Abstract
Covalent modification of proteins with ubiquitin is essential for the majority of biological processes in mammalian cells. Numerous proteins are conjugated with single or multiple ubiquitin molecules or chains in a dynamic fashion, often determining protein half‐lives, localization or function. Experimental approaches to study ubiquitination have been dominated by genetic and biochemical analysis of enzyme structure–function relationships, reaction mechanisms and physiological relevance. Here, we provide an overview of recent developments in microscopy‐based imaging of ubiquitination, available reagents and technologies. We discuss the progress in direct and indirect imaging of differentially linked ubiquitin chains in fixed and living cells using confocal fluorescence microscopy and super‐resolution microscopy, illustrated by the role of ubiquitin in antibacterial autophagy and pro‐inflammatory signalling. Finally, we speculate on future developments and forecast a transition from qualitative to quantitative super‐resolution approaches to understand fundamental aspects of ubiquitination and the formation and distribution of functional E3 ligase protein complexes in their native environment.
Keywords: ubiquitination, LUBAC, OTULIN, Salmonella, super‐resolution microscopy
Subject Categories: Post-translational Modifications, Proteolysis & Proteomics
Glossary
- ACTL8
actin‐like protein 8
- ARIH1
E3 ubiquitin‐protein ligase protein ariadne‐1 homolog
- BiFC
bimolecular fluorescence complementation
- CALCOCO2/NDP52
calcium‐binding and coiled‐coil domain‐containing protein 2/Nuclear domain 10 protein NDP52
- CCCP
carbonylcyanid‐m‐chlorphenylhydrazon
- Cdt1
DNA replication factor Cdt1
- CHIP
caryl terminus of Hsp70‐interacting protein
- CLSM
confocal laser scanning microscopy
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats
- CRL3KLHL21
cullin RING E3 ligase with the KLHL21 adaptor protein
- Cy5
cyanine dye 5
- deGradFP
proteasomal degradation of GFP fusions
- Dha
dehydroalanine
- diGly
lys‐ϵ‐Gly‐Gly
- dSTORM
direct stochastic optical reconstruction microscopy
- DUB
deubiquitinating enzyme
- E1
ubiquitin‐activating enzyme
- E2
ubiquitin‐conjugating enzyme
- E3
ubiquitin protein ligase
- E6AP
human papillomavirus E6‐associated protein
- FC(C)S
fluorescence (cross) correlation spectroscopy
- FCCP
carbonyl cyanide‐4 (trifluoromethoxy) phenylhydrazone
- FK1
anti‐ubiquitin antibody FK1
- FK2
anti‐ubiquitin antibody FK2
- FRAP
fluorescence recovery after photobleaching
- FRET
Förster resonance energy transfer
- FUCCI
fluorescence ubiquitination cell cycle indicator
- GFP
green fluorescent protein
- GFPu
16‐residue CL1 degron fused to GFP
- HECT
homologous to the E6‐AP carboxyl terminus
- HOIL1
RanBP‐type and C3HC4‐type zinc finger‐containing protein 1
- HUWE1
HECT, UBA and WWE domain‐containing protein 1
- IĸBα
NF‐kappa‐B inhibitor alpha
- IKK
inhibitor of nuclear factor kappa‐B kinase
- INT‐Ub.7KR
lysine‐less, internally tagged ubiquitin
- ISG15
interferon stimulated gene 16
- KG
Kusabira‐Green
- K
lysine
- LRSAM1
E3 ubiquitin‐protein ligase leucine‐rich repeat and sterile alpha motif‐containing protein 1
- LUBAC
linear ubiquitin chain assembly complex
- M(et)
methionine
- M1‐SUB
Met1‐linkage‐specific Ub‐binder
- mKO2
monomeric Kusabira‐Orange 2
- NEDD8
neural precursor cell expressed, developmentally down‐regulated 8
- NEMO
NF‐kappa‐B essential modulator
- NF‐ĸB
nuclear factor NF‐kappa‐B
- nm
nanometre
- NSlmb‐vhhGFP4
F‐box‐anti‐GFP nanobody fusion protein
- N‐terminal
amino‐terminal
- NZF
Npl4 zinc finger
- OPTN
optineurin
- OTULIN
OTU domain‐containing deubiquitinase with linear linkage specificity
- OUT
orthogonal Ub transfer (OUT) method
- p65/RelA
nuclear factor NF‐kappa‐B p65 subunit
- PAGFP
photoactivatable GFP
- PAINT
point accumulation for imaging in nanoscale topography
- PALM
photoactivation localization microscopy
- PARKIN
E3 ubiquitin‐protein ligase parkin
- PINK1
serine/threonine‐protein kinase PINK1, mitochondrial
- PML
promyelocytic leukaemia protein
- PolyUb‐FC
polyubiquitin‐mediated fluorescence complementation
- PROTACs
proteolysis‐targeting chimeric molecules
- PSF
point‐spread function
- Pup
prokaryotic ubiquitin‐like protein
- RBR
RING‐in‐between‐RING
- RFP
red fluorescent protein
- RING
really interesting new gene
- SCF
SKP1‐CUL1‐F‐Box
- SCV
salmonella‐containing vacuole
- SIM
structured illumination microscopy
- SLBP
histone RNA hairpin‐binding protein
- SMLM
single‐molecule localization microscopy
- Smurf2
E3 ubiquitin‐protein ligase SMAD ubiquitination regulatory factor 2
- SQSTM1/p62
sequestosome‐1
- SRM
super‐resolution microscopy
- STED
stimulated emission depletion
- SUMO
small ubiquitin‐like modifier
- TAB 2
TGF‐beta‐activated kinase 1 and MAP3K7‐binding protein 2
- TNFR1
tumour necrosis factor receptor superfamily member 1A
- TOM20
mitochondrial import receptor subunit TOM20 homolog
- TUBE
tandem ubiquitin‐binding entity
- UBact
ubiquitin bacterial
- UBAIT
ubiquitin‐activated interaction trap
- UBAN
ubiquitin‐binding in ABIN and NEMO
- UbDha
Ub‐dehydroalanine
- UBD
ubiquitin‐binding domain
- UBE1
ubiquitin‐activating enzyme 1
- UBE2J2
ubiquitin‐conjugating enzyme J2
- UblA‐MS
ubiquitin interactor affinity enrichment‐mass spectrometry
- UBL
ubiquitin‐like
- Ub‐ProT
ubiquitin chain protection from trypsinization
- Ub
ubiquitin
- UCHL3
ubiquitin carboxyl‐terminal hydrolase isozyme L3
- UiFC
ubiquitination‐induced fluorescence complementation
- UIM
ubiquitin‐interacting motif
- UPS
ubiquitin proteasome system
- USP30
ubiquitin carboxyl‐terminal hydrolase 30
- VHH
single‐domain antibody fragments
- VPS27
vacuolar protein sorting‐associated protein 27
- xE1, xE2 and xE3
engineered E1, E2 and E3 OUT enzymes
- YFP
yellow fluorescent protein
Ubiquitination
Major parts of eukaryotic proteomes are controlled and regulated by post‐translational modifications and among the most prominent is the covalent modification with the strictly conserved protein ubiquitin (Ub) (ubiquitination or ubiquitylation). Originally identified as a trigger for protein degradation by the 26S proteasome, ubiquitination serves many more proteasome‐independent functions, including signal transduction and selective autophagy 1, 2, 3. Ubiquitination thus influences and controls the majority of cellular processes and is implicated in a wide variety of pathophysiological states and diseases, ranging from cancer to infections and hereditary disorders 4.
Ubiquitin is a small, 76‐residue regulatory protein that is universally expressed in eukaryotic organisms 2, 3. Ubiquitin is a member of the ubiquitin‐like (UBL) protein family and shares sequence and structural homology with proteins like Small Ubiquitin‐like Modifier (SUMO) 5, 6, Interferon Stimulated Gene 15 (ISG15) 7 and Neural precursor cell Expressed, Developmentally Down‐regulated 8 (NEDD8) 8. Although for many years believed to be strictly expressed in eukaryotes, ubiquitin‐like proteins with some similarities in structure and conjugation systems have now been identified in Mycobacterium tuberculosis (Pup: prokaryotic ubiquitin‐like protein) 9 and in some Gram‐negative bacteria (UBact: Ubiquitin Bacterial) 10.
Ubiquitination is mediated by the sequential action of an ubiquitin‐activating enzyme (E1), an ubiquitin‐conjugating enzyme (E2) and an ubiquitin protein ligase (E3) (Fig 1A and B) 3, 11, 12, 13, 14. The substrate can be modified with a single ubiquitin (mono‐ubiquitination) or with polymeric Ub chains. Depending on which internal lysine (K6, K11, K27, K29, K33, K48, K63) or whether the N‐terminal methionine residue (M1, linear or head‐to‐tail chains) of Ub is used for linkage to the distal Ub different chain types can be generated (Fig 1C and D; Box 1) 3, 15, 16. To add complexity, the differential use of Ub lysine residues can generate homotypic chains (linked through one type of residues) or heterotypic or branched chains, such as K63‐linear and K48‐K11 hybrid polymers, respectively 17, 18. Importantly, the type of ubiquitin signal determines the biological effects of these modifications; for example, K48 and heterotypic K11/K48 chains generally target substrates for degradation by the 26S proteasome. In contrast, chains linked through other residues, like K6, K27, K33, K63 and linear ubiquitin chains, are often involved in non‐degradative purposes, like selective autophagy, DNA damage repair and innate immunity 3. This information is decoded by proteins containing ubiquitin‐binding domains (UBDs) that recognize chain‐specific residues exposed on proximal and distal ubiquitin molecules and within the linker regions connecting two ubiquitin molecules (Fig 1B) 19, 20, 21, 22. Deubiquitinating enzymes (DUBs) counterbalance chain‐growing capacities by removing ubiquitin modifications (Fig 1B) 23, 24. The concerted interplay of chain/linkage formation, recognition by UBDs and Ub hydrolysis creates dynamic networks that control the distribution of different ubiquitin signals, which in turn regulate a plethora of biological processes within the cell.
Figure 1. The complexity of ubiquitin conjugation.
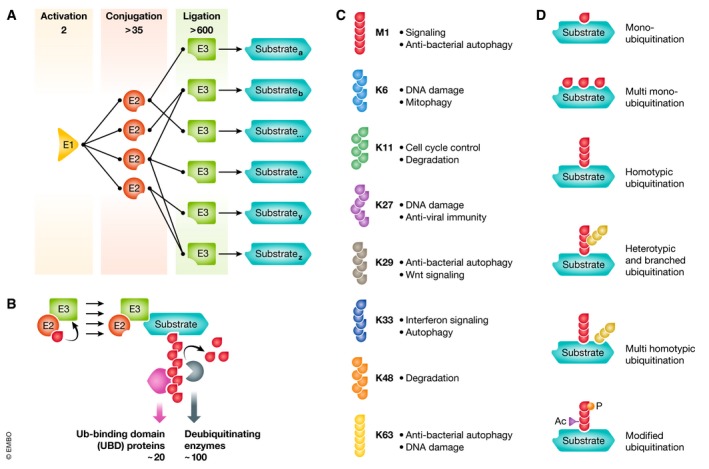
(A) Schematic representation of the abundance and interactions of human ubiquitin‐activating enzyme (E1s), ubiquitin‐conjugating enzymes (E2s) and ubiquitin protein ligases (E3s) involved in ubiquitination. (B) E3 ubiquitin‐protein ligases (like for example RING E3s) recruit ubiquitin‐loaded E2 enzymes and substrates and mediate the formation of ubiquitin chains. These chains can be recognized by ubiquitin‐binding domain (UBD) proteins and/or degraded by deubiquitinating enzymes in a chain‐selective manner. (C) The repertoire of ubiquitin chains, linked through methionine (M) 1 (linear/head‐to‐tail) or through the internal lysine (K) residues 6, 11, 27, 29, 33, 48 and 63 with a short description of their cellular function. (D) Overview of several modes of substrate ubiquitination including different forms of mono‐ and polyubiquitination and the post‐translational modification of ubiquitin itself by acetylation (Ac) and phosphorylation (P).
Box 1: Ubiquitin mutants and derivatives for microscopic analysis of cellular ubiquitination.
