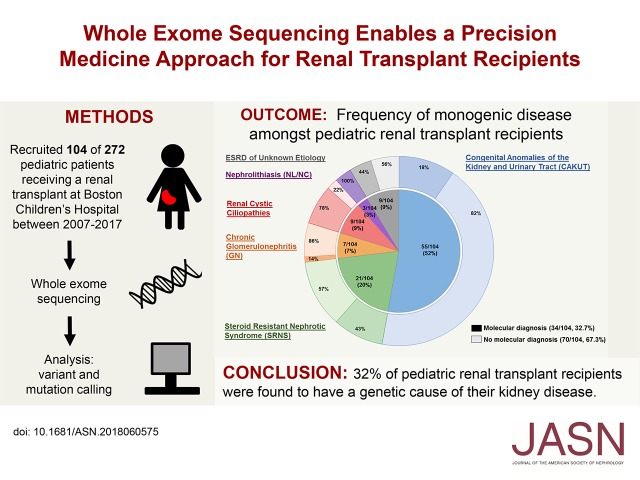
Significance Statement
Case reports describe scenarios in which previously undiagnosed genetic disorders, such as primary hyperoxaluria type 1, caused early allograft failure in kidney transplant recipients. Whole-exome sequencing (WES) has found that approximately 20% of pediatric patients with CKD have a relevant mutation, but the diagnostic yield of WES in kidney transplant recipients is not known. In this study of 104 kidney transplant recipients at a single center, use of WES provided a molecular genetic diagnosis for 34 out of 104 (32.7%) patients. Such diagnoses enabled identification of potential prospective consequences for many patients; in others, receiving the diagnosis earlier in the course of their disease might have mitigated negative consequences. The authors propose considering WES for any child or young adult with CKD.
Keywords: transplantation, end-stage renal disease, genetic renal disease, chronic kidney disease, human genetics
Visual Abstract
Abstract
Background
Whole-exome sequencing (WES) finds a CKD-related mutation in approximately 20% of patients presenting with CKD before 25 years of age. Although provision of a molecular diagnosis could have important implications for clinical management, evidence is lacking on the diagnostic yield and clinical utility of WES for pediatric renal transplant recipients.
Methods
To determine the diagnostic yield of WES in pediatric kidney transplant recipients, we recruited 104 patients who had received a transplant at Boston Children’s Hospital from 2007 through 2017, performed WES, and analyzed results for likely deleterious variants in approximately 400 genes known to cause CKD.
Results
By WES, we identified a genetic cause of CKD in 34 out of 104 (32.7%) transplant recipients. The likelihood of detecting a molecular genetic diagnosis was highest for patients with urinary stone disease (three out of three individuals), followed by renal cystic ciliopathies (seven out of nine individuals), steroid-resistant nephrotic syndrome (nine out of 21 individuals), congenital anomalies of the kidney and urinary tract (ten out of 55 individuals), and chronic glomerulonephritis (one out of seven individuals). WES also yielded a molecular diagnosis for four out of nine individuals with ESRD of unknown etiology. The WES-related molecular genetic diagnosis had implications for clinical care for five patients.
Conclusions
Nearly one third of pediatric renal transplant recipients had a genetic cause of their kidney disease identified by WES. Knowledge of this genetic information can help guide management of both transplant patients and potential living related donors.
CKD is an important cause of morbidity and mortality, often progressing to ESRD and necessitating dialysis and renal transplantation. Causes of CKD in children and young adults differ greatly from those in older individuals. Data from the 2014 North American Pediatric Renal Transplant Cooperative Study (NAPRTCS) show that the most common causes of ESRD in the pediatric population include congenital anomalies of the kidney and urinary tract (CAKUT) (39%), chronic glomerulonephritis (GN) (16%), steroid-resistant nephrotic syndrome (SRNS) (15%), renal cystic ciliopathies (6%), and nephrolithiasis/nephrocalcinosis (2.5%). In 6% of patients, the cause of kidney disease is unknown.1
In recent years, we have gained a better understanding of the genetic landscape of CKD in children. In fact, it has been shown that a monogenic disease-causing mutation in one of approximately 220 genes can be identified in up to 20% of patients who develop CKD before 25 years of age.2 Specifically, a monogenic cause can be identified in 5%–14% of patients with CAKUT,3–5 11%–30% of patients with SRNS,6–10 14% of patients with chronic GN,11 33%–63% of patients with a renal cystic ciliopathy,12–15 and 15%–29% of patients with urinary stone disease.16–19 This has important implications for the clinical management of children and young adults with CKD. For example, patients with SRNS who harbor mutations in COQ2, COQ6, or ADCK4 may respond to administration of coenzyme Q10, which provides a therapeutic option in an otherwise untreatable disease.20–23
Establishing a molecular genetic diagnosis can be of particular importance in the renal transplant population. There are case reports describing situations in which, during the work-up for allograft dysfunction, an underlying genetic diagnosis is identified in a transplant recipient, their living donor, or the deceased donor kidney.24,25 M’dimegh et al. describe a 23-year-old man who developed early allograft dysfunction after renal transplantation. It was not until calcium oxalate deposits were seen on allograft biopsy that a diagnosis of primary hyperoxaluria type 1 (PH1) was suspected.24 In another example, several case reports describe patients with allograft dysfunction in whom subsequent work-up demonstrated that their donors had clinically occult Fabry disease.25–27 In all scenarios, identification of a precise molecular diagnosis before transplantation would have helped to guide peri-transplant management and donor ascertainment, thereby avoiding early allograft failure.
To our knowledge, there is currently no systematic study on the prevalence of monogenic disease in pediatric renal transplant recipients, nor on the utility of whole-exome sequencing (WES) in guiding the management of these patients. We therefore hypothesized that by WES, we can identify a causative mutation in at least 20% of renal transplant recipients and that important consequences for clinical management may result. Here, we report on the results from WES in 104 renal transplant recipients who manifested with CKD before 25 years of age and who received a renal transplant at Boston Children’s Hospital within a continuous 10-year period from 2007 to 2017.
Methods
Human Patients
This study was approved by the Institutional Review Board of Boston Children’s Hospital. Patients who developed CKD before 25 years of age and who received a renal transplant between 2007 and 2017 at Boston Children’s Hospital were enrolled after obtaining informed consent. Our recruitment process is summarized in Supplemental Figure 1.
A total of 104 of 272 probands who met our inclusion criteria were enrolled for WES. Of the 168 probands who were not enrolled, 41 had transitioned care to a different hospital, 18 were unable to provide consent due to guardianship issues, and two were deceased. We excluded 23 probands who developed ESRD secondary to nonintrinsic renal disease (e.g., septic shock). 45 probands declined to participate and 39 remain to be approached.
A small subset of patients had been referred for clinical genetic testing before initiation of this study because of a family history of renal disease or the presence of clinical features suggestive of an underlying genetic disorder. These patients were included in our study as our goal was to determine the diagnostic utility of WES for all kidney transplant recipients with a primary renal disease.
The primary clinical diagnosis of each patient was determined via chart review, and categorized as either CAKUT, SRNS, chronic GN, renal cystic ciliopathy, nephrolithiasis/nephrocalcinosis, or ESRD of unknown etiology. CAKUT was defined as the demonstration of any abnormality of number, size, shape, or anatomic position of the kidneys or urinary tract, and included at least one of the following: renal agenesis, renal hypoplasia/dysplasia, multicystic dysplastic kidneys, obstructive uropathy, or reflux nephropathy. SRNS was defined by the lack of response to steroid treatment in a patient with nephrotic syndrome, and included biopsy findings of FSGS and diffuse mesangial sclerosis. Chronic GN encompassed Alport syndrome, membranoproliferative GN, crescentic GN, IgA nephropathy, and hemolytic uremic syndrome. Renal cystic ciliopathies included patients diagnosed with nephronophthisis, medullary cystic disease, or other renal cystic diseases. Nephrolithiasis/nephrocalcinosis included patients with urinary stone disease.
