDD | Whole blood | Bisulfite conversion using EZ DNA methylation Kit (ZYMO research), Infinium Human Methylation 450 Beadchips | Genome-wide | 363 (313 CGIs) | 363 CpG sites demonstrated lower DNA methylation in MDD patients than in controls. 18 MDD-associated DNA methylation markers to discriminate cases from controls |
| 42 | Haghighi et al. | 2015 | USA | 53 | MDD and suicide cases and controls | Case–control | MDD | Whole blood | Bisulfite conversion using Illumina Infinium HumanMethylation27 BeadChip | Genome-wide | Not mentioned | Increased age-related DNA methylation perturbations in the prefrontal cortex in major depression suicide compared with nonpsychiatric controls |
| 43 | Tadic et al. | 2014 | Germany | 39 | MDD inpatients | Cohort | MDD | Leukocytes | Bisulfite conversion, PCR, and sequencing using BigDye Terminator v3.1 Cycle Sequencing Kit | Treatment response on BDNF | Promoters, CpG islands | Antidepressant treatment did not significantly affect the methylation at BDNF promoter IV; thus, changes in the methylation status in this DNA region seem not to be involved in the response to antidepressant treatment |
| 44 | Khulan et al. | 2014 | Finland | 166 | Senior cases and controls | Case–control | Depressive symptoms | Whole blood | Bisulphite conversion using EZ DNA methylation kit, bead array using Illumina methylation 450k beadchip and Infinium chemistry | Genome-wide | CpG islands, shores, and TSS | Hypomethylation was associated with depressive symptoms. The results supported that DNA methylation differences may be important in the pathogenesis of psychiatric disease |
| 45 | Domschke et al. | 2014 | Germany | 94 | Caucasian cases with antidepressants | Cohort | MDD | Whole blood | Sodium bisulfite converted using EZ-96 DNA methylation Kit, PCR, and sequencing using BigDye Terminator | Treatment response on 5-HTT | Gene body, CpG islands | Hypomethylation of the 5-HTT transcriptional control region might impair antidepressant treatment response in Caucasian patients with MDD |
| 46 | Kaminsky et al. | 2014 | USA | Not mentioned | Not mentioned | Longitudinal | PPD | Whole blood | Not mentioned | HP1BP3 and TTC9B | Not mentioned | HP1BP3 and TTC9B (hypermethylation) predicted PPD with an area under the receiver operator characteristic curve (AUC) of 0.87 |
| 47 | Guintivano et al. | 2014 | USA | 93 | Caucasian women | Longitudinal | PPD | Whole blood | Illumina’s Infinium Human Methylation450 Beadchip Kit | Genome-wide | Two loci within the HP1BP3 and TTC9B genes | CpG methylation levels at two loci within the HP1BP3 and TTC9B genes were identified as biomarkers predictive of PPD |
| 48 | Tseng et al. | 2014 | China (Taiwan) | 74 | MDD cases and controls | Case–control | MDD | Leukocytes | ELISA-based for global DNA methylation profiling. MethylFlash methylated DNA quantification kit (for 5-mc), MethylFlash hydroxymethylated DNA quantification kit (for 5-hmc) | Genome-wide | Gobal methylation levels, no site mentioned | Lower levels of 5-hmc and 5-mc in severe MDD than controls, no difference among severe and remitted patients |
| 49 | Okada et al. | 2014 | Japan | 100 | Untreated cases or cases without a history of depressive episodes | Case–control | MDD | Whole blood | Bisulfite conversion using EZ DNA methylation kits; analyzed using a MassARRAY | SLC6A4 | Promoters, CpG islands | The pre-treatment-methylation rate(CpG3) of SLC6A4 is associated with therapeutic responses to antidepressants in un-medicated patients with MD |
| 50 | Na et al. | 2014 | South Korea | 117 | Untreated cases (no history of antidepressants) | Case–control | MDD | Whole blood | Bisulfite conversion, pyrosequencing using PyroMark ID system with the Pyro Gold reagent kit (Qiagen, Valencia, CA, USA) | NR3C1 | Promoters, CpG islands | MDD had significantly lower methylation than healthy controls at two CPG sites (CpG3, -4) |
| 51 | Davies et al. | 2014 | UK | 454 (50 twins, 354 case–control) | Monozygotic twins, discordant for depression | Twin study and case–control | MDD | Whole blood | Methylated DNA immunoprecipitation combined with ultra-deep sequencing (MeDIP-seq) (enrichment for methylated regions) | Genome-wide | Coding region of ZBTB20 gene | Both AU and UK did not identify DMR of genome-wide significance. MDD was associated with hypermethylation on the coding region of ZBTB20 |
| 52 | Carlberg et al. | 2014 | Austria | 554 | Unrelated in- and outpatients of White European origin | Case-Control | MDD | Peripheral blood mononuclear cells (PBMCs) | Bisulfite conversion using EZ-96 DNA Methylation Kit. Used methylation-specific quantitative PCR following the MethyLight protocol using SYBR green | BDNF, treatment response | Promoters, CpG islands | BDNF exon I promoter was significantly increased in MDD. Current antidepressant therapy was associated with increased methylation |
| 53 | Dell’Osso et al. | 2014 | Italy | 87 | Stable, pharmacological treated MDD and matched controls | Case–control | MDD | Peripheral blood mononuclear cells (PBMCs) | Bisulfite conversion, PCR, and sequencing | BDNF, treatment response | Promoters, CpG islands | Overall lithium and valproate tend to decrease the DNA methylation level at BDNF gene promoter, when compared to other classes of medications. However, within each different disorder, mood stabilizers did not seem to affect DNA methylation, suggesting that such an alteration was likely not due to treatment use |