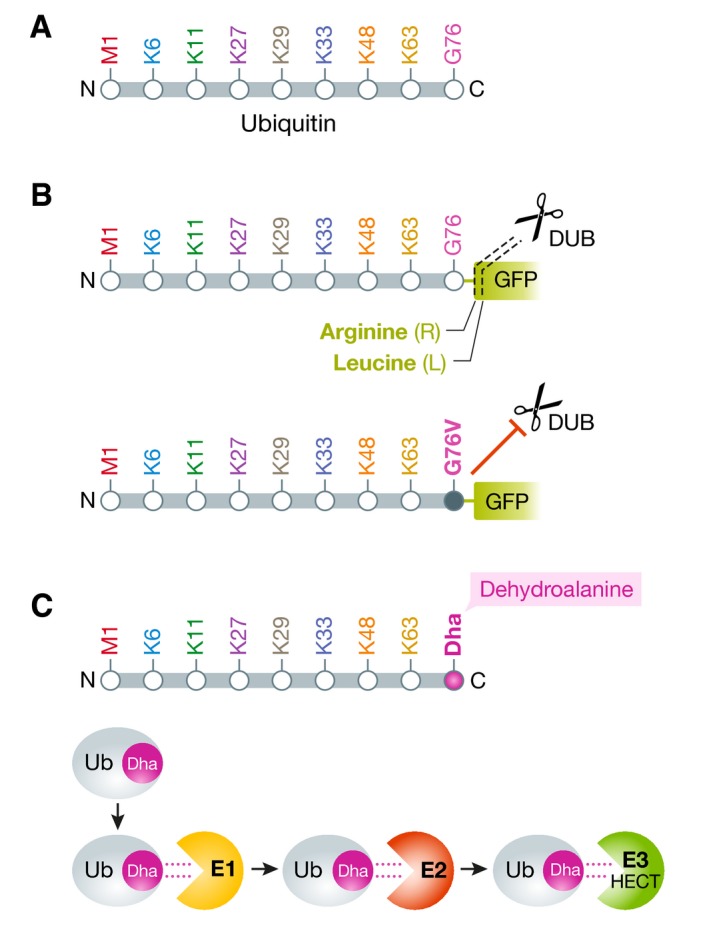
Schematic representation of the ubiquitin molecule. (A) Depicted are the N‐ and C‐termini, the initiator methionine (M1) for linear ubiquitination, the seven internal lysine residues and the C‐terminal glycine‐76. (B) Two exemplary ubiquitin‐green fluorescent protein (GFP) fusion protein reporters, used to image ubiquitin/proteasome‐dependent proteolysis and the degradative functions of ubiquitin. DUB‐mediated cleavage of ubiquitin‐(R)‐GFP or ubiquitin‐(L)‐GFP give rise to GFP molecules with arginine or leucine at the N‐terminus that determine the half‐lives of the GFP molecules by the N‐end rule pathway (upper). The deubiquitinating enzyme (DUB)‐resistant ubiquitin G76V mutant becomes modified with ubiquitin chains that mediate subsequent proteasomal degradation of the reporter, leading to a decrease in GFP signals (lower). (C) Ubiquitin derivatives with dehydroalanine (Dha) at position 76 can be used as cascade probes to investigate the cellular paths of ubiquitin, including the E1, E2 and HECT E3 ligase (see text for info).
Methods to study ubiquitination
Ever since its discovery, biochemistry‐ and imaging‐based approaches to study ubiquitination are continuously evolving, improving and adapting. In recent years, ubiquitin biochemistry has profited from the identification of novel key enzymes, new heterotypic/branched chain types and novel pathways relying on ubiquitin. Imaging‐based approaches are now more and more complementing biochemical methods due to novel developments and applications in reagents to visualize ubiquitin chains with confocal and super‐resolution microscopy.
Biochemical approaches to study ubiquitination
Ubiquitination is classically studied by resolving ubiquitin chains and/or ubiquitinated substrates on Western blot, and biochemical experiments are the method of choice for substrate identification. Antibody‐/affinity reagent‐based substrate/chain enrichment allows mass spectrometry to further discover novel aspects of ubiquitination. Since these methods have been extensively reviewed elsewhere (see, for example, the reviews of 25 and 26), here we only want to highlight some of the most useful tools that were developed in recent years. In particular, the antibody‐based enrichment of Ub Gly‐Gly‐Lysine substrate peptides upon trypsinization ((diGly) ubiquitin remnant proteomics) has enabled powerful and versatile substrate identification in complex biological specimens 26. Furthermore, the development of tandem ubiquitin‐binding entities (TUBEs), in which one or multiple UBDs are fused, has proven to be powerful for chain enrichment and substrate identification 27, 28, 29. Since then, application‐specific TUBE adjustments have been introduced, such as ubiquitin chain protection from trypsinization (Ub‐ProT) to determine ubiquitin chain length 30 and sensor‐based chain‐specific TUBEs, like the linear ubiquitin‐specific M1‐specific ubiquitin binder (SUB) 31. Interestingly, TUBE‐like chain‐binding sensors are used in cellular imaging‐based experiments as well (discussed in more detail later). Genetic trapping approaches like ubiquitin ligase trapping 32, 33 and ubiquitin‐activated interaction traps (UBAITs) further facilitated substrate identification 34. Additionally, orthogonal Ub transfer (OUT) relies on the expression of engineered E1, E2 and E3 enzymes (xE1, xE2 and xE3) that possess reactivity towards an affinity tagged ubiquitin mutant (xUb), but not to endogenous Ub, leading to the identification of selective substrates 35, 36. The applicability of this elegant approach has been demonstrated successfully by the identification of novel E6AP and CHIP E3 ligase substrates 37, 38. Although these approaches have not been adapted to microscopy‐based settings yet, it should theoretically be possible to image substrate ubiquitination with known, tagged E3 ligase and substrate pairs and labelled ubiquitin. Finally, chemical biology, combined with structural information, has yielded a wealth of activity‐based probes that can be used to manipulate and study key enzymes in ubiquitin research (see for an overview for example 39, 40, 41). Chemical and semi‐chemical probes have been developed that target E1, E2 and E3 enzymes and E3‐substrate interactions 42, 43, 44, DUBs 45, 46, 47, 48, UBDs 49 or can be applied for the induction of protein degradation, such as proteolysis‐targeting chimeras (PROTACs) 50, 51, 52, 53, 54.
In conclusion, biochemistry‐based methods are ideally suited for substrate identification, the verification of chain specificity, the differentiation between mono‐ and polyubiquitination and can be applied on a wide range of biological materials, ranging from in vitro ubiquitination reactions, cellular lysates to whole tissues and organisms. However, biochemical measurements often occur post‐lysis and can potentially increase the incidence of artefacts. Moreover, protein interactions might be too weak to be detected by immunoprecipitation and Western blotting. Furthermore, restriction of Ub reactions to specific cellular compartments or subsets of targets often require cell fractionation to enrich specific substrates or chain types. Scaling‐up to high‐throughput or high‐content settings is also difficult to achieve and provides limited spatial‐temporal resolution (Table 1).
Table 1.
Comparative advantages and disadvantages of biochemistry‐ and imaging‐based approaches to study cellular aspects of ubiquitination
| Advantages | Disadvantages | |
|---|---|---|
| Biochemistry‐based |
|
|
| Imaging‐based |
|
|
Strategies to monitor ubiquitination in mammalian cells using microscopy
Complementing the abovementioned approaches, recent developments in optical microscopy have opened the door to image, visualize and trace ubiquitin‐related processes directly in native and live‐cell settings. In particular, the development of microscopy techniques that achieve a spatial resolution approaching the size of single proteins allows functional studies on how proteins organize and interact at the molecular level in their physiological environment. In the following paragraphs, we highlight three important strategies to image cellular ubiquitination and discuss specific advantages and disadvantages. The first approach utilizes fluorescently labelled reporter and model substrates to indirectly image the degradative functions of ubiquitination and the UPS. The second approach applies tagged ubiquitin reagents, chain‐specific antibodies and chain‐specific sensors to directly image ubiquitination in cellular compartments and biological processes. Finally, the application of super‐resolution microscopy allows to image ubiquitin signals in mammalian cells with unprecedented spatial resolution.
Indirect imaging of ubiquitination in protein degradation
The role of ubiquitination in proteasomal degradation has been extensively studied by imaging the stability and degradation of artificial reporter proteins and physiological model substrates. Certain ubiquitin signals like ubiquitin chains linked through K11 and K48 serve as recognition signals for degradation by the 26S proteasome 2, 17, 55. Fluorescently labelled, degradation‐sensitive reporters have been developed, often based on green fluorescent protein (GFP) or derivatives, which are stabilized or degraded when expressed in isolated cells or intact organisms (Fig 2A) 56, 57. Monitoring changes in fluorescence intensity serves as indirect read‐out for ubiquitination and proteasome function and has for example facilitated the development and evaluation of proteasome inhibiting compounds 56, 57.
Figure 2. Indirect imaging of the degradative functions of ubiquitin.
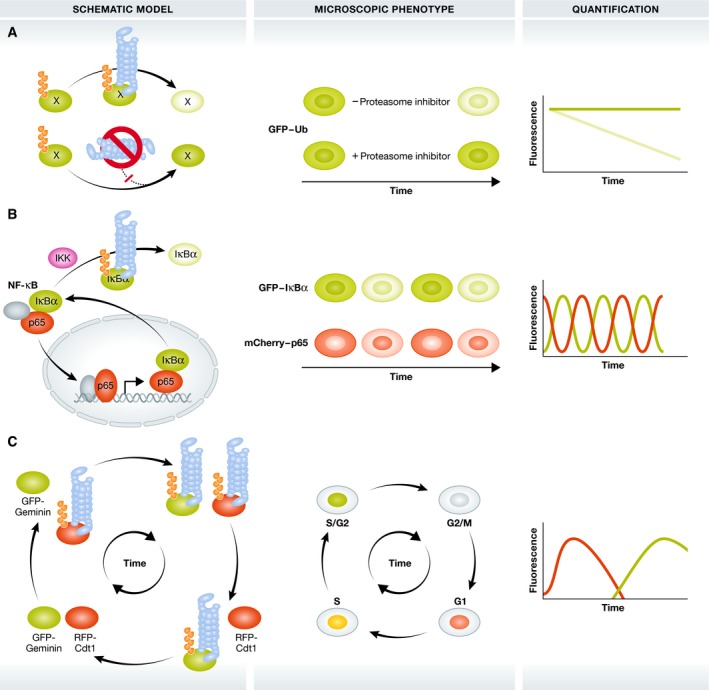
Approaches to image the proteasome‐related functions of Ub in the control of protein stability and breakdown. Darker and lighter green colours indicate the accumulation and breakdown of proteins, respectively. (A) GFP‐labelled Ub or model substrates are modified with degradative Ub signals and degraded by the 26S proteasome (blue barrel). Proteasome inhibition induces stabilization and accumulation of these GFP reporters as ubiquitinated forms. (B) Upon IKK activation, GFP‐tagged IκBα becomes modified with K48‐linked polyubiquitin chains and degraded by the 26S proteasome. This releases mCherry‐RelA/p65 that subsequently translocates in the nucleus to control NF‐κB‐dependent gene expression. IĸBα is among these NF‐κB target genes and shuttles back into the cytosol, creating dynamic NF‐κB degradation/translocation loops. Grey ovals: additional NF‐κB transcription factors (C) FUCCI: co‐expression of the UPS substrates GFP‐Geminin and RFP‐Cdt1 allows microscopic analysis of cell cycle phases by phase‐dependent Ub‐dependent degradation of GFP‐Geminin and RFP‐Cdt1.
One of the first GFP‐based UPS reporter (GFPu) was generated by fusing GFP to the 16‐residue CL1 degron that becomes degraded by the 26S proteasome in an ubiquitin‐dependent manner 58, 59. This GFP‐based reporter has been applied to indirectly study UPS function in isolated cells and intact organisms and can be used to investigate protein degradation in specific cellular compartments, like cytosol and the nucleus 60. Another reporter that probes the activity of DUBs consists of a direct fusion of ubiquitin to GFP. DUB‐dependent cleavage of the ubiquitin molecule generates free GFP with different N‐terminal residues that serve as indicators for N‐end rule pathways degradation since the N‐terminal residue determines the protein half‐live 61, 62. In addition, the non‐cleavable UbG76V‐GFP and –Dendra2 fusion reporters become modified with polyubiquitin chains that subsequently target the complete fusion protein for 26S proteasomal degradation (Fig 2A) and (Box 1) 61, 63, 64. These reporters have been applied in isolated cells and transgenic UbG76V‐GFP reporter mice to study proteasome activity 64.
