Abstract
Comparative genomic studies of the bacterial MFS-type copper importer CcoA, required for cbb3-type cytochrome c oxidase (cbb3-Cox) biogenesis, revealed a widespread CcoA-like transporters (CalT) family, containing the conserved CcoA Cu-binding MxxxM and HxxxM motifs. Surprisingly, this family also included the RfnT-like proteins, earlier suggested to transport riboflavin. However, presence of the Cu-binding motifs in these proteins raised the possibility that they might be Cu transporters. To test this hypothesis, the genomic context of the corresponding genes was examined, and three of such genes from Ochrobactrum anthropi, Rhodopseudomonas palustris and Agrobacterium tumefaciens were expressed in Escherichia coli (ΔribB) and Rhodobacter capsulatus (ΔccoA) mutants. Copper and riboflavin uptake abilities of these strains were compared with those expressing R. capsulatus CcoA and Rhizobium leguminosarum RibN as bona fide copper and riboflavin importers, respectively. Overall data demonstrated that the “RfnT-like” CalT proteins are unable to efficiently transport riboflavin, but they import copper like CcoA. Nevertheless, even though expressed and membrane-localized in a R. capsulatus mutant lacking CcoA, these transporters were unable to accumulate Cu or complement for cbb3-Cox defect. This lack of functional exchangeability between the different subfamilies of CalT homologs suggests that MFS-type bacterial copper importers might be species-specific.
Introduction
The major facilitator superfamily (MFS) transporters are ubiquitous to all branches of life, and play central roles in various biological processes. This superfamily is one of the largest group of secondary active transporters that selectively transport a remarkable variety of substrates across cell membranes (http://www.tcdb.org)1. Their substrates include mono- and oligo-saccharides, peptides, drugs, siderophores, metal ions as well as inorganic anions and cations2,3. Based on the nature of the substrates and modes of transport (uniporters, symporters and antiporters), the MFS superfamily members are divided into several families, according to the Transporter Classification Database (TCDB, http://www.tcdb.org). The primary amino acid sequences of these transporters are not very similar, yet they all share universally conserved (GxxxDxxxxxRxGRR and RxxxG) motifs4. Although the number of transmembrane (TM) helices varies among members of the superfamily5, most MFS transporters are composed of 12 TM helices6–8 resulting from two successive duplications of a three-TM-core. It has been hypothesized that these proteins function via an “alternating access” mechanism9,10, which postulates that the transporter switches between two conformations, allowing the substrate to access the binding site embedded in one side of the membrane. Substrate binding to the transporter in an outward-open conformation induces structural changes, switching it to an inward-open form, which allows release of the substrate to the opposite side of the membrane10,11.
The first bacterial copper (Cu) importer of MFS-type, CcoA (RCC02192) was discovered in the facultative photosynthetic model organism Rhodobacter capsulatus12,13 and since then has become the prototype of the newly defined Copper Uptake Porter Family (TCDB: 2.A1.81.11). Earlier genetic and biochemical studies unequivocally established that CcoA imports Cu destined to the catalytic site (CuB center) of the cbb3-type cytochrome c oxidases (cbb3-Cox), where O2 is reduced to H2O during aerobic respiration14–16. Subsequent studies identified putative Cu-binding sites of CcoA, involving the MxxxM and HxxxM motifs located at its TM7 and TM816. Our previous comparative genomics of CcoA homologs revealed the existence of an extensive family of CcoA-like MFS-type transporters, ubiquitous among bacteria and also present in a few microbial eukaryotes17. Moreover, the corresponding genes often co-occur with genes encoding aa3- and cbb3-Cox in alpha-proteobacterial genomes, but they do not appear to provide Cu to all cupro-enzymes. In Rhodobacter sphaeroides, CcoA is required only for cbb3- but not for aa3-Cox biogenesis even though both enzymes have quasi-identical CuB centers17. In the present study, we refer to this large family of proteins as the CcoA-like transporters or CalT.
Unexpectedly, the protein similarity network analysis of the CalT family identified a subgroup of organisms that contain protein sequences17 previously annotated as riboflavin transporters (RfnT)18. The amino acid sequences from this “RfnT-like” CalT subgroup are highly similar to R. capsulatus and R. sphaeroides CcoA and also contain the putative Cu binding motifs (MxxxM and HxxxM)16. In some species, like Mesorhizobium loti, Sinorhizobium meliloti and Agrobacterium tumafeciens, the rfnT gene is the last open reading frame (ORF) of a hypothetical riboflavin biosynthesis pathway (RBP) operon (nrdR-ribDEH-nusB-rfnT)18. Based on the positional clustering of rfnT with the RBP genes and its homology to membrane transporters, these proteins were suggested to be riboflavin-related transporters. Later, Ochrobactrum anthropi rfnT was introduced into an E. coli riboflavin auxotrophic mutant (ΔribB) conferring the ability to grow in the presence of low amounts (2.5 μM) of riboflavin, which led to the suggestion that O. anthropi RfnT was a riboflavin transporter19,20. Surprisingly, the presence of a riboflavin-responsive riboswitch upstream of nrdR-ribDEH-nusB-rfnT has not been found so far in analyzed genomes19–21. Also, two genes of this cluster, nrdR and nusB, which function as a transcriptional repressor of ribonucleotide reductases and a factor of the bacterial anti-terminator complex, respectively, are not related to riboflavin biosynthesis, further bringing into question the role of RfnT proteins in riboflavin transport.
In this work, we performed a phylogenomic analysis to determine whether RfnT defines a functionally different subgroup of the CalT family, with distinct substrate specificity. To this end, we examined the genomic context of CalT genes, and expressed three RfnT-like CalT homologs from O. anthropi (calT-O: OANT_22955, formerly called RfnT19), A. tumefaciens (calT-A: ATU_1173) and Rhodopseudomonas palustris (calT-R: TX73-RS24555) in E. coli ΔribB and R. capsulatus ΔccoA mutants, and investigated their ability to transport Cu or riboflavin. The overall data supported the conclusion that the RfnT-like CalT subfamily members imported Cu like CcoA, and not riboflavin unlike RibN. The findings showed that the conserved MxxxM and HxxxM motifs are strong predictors of Cu-transport activity, and suggested that members of the CalT family may exhibit species-specificity for Cu uptake, illustrating the ability of different organisms to use dedicated Cu uptake and delivery pathways to channel Cu to target proteins.