All patients completed clinical questionnaires, which included information regarding age of disease onset, family history of renal disease, and presence of consanguinity. Information regarding extrarenal manifestations was obtained from each patient’s clinical chart.
DNA Isolation and WES
Research-based WES was performed as previously described.28 In brief, genomic DNA was isolated from blood lymphocyte or saliva samples and subjected to exome capture using Agilent SureSelect human exome capture arrays (Life Technologies), followed by next-generation sequencing on the Illumina HighSeq sequencing platform. Sequence reads were mapped to the human reference genome assembly (National Center for Biotechnology Information build 37/hg19) using the CLC Genomics Workbench software (version 6.5.2; CLC bio, Aarhus, Denmark).
Mutation Calling in Approximately 400 Genes Known to Cause CKD
After alignment to the human reference genome, variants were filtered as previously described and as summarized in Supplemental Figure 2.6,29 In brief, variant filtering on the basis of population frequency was performed using population databases (Exome Sequencing Project [http://evs.gs.washington.edu/EVS], Exome Aggregation Consortium [http://exac.broadinstitute.org], Genome Aggregation Database [http://gnomad.broadinstitute.org], and 1000 Genomes Project [http://www.internationalgenome.org/1000-genomes-browsers]) to include only rare alleles (i.e., minor allele frequency <1%). The exception to this was the NPHS2 R229Q allele, which, despite having a minor allele frequency >1%, has been reported to be pathogenic when in trans with other specific NPHS2 alleles.30 Synonymous and intronic variants that were not located within splice site regions were excluded.
We then evaluated WES data for causative mutations in 396 genes associated with renal disease (Supplemental Table 1, A–G). This included 41 genes that cause isolated CAKUT, 50 genes for SRNS, 17 genes for chronic GN, 95 genes for renal cystic ciliopathies, and 37 genes for urinary stone disease. We also evaluated WES data for variants in an additional 145 genes that have been reported to cause syndromic CAKUT and 11 genes that have been reported to cause unspecified CKD.
A mean depth coverage of 58× was achieved in our cohort. Six of the 396 known CKD genes did not achieve a mean coverage of at least 30× (Supplemental Table 2).
Surviving variants were then ranked based on their likelihood of being disease-causing, taking into consideration evolutionary conservation among orthologs using the ENSEMBL Genome Browser (http://www.ensembl.org) and assembled using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), as well as prediction scores from the web-based prediction programs PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2), SIFT (http://sift.bii.a-star.edu.sg/), and MutationTaster (http://www.mutationtaster.org). Remaining variants were further evaluated by review of the existing literature and determination of phenotypic match. Clinician-scientists and geneticists who had knowledge of the clinical phenotypes and pedigree structure as well as experience with exome evaluation performed mutation calling. In addition, the American College of Medical Genetics and Genomics guidelines for variant classification were applied and variants were considered to be disease-causing if they were classified as pathogenic or likely pathogenic (Supplemental Table 3).31–33 All variants were confirmed in original patient DNA by Sanger sequencing.
CNV Analysis
For probands in whom clinical SNP arrays revealed a pathogenic copy number variant (CNV) and WES evaluation for single nucleotide variants and small insertions/deletions was negative, we performed CNV analysis on WES data using CoNIFER software in order to verify the clinical findings.34 WES was not utilized to identify novel CNVs because of the relatively low sensitivity of this technique.
Results
Clinical Characteristics
Of 272 probands who received a renal transplant at Boston Children’s Hospital between 2007 and 2017, we recruited 104 individuals for WES (Supplemental Figure 1). Fifty five (52.9%) patients had a clinical diagnosis of CAKUT, which was the most common cause of ESRD in our cohort. Twenty one (20.2%) patients were clinically diagnosed with SRNS, seven (6.8%) with chronic GN, nine (8.6%) with a renal cystic ciliopathy, and three (2.9%) with nephrolithiasis. In nine out of 104 (8.6%) patients, the cause of ESRD was unknown. Sixty two (59.6%) patients were male, nine (8.7%) were from consanguineous families, and 23 (22.1%) had a family history of renal disease. Clinical characteristics of the 104 probands enrolled for WES are compared with those of the 272 total probands who were eligible for the study and 2,196 patients included in the NAPRTCS registry between 2007 and 2014 (Supplemental Table 4). The 104 enrolled probands are fairly representative of the general pediatric renal transplant cohort.
A Monogenic Cause of CKD Is Identified in 32.7% of Renal Transplant Recipients
We performed WES in 104 renal transplant recipients and identified a likely causative mutation in one of approximately 400 CKD genes in 34 (32.7%) individuals (Figure 1, Table 1). Six patients had previously undergone clinical genetic testing and already carried a molecular genetic diagnosis. In five families, targeted gene sequencing performed in our laboratory had identified a pathogenic mutation previously.8 In both of these instances, previously identified mutations were confirmed by WES. For 23 individuals, WES provided a molecular genetic diagnosis for the first time.
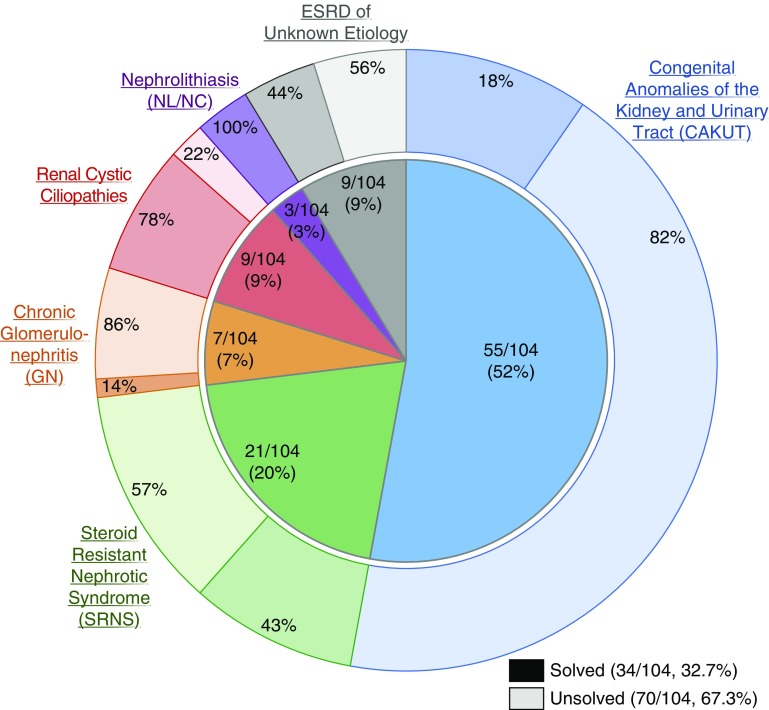
Figure 1.
A monogenic cause of ESRD is identified in 32.7% of renal transplant recipients with onset of CKD before 25 years of age. Probands are categorized by clinical diagnostic group. Inner segments represent the numbers and fractions (in %) of transplant recipients that fall into one of six clinical diagnostic groups, as follows: CAKUT, 55 out of 104 (52%); SRNS, 21 out of 104 (20%); chronic GN, seven out of 104 (7%); renal cystic ciliopathies, nine out of 104 (9%); nephrolithiasis or nephrocalcinosis (NL/NC), three out of 104 (3%); and ESRD of unknown etiology, nine out of 104 (9%). Outer segments represent for each diagnostic group the relative fraction of patients in whom a molecular genetic diagnosis was established (darker colors). A disease-causing mutation was identified in 34 out of 104 families (32.7%). The distribution by clinical diagnostic group is as follows: a molecular diagnosis was established in ten out of 55 (18%) patients with CAKUT, nine out of 21 (43%) patients with SRNS, one out of seven (14%) patients with GN, seven out of nine (78%) patients with a renal cystic ciliopathy, three out of three (100%) patients with nephrolithiasis, and four out of nine (44%) patients with ESRD of unknown etiology.
Table 1.