The above listed reporters either employ artificial substrates or indirectly monitor DUB and proteasome activity. An alternative approach is the direct fluorescent labelling of the physiological ubiquitin substrate itself. This strategy has been applied to NF‐κB signalling (Fig 2B). Under basal conditions, the transcription factor p65/RelA is retained in the cytoplasm by its interaction with IκBα. Pathway activation induces IKK‐dependent IκBα phosphorylation and its ubiquitin‐dependent degradation leading to the liberation and nuclear translocation of p65 65. The co‐imaging of the nucleo‐cytoplasmic shuttling of GFP‐labelled p65 with the oscillatory accumulation/degradation of mCherry‐tagged IĸBα protein has allowed to draw conclusions on NF‐κB kinetics, dynamics and oscillations 66, 67, 68.
Another prominent example is the cell cycle indicator FUCCI (fluorescence ubiquitination cell cycle indicator). Here, two proteins known to be degraded in specific phases of the cell cycle are tagged with two different fluorescent reporters: RFP‐Cdt1 and GFP‐Geminin (Fig 2C) 69. During S, G2 and M phases of the cell cycle, RFP‐Cdt is degraded by the UPS, giving rise to GFP‐positive nuclei, whereas GFP‐tagged geminin is degraded in G1 with the red signal remaining. The G1/S transition phase shows yellow fluorescent nuclei, due to the green and red overlay, since Cdt1 levels are decreasing and geminin levels increase 69. A variant of FUCCI, called FUCCI4, combines four different fluorescent substrates (mKO2‐Cdt (30–120), mTurquoise2‐SLBP (18–126), Clover‐Geminin (1–110) and H1.0‐Maroon) to allow visualization of each cell cycle phase 70. In addition, Fly‐FUCCI has been developed to monitor proliferation in tissues, based on the FUCCI principle 71. The FUCCI principle allows thus a dynamic, indirect imaging of the degradative functions of ubiquitination during cell cycle progression and division.
An interesting development is the application of nanobodies. These are single‐chain VHH antibody regions specifically designed to recognize a specific epitope. These small, monomeric and stable reagents, derived from immunized Camelidae sp., can be labelled with fluorophores and expressed in cells 72, 73. Expression of these so‐called chromobodies can be achieved by conventional transient expression methods or through stable integration using viral transduction in a constitutive or inducible manner, depending on experimental constraints. Chromobodies against a wide variety of endogenous epitopes 72, 73 including GFP are available. GFP nanobodies can have GFP‐quenching or GFP‐stimulating properties 74 and can be genetically tagged to be used as biochemical matrices to enrich GFP‐tagged proteins 74. In an elegant approach, GFP nanobodies have been employed to achieve the selective (ubiquitin‐dependent) degradation of any GFP‐tagged protein of interest 75. The authors fused the Drosophila F‐box protein Slmb to GFP nanobodies (NSlmb‐vhhGFP4). Since F‐box proteins are the substrate specifying determinants of large multimeric E3 ligase complexes, called SKP1‐CUL1‐F‐Box (SCF) complexes 76, recognition of GFP‐tagged proteins by NSlmb‐vhhGFP4 induces their degradation by the SCF complex. The authors demonstrated the applicability of deGradFP (degrade green fluorescent protein) in isolated cells and intact organisms 75. It would be interesting to apply this GFP knockout technique on the degradation of GFP‐tagged ubiquitin chain‐specific sensors proteins, like GFP‐UBAN, that selectively bind linear Ub chains. Theoretically, one would expect proteasomal degradation of the GFP chain sensors and perhaps of the endogenous linear Ub chains as well. This would imply novel modes of manipulation of ubiquitin signalling that can easily be combined with imaging‐based experiments.
In conclusion, several reporters, methods and reagents have been developed and applied that are useful for studying Ub and protein breakdown by the 26S proteasome in imaging‐based set‐ups. Although these techniques do not allow direct imaging of the degradative Ub signals, the focus is on imaging the functional consequences of these signals on reporter stability. For this reason, this approach is only suitable for imaging degradative functions of Ub in well‐characterized biological scenarios that rely on known and well‐studied substrates, E3 ligases and molecular mechanisms. These reporter‐based read‐outs are less suitable for monitoring proteasome‐independent functions of ubiquitin and do not answer questions about the specific type of ubiquitin modification.
Direct imaging of ubiquitination in fixed and living cells
Microscopy‐based imaging of ubiquitination in cells is mostly focused on the visualization of local accumulations of different ubiquitin signals. In contrast to diffuse ubiquitination reactions, that might take place freely in certain organelles, accumulated ubiquitinated structures, like aggregates, foci or puncta, provide assemblies that can be imaged easily. These structures are in most cases ensembles of mixed types of ubiquitin chains, likely combined with (multiple) mono‐ubiquitination and/or branched chains. Two main approaches are currently in use to image ubiquitin, a direct one in which genetically labelled ubiquitin is used and an indirect one using reagents that recognize certain types of ubiquitin signals (Table 2).
Table 2.
Useful reagents for monitoring cellular ubiquitination using microscopy
| Application | Reagent | Advantages | Disadvantages |
|---|---|---|---|
| Degradative proteasomal functions of Ub in biological processes | GFP‐tagged wild‐type ubiquitin and mutants | Robust and applicable in intact animals | Transfection required Potential influence on Ub homeostasis Limited to degradative functions of the ubiquitin/proteasome system |
| GFP‐tagged physiological Ub substrates | Pathways often characterized in detail |
Transfection required Limited to degradative functions of the ubiquitin/proteasome system |
|
| Degron‐tagged nanobodies against GFP | Target flexibility |
Transfection required Limited to degradative functions of the ubiquitin/proteasome system |
|
| Visualization of Ub chains, substrates and processes in cellular structures and processes | Ectopic expression of tagged Ub |
Powerful in combination with biochemistry Ubiquitin mutants for chain characterization |
Transfection required Competition with endogenous Ub Potential influence on Ub homeostasis Tags at Ub might interfere with function Ectopic expression of Ub mutants might influence native chain patterns N‐terminal tagging interferes with linear chain formation |
| Ub chain sensors |
Chain selective Applicable in living cells Compatible with BiFC Potential influences of sensors of process of interest can be used to discover novel chain functions |
Transfection required Prone to influencing process of interest by interfering with endogenous chain recognition Prone to high background signals Target binding potentially susceptible to post‐translational modification of Ub chains |
|
| Ub antibodies |
Easy to use in immunofluorescence Characterized in detail Compatible with library screening and affinity maturation and optimization |
Post‐fixation Only available against selected Ub chain targets |
|
| Ub affimers |
Versatile, especially in combination with library screening Small and physiochemically robust Compatible with genetic tagging and ectopic expression |
Post‐fixation | |
| Ub(Dha) derivatives |
Promising reagents for imaging Ub flux Compatible with live‐cell imaging and in principle with FRAP/FRET |
Electroporation required Optimization required |
The most straightforward way of direct imaging of ubiquitination includes the transient or stable overexpression of tagged or fluorescent labelled ubiquitin in mammalian cells 77. These ubiquitin molecules are recognized by the E1‐E2‐E3 machinery and incorporated into chains. Direct visualization of GFP or immunofluorescence with antibodies against specific tag proteins allows selective imaging. Although this method seems straightforward, a few points have to be considered. First, the ectopically expressed ubiquitin has to compete with the endogenous ubiquitin molecules for incorporation, so sufficient levels of (over)expression need to be achieved. However, ectopic expression of ubiquitin on itself might already affect cellular processes and the (de)ubiquitination balance. Second, it cannot be excluded that tags, especially bulky ones, like GFP, interfere with chain growth and accessibility by UBD proteins. Third, expression of labelled wild‐type ubiquitin does not provide information about the type of ubiquitin chain. To solve this, lysine‐to‐arginine mutation of the internal residues within ubiquitin can be used, but one needs to be cautious with interpreting the obtained results since the mutations might force the generation of specific chains and require sufficient levels of expression in order to function adequately 77. Finally, placing a tag at the N‐terminus of ubiquitin interferes with the ability to form linear chains 78. A strategy to overcome this negative effect of tagging the N‐terminus of ubiquitin is the recent development of lysine‐less, internally STREP II‐tagged ubiquitin (INT‐Ub.7KR) 78. INT‐Ub.7KR can be used as mono‐Ub or as part of linear Ub chains, but INT‐Ub.7KR is expected to be incorporated in virtually every chain type as terminal ubiquitin, so care must be taken concerning this compromised specificity. Up to now, INT‐Ub.7KR has only been applied in mass spectrometry, but internally tagged Ub should allow imaging ubiquitination with microscopy techniques.
Apart from tracing ubiquitin mutants and variants, expression of isolated, chain‐specific UBDs fused to GFP enable visualization and tracking of specific ubiquitin chains in cells (Fig 3). Several UBD‐based sensors have been generated and applied. Among the first is the GFP‐labelled, triple fused ubiquitin‐interacting motif (UIM) domain of VPS27 that specifically monitors K63‐linked chains in vitro and in cells 79. Other sensors use a single GFP‐tagged UBAN domain of NEMO to visualize M1 ubiquitination or the TAB2 Npl4 zinc finger (NZF) domain to detect K63‐linked chains 80. These entities allowed imaging of Ub signals in DNA damage responses, mitochondrial damage and pathogen invasion in fixed and living cells (Fig 3A and B) 79, 80. The current repertoire of UBD‐based sensors is limited to linear, K33, K48 and K63 chains. Recently, a global chain‐interaction screen using immobilized, non‐hydrolysable di‐Ub molecules covering the complete spectrum of homotypic linkages was performed using ubiquitin interactor affinity enrichment‐mass spectrometry (UblA‐MS) 81. This screen identified many putative interactors for Ub chains, like UCHL3 for K27 chains and ACTL8 as K6 interactor that could be further developed into chain‐specific sensors. Bimolecular fluorescence complementation (BiFC) represents an interesting alternative to the application of single or tandem fused UBDs. BiFC is based on the reconstitution of fluorescence of two non‐fluorescent fragments of a normally fluorescent protein, like Venus, a yellow fluorescent protein (YFP) variant 82. When proteins fused to these fragments come in close proximity, the two inert halves reconstitute a functional fluorescent protein 82. Ubiquitination‐induced fluorescence complementation (UiFC) is a variant of BiFC that has been used to image K48‐linked chains in cells. Three tandem K48‐specific UIM domains of epsin1 fused to the N‐ and C‐terminal fragments of Venus 83 indeed detected K48‐linked polyubiquitin chains in vitro and in fixed and living cells. The principle has been applied to image K48 ubiquitin chain dynamics in presynaptic assembly and differentiation 83, 84. In addition, direct fusion of certain ubiquitin mutants to two split fragments of the Kusabira‐Green (KG) fluorescent protein generated a platform (polyubiquitin‐mediated fluorescence complementation, polyUb‐FC) that allowed the imaging of K33 polyubiquitination chains and linked K33 chains to autophagy signalling 85. The specific advantage of this approach is that background fluorescence is strongly reduced, since the Kusabira‐Green protein only emits when the two Kusabira fragments are incorporated into a chain, i.e. as intact fluorescent protein.
Figure 3. Imaging ubiquitination in selective autophagy.
