In this study, Verma et al. show that distinct dependencies exist for telomere synthesis in response to replication stress or DNA double-strand breaks (DSBs). Their findings establish that RAD52 and SLX4 mediate distinct postreplicative DNA repair processes that maintain ALT telomere stability and cancer cell viability.
Keywords: ALT, FANCD2, homologous recombination, RAD52, SLX4, telomere, replication stress
Abstract
Approximately 15% of cancers use homologous recombination for alternative lengthening of telomeres (ALT). How the initiating genomic lesions invoke homology-directed telomere synthesis remains enigmatic. Here, we show that distinct dependencies exist for telomere synthesis in response to replication stress or DNA double-strand breaks (DSBs). RAD52 deficiency reduced spontaneous telomeric DNA synthesis and replication stress-associated recombination in G2, concomitant with telomere shortening and damage. However, viability and proliferation remained unaffected, suggesting that alternative telomere recombination mechanisms compensate in the absence of RAD52. In agreement, RAD52 was dispensable for DSB-induced telomere synthesis. Moreover, a targeted CRISPR screen revealed that loss of the structure-specific endonuclease scaffold SLX4 reduced the proliferation of RAD52-null ALT cells. While SLX4 was dispensable for RAD52-mediated ALT telomere synthesis in G2, combined SLX4 and RAD52 loss resulted in elevated telomere loss, unresolved telomere recombination intermediates, and mitotic infidelity. These findings establish that RAD52 and SLX4 mediate distinct postreplicative DNA repair processes that maintain ALT telomere stability and cancer cell viability.
Homology-directed DNA repair (HDR) can be achieved through multiple pathways that use distinct molecular machineries (Ceccaldi et al. 2016; Verma and Greenberg 2016). Canonical HDR proceeds through a RAD51-dependent search and capture of a homologous sequence followed by templated DNA synthesis. However, noncanonical forms of HDR that occur independent of RAD51 have been described extensively in yeast and are responsible for a subset of break-induced replication (BIR) and telomere maintenance in telomerase-negative survivors (Lundblad and Blackburn 1993; Malkova et al. 1996; Le et al. 1999; Teng and Zakian 1999; Teng et al. 2000; Chen et al. 2001; Lydeard et al. 2007; Anand et al. 2013). Analogous mechanisms in human cells are increasingly believed to underlie the repair of damaged replication forks and the alternative lengthening of telomeres (ALT) pathway of telomere maintenance (Bryan et al. 1995; Cesare and Reddel 2010; Costantino et al. 2014; Dilley et al. 2016). The relative contribution of canonical versus noncanonical HDR to these pathways in humans is poorly understood. We demonstrated recently that human break-induced telomere synthesis (BITS) and spontaneous ALT telomere synthesis are independent of RAD51 (Dilley et al. 2016). ALT involves long-tract conservative DNA synthesis, suggestive of a RAD51-independent BIR mechanism (Dilley et al. 2016; Roumelioti et al. 2016). The molecular control of this alternative form of HDR remains a fundamental question.
Survivors of telomerase mutation in yeast rely on either RAD51-dependent (type I survivors) or RAD51-independent (type II survivors) HDR mechanisms to maintain telomere length. However, both types I and II require the repair protein RAD52 and the DNA polymerase δ subunit Pol32 (Lundblad and Blackburn 1993; Lydeard et al. 2007). Drawing parallels between yeast and human RAD52 may prove challenging, since the cellular function does not appear to be fully conserved. While yeast RAD52 is an important mediator of RAD51 nucleofilament formation, this function has largely been taken over by BRCA2 in human cells (Rijkers et al. 1998; Hanamshet et al. 2016). Biochemically, human RAD52 binds dsDNA and ssDNA and promotes the annealing between complementary ssDNA strands as well as D-loop formation in vitro (Benson et al. 1998; Kagawa et al. 2001, 2002; Van Dyck et al. 1999, 2001). In vivo, mammalian RAD52 was thought to participate mainly in alternative, backup repair pathways (Feng et al. 2011; Lok et al. 2013). However, recent studies identified an essential role for RAD52 in DNA repair synthesis after replication stress in human cells (Bhowmick et al. 2016). These RAD52-dependent repair pathways are independent of RAD51 and occur in mitosis (mitotic DNA synthesis [MiDAS]), a time when canonical homologous recombination (HR) and nonhomologous end joining (NHEJ) are repressed. Recruitment of RAD52 to the sites of MiDAS was reported to be SLX4-dependent (Bhowmick et al. 2016). RAD52-dependent mitotic repair synthesis was also observed at telomeres (Min et al. 2017; Özer et al. 2018). However, the physiological importance of RAD52-mediated synthesis to ALT telomere maintenance remains enigmatic, as is the nature of initiating telomere lesions to which it can effectively respond to direct HDR.
In this study, we set out to investigate the contributions of RAD52 to ALT telomere maintenance. Our findings reveal a SLX4-independent role for RAD52 in spontaneous ALT telomere synthesis in interphase cells. Simultaneous loss of RAD52 and SLX4 leads to telomere abnormalities, loss of mitotic fidelity, and impaired viability, thus establishing their nonepistatic interaction in maintaining genomic integrity.