Disease-causing mutations identified in 34 out of 104 (32.7%) renal transplant recipients with onset of CKD at < 25 years of age
| Familya | A Priori Clinical Diagnosis | Post-WES Diagnosis | Extrarenal Manifestations | Family History | Homozygosity (>75 MB) | Gene | Zygosity | c.Change p.Change Segregation (m, p)b | Conservation | PP2c SIFTd/MutationTastere | Allele Frequency in gnomADf | HGMDg (ACMGh) | PMID (if previously reported) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAKUT | |||||||||||||
| B910i | Wolf–Hirschhorn syndrome | Wolf–Hirschhorn syndrome | Growth failure, seizures, developmental delay | No | No | 4p16.3 deletion | Heterozygous deletion (NA) | NA | #10995514 | ||||
| B643i | L UPJO R MCDK | RCAD | Autism | No | No | 17q12 deletion | Heterozygous deletion (NA) | NA | #25536396 | ||||
| B849i | BOR | BOR | Cleft palate, brachial pit, hearing loss | No | No | EYA1 | Het | c.966+1G>C | N/A | N/A | None | Gene (P) | Novel |
| Splice (NA) | |||||||||||||
| B1162 | BOR | BOR | Malformed ears, hearing loss, polydactyly | Yes (mother) | No | EYA1 | Het | c.1319G>A | C.i. | 0.786 Tol/DC | None | Variant (P) | #10464653 |
| p.R440Q (NA) | |||||||||||||
| B664 | B/L MCDK | RCAD | None | No | No | HNF1B | Het | c.494G>A | C.e. | 0.999 Del/— | None | Variant (LP) | #24254850 |
| p.R165H (NA) | |||||||||||||
| B1137 | VUR | Alagille syndrome | ADHD, scoliosis, heart murmur | Yes (mother) | No | JAG1 | Het | c.2638T>C | D.m. | 0.99 Del/DC | 0/1/251,430 | Gene (LP) | Novel |
| p.C880R (NA) | |||||||||||||
| B848 | Alagille syndrome | Alagille syndrome | Tetralogy of Fallot, liver failure | No | No | JAG1 | Het | c.2957_2958insTT | N/A | N/A | None | Gene (P) | Novel |
| p.L986Ffs*2 (NA) | |||||||||||||
| B1142 | B/L renal agenesis | Feingold syndrome | Duodenal web | Yes (father) | No | MYCN | Het | c.1178G>A | D.m. | 1.00 Del/DC | None | Variant (LP) | #15821734 |
| p.R393H (NA) | |||||||||||||
| B934 | BOR | Townes Brocks syndrome | Malformed ears, hearing loss, Duane syndrome, VSD, polydactyly | No | No | SALL1 | Het | c.826C>T | N/A | N/A | None | Variant (P) | #9973281 |
| p.R276* (NA, WT) | |||||||||||||
| B625 | B/L renal dysplasia | EEC syndrome | Hypergonadotropic hypogonadism, sandal gap deformity | Yes (pat gpa) | No | TP63 | Het | c.1012C>T | C.i. | 0.99 Del/DC | 0/1/251,202 | Gene (LP) | Novel |
| p.R338C (NA; het) | |||||||||||||
Table 1.
Continued
| Familya | A Priori Clinical Diagnosis | Post-WES Diagnosis | Extrarenal Manifestations | Family History | Homozygosity (>75 MB) | Gene | Zygosity | c.Change p.Change Segregation (m, p)b | Conservation | PP2c SIFTd/MutationTastere | Allele Frequency in gnomADf | HGMDg (ACMGh) | PMID (if previously reported) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SRNS | |||||||||||||
| B284j | FSGS | FSGS | Charcot-Marie-Tooth | Yes (mother) | No | INF2 | Het | c.542T>G | H.s. | 0.98 Del/DC | None | Variant (P) | #23014460 |
| p.V181G (het, NA) | |||||||||||||
| B1273i | CNS | CNS | Autism, seizures | No | No | NPHS1 | Cpd het | c.139delG | N/A | N/A | 0/3/277,638 | Variant (P) | #18503012 |
| p.A47Pfs*81 (NA)k | |||||||||||||
| c.1701C>A | N/A | N/A | 0/10/282,168 | Variant (P) | #11317351 | ||||||||
| p.C567* (NA)k | |||||||||||||
| B144j | CNS | CNS | Aortic and pulmonary valve stenosis, microcephaly | Yes (sister) | Yes | NPHS1 | Hom | c.728C>T | D.r. | 1.00 Del/DC | None | Gene (LP) | Novel |
| p.P243L (het; het) | |||||||||||||
| B801i | CNS | CNS | Hypotonia, developmental delay | No | Yes | NPHS1 | Hom | c.1379G>A | M.m. | 0.48 Tol/Poly | 0/1/250,476 | Variant (LP) | #11317351 |
| p.R460Q (NA) | |||||||||||||
| B350j | CNS | CNS | Aortic stenosis | No | No | NPHS1 | Hom | c.1868G>T | D.r. | 1.00 Del/DC | 0/10/282,168 | Variant (P) | #9915943 |
| p.C623F (NA) | |||||||||||||
| B1395i | CNS | CNS | Polymicrogyria, arthrogryposis, developmental delay | Yes (brother) | Yes | NPHS2 | Hom | c.503G>A | D.m. | 0.99 Del/DC | 0/3/221,108 | Variant (P) | #15253708 |
| p.R168H (het; het) | |||||||||||||
| B188j | FSGS | FSGS | None | No | No | NPHS2 | Hom | c.855_856delAA | N/A | N/A | 0/20/281,474 | Variant (P) | #10742096 |
| p.R286Tfs*17 (NA) | |||||||||||||
| B354 | FSGS | FSGS | None | Yes (brother) | Yes | PLCE1 | Hom | c.4978_4981delCAGA | N/A | N/A | None | Variant (P) | #25349199 |
| p.Q1660Lfs*9 (het; het) | |||||||||||||
| B92j | Frasier syndrome | Frasier syndrome | Gonadal dysgenesis, hereditary spherocytosis | Yes (mother, sister) | No | WT1 | Het | c.1432+4C>T | N/A | N/A | None | Variant (P) | #9398852 |
| Splice (het; NA) | |||||||||||||
| Chronic GN | |||||||||||||
| B2440 | Alport syndrome | Alport syndrome | Hearing loss | Yes (mother) | No | COL4A5 | Hemi | c.4791T>G | N/A | N/A | None | Variant (P) | #9848783 |
| p.Y1597* (het; NA) | |||||||||||||
Table 1.