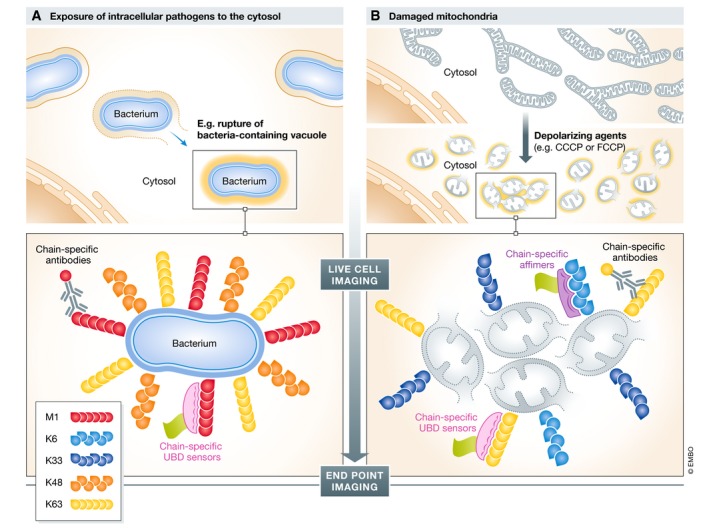
Schematic overview of live‐cell and end‐point Ub imaging applications in antibacterial autophagy (xenophagy) and autophagic breakdown of damaged mitochondria (mitophagy). (A) Intracellular pathogens, like Salmonella, reside mostly in Salmonella‐containing vacuoles. In some cases, vacuolar membrane rupture and pathogens are exposed to the host cytosol, inducing prominent multi‐type Ub deposition at the bacterial surface. These Ub structures serve as non‐self “eat me” signals that trigger bacterial autophagy. (B) Damaged mitochondria (grey circles), for example induced by depolarizing agents like CCCP/FCCP, trigger PINK1/PARKIN‐dependent ubiquitination at the surface of these organelles, thereby recruiting the autophagy machinery. In both examples, Ub chain‐specific antibodies, UBD‐based chain‐specific sensors and chain‐specific affimers were applied to understand the mechanisms and functional relevance of ubiquitination. Substantial overlap in similarities of Ub patterns exists in these two mechanistically distinct forms of ubiquitin‐accumulated structures and the contribution of Ub for selective autophagy.
Although ectopically expressed UBD‐based sensors have been useful in monitoring cellular ubiquitination in fixed and living cells, there are several aspects that need to be taken into account. First, due to their selective binding capacities, often combined with high affinities, sensors may act as interfering reagents that hinder access of endogenous UBD proteins and DUBs. This may lead to artificial stabilization of sensor‐bound chains or disturbances in Ub‐based signalling, especially concerning chain types that are less abundant. Furthermore, post‐translational modification of ubiquitin in chains and/or branched chain types might interfere with sensor recognition and binding. Finally, introduction of sensor molecules requires transfection procedures, which need to be optimized for individual cell types. On the other hand, these inhibitory characteristics can also be employed to resolve the contribution of certain ubiquitin linkages to biological processes, such as the significance of M1/K63‐linked chains for TNFR1‐mediated NF‐κB and interleukin signalling 79, 80. Ideally, the delivery of chain‐specific sensors should be done using inducible expression systems and careful sensor titrations combined with phenotypic assessments.
Besides the abovementioned approaches that rely on the ectopic expression of modified forms of ubiquitin or UBDs, general and chain‐specific ubiquitin antibodies have been developed that can be used for immunofluorescence 86, 87, 88, 89, 90. Although the majority of these antibodies were originally used for enrichment procedures using immunoprecipitation, some of these, like for example FK1 and FK2, do efficiently recognize ubiquitinated structures in fixed and permeabilized cells prepared for fluorescence microscopy 86, 91. The monoclonal antibody FK1 recognizes polyubiquitinated proteins but not monoubiquitinated or free ubiquitin. FK2 on the other hand recognizes both mono‐ and polyubiquitinated substrates 86, 91. Care must be taken since the binding properties of these antibodies have been validated using Western blotting with purified and isolated ubiquitin reagents and might depend on experimental settings, like immobilization of the antibody. Indeed, it has been shown recently that the FK2 monoclonal antibody exhibits some preference for certain linkages, at least in recognizing the eight types of di‐ubiquitin on Western blot 92.
To specifically detect linkage‐specific ubiquitin chains, phage display technologies combined with binding optimization have generated antibodies that selectively recognize linear, K11, K48, K63 and a bispecific antibody recognizing K11/K48 heterotypic chains 17, 87, 88, 89, 90. These antibodies have been applied successfully in immunoprecipitation experiments as well as confocal fluorescence and super‐resolution microscopy (see below) 89.
Affimers are novel and alternative chain‐specific reagents, often based on a 12‐kDa cystatin fold scaffold that contains an alpha‐helix and an anti‐parallel beta‐sheet connected through loop regions 93. These loop regions can be randomized and screened from phage display libraries to bind a protein of interest with high affinity and specificity 93. Affimers are generally much smaller as antibodies or isolated UBDs and are pH stable and thermally robust and allow versatile affimer applications as well as chemical and genetic modification and enable imaging experiments 94, 95. Ub chain‐specific affimers have been developed by screening of affimer variant libraries for binders of K6 and K33/K11 linkages 96, 97. Structure‐based optimization including dimerization improved the performance of these ubiquitin chain affimers and increased target chain affinities 97. K6‐specific affimers were used to study mechanistic aspects of K6 chain generation in a broad array of biochemical and cell biological approaches and led to the identification of HUWE1 as K6‐specific E3 ligase in cells 97. In addition, chemical labelling of K6 affimers with AlexaFluor488 allowed the detection of K6‐linked polyubiquitinated structures on damaged mitochondria and identified USP30 as DUB for K6‐ubiquitinated TOM20 by fluorescence microscopy (Fig 3B) 97. A major advantage of affimers as chain‐specific antibody‐like reagents compared to overexpression other ubiquitin‐binding entities is the lack of confounding inhibitory effects due to ectopic expression. At present affimers are not expressed in cells, but are being used as antibodies after specimen fixation. However, intracellular expression of chain‐specific affimers could potentially be used to understand the biological function of uncommon ubiquitin chains by competing for chain access with endogenous UBDs and/or DUBs. Recently, affimers against SUMO1 and SUMO2/3 have been described as well, highlighting the potential of this technology for other types of post‐translation modifications 98.
A promising contribution towards cellular imaging of Ub functions comes from a remarkable type of activity‐based probe, called UbDHa, that has been developed as synthetic cascading probe that enables monitoring of sequential E1, E2 and HECT‐type E3 ligase activity 43 (Box 1). Inspired by the observation that the G76A Ub variant still can be used by the E1‐E2‐E3 machinery, although with decreased efficiency 99, 100, UbDHA is composed of residues 1–75 of Ub G76A in which the C‐terminal alanine is replaced by dehydroalanine (Dha). Upon activation of UbDha by the E1 enzyme, Dha can either be covalently linked to the E1 enzyme in a E1‐UbDha‐thioether adduct or follow the native ligation pathway to E2 and E3 enzymes. At the level of E2 enzymes, UbDha can again form E2‐thioether adducts or travel further to active site cysteine E3 ligases, i.e. HECT and RBR‐type E3s. This probe has been validated thoroughly in vitro and in vivo 43. Intriguingly, Cy5‐labelled UbDha has been introduced in cells by electroporation together with ectopic expression of GFP‐tagged UBE1, the main mammalian E1 enzyme. Upon co‐expression with GFP‐UBE1, Cy5‐UbDha was found enriched within the nucleus, colocalizing with GFP‐UBE1. Furthermore, Cy5‐UbDha colocalized with the wild‐type GFP‐UBE2J2 E2 enzyme, but not with the catalytically inactive C91S mutant. In addition, this colocalization was dependent on the upstream E1, since E1 inhibition with PYR‐41 inhibited colocalization. The imaging‐based experiments of the UbDha probe highlight the possibilities of applying activity‐based probes for ubiquitination to measure ubiquitin “flux” in cells. Introducing these probes in living cells would allow Fluorescence Recovery After Photobleaching (FRAP)‐like experiments (see below). For this, probe fluorescence could be bleached in certain cellular regions of interest and the redistribution of fluorescence could be monitored in time and space and provide information on Ub dynamics in cells. Combining this analysis with measuring colocalization of specific UPS components, like GFP‐UBE1, this approach should enable measurement of UPS flux. At the moment, Cy5‐UbDha imaging is limited to co‐imaging the probe with GFP‐labelled UPS enzymes and more insights concerning the UbDha transfer efficiencies in cells would be needed. It would be extremely interesting to further test and develop probes to image ubiquitination in cells. However, one potential disadvantage of chemical probes is that these entities are introduced into cells using electroporation that potentially might influence ubiquitination and signalling cascades.
The dynamic interchange of ubiquitin has been studied using FRAP in combination with GFP‐ and photoactivatable GFP (paGFP)‐labelled Ub in different cellular compartments and with proteasome inhibition 101, disease‐associated protein aggregates 102, 103 and autophagy 104. Furthermore, Förster Resonance Energy Transfer (FRET) has been applied for studying example DUB selectivity 105, 106 and regulation of E3 function 107, although mostly applied in in vitro applications.
In conclusion, the continuous development of new and innovative reagents to image ubiquitination has clearly facilitated ubiquitin research. The application of structure‐guided optimization of new methodologies is expected to open new avenues that will further increase our understanding of ubiquitination in cell biology. One of the major remaining problems is that current conventional ways to image ubiquitination focus on imaging dense and accumulated ubiquitinated structures within cells. In other words, current techniques image larger ensembles of ubiquitin signals, including multiple substrates, often modified on multiple residues, modified with multiple types of homo‐ and heterotypic ubiquitin chains of different lengths and different stoichiometry. Diffraction‐limited optical microscopy provides insufficient spatial resolution to uncover molecular details concerning ubiquitination in cells, and therefore, the application of super‐resolution microscopy to study ubiquitination might be a promising approach to overcome these problems 108.
Super‐resolution imaging of ubiquitination
Super‐resolution microscopy (SRM) methods can provide unprecedented levels of spatial resolution into the distribution of biological molecules and processes, such as the molecular architecture of protein complexes, membrane nanostructures and protein aggregates 108. Although the majority of ubiquitin imaging applications use conventional fluorescence microscopy, ectopic expression of labelled ubiquitin and antibody staining of ubiquitin chains is ideally suitable for SRM. While the spatial resolution in conventional fluorescence microscopy is restricted to about 200 nm laterally and 500 nm axially, common SRM techniques reach a spatial resolution of 10–20 nm, thus approaching the size of proteins. In addition, single‐molecule super‐resolution methods can provide quantitative information on protein copy numbers and protein complex stoichiometry. The repertoire of super‐resolution methodologies, technologies and applications is growing and was comprehensively reviewed before 108, 109, 110.
At present, SRM methods that have been successfully used to image ubiquitination are patterned light illuminated techniques, such as structured illumination microscopy (SIM) 111 and localization‐based technologies, like direct stochastic optical reconstruction microscopy (dSTORM) 112 and photoactivated localization microscopy (PALM) 113 (Box 2). Apart from these, stimulated emission depletion (STED) microscopy 114 and (lattice) light‐sheet microscopy 115 can in principle also be used for ubiquitin SRM imaging. SIM uses a finely stripped, patterned illumination (structured illumination) to image a sample, which will generate interference patterns (moiré patterns). These patterns are coarser than the original and can be detected by optical microscopes. SIM captures these patterns with slightly varying the orientation of the structured illumination and processes these images to mathematically reconstruct a high‐resolution structure of the specimen. dSTORM and PALM are examples of single‐molecule localization microscopy (SMLM) and rely on sequential activation and localization of only a subset of fluorophores within a sample. The exact fluorophore position can be determined by finding the centre of the point‐spread function of individual fluorophores. By iterative cycles of activation and deactivation, a super‐resolution image can be constructed 116. SMLM can be realized with organic fluorophores or with photoconvertible fluorescent proteins. An alternative strategy to achieve labelling of a subset of targets is to use reversible duplex formation of DNA, as in point accumulation for imaging in nanoscale topography (DNA‐PAINT) 117, 118. SMLM has been applied for three‐dimensional imaging, for example by using optical astigmatism in which an elliptical shape of the point‐spread function (PSF) provides information concerning the x‐y‐z directions of the fluorophores 119.
Box 2: Three concepts of optical super‐resolution microscopy.