| 54 | Zhao et al. | 2013 | USA | 84 | MZ twins (male veterans) for lifetime and concurrent MDD | Twin study | MDD | Leukocytes | Bisulfite conversion using EZ DNA methylation kit, pyrosequencing using PSQ 96 HS System | SLC6A4 | Promoters, CpG islands | Variation in methylation level within the promoter region of SLC6A4 was associated with variations in depressive symptoms. A 10% increase in the difference in mean DNA methylation level was associated with a 4.4-fold increase in the difference in BDI scores. The 5-HTTLPR genotype did not modulate this association. The use of antidepressants did not affect the relationship between SLC6A4 methylation and depressive symptoms |
| 55 | Melas et al. | 2013 | Sweden | 174 | Female cases and controls | Case–control | Depression in general | Saliva | Bisulfite conversion using EZ-96 DNA Methylation-Gold Kit, PCR, and sequencing, EpiTyper software | MAOA | Gene body, other CpG sites | Overall MAOA methylation levels were decreased in depressed females compared to controls |
| 56 | Byrne et al. | 2013 | Australia | 48 | Queenland twin study (discordant MDD and concordant no MDD) | Twin study | MDD | White blood cells | Bisulphite conversion, Illumina Human Methylation 450 BeadChip | Genome-wide | 17 sites (6 CpG islands) | The difference in mean methylation was significant in females within discordant pairs, but not in males |
| 57 | Kim et al. | 2013 | South Korea | 286 | Patients with a recent ischemic stroke | Longitudinal | Post-stroke depression (both major and minor) | Leukocytes | Bisulfite conversion using EpiTech Bisulfite Kit, pyrosequencing using PSQ 96M System | SLC6A4 | Promoters, CpG islands | Higher SLC6A4 methylation status was independently associated with a major post-stroke depression at both baseline and follow-up |
| 58 | Kim et al. | 2013 | South Korea | 286 | Patients with a recent ischemic stroke | Longitudinal | Post-stroke depression (both major and minor) | Leukocytes | Bisulfite conversion using EpiTech Bisulfite Kit, pyrosequencing using PSQ 96M System | BDNF | Promoters, CpG islands | Prevalent, persistent, and incident PSD had a higher BDNF methylation status. CpG site 6 was significantly associated with incident post-stroke depression |
| 59 | Kang et al. | 2013 | South Korea | 108 | Patients with MDD only | Longitudinal | MDD | Leukocytes | Bisulfite conversion using EpiTech Bisulfite Kit, pyrosequencing using PSQ 96M System | SLC6A4, treatment response | Promoters, CpG islands | SLC6A4 methylation status as a marker for childhood adversities among MDD, but was not associated with treatment outcomes |
| 60 | Bayles et al. | 2013 | Australia | 106 | Newly diagnosed or currently untreated and have not been receiving antidepressants for at least 4 weeks | Case–control | MDD | Leukocytes | Bisulfite conversion, PCR, and sequencing; EpiTYPER methylation analysis | SLC6A2 or NET | Promoters, CpG islands | There were no significant differences between MDD cases and controls in terms of the pattern of methylation of the SLC6A2 promoter. Antidepressant treatment did not change the result |
| 61 | Zill et al. | 2012 | Germany | 162 | Caucasian cases and controls | Case–control | MDD | Leukocytes | Bisulfite conversion, PCR, and sequencing, EpiTect Bisulfite Kit | ACE | Promoters, CpG islands | MDD patients showed a hypermethylation pattern at all the CpG sites compared to healthy controls |
| 62 | Sabunciyan et al. | 2012 | USA | 154 | MDD and controls | Replication | MDD | Postmortem frontal cortex, lymphoblastoid cell lines, postmortem brain | CHARM assay platform | Genome-wide | No site identified | PRIMA1 significantly increased the methylation in MDD in pilot, but not in replication |
| 63 | Uddin et al. | 2011 | USA | 100 | Lifetime depression cases and non-depressed controls | Case–control | Depression in general | Whole blood | Bisulfite conversion using EZ-96 DNA Methylation Kit, bead array using HumanMethylation27 (HM 27) DNA Analysis BeadChip | Genome-wide | 21 uniquely methylated and 107 uniquely unmethylated sites with depression | Uniquely unmethylated gene sets distinguished between those with versus without lifetime depression. In particular, some processes (e.g., brain development, tryptophan metabolism) showed patterns suggestive of increased methylation among individuals with depression whereas others (e.g., lipoprotein) showed patterns suggestive of decreased methylation among individuals with depression |
| 64 | Fuchikami et al. | 2011 | Japan | 38 | Japanese adults | Case–control | MDD | Whole blood | Bisulfite conversion using EZ DNA methylation kit | BDNF | Promoters, CpG islands | Significant methylation difference was found in CpGI, but not in -IV |