Continued
| Familya | A Priori Clinical Diagnosis | Post-WES Diagnosis | Extrarenal Manifestations | Family History | Homozygosity (>75 MB) | Gene | Zygosity | c.Change p.Change Segregation (m, p)b | Conservation | PP2c SIFTd/MutationTastere | Allele Frequency in gnomADf | HGMDg (ACMGh) | PMID (if previously reported) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Renal cystic ciliopathies | |||||||||||||
| A4037l | NPHP | NPHP | Retinal degeneration, pseudotumor cerebri, macrocephaly, XXX karyotype | No | No | CEP83 | Hom | c.2007delA | N/A | N/A | None | Variant (P) | #24882706 |
| p.E669Dfs*14 (het, het) | |||||||||||||
| B659 | Senior Loken syndrome | Senior Loken syndrome | Retinal degeneration | Yes (sister) | No | CEP83 | Hom | c.2007delA | N/A | N/A | None | Variant (P) | #24882706 |
| p.E669Dfs*14 (NA) | |||||||||||||
| B06 | Short-rib thoracic dysplasia | Short-rib thoracic dysplasia | Restrictive lung disease, cholestatic liver disease | No | No | DYNC2H1 | Cpd het | c.9638A>G | D.m. | 1.00 Del/DC | 0/1/31,366 | Gene (LP) | Novel |
| p.Y3213C (WT; het) | |||||||||||||
| c.12431C>G | G.g. | 0.97 Del/DC | 0/3/279,946 | Gene (LP) | Novel | ||||||||
| p.P4144R (het; WT) | |||||||||||||
| B1233 | NPHP | RCAD | None | No | No | HNF1B | Het | c.857T>C | C.i. | 1.00 Del/— | None | Gene (LP) | Novel |
| p.L286P (NA) | |||||||||||||
| B367 | NPHP | NPHP | None | No | No | NPHP1 | Hom | Homozygous deletion (NA) | Variant (P) | #9326933 | |||
| B950 | NPHP | NPHP | Oculomotor apraxia | No | No | NPHP1 | Hom | Homozygous deletion (NA) | Variant (P) | #9326933 | |||
| B375 | ARPKD | ARPKD | Increased heterogeneity of liver, splenomegaly | No | No | PKHD1 | Cpd het | c.10452dupT | N/A | N/A | 0/1/250,588 | Variant (P) | #15108281 |
| p.L3485Sfs*18 (NA)k | |||||||||||||
| c.11452G>T | X.t. | 0.6 Del/DC | 0/1/251,398 | Gene (LP) | Novel | ||||||||
| p.V3818F (NA)k | |||||||||||||
| Nephrolithiasis/nephrocalcinosis | |||||||||||||
| B949 | PH1 | PH1 | None | No | No | AGXT | Cpd het | c.33dupC | N/A | N/A | 0/43/270,822 | Variant (P) | #10394939 |
| p.K12Qfs*156 (NA)k | |||||||||||||
| c.508G>A | D.m. | 1.00 Del/DC | 1/136/252,084 | Variant (P) | #23229545 | ||||||||
| p.G170R (NA)k | |||||||||||||
| B2404 | PH1 | PH1 | Hypothyroidism | No | No | AGXT | Hom | c.245G>A | S.c. | 1.00 Del/DC | 0/5/251,218 | Variant(P) | #1349575 |
| p.G82E (het, het) | |||||||||||||
| B942 | PH1 | PH1 | None | No | No | AGXT | Hom | c.473C>T | D.r. | 0.99 Del/DC | 0/1/226,530 | Variant (P) | #15849466 |
| p.S158L (NA) | |||||||||||||
Table 1.
Continued
| Familya | A Priori Clinical Diagnosis | Post-WES Diagnosis | Extrarenal Manifestations | Family History | Homozygosity (>75 MB) | Gene | Zygosity | c.Change p.Change Segregation (m, p)b | Conservation | PP2c SIFTd/MutationTastere | Allele Frequency in gnomADf | HGMDg (ACMGh) | PMID (if previously reported) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ESRD of unknown etiology | |||||||||||||
| B797 | ESRD | NPHP | Learning disability, cerebellar hypoplasia | No | Yes | NPHP1 | Hom | Homozygous deletion (NA) | Variant (P) | #9326933 | |||
| B238 | ESRD | SRNS | None | No | Yes | NUP93 | Hom | c.575A>G | X.t. | 0.03 Del/DC | 0/7/282,600 | Gene (LP) | Novel |
| p.Y192C (NA) | |||||||||||||
| B2559 | ESRD | SRNS | None | No | No | TRPC6 | Het | c.2684G>T | D.r. | 0.90 Del/DC | None | Variant (P) | #21734084 |
| p.R895L (WT, WT) | |||||||||||||
| B1873 | ESRD | NPHP | Neonatal stroke, mild autism spectrum disorder | No | No | TTC21B | Cpd het | c.986A>T | C.i. | 0.99 Del/DC | 0/1/251,074 | Gene (LP) | |
| p.E329V (het, WT)m | Novel | ||||||||||||
| c.1038G>A | N/A | N/A | None | Gene (P) | Novel | ||||||||
| p.W346* (WT, WT)m | |||||||||||||
Patients are grouped according to their a priori clinical diagnosis. The patients in whom WES clarified the clinical diagnosis or lead to a diagnosis for the first time are underlined. m, p, maternal allele, paternal allele; ACMG, American College of Medical Genetics and Genomics; PMID, PubMed ID; NA, not available; L, left; UPJO, ureteropelvic junction obstruction; R, right; MCDK, multicystic dysplastic kidney; RCAD, renal cysts and diabetes; BOR, branchio-oto-renal syndrome; Het, heterozygous; P, pathogenic; C.i., Ciona intestinalis; Tol, tolerated (SIFT); DC, disease causing (MutationTaster); B/L, bilateral; C.e., Caenorhabditis elegans; Del, deleterious (SIFT); LP, likely pathogenic; VUR, vesicoureteral reflux; ADHD, attention deficient hyperactivity disorder; D.m., Drosophila melanogaster; VSD, ventricular septal defect; EEC, ectrodactyly, ectodermal dysplasia, and cleft lip/palate syndrome; pat gpa, paternal grandfather; H.s., Homo sapiens; CNS, congenital nephrotic syndrome; Cpd het, compound heterozygous; Hemi, hemizygous; Hom, homozygous; D.r., Danio rerio; M.m., Mus musculus; Poly, polymorphism (MutationTaster); NPHP, nephronophthisis; WT, wild type; G.g., Gallus gallus; ARPKD, autosomal recessive polycystic kidney disease; X.t., Xenopus tropicalis; S.c., Saccharomyces cerevisiae.
Family number is underlined: WES altered or further clarified the a priori clinical diagnosis.
Segregation listed as (maternal allele, paternal allele). If maternal and paternal DNA are unavailable, segregation is listed as NA.
PolyPhen-2 score, which predicts potential effect of an amino acid change on the structure and function of a protein (http://genetics.bwh.harvard.edu/pph2). More deleterious mutations are closer to 1.000, whereas tolerant changes are closer to 0.000.
SIFT, which predicts whether an amino acid change will affect protein function (http://sift.bii.a-star.edu.sg/).
MutationTaster, prediction tool to determine deleteriousness of an amino acid substitution (http://www.mutationtaster.org).
Genome Aggregation Database (gnomAD; http://www.gnomad.broadinstitute.org).
Human Gene Mutation Database (HGMD; https://portal.biobase-international.com), listed as “Variant” if mutation is reported in HGMD or “Gene” if gene, but not specific allele is reported.
ACMG classifications as described previously.31
Proband underwent clinical genetic testing before enrollment in this study. Mutation was confirmed via WES.
Proband underwent panel sequencing previously in our laboratory before enrollment in this study. Mutation was confirmed via WES.
Probands with compound heterozygous mutations in whom parental DNA was unavailable for segregation. These were only considered to be molecularly solved if there was a clear genotype-phenotype correlation.
Index case published previously (PMID 24882706).
Inspection of WES reads demonstrate that the two alleles are in trans (Supplemental Figure 5).
In the 34 individuals in whom a molecular genetic diagnosis was established, 29 patients had monogenic mutations in 19 different genes, three patients had homozygous deletions of NPHP1, and two patients harbored pathogenic CNVs (Table 1). There were three patients with compound heterozygous mutations in whom segregation was not definitively established. In these situations, patients were considered to be molecularly solved only if there was a clear genotype-phenotype match.
Clinical Determinants of Establishing a Molecular Diagnosis
The percentage of patients in whom we established a molecular genetic diagnosis differed across clinical diagnostic groups (Supplemental Figure 3). Three patients with urinary stone disease were clinically diagnosed with PH1, and all three were found to have mutations in AGXT. A molecular genetic diagnosis was identified in seven out of nine (78%) patients with a renal cystic ciliopathy, four out of nine (44%) patients with ESRD of unknown etiology, nine out of 21 (43%) patients with SRNS, ten out of 55 (18%) patients with CAKUT, and one out of seven (14%) patients with chronic GN. The broadest genetic heterogeneity was seen among patients with CAKUT, in whom mutations in six different genes and two CNVs were found in ten families.
The likelihood of establishing a molecular genetic diagnosis was also higher for patients who reported a history of consanguinity (67% compared with 30%), patients who had extrarenal manifestations (45% compared with 18%), and patients with a positive family history of renal disease (48% compared with 28%) (Supplemental Figure 4).