Results
Amino acid sequence similarity analyses of CcoA-like transporters (CalT)
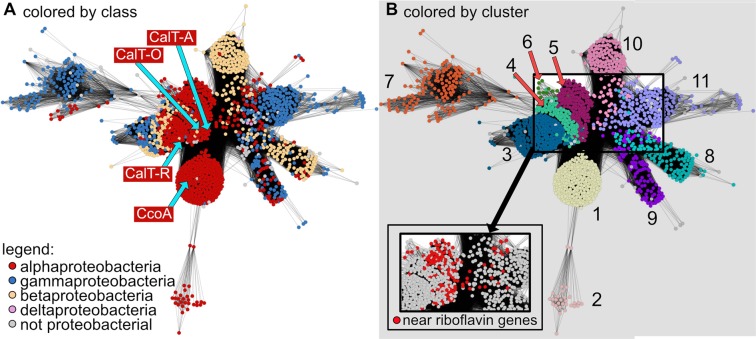
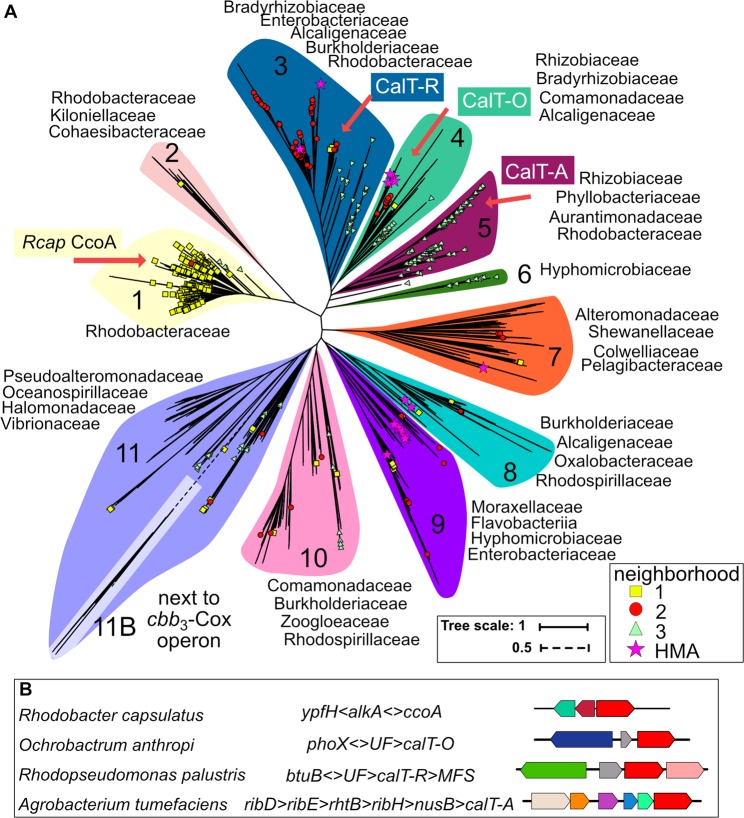
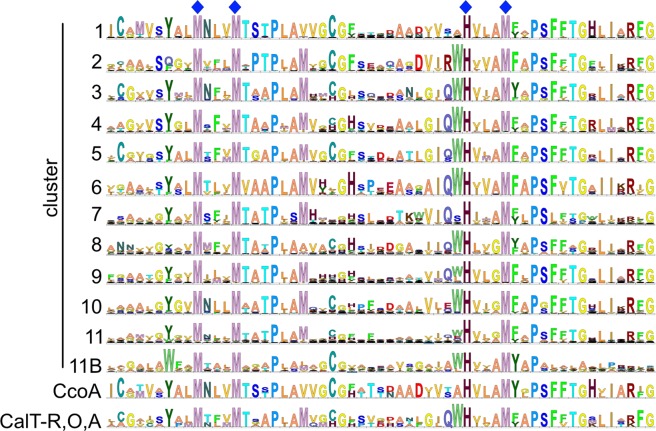
CcoA homologs, referred here as CalT, are found throughout the bacterial kingdom and also encoded in the genomes of some microbial eukaryotes17. The vast majority of proteins from each taxonomically distinct subfamily of CalT contain the motifs MxxxM in TM7 and HxxxM in TM8, which are required for Cu uptake and cbb3-Cox biogenesis in R. capsulatus16. These findings suggest that Cu import might be a ubiquitous function for this family of MFS transporters. As a first step in addressing this hypothesis, we performed a phylogenetic and genomic context analysis on the CalT subfamily members that are mainly from other Proteobacteria and exhibit highest similarity to CcoA from R. capsulatus and CalT-O (formerly RfnT) from O. anthropi, Based on the protein similarity network (Fig. 1) and the phylogenetic tree (Fig. 2A), 11 distinct clusters (numbered 1 to 11, Figs 1B and 2A) were identified, and the amino acid contexts of their conserved MxxxM and HxxxM motifs are shown in Fig. 3. The three largest subunits of cbb3-Cox (CcoN, CcoO and CcoP) were found encoded in most, but not all, of these proteobacterial genomes (SI Fig. 1), suggesting that not all CalT are involved in supplying Cu for cbb3-Cox biogenesis. The CcoA from R. capsulatus13 and R. sphaeroides17 were found in cluster 1, which is shared with orthologous proteins from the Rhodobacteraceae family, whereas CalT-O was found in cluster 4 (Fig. 2A). Due to sequence divergence, the Rhizobiales CalT proteins which are truncated at the C-terminus and whose corresponding genes are located next to the cbb3-Cox biogenesis (ccoNOQP-ccoGHIS) cluster17, were not connected to the network. However, when these sequences were included in the phylogenetic analyses, they were found most closely related to proteins within cluster 11 (Fig. 2A, cluster 11B), instead of cluster 1, which contains members experimentally shown to be required for cbb3-Cox biogenesis.
Figure 1.
CcoA-like transporter (CalT) Protein Similarity Network. A protein similarity network of MFS members that are homologous to CcoA is shown. Each node (circle) represents one or more protein sequences, and each edge (solid line) represents similarity between two proteins (threshold set at an alignment score of 75). (A) Nodes are colored by taxonomic classes as indicated. The locations of nodes representing proteins examined in this study are indicated with blue arrows. (B) Nodes are colored by clusters as found in, and numbered according to Fig. 2A. Nodes that represent proteins encoded by genes found near a riboflavin biosynthetic pathway (RBP) cluster are colored red in the pop-out.
Figure 2.
Phylogenetic distribution of CalT in Proteobacteria. (A) A phylogenetic tree of proteins from the sequence similarity network, plus 12 Rhizobiales sequences that are encoded next to the cbb3-Cox biogenesis gene cluster, is shown. Background shading corresponds to separate clusters (1 to 11), and leaves corresponding to the proteins experimentally examined in this study are indicated by a red arrow. Whether a CalT is encoded by a gene found in one of the three main genomic neighborhoods is indicated with either a yellow square (N1), a red circle (N2) or a green triangle (N3) according to the legend. A star (Heavy Metal Associated, HMA) indicates that the corresponding gene is found near a putative Cu homeostasis gene. A full list of all proteins analyzed and the related information can be found in SI Table S1. (B) Genomic neighborhoods of the RfnT-like CalT proteins examined in this study are compared to R. capsulatus CcoA. All gene abbreviations are listed in SI Table S4.
Figure 3.
Sequence logos for each CalT cluster. Sequence logo “11B” was constructed using only the MFS proteins that are encoded by the calT genes found next to the cbb3-Cox biogenesis genes cluster. The sequence logo “CcoA” was constructed using CcoA from R. capsulatus and R. sphaeroides, and the sequence logo “CalT-R, O, A” was built using CalT-O, CalT-R and CalT-A. Dark blue diamond symbols indicate the putative Cu-binding motifs MxxxM and HxxxM in TM7 and TM8 of CcoA.
Genomic context of CalT
Next, a neighborhood analysis was performed to identify proteins other than cbb3-Cox that might be functionally linked to CalT. Functionally coupled genes tend to cluster physically in bacterial genomes, and both frequency and conservation of gene clustering across evolutionarily distant genomes can be used to detect functional coupling22. We used a window of three genes upstream and downstream of each calT gene encoding a CalT protein from the similarity network to analyze the extent to which the neighboring genes are conserved at the genus, family, and order levels of taxonomy. At the genus level, we identified 605 protein family (Pfam) domains or domain fusions (referred to as neighbors) that were seen in at least two different genera. We ranked these domains by number of genera, excluded putative transcription factors and transporters, and further analyzed the top 17 neighbors (each found in 30 or more genera) (Methods). These neighbors could be arranged into three main neighborhoods (SI Table S1). The first neighborhood N1 (yellow squares in Fig. 2A), which contains CcoA from Rhodobacter species, was composed of one or more of nine genes including the putative DNA repair (alkA, PF00730) and esterase (ypfH, PF02230) proteins (Fig. 2B). The second neighborhood N2 (red circles in Fig. 2A) contained the genes encoding a FabG-like reductase (PF13561) and/or a putative Zn-dependent dehydrogenase (PF00107-PF08240). The third neighborhood N3 (green triangles in Fig. 2A) contained genes encoding a BamE-like outer membrane protein assembly factor (PF04355), a putative ubiquinol-cytochrome c oxidoreductase (cytochrome bc1 complex) chaperone (PF03981), a putative thiamine-monophosphate kinase (PF00586-PF02769) and/or a putative 6,7-dimethyl-8-ribityllumazine synthase (ribH involved in riboflavin biosynthesis, PF00885). These main neighborhoods (yellow squares, red circles and green triangles) are indicated in Fig. 2A, and all neighboring genes are listed in SI Table S1 (Tree and Neighborhood sheets). Of the neighborhoods, only N2 and a putative methyl transferase from N1 were enriched at the family and order levels. The RBP protein RibH, BamE-like outer membrane protein assembly factor, and ubiquinol-cytochrome c oxidoreductase chaperone from N3 were enriched at the family, but not at the order level (SI Table S1).