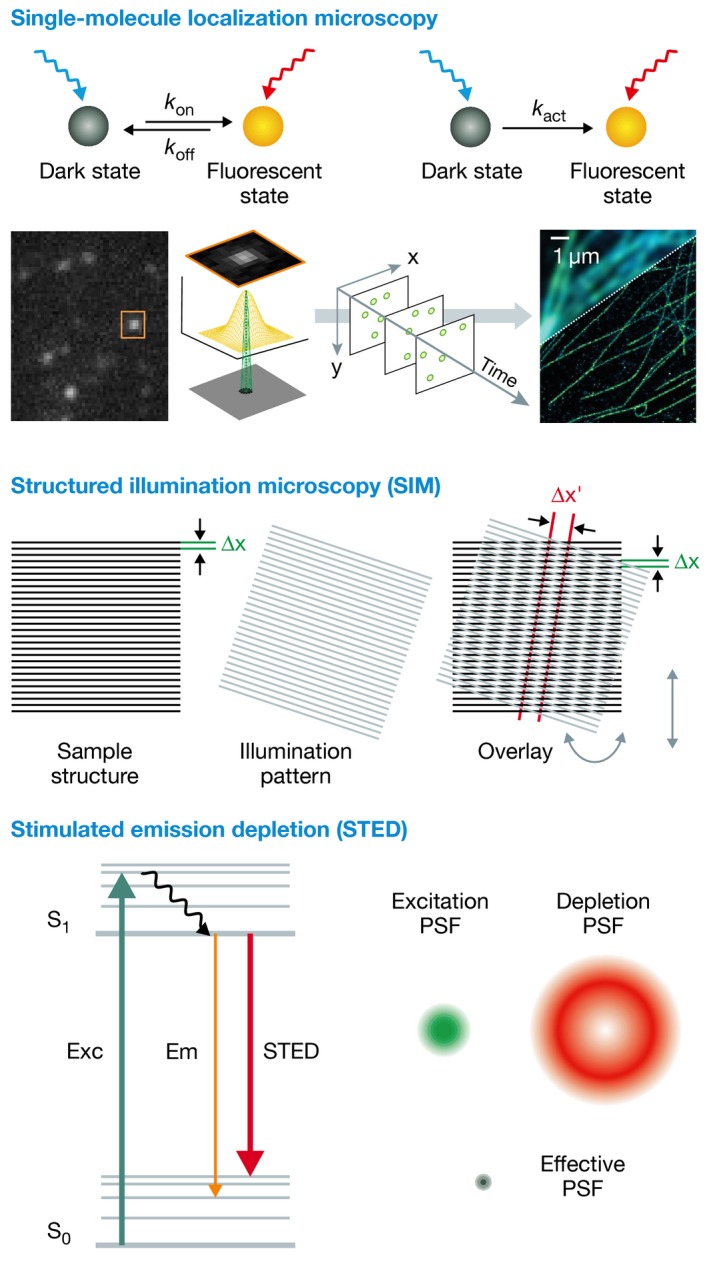
Three concepts of optical super‐resolution microscopy. In single‐molecule localization microscopy (SMLM), an image is generated by determining the precise position of single fluorophore labels. This requires a separation of fluorophores during imaging, which is either achieved by photoswitching/photoactivation, or by fluorophore labels that transiently bind to a target. In structured illumination microscopy (SIM), a sample is illuminated with a periodic pattern, generating moiré fringes. From various translations and rotations of this pattern, a set of images is recorded, and a high‐resolution image is reconstructed computationally. Stimulated emission depletion (STED) employs targeted switching of fluorescence achieved by overlaying an excitation laser with a doughnut‐shaped (null intensity in the centre) de‐excitation beam, generating a sub‐diffraction sized, effective excitation spot. Figures were adapted from 153 and 154.
Multi‐wavelength super‐resolution imaging of ubiquitination as well as SRM of living cells is still challenging. As for all microscopy‐based approaches to study ubiquitination, SRM is dependent on the level of fluorescence intensity compared to background to allow proper image acquisition and detection. Ubiquitin antibodies are generally working well for immunofluorescence, but if the ubiquitin structures of interest are less abundant, adequate detection might be hampered. This becomes especially valid when there is insufficient contrast between the region of interest and adjacent structures. Therefore, SRM‐based techniques are ideally suited for imaging ubiquitination at accumulated structures, like aggregates, organelles or invading pathogens. This is illustrated by 3D SIM imaging of the SUMO 2/3 distribution at arsenic‐induced PML nuclear bodies that provide information on how PML becomes degraded in an ubiquitin‐ and SUMO‐dependent manner 120. Another example is the application of single‐molecule localization microscopy to study the localization of E3 ligases, like Smurf2, STUB1/CHIP, Cullin RING E3 ligase with the KLHL21 adaptor protein (CRL3KLHL21), proteasome storage granules in yeast and proteins closely colocalizing with general and chain‐specific ubiquitin signals 121, 122, 123, 124, 125. Therefore, dense structures and ubiquitination occurring at the surfaces of these structures serve as excellent platforms to image ubiquitin homeostasis and to understand the functions of E3 ligases and DUBs.
Imaging ubiquitination in bacterial autophagy and pro‐inflammatory signalling
The surface of intracellular bacteria, such as Salmonella, is a prototypic example of such dense structures and provides an ideal model system to image many aspects of ubiquitination. Exposure to pathogenic bacteria requires host defence strategies to restrict bacterial proliferation and spread. Salmonella enterica subs. enterica Typhimurium (hereafter referred to as Salmonella) invade host cells by inducing rearrangements in the actin network and the cytoskeleton 126. Most intracellular bacteria reside and replicate in membrane‐derived, dedicated Salmonella‐containing vacuoles (SCVs) that provide shielding against host immune surveillances 126. Due to the nutritional status of the mammalian cytosol, certain subpopulations of vacuolar Salmonella escape and translocate into the cytoplasm 127. These bacteria are covered with dense structures of ubiquitin 127, 128, 129. Salmonella Ub coats act as potent stimulator of bacterial clearance through selective autophagy by recruiting autophagy receptors like SQSTM1/p62, CALCOCO2/NDP52 and optineurin (OPTN) 130, 131, 132. These proteins mediate the formation of double‐membraned autophagosomes that target bacteria for lysosomal destruction in a process called xenophagy 129.
Ubiquitinated intracellular bacteria provide excellent models to image ubiquitin homeostasis for several reasons. First, bacteria are rather large and can be very easily imaged with confocal fluorescence microscopy 133. Second, the onset of ubiquitination is timed and only occurs when bacteria are exposed to the host cytosol, which is relatively well described for Salmonella 128. Third, antibodies against bacterial cell wall markers and immunofluorescent markers for the autophagic machinery are available that allow co‐staining with fluorescent bacteria. Fourth, multiple types of ubiquitin chains can be found in the Salmonella ubiquitin coat. For example, immunofluorescence staining of Salmonella‐infected human epithelial cells with FK1, FK2 and chain‐specific antibodies and imaging of the recruitment of GFP‐tagged linear and K63‐specific UBDs expressed in infected cells confirmed the presence of linear, K11‐, K63‐ and K48‐linked chains around the cytosolic Salmonella 80, 92, 128, 133, 134, 135. These approaches contributed to the identification and characterization of key E3 ligases, like LUBAC 92, PARKIN 135, LRSAM1 136 and ARIH1 137, of the DUB OTULIN 138 and several UBD proteins that recruit the autophagy machinery (like SQSTM1/p62, NDP52 and OPTN) 130, 131, 132, 139, 140. In addition, multiscale imaging has been applied to explore the role of Ub in xenophagy of other intracellular microorganisms, like Shigella and Mycobacterium 135, 141 and even internalized latex beads that mimic cytosolic bacteria 142.
Recently, SIM and dSTORM imaging was performed on cytosolic Salmonella total, M1, K63 and K48 ubiquitin and revealed unprecedented levels of resolution 92, 138. SRM confirmed non‐uniform distributions of several of these ubiquitin structures within the coat in which some chain‐linkages accumulated in micro domain‐like structures 92, 138. In addition, SIM was also used to image the distribution of the autophagic adaptor proteins p62 and NDP52 on the type II Toxoplasma gondii parasitophorous vacuole 140. Quantitative dSTORM imaging of M1 signals in the Salmonella Ub coat revealed striking alterations in M1 antibody distribution and intensity in infected OTULIN‐depleted cells, without affecting K63 Ub, suggesting OTULIN as crucial regulator of linear chain deposition within the ubiquitin coat (Fig 4A and B) 138. These quantitative measurements highlight the potential of applying SRM in monitoring ubiquitination in cells. At present, SRM cannot discriminate between changes in M1 chain lengths or the number of M1 chains within the bacterial Ub surface upon loss of OTULIN expression. Therefore, changes in M1 antibody intensity could indicate OTULIN‐dependent increases in the length of linear chains or M1 modification of novel substrates by LUBAC (Fig 4C).
Figure 4. Super‐resolution imaging‐based differentiation of novel roles for differentially linked polyUb in bacterial autophagy.
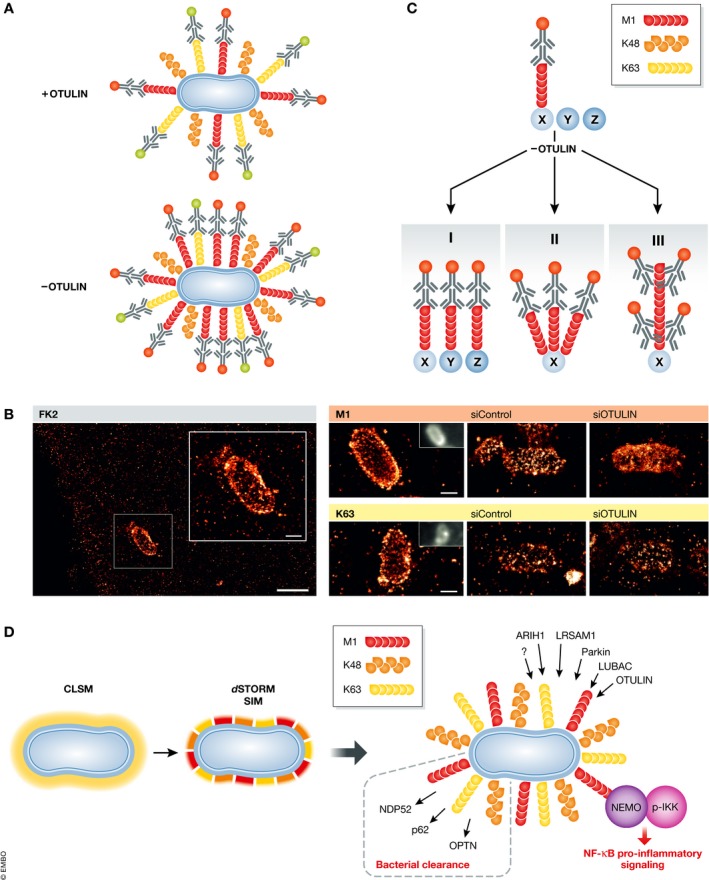
(A) Schematic depiction of dSTORM‐based imaging using chain‐specific Ub antibodies to analyse the role of differential ubiquitination on the surface of cytosolic Salmonella. Ubiquitinated cytosolic Salmonella (blue) labelled with Ub chain‐specific antibodies against M1 (red chains) and K63 (yellow chains) combined with secondary antibodies carrying dSTORM‐compatible fluorescent labels (red and green dots). Locating single, antibody‐associated fluorophores allows reconstruction of super‐resolution images and mathematical modelling of the fluorophore distribution and accumulation. (B) dSTORM imaging of the nanoscale distribution of total ubiquitin and chain‐specific ubiquitin patterns around cytosolic Salmonella in human epithelial cells reveals the importance of the linear deubiquitinating enzyme OTULIN for controlling the distribution of linear ubiquitin patterns at ubiquitin‐coated intracellular bacteria (reproduced with permission in adapted form from 138). (C) dSTORM SRM imaging‐based quantification detected an increased density and altered M1 distribution patterns on cytosolic Salmonella upon loss of the DUB OTULIN. This could indicate (I) modification of novel substrates Y and Z with M1 chains by LUBAC that are normally deubiquitinated by OTULIN, (II) more M1 chains on native substrate X that are removed in the presence of OTULIN or (III) longer M1 chains on native substrate X that are normally trimmed by OTULIN. (D) The higher spatial resolution of dSTORM and SIM imaging compared to confocal laser scanning microscopy (CLSM) reveals the spatial organization of different ubiquitin chains on the surface of Salmonella. NEMO is recruited to M1 chains at the bacterial surface resulting in the accumulation of phosphorylated IKK and the activation of NF‐κB pro‐survival signalling. See text for more details.