WES Can Provide a Precise Etiologic Diagnosis for Renal Transplant Recipients
Nine patients in our cohort were given an a priori clinical diagnosis of ESRD of unknown etiology, and in four of these patients, we made a molecular diagnosis through WES (Figure 1, Table 1). All four patients presented in childhood or early adolescence with advanced renal disease, and biopsies were either deferred or nondiagnostic. WES revealed mutations in the following genes: (1) NPHP1 (homozygous deletion), which causes juvenile NPHP; (2) TRPC6 (c.2684G>T, p.Arg895Leu; de novo), which is a cause of autosomal dominant FSGS;35,36 (3) TTC21B (c.1038G>A, p.Trp346*; c.986A>T, p.Glu328Val), which has been reported to cause both glomerular and cystic renal diseases;37,38 and (4) NUP93 (c.575A>G, p.Tyr192Cys), which is a cause of SRNS.28 Examination of WES reads confirmed that the compound heterozygous TTC21B alleles were inherited in trans (Supplemental Figure 5). Thus, for all four individuals, WES provided a precise diagnosis for the first time.
In addition to these four patients with ESRD of unknown etiology, WES provided a more specific molecular etiologic diagnosis for seven additional patients (underlined in Table 1). This included one patient who was initially diagnosed with a renal cystic ciliopathy, but ultimately was found to have a mutation in HNF1B. This patient did not have any known family history of renal disease and received a deceased donor renal transplant. There were also six patients initially thought to have isolated CAKUT, but were found to have mutations in genes that cause syndromic disease. Upon further chart review, it was noted that five of these six patients had subtle extrarenal manifestations. In total, identification of a molecular diagnosis provided clarification of a patient’s clinical diagnosis for 11 out of 34 probands (32%).
Implications of Establishing a Molecular Genetic Diagnosis for Clinical Management
Because of the retrospective nature of this study, virtually all recruited patients underwent genetic testing after kidney transplantation (Figure 2). Several patients did carry an accurate clinical diagnosis, which retrospectively was confirmed by WES. However, in many cases, the clinical diagnosis was made several years after a patient’s initial presentation, and early initiation of genetic testing could have had important implications for clinical care. We identified five probands, four with correct clinical diagnoses, in whom identification of a molecular genetic etiology of CKD could have had clinical consequences (Table 2). Here, we highlight several examples.
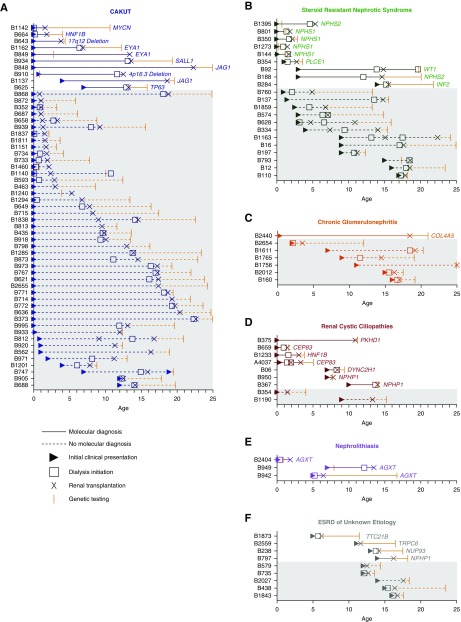
Figure 2.
Relationship between the time point at which WES was performed and relevant diagnostic and treatment events in 104 renal transplant recipients. Each patient is denoted as a separate entry on the y-axis. Age is represented on the x-axis. For each patient, age at clinical presentation (triangles), dialysis initiation (open squares), renal transplantation (X), and genetic testing (orange vertical hatch) are depicted. Patients in whom a molecular genetic diagnosis was made are on the top, and depicted as solid lines on a white background. Patients in whom no genetic mutations were identified are shown as dashed lines on a gray background. Some patients received a renal transplant preemptively and never required dialysis. For most patients, genetic testing on a research basis was completed after kidney transplantation because of the retrospective nature of this study. Patient data are grouped by clinical diagnosis within the same categories depicted in Figure 1, i.e., (A) CAKUT, (B) SRNS, (C) chronic GN, (D) renal cystic ciliopathies, (E) nephrolithiasis/nephrocalcinosis, and (F) ESRD of unknown etiology.
Table 2.
Five probands in whom early genetic diagnosis could have affected clinical care
| Family | Gene | Gender, Ethnicity | Age at Presentation/Age at ESRD | Clinical Details | Biopsy (if done) | Retrospective or Prospective Implications after WES |
|---|---|---|---|---|---|---|
| B2404 | AGXT | Female, Indian | 2 mo/2.5 mo | • Presented with advanced CKD in infancy | None | • Early initiation of daily hemodialysis to decrease risk for systemic oxalosis |
| • Initial work-up included normal serum oxalate level | • Earlier listing for combined liver-kidney transplantation | |||||
| • Presumed diagnosis of renal dysplasia | ||||||
| • PH1 diagnosis made at 1 yr of age during evaluation for LRD transplant | ||||||
| B188 | NPHS2 | Female, white | 3 yr/12 yr | • Presented with edema, proteinuria, and hypoalbuminemia | FSGS | • Avoidance of pretransplant IS and pheresis catheter placement |
| • Treatment with steroids, cyclophosphamide, cyclosporine, and ACE inhibitors | • Using a lower-risk (e.g., steroid minimization) protocol for IS given low risk for recurrence | |||||
| • Received plasmapheresis before transplantation to reduce risk of FSGS recurrence8,39-41 | ||||||
| B354 | PLCE1 | Male, Pakistani | 12 mo/15 mo | • Presented with edema and proteinuria | Biopsy 1: mesangial proliferative GN | • Avoidance of pretransplant IS (steroids and cyclosporine) |
| • No response to steroids or calcineurin inhibitors | Biopsy 2: DMS | |||||
| B2559 | TRPC6 | Female, white | 11 yr/11 yr | • Presented with malignant hypertension, anemia, thrombocytopenia, and elevated LDH | Global and segmental sclerosis, tubular atrophy, interstitial fibrosis | • No need for treatment with eculizumab |
| • Received treatment with eculizumab because of concern for atypical HUS and initiated on hemodialysis | • Genetic counseling given autosomal dominant inheritance and 50% risk of passing along mutant allele | |||||
| B92 | WT1 | Female, white | 4 yr/13 yr | • Treatment with steroids, cyclophosphamide, and ACE inhibitors | FSGS | • Earlier initiation of hormone therapy and prophylactic gonadectomy |
| • Gonadal dysgenesis diagnosed at 18 yr of age during work-up for primary amenorrhea |
LRD, living related donor; ACE, angiotensin-converting enzyme; IS, immunosuppression; DMS, diffuse mesangial sclerosis; LDH, lactate dehydrogenase; HUS, hemolytic uremic syndrome.
We established a molecular genetic diagnosis in nine out of the 21 patients clinically diagnosed with SRNS. Three out of these nine patients received treatment with steroids or other immunosuppressive therapy, and one patient also received plasmapheresis before transplantation, as some studies suggest that prophylactic plasmapheresis can reduce the likelihood of FSGS recurrence for high-risk patients.39–41 It was not until after kidney transplantation that WES was performed for all three individuals. Identification of a molecular genetic diagnosis earlier in the patients’ disease courses could minimize the exposure to intensive immunosuppressive medications both pre- and post-transplant.42 In addition, it could also have obviated the need for invasive procedures, such as renal biopsy or catheter placement for plasmapheresis.