Positional clustering of RfnT-like CalT proteins with RBP genes
The neighborhood N3 captured the positional clustering that originally led to the identification of RfnT in M. loti, S. meliloti and A. tumefaciens, and prediction that these might be riboflavin transporters18. Many more bacterial genomes have been sequenced since that original analysis, and our data show that positional clustering between RBP genes and those encoding RfnT-like CalT proteins is conserved only in Rhizobiales, in a small subset of Rhodobacterales, and in Rhodospirillales (SI Figs S1A and S2A). Current data indicate that the proximity of these rfnT-like calT to ribH is mainly observed in clusters 5 and 6, and near the base of the clusters 3, 4, 10 and 11. The core unit, seen in cluster 5, is composed of the RBP gene ribH, followed by nusB encoding a subunit of the global transcriptional antitermination complex, and finally by calT. In addition, the thiL gene encoding thiamine-monophosphate kinase (vitamin B1 biosynthesis) and some other presumably functionally unrelated genes separating rfnT from ribH-nusB (clusters 3, 4, 10, and 11) were frequently seen (SI Fig. S2B). In most cases, RBP genes other than ribH are also conserved upstream of the core ribH-nusB-rfnT unit (SI Fig. S2B). Thus, the genomic proximity of the genes encoding the RfnT-like CalT to RibH-related proteins is not general, but is only seen in a subset of the clusters and in relatively closely related bacteria. Similarly, the previously identified group of Rhizobiales CalT is the only example where calT was located next to the cbb3-Cox biogenesis genes (ccoNOQP-ccoGHIS)17.
Cu-related proteins found in neighborhoods containing CalT
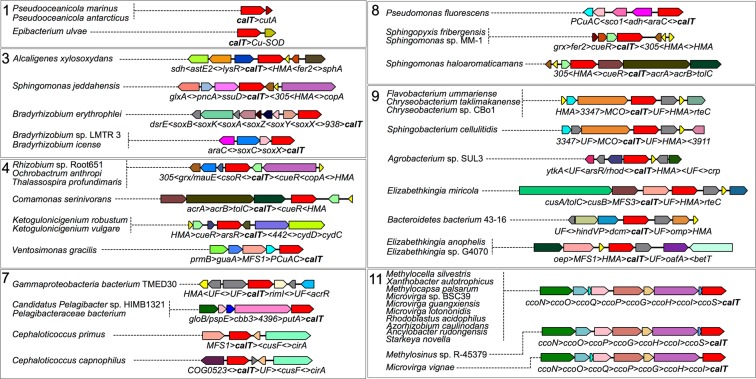
Given the experimentally defined role of CcoA as being a Cu transporter13,14, we searched within genomic neighborhoods for genes encoding either cuproproteins or other proteins involved in Cu homeostasis. Noticeably, the previously identified group of Rhizobiales CalT (cluster 11B) was an example where calT could be found located next to the cbb3-Cox biogenesis genes (ccoNOQP-ccoGHIS)17. In addition, calT homologs were observed next to a gene containing a cytochrome_CBB3 domain (PF13442), similar to subunit III of cbb3-Cox, in two unclassified Pelagibacteraceae bacteria and Pelagibacter sp. HIMB1321 (cluster 7), and Bradyrhizobium sp. LMTR 3, Bradyrhizobium icense and Bradyrhizobium erythrophlei (cluster 3 in Fig. 4). In the case of Bradyrhizobium spp., this gene putatively encodes SoxX and is found in a putative sulfur-oxidizing gene cluster. This finding is significant as the SoxAX from Starkeya novella was shown to contain a mononuclear Cu2+ center23. Putative Cu chaperones containing a Heavy Metal Associated (HMA, PF00403) domain were found in the top 15 neighbors at the family and order levels, with the corresponding CalT proteins being in clusters 3, 4, 7, 8 and 9 (45 of them are shown in Fig. 4). Interestingly, the O. anthropi genome contains two calT genes in cluster 4; one corresponds to the earlier described rfnT19, and a paralog is located near a putative Cu chaperone gene. In addition, several clusters contained copA, csoR, cueR or mco genes that encode proteins involved in Cu-detoxification (Fig. 4). Out of the 1635 calT genes analyzed, only a few were observed proximal to additional genes also encoding Cu-responsive or Cu-homeostasis related proteins, such as CusF (Cephaloticoccus primus and Cephaloticoccus capnophilus), PCuAC (Ventosimonas gracilis), SCO1 (Pseudomonas tolaasii and Pseudomonas fluorescens), CutA1 (divalent ion tolerance protein, PFAM3091) (Pseudooceanicola marinus and Pseudooceanicola antarcticus) and Cu/Zn superoxide dismutase (Epibacterium ulvae).
Figure 4.
Genomic neighborhoods containing CalT near putative Cu-related proteins. Close proximity between CalT-encoding genes and putative Cu-homeostasis genes was observed for various genomes in clusters 1, 3, 4, 7, 8, 9 and 11. A cartoon for each neighborhood is shown with corresponding genomes (on the left) in which the neighborhood was found. All gene abbreviations are listed in SI Table S4.
Heterologous expression of RfnT-like CalT proteins in E. coli
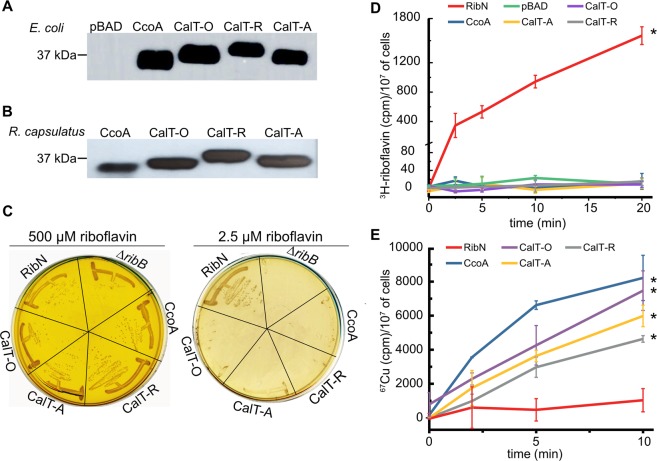
Although in our analysis 190 (out of 1635) CalT members from Proteobacteria are found near ribH, the phylogenomic analysis alone could not definitively distinguish putative Cu transporters from putative riboflavin transporters. Indeed, phylogenetic clusters that contained calT genes clustering with the RBP genes also contained homologs nearby the Cu homeostasis genes (cluster 3, 4 and 11). Thus, to further define the functions of CalT family members, we tested experimentally the ability of CcoA-like and RfnT-like members to transport riboflavin and Cu, respectively. We chose three RfnT-like CalT-encoding genes: O. anthropi calT (CalT-O, WP_010660541, previously called RfnT) from cluster 4, R. palustris calT (CalT-R, WP_011160333) from cluster 3, and A. tumefaciens calT (CalT-A, WP_010971445) from cluster 5. These calT homologs were cloned into a L-Ara inducible plasmid, as done earlier with R. capsulatus CcoA16 (Methods). Upon L-Ara induction, E. coli K12 strains (LMG194, ΔaraC) harboring plasmids carrying calT-O, calT-R and calT-A (SI Table S2) expressed Myc-tagged CalT-O, CalT-R and CalT-A (Mr ranging from 37 to 40 kDa) proteins, as detected by immunoblot analysis of whole cell extracts using anti-Myc antibodies (Fig. 5A). The same E. coli strain producing R. capsulatus CcoA (running as ~37 kDa)16 was used as a control. Similarly, whole cells and chromatophore membranes of a R. capsulatus strain lacking CcoA (ΔccoA) harboring appropriate plasmids with ccoA, calT-O, calT-R and calT-A (SI Table S2) also contained comparable amounts of the respective proteins (Fig. 5B), indicating that they were expressed and inserted in the cytoplasmic membrane in these species.
Figure 5.
