ABSTRACT
Metazoans contain two homologs of the Gcn5-binding protein Ada2, Ada2a and Ada2b, which nucleate formation of the ATAC and SAGA complexes, respectively. In Drosophila melanogaster, there are two splice isoforms of Ada2b: Ada2b-PA and Ada2b-PB. Here, we show that only the Ada2b-PB isoform is in SAGA; in contrast, Ada2b-PA associates with Gcn5, Ada3, Sgf29 and Chiffon, forming the Chiffon histone acetyltransferase (CHAT) complex. Chiffon is the Drosophila ortholog of Dbf4, which binds and activates the cell cycle kinase Cdc7 to initiate DNA replication. In flies, Chiffon and Cdc7 are required in ovary follicle cells for gene amplification, a specialized form of DNA re-replication. Although chiffon was previously reported to be dispensable for viability, here, we find that Chiffon is required for both histone acetylation and viability in flies. Surprisingly, we show that chiffon is a dicistronic gene that encodes distinct Cdc7- and CHAT-binding polypeptides. Although the Cdc7-binding domain of Chiffon is not required for viability in flies, the CHAT-binding domain is essential for viability, but is not required for gene amplification, arguing against a role in DNA replication.
KEY WORDS: Dbf4, Gcn5, Cell cycle, Gene expression, Histone acetylation
Highlighted Article: The Drosophila ortholog of Dbf4, Chiffon, binds Gcn5 to form a novel histone acetyltransferase complex that is essential for viability in flies, but is not required for DNA replication.
INTRODUCTION
Chromatin modifications impact both transcription and cell cycle events such as DNA replication (Li et al., 2007; Ma et al., 2015). In particular, histone acetylation contributes to transcription and correlates with the timing of the initial step in DNA replication, origin firing. The histone acetyltransferase Gcn5 stimulates transcription by generating a permissive chromatin environment that facilitates chromatin remodeling by complexes such as SWI/SNF (Hassan et al., 2002; Weake and Workman, 2010). Gcn5 also stimulates origin firing when tethered to a late-firing origin in yeast (Vogelauer et al., 2002) and enhances the rate of DNA synthesis from a chromatin template in vitro (Kurat et al., 2017). In Saccharomyces cerevisiae, Gcn5 function in transcription is mediated predominantly through the Spt–Ada–Gcn5 acetyltransferase (SAGA) complex (Grant et al., 1997). During evolution, there has been an expansion in the diversity of Gcn5-containing complexes (Spedale et al., 2012). In metazoans, including Drosophila, there are two homologs of the Gcn5-binding protein Ada2, Ada2a and Ada2b, which nucleate formation of the Ada2a-containing (ATAC) or SAGA transcription coactivator complexes, respectively (Kusch et al., 2003; Wang et al., 2008). All Gcn5 complexes share Sgf29 and Ada3 subunits, which, together with their respective Ada2 homolog, enable nucleosomal histone acetyltransferase activity (Balasubramanian et al., 2002; Grant et al., 1997; Spedale et al., 2012). The SAGA and ATAC complexes activate transcription during development and in response to signaling pathways or external stimuli (Spedale et al., 2012). In addition, ATAC has roles in cell cycle progression via acetylation of cyclin A, which promotes progression through mitosis (Orpinell et al., 2010).
In Drosophila, Ada2b has two splice isoforms that differ in their C-terminal regions but share a common N-terminal region (Pankotai et al., 2013; Qi et al., 2004). The short Ada2b-PA isoform binds Gcn5 and is necessary for histone H3 acetylation in vivo (Pankotai et al., 2013; Qi et al., 2004). Both Ada2b isoforms are required to fully complement ada2b mutations, although expression of the short Ada2b-PA isoform alone can support development and partially restore histone H3 acetylation (Pankotai et al., 2013). Although it has been assumed that the short Ada2b-PA isoform functions as part of the SAGA complex, our previous studies showed that the long Ada2b-PB splice isoform is associated with Drosophila SAGA (Weake et al., 2009). In this study, we describe a novel Gcn5-containing complex nucleated by the short Ada2b-PA isoform that contains the cell cycle regulatory protein, Chiffon: the Chiffon histone acetyltransferase (CHAT) complex. Chiffon is the Drosophila ortholog of Dbf4 (Landis and Tower, 1999), and it binds and activates the cell cycle kinase Cdc7 (Stephenson et al., 2015). In yeast, Dbf4 and Cdc7 form the Dbf4-dependent kinase (DDK) complex that phosphorylates the Mcm helicase to initiate DNA replication (Lei et al., 1997; Weinreich and Stillman, 1999). Although Dbf4 is essential for DNA replication in most organisms, previous studies found that chiffon null mutants were viable (Landis and Tower, 1999). Here, we show that the Cdc7-binding activity of Chiffon is dispensable in flies. However, the C-terminal insect-specific domain of Chiffon that nucleates formation of the CHAT complex is required in flies for histone H3 acetylation and viability, but not for DNA replication. Our data demonstrate that the DNA replication and histone acetylation activities of Chiffon can be genetically separated, raising the question of why these two activities are encoded by the same gene. One possibility, although not tested in this study, is that the DDK and CHAT complexes are encoded as part of the same gene to coordinate their expression and/or levels during the cell cycle or development. This could provide a mechanism to coordinate histone acetylation with DNA replication, potentially during particular developmental stages in flies.
RESULTS
Identification of a novel Chiffon–Gcn5 complex in Drosophila
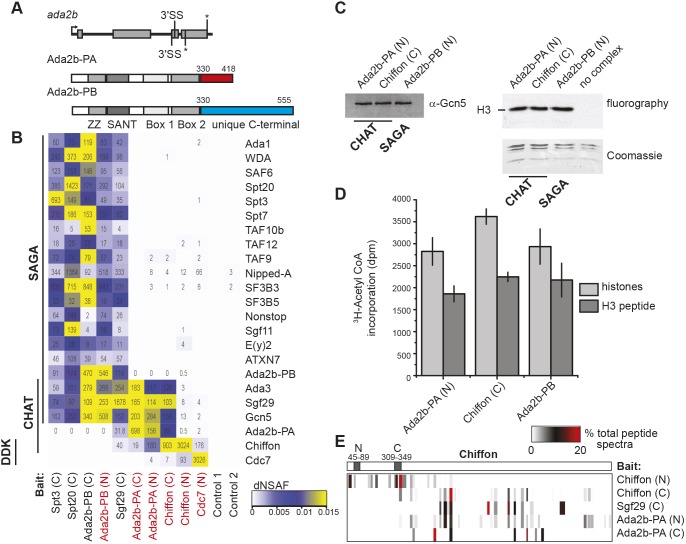
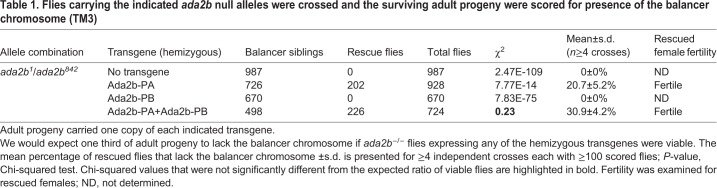
Drosophila Ada2b has two splice isoforms that differ in their C-terminal regions but share a common N-terminal region containing the zinc finger-like ZZ, SANT and two of the three previously described ADA box domains (Pankotai et al., 2013; Qi et al., 2004) (Fig. 1A). Similar to published observations (Pankotai et al., 2013), we observed that expression of Ada2b-PA alone partially rescued adult viability in ada2b null mutants, although both Ada2b isoforms were required to fully complement lethality of the null ada2b1/ada2b842 allele combination (Table 1). Expression of single-copy transgenes for both Ada2b-PA and Ada2b-PB restored viability to the expected one third of ada2b1/ada2b842 progeny (30.9±4.2%, χ2=0.23) compared with 20.7±5.2% with expression of Ada2b-PA alone. In contrast, expression of Ada2b-PB alone did not restore viability to ada2b1/ada2b842 progeny. These data suggested that the Ada2b isoforms have nonredundant functions in flies, and that Ada2b-PA alone can partially support development. Thus, we sought to determine whether these Ada2b isoforms were both required in SAGA, or alternatively, like Ada2a, each Ada2b isoform nucleated formation of a distinct Gcn5-containing complex. To distinguish between these alternatives, we purified the Ada2b-PA and Ada2b-PB isoforms from cultured S2 cells using tandem FLAG-HA affinity chromatography and examined the co-purifying proteins using multidimensional protein identification technology (MudPIT). Using this approach, the Ada2b isoforms could be distinguished by peptide spectra that mapped to their unique C-terminal regions. Ada2b-PB co-purified all other 19 SAGA subunits (Stegeman et al., 2016), but did not co-purify any peptide spectra specific to the short Ada2b-PA isoform (Fig. 1B). Similarly, SAGA-specific purifications using bait proteins such as Spt3 and Spt20 contained peptide spectra specific to Ada2b-PB, but not Ada2b-PA. Instead, Ada2b-PA co-purified Gcn5, Ada3 and Sgf29, but not Ada2b-PB or other SAGA subunits. Ada2b-PA also did not co-purify ATAC-specific subunits such as Atac1, Atac2 or D12 (Table S1). Epitope tagging of Ada2b-PA did not disrupt its interaction with SAGA because similar results were observed with Ada2b isoforms tagged at their shared N- or unique C-termini. These data suggest that Ada2b-PA associates with Gcn5, Sgf29 and Ada3 in a complex that is distinct from either ATAC or SAGA.