Results
BITS escapes mitotic inhibition
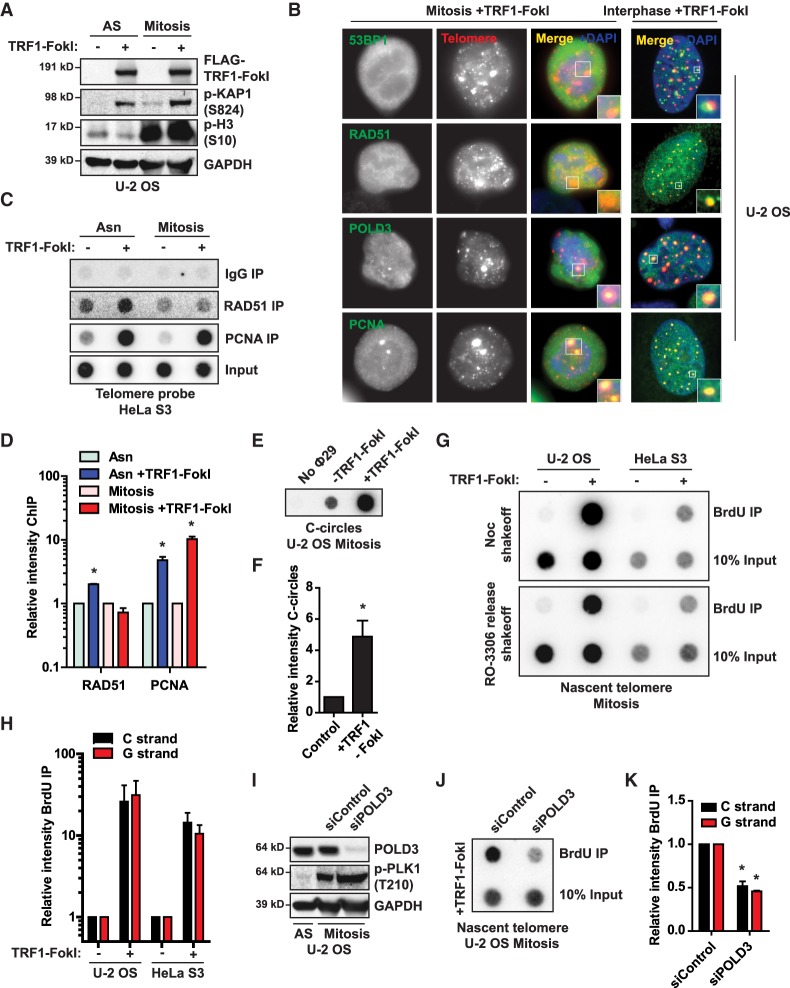
DNA double-strand break (DSB) repair is controlled throughout the cell cycle (Hustedt and Durocher 2017). In mitosis, a series of Cdk1-mediated phosphorylation events is thought to effectively inhibit canonical HR and NHEJ, whereas alternative forms of repair such as MiDAS are permitted (Giunta et al. 2010; Orthwein et al. 2014; Minocherhomji et al. 2015). Since BITS occurs independent of RAD51, we hypothesized that it would escape mitotic inhibition (Dilley et al. 2016). To test this, we isolated mitotic cells using nocodazole shakeoff or RO-3306 release followed by shakeoff and subsequently turned on telomere restriction fragment 1 (TRF1)-FokI to create telomere breaks in mitosis. TRF1-FokI induction for 2 h resulted in activation of ATM signaling in mitotic cells, as indicated by increased p-KAP1 (Fig. 1A). During this time frame, break-induced replisome components POLD3 and PCNA localized to damaged mitotic telomeres, whereas RAD51 and 53BP1 were excluded (Fig. 1B–D). As seen for asynchronous cells, mitotic telomere damage resulted in the generation of C circles as well as a dramatic increase in POLD3-dependent nascent telomere synthesis, quantified using a BrdU pull-down assay that allows for detection of BITS on nitrocellulose membranes when hybridized with a radiolabeled telomeric oligonucleotide (Fig. 1E–K; Supplemental Fig. S1A,B; Cho et al. 2014; Dilley et al. 2016; Verma et al. 2018). Spontaneous mitotic telomere synthesis was present at higher levels in several ALT cell lines compared with telomerase lines (Supplemental Fig. S1C–E), consistent with recent reports (Min et al. 2017; Özer et al. 2018). However, the baseline synthesis was negligible compared with that seen after TRF1-FokI in both ALT and telomerase lines (Fig. 1G,H). Taken together, our data demonstrate that telomere repair synthesis escapes mitotic inhibition and can occur in direct response to breaks generated in mitosis. In addition to highlighting the noncanonical nature of this HDR and the importance of the POLD3-containing replisome, our data suggest that mitosis could serve as a window to identify repair factors that are critical to telomere synthesis and repair.
Figure 1.
BITS escapes mitotic inhibition. (A) Western blot from asynchronous and mitosis-arrested U-2 OS induced with TRF1-FokI for 2 h. p-KAP1 was used to examine whether telomeric DSBs lead to the activation of ATM signaling. (B) Immunofluorescence images of repair factor (green) recruitment to damaged mitotic and interphase telomeres (red) in U-2 OS cells. (C,D) Chromatin immunoprecipitation (ChIP) dot blot from HeLa S3 (C) with quantification (D). (E,F) C-circle dot blot from mitosis arrested U-2 OS induced with TRF1-FokI (E) with quantification (F). (G,H) BrdU pull-down dot blots for telomere content from mitosis-arrested U-2 OS and HeLa S3 induced with TRF1-FokI (G), with quantification of nascent telomeric C and G strands (H). (I) Western blot from asynchronous and mitosis-arrested U-2 OS treated with the indicated siRNAs. p-Plk1 served as a marker for the mitotic enriched population. (J,K) BrdU pull-down dot blot for telomere content from mitotis-arrested U-2 OS induced with TRF1-FokI (J), with quantification of nascent telomeric C and G strands (K). (AS) Asynchronous; (Noc) nocodazole. Data represent mean values of at least two independent experiments. (*) P ≤ 0.05, Student's t-test. Error bars indicate SEM.
