Abstract
Aberrant synaptic function is thought to underlie social deficits in neurodevelopmental disorders such as autism and schizophrenia. Although microRNAs have been shown to regulate synapse development and plasticity, their potential involvement in the control of social behaviour in mammals remains unexplored. Here, we show that deletion of the placental mammal‐specific miR379‐410 cluster in mice leads to hypersocial behaviour, which is accompanied by increased excitatory synaptic transmission, and exaggerated expression of ionotropic glutamate receptor complexes in the hippocampus. Bioinformatic analyses further allowed us to identify five “hub” microRNAs whose deletion accounts largely for the upregulation of excitatory synaptic genes observed, including Cnih2, Dlgap3, Prr7 and Src. Thus, the miR379‐410 cluster acts a natural brake for sociability, and interfering with specific members of this cluster could represent a therapeutic strategy for the treatment of social deficits in neurodevelopmental disorders.
Keywords: autism, glutamate receptor, hippocampus, microRNA, sociability
Subject Categories: Neuroscience, RNA Biology
Introduction
Social interactions with conspecifics are important for physical and mental health, and abnormal social functioning is one of the core symptoms of several neurodevelopmental disorders. Whereas autism spectrum disorder (ASD) and schizophrenia are characterized by social withdrawal, patients suffering from Williams–Beuren syndrome (WBS) or Angelman syndrome (AS) typically display enhanced sociability, pointing to the existence of endogenous pathways that restrict sociability in the healthy brain. Several brain areas have been implicated in the regulation of diverse social behaviours, including the prefrontal cortex, the midbrain, the amygdala and the hippocampus 1. The latter, especially the ventral part, appears to play a particularly important role in the control of social interactions 2, 3. The molecular mechanisms underlying the development of neural circuitries governing social behaviour in the hippocampus however remain largely mysterious. Recent work demonstrated an important role of microRNAs (miRNAs) in several aspects of neuronal development, including neuronal differentiation, polarization, axonogenesis, dendritogenesis and dendritic spine development 4. Specific miRNAs are enriched in the synapto‐dendritic compartment of postmitotic cortical and hippocampal neurons, where they regulate the activity‐dependent local translation of mRNAs encoding for key synaptic proteins 5. Among those, members of a large placental mammal‐specific miRNA cluster (miR379‐410), encompassing 38 miRNAs, appear to play a particularly important role 6. The non‐coding Mirg RNA, which functions as a host gene for multiple miR‐379‐410 members, is expressed in multiple brain areas of adult mice, including the cortex and hippocampus (Allen Brain Atlas; http://mouse.brain-map.org/experiment/show?id=70946557). Arguably the most intensively studied miRNA within miR379‐410 is miR‐134. miR‐134 is broadly expressed during embryonic development, but largely restricted to the brain postnatally 7, 8. Within the brain, miR‐134 is strongly expressed in pyramidal neurons of the hippocampus 7, 9, but not in cell types of glial origin 8. miR‐134 controls cortical neuron migration 10, activity‐dependent dendritogenesis 11, dendritic spine development 7, long‐term potentiation 12 and homeostatic synaptic downscaling 13. In addition to miR‐134, other members of miR379‐410 have been shown to regulate neuroplasticity. For example, miR‐381 and miR‐329 are required for activity‐dependent dendritogenesis 11. miR‐369, miR‐496 and miR‐543 regulate neurogenesis and neuronal migration by fine‐tuning the levels of N‐cadherin 14. miR‐485 is required for homeostatic synaptic plasticity by inhibiting the synaptic vesicle protein SV2A 15. Furthermore, links between miR379‐410 and neuropsychiatric disorders have been reported. Genome‐wide copy number variation analysis in Han Chinese identified duplication of a genomic segment spanning miR379‐410 in an autistic patient 16. Several miR‐379‐410 members are dysregulated in peripheral blood of schizophrenic patients 17 as well as post‐mortem brain samples from both schizophrenia 18 and ASD subjects 19. Loss of the ASD risk gene MeCP2 in mice leads to upregulation of miR379‐410 members, including miR‐134 20, 21. On the other hand, a transcript variant of Ube3a, which is mutated in Angelman syndrome and upregulated in ASD, acts as a competing endogenous RNA for miR379‐410 in dendrites of rat hippocampal neurons 22. A behavioural characterization of a different miR379‐410 knockout (ko) mouse model recently revealed enhanced anxiety‐like behaviour in the absence of miR379‐410 23, further suggesting a role for miR379‐410 in emotional processing.
Based on these observations, we hypothesized that miR379‐410‐dependent regulation of gene expression in the hippocampus could be involved in the control of social behaviour. To test this, we employed a combination of behavioural phenotyping, neuromorphological analysis, electrophysiology and transcriptomic profiling in miR379‐410‐deficient mice. Therefore, we uncovered a negative regulatory role of miR379‐410 in social behaviour, which was accompanied by increased excitatory drive in the hippocampus.
Results and Discussion
miR379‐410 deficiency in mice leads to hypersocial behaviour
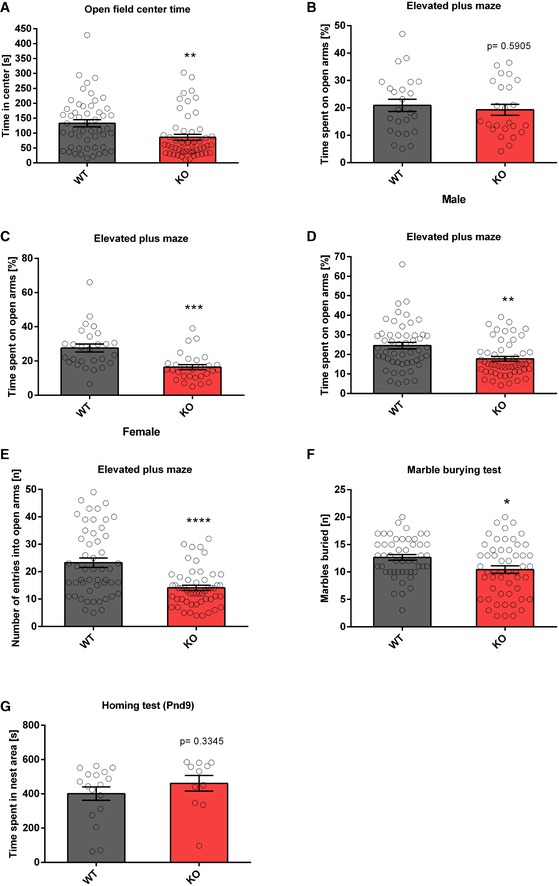
To study the role of the miR379‐410 cluster in social behaviour, we performed behavioural testing in constitutive miR379‐410 knockout (ko) mice 22, (Appendix Fig S1; refer also to Appendix Supplementary Methods for detailed description). To obtain mice deficient for the miR379‐410 cluster, we bred wild‐type (wt) male mice with miR379‐410 heterozygous mutant female mice. Due to paternal imprinting of the miR379‐410 locus 24, the resulting miR379‐410 heterozygous offspring does not detectably express miR379‐410 miRNAs, as previously shown 22. We therefore refer to these mice as miR379‐410 ko mice. Wt mice obtained from these breedings were used as littermate controls in all subsequent experiments. miR379‐410 ko mice display normal macroscopic brain development 22 and are indistinguishable from wild‐type (wt) littermates in the majority of developmental milestones tested (Appendix Fig S2). However, several lines of evidence indicate that lack of miR379‐410 expression increases sociability. First, upon isolation from their mother, miR‐379‐410 ko pups (P3‐12) displayed increased ultrasonic vocalizations (USVs), important communicative signals known to stimulate maternal search and retrieval behaviour (Fig 1A). Second, juvenile (P23) miR379‐410 knockout pairs spent significantly more time interacting with each other compared to pairs of wild‐type mice (Fig 1B). Exaggerated reciprocal social interaction behaviour was accompanied by a strong increase in the emission of pro‐social calls based on USV serving an affiliative function (Fig 1C). Third, adolescent miR379‐410 ko mice displayed increased social approach behaviour compared to controls (Fig 1D). Together, this demonstrates that lack of miR379‐410 increases sociability across the lifespan. In contrast, social recognition and object recognition were not affected in miR379‐410 ko mice (Appendix Fig S3), indicating intact memory function. In addition, miR379‐410 ko mice show reduced repetitive behaviour in the marble burying test (Fig EV1F), and, in agreement with a previous study 23, increased anxiety‐related behaviour in the open field and elevated plus maze (EPM) tests (Fig EV1A–E). No differences in the homing test were observed (Fig EV1G), suggesting intact olfaction in miR379‐410 ko mice. Taken together, our results indicate that the miR379‐410 cluster acts as a natural brake for the development of sociability in the rodent brain.