| 65 | Olsson et al. | 2010 | Australia | 150 | Australian adolescents (cases and controls) | Case–control | MDD | Buccal cells | Bisulfite conversion, Sequenom MassARRAY EpiTyping | SLC6A4 | Promoters, CpG islands | There was no association between depressive symptoms and either buccal cell 5-HTT methylation or 5-HTTLPR. Depressive symptoms were more common among those with elevated buccal cell 5-HTT methylation who carried a 5-HTTLPR short allele |
| 66 | Alt et al. | 2010 | The Netherlands | 12 | Depression and control groups matched for sex, age, brain weight, and postmortem delay | Case–control | MDD | Brain tissues | Bisulphite conversion, pyrosequencing using PyroMark ID | NR3C1 | Promoters, CpG islands | No significant difference in methylation pattern was found between case and control groups |
| 67 | Philibert et al. | 2008 | USA | 192 | Lifetime MDD and controls | Longitudinal | MDD | Lymphoblast cell lines | Bisulfite conversion, methylation ratios calculated by usingMassARRAY | SLC6A4 | Promoters, CpG islands | Greater amounts of methylation in females vs males, and a trend of higher methylation was associated with greater vulnerability of lifetime MDD |
MDD = major depressive disorder, PPD = postpartum depression, PFC = prefrontal cortex, BD = bipolar disorder, HIP = hippocampus, SSRI = selective serotonin reuptake inhibitors, DMR = differentially methylated regions, PSD = poststroke depression
This review was designed to apply evidence-based approaches to summarize the findings between DNA methylation and depression. High heterogeneity was identified among the studies reviewed. The Cochrane guidelines do not recommend using quantitative methods, such as meta-analysis, to synthesize the research findings. Thus, qualitative methods were then used to summarize the overview of the research findings. We present the results in two categories based on the research objectives of these selected studies, namely etiological (genome-wide and candidate-gene) and treatment studies. Supplementary Appendix 3 provides a detailed description of each subgroup and its results.
Etiological studies: genome-wide
Although all genome-wide studies found significant methylation modifications associated with depression, both hyper-and hypomethylation correlations were reported. Inconsistent results were also noted. For instance, in one study, hypermethylation was previously found in a pilot study, but was not present on its replication15; a significant decrease in mean methylation was observed among females, but not among males16; lower methylation levels were found among severe MDD patients vs healthy controls, but no difference between severe vs remitted patients17; and one study found both hypermethylations in some processes (e.g., brain development and tryptophan metabolism), and hypomethylations in other tissues (e.g., lipoprotein)18. Generally, sample sizes were not associated with study designs or major findings. However, studies with large sample sizes were more likely to use DNA purification methods and examine gene expression than those with smaller samples. Results from studies with large sample sizes are considered to be more reliable.
Etiological studies: candidate-gene
Generally, most studies found BDNF and SLC6A4 hypermethylation to be associated with MDD or depression. Studies on NR3C1, OXTR, and the rest of candidate genes, which were tested by only a few studies, reported mixed findings (hyper- and hypomethylation modifications and non-significant differences). The promoter regions and CpG islands were frequently targeted in these studies. The sample size in each group varied dramatically from 12 to 1024. Some of these studies also had gene expression for significant findings. Replications of findings were better in BDNF and SLC6A4 than in other studied genes. Studies with a longitudinal study design, reliable laboratory arrays, and statistical analyses were more likely to provide robust results.
Treatment studies
Findings in this group are more inconsistent compared to those in etiological studies. Half of the studies did not identify any significant methylation sites associated with antidepressant responses, and the rest had mixed significant findings (hyper- and hypomethylations) on different candidate genes. Again, the promoter regions and CpG islands were the major targets. This group of studies had a higher level of heterogeneity compared to other subgroups, as treatment history and stages of treatments may influence methylation modifications.
Discussion
This review firstly explored the role of DNA methylation in depression considering both the laboratory and analytic factors that could potentially confound the findings. A total of 67 articles were included in this review. The majority of the selected studies were recently published and were from developed countries. Whole blood was the most common tissue used in these analyses. Bisulfite conversion, along with pyrosequencing, was widely used to test DNA methylation level. There was a high heterogeneity among the studies in terms of the laboratory and statistical methodologies used and study designs. Large sample size and laboratory verification (DNA purification and DNA methylation validation) are the major characteristics important for accurate results.