RAD52 is recruited to ALT telomeres to resolve replication stress
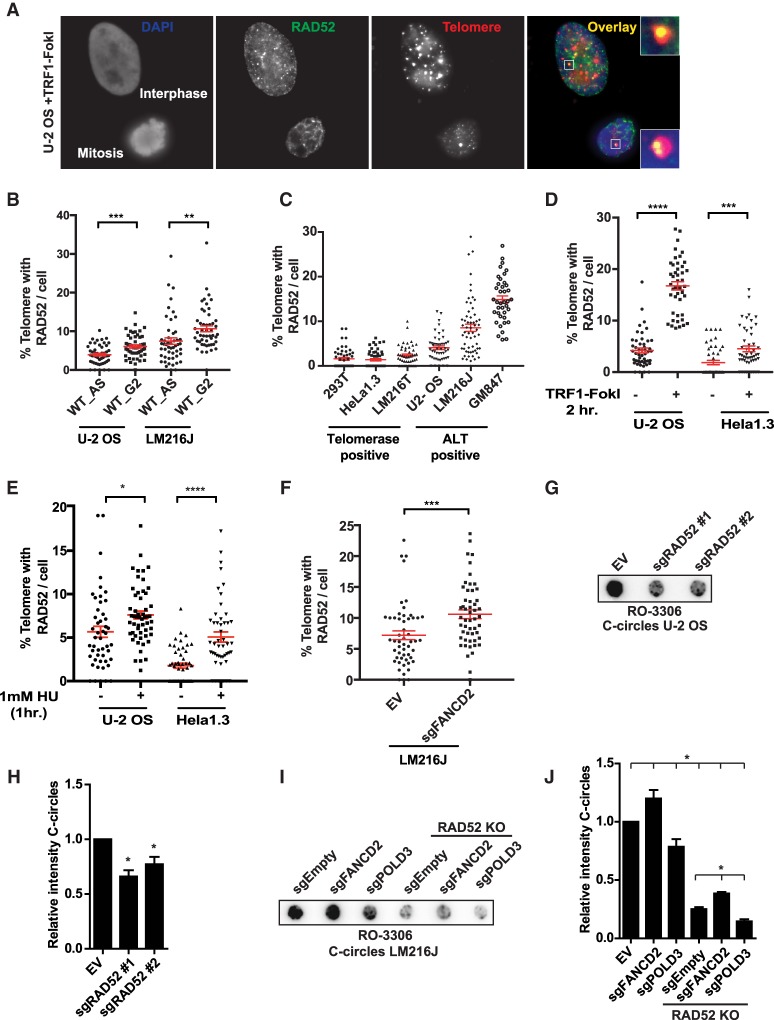
Human RAD52 participates in alternative repair pathways independent of RAD51 (Bhowmick et al. 2016; Sotiriou et al. 2016). RAD52 has been visualized at ALT telomeres and contributes to telomeric MiDAS, but its functional role in human ALT remains poorly understood (Yeager et al. 1999; Min et al. 2017; Özer et al. 2018). In particular, its contributions to telomere synthesis outside of mitosis has not been investigated. Taking advantage of the mitotic repression of canonical repair pathways, we found that RAD52 localized to damaged mitotic as well as interphase telomeres (Fig. 2A). RAD52 telomere localization increased in G2-arrested cells and was more prominent in ALT cell lines compared with telomerase-positive lines, suggesting a damage dependence (Fig. 2B,C; Supplemental Fig. S2A,B; Cesare et al. 2009; Cesare and Reddel 2010). Given that HeLa1.3 cells have telomere lengths comparable with ALT cells, this rules out the possibility that the increased localization observed was related to telomere length. Generation of telomere DSBs using TRF1-FokI resulted in further elevation of RAD52 localization (Fig. 2D). Additionally, replication stress due to hydroxyurea (HU) treatment or collapsed replication forks following FANCD2 depletion also increased RAD52 recruitment to telomeres (Fig. 2E,F; Schlacher et al. 2012).
Figure 2.
RAD52 is enriched at damaged telomeres and is required for C-circle generation. (A) Representative immunofluorescence image of a U-2 OS interphase and mitotic cell showing localization of RAD52 (green) at damaged telomeres (red). DNA was stained using DAPI (blue). (B–F) Quantification of RAD52 localization at telomeres in asynchronous and Cdk1 inhibitor (Cdk1i)-arrested G2-phase cells (B) and telomerase-positive and ALT-positive cells (C) after 2 h of TRF1-FokI induction (D) and 1 h of treatment with 1 mM HU (E) and in LM216J cells expressing sgFANCD2 (F). (G–J) Representative image (G,I) and quantification (H,J) of C circles in U-2 OS (G,H) and LM216J (I,J) cells arrested in G2 phase using Cdk1 inhibitor RO-3306 for 16 h. (EV) Empty vector; (WT) wild type; (KO) knockout; (AS) Asynchronous. Data represent mean values of at least two independent experiments. (*) P≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.0001; (****) P ≤ 0.0001, Student's t-test. Error bars indicate SEM.
Replication stress-associated telomere localization in interphase suggests that RAD52 is part of a repertoire of repair proteins that can contribute to postreplicative synthesis during ALT. C circles are a marker of ALT activity that are proposed to arise from the resolution of replication stress intermediates (Henson et al. 2009; O'Sullivan et al. 2014; Rivera et al. 2017). FANCD2 loss increased C circles in ALT cells, consistent with previous reports (Supplemental Figs. S3A,B, S4A; Fan et al. 2009; Root et al. 2016). Nascent C-circle generation was highest in G2-arrested cells and decreased dramatically as cells entered mitosis in both control and FANCD2-depleted cells (Supplemental Fig. S3C). These data imply that ALT cells may resolve most replication stress during G2. Interestingly, RAD52 localization to ALT telomeres is increased during this phase of the cell cycle (Fig. 2B). This observation may relate to the known recruitment of DNA damage response factors to telomeres after completion of DNA replication (Verdun et al. 2005). CRISPR guides targeting RAD52 (sgRAD52) reduced C circles in G2-arrested and asynchronous cells as well as mitotic cells induced with TRF1-FokI (Fig. 2G,H; Supplemental Figs. S3D–F, S4B). Notably, this C-circle reduction was consistent across two independent guides and in both cell populations and pooled knockout clones (Fig. 2G,H; Supplemental Fig. S3D,E). While sgFANCD2 and sgPOLD3 had opposing effects on C circles in G2, RAD52 knockout had a dominant effect, leading to a reduction in all genetic backgrounds (Fig. 2I,J; Supplemental Figs. S2C, S4C). Taken together, our data demonstrate that RAD52 acts in response to replication stress at telomeres and promotes features of ALT.