L-Ara inducible production of CcoA and RfnT-like CalT and radioactive 67Cu uptake activities of E. coli strains expressing them. (A) Immunoblot analysis of CcoA from R. capsulatus and CalT from O. anthropi (CalT-O, formerly called RfnT), R. palustris (CalT-R) and A. tumefaciens (CalT-A) expressed in E. coli. (B) Immunoblot analysis of CcoA, CalT-O, CalT-R, CalT-A expressed in R. capsulatus. The E. coli (LMG194) and R. capsulatus ∆ccoA (SE8, not shown) strains harboring the empty expression vector pBAD/Myc-His A were used as a negative control and referred to as pBAD. Full-length blots are presented in SI Fig. S2. (C) The ΔribB mutants expressing CalT from O. anthropi (CalT-O), R. palustris (CalT-R) and A. tumefaciens (CalT-A), R. capsulatus CcoA or R. leguminosum RibN were grown at 37 °C on LB plates containing either high (500 µM) (left with yellow background due to high amount of riboflavin) or low (2.5 µM) (right) concentrations of riboflavin. In the presence of 2.5 µM riboflavin, the auxotrophic phenotype of the E. coli ΔribB mutant was restored only when cells expressed the bona fide riboflavin transporter RibN, but not any one of the CalT proteins. (D) Radioactive 3H-riboflavin uptake assays using appropriate E coli cells (strain LMG194 derivatives, SI Table S2) expressing either CcoA or RibN or the CalT-O, -R or -A. pBAD corresponds to the same cells carrying an empty expression vector (SI Table S2). In each case, the uptake assays were repeated at least three times using at least two independently grown cultures, and statistical analysis was performed using the Student’s t test, with p < 0.01 as the level of significance between RibN (*) and the other strains. (E) 67Cu uptake kinetics were performed using E. coli strain LMG194 expressing the RfnT-like CalT proteins from R. capsulatus CcoA, O. anthropi (CalT-O, formerly called RfnT), R. palustris (CalT-R) and A. tumefaciens (CalT-A), or the riboflavin transporter RibN. All uptake assays were performed at 37 °C and on ice as described in Methods, and in each case the activities detected with cells kept on ice were subtracted from those incubated at 37 °C. Of these corrected values the background activity measured with the E. coli strain carrying pBAD/Myc-His (pBAD) were subtracted and plotted in function of time. Each assay was repeated at least three times using multiple independently grown cultures, and statistical analysis was performed using the Student’s t test, with p < 0.01 as the level of significance between RibN and the other strains (*).
CalT-O, CalT-R and CalT-A do not complement E. coliΔribB mutant for riboflavin auxotrophy
E. coli has no known riboflavin transporter, but produces riboflavin via its endogenous RBP, which includes the ribB gene encoding the 3,4-dihydroxy-2-butanone-4-phosphate synthase20,24. Thus, an E. coli ΔribB mutant cannot grow on LB medium unless supplemented with a large amount (500 µM) of riboflavin, which is thought to diffuse passively across the membrane20. In contrast, heterologous expression of an efficient riboflavin uptake transporter, such as the Rhizobium leguminosarum RibN, enables growth of an E. coli ΔribB mutant on LB medium containing low amounts (2.5 µM) of riboflavin25.
In order to assess whether heterologous expression of CalT-R, -O and -A could confer riboflavin uptake activity in E. coli, plasmids encoding these orthologs were transformed into the E. coli ΔribB mutant (BW25141::ΔribB, SI Table S2) using LB plates containing 500 µM riboflavin. These transformants were then tested for growth on LB plates with low concentration of riboflavin (2.5 µM), in the absence and presence (0 to 2%) of L-Ara. A plasmid expressing the R. leguminosarum RibN bona fide riboflavin importer (SI Table S2) was used as a positive control25. Neither the E. coli ΔribB mutant, nor its derivatives carrying the calT- O, -R and -A genes were able to grow on 2.5 to 10 µM of riboflavin containing plates, irrespective of the presence of L-Ara, unlike those carrying ribN (Fig. 5C). As these CalT proteins were expressed in E. coli (Fig. 5A), their inability to rescue the growth on low concentration of riboflavin suggested that they could not confer efficient riboflavin uptake to sustain growth of E. coli, unlike RibN. Similar results were also obtained with a plasmid (pBK68) carrying R. capsulatus ccoA, indicating that CcoA also did not have such uptake activity (Fig. 5C).
During these experiments we observed that the E. coli ΔribB mutant (SI Table S2), and its derivatives expressing various CalT yielded spontaneous revertants that regained riboflavin-independent growth ability on LB medium in the absence, or presence of 2.5 µM of riboflavin (Fig. 5C, e.g., ΔribB expressing CcoA or CalT-O). These observations suggested that similar events might have occurred during the earlier work with O. arthropi gene19.
Neither CcoA nor RfnT-like CalT exhibit riboflavin uptake activity in E. coli
E. coli cells producing CcoA or RfnT-like CalT were tested for their ability to take up radioactive 3H-riboflavin. The data showed that 3H-riboflavin was taken up readily by the E. coli cells expressing RibN, but not by those expressing the three CalT homologs or CcoA (Fig. 5D). Moreover, in the case of CcoA, which is known to transport Cu, addition of Cu (100 μM) did not affect its inability to take up 3H-riboflavin. We concluded that neither CcoA nor the RfnT-like CalT members exhibited any efficient riboflavin uptake activity in E. coli, in agreement with their lack of complementation of the E. coli ΔribB strain for auxotrophy at low riboflavin amounts, suggesting that the earlier observed growth with O. anthropi rfnT19 (i.e., calT-O) might have been due to spontaneous reversion.
The RfnT-like CalT proteins mediate 67Cu uptake activity in E. coli cells
The conservation of the CcoA Cu-binding motifs (MxxxM and HxxxM) in the RfnT-like CalT proteins led us to investigate whether they could import Cu into E. coli cells, like R. capsulatus CcoA15,16. Time dependent 67Cu uptake activities of appropriate strains were measured using whole cells grown in the presence of 0.5% L-Ara. As a control, E. coli cells expressing wild-type R. capsulatus CcoA exhibited significantly higher amounts of 67Cu uptake than the same cells lacking CcoA (i.e., CcoA-independent 67Cu uptake background, Methods), as reported earlier16. Remarkably, E. coli cells expressing CalT-O or -R or -A also showed robust 67Cu uptake activities, whereas the same E. coli (or a ΔribB derivative) cells expressing RibN had no detectable 67Cu uptake activity (Fig. 5E). Therefore, we concluded that RfnT-like CalT proteins have Cu, but not riboflavin, uptake activity when expressed in E. coli, similar to CcoA. We note that the amounts of 67Cu accumulated in E. coli cells expressing different CalT proteins were slightly different. This point being out of the scope of this work, the amounts and affinities for Cu of those transporters were not studied further.
RfnT-like CalT proteins do not complement the R. capsulatusΔccoA mutant for cbb3-Cox defect
Considering that CcoA is a Cu importer required for cbb3-Cox biogenesis in R. capsulatus14,15 and R. sphaeroides17, and that the RfnT-like CalT proteins can also import Cu, their ability to complement the R. capsulatus ΔccoA mutant for its cbb3-Cox biogenesis defect was tested. Appropriate plasmids expressing CalT-O or -R or -A were conjugated into a R. capsulatus strain lacking CcoA. In parallel, a similar plasmid (pBK69) expressing wild-type R. capsulatus CcoA was used as a control (SI Table S2). The trans-conjugants were first tested for the presence of cbb3-Cox activity using the Nadi staining procedure (Cox activity dependent conversion of α-naphthol to indigo blue26). Colonies containing CcoA turned blue (i.e., Nadi+ phenotype) immediately (<30 sec), while those with the RfnT-like CalT proteins remained unstained upon longer (>10 min) exposure times (Fig. 6A). In addition, supplementation of the growth medium with 1 to 500 nM Cu, to increase Cu availability (in case of the lower uptake activities, or Cu affinities of CalT’s tested) was not efficient. Unfortunately, use of higher amounts of Cu supplementation was not informative because of the phenotypic suppression of a ΔccoA mutant for cbb3-Cox activity caused by μM amounts of external Cu13,15. However, immunoblot analyses of membrane preparations from the trans-conjugants using anti-myc antibodies showed that they all contained membrane-bound RfnT-like CalT proteins at levels comparable to those of CcoA (Fig. 5B). These findings suggested that, although produced and inserted into the membrane, the RfnT-like CalT proteins were unable to yield any active cbb3-Cox. Indeed, the trans-conjugants expressing CalT-O or -R or -A had very low levels of cbb3-Cox activity (~3–5%) compared with the R. capsulatus ΔccoA complemented with CcoA (100%) (Fig. 6B). Moreover, determination of the total cellular amounts of Cu associated with cells expressing CalT-A showed no accumulation of cellular Cu, unlike those containing CcoA (Fig. 6C) (see also SI Fig. S3 for the metal contents of these cells), suggesting that it was inactive in R. capsulatus membranes. Overall data showed that although the RfnT-like CalT proteins exhibited Cu uptake activity in E. coli cells, they were unable to complement a R. capsulatus strain lacking CcoA for cbb3-Cox biogenesis.
Figure 6.