Figure 1. Increased sociability in miR379‐410 ko mice.
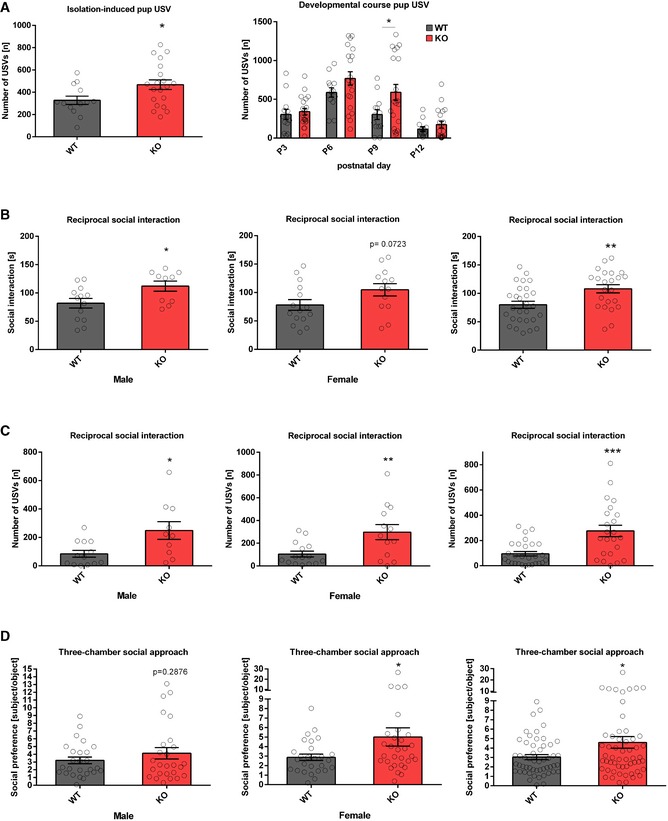
- Ultrasonic vocalizations (USV) in isolated miR379‐410 ko mouse pups emitted on postnatal day P3, P6, P9 and P12. Left panel: total number of ultrasonic vocalizations (USV) in isolated miR379‐410 wt and ko mouse pups emitted on average (P 3, 6, 9 and 12) when isolated from the mother, wt n = 13 (male n = 9, female n = 4), ko n = 20 (male n = 11, female n = 9), t 31 = 2.274, *P = 0.0301, unpaired Student's t‐test. Right panel: developmental course for total number of USV, wt n = 13 (male n = 9, female n = 4), ko n = 20 (male n = 11, female n = 9; development: F 3,87 = 25.463, P < 0.001; genotype: F 1,29 = 4.979, P = 0.03; development × genotype: F 3,87 = 1.403, P = 0.22, rm‐ANOVA); P 9: t 31 = 2.096, *P = 0.044, unpaired Student's t‐test.
- Reciprocal social interaction in juvenile miR379‐410 wt and ko mice pairs (P23). Aberrant time spending in social interaction activity in miR379‐410 ko pairs compared to their control wt littermate pairs. Left panel: wt pairs n = 13 (male), ko pairs n = 10 (male), t 21 = 2.433, *P = 0.0240, unpaired Student's t‐test. Middle panel: wt pairs n = 15 (female), ko pairs n = 13 (female), t 26 = 1.873, ns P = 0.0723, unpaired Student's t‐test. Right panel: pooled data of wt pairs n = 28 (male n = 13 and female n = 15), ko pairs n = 23 (male n = 10 and female n = 13), t 49 = 2.969, **P = 0.005, unpaired Student's t‐test.
- Emitted USVs during social interaction activity in genotype‐matched pairs in juvenile mice (P23). Left panel: wt pairs n = 13 (male), ko pairs n = 15 (male), t 21 = 2.704, *P = 0.0133, unpaired Student's t‐test. Middle panel: wt pairs n = 15 (female), ko pairs n = 13 (female), t 26 = 2.869, **P = 0.0081, unpaired Student's t‐test. Right panel: pooled data of wt pairs n = 28 (male n = 13 and female n = 15), ko pairs n = 23 (male n = 10 and female n = 13), t 49 = 3.992, ***P < 0.001, unpaired Student's t‐test.
- Social preference index in adolescent mice—defined as the ratio of time sniffing a stranger mouse vs. an object—is shown. Left panel: wt n = 26 (male), ko n = 26 (male), t 50 = 1.075, ns P = 0.2876, unpaired Student's t‐test. Middle panel: wt n = 27 (female), ko n = 30 (female), t 55 = 2.017, *P = 0.0486, unpaired Student's t‐test. Right panel: pooled data of wt n = 53 (male n = 26 and female n = 27), ko n = 56 (male n = 26 and female n = 30), t 107 = 2.283, *P = 0.0244, unpaired Student's t‐test.
Figure EV1. miR379‐410 ko mice demonstrated increased anxiety‐related behaviour.
-
AOpen field test in adult mice. Time spent in the central arena was examined, wt n = 51 (male n = 24 and female n = 27), ko n = 53 (male n = 24 and female n = 29), t 102 = 3.020, **P = 0.0032, unpaired Student's t‐test.
-
B–EElevated plus maze test in adult mice. (B) Time spent on open arms (percentage) are shown, wt n = 24 (male), ko n = 24 (male), t 46 = 0.5419, ns P = 0.5905. unpaired Student's t‐test. (C) Time spent on open arms (percentage) is shown, wt n = 27 (female), ko n = 29 (female), t 54 = 4.099, ***P < 0.001, unpaired Student's t‐test. (D) Time spent on open arms (percentage) is shown for pooled data of male and female; wt n = 51 (male n = 24 and female n = 27), ko n = 53 (male n = 24 and female n = 29), t 102 = 3.265, **P = 0.0015, unpaired Student's t‐test. Please notice that the genotype effect is mainly in females (P < 0.001) but not in males (P = 0.5905); Genotype × Sex: F 1,100 = 5.551, P = 0.020, ANOVA. (E) Number of open arm entries is shown; wt n = 51 (male n = 24 and female n = 27), ko n = 53 (male n = 24 and female n = 29), t 102 = 4.679, ****P < 0.0001, unpaired Student's t‐test.
-
FMarble burying test as a measure for repetitive behaviour. Total number of marbles (half and completely) buried by adult miR379‐410 wt and ko mice is shown; wt n = 49 (male n = 23 and female n = 26), ko n = 53 (male n = 24 and female n = 29), t 100 = 2.569, *P = 0.0117, unpaired Student's t‐test.
-
GPup homing test performed at P9 during developmental milestones. Time spent in the nest area (containing nest litter) is shown, wt n = 17 (male n = 8 and female n = 9); ko n = 11 (male n = 5 and female n = 6), t 26 = 0.9834, ns P = 0.3345, unpaired Student's t‐test.