RAD52 is required for spontaneous ALT telomere synthesis and maintenance
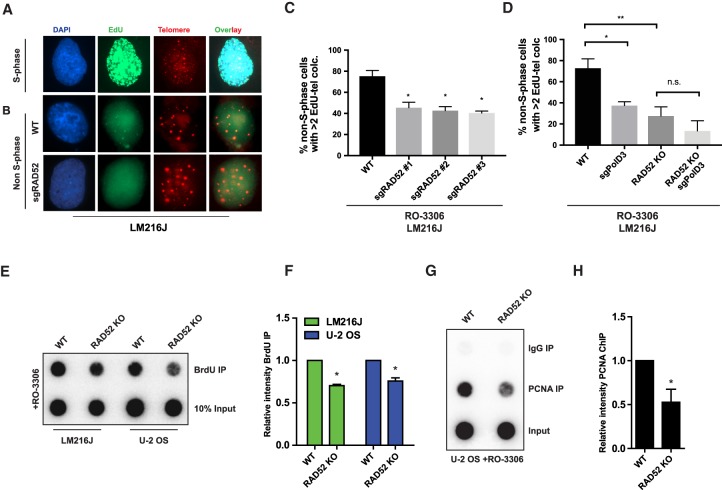
Deficiency in factors that promote telomere synthesis during ALT has been reported to decrease C circles (Dilley et al. 2016). Since RAD52 loss reduced C circles, we hypothesized that it might be involved in directing DNA repair synthesis at ALT telomeres downstream from replication stress. To test this directly, we analyzed the effect of RAD52 depletion on non-S-phase telomere synthesis, a characteristic found only in ALT cells that can be visualized by EdU incorporation selectively at telomeres (Fig. 3A,B; Nabetani et al. 2004; Cho et al. 2014; Dilley et al. 2016). RAD52 colocalized with EdU-positive telomeres in non-S-phase cells (Supplemental Fig. S3G). ALT-positive LM216J cells treated with three independent sgRAD52 guides displayed a significant reduction in non-S-phase telomere synthesis, as measured by EdU incorporation (Fig. 3B,C; Supplemental Fig. S3H). Notably, depletion of RAD52 and POLD3 resulted in similar reductions in telomere synthesis, with no obvious additive effect when combined (Fig. 3D). These results were validated using BrdU pull-downs of nascent telomeres in undamaged cells arrested in G2. Using this approach, RAD52 knockout also decreased spontaneous telomere synthesis in LM216J and U-2 OS cells (Fig. 3E,F). This reduction in synthesis corresponds to diminished PCNA loading onto ALT telomeres in G2 in RAD52 knockout cells (Fig. 3G,H). In contrast, RAD51 loss did not have an effect on this RAD52-mediated ALT synthesis (Supplemental Figs. S3I, S4D). Taken together, our data implicate a RAD51-independent role of RAD52 in ALT telomere synthesis during G2.
Figure 3.
RAD52 is required for spontaneous ALT telomere synthesis. (A) Representative immunofluorescence image of EdU (green) incorporation in a S-phase cell. (B–D) Representative immunofluorescence image (B) and quantification (C,D) of EdU (green) incorporation at telomeres (red) in the indicated LM216J lines arrested in G2 phase using Cdk1 inhibitor (Cdk1i) RO-3306 for 16 h. DNA was stained using DAPI (blue). Error bars indicate SD. (E,F) Representative dot blot (E) and quantification (F) of BrdU immunoprecipitation for telomeric DNA using a 32P-labeled telomeric probe in G2-arrested U-2 OS and LM216J lines. Error bars indicate SEM. (G,H) Representative dot blot (G) and quantification (H) of PCNA immunoprecipitation for telomere content using a 32P-labeled telomeric probe from U-2 OS cells arrested in G2 phase using Cdk1 inhibitor RO-3306 for 16 h. Error bars indicate SEM. Data are mean values of at least two independent experiments. (WT) Wild type; (KO) knockout. (*) P ≤ 0.05; (**) P ≤ 0.01; (n.s.) nonsignificant, Student's t-test.