Modulating M1 homeostasis by altering LUBAC or OTULIN levels allowed visualizing of the balance between linear ubiquitin conjugation and deconjugation in cells 92, 138. Apart from mediating bacterial autophagy, increased M1 Ub also recruits NEMO, an M1‐specific UBD with well‐described roles in TNF‐mediated NF‐κB signalling 143, and the IKK complex 143, 144, 145, 146. Imaging IKK activation by quantifying IKK serine 176 and serine 180 phosphorylation using immunofluorescence at the bacterial Ub coat indicate local activation of canonical NF‐κB activation directly on the bacterial Ub surface (Fig 4D) 92, 138.
Taken together, these observations emphasize the usefulness and relevance of Ub CSLM and SRM to discover novel parallels between antibacterial autophagy and pro‐inflammatory signalling in host cell autonomous signalling. Further application and combination of Ub imaging with existing biochemical methodologies is expected to identify novel roles of ubiquitination at different levels of resolution.
Conclusions
In recent years, advances in microscopic methodologies to study ubiquitination contributed immensely to our understanding of the cellular roles of ubiquitin. Application and improvements of microscopic Ub imaging in a wide variety of physiologically relevant scenarios have increased our insights in this fundamental type of post‐translational modification. Conventional fluorescence microscopy and super‐resolution imaging emerge as valuable supplements for existing biochemical and mass‐spectrometric methodologies and open novel possibilities to image ubiquitination in real time in living cells. Multi‐disciplinary approaches to monitor ubiquitination will open new areas of ubiquitin research, especially quantitative cellular aspects with single‐molecule resolution. Ongoing developments in biochemical reagents and mass spectrometry‐assisted ubiquitinome profiling, CRISPR/Cas9 and label‐free proteomics already allowed to map ubiquitination sites and substrates in an organismal setting with unprecedented depth. Finding ways to combine reagents that specifically label Ub chains, based on recombinant affimers and chain‐selective binding moieties, is expected to further reveal important insights in cellular Ub homeostasis and functions.
Cellular imaging with super‐resolution will increase our understanding of functional aspects of ubiquitination. A broad repertoire of probes, UBD sensors, affimers and antibodies are available and compatible with various super‐resolution techniques. In order to answer fundamental questions in the ubiquitin research field, such as measuring chain length in cells or visualize substrate modification in native environments, quantitative methods will be beneficial, such as single‐molecule counting approaches 147, 148, 149 and advanced biophysical techniques, like fluorescence (cross) correlation spectroscopy (FC(C)S) 150. For live‐cell studies, novel approaches that employ exchangeable fluorophores allow for long‐term super‐resolution imaging with minimal photobleaching 151, 152.
Imaging ubiquitination with increased spatial resolution will unravel novel quantitative aspects of ubiquitination and ubiquitin chains in mammalian cells and permits quantitative measurements of ubiquitin chain parameters and associated protein networks in the future. Finally, current Ub imaging focus on larger ubiquitinated cellular structures, like protein aggregates, organelles and intracellular pathogens as ideal reaction platforms to study ubiquitin homeostasis, chain conformation and E3/DUB selectivity in cells. Application of multi‐level ubiquitin imaging approaches in structurally not well‐defined cellular compartments, like membranes or diffuse cytosolic ubiquitination reactions, will be a major future challenge for ubiquitin research.
In need of answers.
Although many basic aspects of cellular ubiquitination have been elucidated in recent years, some central questions in the ubiquitin research field remain unanswered.
-
Development of labelling and microscopic tools to image the complete “ubiquitin code” with molecular resolution in cellular settings, especially:
Robust and versatile methods for real‐time analysis of chain formation and/or breakdown using sensors and FRET/FRAP analysis would be extremely useful in the analysis of certain E3/DUBs on cellular ubiquitination reactions.
Versatile chemical probes to specifically monitor the ubiquitin flux over E1, E2, E3 enzymes, DUBs or structural compartments would also contribute to our understanding of cellular ubiquitination events, especially those that can be applied in (live‐cell) imaging‐based applications.
What are the spatial molecular patterns of E3 ligases and DUBs and how is ubiquitin chain distribution and length controlled at the molecular level in a cell?
How is ubiquitin and substrate degradation organized by the 26S proteasome in cells at the molecular level?
How is the ubiquitin exchange and flux regulated on ubiquitinated surfaces of (dense) cellular structures?
What are the molecular mechanisms of diffuse ubiquitination reactions in not well‐defined compartments?
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
The authors acknowledge Christina Hugenberg for expert proofreading. This work was supported by grants from the DFG (SFB1177) to ID, SF and MH, BMBF to SF, LOEWE Research Initiative Network Ub‐Net to ID, SF, MH and SJLvW and DFG (WI 5171/1_1) to SJLvW.
EMBO Reports (2019) 20: e46520
See the Glossary for abbreviations used in this article.
Contributor Information
Sjoerd JL van Wijk, Email: s.wijk@kinderkrebsstiftung-frankfurt.de.
Mike Heilemann, Email: heilemann@chemie.uni-frankfurt.de.
References
- 1. Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Dikic I (2017) Proteasomal and autophagic degradation systems. Annu Rev Biochem 86: 193–224 [DOI] [PubMed] [Google Scholar]
- 3. Behrends C, Harper JW (2011) Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol 18: 520–528 [DOI] [PubMed] [Google Scholar]
- 4. Rape M (2017) Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol, 19: 59–70 [DOI] [PubMed] [Google Scholar]
- 5. Mahajan R, Delphin C, Guan T, Gerace L, Melchior F (1997) A small ubiquitin‐related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88: 97–107 [DOI] [PubMed] [Google Scholar]
- 6. Matunis MJ, Coutavas E, Blobel G (1996) A novel ubiquitin‐like modification modulates the partitioning of the Ran‐GTPase‐activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loeb KR, Haas AL (1992) The interferon‐inducible 15‐kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem 267: 7806–7813 [PubMed] [Google Scholar]
- 8. Kamitani T, Kito K, Nguyen HP, Yeh ET (1997) Characterization of NEDD8, a developmentally down‐regulated ubiquitin‐like protein. J Biol Chem 272: 28557–28562 [DOI] [PubMed] [Google Scholar]
- 9. Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH (2008) Ubiquitin‐like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322: 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lehmann G, Udasin RG, Livneh I, Ciechanover A (2017) Identification of UBact, a ubiquitin‐like protein, along with other homologous components of a conjugation system and the proteasome in different gram‐negative bacteria. Biochem Biophys Res Commun 483: 946–950 [DOI] [PubMed] [Google Scholar]
- 11. Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC (1999) The tyrosine kinase negative regulator c‐Cbl as a RING‐type, E2‐dependent ubiquitin‐protein ligase. Science 286: 309–312 [DOI] [PubMed] [Google Scholar]
- 12. Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP (1999) Structure of an E6AP‐UbcH7 complex: insights into ubiquitination by the E2‐E3 enzyme cascade. Science 286: 1321–1326 [DOI] [PubMed] [Google Scholar]
- 13. Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT (2007) The ring between ring fingers (RBR) protein family. Genome Biol 8: 209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schulman BA (2011) Twists and turns in ubiquitin‐like protein conjugation cascades. Protein Sci 20: 1941–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikeda F, Dikic I (2008) Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: beyond the Usual Suspects’ review series. EMBO Rep 9: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez‐Martin N, Matsumoto ML, Dixit VM et al (2017) Assembly and function of heterotypic ubiquitin chains in cell‐cycle and protein quality control. Cell 171: 918–933.e920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P (2013) Activation of the canonical IKK complex by K63/M1‐linked hybrid ubiquitin chains. Proc Natl Acad Sci USA 110: 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trempe JF (2011) Reading the ubiquitin postal code. Curr Opin Struct Biol 21: 792–801 [DOI] [PubMed] [Google Scholar]
- 20. Husnjak K, Dikic I (2012) Ubiquitin‐binding proteins: decoders of ubiquitin‐mediated cellular functions. Annu Rev Biochem 81: 291–322 [DOI] [PubMed] [Google Scholar]
- 21. Rahighi S, Dikic I (2012) Selectivity of the ubiquitin‐binding modules. FEBS Lett 586: 2705–2710 [DOI] [PubMed] [Google Scholar]
- 22. Randles L, Walters KJ (2012) Ubiquitin and its binding domains. Front Biosci 17: 2140–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mevissen TET, Komander D (2017) Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86: 159–192 [DOI] [PubMed] [Google Scholar]
- 24. Nijman SM, Luna‐Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- 25. Iconomou M, Saunders DN (2016) Systematic approaches to identify E3 ligase substrates. Biochem J 473: 4083–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ordureau A, Munch C, Harper JW (2015) Quantifying ubiquitin signaling. Mol Cell 58: 660–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoshida Y, Saeki Y, Murakami A, Kawawaki J, Tsuchiya H, Yoshihara H, Shindo M, Tanaka K (2015) A comprehensive method for detecting ubiquitinated substrates using TR‐TUBE. Proc Natl Acad Sci USA 112: 4630–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hjerpe R, Aillet F, Lopitz‐Otsoa F, Lang V, England P, Rodriguez MS (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin‐binding entities. EMBO Rep 10: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emmerich CH, Cohen P (2015) Optimising methods for the preservation, capture and identification of ubiquitin chains and ubiquitylated proteins by immunoblotting. Biochem Biophys Res Commun 466: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsuchiya H, Burana D, Ohtake F, Arai N, Kaiho A, Komada M, Tanaka K, Saeki Y (2018) Ub‐ProT reveals global length and composition of protein ubiquitylation in cells. Nat Commun 9: 524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd‐Hansen M, Krappmann D et al (2013) OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1‐linked polyubiquitin. Cell 153: 1312–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mark KG, Simonetta M, Maiolica A, Seller CA, Toczyski DP (2014) Ubiquitin ligase trapping identifies an SCF(Saf1) pathway targeting unprocessed vacuolar/lysosomal proteins. Mol Cell 53: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mark KG, Loveless TB, Toczyski DP (2016) Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase‐polyubiquitin‐binding domain fusions (ligase traps). Nat Protoc 11: 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Connor HF, Lyon N, Leung JW, Agarwal P, Swaim CD, Miller KM, Huibregtse JM (2015) Ubiquitin‐activated interaction traps (UBAITs) identify E3 ligase binding partners. EMBO Rep 16: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao B, Bhuripanyo K, Zhang K, Kiyokawa H, Schindelin H, Yin J (2012) Orthogonal ubiquitin transfer through engineered E1‐E2 cascades for protein ubiquitination. Chem Biol 19: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu X, Zhao B, Sun L, Bhuripanyo K, Wang Y, Bi Y, Davuluri RV, Duong DM, Nanavati D, Yin J et al (2017) Orthogonal ubiquitin transfer identifies ubiquitination substrates under differential control by the two ubiquitin activating enzymes. Nat Commun 8: 14286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y, Liu X, Zhou L, Duong D, Bhuripanyo K, Zhao B, Zhou H, Liu R, Bi Y, Kiyokawa H et al (2017) Identifying the ubiquitination targets of E6AP by orthogonal ubiquitin transfer. Nat Commun 8: 2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bhuripanyo K, Wang Y, Liu X, Zhou L, Liu R, Duong D, Zhao B, Bi Y, Zhou H, Chen G et al (2018) Identifying the substrate proteins of U‐box E3s E4B and CHIP by orthogonal ubiquitin transfer. Sci Adv 4: e1701393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Witting KF, Mulder MPC, Ovaa H (2017) Advancing our understanding of ubiquitination using the Ub‐Toolkit. J Mol Biol 429: 3388–3394 [DOI] [PubMed] [Google Scholar]