miR379‐410 deficiency leads to increased excitatory synaptic transmission and spine density in the hippocampus
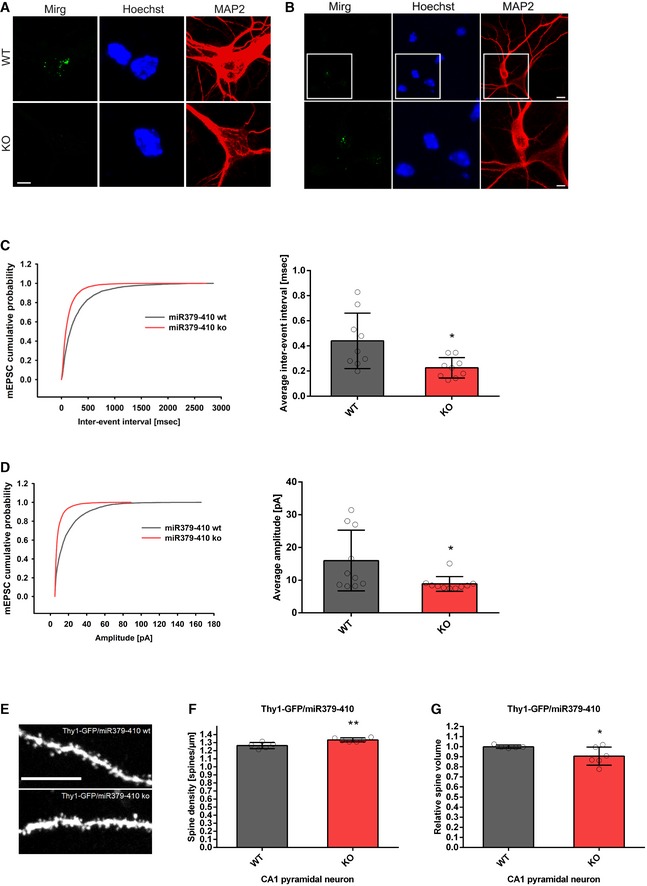
We next investigated whether behavioural alterations observed upon miR379‐410 deletion are accompanied by changes in neuronal physiology and/or morphology. Therefore, we first explored expression of the miR379‐410 encoding Mirg in mouse hippocampal neuron cultures using single‐molecule fluorescent in situ hybridization (smFISH). Thereby, we observed intense nuclear Mirg localization in wt, but not miR379‐410 ko neurons (Fig 2A). Importantly, Mirg expression was restricted to MAP2‐positive neurons and absent from non‐neuronal cells present in our cultures, such as glial cells (Fig 2B). In addition, we could validate the specific lack of several miR379‐410 miRNAs in miR379‐410 ko mice across the lifespan by qPCR (Fig EV2). Using patch‐clamp electrophysiological recordings, we observed a significant increase in mEPSC frequency (Fig 2C), along with a decrease in average mEPSC amplitudes (Fig 2D) in miR379‐410 ko neuron cultures compared to controls. In contrast, no genotype‐dependent differences in mEPSC decay times were observed (Appendix Fig S4). Consistent with our electrophysiology data, CA1 pyramidal neurons in adult Thy1‐GFP/miR379‐410 ko mice displayed a significant increase in dendritic spine density and a significant reduction in dendritic spine volume, as judged by multiphoton imaging (Fig 2E–G). Neuronal health and dendritic complexity were not affected by miR379‐410 deficiency (Appendix Fig S5) 22. Together, our results suggest increased excitatory synaptic transmission due to the loss of the miR379‐410 cluster in CA1 hippocampal pyramidal neurons.
Figure 2. Increased excitatory synaptic transmission and spine density in miR379‐410 ko hippocampal neurons.
-
A, BSingle‐molecule FISH (smFISH) on mouse hippocampal neurons. (A) Representative pictures of smFISH on BDNF‐treated WT (top) and KO (bottom) mouse hippocampal neurons (DIV6) using a probe directed against the Mirg transcript (green). (Middle) Hoechst (blue). (Right) MAP2 immunostaining (red). Pictures show a zoom‐in on the cell body, since the signal detected in WT neurons was restricted to the nuclei (scale bar: 5 μm). (B) Representative pictures of smFISH on mouse wild‐type hippocampal neuron cultures (DIV6; basal) using a probe specific for the Mirg transcript (green). (Middle) Hoechst (blue). (Right) MAP2 immunostaining (red). Top: overview, scale bar: 10 μm; bottom: zoom‐in, scale bar: 5 μm.
-
C, DPatch‐clamp electrophysiological recordings in cultured mouse hippocampal neurons (DIV 8–10) isolated from wt or miR379‐410 ko animals (n = 3 per group). (C) Cumulative distribution (left panel, D = 0.25, P < 0.0001, KS test) and mean (right panel, t 16 = 2.736, *P = 0.0146, unpaired Student's t‐test) of mEPSC inter‐event intervals; wt n = 9, ko n = 9 cells analysed. (D) Cumulative distribution (left panel, D = 0.34741, P < 0.0001, KS test) and mean (right panel, t 18 = 2.357, *P = 0.0299, unpaired Student's t‐test) of mEPSC amplitudes; wt n = 10, ko n = 10 cells analysed.
-
ERepresentative confocal images of CA1 pyramidal neuron apical dendrites from adult Thy1‐GFP/miR379‐410 wt and Thy1‐GFP/miR379‐410 ko mouse tissue. Scale bar: 10 μm.
-
FSpine density in CA1 pyramidal neurons of adult male Thy1‐GFP/miR379‐410 wt and ko mice; wt n = 5, ko n = 6 animals. Mean of basal and apical neurons per mouse is shown, t 9 = 3.662, **P = 0.0052, unpaired Student's t‐test.
-
GSpine volume in CA1 pyramidal neurons of adult Thy1‐GFP/miR379‐410 wt and ko mice; wt n = 5, ko n = 6 animals. Mean of basal and apical neurons per mouse is shown, t 9 = 2.265, *P = 0.0497, unpaired Student's t‐test.
Figure EV2. qPCR expression levels of RNA samples in different brain regions and ages of miR379‐410 wt/ko mice.
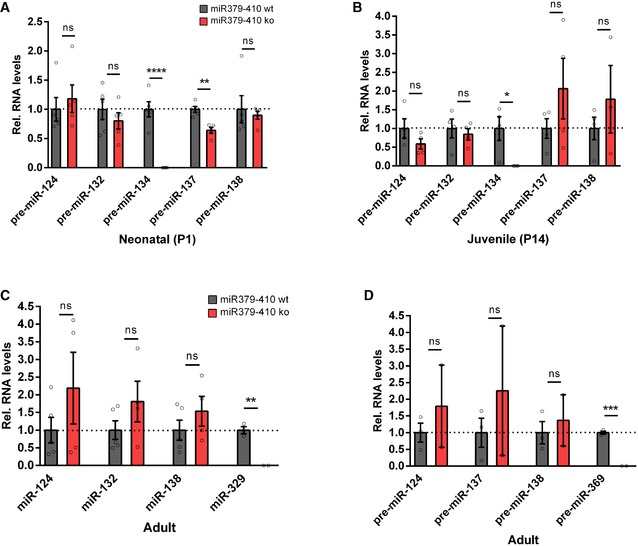
- Expression of precursor miRNAs in hippocampal neonatal (P1) miR379‐410 wt/ko pups: pre‐miR‐124 (wt n = 5, ko n = 5, t 8 = 0.5848, ns P = 0.5748), pre‐miR‐132 (wt n = 5, ko n = 5, t 8 = 0.8740, ns P = 0.4076), pre‐miR‐137 (wt n = 5, ko n = 5, t 8 = 5.037, **P = 0.0010), pre‐miR‐138.2 (wt n = 5, ko n = 5, t 8 = 0.4145, ns P = 0.6894) and from the miR379‐410 cluster pre‐miR‐134 (wt n = 5, ko n = 5, t 8 = 7.637, ****P < 0.0001), unpaired Student's t‐test.
- Expression of precursor miRNAs in hippocampal juvenile (P14) miR379‐410 wt/ko mice: pre‐miR‐124 (wt n = 4, ko n = 4, t 6 = 1.406, ns P = 0.2093), pre‐miR‐132 (wt n = 4, ko n = 4, t 6 = 0.5348, ns P = 0.6120), pre‐miR‐137 (wt n = 4, ko n = 4, t 6 = 1.250, ns P = 0.2578), pre‐miR‐138.2 (wt n = 4, ko n = 3, t 5 = 0.9339, ns P = 0.3932) and from the miR379‐410 cluster pre‐miR‐134 (wt n = 4, ko n = 4, t 6 = 3.150, *P = 0.0198); unpaired Student's t‐test.
- Expression of mature miRNAs in cerebellum of adult miR379‐410 wt/ko mice: miR‐124 (wt n = 5, ko n = 4, t 7 = 1.214, ns P = 0.2642), miR‐132 (wt n = 5, ko n = 4, t 7 = 1.382, ns P = 0.2095), miR‐138 (wt n = 5, ko n = 4, t 7 = 1.089, ns P = 0.3120) and from the miR379‐410 cluster miR‐329 (wt n = 3, ko n = 2, t 3 = 7.720, **P = 0.0045); unpaired Student's t‐test.