Fig. 1.
Identification of a novel Chiffon–Gcn5 complex in Drosophila. (A) The ada2b gene encodes two splice isoforms, Ada2b-PA and Ada2b-PB, resulting from alternative use of 3′ splice acceptor sites in exon 3 (splice site, SS) that generate a frameshift following amino acid 330 (asterisks indicate stop codons). Ada2b isoforms differ only in their highlighted C-terminal regions (red/blue). (B) Heat map showing the relative spectral abundance of SAGA subunits, Chiffon and Cdc7 expressed as distributive normalized spectral abundance factor (dNSAF) in tandem FLAG-HA purifications from S2 cells using the indicated bait proteins (N/C epitope tag shown in brackets). Control 1, untagged; Control 2, CG6459 (nonspecific bait). Bait proteins new to this study are highlighted in red. The dNSAF scale represents abundance of subunits on a scale from yellow (high) to blue (low), with subunits that were not identified shown in white. dNSAF values used to generate the heat map are provided in Table S1. The numbers of spectra specific to each protein isoform (distributed spectra, dS) are shown in each box. Data for each bait protein represent the sum of two technical MudPIT experiments. (C) The histone acetyltransferase activity of FLAG-purified CHAT (via Ada2b-PA or Chiffon) or SAGA (Ada2b-PB) complexes containing equivalent amounts of Gcn5 as determined by western blotting with anti-Gcn5 antibody (left) were assayed using core histones as substrate. Incorporation of 3H-acetyl CoA was assayed by fluorography (top right) and the migration of histone H3 was determined by Coomassie staining (bottom right). The negative control lane consists of histones and 3H-acetyl CoA with no complex added. (D) Histone acetyltransferase activity of the indicated complexes was quantified by scintillation counting of 3H-acetyl CoA incorporated into core histones or H3 tail peptides as in C. Mean±s.d. is shown for three independent histone acetyltransferase assays relative to no complex control. (E) Heat map showing the percentage of total spectra mapping to each region of full-length Chiffon (1695 aa) purifications using the indicated bait proteins as in B. The conserved Dbf4 N and C motifs in Chiffon are indicated by the gray shaded boxes.
Table 1.
Flies carrying the indicated ada2b null alleles were crossed and the surviving adult progeny were scored for presence of the balancer chromosome (TM3)
To identify other proteins in this Ada2b-PA complex, we examined the MudPIT data to find proteins that co-purified specifically with Ada2b-PA, but not with SAGA-specific subunits. A single protein, Chiffon (CG5813; FBgn0000307), co-purified with Ada2b-PA or Sgf29, but not with other SAGA subunits or with the negative controls (Fig. 1B; Table S1). Moreover, reciprocal purifications of C-terminally tagged Chiffon co-purified Gcn5, Ada3, Sgf29 and Ada2b-PA, but not Ada2b-PB. Chiffon is the Drosophila ortholog of Dbf4, which binds and activates the cell cycle kinase Cdc7 to phosphorylate the Mcm helicase, initiating DNA replication (Landis and Tower, 1999; Stephenson et al., 2015). The Chiffon-purified or Ada2b-PA-purified complexes exhibited similar levels and specificity of in vitro histone acetyltransferase activity to Drosophila SAGA purified via Ada2b-PB, with predominant activity on histone H3 in core histones (Fig. 1C) and ability to acetylate histone H3 tail peptides (Fig. 1D). Thus, we conclude that Chiffon is a bona fide subunit of a novel histone acetyltransferase complex containing Ada2b-PA, Gcn5, Sgf29 and Ada3 that we named the CHAT complex.
Most CHAT complexes do not contain Cdc7
Because Chiffon is the regulatory subunit of the cell cycle kinase Cdc7 (Stephenson et al., 2015), we next asked whether Cdc7 was present in the CHAT complex. Only seven peptide spectra were identified for Cdc7 using C-terminally tagged Chiffon as bait, which co-purified 89–158 peptide spectra for each of the other subunits of the CHAT complex: Ada2b-PA, Gcn5, Sgf29 and Ada3 (Fig. 1B). Thus, we next asked whether Chiffon did, in fact, bind Cdc7 in vivo. We previously showed that the N-terminal domain of Chiffon [1–400 amino acids (aa)] is sufficient to bind and stimulate Cdc7 kinase activity in vitro (Stephenson et al., 2015). Indeed, Cdc7 and Chiffon interact in vivo because N-terminally tagged Chiffon co-purified 93 peptide spectra for Cdc7, and Cdc7 reciprocally co-purified 176 peptide spectra for Chiffon. In contrast, N-terminally tagged Chiffon or Cdc7 co-purified fewer than 13 peptide spectra for other components of the CHAT complex such as Gcn5. There are two possibilities for the mutually exclusive binding of Cdc7 and CHAT subunits with Chiffon; first, Cdc7 blocks binding of CHAT subunits to Chiffon; or second, chiffon encodes two separate polypeptides that interact with Cdc7 or CHAT independently. Our data support the latter possibility because most peptide spectra for C-terminally tagged Chiffon map to its C-terminal region, whereas most peptide spectra for N-terminally tagged Chiffon map to its N-terminal region (Fig. 1E). These data suggest that very little full-length Chiffon exists in asynchronous cultured cells. However, a small fraction of Chiffon might interact with both Cdc7 and CHAT because we identified a few peptide spectra corresponding to Cdc7 in CHAT purifications (Ada2b-PA and Chiffon-C). Similarly, we also observed a few peptide spectra for CHAT subunits in Cdc7 or Chiffon-N purifications. Thus, we conclude that although a small fraction of Chiffon protein might interact with Cdc7 and CHAT simultaneously, most Chiffon interacts separately with either Cdc7 or CHAT, likely as two independent Chiffon polypeptides.
The insect-specific C-terminal domain of Chiffon directly binds Gcn5
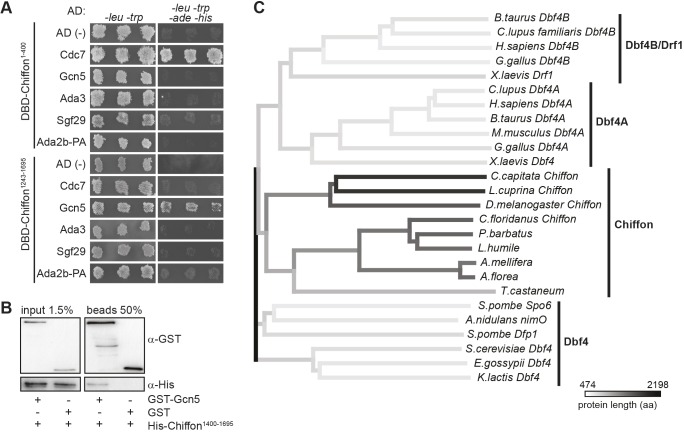
To test whether the N- and C-terminal domains of Chiffon could interact independently with Cdc7 and CHAT subunits, as suggested by the mass spectrometry data, we used a yeast two-hybrid approach to screen for interactions between different domains of Chiffon and each CHAT subunit (Fig. S1). Using this approach, we identified a strong reciprocal interaction between the N-terminal Chiffon domain (1–400 aa) and Cdc7 (Fig. 2A). This is consistent with our MudPIT data and with the previous observation that the N-terminal 400 aa of Chiffon are sufficient to bind Cdc7 in vitro (Stephenson et al., 2015). We also observed a weak unidirectional interaction between Gcn5 and the C-terminal Chiffon domain (1243–1695 aa) (Fig. 2A). We were able to co-immunoprecipitate the recombinant C-terminal region (1400–1695 aa) of Chiffon and Gcn5 under low-salt (150 mM NaCl) conditions in vitro (Fig. 2B), further suggesting that Chiffon and Gcn5 interact directly, albeit weakly.
Fig. 2.