Three patients were clinically diagnosed with PH1. Although all three patients were clinically diagnosed before WES, the diagnosis was delayed by almost 1 year for patient B2404, who was initially thought to have renal dysplasia. This patient developed anuric renal failure as an infant, and required RRT shortly after birth. She did undergo biochemical evaluation for PH1 at the time of her initial presentation, but her serum oxalate levels returned normal, potentially because she was receiving intermittent hemodialysis at the time. The patient was ultimately found to have a markedly elevated serum oxalate level several months later while on peritoneal dialysis and undergoing evaluation for kidney transplantation. After her clinical diagnosis of PH1, she was initiated on a more frequent dialysis regimen to decrease her risk for systemic oxalosis and was listed for a combined liver-kidney transplant, which is the treatment of choice.43 Performing WES at the time of her initial presentation may have led to a more rapid diagnosis and earlier initiation of these treatment measures. In particular, WES has an advantage over traditional screening methods, such as urinary oxalate levels, which can vary depending on age and diet, and can also be inaccurate in advanced CKD.43 Furthermore, WES would reduce the need for more invasive procedures, such as renal or liver biopsy.
Finally, one patient with ESRD of unknown etiology was found to have a de novo mutation in TRPC6. This patient initially presented in adolescence with hypertensive crisis, severely depressed renal function, and laboratory studies suggestive of a thrombotic microangiopathy. There was concern for atypical hemolytic uremic syndrome, and she received treatment with eculizumab for several months. After thoughtful consideration, eculizumab was discontinued before renal transplantation, and there was no recrudescence of the prior microangiopathic process. However, WES at the time of the patient’s initial presentation may have prevented the need for prolonged treatment with eculizumab, which increases the risk for severe infection. Additionally, identification of a genetic cause for this patient’s renal disease provides closure for the family and will also be important for future genetic counseling.
A summary of the potential implications for each of the genes in which a mutation was found in our cohort is provided in Supplemental Table 5.
Discussion
In this study, we performed WES in 104 probands who developed CKD before 25 years of age and who received a renal transplant at Boston Children’s Hospital between 2007 and 2017. We show that a molecular genetic diagnosis can be established in 32% of pediatric renal transplant recipients.
It has been demonstrated previously that a causal mutation can be detected in approximately 20% of all children and young adults who present with CKD before 25 years of age.2 To our knowledge, this is the first study to systematically assess the diagnostic yield of WES in pediatric renal transplant recipients. We determined that the percentage of patients with a molecular diagnosis is slightly higher than that for patients with milder degrees of CKD. This suggests that patients with genetic forms of CKD may have a more severe disease course, although future studies with more patients will be needed for definitive conclusions to be made. In addition, consistent with prior literature, the likelihood of identifying a genetic mutation depended upon a patient’s clinical disease group, as well as the presence of consanguinity or extrarenal manifestations.3,13 With the exception of the cohort of patients with urinary stone disease, in which all three patients had mutations in AGXT, the molecular diagnostic rate for each clinical group in our study was slightly higher than, but overall similar to, those that have been previously reported.3–15,19
Although research-based genetic testing for most of our patients was performed after kidney transplantation, we identified both retrospective and prospective clinical implications for five patients in whom a molecular diagnosis was established. In 11 cases, identification of a genetic mutation provided a more precise etiological cause for the patient’s CKD. This is perhaps most salient for the four patients with ESRD of unknown etiology in whom a genetic mutation was identified. Additionally, genetic testing may also lead to more rapid diagnoses for certain patients, such as the patient with PH1 in whom an accurate diagnosis was delayed for 1 year. In fact, it is not uncommon for diagnoses of PH1 to be delayed or even missed entirely, as reports suggest that up to 10% of patients with PH1 are diagnosed only after allograft failure after isolated kidney transplantation.44 A timely diagnosis of PH1 is paramount for patient care, as increased frequency of dialysis, combined liver-kidney transplantation, and, for some patients, a trial of pyridoxine, can all help to reduce the systemic oxalate burden.45–47
Establishing a precise molecular genetic diagnosis can also allow for preemptive screening for extrarenal manifestations. Recent studies suggest that patients clinically diagnosed with isolated CAKUT can have mutations in genes that cause syndromic disease.48 In some cases, this can be attributed to the differential effects of hypomorphic mutations, as compared with null mutations, or to a gene’s variable expressivity.49 Family members with the same JAG1 mutation, for instance, can have varying clinical manifestations, ranging from isolated renal disease to severe cholestatic liver disease.33,50 In other situations, extrarenal manifestations may manifest later in life or, alternatively, subtle phenotypes may be initially overlooked and only identified retrospectively once a genetic diagnosis is made.48 In these cases of reverse phenotyping, identification of a genetic mutation can lead to preemptive screening for extrarenal manifestations and earlier treatment when available.
Establishing a molecular genetic diagnosis also allows for tailoring of pre- and post-transplant treatment. As another example to the specific therapy for PH1 discussed above, immunosuppression regimens can also be adjusted for patients found to have genetic causes of SRNS. For example, reports suggest that up to 20%–30% of patients with SRNS experience disease-recurrence after kidney transplantation; however, this risk is only 4%–8% in patients who have genetic forms of SRNS.10,42,51 Certain patients with hereditary SRNS, therefore, could be candidates for a steroid minimization protocol at the time of transplantation given their low risk for immunologic disease.52 This is particularly pertinent in the pediatric population, in whom corticosteroids can have significant adverse effects on linear growth, body habitus, and self-image, and in whom compliance is a large issue. One exception is in patients who have the Fin(major) NPHS1 allele (c.121delCT), who have been reported to develop anti-nephrin antibodies post-transplantation, and are at slightly increased risk for SRNS recurrence.53 It is likely that future studies will elucidate further genotype-phenotype correlations, which would allow us to provide more personalized care for each patient.
Finally, there are examples in which establishing a causal molecular genetic diagnosis helps to guide living donor ascertainment. It has been established that women with heterozygous mutations in COL4A5, which causes X-linked Alport syndrome, are at a higher risk for developing CKD later in life. Thus, appropriate genetic counseling should be provided for potential donors who are found to harbor deleterious heterozygous COL4A5 mutations.54,55 Similarly, autosomal dominant causes of FSGS often manifest later in life and can be associated with variable expressivity. Family members of an individual found to carry a mutation in one of these genes should be offered genetic testing, and those in whom a mutation is identified should potentially be precluded from living kidney donation, given their risk for developing disease. Finally, although allograft survival is improved after living renal donation when compared with deceased donation, there often may be hesitancy in pursuing living donor transplantation in patients with FSGS because of the risk of recurrent disease. If a monogenic cause of FSGS is discovered in a patient, concerns regarding living donation may be alleviated because of the reduced risk of recurrence.56
Limitations of our study include a relatively small sample size and single center design. Because our study was conducted at a large, tertiary referral center, our results may not be generalizable to all medical centers worldwide. In addition, although we took a nonbiased approach in recruiting patients with a primary renal disease who received a transplant at our center, there may be potential ascertainment bias because of clinical differences between the patients who declined to participate and those who were enrolled. Finally, WES may miss mutations in introns and promotor regions, certain CNVs, and mutations in exons with low coverage. We additionally predict that the percentage of individuals in whom a causal diagnosis can be made would be higher if formal CNV analysis is added. However, given the progressively declining costs of WES and utility demonstrated in many clinical scenarios,57,58 it is becoming an efficient and cost-effective diagnostic study. Thus, given the effect that identification of a genetic mutation can have on pre- and post-transplant care for renal transplant recipients, we propose that WES be considered for patients who develop CKD at 25 years of age or younger.
Disclosures
Dr. Hildebrandt is a cofounder, SAC member, and holds stock in Goldfinch-Bio.
Supplementary Material
Acknowledgments
We thank the affected individuals, their families, and their physicians who contributed to this study.
N.M., K.A., L.S., M.J.G.S., M.A.F., A.Z.T., D.R.S., M.A.B., G.H.D., S.M., K.V., H.B.K., N.M.R., and F.H. recruited patients and gathered detailed clinical information for this study. S.S., S.M.M., and R.P.L. performed whole-exome sequencing and downstream data analyses. N.M., D.A.B., W.T., S.S., D.M.C., M.N., R.S., T.M.K., A.T.v.d.V., J.C., H.I., A.V., A.J.M., A.D., J.K.W., S.L., S.A., T.J.-S., E.W., H.H., S.M., and F.H. evaluated whole-exome sequencing data and performed variant calling. F.H. conceived of and directed the study with N.M. N.M. and F.H. wrote the manuscript. All authors approved the final version. F.H. is the William E. Harmon Professor of Pediatrics.