- 40. Ovaa H, Vertegaal ACO (2018) Probing ubiquitin and SUMO conjugation and deconjugation. Biochem Soc Trans 46: 423–436 [DOI] [PubMed] [Google Scholar]
- 41. Hewings DS, Flygare JA, Bogyo M, Wertz IE (2017) Activity‐based probes for the ubiquitin conjugation‐deconjugation machinery: new chemistries, new tools, and new insights. FEBS J 284: 1555–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Scott DC, Hammill JT, Min J, Rhee DY, Connelly M, Sviderskiy VO, Bhasin D, Chen Y, Ong SS, Chai SC et al (2017) Blocking an N‐terminal acetylation‐dependent protein interaction inhibits an E3 ligase. Nat Chem Biol 13: 850–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mulder MP, Witting K, Berlin I, Pruneda JN, Wu KP, Chang JG, Merkx R, Bialas J, Groettrup M, Vertegaal AC et al (2016) A cascading activity‐based probe sequentially targets E1‐E2‐E3 ubiquitin enzymes. Nat Chem Biol 12: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pao KC, Stanley M, Han C, Lai YC, Murphy P, Balk K, Wood NT, Corti O, Corvol JC, Muqit MM et al (2016) Probes of ubiquitin E3 ligases enable systematic dissection of parkin activation. Nat Chem Biol 12: 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weber A, Elliott PR, Pinto‐Fernandez A, Bonham S, Kessler BM, Komander D, El Oualid F, Krappmann D (2017) A linear diubiquitin‐based probe for efficient and selective detection of the deubiquitinating enzyme OTULIN. Cell Chem Biol 24: 1299–1313.e1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Flierman D, van der Heden van Noort GJ, Ekkebus R, Geurink PP, Mevissen TE, Hospenthal MK, Komander D, Ovaa H (2016) Non‐hydrolyzable diubiquitin probes reveal linkage‐specific reactivity of deubiquitylating enzymes mediated by S2 pockets. Cell Chem Biol 23: 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tan XD, Pan M, Gao S, Zheng Y, Shi J, Li YM (2017) A diubiquitin‐based photoaffinity probe for profiling K27‐linkage targeting deubiquitinases. Chem Commun (Camb) 53: 10208–10211 [DOI] [PubMed] [Google Scholar]
- 48. de Jong A, Merkx R, Berlin I, Rodenko B, Wijdeven RH, El Atmioui D, Yalcin Z, Robson CN, Neefjes JJ, Ovaa H (2012) Ubiquitin‐based probes prepared by total synthesis to profile the activity of deubiquitinating enzymes. ChemBioChem 13: 2251–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liang J, Zhang L, Tan XL, Qi YK, Feng S, Deng H, Yan Y, Zheng JS, Liu L, Tian CL (2017) Chemical synthesis of diubiquitin‐based photoaffinity probes for selectively profiling ubiquitin‐binding proteins. Angew Chem Int Ed Engl 56: 2744–2748 [DOI] [PubMed] [Google Scholar]
- 50. Uehara T, Minoshima Y, Sagane K, Sugi NH, Mitsuhashi KO, Yamamoto N, Kamiyama H, Takahashi K, Kotake Y, Uesugi M et al (2017) Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat Chem Biol 13: 675–680 [DOI] [PubMed] [Google Scholar]
- 51. Mot AC, Prell E, Klecker M, Naumann C, Faden F, Westermann B, Dissmeyer N (2018) Real‐time detection of N‐end rule‐mediated ubiquitination via fluorescently labeled substrate probes. New Phytol 217: 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe‐Paganon S, Bradner JE (2015) Drug Development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348: 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ (2001) Protacs: chimeric molecules that target proteins to the Skp1‐Cullin‐F box complex for ubiquitination and degradation. Proc Natl Acad Sci USA 98: 8554–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bondeson DP, Mares A, Smith IE, Ko E, Campos S, Miah AH, Mulholland KE, Routly N, Buckley DL, Gustafson JL et al (2015) Catalytic in vivo protein knockdown by small‐molecule PROTACs. Nat Chem Biol 11: 611–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meyer HJ, Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157: 910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Matilainen O, Jha S, Holmberg CI (2016) Fluorescent tools for in vivo studies on the ubiquitin‐proteasome system. Methods Mol Biol 1449: 215–222 [DOI] [PubMed] [Google Scholar]
- 57. Menendez‐Benito V, Heessen S, Dantuma NP (2005) Monitoring of ubiquitin‐dependent proteolysis with green fluorescent protein substrates. Methods Enzymol 399: 490–511 [DOI] [PubMed] [Google Scholar]
- 58. Gilon T, Chomsky O, Kulka RG (1998) Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J 17: 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bence NF, Sampat RM, Kopito RR (2001) Impairment of the ubiquitin‐proteasome system by protein aggregation. Science 292: 1552–1555 [DOI] [PubMed] [Google Scholar]
- 60. Bence NF, Bennett EJ, Kopito RR (2005) Application and analysis of the GFPu family of ubiquitin‐proteasome system reporters. Methods Enzymol 399: 481–490 [DOI] [PubMed] [Google Scholar]
- 61. Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG (2000) Short‐lived green fluorescent proteins for quantifying ubiquitin/proteasome‐dependent proteolysis in living cells. Nat Biotechnol 18: 538–543 [DOI] [PubMed] [Google Scholar]
- 62. Bachmair A, Finley D, Varshavsky A (1986) In vivo half‐life of a protein is a function of its amino‐terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- 63. Stack JH, Whitney M, Rodems SM, Pollok BA (2000) A ubiquitin‐based tagging system for controlled modulation of protein stability. Nat Biotechnol 18: 1298–1302 [DOI] [PubMed] [Google Scholar]
- 64. Hamer G, Matilainen O, Holmberg CI (2010) A photoconvertible reporter of the ubiquitin‐proteasome system in vivo . Nat Methods 7: 473–478 [DOI] [PubMed] [Google Scholar]
- 65. Baeuerle PA, Baltimore D (1988) I kappa B: a specific inhibitor of the NF‐kappa B transcription factor. Science 242: 540–546 [DOI] [PubMed] [Google Scholar]
- 66. Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG et al (2004) Oscillations in NF‐kappaB signaling control the dynamics of gene expression. Science 306: 704–708 [DOI] [PubMed] [Google Scholar]
- 67. Nelson G, Paraoan L, Spiller DG, Wilde GJ, Browne MA, Djali PK, Unitt JF, Sullivan E, Floettmann E, White MR (2002) Multi‐parameter analysis of the kinetics of NF‐kappaB signalling and transcription in single living cells. J Cell Sci 115: 1137–1148 [DOI] [PubMed] [Google Scholar]
- 68. Kovalenko A, Wallach D (2006) If the prophet does not come to the mountain: dynamics of signaling complexes in NF‐kappaB activation. Mol Cell 22: 433–436 [DOI] [PubMed] [Google Scholar]
- 69. Sakaue‐Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H et al (2008) Visualizing spatiotemporal dynamics of multicellular cell‐cycle progression. Cell 132: 487–498 [DOI] [PubMed] [Google Scholar]
- 70. Bajar BT, Lam AJ, Badiee RK, Oh YH, Chu J, Zhou XX, Kim N, Kim BB, Chung M, Yablonovitch AL et al (2016) Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat Methods 13: 993–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zielke N, Korzelius J, van Straaten M, Bender K, Schuhknecht GF, Dutta D, Xiang J, Edgar BA (2014) Fly‐FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep 7: 588–598 [DOI] [PubMed] [Google Scholar]
- 72. Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H (2008) A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7: 282–289 [DOI] [PubMed] [Google Scholar]
- 73. Rothbauer U, Zolghadr K, Tillib S, Nowak D, Schermelleh L, Gahl A, Backmann N, Conrath K, Muyldermans S, Cardoso MC et al (2006) Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods 3: 887–889 [DOI] [PubMed] [Google Scholar]
- 74. Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas‐Delucchi CS, Cardoso MC et al (2010) Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17: 133–138 [DOI] [PubMed] [Google Scholar]
- 75. Caussinus E, Kanca O, Affolter M (2011) Fluorescent fusion protein knockout mediated by anti‐GFP nanobody. Nat Struct Mol Biol 19: 117–121 [DOI] [PubMed] [Google Scholar]
- 76. Skaar JR, Pagan JK, Pagano M (2013) Mechanisms and function of substrate recruitment by F‐box proteins. Nat Rev Mol Cell Biol 14: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Volk S, Wang M, Pickart CM (2005) Chemical and genetic strategies for manipulating polyubiquitin chain structure. Methods Enzymol 399: 3–20 [DOI] [PubMed] [Google Scholar]
- 78. Kliza K, Taumer C, Pinzuti I, Franz‐Wachtel M, Kunzelmann S, Stieglitz B, Macek B, Husnjak K (2017) Internally tagged ubiquitin: a tool to identify linear polyubiquitin‐modified proteins by mass spectrometry. Nat Methods 14: 504–512 [DOI] [PubMed] [Google Scholar]
- 79. Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE (2012) Polyubiquitin‐sensor proteins reveal localization and linkage‐type dependence of cellular ubiquitin signaling. Nat Methods 9: 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I (2012) Fluorescence‐based sensors to monitor localization and functions of linear and K63‐linked ubiquitin chains in cells. Mol Cell 47: 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang X, Smits AH, van Tilburg GB, Jansen PW, Makowski MM, Ovaa H, Vermeulen M (2017) An interaction landscape of ubiquitin signaling. Mol Cell 65: 941–955.e948 [DOI] [PubMed] [Google Scholar]
- 82. Kerppola TK (2009) Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev 38: 2876–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen Z, Zhong Y, Wang Y, Xu S, Liu Z, Baskakov IV, Monteiro MJ, Karbowski M, Shen Y, Fang S (2013) Ubiquitination‐induced fluorescence complementation (UiFC) for detection of K48 ubiquitin chains in vitro and in live cells. PLoS One 8: e73482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pinto MJ, Pedro JR, Costa RO, Almeida RD (2016) Visualizing K48 ubiquitination during presynaptic formation by ubiquitination‐induced fluorescence complementation (UiFC). Front Mol Neurosci 9: 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nibe Y, Oshima S, Kobayashi M, Maeyashiki C, Matsuzawa Y, Otsubo K, Matsuda H, Aonuma E, Nemoto Y, Nagaishi T et al (2018) Novel polyubiquitin imaging system, PolyUb‐FC, reveals that K33‐linked polyubiquitin is recruited by SQSTM1/p62. Autophagy, 14: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fujimuro M, Yokosawa H (2005) Production of antipolyubiquitin monoclonal antibodies and their use for characterization and isolation of polyubiquitinated proteins. Methods Enzymol 399: 75–86 [DOI] [PubMed] [Google Scholar]
- 87. Matsumoto ML, Dong KC, Yu C, Phu L, Gao X, Hannoush RN, Hymowitz SG, Kirkpatrick DS, Dixit VM, Kelley RF (2012) Engineering and structural characterization of a linear polyubiquitin‐specific antibody. J Mol Biol 418: 134–144 [DOI] [PubMed] [Google Scholar]
- 88. Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M et al (2010) K11‐linked polyubiquitination in cell cycle control revealed by a K11 linkage‐specific antibody. Mol Cell 39: 477–484 [DOI] [PubMed] [Google Scholar]
- 89. Newton K, Matsumoto ML, Ferrando RE, Wickliffe KE, Rape M, Kelley RF, Dixit VM (2012) Using linkage‐specific monoclonal antibodies to analyze cellular ubiquitylation. Methods Mol Biol 832: 185–196 [DOI] [PubMed] [Google Scholar]
- 90. Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA et al (2008) Ubiquitin chain editing revealed by polyubiquitin linkage‐specific antibodies. Cell 134: 668–678 [DOI] [PubMed] [Google Scholar]
- 91. Fujimuro M, Sawada H, Yokosawa H (1994) Production and characterization of monoclonal antibodies specific to multi‐ubiquitin chains of polyubiquitinated proteins. FEBS Lett 349: 173–180 [DOI] [PubMed] [Google Scholar]
- 92. Noad J, von der Malsburg A, Pathe C, Michel MA, Komander D, Randow F (2017) LUBAC‐synthesized linear ubiquitin chains restrict cytosol‐invading bacteria by activating autophagy and NF‐kappaB. Nat Microbiol 2: 17063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tiede C, Bedford R, Heseltine SJ, Smith G, Wijetunga I, Ross R, AlQallaf D, Roberts AP, Balls A, Curd A et al (2017) Affimer proteins are versatile and renewable affinity reagents. Elife 6: e24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schlichthaerle T, Eklund AS, Schueder F, Strauss MT, Tiede C, Curd A, Ries J, Peckham M, Tomlinson DC, Jungmann R (2018) Site‐specific labeling of affimers for DNA‐PAINT microscopy. Angew Chem Int Ed Engl, 57: 11060–11063 [DOI] [PubMed] [Google Scholar]