The insect-specific C-terminal domain of Chiffon directly binds Gcn5. (A) Yeast two-hybrid assay was performed to test the pairwise interaction of each CHAT subunit with Chiffon. The Gal4 activating domain (AD) was fused to Cdc7, Gcn5, Ada3, Sgf29 or Ada2b-PA, and the Gal4 DNA-binding domain (DBD) was fused to either the N-terminal (1–400 aa) or C-terminal (1243–1695 aa) domains of Chiffon. Empty plasmids expressing only the AD or DBD were used to test for auto-activation of each protein. Three independent transformed yeast colonies were patched on media lacking leucine and tryptophan to test for the presence of the AD and DBD plasmids, and on media lacking leucine, tryptophan, adenine and histidine to test for interaction. (B) Glutathione-sepharose pull-down of recombinant GST–Gcn5 and the C-terminal domain of Chiffon (1400–1695 aa) tagged with His followed by western blotting with antibodies against GST and His. Representative data from three experiments are shown. (C) Phylogenetic tree constructed using neighbor-joining method showing Dbf4 homologs from fungi, insects and vertebrates based on Clustal-Omega multiple sequence alignment of full-length proteins. Shading represents protein length (aa, amino acids).
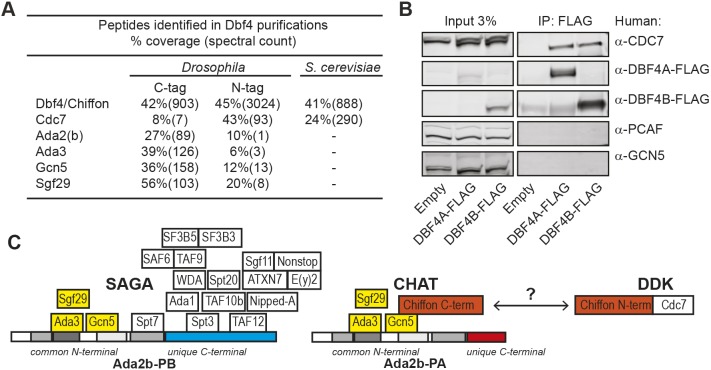
The C-terminal domain of Chiffon in Drosophila and other insects is much longer than that of other Dbf4 homologs and is not present in yeast or vertebrate Dbf4 (Landis and Tower, 1999; Tower, 2004) (Fig. 2C). Moreover, the C-terminal region of Chiffon that binds Gcn5 shares several highly conserved regions with other insects (Fig. S2). Because this insect-specific C-terminal domain interacted with Gcn5, we predicted that yeast or mammalian Dbf4 would be unlikely to interact with Gcn5. Indeed, we did not observe any peptides for Ada2, Gcn5, Sgf29 or Ada3 in TAP-purified Dbf4 from yeast cells (Fig. 3A). Moreover, human DBF4A and DBF4B co-immunoprecipitated CDC7, but not GCN5 (also known as KAT2A) or its paralog, PCAF (also known as KAT2B), from human embryonic kidney (HEK) 293T cells (Fig. 3B). Thus, the insect-specific C-terminal domain of Chiffon interacts directly with Gcn5, while the conserved N-terminal domain of Chiffon binds Cdc7.
Fig. 3.
Dbf4 does not bind Gcn5 in yeast or humans. (A) Table showing proteins identified in Chiffon and Dbf4 purifications from Drosophila melanogaster (tandem FLAG-HA) or Saccharomyces cerevisiae (TAP-tagged). Sequence coverage (percentage) and number of spectra are shown for each protein. (B) FLAG-tagged human DBF4A or DBF4B were immunoprecipitated from HEK293T cell extracts using anti-FLAG antibodies, and analyzed by western blotting using the indicated antibodies. Control, empty vector. Representative data from three experiments are shown. (C) Schematic showing subunit composition of the SAGA, CHAT and DDK complexes. Interactions between subunits are based on the yeast two-hybrid analysis from Figs S1 and S3, which suggest that Ada2b-PB binds Spt3 and TAF12 via its unique C-terminal domain to nucleate SAGA formation. In contrast, CHAT formation is nucleated by the binding of Chiffon's C-terminal to Gcn5, which precludes association of other SAGA subunits. Chiffon interacts with Cdc7 via its N-terminal domain to form the DDK complex, and the DDK and CHAT complexes appear to be largely separate in vivo.
Because Gcn5 is a component of all three of the SAGA, ATAC and CHAT complexes in flies, and because Gcn5 binds the C-terminal domain of Chiffon, we wondered why Chiffon did not associate with either the SAGA or ATAC complexes (Fig. 1B; Table S1). To answer this question, we examined the interaction of each Ada2b isoform with all SAGA subunits except Nipped-A (also known as Tra1) using yeast two-hybrid analysis (Fig. S3). Using this approach, we found that Ada2b-PB, but not Ada2b-PA, auto-activated when fused to the Gal4 DNA-binding domain. This suggests that Ada2b-PB, but not Ada2b-PA, might interact with yeast transcriptional coactivators like SAGA to activate expression of the reporter genes in this assay. We also observed that Ada2b-PA interacted with the CHAT subunits Gcn5 and Ada3, and surprisingly also with the SAGA-specific subunit Spt7 (Fig. S3A). Further, Ada2b-PB fused to the Gal4 activating domain interacted with two additional SAGA-specific subunits that did not interact with Ada2b-PA: Spt3 and TAF12 (Fig. S3B). These data suggest a model in which the unique C-terminal region of the Ada2b-PB isoform binds SAGA through Spt3 and TAF12, enhancing binding of Spt7 to the Ada2b-PB N-terminal, which precludes Gcn5 binding to Chiffon (Fig. S3C). The Ada2b-PA isoform lacks the C-terminal region necessary for binding Spt3 and TAF12, preventing stable binding of Spt7 to the N-terminal of Ada2b-PA, and instead enabling Gcn5 to bind Chiffon. This model is partially based on the observation that Ada2b-PA did not interact with Spt7 in our mass spectrometry data, even though it is capable of binding Spt7 by yeast two-hybrid assay. We further suggest that the Ada2b-PB C-terminal domain might also be capable of interacting with yeast SAGA, potentially via yeast Spt3 and TAF12. Although Ada2b-PA and Ada2b-PB interacted in one direction by yeast two-hybrid assay (Fig. S3B), our MudPIT data indicate that the Ada2b isoforms are not present in the same complex in vivo (Fig. 1B; Table S1). We conclude that the unique C-terminal regions of the Ada2b isoforms control protein–protein interactions that determine formation of the SAGA or CHAT complexes, and that the extended C-terminal domain in Ada2b-PB is necessary for SAGA formation (Fig. 3C).
Chiffon is necessary for histone H3 acetylation in vivo
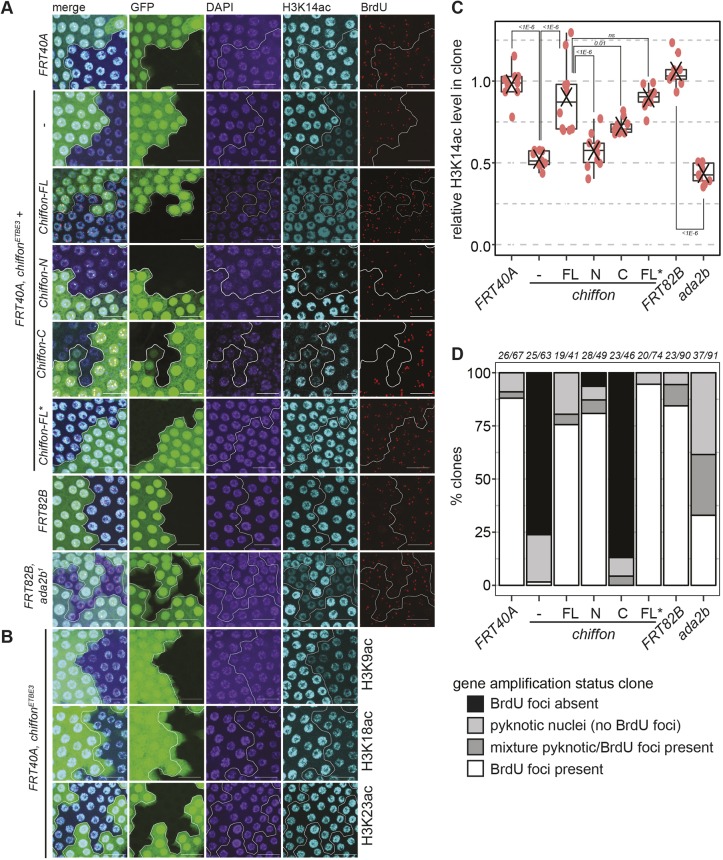
Since the CHAT complex exhibits in vitro histone acetyltransferase activity against histone H3, we next asked whether Chiffon was necessary for proper histone H3 acetylation in vivo. To do this, we used Drosophila ovary follicle cells in which specific regions of the genome undergo repeated bidirectional replication initiation to increase DNA copy number (Spradling and Mahowald, 1980). Chiffon is necessary for gene amplification in these cells (Landis and Tower, 1999; Stephenson et al., 2015; Zhang and Tower, 2004). We generated somatic mosaics for the chiffonETBE3 null allele in ovaries using the FLP/FRT system and examined levels of different histone H3 acetyl marks by immunostaining. Notably, chiffonETBE3 mutant cells showed decreased levels of histone H3 acetylated at lysine 14 (H3K14ac) relative to the adjacent GFP-positive cells (Fig. 4A). We also observed decreased levels of H3K9ac and H3K18ac, but not H3K23ac, in chiffonETBE3 mutant cells (Fig. 4B). H3K18ac levels were only modestly reduced in chiffonETBE3 clones, consistent with data showing that p300/CBP (encoded by nejire) is the major histone acetyltransferase for H3K18 in Drosophila and in mammalian cells (Jin et al., 2011; Tie et al., 2009). H3K14ac levels within chiffonETBE3, but not control FRT40A, clones were reduced to ∼50% of the surrounding tissue (Fig. 4A,C). The nuclei in some chiffonETBE3 clones appeared slightly more condensed using 4′,6-diamidino-2-phenylindole (DAPI) staining, suggesting that the reduced histone acetylation could be due to decreased DNA content in these cells. However, we and others have previously shown that chiffon is not essential for endoreplication, which determines the ploidy of follicle cells (Landis and Tower, 1999; Stephenson et al., 2015; Zhang and Tower, 2004). Moreover, some H3 acetyl marks such as H3K23ac were not reduced in chiffonETBE3 clones (Fig. 4B). Together, these data demonstrate that Chiffon is required for full levels of histone H3 acetylation at lysines 9, 14 and 18 in vivo, suggesting that the CHAT complex contributes to bulk levels of histone H3 acetylation