This research was supported by grants from the National Institutes of Health to F.H. (DK076683, DK088767, and DK068306) and from the Yale Center for Mendelian Genomics (grant U54HG006504, to R.P.L.). N.M. is supported by funding from the National Institutes of Health T32-DK007726-33 grant at the Boston Children’s Hospital and by a Fred Lovejoy Resident Research and Education Award. D.M.C. is funded by the Health Research Board, Ireland (HPF-206-674), the International Pediatric Research Foundation Early Investigators’ Exchange Program, and the Amgen Irish Nephrology Society Specialist Registrar Bursary. M.N. is supported by a grant from the Japan Society for the Promotion of Science. T.M.K. is supported by a Postdoctoral Fellowship award from the KRESCENT Program, a national kidney research training partnership of the Kidney Foundation of Canada, the Canadian Society of Nephrology, and the Canadian Institutes of Health Research. A.T.v.d.V. is supported by postdoctoral research fellowship VE969-7 from the German Research Foundation (DFG). A.J.M. is supported by funding from the National Institutes of Health T32-DK007726-31 grant at the Boston Children’s Hospital, the 2017 Harvard Stem Cell Institute Kidney Group Interlaboratory Postdoctoral Fellowship Grant, and the 2018 Polycystic Kidney Disease Foundation Jared J. Grantham Research Fellowship from the American Society of Nephrology Ben J. Lipps Research Fellowship Program. T.J.S. is supported by a grant of the Deutsche Forschungsgemeinschaft (Jo 1324/1-1). E.W. is supported by the German National Academy of Sciences Leopoldina (LPDS-2015-07). The Broad Center for Mendelian Genomics (UM1 HG008900) is funded by the National Human Genome Research Institute with supplemental funding provided by the National Heart, Lung, and Blood Institute under the Trans-Omics for Precision Medicine (TOPMed) program and the National Eye Institute. This research was also supported by the Isabella Forrest Julian Research Fund for Pediatric Post Kidney Transplant Research.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2018060575/-/DCSupplemental.
Supplemental Table 1A. Forty one genes that represent monogenic causes of human isolated CAKUT, if mutated.
Supplemental Table 1B. Fifty genes that represent monogenic causes of human nephrotic syndrome, if mutated.
Supplemental Table 1C. Seventeen genes that represent monogenic causes of human nephritis, if mutated.
Supplemental Table 1D. Ninety five genes that represent monogenic causes of human nephronophthisis or cystic kidney disease, if mutated.
Supplemental Table 1E. Thirty seven genes that represent monogenic causes of human nephrolithiasis or nephrocalcinosis, if mutated.
Supplemental Table 1F. One hundred forty five genes that represent monogenic causes of human syndromic CAKUT, if mutated.
Supplemental Table 1G. Eleven genes that represent rare monogenic causes of human kidney disease (miscellaneous category), if mutated.
Supplemental Table 2. Information on six out of 396 known CKD genes that did not achieve a mean coverage of at least 30×.
Supplemental Table 3. ACMG guidelines for variant calling.
Supplemental Table 4. Clinical characteristics of probands at Boston Children’s Hospital who underwent renal transplantation between 2007 and 2017 compared with patients in the NAPRTCS registry.
Supplemental Table 5. Clinical consequences after establishment of a molecular genetic diagnosis for 21 genes identified in 104 families.
Supplemental Figure 1. Recruitment strategy and likelihood of detecting a monogenic cause of CKD in 104 renal transplant recipients.
Supplemental Figure 2. Variant filtering process for the identification of causative mutations in genes known to cause CKD.
Supplemental Figure 3. Percentage of patients with a molecular genetic diagnosis varies by clinical diagnostic group.
Supplemental Figure 4. The likelihood of identifying a molecular diagnosis is higher when there is consanguinity, extrarenal manifestations, or a positive family history.
Supplemental Figure 5. TTC21B compound heterozygous alleles are inherited in trans for patient B1873.
References
- 1.North American Pediatric Renal Trials and Collaborative Studies: NAPRTCS Annual Transplant Report, 2014. Available at: https://web.emmes.com/study/ped/annlrept/annualrept2014.pdf. Accessed May 1, 2018
- 2.Vivante A, Hildebrandt F: Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol 12: 133–146, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Ven AT, Connaughton DM, Ityel H, Mann N, Nakayama M, Chen J, et al.: Whole-exome sequencing identifies causative mutations in families with congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 29: 2348–2361, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber S, Moriniere V, Knüppel T, Charbit M, Dusek J, Ghiggeri GM, et al.: Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: Results of the ESCAPE study. J Am Soc Nephrol 17: 2864–2870, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Sanna-Cherchi S, Westland R, Ghiggeri GM, Gharavi AG: Genetic basis of human congenital anomalies of the kidney and urinary tract. J Clin Invest 128: 4–15, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, et al.: SRNS Study Group : A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 26: 1279–1289, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warejko JK, Tan W, Daga A, et al.: Whole exome sequencing of patients with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 13: 53–62, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W, Lovric S, Ashraf S, Rao J, Schapiro D, Airik M, et al.: Analysis of 24 genes reveals a monogenic cause in 11.1% of cases with steroid-resistant nephrotic syndrome at a single center. Pediatr Nephrol 33: 305–314, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown EJ, Pollak MR, Barua M: Genetic testing for nephrotic syndrome and FSGS in the era of next-generation sequencing. Kidney Int 85: 1030–1038, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trautmann A, Bodria M, Ozaltin F, Gheisari A, Melk A, Azocar M, et al.: PodoNet Consortium : Spectrum of steroid-resistant and congenital nephrotic syndrome in children: The PodoNet registry cohort. Clin J Am Soc Nephrol 10: 592–600, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schapiro D, Daga A, Lawson JA, Majmundar AJ, Lovric S, Tan W, et al.: Panel sequencing distinguishes monogenic forms of nephritis from nephrosis in children. Nephrol Dial Transplant gfy050, 2018. doi: 10.1093/ndt/gfy050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halbritter J, Porath JD, Diaz KA, Braun DA, Kohl S, Chaki M, et al.: GPN Study Group : Identification of 99 novel mutations in a worldwide cohort of 1,056 patients with a nephronophthisis-related ciliopathy. Hum Genet 132: 865–884, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun DA, Schueler M, Halbritter J, Gee HY, Porath JD, Lawson JA, et al.: Whole exome sequencing identifies causative mutations in the majority of consanguineous or familial cases with childhood-onset increased renal echogenicity. Kidney Int 89: 468–475, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun DA, Hildebrandt F. Ciliopathies. Cold Spring Harb Perspect Biol 9: a028191, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava S, Molinari E, Raman S, Sayer JA: Many genes-one disease? Genetics of nephronophthisis (NPHP) and NPHP-associated disorders. Front Pediatr 5: 287, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halbritter J, Baum M, Hynes AM, Rice SJ, Thwaites DT, Gucev ZS, et al.: Fourteen monogenic genes account for 15% of nephrolithiasis/nephrocalcinosis. J Am Soc Nephrol 26: 543–551, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun DA, Lawson JA, Gee HY, Halbritter J, Shril S, Tan W, et al.: Prevalence of monogenic causes in pediatric patients with nephrolithiasis or nephrocalcinosis. Clin J Am Soc Nephrol 11: 664–672, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daga A, Majmundar AJ, Braun DA, et al.: Whole exome sequencing frequently detects a monogenic cause in early onset nephrolithiasis and nephrocalcinosis. Kidney Int 93: 204–213, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halbritter J, Seidel A, Müller L, Schönauer R, Hoppe B: Update on hereditary kidney stone disease and introduction of a new clinical patient registry in Germany. Front Pediatr 6: 47, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montini G, Malaventura C, Salviati L: Early coenzyme Q10 supplementation in primary coenzyme Q10 deficiency. N Engl J Med 358: 2849–2850, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, Ji Z, et al.: COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest 121: 2013–2024, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashraf S, Gee HY, Woerner S, Xie LX, Vega-Warner V, Lovric S, et al.: ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest 123: 5179–5189, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozaltin F: Primary coenzyme Q10 (CoQ 10) deficiencies and related nephropathies. Pediatr Nephrol 29: 961–969, 2014 [DOI] [PubMed] [Google Scholar]