- Expression of precursor miRNAs in cerebellum of adult miR379‐410 wt/ko mice: pre‐miR‐124 (wt n = 3, ko n = 2, t 3 = 0.8014, ns P = 0.4815), pre‐miR‐137 (wt n = 3, ko n = 2, t 3 = 0.8107, ns P = 0.4769), pre‐miR‐138.2 (wt n = 3, ko n = 2, t 3 = 0.5169, ns P = 0.6409) and from the miR379‐410 cluster pre‐miR‐369 (wt n = 3, ko n = 2, t 3 = 16.78, ***P = 0.0005); unpaired Student's t‐test. Data are presented as mean ± s.e.m.
miR379‐410 deficiency leads to increased expression of ionotropic glutamate receptor components in the hippocampus
To pinpoint the molecular pathways responsible for the observed behavioural and cellular alterations in miR379‐410 mice, we performed comparative transcriptome analysis in the hippocampus of adult miR379‐410 ko and control mice using polyA‐RNAseq. miR379‐410 deficiency caused extensive changes in the hippocampal transcriptome [differentially expressed genes (DEGs): n = 3,068; upregulated in miR379‐410 ko: n = 2,170; downregulated: n = 898; q < 0.05; Fig 3A and Appendix Fig S6]. Consistent with our cellular data, GO term enrichment analysis revealed a number of significant terms associated with synaptic transmission (Fig 3B and Appendix Figs S7 and S8). Further clustering of synaptic GO terms led to the identification of an extensive network comprising main ionotropic receptor complexes, i.e. glutamate and GABA receptor complexes (Figs 3C and EV3). Notably, a large number of known positive regulators of ionotropic glutamate receptor complexes (e.g. Grin2c/d, Shank1/3, Cacng4/7/8, Homer3) were upregulated in the absence of miR‐379‐410 (Fig 3C), whereas negative regulators (e.g. Homer1, Fmr1) were primarily downregulated. Given the well‐established inhibitory role of miRNAs, we considered upregulated genes as potential direct miR379‐410 targets. Surprisingly, motif analysis revealed that binding sites for only five cluster miRNAs (miR‐485‐5p, miR‐134‐5p, miR‐377‐3p, miR‐381‐3p and miR‐329‐3p) were overrepresented in upregulated genes (Fig 3D). Among this miR379‐410 subset, miR‐134‐5p/miR‐485‐5p and miR‐377‐3p/miR‐329‐3p share similar seed sequences (Fig EV4A), suggesting that the target spectrum of these miRNAs might be at least partially overlapping.
Figure 3. Coordinated upregulation of ionotropic glutamate receptor complexes in the miR379‐410 ko hippocampus.
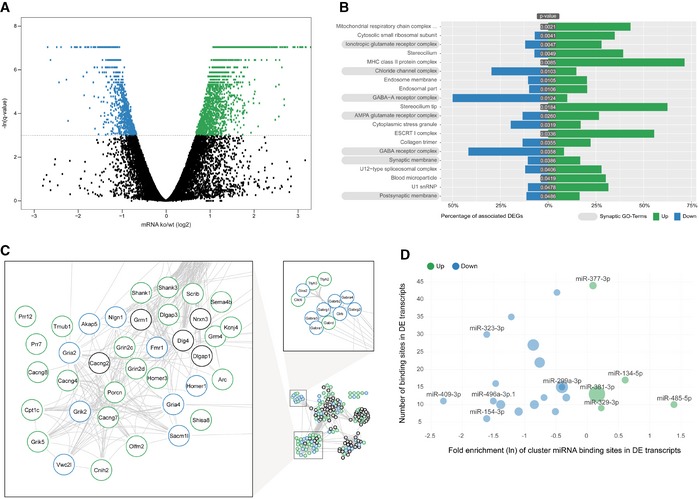
- RNAseq differentially expressed gene (DEG) analysis in the adult hippocampus of miR379‐410 ko and wt mice. Upregulated (green) and downregulated (blue) DEGs are indicated by average changes in mRNA levels (ko/wt; log2) (n = 3; q‐values represent FDR‐corrected P‐values; q < 0.05); 2,170/898 genes were significantly up‐ and downregulated in miR379‐410 mice, respectively.
- GO term enrichment analysis of DEG reveals several synaptic cellular components (highlighted in grey). Green and blue bars indicate the percentage of significantly up‐ and downregulated genes associated with specific GO terms, respectively. GO terms with more than 300 annotated genes are not shown.
- String database protein–protein interaction network. The network is built including String‐DB parameters Experiments, Databases and Textmining. Clusters associated with synaptic transmission are enlarged (for the complete network, see Fig EV3B). Green and blue borders indicate significantly up‐ and downregulated genes, respectively.
- Enrichment analysis of cluster miRNA binding motifs (TargetScan v7.1). miRNAs overrepresented in differentially expressed (DE) transcripts are highlighted in green and blue, respectively. Shown is the natural logarithm (ln) of the wt/ko ratio of binding sites in DE transcripts. Bubble sizes correspond to RPM values in the mouse brain.
Figure EV3. Clustering of synaptic GO terms and String database protein–protein interaction network.
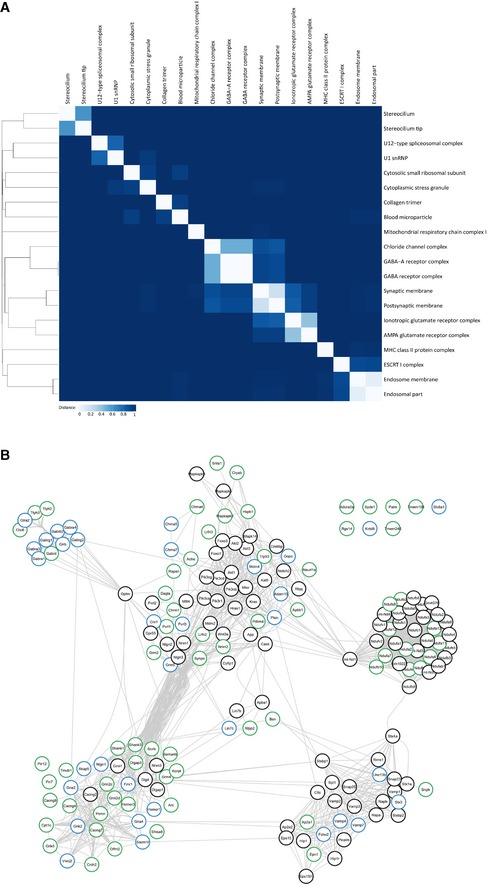
- Binary distance matrix of detected GO terms (cellular component). Hierarchical clustering has been performed by the unweighted pair group method with arithmetic mean. GO terms with more than 300 annotated genes are not shown.
- Complete String database protein–protein interaction network.
Figure EV4. miRNA binding site analysis and target validation.
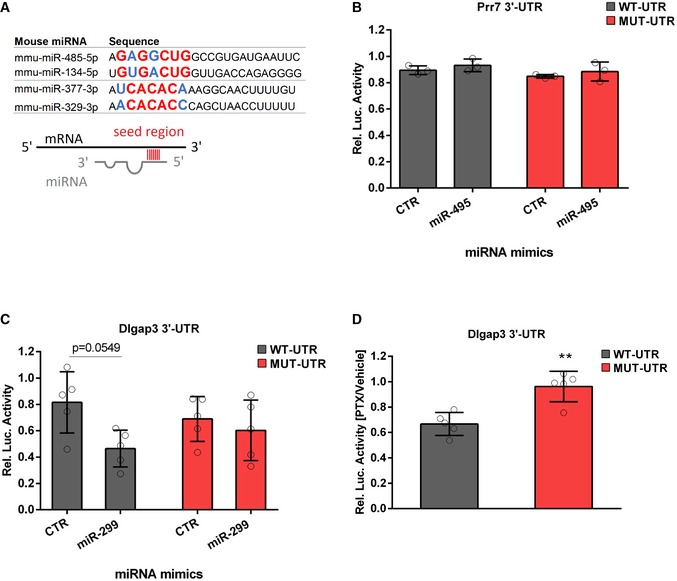
- miRNAs of the miR379‐410 cluster sharing similar seed regions: miR485‐5p/miR‐134‐5p and miR‐377‐3p/miR‐329‐3p. Seed regions (nucleotide 2–8) are highlighted in bigger letters, conserved nucleotides in red and those that allow wobble base pairing in blue.