in flies. Interestingly, ada2b1 clones showed only slightly lower levels of H3K14ac (not significant) when compared with chiffonETBE3 clones, despite the fact that the ada2b1 allele removes both Ada2b isoforms (Fig. 4A,C). This suggests that the CHAT complex, rather than SAGA, might contribute to the majority of histone H3 acetylation in ovary follicle cells. To test whether CHAT was also required in other cell types for histone H3 acetylation, we compared H3K14ac levels in chiffonETBE3 and ada2b1 clones from imaginal discs. Similar to ovary follicle cells, most chiffonETBE3 and ada2b1 clones from imaginal discs showed decreased levels of H3K14ac (Fig. S4). Although some large chiffonETBE3 clones appeared to have little or no H3K14ac staining, other chiffonETBE3 clones showed only moderate decreases in H3K14ac more similar to those observed in ovary follicle cells. Notably, some chiffonETBE3 clones also appeared to contain fewer nuclei, suggesting that Chiffon might also contribute to cell number, potentially through DNA replication, in this cell type. While some ada2b1 clones showed only slight decreases in H3K14ac, other clones showed similar levels of H3K14ac to those observed in ada2b1 ovary follicle clones. Previous studies showed that mutations in the SAGA-specific subunit, wda, strongly reduced H3K9ac levels in Drosophila embryos (Guelman et al., 2006), suggesting that SAGA is necessary for full levels of histone H3 acetylation in embryos. It remains unclear whether SAGA and CHAT have overlapping or specialized functions with regard to histone H3 acetylation in Drosophila. However, our observation that the CHAT-specific Ada2b-PA isoform is sufficient to partially restore viability to ada2b null flies suggests that CHAT might compensate for some of SAGA's essential functions during development. Overall, these data indicate that chiffon is required for histone H3 acetylation in vivo, and that the CHAT complex contributes to histone H3 acetylation in several tissues in flies.
Fig. 4.
Chiffon is necessary for histone H3 acetylation in vivo. (A) Mosaic egg chambers were generated using the FLP/FRT system for chiffonETBE3 and ada2b1, their respective controls, FRT40A and FRT82B, and for chiffonETBE3 clones expressing single copies of the indicated Chiffon rescue transgenes. Maximum-intensity projection images showing BrdU incorporation, α-H3K14ac and DAPI staining from amplification-stage egg chamber follicle cells containing representative clones, marked by the absence of GFP and outlined in white. Scale bars: 20 µm. (B) Mosaic egg chambers for chiffonETBE3 were examined for H3K9ac (n=5), H3K18ac (n=6) or H3K23ac (n=7) as in A. Representative images are shown for each histone modification. Scale bars: 20 µm. (C) Box plots showing relative H3K14ac levels in mutant clones versus GFP-positive control regions. 10–30 nuclei were quantified per region for 9–10 independent animals (red dots indicate clone analyzed from an individual animal; X, mean). P-values for the indicated comparisons were determined by ANOVA+Tukey-HSD; ns, not significant. (D) The percentage of clones undergoing gene amplification (BrdU-positive foci) in amplification-stage egg chambers from the indicated genotypes was determined. Several genotypes showed clones that were composed entirely or partially of pyknotic nuclei, which did not undergo gene amplification. The number of independent animals and clones examined for each genotype is shown above the plot (animals/clones).
CHAT-mediated histone acetylation is not required for gene amplification
Histone acetylation correlates with and contributes to localized replication at the amplified follicle cell origins (Aggarwal and Calvi, 2004; Liu et al., 2012; McConnell et al., 2012). Moreover, mutations in chiffon eliminate gene amplification in follicle cells (Landis and Tower, 1999; Stephenson et al., 2015; Zhang and Tower, 2004). Therefore, we asked whether CHAT-mediated histone acetylation was also necessary for gene amplification. As observed previously, chiffonETBE3 clones lack the characteristic 5-bromo-2-deoxyuridine (BrdU) foci indicative of chorion gene re-replication that are present in the wild-type cells adjacent to the clone or in the FRT40A control clone (Fig. 4A,D). To test whether CHAT-mediated histone acetylation was required for gene amplification, we examined ada2b1 somatic ovary mosaics, which exhibit decreased levels of histone H3 acetylation, similar to that observed in chiffonETBE3 mutant cells (Fig. 4A,C). In contrast to chiffonETBE3 clones that lack detectable gene amplification, we observed multiple ada2b1 clones undergoing gene amplification (Fig. 4A,D). We note that there were more pyknotic nuclei in ada2b1 clones, suggesting that loss of both Ada2b isoforms increased cell death in follicle cells, potentially due to pleiotropic effects resulting from loss of both the SAGA and CHAT complexes. Supporting this, loss of ada2b in the germline cells of female flies results in arrested oogenesis and increased apoptosis, suggesting that proper histone acetylation is necessary for other aspects of egg development (Li et al., 2017). Despite this, these data suggest that Ada2b-PA, which is necessary for histone acetyltransferase activity of the CHAT complex, is not required for gene amplification.
Because our MudPIT data and binding studies suggested that the N- and C-terminal domains of Chiffon interacted independently with Cdc7 and the CHAT complex, respectively, we wondered whether expression of these domains would restore either gene amplification or histone acetylation in chiffon mutants. To test this, we generated flies expressing either full-length Chiffon (1–1695 aa, Chiffon-FL), or its N-terminal (1–375 aa, Chiffon-N) or C-terminal (401–1695 aa, Chiffon-C) domains. Each Chiffon construct was expressed under the control of chiffon genomic regulatory elements from transgenes inserted in the third chromosome attP2 site (Fig. 5, Materials and Methods). Both H3K14ac levels and gene amplification were restored in chiffonETBE3 mutant clones by expression of a single copy of full-length Chiffon (Fig. 4A,C,D). In contrast, the N-terminal Chiffon transgene rescued gene amplification, but not histone acetylation, in chiffonETBE3 clones. Further, the C-terminal Chiffon transgene partially rescued histone acetylation, but did not restore gene amplification, in chiffonETBE3 clones. To our surprise, a full-length Chiffon transgene that contained a stop codon at position 376 (Chiffon-FL*), separating the N-terminal Cdc7-binding domain from the C-terminal Gcn5-binding domain, fully restored both gene amplification and histone acetylation in chiffonETBE3 clones. As histone acetyltransferases function redundantly to stimulate follicle cell gene amplification (McConnell et al., 2012), we cannot exclude the possibility that CHAT functions redundantly with other histone acetyltransferases to stimulate origin activity. Indeed, although bulk H3K14ac was reduced in chiffonETBE3 clones expressing the N-terminal Chiffon transgene, we observed residual H3K14ac foci that co-localized with the BrdU foci in half of the images (five of the ten images) analyzed for acetylation in this genotype. We also observed H3K14ac foci that co-localized with the BrdU foci in some of the ada2b clones (three of the nine images), but these were much fainter than those present in chiffonETBE3 clones expressing the N-terminal Chiffon transgene. This suggests that other histone acetyltransferases target the amplified follicle cell origins in the absence of CHAT, likely including SAGA. Thus, we conclude that the histone acetyltransferase activity of the CHAT complex alone is not essential for the specialized gene amplification form of DNA replication that occurs in follicle cells.
Fig. 5.