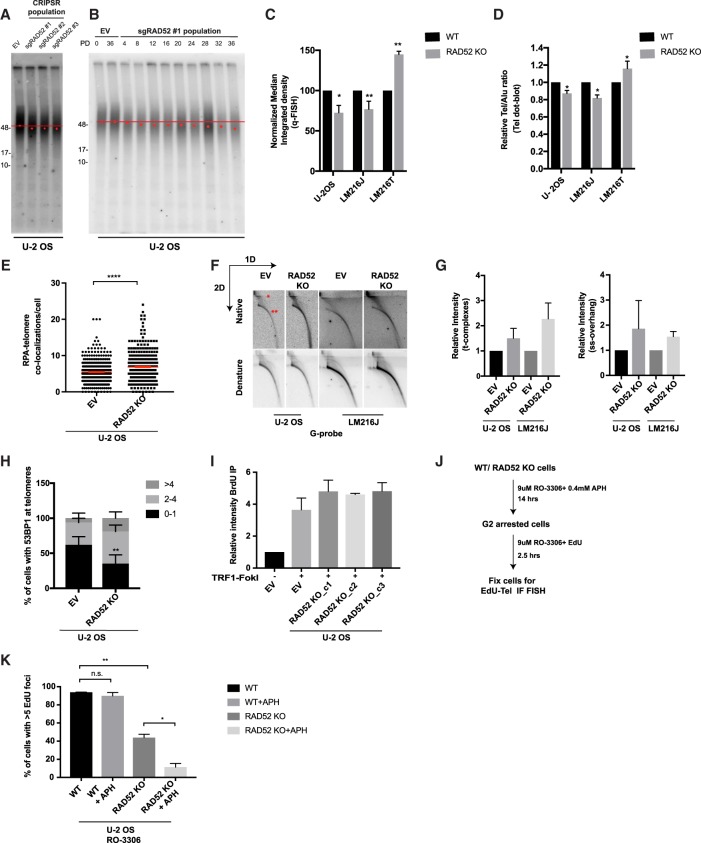
To determine the importance of RAD52 for ALT telomere maintenance, we analyzed telomere length and content in sgRAD52 populations and RAD52 knockout clones. All three sgRNAs targeting RAD52 resulted in observable telomere shortening in populations of U-2 OS cells, as determined by telomere restriction fragment (TRF) analysis (Fig. 4A,B). Multiple RAD52 knockout clones pooled together also displayed a significant loss of telomere content, as quantified by quantitative telomere fluorescence in situ hybridization (Q-FISH) and dot blot in U-2 OS and LM216J but not in the matched telomerase-positive LM216T line (Fig. 4C,D; Supplemental Fig. S4E). In accordance with a reduced ability to resolve replication stress, RAD52 knockout cells accumulated RPA, ssDNA, and unresolved complexes at telomeres (Fig. 4E–G). Despite complete loss of RAD52, cell viability was not significantly compromised, raising the possibility that other repair factors compensate for RAD52 loss or act in parallel to maintain ALT telomeres. Loss of RAD52 resulted in increased 53BP1 localization at telomeres, suggesting a conversion of replication intermediates to DSBs (Fig. 4H). Given that DSB lesions are a potent inducer of BITS, we speculated that RAD52 knockout ALT cells may become increasingly reliant on alternative repair synthesis for survival. Indeed, RAD52 was dispensable for BITS following TRF1-FokI induction (Fig. 4I). On the other hand, residual G2 synthesis in RAD52 knockout cells was hypersensitive to low-dose aphidicolin treatment, consistent with a reported role of RAD52 in replication fork restart (Fig. 4J,K; Sotiriou et al. 2016). These findings implicate RAD52 downstream from replication stress in directing POLD3-dependent telomere maintenance. On the other hand, RAD52-independent pathways can function in response to DSBs to execute BITS.
Figure 4.
RAD52 is required for maintenance of ALT telomere. (A) TRF analysis using pulse-field electrophoresis from a population of U-2 OS cells expressing either empty vector or a sgRNA for RAD52 at ∼25 population doublings. Peak intensity of telomere signal is indicated by a red dot. (B) TRF analysis using pulse-field electrophoresis from a population of U-2 OS cells expressing either empty vector or sgRAD52 #1. After transduction with either empty vector or sgRAD52 #1, cells were collected after every four population doublings. Peak intensity of telomere signal is indicated by a red dot. (C) Quantification of relative telomere length by Q-FISH of metaphase spreads from the indicated cell lines. Error bars indicate SD. (D) Quantification of relative telomere content by dot blot from the indicated cell lines. Error bars indicate SD. (E) Quantification of RPA localization at telomeres. Error bars indicate SEM. (F,G) Native and denatured two-dimensional (2D) gels showing telomeric DNA from U-2 OS and LM216J cells. A single asterisk indicates t-complexes, and two asterisks indicate single-strand overhang (F) and quantification (G). Error bars indicate SD. (H) Quantification of 53BP1 localization at telomeres. Error bars indicate SD. (I) Quantification of BrdU pull-down dot blots for telomere content from the indicated cell lines following TRF1-FokI induction. C1–C3 represent individual CRISPR clones isolated from a population expressing sgRAD52 sg#1. Error bars indicate SD. (J,K) Experimental outline (J) and quantification (K) of telomere synthesis in G2 after low-dose APH treatment. Error bars indicate SD. (EV) Empty vector; (WT) wild type; (KO) knockout; (PD) population doubling; (APH) aphidicolin. Data represent mean values of at least two independent experiments. (n.s.) Nonsignificant; (*) P ≤ 0.05; (**) P ≤ 0.01; (****) P ≤ 0.0001, Student's t-test.
CRISPR screening reveals sensitivities of RAD52-deficient cells
We reasoned that RAD52-deficient ALT cells should be particularly sensitive to depletion of DNA repair factors that mediate RAD52-independent telomere maintenance. We performed a targeted CRISPR screen of candidate DNA repair proteins that have been implicated previously in different aspects of recombination (Fig. 5A,B). The screen was performed in wild-type and RAD52 knockout ALT-dependent LM216J cells. A sgRNA library consisting of 56 guides targeting 15 DNA repair proteins with distinct functionalities as well as three controls were cloned into a lentiviral vector that expresses GFP under the control of an internal ribosome entry sequence (IRES) (Fig. 5B; Tarumoto et al. 2018). The number of sgRNAs designed per gene was roughly based on gene size and the number of functional domains. Targeting functional domains has been documented to yield increased CRISPR Cas9-mediated knockout efficiency (Shi et al. 2015). Wild-type and RAD52 knockout LM216J ALT cells stably expressing Cas9 were transduced with lentivirus expressing sgRNAs and GFP as a tracker at a multiplicity of infection (MOI) of <0.6 to allow competition between the transduced GFP-positive and the nontransduced GFP-negative cells. These cells were subsequently monitored for changes in the percentage of GFP positivity every two doubling times for seven passages as a measure of cell proliferation (Fig. 5A). While GFP percent remained constant with each passage in cells targeted with the negative control ROSA26 (Fig. 5C), there was a total reduction in GFP positivity over the course of the experiment in both wild-type and RAD52 knockout cells treated with positive controls targeting the essential proteins PCNA and RPA3. However, we observed significantly reduced editing efficiency in RAD52 knockout cells compared with wild-type cells, biasing our screen against identifying hits that would be selectively toxic in cells lacking RAD52 (Fig. 5C).