- 95. Bedford R, Tiede C, Hughes R, Curd A, McPherson MJ, Peckham M, Tomlinson DC (2017) Alternative reagents to antibodies in imaging applications. Biophys Rev 9: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gersch M, Gladkova C, Schubert AF, Michel MA, Maslen S, Komander D (2017) Mechanism and regulation of the Lys6‐selective deubiquitinase USP30. Nat Struct Mol Biol 24: 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Michel MA, Swatek KN, Hospenthal MK, Komander D (2017) Ubiquitin linkage‐specific affimers reveal insights into k6‐linked ubiquitin signaling. Mol Cell 68: 233–246.e235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hughes DJ, Tiede C, Penswick N, Tang AA, Trinh CH, Mandal U, Zajac KZ, Gaule T, Howell G, Edwards TA et al (2017) Generation of specific inhibitors of SUMO‐1‐ and SUMO‐2/3‐mediated protein‐protein interactions using Affimer (Adhiron) technology. Sci Signal 10: eaaj2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hodgins RR, Ellison KS, Ellison MJ (1992) Expression of a ubiquitin derivative that conjugates to protein irreversibly produces phenotypes consistent with a ubiquitin deficiency. J Biol Chem 267: 8807–8812 [PubMed] [Google Scholar]
- 100. Pickart CM, Kasperek EM, Beal R, Kim A (1994) Substrate properties of site‐specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin‐activating enzyme (E1). J Biol Chem 269: 7115–7123 [PubMed] [Google Scholar]
- 101. Dantuma NP, Groothuis TA, Salomons FA, Neefjes J (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol 173: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ben Yehuda A, Risheq M, Novoplansky O, Bersuker K, Kopito RR, Goldberg M, Brandeis M (2017) Ubiquitin accumulation on disease associated protein aggregates is correlated with nuclear ubiquitin depletion, histone De‐ubiquitination and impaired DNA damage response. PLoS One 12: e0169054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Juenemann K, Jansen AHP, van Riel L, Merkx R, Mulder MPC, An H, Statsyuk A, Kirstein J, Ovaa H, Reits EA (2018) Dynamic recruitment of ubiquitin to mutant huntingtin inclusion bodies. Sci Rep 8: 1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sun D, Wu R, Zheng J, Li P, Yu L (2018) Polyubiquitin chain‐induced p62 phase separation drives autophagic cargo segregation. Cell Res 28: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Geurink PP, van Tol BD, van Dalen D, Brundel PJ, Mevissen TE, Pruneda JN, Elliott PR, van Tilburg GB, Komander D, Ovaa H (2016) Development of diubiquitin‐Based FRET probes to quantify ubiquitin linkage specificity of deubiquitinating enzymes. ChemBioChem 17: 816–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ye Y, Blaser G, Horrocks MH, Ruedas‐Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D (2012) Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bowman J, Rodgers MA, Shi M, Amatya R, Hostager B, Iwai K, Gao SJ, Jung JU (2015) Posttranslational modification of HOIP blocks toll‐like receptor 4‐mediated linear‐ubiquitin‐chain formation. MBio 6: e01777–01715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sauer M, Heilemann M (2017) Single‐molecule localization microscopy in eukaryotes. Chem Rev 117: 7478–7509 [DOI] [PubMed] [Google Scholar]
- 109. Sahl SJ, Hell SW, Jakobs S (2017) Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol 18: 685–701 [DOI] [PubMed] [Google Scholar]
- 110. Betzig E (2015) Single molecules, cells, and super‐resolution optics (Nobel Lecture). Angew Chem Int Ed Engl 54: 8034–8053 [DOI] [PubMed] [Google Scholar]
- 111. Gustafsson MG (1999) Extended resolution fluorescence microscopy. Curr Opin Struct Biol 9: 627–634 [DOI] [PubMed] [Google Scholar]
- 112. Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M (2008) Subdiffraction‐resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl 47: 6172–6176 [DOI] [PubMed] [Google Scholar]
- 113. Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott‐Schwartz J, Hess HF (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313: 1642–1645 [DOI] [PubMed] [Google Scholar]
- 114. Hell SW, Wichmann J (1994) Breaking the diffraction resolution limit by stimulated emission: stimulated‐emission‐depletion fluorescence microscopy. Opt Lett 19: 780–782 [DOI] [PubMed] [Google Scholar]
- 115. Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA III, Liu Z et al (2014) Lattice light‐sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346: 1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Furstenberg A, Heilemann M (2013) Single‐molecule localization microscopy‐near‐molecular spatial resolution in light microscopy with photoswitchable fluorophores. Phys Chem Chem Phys 15: 14919–14930 [DOI] [PubMed] [Google Scholar]
- 117. Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R (2017) Super‐resolution microscopy with DNA‐PAINT. Nat Protoc 12: 1198–1228 [DOI] [PubMed] [Google Scholar]
- 118. Deussner‐Helfmann NS, Auer A, Strauss MT, Malkusch S, Dietz MS, Barth HD, Jungmann R, Heilemann M (2018) Correlative single‐molecule FRET and DNA‐PAINT imaging. Nano Lett 18: 4626–4630 [DOI] [PubMed] [Google Scholar]
- 119. Huang B, Wang W, Bates M, Zhuang X (2008) Three‐dimensional super‐resolution imaging by stochastic optical reconstruction microscopy. Science 319: 810–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hands KJ, Cuchet‐Lourenco D, Everett RD, Hay RT (2014) PML isoforms in response to arsenic: high‐resolution analysis of PML body structure and degradation. J Cell Sci 127: 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hausmann M, Ilic N, Pilarczyk G, Lee JH, Logeswaran A, Borroni AP, Krufczik M, Theda F, Waltrich N, Bestvater F et al (2017) Challenges for super‐resolution localization microscopy and biomolecular fluorescent nano‐probing in cancer research. Int J Mol Sci 18: E2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yonezawa T, Takahashi H, Shikata S, Liu X, Tamura M, Asada S, Fukushima T, Fukuyama T, Tanaka Y, Sawasaki T et al (2017) The ubiquitin ligase STUB1 regulates stability and activity of RUNX1 and RUNX1‐RUNX1T1. J Biol Chem 292: 12528–12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Courtheoux T, Enchev RI, Lampert F, Gerez J, Beck J, Picotti P, Sumara I, Peter M (2016) Cortical dynamics during cell motility are regulated by CRL3(KLHL21) E3 ubiquitin ligase. Nat Commun 7: 12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gu ZC, Wu E, Sailer C, Jando J, Styles E, Eisenkolb I, Kuschel M, Bitschar K, Wang X, Huang L et al (2017) Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol Biol Cell 28: 2479–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Keller KE, Yang YF, Sun YY, Sykes R, Acott TS, Wirtz MK (2013) Ankyrin repeat and suppressor of cytokine signaling box containing protein‐10 is associated with ubiquitin‐mediated degradation pathways in trabecular meshwork cells. Mol Vis 19: 1639–1655 [PMC free article] [PubMed] [Google Scholar]
- 126. LaRock DL, Chaudhary A, Miller SI (2015) Salmonellae interactions with host processes. Nat Rev Microbiol 13: 191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Randow F, Youle RJ (2014) Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe 15: 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH (2004) Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol 14: 806–811 [DOI] [PubMed] [Google Scholar]
- 129. Herhaus L, Dikic I (2017) Regulation of Salmonella‐host cell interactions via the ubiquitin system. Int J Med Microbiol 308: 176–184 [DOI] [PubMed] [Google Scholar]
- 130. Cemma M, Kim PK, Brumell JH (2011) The ubiquitin‐binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria‐associated microdomains to target Salmonella to the autophagy pathway. Autophagy 7: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C et al (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin‐coated bacteria. Nat Immunol 10: 1215–1221 [DOI] [PubMed] [Google Scholar]
- 133. Lork M, Delvaeye M, Goncalves A, Van Hamme E, Beyaert R (2016) Monitoring ubiquitin‐coated bacteria via confocal microscopy. Methods Mol Biol 1449: 243–250 [DOI] [PubMed] [Google Scholar]
- 134. van Wijk SJ, Fiskin E, Dikic I (2013) Selective monitoring of ubiquitin signals with genetically encoded ubiquitin chain‐specific sensors. Nat Protoc 8: 1449–1458 [DOI] [PubMed] [Google Scholar]
- 135. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS (2013) The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501: 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ (2012) The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin‐dependent autophagy of intracellular salmonella typhimurium. Cell Host Microbe 12: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Polajnar M, Dietz MS, Heilemann M, Behrends C (2017) Expanding the host cell ubiquitylation machinery targeting cytosolic Salmonella. EMBO Rep 18: 1572–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. van Wijk SJL, Fricke F, Herhaus L, Gupta J, Hotte K, Pampaloni F, Grumati P, Kaulich M, Sou YS, Komatsu M et al (2017) Linear ubiquitination of cytosolic salmonella typhimurium activates NF‐kappaB and restricts bacterial proliferation. Nat Microbiol 2: 17066 [DOI] [PubMed] [Google Scholar]
- 139. Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, Zaffagnini G, Wild P, Martens S, Wagner SA et al (2016) Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA 113: 4039–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Clough B, Wright JD, Pereira PM, Hirst EM, Johnston AC, Henriques R, Frickel EM (2016) K63‐linked ubiquitination targets toxoplasma gondii for endo‐lysosomal destruction in IFNgamma‐stimulated human cells. PLoS Pathog 12: e1006027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zhou Y, Dong N, Hu L, Shao F (2013) The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. PLoS One 8: e57558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S et al (2013) Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol 203: 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D et al (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF‐kappaB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- 144. Emmerich CH, Schmukle AC, Walczak H (2011) The emerging role of linear ubiquitination in cell signaling. Sci Signal 4: re5 [DOI] [PubMed] [Google Scholar]
- 145. Fiil BK, Gyrd‐Hansen M (2014) Met1‐linked ubiquitination in immune signalling. FEBS J 281: 4337–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Hacker H, Karin M (2006) Regulation and function of IKK and IKK‐related kinases. Sci STKE 2006: re13 [DOI] [PubMed] [Google Scholar]
- 147. Fricke F, Beaudouin J, Eils R, Heilemann M (2015) One, two or three? Probing the stoichiometry of membrane proteins by single‐molecule localization microscopy. Sci Rep 5: 14072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Hummer G, Fricke F, Heilemann M (2016) Model‐independent counting of molecules in single‐molecule localization microscopy. Mol Biol Cell 27: 3637–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Karathanasis C, Fricke F, Hummer G, Heilemann M (2017) Molecule counts in localization microscopy with organic fluorophores. ChemPhysChem 18: 942–948 [DOI] [PubMed] [Google Scholar]
- 150. Elson EL (2011) Fluorescence correlation spectroscopy: past, present, future. Biophys J 101: 2855–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Spahn C, Grimm JB, Lavis LD, Lampe M, Heilemann M (2018) Whole‐cell, 3D and multi‐color STED imaging with exchangeable fluorophores. Nano Lett 19: 500–505 [DOI] [PubMed] [Google Scholar]
- 152. Spahn CK, Glaesmann M, Grimm JB, Ayala AX, Lavis LD, Heilemann M (2018) A toolbox for multiplexed super‐resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels. Sci Rep 8: 14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Heilemann M (2012) Elucidating cellular structures – superresolution microscopy In Comprehensive Biophysics, Egelman E. (ed). Amsterdam: Elsevier; ISBN 9780123749208. [Google Scholar]
- 154. Sauer M, Heilemann M (2017) Localization‐based super‐resolution microscopy In Fluorescence Microscopy, Kubitscheck U. (ed), pp 267–290. Weinheim: Wiley‐Blackwell; [Google Scholar]