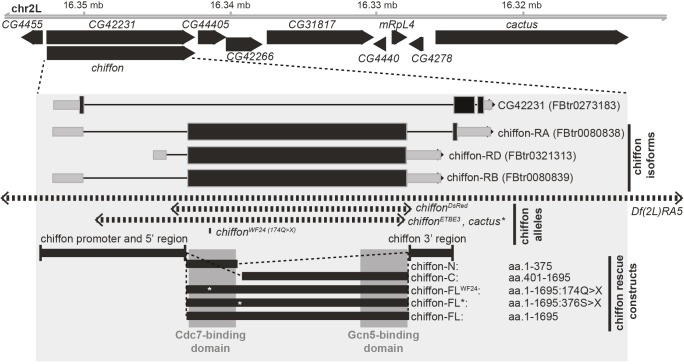
Map of chiffon gene structure. Schematic of chiffon locus showing nearby genes including cactus. The gene structure for chiffon is shown in the inset shaded box as coding sequences (black boxes), untranslated regions (gray boxes), and introns (lines). There are three annotated splice isoforms for chiffon: RA encodes a 1711-aa protein; RB and RD encode 1695-aa proteins from a single ∼5 kb exon. An overlapping gene, CG42231, shares a promoter with chiffon but differs in its reading frame and encodes a separate polypeptide. The genomic regions deleted/mutated in each of the indicated chiffon alleles (chiffonDsRed, chiffonETBE3 and chiffonWF24) are shown by the dashed line arrows. The chiffon rescue transgenes are shown by the black boxes at the base of the panel. Rescue constructs contain the indicated chiffon 5′ and 3′ regulatory regions (black boxes) and the chiffon coding sequences. The conserved Dbf4 N-terminal domain and C-terminal insect-specific Gcn5-binding domain are indicated by the shaded boxes overlaying the rescue constructs, and the position of each nonsense mutation in the rescue constructs is indicated by an asterisk.
CHAT-mediated histone acetylation is essential for viability in flies
To our surprise, a premature stop codon in one of the chiffon transgenes (Chiffon-FL*), that should have truncated the protein prior to the Gcn5-binding region, fully rescued histone acetylation in chiffon mutant cells. These data implied that there could be an internal translation start site within the single, large exon in the chiffon gene (Fig. 5). Indeed, we identified a potential consensus initiation codon sequence 393 aa from the end of the chiffon coding region that would be expected to generate a ∼43 kDa polypeptide. Although chiffon has been reported to be dispensable for viability in flies, these conclusions were based largely on an allele containing a nonsense mutation at position 174, chiffonWF24 (Landis and Tower, 1999). This mutation disrupts the Cdc7-binding domain and results in viable flies with phenotypes indicative of partially disrupted DNA replication, such as rough eyes and female infertility. Although these data suggested that the Cdc7-binding activity of Chiffon was not essential for viability in flies, we wondered whether this was also the case for the CHAT complex. Because Ada2b-PA was sufficient to partially restore viability to ada2b mutants (Table 1), we hypothesized that the CHAT complex is essential for development in flies.
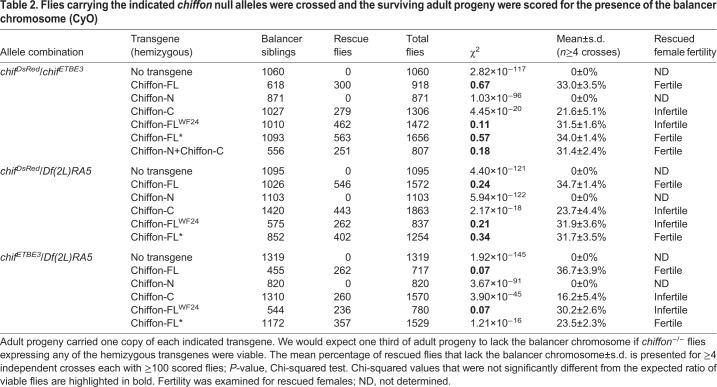
To test this, we used CRISPR-Cas9 technology to generate a new null chiffon allele in which the entire chiffon coding region was replaced with a visible eye marker, 3xP3-DsRed (Fig. 5, chiffonDsRed). We then crossed these chiffonDsRed flies with the chiffonETBE3 null allele generated by Landis and Tower (1999), or the Df(2L)RA5 deficiency that spans the chiffon gene and removes several adjacent genes. Lethality in the chiffonETBE3 flies was previously attributed to a secondary mutation in the nearby cactus gene, which is also missing in the Df(2L)RA5 deficiency. However, we found that combinations of any of these three chiffon alleles resulted in complete adult lethality (Table 2). We then expressed single-copy chiffon rescue transgenes expressing full-length Chiffon (Chiffon-FL) with or without the chiffonWF24 mutation (174Q>X). If the chiffon rescue transgene fully restored Chiffon function, we would expect to observe one third of adult progeny lacking the balancer chromosome. Moreover, we would expect that female adult progeny with restored Chiffon function would be fertile due to restoration of Chiffon activity in ovary follicle cells. Indeed, expression of the wild-type full-length Chiffon transgene fully restored both viability and female fertility in all three allele combinations (Table 2). Moreover, similar to Landis and Tower (1999), the Chiffon-FLWF24 transgene fully restored viability, but not female fertility, in the chiffonDsRed/chiffonETBE3 progeny; similar results were observed with the other chiffon allele combinations. Thus, the chiffonWF24 mutation, which disrupts the Cdc7-binding domain of Chiffon, eliminates Chiffon function with respect to female fertility, but does not disrupt Chiffon's role in adult viability. Next, we asked whether expression of the Chiffon-C domain, which partially rescued histone acetylation in chiffon clones but did not restore gene amplification, could restore adult viability. Indeed, consistent with the observations for the Chiffon-FLWF24 transgene, expression of the Chiffon-C domain restored viability, although not to the same extent as Chiffon-FL, but the resulting females were infertile. In contrast, expression of the Chiffon-N transgene did not restore viability, even though this transgene did rescue gene amplification in chiffon clones (Fig. 4A,D). Further supporting the possibility that chiffon contains an internal translation start site, a full-length Chiffon transgene that contained a stop codon at position 376 (Chiffon-FL*), separating the N-terminal Cdc7-binding domain from the C-terminal insect-specific region, fully complemented both viability and fertility in chiffon mutants. Thus, we asked whether the Cdc7- and Gcn5-binding domains of Chiffon could function in trans. To test this, we expressed single copies of the Chiffon-N and Chiffon-C in combination, and found that this fully restored both viability and female fertility to chiffon mutants (Table 2). These data demonstrate that Chiffon, like Ada2b-PA, is essential for viability in flies. Moreover, the essential function of Chiffon relates to its histone acetyltransferase activity rather than Cdc7 activation.
Table 2.
Flies carrying the indicated chiffon null alleles were crossed and the surviving adult progeny were scored for the presence of the balancer chromosome (CyO)
Our genetic observations support the possibility that chiffon is a dicistronic gene that encodes two distinct polypeptides; although this type of gene structure is relatively rare in Drosophila, there are several examples of dicistronic genes in flies, including stoned A and stoned B, and Adh (Andrews et al., 1996; Brogna and Ashburner, 1997; Komonyi et al., 2009). The coding sequence for the 1695-aa Chiffon protein encoded by the RD or RB transcripts lies within a single exon (Fig. 5), and northern blot analysis previously identified a single 6.5 kb chiffon transcript (Landis and Tower, 1999), suggesting that alternative splicing is unlikely to account for our observations. To test whether a C-terminal product was generated from either of the Chiffon transgenes that contained premature stop codons, we immunoprecipitated the Chiffon-FL* and Chiffon-FLWF24 proteins via their C-terminal FLAG epitope tags, and performed western blotting with anti-FLAG antibodies. We observed a ∼48 kDa product that was recognized by anti-FLAG antibodies in both the immunoprecipitations from Chiffon-FL* and Chiffon-FLWF24 lysates, but not those from untagged embryo lysates (Fig. 6A). Further, both Chiffon-FL* and Chiffon-FLWF24 co-immunoprecipitated Gcn5, suggesting that the C-terminal product expressed by these transgenes interacted with the CHAT complex. Thus, the Chiffon C-terminal domain nucleates CHAT formation, can be expressed from an alternative translation start site in the chiffon gene and is essential for viability in Drosophila (Fig. 6B).
Fig. 6.