The findings of our study are as follows. (1) For studies using candidate-gene approaches, BDNF, NR3C1, SLC6A4, and OXTR genes were the most frequently studied genes. Promoters and CpG islands were the common targeted regions. Overall, most of the studies found that BDNF and SLC6A4 hypermethylations were associated with depression. Studies on NR3C1, OXTR, and other candidate genes reported mixed findings in terms of methylation modification and depression. Again, promoters and CpG islands still were the focus. (2) All genome-wide studies found significant methylation sites, including hyper- and hypomethylations. (3) For studies that explored antidepressant treatment responses, their results were inconsistent as they targeted on a number of different genes and different stages of treatment. (4) Large-sample size studies were more likely to use DNA purification methods, examine gene expression in their analyses, and provide more reliable results.
Findings on etiological genome-wide studies
All genome-wide studies reported that DNA methylation was significantly associated with depression. Hypermethylations were observed in six studies on the following genes: zinc finger and BTB domain containing 20 (ZBTB20), heterochromatin protein 1-binding protein 3 (HP1BP3), tetratricopeptide repeat domain 9B (TTC9B), and glutamate ionotropic receptor NMDA type subunit 2A (GRIN2A)19–24.
ZBTB20 exists in the hippocampal neurons and cerebellum granule cells25, and plays a role in many processes, including neurogenesis, glucose homeostasis, and postnatal growth26. It may also have an impact on the development and regionalization of the human hippocampus, which has been found to be related to depression27–29.
Both HP1BP3 and TTC9B are linked to estrogen signaling. HP1BP3 is highly expressed in the brain and is related to a number of physical and behavioral phenotypes in mice, such as dwarfism, impaired bone mass, impaired maternal behavior, and anxiety30,31. Lower HP1BP3 has been found to be associated with postpartum depression and Alzheimer’s disease in humans21,32. TTC9B has been identified to be related to gonadal hormones33 and may be linked to hippocampal synaptic plasticity, which is critical for hippocampal long-term potentiation and depression34. These markers in peripheral blood may indicate estrogen-mediated epigenetic changes in the hippocampus and in turn, potentially, raise the vulnerable phenotypes based on their actions in brain21.
The GRIN2A gene provides the instructions for making a protein called glutamate receptor subunit epsilon-1 in human encoded GluN2A, which is one of the components (subunit) of a subset of N-methyl-D-aspartate (NMDA) receptors. They are involved in normal brain development and are responsible for changes in the brain in response to experience (synaptic plasticity), learning, and memory26. Methylation modifications in GRIN2A may play a key role in determining the function of NMDA receptors. Generally, gene promoter-region methylation could repress the gene expression, but the methylation on gene body can be positively correlated with expression activity35. This suggests that the hypermethylation of the GRIN2A gene body may result in the overexpression of NR2A and, thus, promote vulnerability for MDD via up-regulating NMDA receptor-dependent glutamatergic signaling36.
Hypomethylations were also observed among depression patients on the following genes: WD repeat domain 26 (WDR26), the promoter region of miRNA4646, 5-hydroxymethylcytosine (5-hmc), and 5-methylcytosine (5-mc)17,23,37–41. Consistent with our findings on WDR26, previous studies have found that the hypomethylation of WDR26 in depressed individuals may be related to lower gene-expression levels42. Additionally, the decreased blood transcription levels of WDR26 were associated with depression-related phenotypes42–45. 5-mc is a methylated form of the DNA base cytosine, which could be involved in the regulation of gene transcription. Its presence is important for the maintenance of the active chromatic state and for neurogenesis at non-promoter CpG islands46, and is associated with stable and long-term transcriptional silencing of promoters47. 5-mc is also found to be involved in the critical mechanism mediating genomic imprinting. This process has been identified as a key for normal development, and its abnormal imprinting can result in disorders such as Prader–Willi, Angelman, and Beckwith–Wiedemann syndrome47.
5-hmc is a product of conversion of 5-mc. It is related to the regulation of gene expression, prompting DNA demethylation. The three ten-eleven translocation (TET) enzymes oxidize each step in the demethylation of 5-mc. 5-mc is first converted to 5-hmc, then to 5-formylcytosine (5fC), and then to 5-carboxylcytosine (5caC), each by TET1-348. Reduced levels of TET1 and, subsequently, 5hmc cause impaired self-renewal of stem cells49.
Notably, inconsistent results were identified within the same studies among different subgroups; for example, different sexes16, processes (e.g., brain development, tryptophan metabolism, and lipoprotein)18, tissues (white blood cells, brain, and sperm)50, or between pilot and replication studies15.