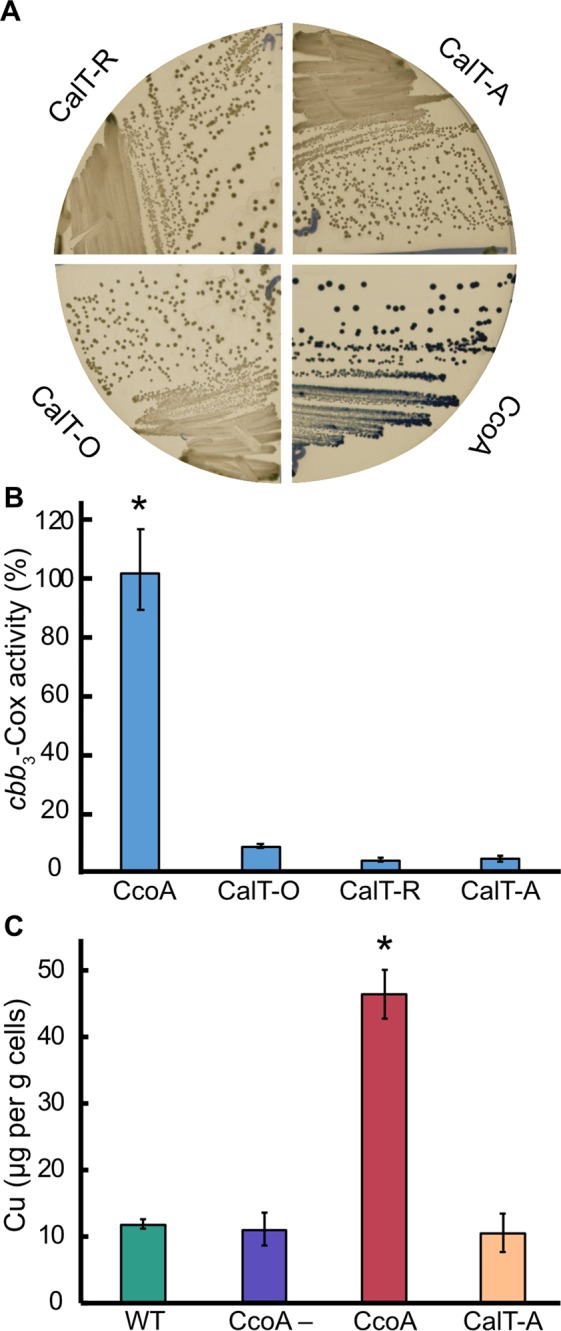
The RfnT-like CalT proteins do not restore the cbb3-Cox defect of R. capsulatus ΔccoA mutant. (A) R. capsulatus ΔccoA mutant (SE8), the ΔccoA mutant expressing either R. capsulatus CcoA (positive control) or the CalT from O. anthropi (CalT-O), R. palustris (CalT-R) or A. tumefaciens (CalT-A) were stained using the NADI procedure (Methods), which detects cbb3-Cox activity in R. capsulatus colonies via the development of a blue color. Note that cbb3-Cox activity was detected only in the presence of CcoA, and not the CalT-O, -R or -A proteins. (B) In vitro cbb3-Cox activities in chromotophore membranes of R. capsulatus SE8 strain expressing either CcoA or CalT-O, -R or -A. Cox assays were initiated by adding 30–150 μg of DDM-dispersed chromatophore membrane proteins to the assay buffer containing 0.1% DDM and 50 μM of reduced cyt c. Decrease in absorbance at 550 nm resulting from oxidation of cyt c was recorded. Addition of 0.1 mM KCN stopped immediately cyt c oxidation confirming the specificity of the cbb3-Cox activity. The cbb3-Cox activity determined for SE8/CcoA strain was 462 nmoles of cyt c oxidized/min/mg protein and set as 100%. Note that the R. capsulatus SE8 strains expressing CalT-O, -R or -A proteins have no cbb3-Cox activity. Mean values of at least three independent measurements with corresponding error bars are shown, and statistical analysis was performed using the Student’s t test, with p < 0.01 as the level of significance between CcoA (*) and the other strains. (C) Whole-cell Cu content measured by ICP-MS for wild-type R. capsulatus (WT), CcoA−, (ΔccoA) or a CcoA– strain expressing either CcoA or CalT-A. Each bar represents the mean values of four individually digested and independently measured samples from each strain and statistical analysis was performed using the Student’s t test, with p < 0.01 as the level of significance between CcoA (*) and the other strains.
Discussion
During our previous comparative genomic study of CcoA required for cbb3-Cox biogenesis13–16 in R. capsulatus and R. sphaeroides17 we noticed that a subgroup of the CcoA homologs (CcoA-like transporters or CalT) included the RfnT proteins previously predicted to transport riboflavin18–20. Moreover, the conserved (MxxxM and HxxxM) motifs of CcoA, which are associated with Cu import and cbb3-Cox biogenesis16,27, were also conserved in this subgroup. This similarity led us to further investigate this subfamily in order to probe whether the different members of the CalT family could transport different substrates such as Cu or riboflavin. We first divided the CalT family into 11 clusters based on the phylogenomic and genomic context analyses. While CcoA from R. capsulatus and R. sphaeroides belongs to a distinct cluster of proteins (cluster 1) shared with orthologs from other Rhodobacteraceae, we were unable to make a clear phylogenetic distinction between putative Cu transporters and putative riboflavin transporters. In the same protein cluster (e.g., clusters 3 and 4) we found calT genes that were located proximal to HMA-domain containing Cu chaperones involved in Cu response or detoxification, in support of a Cu-related function, whereas in other genomes their orthologs were next to RBP gene clusters. Thus, to establish the substrate specificity of different CalT subfamilies with respect to Cu and riboflavin we used an empirical approach. Three RfnT-like CalT proteins from three cbb3-Cox encoding proteobacterial species were introduced into appropriate E. coli and R. capsulatus mutants. Protein expression, phenotypic complementation and radiolabeled Cu and riboflavin uptake kinetics data showed that CalT-O, -R and -A from O. anthropi (cluster 4), R. palustris (cluster 3) and A. tumefaciens (cluster 5), respectively, were MFS-type Cu transporters just like R. capsulatus CcoA, and not efficient riboflavin transporters. Conceivably, currently unknown link(s) between Cu and riboflavin might exist, and these proteins may transport Cu and/or riboflavin at much higher concentrations or under specific conditions that are different than those used here. In any event, our findings validated the conservation of the MxxxM and HxxxM motifs in these CalT subfamily members, and suggested that this motif may be a good predictor of Cu importers among the MFS transporters. Currently, this point is further pursued using appropriate strains and species.
Most bacteria have an active RBP and are able to synthesize riboflavin de novo20,24, yet some species can also take up riboflavin from their environment via specific riboflavin uptake transporters20. Several such transporters have been described, and among them the energy coupling factor (ECF)-type RibU28,29, PnuX/RibM30–32, and RibN19,25 have been shown to transport riboflavin or its derivatives, whereas some others (e.g., ImpX and RibXY) are less studied. With the exception of the well characterized ECF-type RibU29, very little is known about the structural properties of bacterial riboflavin transporters and the specific motifs involved in substrate binding. Initially, rfnT was proposed to encode another riboflavin transporter based on its physical proximity to the RBP genes in Rhizobiales genomes18,19. However, neither the expression of CalT-O in the ΔribB mutant, nor an ability to take up riboflavin was examined19. During our analyses, we found that the E. coli ΔribB mutant (BW25141::ΔribB) used in previous studies reverted spontaneously to riboflavin protrophy. Similarly, the ΔribB derivatives expressing CalT-O, -R and -A yielded riboflavin prototrophic revertants, raising the issue of whether the RfnT-like CalT proteins were efficient riboflavin transporters. Indeed, 3H-riboflavin uptake experiments showed that cells harboring these proteins (and even CcoA) were unable to take up riboflavin, unlike a bone fide riboflavin transporter (e.g., R. leguminosarum RibN25). Instead, these transporters also exhibited Cu-transport activity in E. coli like R. capsulatus CcoA.
The unusual association of some calT subfamilies with RBP genes might suggest a possible, but currently unknown role for riboflavin in Cu homeostasis or Cu in riboflavin biosynthesis, or even cytochrome biogenesis in bacteria. Notably, some calT genes located in neighborhood N3 were found to be associated with a gene encoding a putative chaperone of ubiquinol-cytochrome c oxidoreductase. Moreover, a recent work using transcriptomics suggested that RibN-imported riboflavin might be involved in c-type cytochrome biogenesis in Vibrio cholerae21.