- 3′UTR luciferase reporter gene assays in cultured rat cortical neurons using the Prr7 3′UTR reporter gene (wt or seed mutant) together with miR‐495 or control mimic, respectively (n = 3 per group; wt: P = 0.7775; mut: P = 0.7797; mimic: F 1,8 = 1.816, P = 0.2147; utr‐construct: F 1,8 = 2.973, P = 0.1229; mimic × utr‐construct: F 1,8 = 9.47e‐006, P = 0.09976); two‐way ANOVA.
- 3′UTR luciferase reporter gene assays in cultured rat cortical neurons using the Dlgap3 3′UTR reporter gene (wt or seed mutant) together with miR‐299 or control mimic, respectively (n = 5 per group; wt: P = 0.0549; mut: P = 0.8982; mimic: F 1,16 = 6.145, P = 0.0247; utr‐construct: F 1,16 = 0.005108, P = 0.9434; mimic × utr‐construct: F 1,16 = 2.246, P = 0.1534); two‐way ANOVA.
- 3′UTR luciferase reporter gene assays in cultured rat hippocampal neurons using the Dlgap3 3′UTR reporter gene (wt or seed mutant) after 48 h PTX stimulation (n = 5 per group, t 8 = 4.397, **P = 0.0023); unpaired Student's t‐test.
Ionotropic glutamate receptor components are direct targets of a specific subset of miR379‐410 miRNAs
When inspecting 3′UTR sequences of DEGs encoding for components of ionotropic glutamate receptor complexes, we found that several of these genes contain at least one conserved binding site for the miR379‐410 subset (Fig 4A). Using qPCR, we were able to confirm upregulated expression of several of these predicted direct targets in the miR379‐410 ko hippocampus (Fig 4B and Appendix Fig S9). Therefore, our findings suggest that loss of a specific subset of miR379‐410 miRNAs might largely explain the coordinated upregulation of ionotropic glutamate receptor components in miR379‐410 ko neurons. We went on to validate direct regulation of candidate targets by specific miR379‐410 miRNAs using 3′UTR luciferase reporter gene assays. In primary rat cortical neurons, transfection of miR‐485‐5p and miR‐329‐3p mimics downregulated expression of the cognate reporter genes (Prr7, Cnih2, Src) in the presence of wt, but not (Cnih2, Src) or to a lesser extent (Prr7) in the presence of mutant 3′UTR binding sites (Fig 4C). In contrast, transfection of an unrelated mimic (miR‐495) did not significantly affect wt or mutant reporter gene activity, further demonstrating miRNA specificity (Fig EV4B). Importantly, transfection efficiencies were comparable between different miRNA mimics (Appendix Fig S10). Together, our findings demonstrate the functionality of the identified miRNA binding sites. To further address the relevance of endogenous miRNAs for target gene regulation, we assessed reporter gene activity in hippocampal neurons treated for 48 h with the GABA‐A receptor blocker picrotoxin (PTX). We used PTX for these experiments, since we previously found that PTX upregulates several members of miR379‐410 and that miR‐134 was required for PTX‐dependent homeostatic synaptic downscaling 13, 25. Accordingly, PTX‐mediated downregulation of the miR‐485‐5p/329‐3p targets Prr7 and Src was abrogated by the transfection of the cognate locked nucleic acid (LNA)‐modified antisense oligonucleotides (“anti‐miRNAs”) in a miRNA binding site‐specific manner (Fig 4D). Similar results were observed for the miR‐299‐3p target Dlgap3 (Fig EV4C and D). Finally, Prr7 protein levels were upregulated in the hippocampus of miR379‐410 ko mice compared to wt mice as judged by Western blot (Fig EV5 and Appendix Figs S11 and S12). Together, these experiments demonstrate that a number of important ionotropic glutamate receptor components are direct targets of a specific subset of endogenous miR379‐410 miRNAs.
Figure 4. Validation of direct miR379‐410 targets.
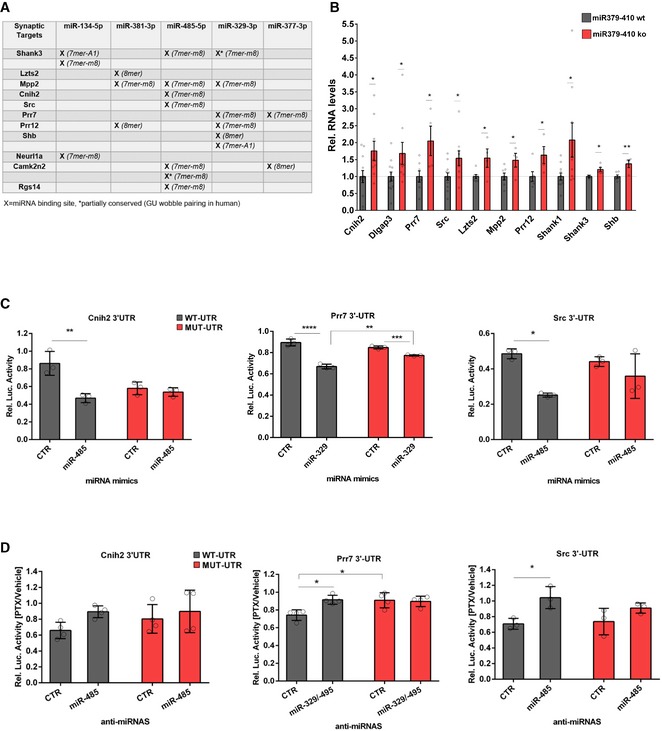
-
AList of synaptic targets among DEG containing conserved binding sites (human and mouse) for indicated miRNAs based on TargetScan v7.1. Category of binding site from TargetScan (v7.1) is shown in brackets.
-
BqPCR validation of predicted direct miR379‐410 targets using RNA from hippocampi of juvenile (P 22–24) mice, except of Shank3 where adult miR‐379‐410 wt and ko animals were measured. If not indicated, both male and female samples were used. Cnih2 (wt n = 12, ko n = 9, t 19 = 2.344, *P = 0.0301), Dlgap3 (wt n = 12, ko n = 9, t 19 = 2.128, *P = 0.0467), Prr7 (male wt n = 7, ko n = 4, t 9 = 2.719, *P = 0.0236), Src (wt n = 12, ko n = 9, t 19 = 2.410, *P = 0.0263), Lzts2 (male wt n = 7, ko n = 4, t 9 = 2.554, *P = 0.0310), Mpp2 (male wt n = 7, ko n = 4, t 9 = 2.369, *P = 0.0420), Prr12 (wt n = 6, ko n = 4, t 8 = 2.375, *P = 0.0449), Shank1 (wt n = 11, ko n = 9, t 18 = 2.310, *P = 0.0329), Shb (male wt n = 6, ko n = 4, t 8 = 3.451, **P = 0.0087) and Shank3 (adult male wt n = 3, ko n = 5, t 6 = 2.538, *P = 0.0442), unpaired Student's t‐test. Data are presented as mean ± s.e.m.