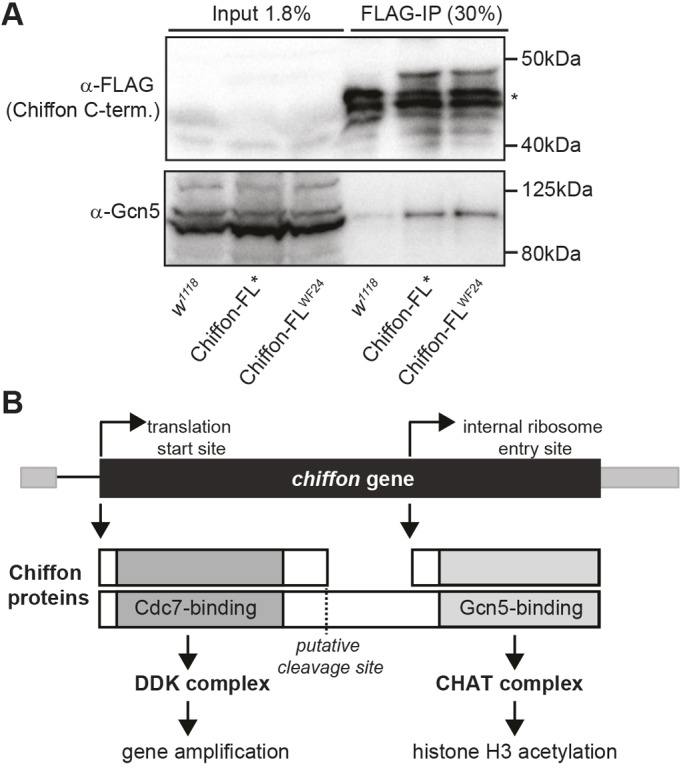
An internal translation start site in chiffon expresses a C-terminal product that binds Gcn5. (A) C-terminally FLAG-tagged Chiffon-FL* or Chiffon-FLWF24 transgenes, that contain premature stop codons at amino acids 376 or 174, respectively, were immunoprecipitated from embryo lysates using anti-FLAG antibodies. Co-immunoprecipitated proteins were analyzed by SDS-PAGE and western blotting with antibodies against FLAG (Chiffon) and Gcn5. The asterisk indicates nonspecific bands present in the w1118 control. Representative data from three experiments are shown. (B) Schematic illustrating the two polypeptides encoded by chiffon. The first start codon encodes full-length Chiffon (1695 aa) with the conserved Dbf4 Cdc7-binding domain in its N-terminal region. The N-terminal Chiffon product binds Cdc7, nucleates DDK formation and is necessary for gene amplification. An alternative internal ribosome entry site generates a C-terminal product containing the insect-specific Gcn5-binding domain that nucleates CHAT formation, and is essential for histone acetylation and development. Our data suggest that two mechanisms might control production of the alternative Chiffon products that nucleate DDK versus CHAT complex formation: (1) translational switching between cap-dependent and IRES-dependent start sites; and/or (2) proteolytic cleavage of full-length Chiffon.
DISCUSSION
Here, we show that the Drosophila Dbf4 ortholog chiffon is a dicistronic gene that encodes two distinct polypeptides from alternative translation start sites. Chiffon's two activities can be separated genetically; its N-terminal domain binds Cdc7 and its C-terminal domain binds the histone acetyltransferase Gcn5 (Fig. 6B). The interaction between Chiffon and Gcn5 forms the CHAT complex that is required for histone H3 acetylation and viability in flies. Thus, in addition to the Gcn5-containing SAGA and ATAC complexes, flies contain a third Gcn5-containing complex: CHAT. The CHAT complex is not present in yeast or human cells, and is likely to be specific to insects because it is nucleated by Chiffon's insect-specific C-terminal domain. Our mass spectrometry data suggest that most Chiffon interacts in a mutually exclusive manner with Cdc7 and CHAT, and transgenes that separate the N- and C-terminal domains of chiffon fully restore both functions. Thus, our data demonstrate that the DDK and CHAT complexes function independently in DNA replication and histone acetylation, respectively.
What might be the function of this CHAT complex in flies? Gcn5 and another histone acetyltransferase, Esa1, stimulate DNA replication in yeast in vitro (Kurat et al., 2017). In addition, several histone acetyltransferases work together to stimulate follicle cell amplification in Drosophila (McConnell et al., 2012). However, our work argues against a role for the CHAT complex in DNA replication; although we cannot exclude the possibility that the CHAT complex functions redundantly with other histone acetyltransferases to stimulate DNA replication, CHAT is not essential for gene amplification in follicle cells. Because SAGA is required for proper gene expression in flies, and because the CHAT-specific Ada2b-PA isoform can restore viability to ada2b mutants, we propose that CHAT, like the SAGA and ATAC complexes, regulates gene expression in flies. In other organisms, Dbf4 levels fluctuate throughout the cell cycle to control activity of Cdc7 (Cheng et al., 1999; Oshiro et al., 1999): Dbf4 protein levels correlate with Cdc7 activity and increase at the G1–S transition, peak in S phase, and then become low during G1 phase, when Dbf4 is degraded by the anaphase-promoting complex (Cheng et al., 1999; Oshiro et al., 1999). One possibility in flies is that Chiffon levels are also cell cycle regulated, and if so, CHAT complex expression could be controlled by Chiffon levels, potentially peaking in S phase. Thus, the DDK and CHAT functions of chiffon could have evolved as part of the same gene structure to coordinate DNA replication with expression of CHAT target genes during the cell cycle in insects.
Although Dbf4 did not interact with Gcn5 in yeast or in human cells, some observations support a potential role for Dbf4 in gene expression in these organisms. For example, the C-terminal domain of human DBF4 (also known as ASK) binds the chromatin-associated protein Lens epithelium-derived growth factor (LEDGF; also known as PSIP1), which is associated with the MLL histone H3 methyltransferase complex (Hughes et al., 2010; Yokoyama and Cleary, 2008). Further, the C-terminus of yeast Dbf4 binds forkhead transcription factors (Fang et al., 2017). In addition, DDK phosphorylates Thr45 of histone H3 in budding yeast (Baker et al., 2010), demonstrating a direct role for DDK complexes in chromatin modification. Thus, although the CHAT complex might be specific to insects, Dbf4 orthologs could have a more general role in gene expression in addition to their essential DNA replication activity.
One unusual feature of Chiffon in flies is that its Cdc7-binding activity is dispensable for viability. Loss of either Dbf4 or Cdc7 disrupts DNA replication and mitosis in organisms from yeast to mammals, leading to growth defects and/or cell death (Labib, 2010). In flies, Cdc7 is also an essential gene (Stephenson et al., 2015), and recent work showed that Cdc7 is required for early embryonic nuclear cycles, consistent with its essential role in DNA replication (Seller and O'Farrell, 2018). However, our data show that Chiffon's Cdc7-binding activity is not essential for DNA replication or viability in flies, although it is required for follicle cell gene amplification in the ovary. These conclusions are consistent with the previous findings of Landis and Tower (1999), and are in stark contrast to the absolute requirement of Cdc7 and Dbf4 for DNA replication and cell viability in organisms from yeast to vertebrates (Labib, 2010; Landis and Tower, 1999). Despite this, chiffon is essential for development in flies but this is due to a requirement for the CHAT complex, likely due to its role in histone acetylation. Thus, our studies raise the question of how Drosophila Cdc7 can function in the absence of its Dbf4 regulatory partner, because flies do not have any other detectable sequence homolog for Dbf4. Whereas budding yeast possesses only one homolog for Dbf4 and Cdc7, several organisms possess paralogs of DDK subunits with specialized functions in meiosis and development. In particular, the vertebrate Dbf4B paralog has specialized roles in early embryogenesis (Collart et al., 2017; Montagnoli et al., 2002; Silva et al., 2006; Yoshizawa-Sugata et al., 2005). If Chiffon, like Dbf4B, has a more specialized developmental role in DNA re-replication in ovary follicle cells, then our data suggest that there might be an alternative mechanism to regulate Cdc7 activity in flies.
MATERIALS AND METHODS
Affinity purification, MudPIT analysis and histone acetyltransferase assays
Tandem FLAG-HA affinity purification and MudPIT analysis was conducted from stable Drosophila S2 cell lines as described previously (Stegeman et al., 2016). TAP purifications from S. cerevisiae was performed as described previously (Lee et al., 2004). To estimate relative protein levels, distributed normalized abundance factors (dNSAFs) were calculated for each nonredundant protein or protein group (Zhang et al., 2010). Briefly, shared spectral counts (sSpC) were distributed based on spectral counts unique to each protein (uSpC). Histone acetyltransferase assays were performed as previously described (Stegeman et al., 2016) using Flag-purified complexes and HeLa core histones or human histone H3 peptide (K5–K23) as substrate.