An unexpected finding was the inability of the RfnT-like CalT proteins to complement the cbb3-Cox defect of a R. capsulatus mutant lacking CcoA. Considering the successful heterologous production and membrane localization of CalT-O, -R and -A in R. capsulatus, and their Cu uptake activities seen in E. coli, the basis of this observation remains unclear. A possibility is that the RfnT-like CalT subfamily members might be inactive for unknown reason(s) for Cu uptake in R. capsulatus despite their competence in E. coli. The ICP-MS data suggested that R. capsulatus cells producing CalT-A do not accumulate Cu unlike those containing CcoA. A different possibility is that the Cu uptake and delivery pathways during cbb3-Cox biogenesis via the CalT family members might be species-specific. If so, these proteins (or the chemical nature of Cu cargo) might be incompatible to interact with their heterologous partner(s) to convey Cu to its ultimate destination, rendering them non-interchangeable for cbb3-Cox biogenesis. Similar diversity occurs with cytoplasmic Cu chaperones in lower eukaryotes33. Ongoing work aiming at inactivating a RfnT-like CalT member (i.e., CalT-A) in a genetically tractable species like A. tumefaciens, and defining its effect(s) on cbb3-Cox biogenesis and Cu transport might shed further light to some of these issues. Moreover, the role of CalT in the provision of Cu to other cuproproteins also remains a possibility as not all CalT-encoding genomes encode a cbb3-Cox.
Finally, the biogenesis of cbb3-Cox is a complex process that is not yet fully understood12,27. It involves an increasing number of Cu chaperones and transporters, including SenC (PrrC/Sco homolog) and PccA (PCuAC homolog)34–36 that work collaboratively37 with the dedicated P1B-type transporter CcoI (also known as CtpA/CopA2)38–40. The spatial and temporal order(s) with which these Cu chaperones handle Cu, and interact with each other, is only now emerging27. In the absence of a three-dimensional structure for a CalT member, it is difficult to speculate about the amino acid residues that might be responsible for the observed differences. Nonetheless, sequence alignments show salient differences located around the cytoplasmic and periplasmic loops between the TM6 - TM7 and TM11 - TM12 of CalT members, respectively (SI Fig. S4). The occurrence of amino acid residues that are conserved in the CcoA and not in the RfnT-like CalT subfamilies, and vice versa, might be important in defining their specificity.
In summary, this study further defined the extended family of CalT in Proteobacteria and demonstrated that the RfnT-like CalT subfamily members are not riboflavin transporters, but they are rather bona fide Cu importer members of the Cu Uptake Porter family of TCDB1. Moreover, the occurrence of the conserved MxxxM and HxxxM motifs among this family appears to be a reliable predictor of Cu import activity. Whether all members of the CalT family exclusively provide Cu to the CuB center of cbb3-Cox, or to other cuproproteins as well remains to be seen.
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in SI Table S2. Standard molecular biology techniques were used41. E. coli strains were grown in LB medium at 37 °C supplemented as needed with ampicillin (Amp), chloramphenicol (Cm), tetracycline (Tet) and kanamycin (Km) at final concentrations of 100, 30, 12.5 and 50 μg/mL, respectively42,43. E. coli ΔribB strains were grown in the presence of 500 μM riboflavin, because they are unable to grow at lower concentrations (e.g., 2.5 μM) unless they express a functional heterologous riboflavin transporter (e.g., RibN)25. Complementation of this auxotrophic growth phenotype of the ΔribB strain was used to assess the ability of a gene product to transport riboflavin upon heterologous expression. E. coli strains containing pBAD/Myc-His A plasmid derivatives were grown overnight in LB medium with 0.5% L-arabinose (L-Ara) to express L-Ara-inducible genes. The R. capsulatus SE8 (ΔccoA) strain derivatives were grown in enriched medium (MPYE) at 35 °C supplemented with 2.5 μg/mL Tet. The L-Ara-inducible pBAD-pRK415 plasmid derivatives were conjugated into R. capsulatus by tri-parental mating using the helper plasmid pRK201342,44 and cells were grown overnight in the presence of L-Ara (0.5% to 2% as needed)16.
Construction of the expression plasmids
R. palustris calT gene (calT-R) was amplified using primers RPA-F and RPA-R (SI Table S3), and the resulting 1248 bp PCR fragment was digested with HindIII and KpnI and cloned into pBAD/Myc-His A vector, yielding plasmid pYZ02 encoding a C-terminally Myc-His-fused CalT-R (SI Table S2). Similarly, a 1224 bp PCR fragment containing the calT gene from A. tumefaciens (calT-A) was amplified using primers Atu-F and Atu-R, and cloned into pBAD/Myc-His A as above, yielding plasmid pYZ03 with a Myc-His tagged CalT-A. The calT gene (previously called rfnT18) from O. anthropi (calT-O) was amplified using primers OanT-F and OanT-R (SI Table S3) resulting in a 1197 bp PCR fragment that was cloned into pBAD/Myc-His A digested with EcoRI and KpnI, yielding the plasmid pYZ09 with a Myc-His tagged CalT-O (SI Table S2). Plasmids pYZ02 and pYZ09 were digested with NsiI, and ligated into the broad-host-range vector pRK415 digested with PstI (compatible cohesive ends with NsiI), yielding pYZ07 and pYZ11, respectively (SI Table S2). As the wild-type calT-A contains an internal NsiI (ATGCAT) site, the adenine of the NsiI site was replaced by a cytosine (ATGCCT) using the Q5 Site-Directed Mutagenesis Kit (NEB, Beverly, MA). Plasmid pYZ03 and primers AtuN-F and AtuN-R were used, yielding plasmid pYZ08 that was digested with NsiI and ligated into pRK415, digested with PstI, yielding plasmid pYZ13 (SI Table S2).
Whole cell lysates and chromatophore membrane preparation, SDS-PAGE and immunoblots
For whole-cell lysates, E. coli (1.5 mL) and R. capsulatus (10 mL) cultures were grown overnight in the presence of 0.5% L-Ara16. Cells were collected by centrifugation, washed and resuspended in CelLytic™ B Cell Lysis Reagent (Sigma-Aldrich, Saint Louis, MO) (200 μL) supplemented with lysozyme (20 μg/mL), DNAse (10 μg/ mL) and phenylmethylsulfonyl fluoride (PMSF, 200 μM), and incubated at room temperature for 15 min with shaking. Lysates were supplemented with EDTA (4 μM) and cleared by centrifugation at 19000 × g for 15 min. Proteins (30 μg of cell lysates or 40–60 μg of chromatophore membranes) were separated using 12% Tris-Glycine SDS-PAGE and transferred to Immobilon-P PVDF membranes. For chromatophore membrane preparations, R. capsulatus strains were grown in 1 L cultures. Cells were collected and resuspended in 25 mM Tris pH 7.5, 150 mM NaCl buffer, 200 μM PMSF (Cox assay buffer) and intracytoplasmic membrane vesicles were prepared as done earlier45. Protein concentrations were determined using the bicinchoninic acid assay (Sigma Inc.; procedure TPRO-562). Immunoblot analysis to detect the presence of the c-Myc epitope using either E. coli or R. capsulatus cell extracts or chromatophore membrane proteins (R. capsulatus) was done as in16. The presence of the CcoA or CalT in cell lysates of E. coli, and in the membrane fraction of R. capsulatus was confirmed by immuno-detection using anti-Myc monoclonal antibody and horseradish peroxidase conjugated anti-mouse IgG. Signal was detected using the Supersignal West Pico chemiluminescence substrate.
In vivo and in vitro cbb3-Cox activity
TetR derivatives of R. capsulatus SE8 (ΔccoA) containing plasmids pYZ07, pYZ11, pYZ13 and pBK69 (SI Table S2) were purified on appropriate MPYE plates under respiratory growth conditions, and their cbb3-Cox activities visualized qualitatively with the Nadi staining procedure26. Staining of the colonies was done as previously described17. The cbb3-Cox activities were measured by monitoring oxidation of reduced horse heart cyt c (Sigma Inc.) using chromatophore membranes according to17,46.