-
C, DCnih2, Prr7 and Src are direct miR‐379‐410 target mRNAs. (C) 3′UTR luciferase reporter gene assays in cultured rat cortical neurons using the indicated 3′UTR reporter genes (wt or seed mutant) together with miRNA mimics. Cnih2 (n = 3 per group; wt: **P = 0.0020, mut: P = 0.9267; mimic: F 1,8 = 19.92, P = 0.0021; utr‐construct: F 1,8 = 4.760, P = 0.0607; mimic × utr‐construct: F 1,8 = 12.99, P = 0.0069), Prr7 (n = 3 per group; wt: ****P < 0.0001, mut: *P = 0.0103; wt vs. mut mimic miR329: **P = 0.0014; mimic: F 1,8 = 153.1, P < 0.0001; utr‐construct: F 1,8 = 5.631, P = 0.0450; mimic × utr‐construct: F 1,8 = 38.54, P = 0.0003); Src (n = 3 per group; wt: *P = 0.0110, mut: P = 0.4724; mimic: F 1,8 = 16.99, P = 0.0033; utr‐construct: F 1,8 = 0.6809, P = 0.4332; mimic vs. utr‐construct: F 1,8 = 3.920, P = 0.0831); two‐way ANOVA. (D) Same as in (C), but in the presence of anti‐miRs (pLNAs) in hippocampal neurons after 48 h of PTX stimulation. Cnih2 (n = 4 per group; wt: P = 0.2688; mut: P = 0.8668; wt ctr vs. mut ctr: P = 0.6463; anti‐miRNA: F 1,12 = 3.629, P = 0.0810; utr‐construct: F 1,12 = 0.7407, P = 0.4063; anti‐miRNA × utr‐construct: F 1,12 = 0.6679, P = 0.4297), Prr7 (n = 4 per group; wt: *P = 0.0123; mut: P = 0.9917; wt ctr vs. mut ctr: *P = 0.0152; anti‐miRNA: F 1,12 = 6.082, P = 0.0297; utr‐construct: F 1,12 = 5.271; P = 0.0405; anti‐miRNA × utr‐construct: F 1,12 = 8.212, P = 0.0142); Src (n = 3 per group; wt: *P = 0.0370; mut: P = 0.3540; wt ctr vs. mut ctr: P = 0.9906; anti‐miRNA: F 1,12 = 13.43, P = 0.0064; utr‐construct: F 1,12 = 0.5672, P = 0.4730; anti‐miRNA x utr‐construct: F 1,12 = 1.362, P = 0.2768), two‐way ANOVA. Data are presented as mean ± s.d.
Figure EV5. Protein expression in hippocampus of 5‐week‐old juvenile miR379‐410 wt and ko mice.
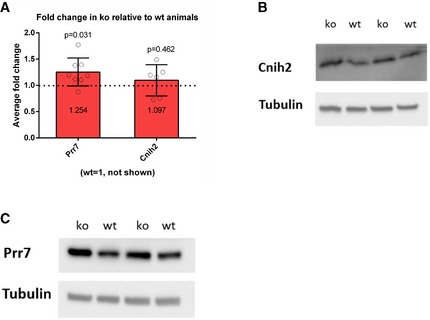
-
AAverage fold change in ko relative to wt animals is presented (wt = 1, not shown), Cnih2 (wt n = 6, ko n = 6, P = 0.4625), Prr7 (wt n = 7, ko = 8, P = 0.0311), one‐sample t‐test (2.3 test in excel). Data are presented as mean ± s.d.
-
B, CRepresentative Western blots of Cnih2 and Prr7 protein expression in hippocampus of juvenile mice.
Conclusions
In this study, we provide first evidence for an important role of miRNA‐dependent post‐transcriptional regulation of excitatory synapse development in the control of rodent sociability. miR‐124 was recently shown to partially restore social deficits associated with frontotemporal dementia 26, but in contrast to our study, this study relied on miRNA overexpression and focussed on the late adult stage. In addition, specific miRNAs, including miR‐124 and the miR379‐410 cluster, were shown to control anxiety‐like behaviour 23, 27, 28, 29. Intriguingly, the unusual combination of hypersocial and anxiety‐like behaviour we observed in miR379‐410 ko mice is present in patients suffering from two rare neurodevelopmental disorders, WBS 30 and AS 31. The candidate disease genes of WBS and AS, LIMK1 and UBE3A, represent validated targets of miR‐134 7, 22, raising the possibility that a common molecular aetiology could underlie phenotypic alterations observed in WBS, AS and miR379‐410 deficiency. Previous reports also suggest an association of the human syntenic genomic region (14q32.2) encompassing the miR379‐410 cluster with neurodevelopmental disease. For example, 15 reports of copy number variations (both deletions and duplications) within 14q32.2 are listed in the Sfari autism gene database (gene.sfari.org). Moreover, the rare imprinting disorders Temple syndrome [uniparental disomy (upd) 14 of maternal origin] and Kagami–Ogata syndrome (upd 14 of paternal origin) frequently display, among other symptoms, intellectual disability 32.
Results from previous in vitro studies and our extensive molecular and cellular characterization presented here point to the hippocampus as a critical site of miR379‐410 function. Moreover, expression of miR379‐410 during postnatal development and in adulthood is mostly restricted to the brain 33. However, since our model encompasses organism‐wide miR379‐410 deficiency throughout development, we cannot completely rule out that deficits outside the brain or in brain regions other than the hippocampus, such as amygdala, prefrontal or entorhinal cortex, might be causally involved in the manifestation of behavioural phenotypes 2, 34, 35.
Our bioinformatics analysis suggests that loss of a small subgroup of related miRNAs from the cluster might explain to a large extent the upregulation of key synaptic proteins. Intriguingly, two of these miRNAs, miR‐134‐5p and miR‐485‐5p, are encoded within a single transcript (Mirg/Meg9) 36 and contain a similar seed sequence (Fig EV4A). Since previous in vitro work showed that miR‐134‐5p and miR‐485‐5p are required for homeostatic synaptic downscaling 13, 15, we hypothesize that enhanced excitation in miR379‐410 ko neurons could be the result of inefficient homeostatic compensation. In agreement, excitatory dysfunction is observed in the hippocampus of a number of autism mouse models, such as MeCP2, FMRP, Shank or Nrxn mutants 37.
In conclusion, our results provide important new insight into the physiological mechanisms that control sociability in the healthy brain. Moreover, we envision that interfering with specific members of the miR379‐410 cluster could represent a promising strategy to counteract social deficits observed in neurodevelopmental disorders.
Materials and Methods
A more detailed description of methods is provided in the Appendix.
Animals and housing
Animal experiments were performed in accordance with the animal protection law of Germany and were approved by the local authorities responsible for the Philipps University Marburg (Regierungspräsidium, Gießen, Germany). For reciprocal social interaction test, P22 juvenile mice were housed in isolation for 24 h before testing. Otherwise, all rodents were housed under standard cage conditions with food and water ad libitum and maintained on a 12‐h/12‐h light/dark cycle. The miR379–410 conditional knockout mouse was generated at Taconic/Artemis (Cologne, Germany) and described previously 22. The Thy1‐GFP reporter mice were provided by Marco Rust (Philipps University Marburg). For three‐chamber social memory test, stimulus C57BL/6 mice were taken, provided by Charles River (Sulzfeld, Germany).
General behavioural procedures
Behavioural assays were performed in pups (P3‐12), juvenile (P23), adolescent (P25‐42) or adult mice (P84‐224) of both sexes in two independent cohorts. To examine developmental milestones, a third independent cohort was used to minimize handling stress. All behavioural tests, except for the elevated plus maze, were performed under dim red light. Behavioural assays were done with all animals in the following order: Pup isolation induced USV, reciprocal social interaction, three‐chamber social and object memory, open field, marble burying, elevated plus maze.
Developmental milestones and somatosensory reflexes
Pups were tested according to a modified Fox battery for developmental milestones and somatosensory reflexes as described previously 38.
Ultrasonic vocalizations in isolated pups
To initiate isolation‐induced USV, the protocol was used as described earlier 39.
Open field locomotion
Mice were placed randomly in one of the corner of a standard open field box (TSE System, Bad Homburg, Germany), and the locomotor activity was recorded for 10 min and automatically collected using the TSE VideoMot2 analyser software (TSE Systems, Bad Homburg, Germany).
Elevated plus maze
The elevated plus maze test was performed as described previously 39.
Marble burying test
The marble burying protocol was performed as described earlier 40.
Reciprocal social interaction and USV analysis
The reciprocal social interaction assay was performed in juvenile mice on P23 as described earlier 41. To observe social interactions, two mice of the same age, sex and genotype that were unfamiliar to each other but with the same early‐life history were put together for 5 min in a novel testing cage with fresh bedding. Social behaviours of each pair were videotaped, and time of social events and USV calls was recorded by an investigator blinded to the genotypes.
Three‐chamber box
The three‐chamber box apparatus was used in 4‐ to 6‐week‐old juvenile animals for social memory and object memory, as previously described in detail 42. One animal was not able to perform the social memory assay since it was outside the test field and had therefore to be excluded from analysis.