Fly stocks and genetics
Genotypes for flies used in this study are described in Table S2. The chiffonETBE3 (Landis and Tower, 1999) and ada2b1 (Qi et al., 2004) null alleles were used for somatic mosaic analysis. The null ada2b1 and ada2b842 (Pankotai et al., 2013) alleles that disrupt both Ada2b isoforms were used to assess adult survival. Ada2b rescue transgenes contain genomic ada2b enhancer sequences that begin −1878 bp from the transcription start site and extend +1782 bp to the end of the second exon. The alternative exon 3 and 4 sequences for each Ada2b isoform are fused directly to the 3′ end of exon 2. Constructs were generated in the pCa4B vector with the addition of the Adh 3′ UTR and polyadenylation signal sequences from the pRmaHa3 vector. Transgenic flies were generated using the phiC31 site-specific integration system in the attP40 site on chromosome 2L. Chiffon rescue transgenes contain genomic chiffon enhancer sequences that span −3480 bp relative to the translation start site of the chiffon-RD transcript, and include the chiffon 3′ UTR sequences that extend 1056 bp past the stop codon of the chiffon-RD transcript. Chiffon domain constructs encode the indicated number of amino acids relative to 1695 aa full-length Chiffon based on the chiffon-RD transcript. Chiffon constructs were N- and C-terminally epitope tagged with 2xHA and FLAG, respectively. Transgenic flies were generated in the attP2 site on chromosome 3L. The chiffonDsRed allele was generated using CRISPR-Cas9 technology (Gratz et al., 2014). The following guide RNAs were used to target the chiffon 5088 bp exon for replacement: 5′-GGAGGGAAACTTTATAGGAGTGG-3′ and 5′-GATGATGATTAGATGACACAGGG-3′. Flanking regions immediately upstream and downstream of the chiffon coding region (chiffon-RD) were cloned into the flyCRISPR vector pHD-DsRed-attP and used as a template for homologous recombination. Flies expressing DsRed were selected, and the insertion position of the 3xP3-DsRed-attP cassette was confirmed by PCR and sequencing. The chiffonETBE3 allele was also confirmed by PCR and sequencing. The genomic positions of the regions deleted in each chiffon allele are as follows: chiffonDsRed chr2L, 16344356–16349852; chiffonETBE3 chr2L, 16344400–16351631.
Immunohistochemistry
Somatic clones were induced in egg chambers, and ovaries were dissected from adult females at 3 days posteclosion, labeled with BrdU, fixed, and immunostained with anti-BrdU (#555627, BD Pharmingen, mouse, 1:20) and either anti-H3K14ac (07-353, Millipore, rabbit, 1:100), anti-H3K9ac (ab10812, Abcam, rabbit, 1:500), anti-H3K18ac (ab1191, Abcam, rabbit, 1:400) or anti-H3K23ac (ab47813, Abcam, rabbit, 1:700) antibodies, followed by Alexa Fluor 568- and Alexa Fluor 633-conjugated secondary antibodies (Life Technologies). They were then imaged as described previously (Stephenson et al., 2015). H3K14ac levels were quantified for 10–30 nuclei in each clone relative to a similar number of nuclei from the surrounding wild-type region of the tissue (GFP positive). Acetylation levels were determined as average sum intensity values for nuclear-localized fluorescence using NIS-Elements Analysis software. Acetylation levels were quantified for four individual frames from a z-stack image of each egg chamber. These four frames were selected based on those images that contained the brightest H3K14ac signal in the wild-type region (GFP positive) of the egg chamber. Somatic clones were induced in imaginal discs by heat shock for 30 min at 37°C 72 h after egg laying. Imaginal discs were dissected from wandering third-instar larvae and immunostained with anti-H3K14ac.
Phylogenetic analysis
The following protein sequences were aligned using Clustal Omega (Sievers et al., 2011) and used to generate a neighbor-joining phylogenetic tree, which was plotted using phytools in R (Revell, 2012): Bos taurus, XP_024836692.1 and XP_015324178.1; Canis lupus familiaris, XP_532451.2 and XP_022278602.1; Homo sapiens, NP_006707.1 and NP_663696.1; Gallus gallus, XP_004939326.1 and XP_004948536.1; Xenopus laevis, ABB16337.1 and BAC76421.1; Mus musculus, NP_001177646.1; Ceratitis capitata, XP_004521831.1; Lucilia cuprina, XP_023301579.1; Drosophila melanogaster, AAD48779.1; Camponotus floridanus, EFN62957.1; Pogonomyrmex barbatus, XP_011633258.1; Linepithema humile, XP_012229084.1; Apis mellifera, XP_016770645.1; Apis florea, XP_003693265.1; Tribolium castaneum, XP_008197891.1; Schizosaccharomyces pombe, CAA19117.1 and CAB39799.1; Aspergillus nidulans AAD01519.1; S. cerevisiae, NP_010337.3; Eremothecium gossypii, NP_986462.1; Kluyveromyces lactis, XP_455609.1.
Yeast two-hybrid assay
Yeast two-hybrid analysis was performed with the Matchmaker Gold Yeast two-hybrid system as per the manufacturer's instructions (Clontech). Three independent transformed colonies were replica plated on the different selective media for each interaction tested.
Co-immunoprecipitation and western blotting analysis
Recombinant proteins (500 ng) were incubated with glutathione-sepharose (16100, Thermo Fisher Scientific) in the following buffer: 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% NP-40. Embryo lysates (4 mg protein) were immunoprecipitated using FLAG-agarose in the following buffer: 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 0.5% NP-40, 10% glycerol. C-terminally FLAG-tagged DBF4A, DBF4B or pENTER empty vector control (CG801040, CH874659, P100001, Vigene Biosciences, Rockville, MD) were transiently transfected into HEK293 T cells, and nuclear lysates (1 mg protein) were immunoprecipitated using Flag M2 antibodies and Protein-G Dynabeads in the following buffer: 50 mM Tris-HCl pH 8.0, 150 mM NaCl and 0.2% IPEGAL with PMSF, aprotinin, leupeptin and pepstatin. The following antibodies were used for western blot analysis: anti-GST (PC53, Millipore, rabbit, 1:1000), anti-His-HRP (MA1-21315, Invitrogen, mouse, 1:1000), anti-FLAG-HRP (A8592, Sigma-Aldrich, mouse, 1:5000), anti-Drosophila Gcn5 (rabbit, 1:3000) (Kusch et al., 2003), anti-Flag M2 (Sigma-Aldrich; 1:1000), anti-human CDC7 (ab108382, Abcam, 1:1000), anti-human PCAF (ab12188, Abcam, 1:500) and anti-human GCN5 (ab153903, Abcam, 1:1000). HEK293T cells were obtained from the American Type Culture Collection and were tested for mycoplasma contamination using a MycoAlert Mycoplasma Detection Kit (Lonza).
Supplementary Material
Acknowledgements
We thank John Tower, Scott Hawley, Mattias Mannervik, Imre Boros and the Bloomington Drosophila Stock Center (NIH P40OD018537) for flies. The following undergraduate students contributed to the yeast two-hybrid experiments: Jose Adorno-Cancel, Evan Baker, William Delacruz, Shohei Fujikawa, Brianna Kennedy, Macey Lee, Emily Olson, Delayna Pagan, Josie Rhodes, Karissa Rulon, Ana Stenstrom and Samantha Villagomez.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.F.T.-Z., V.M.W.; Methodology: M.P.W., V.M.W.; Formal analysis: E.F.T.-Z., V.M.W.; Investigation: E.F.T.-Z., R.E.S., A.A., B.D.A., S.K.S., L.F., V.M.W.; Resources: M.P.W., V.M.W.; Data curation: E.F.T.-Z., L.F., V.M.W.; Writing - original draft: E.F.T.-Z., V.M.W.; Writing - review & editing: E.F.T.-Z., V.M.W.; Visualization: E.F.T.-Z., R.E.S., A.A., V.M.W.; Supervision: L.F., E.C.D., M.P.W., V.M.W.; Project administration: V.M.W.; Funding acquisition: V.M.W.
Funding
This work was funded by grants to Purdue University Center for Cancer Research from the American Cancer Society [58-006-53] and National Institutes of Health (NIH) [P30 CA023168]. The work was initiated with support from the National Institute of General Medical Sciences [GM99945-01]. Microscopy studies were performed in the Bindley Bioscience Imaging Facility, funded by the NIH-funded Indiana Clinical and Translational Sciences Institute. Deposited in PMC for release after 12 months.