- 24.M’dimegh S, Omezzine A, Hamida-Rebai MB, Aquaviva-Bourdain C, M’barek I, Sahtout W, et al.: Identification of a novel AGXT gene mutation in primary hyperoxaluria after kidney transplantation failure. Transpl Immunol 39: 60–65, 2016 [DOI] [PubMed] [Google Scholar]
- 25.Taneda S, Honda K, Nakajima I, Huchinoue S, Oda H: Renal transplantation between siblings with unrecognized Fabry disease. Transplant Proc 45: 115–118, 2013 [DOI] [PubMed] [Google Scholar]
- 26.Basic-Jukic N, Coric M, Kes P, Bubic-Filipi LJ, Pasini J, Mokos I: Anderson-Fabry disease in kidneys from deceased donor. Am J Transplant 7: 2829–2833, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Kochar O, Wick MR, Kerr SE, Oglesbee D, Cathro HP: Unexpected Fabry disease in a renal allograft kidney: An underrecognized cause of poor allograft function. Ultrastruct Pathol 35: 92–96, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Braun DA, Sadowski CE, Kohl S, Lovric S, Astrinidis SA, Pabst WL, et al.: Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet 48: 457–465, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gee HY, Otto EA, Hurd TW, Ashraf S, Chaki M, Cluckey A, et al.: Whole-exome resequencing distinguishes cystic kidney diseases from phenocopies in renal ciliopathies. Kidney Int 85: 880–887, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tory K, Menyhárd DK, Woerner S, Nevo F, Gribouval O, Kerti A, et al.: Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat Genet 46: 299–304, 2014 [DOI] [PubMed] [Google Scholar]
- 31.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al.: ACMG Laboratory Quality Assurance Committee : Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, et al.: Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: Recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med 20: 351–359, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamath BM, Bason L, Piccoli DA, Krantz ID, Spinner NB: Consequences of JAG1 mutations. J Med Genet 40: 891–895, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, et al.: NHLBI Exome Sequencing Project : Copy number variation detection and genotyping from exome sequence data. Genome Res 22: 1525–1532, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gigante M, Caridi G, Montemurno E, Soccio M, d’Apolito M, Cerullo G, et al.: TRPC6 mutations in children with steroid-resistant nephrotic syndrome and atypical phenotype. Clin J Am Soc Nephrol 6: 1626–1634, 2011 [DOI] [PubMed] [Google Scholar]
- 36.Reiser J, Polu KR, Möller CC, Kenlan P, Altintas MM, Wei C, et al.: TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat Genet 37: 739–744, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huynh Cong E, Bizet AA, Boyer O, Woerner S, Gribouval O, Filhol E, et al.: A homozygous missense mutation in the ciliary gene TTC21B causes familial FSGS. J Am Soc Nephrol 25: 2435–2443, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, et al.: NISC Comparative Sequencing Program : TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43: 189–196, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohta T, Kawaguchi H, Hattori M, Komatsu Y, Akioka Y, Nagata M, et al.: Effect of pre-and postoperative plasmapheresis on posttransplant recurrence of focal segmental glomerulosclerosis in children. Transplantation 71: 628–633, 2001 [DOI] [PubMed] [Google Scholar]
- 40.Gohh RY, Yango AF, Morrissey PE, Monaco AP, Gautam A, Sharma M, et al.: Preemptive plasmapheresis and recurrence of FSGS in high-risk renal transplant recipients. Am J Transplant 5: 2907–2912, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Bacchetta J, Cochat P: Primary disease recurrence—effects on paediatric renal transplantation outcomes. Nat Rev Nephrol 11: 371–384, 2015 [DOI] [PubMed] [Google Scholar]
- 42.Ruf RG, Lichtenberger A, Karle SM, Haas JP, Anacleto FE, Schultheiss M, et al.: Arbeitsgemeinschaft Für Pädiatrische Nephrologie Study Group : Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol 15: 722–732, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Cochat P, Hulton SA, Acquaviva C, Danpure CJ, Daudon M, De Marchi M, et al.: OxalEurope : Primary hyperoxaluria Type 1: Indications for screening and guidance for diagnosis and treatment. Nephrol Dial Transplant 27: 1729–1736, 2012 [DOI] [PubMed] [Google Scholar]
- 44.Cochat P, Rumsby G: Primary hyperoxaluria. N Engl J Med 369: 649–658, 2013 [DOI] [PubMed] [Google Scholar]
- 45.Hoppe B: An update on primary hyperoxaluria. Nat Rev Nephrol 8: 467–475, 2012 [DOI] [PubMed] [Google Scholar]
- 46.Monico CG, Olson JB, Milliner DS: Implications of genotype and enzyme phenotype in pyridoxine response of patients with type I primary hyperoxaluria. Am J Nephrol 25: 183–188, 2005 [DOI] [PubMed] [Google Scholar]
- 47.van Woerden CS, Groothoff JW, Wijburg FA, Annink C, Wanders RJ, Waterham HR: Clinical implications of mutation analysis in primary hyperoxaluria type 1. Kidney Int 66: 746–752, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Vivante A, Hwang DY, Kohl S, Chen J, Shril S, Schulz J, et al.: Exome sequencing discerns syndromes in patients from consanguineous families with congenital anomalies of the kidneys and urinary tract. J Am Soc Nephrol 28: 69–75, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kohl S, Hwang DY, Dworschak GC, Hilger AC, Saisawat P, Vivante A, et al.: Mild recessive mutations in six Fraser syndrome-related genes cause isolated congenital anomalies of the kidney and urinary tract. J Am Soc Nephrol 25: 1917–1922, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamath BM, Spinner NB, Rosenblum ND: Renal involvement and the role of Notch signalling in Alagille syndrome. Nat Rev Nephrol 9: 409–418, 2013 [DOI] [PubMed] [Google Scholar]
- 51.Karle SM, Uetz B, Ronner V, Glaeser L, Hildebrandt F, Fuchshuber A: Novel mutations in NPHS2 detected in both familial and sporadic steroid-resistant nephrotic syndrome. J Am Soc Nephrol 13: 388–393, 2002 [DOI] [PubMed] [Google Scholar]
- 52.Nehus EJ, Liu C, Lu B, Macaluso M, Kim MO: Graft survival of pediatric kidney transplant recipients selected for de novo steroid avoidance-a propensity score-matched study. Nephrol Dial Transplant 32: 1424–1431, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holmberg C, Jalanko H: Congenital nephrotic syndrome and recurrence of proteinuria after renal transplantation. Pediatr Nephrol 29: 2309–2317, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savige J, Gregory M, Gross O, Kashtan C, Ding J, Flinter F: Expert guidelines for the management of Alport syndrome and thin basement membrane nephropathy. J Am Soc Nephrol 24: 364–375, 2013 [DOI] [PubMed] [Google Scholar]
- 55.Kashtan CE: Renal transplantation in patients with Alport syndrome. Pediatr Transplant 10: 651–657, 2006 [DOI] [PubMed] [Google Scholar]
- 56.Baum MA, Ho M, Stablein D, Alexander SR; North American Pediatric Renal Transplant Cooperative Study : Outcome of renal transplantation in adolescents with focal segmental glomerulosclerosis. Pediatr Transplant 6: 488–492, 2002 [DOI] [PubMed] [Google Scholar]
- 57.Meng L, Pammi M, Saronwala A, Magoulas P, Ghazi AR, Vetrini F, et al.: Use of exome sequencing for infants in intensive care units: Ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr 171: e173438, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan TY, Dillon OJ, Stark Z, Schofield D, Alam K, Shrestha R, et al.: Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr 171: 855–862, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.