Cell culture
Primary hippocampal and cortical neuron cultures were prepared from rats (E18, Sprague Dawley, Charles River Laboratories, Sulzfeld, Germany) or mice (P1, C57BL/6 wt or miR379‐410 ko) as described previously with minor modifications 7.
smFISH
smFISH was performed on DIV6 mouse hippocampal neurons using a type 4 Alexa 488 probe against the mouse Mirg transcript (Assay‐ID: VB4‐3140541, Thermo Fisher) and the QuantiGene ViewRNA ISH Cell Assay kit as previously described 22, but omitting the protease treatment.
Electrophysiology in primary mouse culture
Spontaneous miniature excitatory postsynaptic currents (mEPSCs) were recorded in in vitro‐cultured hippocampal cells (DIV 8–10, genotypes were equally distributed across the days) as described previously 43.
Luciferase reporter assay
Primary neurons were transfected in triplicate per independent experiment (“n”) in 24‐well plates using for each well 100 ng (for CTX cells) or 50 ng (for HC cells) of pmirGLO dual‐luciferase expression vector reporter (Promega, Madison, WI, USA) with respective 10 pmol miRNA mimics (pre‐miR miRNA precursor, Ambion, Thermo Fisher Scientific, Darmstadt, Germany) or 20 pmol anti‐miRs (pLNAs, Exiqon, Vedbaek, Denmark). Control transfections (CTR) were performed with scrambled miRNA mimics (pre‐miR negative control#1, Ambion cat#4464058) or anti‐miRNAs (negative control A, Exiqon cat#199006‐100), respectively. Luciferase assays were performed using the dual‐luciferase reporter assay system on a GloMax R96 Microplate Luminometer (Promega).
DNA constructs
3′ UTRs of Prr7 (NM_001030296.4), Src (NM_009271.3), Cnih2 (NM_009920.4) and Dlgap3 (NM_198618.5) were amplified either from mouse genomic DNA or from mouse cDNA and cloned into the pmirGLO dual‐luciferase expression vector. Mutants of conserved miRNA binding sites were produced by site‐directed mutagenesis using Pfu Plus! DNA Polymerase (Roboklon) or with the QuikChange Site‐Directed Mutagenesis kit (Stratagene) according to manufacturer's instructions.
Tissue preparation
After perfusion fixation with 4% paraformaldehyde, brains were taken out immediately and 50‐μm‐thick coronal brain sections were prepared using a VT1000S vibratome (Leica, Wetzlar, Germany). Brain slices were mounted on microscope slides using Aqua‐Poly/Mount (Polysciences, Inc., Valley Road, PA, USA). For RNA extraction (qPCR and RNAseq), brains were rapidly removed after cervical dislocation of the animals and hippocampi were dissected on ice. Afterwards, RNA was isolated with the mirVana miRNA isolation kit (Thermo Fisher Scientific).
Protein extracts for Western blot
Hippocampal tissue from 5‐week‐old juvenile mouse brain was removed quickly after cervical dislocation and homogenized on ice with a small plastic stamper in modified RIPA buffer (50 mM Tris–HCL, 150 mM NaCl, 0.5% NP‐40, 0.1% SDS, pH 7.4) containing Complete Protease Inhibitor Cocktail EDTA‐free (Merck, Darmstadt, Germany) to extract proteins. Please see Appendix Supplementary Methods for further details.
Image analysis
For spine analysis of Thy1‐GFP/miR379‐410 mice brain sections, images were taken with a multiphoton microscopy BX36 (Olympus, Hamburg, Germany) equipped with a 25× objective and the numerical aperture was set to 1.05. An excitation wavelength of 880–940 nm (HV: 500–600 V) was chosen for imaging GFP tissue fluorescence. Images (1,024 × 1,024 pixels) were recorded in time series of z‐stacks (with 0.5 μm step size) using the software FluoView FV30S‐SW (Olympus). Morphological changes in hippocampal CA1 basal and apical dendrites were analysed as described earlier 44. Spine density and volumes were subsequently analysed with ImageJ software (NIH, Bethesda, MD, USA) as previously described 13.
Quantitative real‐time PCR
qPCR was performed as described earlier 22. The geometric mean of U6, Rp2 and Ywhaz was used for normalization. One outlier was detected for each Prr12, Shank1 and Shb and removed from the analysis.
RNAseq and bioinformatic analysis
Total RNA was isolated from three miR379‐410 wt and three ko adult (8‐month‐old) male hippocampi. Stranded polyA+ enriched RNA sequencing libraries were prepared at the GENCORE (EMBL, Genomics Core Facility, Heidelberg, Germany) and sequenced on an Illumina HiSeq 2000 machine using a 50nt paired‐end protocol.
Further details on the processing of RNAseq data as well as GO term, miRNA binding site and network analysis are provided in the Appendix Supplementary Methods.
Statistical analysis
Normal distribution was tested by using the Kolmogorov–Smirnov test. Statistical significance was determined using either two‐tailed, unpaired Student's t‐test for two population comparisons or paired Student's t‐test for comparing identical population in the three‐chamber assay. For isolation‐induced pup USV and developmental milestones, a repeated‐measures ANOVA with within‐subject factor “postnatal day” and between‐subject factor “genotype” was performed. For the three‐chamber box assays (social approach/recognition and object recognition), a repeated‐measures ANOVA with within‐subject factor “side” and between‐subject factors “sex” and “genotype” was performed. For the remaining behavioural data, a two‐way ANOVA with between‐subject factors “genotype” and “sex” was performed. For the luciferase assay, a two‐way ANOVA was used with two independent variables followed by the Tukey post hoc test. Data were analysed using GraphPad Prism and SPSS.
Author contributions
ML performed behavioural assays, multiphoton microscopy, qPCR, luciferase assays and related data analysis. AÖS performed and analysed behavioural assays. RD performed and analysed patch‐clamp recordings and helped with hippocampus isolation and blotting for the WB experiments. MS performed integrative bioinformatic analysis of RNAseq data and cloned reporter gene plasmids. SB performed smFISH. LS analysed the USV data. RF established and characterized the miR379–410 knockout mouse colony and performed WB experiments. TW performed WB and qPCR experiments. CD analysed RNAseq data and performed differential gene expression analysis. RKWS and MW designed and supervised behavioural studies. GS designed the study, wrote the manuscript and coordinated the entire project.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Marco Rust for providing Thy1‐GFP reporter mice and Tim Plant for generously granting access to a patch‐clamp electrophysiology set‐up. We thank Robert Große for assistance in behavioural assays. We thank Eva Becker for excellent technical support and Helena Martins for assistance in mouse cell culture preparation. This work was supported by grants from the Deutsche Forschungsgemeinschaft to GS (DFG SCHR 1136/8‐1, 1136/4‐1), RF (DFG FI215/2‐1), CD (DFG DI1501/5‐1) and MW (DFG WO 1732/1‐1, 1732/4‐1).
EMBO Reports (2019) 20: e46429
See also: MJ Cairns (February 2019)
Data availability
All primary data are freely available from the corresponding author upon request. RNAseq data were submitted to the NCBI BioProject database, accession no. PRJNA494281.