Whole cells 67Cu and 3H-riboflavin uptake assays
The Cu uptake assays were performed according to15. Radioactive 67Cu (half-life of ~62 hours) was obtained from the DOE-Brookhaven National Laboratory (NY). E. coli strain LMG194 containing the pBAD/Myc-His derivatives encoding R. capsulatus ccoA (pBK68) or various calT (pYZ02, pYZ03 and pYZ09), or R. leguminasorum riboflavin transporter RibN (pGRibN)25 (SI Table S2) were grown in 10 mL of LB supplemented with 0.5% L-Ara until an OD600 of 0.5. Similarly, the E. coli strains BW25141::ΔribB (ΔribB derivative of BW25141)19 and BW25141::ΔribB/pGRibN were grown in the presence of appropriate amounts of riboflavin as control strains. Cells were collected, washed with 50 mM sodium citrate, pH 6.5, 5% glucose buffer (uptake assay buffer) and re-suspended in 1 mL of the uptake assay buffer. Optical density at 600 nm was determined. For each assay, a total of 7.5 × 108 cells per 500 µL of total assay mixture (1.0 A600 = 5 × 108 cells/mL) were used. Cells were incubated for 10 min either at 35 °C or on ice, before each assay. Cu uptake was initiated by addition of 106 cpm of 67Cu (determined immediately before use) to the cell suspension. At each time point (0, 1, 2, 5, and 10 min), aliquots of 50 µL of assay mixture were collected and combined with 50 µl of CuCl2 (1 mM) and 50 µL of EDTA (50 mM, pH 6.5) to stop the uptake activity, and stored on ice. The aliquots were then centrifuged, and cells washed twice with 100 µL of ice-cold EDTA (50 mM, pH 6.5) solution. Cells were re-suspended in 1 mL of scintillation liquid (Bio-Safe II, RPI, Mt. Prospect, IL) and counted using a scintillation counter with a wide-open window. Background uptake activities determined using cell mixtures kept on ice during the assays were subtracted from those obtained at 35 °C, and plotted in function of time.
For 3H-riboflavin (Moravek Inc., Brea, CA) uptake assays, E. coli strains were grown to an OD600 of 0.4–0.6, washed with LB medium and re-suspended in LB medium to a final OD600 of 12. A total of 7.5 × 108 cells were diluted with uptake assay buffer to a final volume of 500 µL. Assay mixtures were pre-incubated either at 37 °C or kept on ice for 10 min before initiating the assay by addition of 2.5 μM riboflavin containing 2 μCi of 3H-riboflavin. At each time point (0, 2, 5, 10, and 20 min), an aliquot of 50 µL was taken and mixed with 50 µL of ice cold stopping solution (100 μM non-radioactive riboflavin in LB medium) and stored on ice. Cells were then pelleted, washed with 500 µL of stopping solution and re-suspended in 1 mL of scintillation liquid, and counted using a scintillation counter (Tri-Carb 2900 TR, Perkin Elmer).
Determination of total cellular Cu contents using ICP-MS
Samples for determination of total cellular Cu contents were prepared as described earlier15. Briefly, R. capsulatus strains were grown by respiration in 1 L of enriched MPYE medium prepared with metal-free water (stirred at room temperature with Chelex100 at a concentration of 5 g/L for 1 hour) to an OD630 of 0.8–0.9. Cells were harvested by centrifugation and washed three times with metal-free 20 mM Tris-HCl pH 8.0 and once with ice cold metal free water. Cell pellets were lyophilized to complete dryness. A total of 50 mg of dry cell powder per sample was digested in 1 ml trace-metal grade nitric acid (Sigma) at 65 °C. To obtain a corresponding blank, the volume of the cell powder was replaced by milli-Q grade water (ultrapure) and treated the same as the samples. The digested samples were then diluted with milli-Q grade water to a final concentration of 1 mg/ml cell powder. Total metal content was measured by ICP-MS (Nexion 350D, Perkin Elmer equipped with an Element Scientific prepFAST M5 autosampler) using quadruplicate digested samples for each strain.
Comparative genomic and phylogenetic analyses
The protein similarity network was constructed using the EFI-EST tool (http://efi.igb.illinois.edu/efi-est/)47 with an alignment score of 75. CalT proteins that were not connected to the main network hub were deleted and not included in further analyses. A full list of identified CalT members was published previously17, and the sequences used in this study are available in SI Table S1. The network was visualized with the yFiles organic layout provided with the Cytoscape software (http://www.cytoscape.org)48. The nodes in the network were colored either by taxonomy as provided by the UniProt database49, by cluster as determined by the phylogenetic analysis, or by the presence of proteins containing CcoN (IPR004677), CcoO (IPR003468) and CcoP (IPR004678 or IPR032858) as determined with the Interpro database50. The phylogenetic analysis was performed using NCBI’s COBALT51 for sequence alignment and IQ Tree52 as implemented on the CIPRES web portal53 with 1000 bootstrap replicates54. In addition to the sequences found in the network, 12 Rhizobiales CalT sequences, which are encoded by genes found near the cbb3-Cox biogenesis cluster were added to the phylogenetic analysis. Before tree building, the multiple-sequence alignment was edited to remove positions with a quality score less than 82655 and those sequences that did not contain the MxxxM and HxxxM motifs. Sequence logos were built with Skylign56 using the same multiple-sequence alignment used for the phylogenetic analysis.
Gene neighborhoods (a window of three genes upstream and downstream of each gene encoding a CalT protein from the similarity network) were retrieved using the EFI-GNT tool (https://efi.igb.illinois.edu/efi-gnt/). At the genus level, we identified 605 protein family (Pfam) domains or domain fusions (referred to as neighbors) that were seen in at least two different genera. We ranked these domains by number of genera and set a threshold at 30 individual genera, which resulted in 19 neighboring PFam domains. Of these, transcription factors, PF07690 (MFS_1) and PF00005 (ABC_tran) were excluded from further analysis because they are particularly large multi-functional families. The remaining 17 neighboring PFam domains could be collapsed into three main neighborhoods (SI Table S1).
Statistics analysis
The data are presented as means ± S.D, and statistical analysis was performed using the Student’s t test, with p < 0.01 as the level of significance and indicated in the figure legends.
Supplementary information
Acknowledgements
This work was supported mainly by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of Department of Energy [DOE DE-FG02-91ER20052] to FD, and partly by the National Institute of Health [NIH GM 38237] to FD. We acknowledge the partial support provided by the Office of Biological and Environmental Research of the Department Of Energy to CBH. Support by the Deutsche Forschungs Gemeinschaft to HGK (IRTG 1478 and RTG 2202) is greatly appreciated. We thank Dr. Chunxia Dong and Wenchu Zhao for preparation of R. capsulatus membrane fragments.
Author Contributions
All authors have given approval to the final version of the manuscript. B.K.-H., C.B.-H., and F.D. designed and performed experiments, analyzed data and wrote the manuscript. Y.Z., S.S. and A.F.V. performed experiments, analyzed data and edited the manuscript. V.G.-A. and H.-G.K. did critical reading and revision of the manuscript. F.D. managed the project and supervised the study.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yang Zhang and Crysten E. Blaby-Haas contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37988-4.