Findings on etiological candidate-gene studies
For candidate-gene studies, the majority (11/12) of studies on BDNF found BDNF hypermethylation were associated with cases suffering from depression. Most of the studies had relatively large sample sizes and examined DNA purification. This is consistent with the recent reviews on BDNF and depression. Chen et al. indicated that more than half of the studies showed an increased BDNF methylation in depressed patients. Bakusic et al. concluded in their review that hypermethylation was consistently found in MDD subjects across the three studies selected10. The BDNF gene provides the instructions for making a protein found in the brain and spinal cord, and promotes the survival of nerve cells (neurons). It is actively involved in the growth, maturation, and maintenance of these neurons, and in the regulation of synaptic plasticity, which is important for learning and memory26,51. It is reported that changes in the methylation level of the BDNF promoter are associated with its lower expression in the prefrontal cortex52 and its activity in the hippocampus in animal studies53. A similar decrease in BDNF levels was also found in the serum and plasma of MDD patients; thus, it is hypothesized that MDD is related to impaired neuronal plasticity53.
Positive associations between SLC6A4 methylation modifications and depression have also been identified in many studies in this review and previous reviews10,11. All longitudinal studies in this review and studies with more comprehensive considerations of lab and statistical work have consistently found that depression patients had SLC6A4 hypermethylation compared to controls. SLC6A4 gives the instructions for making a protein in the brain that is involved in the regulation of serotonergic signaling by transporting serotonin or 5-hydroxytryptamine (5-HT) from synaptic spaces into presynaptic neurons54 and in the regulation of emotional behaviors55. The alterations of SLC6A4 play an important role in brain development and function in humans56. It has been hypothesized that DNA hypermethylation may result in the reduction of SLC6A4 expression and 5-HT reuptake, which in turn may increase the vulnerability to affective disorders at critical stages of development57,58.
Findings on NR3C1, OXTR, and other genes were less coherent. Both hypo- and hypermethylation levels were noted in depressive patients compared to controls. No significant associations between DNA methylation on these genes and depression were also reported by some studies. Similar findings were also found by recent reviews10,11. NR3C1 is the receptor to which cortisol and glucocorticoids bind. It regulates gene transcriptions and is linked to development, metabolism, and immune response59,60. OXTR is a receptor of the hormone and neurotransmitter oxytocin61,62. It presents in the central nervous system and plays an important role in modulating various behaviors, such as stress and anxiety, social memory and recognition, sexual and aggressive behaviors, bonding/affiliation, and maternal behavior63–65. We found that some of the selected studies had certain limitations in terms of the type of study design, sample size, and range of laboratory work and statistical analyses. Due to the high heterogeneity across the selected studies, this review could not provide more conclusive results on these genes in terms of relationships between DNA methylation modifications on these genes and depression.
Findings on treatment studies
Findings of this subgroup were less consistent than those of the other two subgroups analyzed. However, this is in line with another recent review on DNA methylation, and clinical response to antidepressants in MDD patients was inadequate to provide any consistent support for such a relationship66. Both the increased and decreased DNA methylation levels on SLC6A4 and BDNF genes were associated with the use of antidepressant medications, whereas MAOA methylation modification was not linked to antidepressant response. The relationship between antidepressant treatment and DNA methylation of certain genes has been reported, i.e., BDNF DNA methylation modification was associated with decreased gene expression, which can lead to MDD67. The use of antidepressants can restore the decreased BDNF to the normal level and alleviate depressive symptoms53,67. Inconsistencies across all these findings may be explained by different ethnicities, duration of treatments, and pharmacogenetic heterogeneities68,69. Investigations on antidepressant response should cover all the different treatment stages, since the level of DNA methylation may be altered during the treatment70.
Strengths and limitations
This review synthesizes the findings on DNA methylation associated with depression and critically appraised the major study characteristics that can significantly impact this association, including study design, study population, targeted genetic variations, methylation arrays, types of tissues, DNA purification, methylation validation, appropriate statistical methods, and the consideration of downstream analyses, e.g., genotyping and gene expression.
However, there are several limitations to be noted. First, this review was designed to provide an overview of the relationship between DNA methylation and depression. Therefore, all eligible studies with a wide range of genomic coverage, i.e., targeted genes or whole genome, and different types of study designs were included. As many study characteristics were heterogeneous, no pooled results were made to simply estimate this relationship. Second, although we used subgroup analyses to synthesize homogeneous studies, different types of tissues, study designs, phenotypes of the outcome, comparison groups, analytic methods, and sample sizes can still lead to inconsistent results. Third, most of studies were cross-sectional. DNA methylation level is dynamic and potentially reversible, and can be affected by a number of environmental factors. Findings from these cross-sectional studies may not be able to reveal the true nature of this complex relationship. Finally, only English databases were searched, which may limit the comprehensiveness of eligible studies.
Overall, we found that hyper- and hypomethylations on promoter regions and CpG islands of a number of genes were significantly associated with the disease. Most of the studies applied the widely acceptable laboratory techniques and statistical analyses, which made the pooled results more likely to reach a consistent finding. Future studies should adopt longitudinal study designs to explore the dynamic change of methylation levels. To allow for a systematic comparison of studies, there should be an agreement upon the consistent set of standards involving a minimum set for the items for the execution and reporting of methylation studies similar to what is required for the reporting of clinical trials, systematic reviews and meta-analysis12,71. Gene expression should also be routinely added into the research to uncover how, when, and what underlying mechanisms link these identified methylation sites to depression. This would advance the field and provide a firm base for the evidence on the relationship between DNA methylation and depression.