References
- 1. Barak B, Feng G (2016) Neurobiology of social behavior abnormalities in autism and Williams syndrome. Nat Neurosci 19: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Felix‐Ortiz AC, Tye KM (2014) Amygdala inputs to the ventral hippocampus bidirectionally modulate social behavior. J Neurosci 34: 586–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. (2014) Natural neural projection dynamics underlying social behavior. Cell 157: 1535–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajman M, Schratt G (2017) MicroRNAs in neural development: from master regulators to fine‐tuners. Development 144: 2310–2322 [DOI] [PubMed] [Google Scholar]
- 5. Schratt G (2009) microRNAs at the synapse. Nat Rev Neurosci 10: 842–849 [DOI] [PubMed] [Google Scholar]
- 6. Winter J (2015) MicroRNAs of the miR379‐410 cluster: New players in embryonic neurogenesis and regulators of neuronal function. Neurogenesis (Austin) 2: e1004970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME (2006) A brain‐specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- 8. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jimenez‐Mateos EM, Engel T, Merino‐Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O'Tuathaigh C, Waddington JL, Prenter S, et al. (2012) Silencing microRNA‐134 produces neuroprotective and prolonged seizure‐suppressive effects. Nat Med 18: 1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P (2011) Stage‐specific modulation of cortical neuronal development by Mmu‐miR‐134. Cereb Cortex 21: 1857–1869 [DOI] [PubMed] [Google Scholar]
- 11. Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G (2009) Mef2‐mediated transcription of the miR379‐410 cluster regulates activity‐dependent dendritogenesis by fine‐tuning Pumilio2 protein levels. EMBO J 28: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH (2010) A novel pathway regulates memory and plasticity via SIRT1 and miR‐134. Nature 466: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fiore R, Rajman M, Schwale C, Bicker S, Antoniou A, Bruehl C, Draguhn A, Schratt G (2014) MiR‐134‐dependent regulation of Pumilio‐2 is necessary for homeostatic synaptic depression. EMBO J 33: 2231–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rago L, Beattie R, Taylor V, Winter J (2014) miR379‐410 cluster miRNAs regulate neurogenesis and neuronal migration by fine‐tuning N‐cadherin. EMBO J 33: 906–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen JE, Lee PR, Chen S, Li W, Fields RD (2011) MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci U S A 108: 11650–11655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yin CL, Chen HI, Li LH, Chien YL, Liao HM, Chou MC, Chou WJ, Tsai WC, Chiu YN, Wu YY, et al. (2016) Genome‐wide analysis of copy number variations identifies PARK2 as a candidate gene for autism spectrum disorder. Mol Autism 7: 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ (2011) Imprinted DLK1‐DIO3 region of 14q32 defines a schizophrenia‐associated miRNA signature in peripheral blood mononuclear cells. Mol Psychiatry, 10.1038/mp.2011.78mp201178 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ (2010) Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry 15: 1176–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu YE, Parikshak NN, Belgard TG, Geschwind DH (2016) Genome‐wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat Neurosci 19: 1463–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng TL, Wang Z, Liao Q, Zhu Y, Zhou WH, Xu W, Qiu Z (2014) MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev Cell 28: 547–560 [DOI] [PubMed] [Google Scholar]
- 21. Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, Ruan X, Ruan Y, Sun YE (2010) Genome‐wide analysis reveals methyl‐CpG‐binding protein 2‐dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci U S A 107: 18161–18166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Valluy J, Bicker S, Aksoy‐Aksel A, Lackinger M, Sumer S, Fiore R, Wust T, Seffer D, Metge F, Dieterich C, et al. (2015) A coding‐independent function of an alternative Ube3a transcript during neuronal development. Nat Neurosci 18: 666–673 [DOI] [PubMed] [Google Scholar]
- 23. Marty V, Labialle S, Bortolin‐Cavaille ML, Ferreira De Medeiros G, Moisan MP, Florian C, Cavaille J (2016) Deletion of the miR‐379/miR‐410 gene cluster at the imprinted Dlk1‐Dio3 locus enhances anxiety‐related behaviour. Hum Mol Genet 25: 728–739 [DOI] [PubMed] [Google Scholar]
- 24. Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson‐Smith AC, Cavaille J (2004) A large imprinted microRNA gene cluster at the mouse Dlk1‐Gtl2 domain. Genome Res 14: 1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rajman M, Metge F, Fiore R, Khudayberdiev S, Aksoy‐Aksel A, Bicker S, Ruedell Reschke C, Raoof R, Brennan GP, Delanty N, et al. (2017) A microRNA‐129‐5p/Rbfox crosstalk coordinates homeostatic downscaling of excitatory synapses. EMBO J 36: 1770–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gascon E, Lynch K, Ruan H, Almeida S, Verheyden JM, Seeley WW, Dickson DW, Petrucelli L, Sun D, Jiao J, et al. (2014) Alterations in microRNA‐124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat Med 20: 1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bahi A (2016) Sustained lentiviral‐mediated overexpression of microRNA124a in the dentate gyrus exacerbates anxiety‐ and autism‐like behaviors associated with neonatal isolation in rats. Behav Brain Res 311: 298–308 [DOI] [PubMed] [Google Scholar]
- 28. Haramati S, Navon I, Issler O, Ezra‐Nevo G, Gil S, Zwang R, Hornstein E, Chen A (2011) MicroRNA as repressors of stress‐induced anxiety: the case of amygdalar miR‐34. J Neurosci 31: 14191–14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fonken LK, Gaudet AD, Gaier KR, Nelson RJ, Popovich PG (2016) MicroRNA‐155 deletion reduces anxiety‐ and depressive‐like behaviors in mice. Psychoneuroendocrinology 63: 362–369 [DOI] [PubMed] [Google Scholar]
- 30. Meyer‐Lindenberg A, Hariri AR, Munoz KE, Mervis CB, Mattay VS, Morris CA, Berman KF (2005) Neural correlates of genetically abnormal social cognition in Williams syndrome. Nat Neurosci 8: 991–993 [DOI] [PubMed] [Google Scholar]
- 31. Stoppel DC, Anderson MP (2017) Hypersociability in the Angelman syndrome mouse model. Exp Neurol 293: 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenfeld JA, Fox JE, Descartes M, Brewer F, Stroud T, Gorski JL, Upton SJ, Moeschler JB, Monteleone B, Neill NJ, et al. (2015) Clinical features associated with copy number variations of the 14q32 imprinted gene cluster. Am J Med Genet A 167A: 345–353 [DOI] [PubMed] [Google Scholar]
- 33. Labialle S, Marty V, Bortolin‐Cavaille ML, Hoareau‐Osman M, Pradere JP, Valet P, Martin PG, Cavaille J (2014) The miR‐379/miR‐410 cluster at the imprinted Dlk1‐Dio3 domain controls neonatal metabolic adaptation. EMBO J 33: 2216–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bicks LK, Koike H, Akbarian S, Morishita H (2015) Prefrontal Cortex and Social Cognition in Mouse and Man. Front Psychol 6: 1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hollins SL, Zavitsanou K, Walker FR, Cairns MJ (2014) Alteration of imprinted Dlk1‐Dio3 miRNA cluster expression in the entorhinal cortex induced by maternal immune activation and adolescent cannabinoid exposure. Transl Psychiatry 4: e452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hagan JP, O'Neill BL, Stewart CL, Kozlov SV, Croce CM (2009) At least ten genes define the imprinted Dlk1‐Dio3 cluster on mouse chromosome 12qF1. PLoS ONE 4: e4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nelson SB, Valakh V (2015) Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron 87: 684–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wohr M, Silverman JL, Scattoni ML, Turner SM, Harris MJ, Saxena R, Crawley JN (2013) Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav Brain Res 251: 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sungur AO, Schwarting RK, Wohr M (2016) Early communication deficits in the Shank1 knockout mouse model for autism spectrum disorder: Developmental aspects and effects of social context. Autism Res 9: 696–709 [DOI] [PubMed] [Google Scholar]
- 40. Sungur AO, Vorckel KJ, Schwarting RK, Wohr M (2014) Repetitive behaviors in the Shank1 knockout mouse model for autism spectrum disorder: developmental aspects and effects of social context. J Neurosci Methods 234: 92–100 [DOI] [PubMed] [Google Scholar]
- 41. Wohr M, Orduz D, Gregory P, Moreno H, Khan U, Vorckel KJ, Wolfer DP, Welzl H, Gall D, Schiffmann SN, et al. (2015) Lack of parvalbumin in mice leads to behavioral deficits relevant to all human autism core symptoms and related neural morphofunctional abnormalities. Transl Psychiatry 5: e525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sungur AO, Jochner MCE, Harb H, Kilic A, Garn H, Schwarting RKW, Wohr M (2017) Aberrant cognitive phenotypes and altered hippocampal BDNF expression related to epigenetic modifications in mice lacking the post‐synaptic scaffolding protein SHANK1: Implications for autism spectrum disorder. Hippocampus 27: 906–919 [DOI] [PubMed] [Google Scholar]
- 43. Saba R, Storchel PH, Aksoy‐Aksel A, Kepura F, Lippi G, Plant TD, Schratt GM (2012) Dopamine‐regulated microRNA MiR‐181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol 32: 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K (2010) Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS ONE 5: e15497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Data Availability Statement
All primary data are freely available from the corresponding author upon request. RNAseq data were submitted to the NCBI BioProject database, accession no. PRJNA494281.