References
- 1.Saier JMH, et al. The Transporter Classification Database (TCDB): Recent Advances. Nucleic Acids Research. 2016;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marger MD, Saier MH. A Major Superfamily of Transmembrane Facilitators that Catalyse Uniport, Symport and Antiport. Trends in Biochemical Sciences. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-W. [DOI] [PubMed] [Google Scholar]
- 3.Pao SS, Paulsen IT, Saier MH. Major Facilitator Superfamily. Microbiology and Molecular Biology Reviews. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang XC, Zhao Y, Heng J, Jiang D. Energy Coupling Mechanisms of MFS Transporters. Protein Science. 2015;24:1560–1579. doi: 10.1002/pro.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newstead S, et al. Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide–Proton Symporters, PepT1 and PepT2. EMBO J. 2011;30:417. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abramson J, et al. Structure and Mechanism of the Lactose Permease of Escherichia coli. Science. 2003;301:610. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Lemieux MJ, Song J, Auer M, Wang D-N. Structure and Mechanism of the Glycerol-3-Phosphate Transporter from Escherichia coli. Science. 2003;301:616. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 8.Deng D, et al. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510:121. doi: 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- 9.Law CJ, Maloney PC, Wang D-N. Ins and Outs of Major Facilitator Superfamily Antiporters. Annual review of microbiology. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan N. Structural Advances for the Major Facilitator Superfamily (MFS) Transporters. Trends in Biochemical Sciences. 2013;38:151–159. doi: 10.1016/j.tibs.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Quistgaard EM, Löw C, Guettou F, Nordlund P. Understanding Transport by the Major Facilitator Superfamily (MFS): Structures Pave the Way. Nature Reviews Molecular Cell Biology. 2016;17:123. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 12.Ekici S, Pawlik G, Lohmeyer E, Koch H-G, Daldal F. Biogenesis of cbb3-type cytochrome c oxidase in Rhodobacter capsulatus. Biochimica et Biophysica Acta. 2012;1817:898–910. doi: 10.1016/j.bbabio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekici S, Yang H, Koch H-G, Daldal F. Novel Transporter Required for Biogenesis of cbb3-Type Cytochrome c Oxidase in Rhodobacter capsulatus. mBio. 2012;3:e00293–00211. doi: 10.1128/mBio.00293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaudoin J, et al. Copper transport and regulation in Schizosaccharomyces pombe. Biochemical Society Transactions. 2013;41:1679–1686. doi: 10.1042/BST2013089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekici S, et al. Intracytoplasmic Copper Homeostasis Controls Cytochrome c Oxidase Production. mBio. 2014;5:e01055–01013. doi: 10.1128/mBio.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalfaoui-Hassani B, Verissimo AF, Koch H-G, Daldal F. Uncovering the Transmembrane Metal Binding Site of the Novel Bacterial Major Facilitator Superfamily-Type Copper Importer CcoA. mBio. 2016;7:e01981–01915. doi: 10.1128/mBio.01981-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalfaoui-Hassani B, et al. Widespread Distribution and Functional Specificity of the Copper Importer CcoA: Distinct Cu Uptake Routes for Bacterial Cytochrome c Oxidases. mBio. 2018;9:e00065–00018. doi: 10.1128/mBio.00065-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of Riboflavin Biosynthesis and Transport Genes in Bacteria by Transcriptional and Translational Attenuation. Nucleic Acids Research. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutiérrez-Preciado A, et al. Extensive Identification of Bacterial Riboflavin Transporters and Their Distribution across Bacterial Species. PLoS ONE. 2015;10:e0126124. doi: 10.1371/journal.pone.0126124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Angulo VA. Overlapping Riboflavin Supply Pathways in Bacteria. Critical Reviews in Microbiology. 2017;43:196–209. doi: 10.1080/1040841x.2016.1192578. [DOI] [PubMed] [Google Scholar]
- 21.Sepulveda-Cisternas I, Lozano Aguirre L, Fuentes Flores A, Vasquez Solis de Ovando I, Garcia-Angulo VA. Transcriptomics reveals a cross-modulatory effect between riboflavin and iron and outlines responses to riboflavin biosynthesis and uptake in Vibrio cholerae. Sci Rep. 2018;8:3149. doi: 10.1038/s41598-018-21302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. The Use of Gene Clusters to Infer functional Coupling. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kappler U, et al. SoxAX cytochromes, a new type of heme copper protein involved in bacterial energy generation from sulfur compounds. J Biol Chem. 2008;283:22206–22214. doi: 10.1074/jbc.M800315200. [DOI] [PubMed] [Google Scholar]
- 24.Fischer M, Bacher A. Biosynthesis of Flavocoenzymes. Nat. Prod. Rep. 2005;22:324–350. doi: 10.1039/B210142B. [DOI] [PubMed] [Google Scholar]
- 25.García Angulo VA, et al. Identification and Characterization of RibN, a Novel Family of Riboflavin Transporters from Rhizobium leguminosarum and Other Proteobacteria. Journal of Bacteriology. 2013;195:4611–4619. doi: 10.1128/JB.00644-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch H-G, Hwang O, Daldal F. Isolation and Characterization of Rhodobacter capsulatus Mutants Affected in Cytochrome cbb3 Oxidase Activity. Journal of Bacteriology. 1998;180:969–978. doi: 10.1128/jb.180.4.969-978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalfaoui-Hassani, B. et al. In Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling (eds William A. Cramer & Toivo Kallas) 527–554 (Springer Netherlands, 2016).
- 28.Karpowich NK, Song JM, Cocco N, Wang D-N. ATP Binding Drives Substrate Capture in an ECF Transporter by a Release and Catch Mechanism. Nature Structural & Molecular Biology. 2015;22:565–571. doi: 10.1038/nsmb.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng Z, Jiawei W, Yigong SHI. Structure and Mechanism of the S Component of a Bacterial ECF Transporter. Nature (Lond.) 2010;468:717–720. doi: 10.1038/nature09488. [DOI] [PubMed] [Google Scholar]
- 30.Grill S, et al. Identification and Characterization of two Streptomyces davawensis Riboflavin Biosynthesis gene Clusters. Archives of Microbiology. 2007;188:377–387. doi: 10.1007/s00203-007-0258-1. [DOI] [PubMed] [Google Scholar]
- 31.Hemberger S, et al. RibM from Streptomyces davawensis is a Riboflavin/Roseoflavin Transporter and may be Useful for the Optimization of Riboflavin Production Strains. BMC Biotechnology. 2011;11:119–119. doi: 10.1186/1472-6750-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogl C, et al. Characterization of Riboflavin (Vitamin B(2)) Transport Proteins from Bacillus subtilis and Corynebacterium glutamicum. Journal of Bacteriology. 2007;189:7367–7375. doi: 10.1128/JB.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swem DL, Swem LR, Setterdahl A, Bauer CE. Involvement of SenC in Assembly of Cytochrome c Oxidase in Rhodobacter capsulatus. Journal of Bacteriology. 2005;187:8081–8087. doi: 10.1128/JB.187.23.8081-8087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson AK, Gray J, Liu A, Hosler JP. The roles of Rhodobacter sphaeroides copper chaperones PCu(A)C and Sco (PrrC) in the assembly of the copper centers of the aa3-type and the cbb3-type cytochrome c oxidases. Biochimica et Biophysica Acta. 2012;1817:955–964. doi: 10.1016/j.bbabio.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trasnea P-I, et al. Cooperation Between Two Periplasmic Copper Chaperones is Required for Full Activity of the cbb3-type Cytochrome c Oxidase and Copper Homeostasis in Rhodobacter capsulatus. Molecular Microbiology. 2016;100:345–361. doi: 10.1111/mmi.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trasnea, P.-I. et al. A Copper Relay System Involving Two Periplasmic Chaperones Drives cbb3-type Cytochrome c Oxidase Biogenesis in Rhodobacter capsulatus. ACS Chem. Biol., 10.1021/acschembio.8b00293 (2018). [DOI] [PMC free article] [PubMed]
- 38.Koch H-G, Winterstein C, Saribas AS, Alben JO, Daldal F. Roles of the ccoGHIS Gene Products in the Biogenesis of the cbb3-type Cytochrome c Oxidase. Journal of Molecular Biology. 2000;297:49–65. doi: 10.1006/jmbi.2000.3555. [DOI] [PubMed] [Google Scholar]
- 39.Hassani BK, Astier C, Nitschke W, Ouchane S. CtpA, a Copper-translocating P-type ATPase Involved in the Biogenesis of Multiple Copper-requiring Enzymes. The Journal of Biological Chemistry. 2010;285:19330–19337. doi: 10.1074/jbc.M110.116020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González‐Guerrero M, Raimunda D, Cheng X, Argüello José M. Distinct Functional Roles of Homologous Cu+ Efflux ATPases in Pseudomonas aeruginosa. Molecular Microbiology. 2010;78:1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory, 2001).
- 42.Atta-Asafo-Adjei E, Daldal F. Size of the amino acid side chain at position 158 of cytochrome b is critical for an active cytochrome bc1 complex and for photosynthetic growth of Rhodobacter capsulatus. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:492–496. doi: 10.1073/pnas.88.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray KA, Davidson E, Daldal F. Mutagenesis of methionine-183 drastically affects the physicochemical properties of cytochrome c1 of the bc1 complex of Rhodobacter capsulatus. Biochemistry. 1992;31:11864–11873. doi: 10.1021/bi00162a027. [DOI] [PubMed] [Google Scholar]
- 44.Darrouzet E, Daldal F. Movement of the iron-sulfur subunit beyond the ef loop of cytochrome b is required for multiple turnovers of the bc1 complex but not for single turnover Qo site catalysis. J Biol Chem. 2002;277:3471–3476. doi: 10.1074/jbc.M107974200. [DOI] [PubMed] [Google Scholar]
- 45.Cooley JW, Ohnishi T, Daldal F. Binding dynamics at the quinone reduction (Qi) site influence the equilibrium interactions of the iron sulfur protein and hydroquinone oxidation (Qo) site of the cytochrome bc1 complex. Biochemistry. 2005;44:10520–10532. doi: 10.1021/bi050571 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onder O, et al. Absence of Thiol-Disulfide Oxidoreductase DsbA Impairs cbb3-Type Cytochrome c Oxidase Biogenesis in Rhodobacter capsulatus. Front Microbiol. 2017;8:2576. doi: 10.3389/fmicb.2017.02576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerlt JA, et al. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GEL), 1–8 (2010).
- 54.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheeler TJ, Clements J, Finn RD. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics. 2014;15:7. doi: 10.1186/1471-2105-15-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.