Supplementary information
Appendix 1 Search strategy for this systematic review
Appendix 2 Data references for selected 67 articles in this systematic review
Appendix 3 A summary of findings on etiological - candidate genes studies
Acknowledgements
This work was supported by a grant from the Canadian Institute of Health Research (PJT-148845) to X.M. and a scholar award from the Fonds de recherche du Québec- Sante, Canada, to X.M.
Authors' contributions
X.L. and M.L. conducted the search and, together with X.M., reviewed the articles returned by the search for eligibility, reviewed all data extraction, and prepared the draft of this manuscript. X.M. and C.D. designed this review. T.Z. and R.J. assisted with the interpretation of the results. X.M. oversaw the project, provided feedback on all steps of the search, data extraction, and interpretation. All authors contributed to the writing and editing of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-019-0412-y).
References
- 1.Levinson DF. The genetics of depression: a review. Biol. Psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 2.Lohoff FW. Overview of the genetics of major depressive disorder. Curr. Psychiatry Rep. 2010;12:539–546. doi: 10.1007/s11920-010-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shyn SI, Hamilton SP. The genetics of major depression: moving beyond the monoamine hypothesis. Psychiatr. Clin. North Am. 2010;33:125–140. doi: 10.1016/j.psc.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saveanu RV, Nemeroff CB. Etiology of depression: genetic and environmental factors. Psychiatr. Clin. North Am. 2012;35:51–71. doi: 10.1016/j.psc.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Cohen-Woods S, Craig IW, McGuffin P. The current state of play on the molecular genetics of depression. Psychol. Med. 2013;43:673–687. doi: 10.1017/S0033291712001286. [DOI] [PubMed] [Google Scholar]
- 6.Dunn EC, et al. Genetic determinants of depression: recent findings and future directions. Harv. Rev. Psychiatry. 2015;23:1–18. doi: 10.1097/HRP.0000000000000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lockwood LE, Su S, Youssef NA. The role of epigenetics in depression and suicide: A platform for gene–environment interactions. Psychiatry Res. 2015;228:235–242. doi: 10.1016/j.psychres.2015.05.071. [DOI] [PubMed] [Google Scholar]
- 8.Uddin M, Sipahi L, Li J, Koenen KC. Sex differences in DNA methylation may contribute to risk of PTSD and depression: a review of existing evidence. Depress. Anxiety. 2013;30:1151–1160. doi: 10.1002/da.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton VS, Kolshus E, McLoughlin DM. Epigenetics and depression: return of the repressed. J. Affect. Disord. 2014;155:1–12. doi: 10.1016/j.jad.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Bakusic J, Schaufeli W, Claes S, Godderis L. Stress, burnout and depression: A systematic review on DNA methylation mechanisms. J. Psychosom. Res. 2017;92:34–44. doi: 10.1016/j.jpsychores.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Meng L, Pei F, Zheng Y, Leng J. A review of DNA methylation in depression. J. Clin. Neurosci. 2017;43:39–46. doi: 10.1016/j.jocn.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.APA. Diagnostic and Statistical Mannual of Mental Disorders. 5th ed. (American Psychiatric Association, Washington, 2013).
- 14.WHO. The ICD-10 Classification of Mental and Behavioral Disorders: Clinical Descriptions and Diagnostic Guidelines. (World Health Organization, Geneva, 1992).