Figure 5.
CRISPR screen reveals new vulnerabilities in RAD52 knockout cells. (A) Schematic of the experimental approach. (B) List of genes in the screen, classified according to their functionalities. (C–E) Growth curves representing the percentage of GFP-positive cells over seven passages (14 population doublings) in controls (C), group II genes (D), and group III genes (E). Each data point represents the mean of two independent experiments. Two-thousand cells were analyzed at most time points. Data points obtained from <500 cells were excluded from the analysis. (EV) Empty vector; (KO) knockout. The error bar indicates SD.
We observed three different proliferation trends, denoted by groups I–III (Fig. 5D,E; Supplemental Fig. S5). Group I sgRNAs did not reduce the GFP-positive population below 40% over 14 passage doublings in both wild-type and RAD52 knockout cells. These genes included RAD54A, RAD54B, Mus81, Gen1, and Polθ (Supplemental Fig. S5). Group II sgRNAs significantly reduced the GFP-positive population to <40% in wild-type cells but not in RAD52 knockout cells. This included RAD51, its paralogs (RAD51A, RAD51C, RAD51D, XRCC2, and XRCC3), and Fen1. We attribute the diminished sensitivity of targeting these essential recombination factors to the reduced editing efficiency in RAD52 knockout cells (Fig. 5D). Group III sgRNAs significantly reduced cell proliferation to <40% in both wild-type and RAD52 knockout cells. sgRNAs against SLX4, FANCD2, and BLM reduced cell proliferation of wild-type and RAD52 knockout cells to a comparable extent. Because of the reduced targeting efficiency in cells lacking RAD52 (Fig. 5E), we propose that the magnitude of the effect of these genetic interactions is underrepresented in RAD52 knockout cells.
To further understand these genetic interactions, we examined interdependencies for telomere localization. BLM and FANCD2 telomere localization was increased after RAD52 loss; however, SLX4, which is constitutively present at the telomere, remained unchanged (Supplemental Fig. S6A–C; Wilson et al. 2013). Conversely, loss of FANCD2 or SLX4 resulted in an increased RAD52 telomere localization (Fig. 2F; Supplemental Fig. S6D). BLM deficiency, on the other hand, reduced RAD52 signal at telomeres, suggesting that BLM acts upstream of RAD52 during ALT telomere synthesis (Supplemental Fig. S6D).
Combined loss of SLX4 and RAD52 causes genomic instability
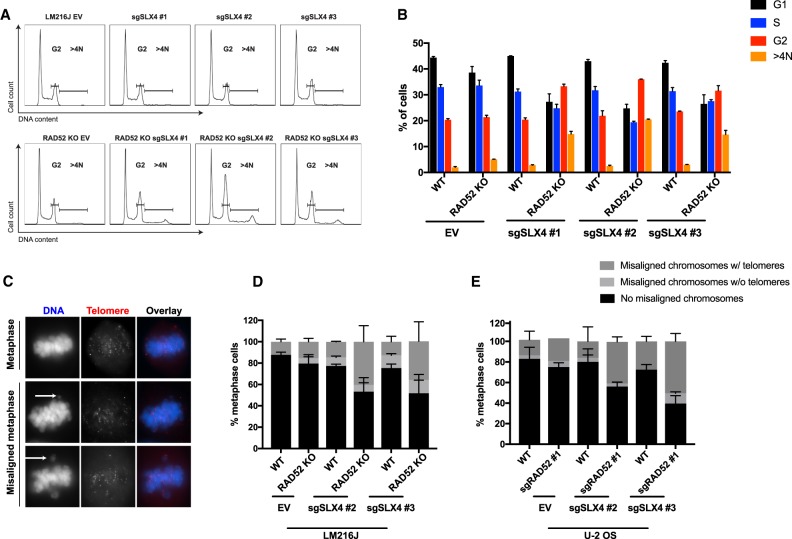
SLX4 has been reported upstream of and epistatic to RAD52 during MiDAS (Bhowmick et al. 2016). However, our results are indicative of nonoverlapping functions of RAD52 and SLX4, prompting us to examine the basis for this genetic interaction. We used three different sgRNAs to deplete the SLX4 protein in wild-type and RAD52 knockout LM216J cells. A blasticidin selection marker on each sgRNA allowed for enrichment of cells expressing SLX4 sgRNA. Consistent with screening data, there was extensive cell death following transduction with lentivirus expressing SLX4 sgRNAs. Immunoblot analysis of the surviving population revealed approximately twofold to fivefold reduction in SLX4 protein (Supplemental Fig. S4F). These pools of cells expressing SLX4 sgRNA were then used for subsequent experiments. Wild-type cells expressing sgRNA for SLX4 and RAD52 knockout cells did not display changes in the cell cycle profile. However, RAD52 knockout cells expressing SLX4 sgRNAs (RAD52 knockout sgSLX4) showed an increase in pChk1 positivity and hence G2/M arrest, indicating higher endogenous DNA damage in their combined absence (Fig. 6A,B; Supplemental Fig. S6E–G). Moreover, an increased fraction of polyploid cells was observed in RAD52 knockout sgSLX4 cells, suggestive of mitotic errors (Fig. 6A,B).
Figure 6.