Data availability
All raw and supporting data including detailed protocols have been deposited at the Purdue University Research Repository (PURR) as a publically available, archived data set and can be accessed using https://doi.org/10.4231/R72V2DD0. Original mass spectrometry data files underlying this paper can be accessed from the Stowers Original Data Repository at libpb-1243, or from the MassIVE/ProteomeXChange databases at MSV000081791 or PXD008391.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.214072.supplemental
References
- Aggarwal B. D. and Calvi B. R. (2004). Chromatin regulates origin activity in Drosophila follicle cells. Nature 430, 372-376. 10.1038/nature02694 [DOI] [PubMed] [Google Scholar]
- Andrews J., Smith M., Merakovsky J., Coulson M., Hannan F. and Kelly L. E. (1996). The stoned locus of Drosophila melanogaster produces a dicistronic transcript and encodes two distinct polypeptides. Genetics 143, 1699-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. P., Phillips J., Anderson S., Qiu Q., Shabanowitz J., Smith M. M., Yates J. R. III, Hunt D. F. and Grant P. A. (2010). Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell Biol. 12, 294-298. 10.1038/ncb2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R., Pray-Grant M. G., Selleck W., Grant P. A. and Tan S. (2002). Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277, 7989-7995. 10.1074/jbc.M110849200 [DOI] [PubMed] [Google Scholar]
- Brogna S. and Ashburner M. (1997). The Adh-related gene of Drosophila melanogaster is expressed as a functional dicistronic messenger RNA: multigenic transcription in higher organisms. EMBO J. 16, 2023-2031. 10.1093/emboj/16.8.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Collyer T. and Hardy C. F. J. (1999). Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol. Cell. Biol. 19, 4270-4278. 10.1128/MCB.19.6.4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C., Smith J. C. and Zegerman P. (2017). Chk1 inhibition of the replication factor Drf1 guarantees cell-cycle elongation at the Xenopus laevis mid-blastula transition. Dev. Cell 42, 82-96 e3. 10.1016/j.devcel.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D., Lengronne A., Shi D., Forey R., Skrzypczak M., Ginalski K., Yan C., Wang X., Cao Q., Pasero P. et al. (2017). Dbf4 recruitment by forkhead transcription factors defines an upstream rate-limiting step in determining origin firing timing. Genes Dev. 31, 2405-2415. 10.1101/gad.306571.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. A., Duggan L., Cote J., Roberts S. M., Brownell J. E., Candau R., Ohba R., Owen-Hughes T., Allis C. D., Winston F. et al. (1997). Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640-1650. 10.1101/gad.11.13.1640 [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M. and O'Connor-Giles K. M. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961-971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S., Suganuma T., Florens L., Weake V., Swanson S. K., Washburn M. P., Abmayr S. M. and Workman J. L. (2006). The essential gene wda encodes a WD40 repeat subunit of Drosophila SAGA required for histone H3 acetylation. Mol. Cell. Biol. 26, 7178-7189. 10.1128/MCB.00130-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A. H., Prochasson P., Neely K. E., Galasinski S. C., Chandy M., Carrozza M. J. and Workman J. L. (2002). Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111, 369-379. 10.1016/S0092-8674(02)01005-X [DOI] [PubMed] [Google Scholar]
- Hughes S., Jenkins V., Dar M. J., Engelman A. and Cherepanov P. (2010). Transcriptional co-activator LEDGF interacts with Cdc7-activator of S-phase kinase (ASK) and stimulates its enzymatic activity. J. Biol. Chem. 285, 541-554. 10.1074/jbc.M109.036491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Yu L.-R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y. and Ge K. (2011). Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249-262. 10.1038/emboj.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komonyi O., Schauer T., Papai G., Deak P. and Boros I. M. (2009). A product of the bicistronic Drosophila melanogaster gene CG31241, which also encodes a trimethylguanosine synthase, plays a role in telomere protection. J. Cell Sci. 122, 769-774. 10.1242/jcs.035097 [DOI] [PubMed] [Google Scholar]
- Kurat C. F., Yeeles J. T. P., Patel H., Early A. and Diffley J. F. X. (2017). Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 65, 117-130. 10.1016/j.molcel.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T., Guelman S., Abmayr S. M. and Workman J. L. (2003). Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23, 3305-3319. 10.1128/MCB.23.9.3305-3319.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K. (2010). How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 24, 1208-1219. 10.1101/gad.1933010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G. and Tower J. (1999). The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development 126, 4281-4293. [DOI] [PubMed] [Google Scholar]
- Lee K. K., Prochasson P., Florens L., Swanson S. K., Washburn M. P. and Workman J. L. (2004). Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem. Soc. Trans. 32, 899-903. 10.1042/BST0320899 [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki Y., Young M. R., Kihara M., Sugino A. and Tye B. K. (1997). Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 11, 3365-3374. 10.1101/gad.11.24.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M. and Workman J. L. (2007). The role of chromatin during transcription. Cell 128, 707-719. 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Li X., Seidel C. W., Szerszen L. T., Lange J. J., Workman J. L. and Abmayr S. M. (2017). Enzymatic modules of the SAGA chromatin-modifying complex play distinct roles in Drosophila gene expression and development. Genes Dev. 31, 1588-1600. 10.1101/gad.300988.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., McConnell K., Dixon M. and Calvi B. R. (2012). Analysis of model replication origins in Drosophila reveals new aspects of the chromatin landscape and its relationship to origin activity and the prereplicative complex. Mol. Biol. Cell 23, 200-212. 10.1091/mbc.e11-05-0409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Kanakousaki K. and Buttitta L. (2015). How the cell cycle impacts chromatin architecture and influences cell fate. Front. Genet. 6, 19 10.3389/fgene.2015.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell K. H., Dixon M. and Calvi B. R. (2012). The histone acetyltransferases CBP and Chameau integrate developmental and DNA replication programs in Drosophila ovarian follicle cells. Development 139, 3880-3890. 10.1242/dev.083576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A., Bosotti R., Villa F., Rialland M., Brotherton D., Mercurio C., Berthelsen J. and Santocanale C. (2002). Drf1, a novel regulatory subunit for human Cdc7 kinase. EMBO J. 21, 3171-3181. 10.1093/emboj/cdf290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orpinell M., Fournier M., Riss A., Nagy Z., Krebs A. R., Frontini M. and Tora L. (2010). The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 29, 2381-2394. 10.1038/emboj.2010.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro G., Owens J. C., Shellman Y., Sclafani R. A. and Li J. J. (1999). Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol. Cell. Biol. 19, 4888-4896. 10.1128/MCB.19.7.4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T., Zsindely N., Vamos E. E., Komonyi O., Bodai L. and Boros I. M. (2013). Functional characterization and gene expression profiling of Drosophila melanogaster short dADA2b isoform-containing dSAGA complexes. BMC Genomics 14, 44 10.1186/1471-2164-14-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D., Larsson J. and Mannervik M. (2004). Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol. Cell. Biol. 24, 8080-8089. 10.1128/MCB.24.18.8080-8089.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217-223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Seller C. A. and O'Farrell P. H. (2018). Rif1 prolongs the embryonic S phase at the Drosophila mid-blastula transition. PLoS Biol. 16, e2005687 10.1371/journal.pbio.2005687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J. et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva T., Bradley R. H., Gao Y. and Coue M. (2006). Xenopus CDC7/DRF1 complex is required for the initiation of DNA replication. J. Biol. Chem. 281, 11569-11576. 10.1074/jbc.M510278200 [DOI] [PubMed] [Google Scholar]
- Spedale G., Timmers H. T. M. and Pijnappel W. W. M. P. (2012). ATAC-king the complexity of SAGA during evolution. Genes Dev. 26, 527-541. 10.1101/gad.184705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C. and Mahowald A. P. (1980). Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77, 1096-1100. 10.1073/pnas.77.2.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman R., Spreacker P. J., Swanson S. K., Stephenson R., Florens L., Washburn M. P. and Weake V. M. (2016). The spliceosomal protein SF3B5 is a novel component of Drosophila SAGA that functions in gene expression independent of splicing. J. Mol. Biol. 428, 3632-3649. 10.1016/j.jmb.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson R., Hosler M. R., Gavande N. S., Ghosh A. K. and Weake V. M. (2015). Characterization of a Drosophila ortholog of the Cdc7 kinase: a role for Cdc7 in endoreplication independent of Chiffon. J. Biol. Chem. 290, 1332-1347. 10.1074/jbc.M114.597948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F., Banerjee R., Stratton C. A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M. O., Scacheri P. C. and Harte P. J. (2009). CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131-3141. 10.1242/dev.037127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J. (2004). Developmental gene amplification and origin regulation. Annu. Rev. Genet. 38, 273-304. 10.1146/annurev.genet.37.110801.143851 [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Rubbi L., Lucas I., Brewer B. J. and Grunstein M. (2002). Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223-1233. 10.1016/S1097-2765(02)00702-5 [DOI] [PubMed] [Google Scholar]
- Wang Y. L., Faiola F., Xu M., Pan S. and Martinez E. (2008). Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 283, 33808-33815. 10.1074/jbc.M806936200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake V. M. and Workman J. L. (2010). Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 11, 426-437. 10.1038/nrg2781 [DOI] [PubMed] [Google Scholar]
- Weake V. M., Swanson S. K., Mushegian A., Florens L., Washburn M. P., Abmayr S. M. and Workman J. L. (2009). A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev. 23, 2818-2823. 10.1101/gad.1846409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M. and Stillman B. (1999). Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18, 5334-5346. 10.1093/emboj/18.19.5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A. and Cleary M. L. (2008). Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14, 36-46. 10.1016/j.ccr.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa-Sugata N., Ishii A., Taniyama C., Matsui E., Arai K. and Masai H. (2005). A second human Dbf4/ASK-related protein, Drf1/ASKL1, is required for efficient progression of S and M phases. J. Biol. Chem. 280, 13062-13070. 10.1074/jbc.M411653200 [DOI] [PubMed] [Google Scholar]
- Zhang H. and Tower J. (2004). Sequence requirements for function of the Drosophila chorion gene locus ACE3 replicator and ori-beta origin elements. Development 131, 2089-2099. 10.1242/dev.01064 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wen Z., Washburn M. P. and Florens L. (2010). Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272-2281. 10.1021/ac9023999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.