- 15.Sabunciyan S, et al. Genome-wide DNA methylation scan in major depressive disorder. PLoS ONE. 2012;7:e34451. doi: 10.1371/journal.pone.0034451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrne EM, et al. Monozygotic twins affected with major depressive disorder have greater variance in methylation than their unaffected co-twin. Transl. Psychiatry. 2013;3:e269. doi: 10.1038/tp.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tseng PT, et al. Age-associated decrease in global DNA methylation in patients with major depression. Neuropsychiatr. Dis. Treat. 2014;10:2105–2114. doi: 10.2147/NDT.S71997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uddin M, et al. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol. Med. 2011;41:997–1007. doi: 10.1017/S0033291710001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker RM, et al. DNA methylation in a Scottish family multiply affected by bipolar disorder and major depressive disorder. Clin. Epigenetics. 2016;8:5. doi: 10.1186/s13148-016-0171-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies MN, et al. Hypermethylation in the ZBTB20 gene is associated with major depressive disorder. Genome Biol. 2014;15:R56. doi: 10.1186/gb-2014-15-4-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guintivano J, Arad M, Gould TD, Payne JL, Kaminsky ZA. Antenatal prediction of postpartum depression with blood DNA methylation biomarkers. Mol. Psychiatry. 2014;19:560–567. doi: 10.1038/mp.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osborne L, et al. Replication of epigenetic postpartum depression biomarkers and variation with hormone levels. Neuropsychopharmacology. 2016;41:1648–1658. doi: 10.1038/npp.2015.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaut O, et al. Aberrant NMDA receptor DNA methylation detected by epigenome-wide analysis of hippocampus and prefrontal cortex in major depression. Eur. Arch. Psychiatry Clin. Neurosci. 2015;265:331–341. doi: 10.1007/s00406-014-0572-y. [DOI] [PubMed] [Google Scholar]
- 24.Haghighi F, et al. Increased DNA methylation in the suicide brain. Dialogues. Clin. Neurosci. 2014;16:430–438. doi: 10.31887/DCNS.2014.16.3/jmann. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchelmore C, et al. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J. Biol. Chem. 2002;277:7598–7609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen JV, Thomassen M, Mollgard K, Noraberg J, Jensen NA. Zbtb20 defines a hippocampal neuronal identity through direct repression of genes that control projection neuron development in the isocortex. Cereb. Cortex. 2014;24:1216–1229. doi: 10.1093/cercor/bhs400. [DOI] [PubMed] [Google Scholar]
- 27.Sheline YI, Mittler BL, Mintun MA. The hippocampus and depression. Eur. Psychiatry. 2002;17:300–305. doi: 10.1016/S0924-9338(02)00655-7. [DOI] [PubMed] [Google Scholar]
- 28.Bremner JD, et al. Hippocampal volume reduction in major depression. Am. J. Psychiatry. 2000;157:115–118. doi: 10.1176/ajp.157.1.115. [DOI] [PubMed] [Google Scholar]
- 29.Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW. Hippocampal atrophy in recurrent major depression. Proc. Natl Acad. Sci. USA. 1996;93:3908–3913. doi: 10.1073/pnas.93.9.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garfinkel BP, et al. Proportionate dwarfism in mice lacking heterochromatin protein 1 binding protein 3 (HP1BP3) is associated with alterations in the endocrine IGF-1 pathway. Endocrinology. 2015;156:4558–4570. doi: 10.1210/en.2015-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garfinkel BP, et al. HP1BP3 expression determines maternal behavior and offspring survival. Genes Brain Behav. 2016;15:678–688. doi: 10.1111/gbb.12312. [DOI] [PubMed] [Google Scholar]
- 32.Neuner SM, et al. Systems genetics identifies Hp1bp3 as a novel modulator of cognitive aging. Neurobiol. Aging. 2016;46:58–67. doi: 10.1016/j.neurobiolaging.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao S, Iyer JK, Lin V. Identification of tetratricopeptide repeat domain 9, a hormonally regulated protein. Biochem. Biophys. Res. Commun. 2006;345:310–317. doi: 10.1016/j.bbrc.2006.04.091. [DOI] [PubMed] [Google Scholar]
- 34.Gerges NZ, et al. Independent functions of hsp90 in neurotransmitter release and in the continuous synaptic cycling of AMPA receptors. J. Neurosci. 2004;24:4758–4766. doi: 10.1523/JNEUROSCI.0594-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 36.Calabrese F, et al. Stress-induced changes of hippocampal NMDA receptors: modulation by duloxetine treatment. PLoS ONE. 2012;7:e37916. doi: 10.1371/journal.pone.0037916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordova-Palomera A, et al. Genome-wide methylation study on depression: differential methylation and variable methylation in monozygotic twins. Transl. Psychiatry. 2015;5:e557. doi: 10.1038/tp.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khulan B, et al. Epigenomic profiling of men exposed to early-life stress reveals DNA methylation differences in association with current mental state. Transl. Psychiatry. 2014;4:e448. doi: 10.1038/tp.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Numata S, et al. Blood diagnostic biomarkers for major depressive disorder using multiplex DNA methylation profiles: discovery and validation. Epigenetics. 2015;10:135–141. doi: 10.1080/15592294.2014.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagy C, et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol. Psychiatry. 2015;20:320–328. doi: 10.1038/mp.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bostrom AE, et al. A MIR4646 associated methylation locus is hypomethylated in adolescent depression. J. Affect. Disord. 2017;220:117–128. doi: 10.1016/j.jad.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Pajer K, et al. Discovery of blood transcriptomic markers for depression in animal models and pilot validation in subjects with early-onset major depression. Transl. Psychiatry. 