Combined loss of SLX4 and RAD52 results in G2 arrest and increased mitotic abnormalities. (A,B) Representative FACS profile (A) and quantification (B) of cell cycle distribution in the indicated cell lines. The error bar indicates SD. (C,D) Representative immunofluorescence-FISH images (C) and quantification (D,E) of metaphase chromosomal abnormalities (blue) with telomeres (red) in the indicated cell lines. The error bar indicates SD. The white arrow indicates the aberration. (EV) Empty vector; (WT) wild type; (KO) knockout. Data represent mean values of two independent experiments.
We therefore examined LM216J and U-2 OS cells expressing sgRNAs for SLX4 and/or RAD52 for mitotic aberrations (Fig. 6C; Supplemental Fig. S4F). While treatment with either RAD52 or SLX4 sgRNA itself did not have a major effect on metaphase alignment, >40% of cells expressing sgRNAs for both RAD52 and SLX4 resulted in a significant increase in misaligned chromosomes. Nearly 80% of the misaligned chromosomes had telomere signal, suggesting that they arise from the distal ends of the chromosomes (Fig. 6D,E). On the basis of these results, we conclude that the combined loss of RAD52 and SLX4 enhances DNA damage and mitotic abnormalities that may in turn lead to reduced proliferation.
SLX4 and RAD52 contribute to distinct pathways in ALT telomere maintenance
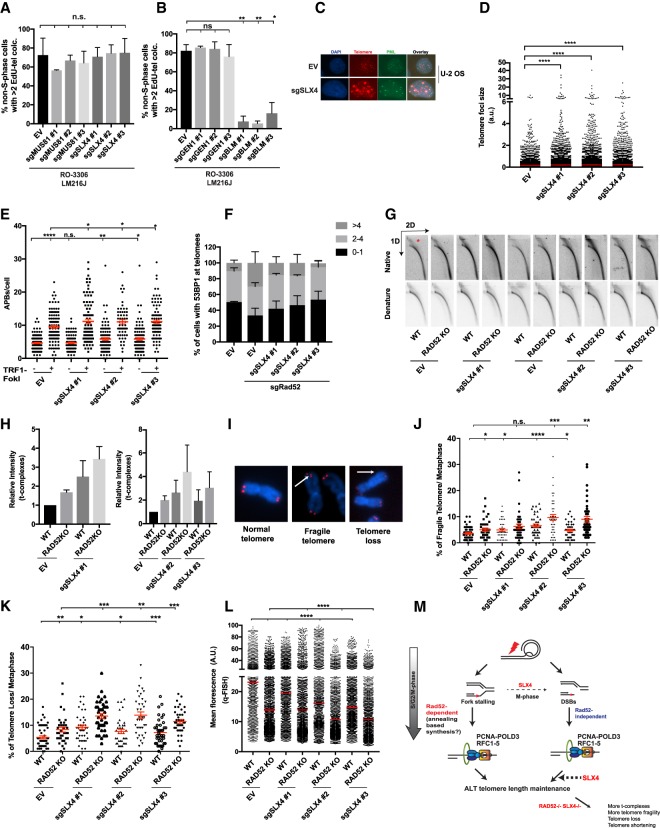
We next investigated the consequences of combined RAD52 and SLX4 loss on ALT telomere synthesis and stability. We first examined whether SLX4 and its associated nuclease, Mus81, contributes to G2-phase synthesis associated with ALT telomeres. None of the three sgRNAs for Mus81 or SLX4 had an effect on EdU incorporation at telomeres in Cdk1 inhibitor-arrested cells (Fig. 7A; Supplemental Figs. S2D, S4F,G). Additionally, Gen1, a nuclease that shares some functional redundancy with SLX4, did not have an effect on G2-phase telomere synthesis (Fig. 7B; Garner et al. 2013; Wyatt et al. 2013). On the other hand, sgBLM significantly reduced G2-phase telomere synthesis, consistent with our RAD52 localization studies and prior reports (Fig. 7B; Supplemental Figs. S2E, S4H, S6D; Sobinoff et al. 2017). These observations reveal that RAD52-mediated telomere synthesis in G2 is SLX4-independent, differing from its reported dependency on SLX4 in mitosis (Bhowmick et al. 2016).
Figure 7.
Nonepistatic interaction between RAD52 and SLX4 during ALT. (A,B) Quantification of EdU incorporation at telomeres in the indicated LM216J cells arrested in G2 phase using Cdk1 inhibitor (Cdk1i) RO-3306 for 16 h. Error bars indicate SD. (C) Immunofluorescence image showing telomere foci (red) colocalizing with promyelocytic leukemia (PML; green) in the indicated U-2 OS lines. DNA was stained using DAPI (blue). (D) Distribution of individual telomere focus size in the indicated U-2 OS lines. Error bars indicate SEM. (E) Quantification of APBs by measuring PMLs colocalizing at telomeres in the indicated U-2 OS lines. TRF1-FokI was induced for 2 h. Error bars indicate SEM. (F) Quantification of 53BP1 localization at the telomeres in the indicated U-2 OS lines. Error bars indicate SD. (G,H) Native and denatured 2D gels showing telomeric DNA from the indicated LM216J cells. An asterisk indicates t complexes (G) and quantification (H). Error bars indicate SD. (I–K) Representative metaphase spreads (I) and quantification of fragile telomeres (J) and telomere loss (K) from the indicated LM216J cell lines. Error bars indicate SEM. (L) Q-FISH analysis of the indicated LM216J cell lines. Error bars indicate SEM. (M) Model for ALT telomere maintenance: RAD52 is required for telomere synthesis ensuing from stalled forks. BITS occurs in a RAD52-independent manner. The major function of SLX4 is to resolve recombination intermediates (bold arrow). In the absence of RAD52, SLX4 may cleave the stalled forks during mitosis (light arrow). Combined loss of RAD52 and SLX4 results in the accumulation of unresolved replication and recombination intermediates, resulting in telomere fragility, loss, and shortening. (a.u.) Arbitrary units; (EV) empty vector; (WT) wild type; (KO) knockout. Data represent mean values of at least two independent experiments. (n.s.) Nonsignificant; (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001; (****) P ≤ 0.0001, Student's t-test.