2012;2:e101. doi: 10.1038/tp.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karanges EA, et al. Hippocampal protein expression is differentially affected by chronic paroxetine treatment in adolescent and adult rats: a possible mechanism of “paradoxical” antidepressant responses in young persons. Front. Pharmacol. 2013;4:86. doi: 10.3389/fphar.2013.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wray NR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol. Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee HC, et al. Gene expression profiling in hypothalamus of immobilization-stressed mouse using cDNA microarray. Brain. Res. Mol. Brain. Res. 2005;135:293–300. doi: 10.1016/j.molbrainres.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butler MG. Genomic imprinting disorders in humans: a mini-review. J. Assist. Reprod. Genet. 2009;26:477–486. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freudenberg JM, et al. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012;40:3364–3377. doi: 10.1093/nar/gkr1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh G, et al. DNA modification study of major depressive disorder: beyond locus-by-locus comparisons. Biol. Psychiatry. 2015;77:246–255. doi: 10.1016/j.biopsych.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malcangio M, Lessmann V. A common thread for pain and memory synapses? Brain-derived neurotrophic factor and trkB receptors. Trends Pharmacol. Sci. 2003;24:116–121. doi: 10.1016/S0165-6147(03)00025-7. [DOI] [PubMed] [Google Scholar]
- 52.Zheleznyakova GY, Cao H, Schioth HB. BDNF DNA methylation changes as a biomarker of psychiatric disorders: literature review and open access database analysis. Behav. Brain. Funct. 2016;12:17. doi: 10.1186/s12993-016-0101-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee BH, Kim YK. The roles of BDNF in the pathophysiology of major depression and in antidepressant treatment. Psychiatry Investig. 2010;7:231–235. doi: 10.4306/pi.2010.7.4.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tao-Cheng JH, Zhou FC. Differential polarization of serotonin transporters in axons versus soma-dendrites: an immunogold electron microscopy study. Neuroscience. 1999;94:821–830. doi: 10.1016/S0306-4522(99)00373-5. [DOI] [PubMed] [Google Scholar]
- 55.Meyer-Lindenberg A. Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum. Brain Mapp. 2009;30:1938–1946. doi: 10.1002/hbm.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Booij L, Wang D, Levesque ML, Tremblay RE, Szyf M. Looking beyond the DNA sequence: the relevance of DNA methylation processes for the stress-diathesis model of depression. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20120251. doi: 10.1098/rstb.2012.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaspar P, Cases O, Maroteaux L. The developmental role of serotonin: news from mouse molecular genetics. Nat. Rev. Neurosci. 2003;4:1002–1012. doi: 10.1038/nrn1256. [DOI] [PubMed] [Google Scholar]
- 58.Olsson CA, et al. Prospects for epigenetic research within cohort studies of psychological disorder: a pilot investigation of a peripheral cell marker of epigenetic risk for depression. Biol. Psychol. 2010;83:159–165. doi: 10.1016/j.biopsycho.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Lu NZ, et al. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol. Rev. 2006;58:782–797. doi: 10.1124/pr.58.4.9. [DOI] [PubMed] [Google Scholar]
- 60.Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids--new mechanisms for old drugs. N. Engl. J. Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 61.Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol. Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 62.Zingg HH, Laporte SA. The oxytocin receptor. Trends Endocrinol. Metab. 2003;14:222–227. doi: 10.1016/S1043-2760(03)00080-8. [DOI] [PubMed] [Google Scholar]
- 63.Caldwell H. & Young W. Handbook of Neurochemistry and Molecular Neurobiology 3rd ed (eds Lajtha, A. & Ramon, L.) Oxytocin and Vasopressin: Genetics and Behavioral Implications (Springer, Berlin, 2006).
- 64.Kiss A, Mikkelsen JD. Oxytocin--anatomy and functional assignments: a minireview. Endocr. Regul. 2005;39:97–105. [PubMed] [Google Scholar]
- 65.Veenema AH, Neumann ID. Central vasopressin and oxytocin release: regulation of complex social behaviours. Prog. Brain. Res. 2008;170:261–276. doi: 10.1016/S0079-6123(08)00422-6. [DOI] [PubMed] [Google Scholar]
- 66.Lisoway AJ, Zai CC, Tiwari AK, Kennedy JL. DNA methylation and clinical response to antidepressant medication in major depressive disorder: A review and recommendations. Neurosci. Lett. 2017;669:14–23. doi: 10.1016/j.neulet.2016.12.071. [DOI] [PubMed] [Google Scholar]
- 67.Duman RS. Pathophysiology of depression: the concept of synaptic plasticity. Eur. Psychiatry. 2002;17(Suppl 3):306–310. doi: 10.1016/S0924-9338(02)00654-5. [DOI] [PubMed] [Google Scholar]
- 68.Domschke K, et al. Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int. J. Neuropsychopharmacol. 2014;17:1167–1176. doi: 10.1017/S146114571400039X. [DOI] [PubMed] [Google Scholar]
- 69.Kang HJ, et al. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2013;44:23–28. doi: 10.1016/j.pnpbp.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Roberts S, et al. Serotonin transporter [corrected] methylation and response to cognitive behaviour therapy in children with anxiety disorders. Transl. Psychiatry. 2014;4:e444. doi: 10.1038/tp.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schulz KF, Altman DG, Moher D, Group C. CONSORT 2010 Statement: Updated guidelines for reporting parallel group randomised trials. J. Clin. Epidemiol. 2010;63:834–840. doi: 10.1016/j.jclinepi.2010.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1 Search strategy for this systematic review
Appendix 2 Data references for selected 67 articles in this systematic review
Appendix 3 A summary of findings on etiological - candidate genes studies