While not affecting G2 telomere synthesis, cells expressing sgRNAs for SLX4 displayed strikingly large telomere foci that often coalesced in APBs. Distribution of focus size revealed an increased number of telomeres with two to 10 times larger focus size in sgSLX4-expressing cells compared with wild type (Fig. 7C,D). However, cells expressing sgRNAs targeting RAD52 showed reduced telomere focus size, consistent with its role in telomeric DNA synthesis (Supplemental Fig. S6H). Moreover, loss of SLX4 increased APB formation, and this was further elevated after induction of DSBs via TRF1-FokI (Fig. 7E). These observations suggest that SLX4 can act downstream from DSBs at telomeres to resolve recombination intermediates. The distinction in phenotypes conferred in the absence of either RAD52 or SLX4 prompted us to examine effects on telomeres after the combined loss of these proteins. Transient expression of sgSLX4 in sgRAD52-expressing cells reduced DSBs at telomeres (Fig. 7F). The data suggest that the replication intermediates generated in the absence of RAD52 may be processed by SLX4 to generate DSBs. Our observations lead to the prediction that concomitant loss of RAD52 and SLX4 would result in accumulation of unresolved stalled forks and recombination intermediates. Indeed, RAD52 knockout cells expressing sgSLX4 showed an increase in t complexes (Fig. 7G,H).
Failure to repair stalled forks has been demonstrated to increase fragile telomeres and loss of telomeric DNA (Sfeir et al. 2009; Pepe and West 2014; Pinzaru et al. 2016). In agreement, RAD52 knockout cells displayed a modest but significant increase in telomere fragility and loss. Consistent with previous literature, SLX4 deficiency increased the percentage of fragile telomeres (Wilson et al. 2013). The combined loss of RAD52 and SLX4 further enhanced both telomere loss and fragility (Fig. 7I–K). Finally, quantitative telomere FISH analysis revealed a significant reduction in telomere length in cells depleted for both RAD52 and SLX4 compared with the individual knockouts, establishing a coordinated role of the two proteins in ALT telomere maintenance (Fig. 7L).
Discussion
This study provides evidence that distinct repair synthesis pathways operate at ALT telomere synthesis depending on the nature of the initiating telomere lesion and cell cycle phase. We demonstrate mechanistic distinctions between the three RAD51-independent pathways that have been documented at ALT telomeres: MiDAS, spontaneous synthesis in G2, and BITS (Dilley et al. 2016; Özer et al. 2018). SLX4 and RAD52 were dispensable for BITS, while each is required for telomeric MiDAS (Fig. 4I; data not shown). RAD52, but not SLX4 or Mus81, contributed to spontaneous telomere synthesis in G2. While MiDAS is restricted to prometaphase, BITS occurred in interphase, prometaphase, and metaphase. TRF1-FokI-directed DSBs may bypass the requirement of structure-specific nucleases at later stages of mitosis, allowing replisome loading and subsequent HDR synthesis. Notably, all three pathways converge on a PCNA–POLD3-based replisome for executing DNA synthesis (Fig. 7M).
Our studies establish a crucial role for RAD52 in resolving replication stress and maintaining telomere length during ALT. The short duration of mitosis and the low frequency of MiDAS, (reported to be 0.5 EdU telomere foci per metaphase spread) suggest that mitotic DNA synthesis is likely not the primary means for telomere length maintenance during ALT (Min et al. 2017). We therefore propose that the progressive telomere shortening over several generations observed in RAD52 knockout cells results from reduced G2-phase synthesis. RAD52-dependent resolution of replication stress is likely activated more frequently and is responsible for a large portion of POLD3-dependent telomere synthesis in G2. However, the balance between RAD52-dependent and RAD52-independent pathways could shift in response to genetic disruption of repair proteins. As such, RAD52-null ALT cells continue to proliferate but are rendered hypersensitive to the loss of SLX4, BLM, and FANCD2. Notably, they were not hypersensitive to loss of RAD51 or its paralogs. Mitotic aberrations, along with increased G2 and polyploidy cell populations, demonstrate that lethal forms of genomic abnormalities accumulate in cells lacking both RAD52 and SLX4. It will be interesting to determine whether this nonepistatic interaction between RAD52 and SLX4 extends beyond telomere damage in cells that experience a high degree of replicative stress.
Several possibilities exist to explain the nonepistatic interaction between RAD52 and SLX4 in telomere maintenance, including (1) SLX4-directed endonucleolytic cleavage of stalled forks in the absence of RAD52 that promotes replication restart and/or (2) RAD52-mediated annealing of single-strand regions during fork remodeling that would mitigate the requirement of nucleolytic processing for fork restart (Ciccia and Symington 2016). Additionally, SLX4 would also be required for resolving recombination intermediates downstream from synthesis involving strand invasion events (Fig. 7M). In accordance, our studies reveal that the combined loss of RAD52 and SLX4 leads to increased accumulation of complex telomeric structures. These unresolved intermediates may serve as an impediment to the DNA synthesis machinery on subsets of telomeres and result in gradual or catastrophic telomere shortening. These events might not be evident in BrdU pull-down and C-circle assays that monitor HDR within a single cell cycle (Supplemental Figs. S2D, S6I–K). Approaches to directly visualize the ensuing telomere recombination intermediates that arise in the context of different DNA repair factor deficiencies may be necessary to address these possibilities.
Materials and methods
Cell culture
HeLa1.3, 293T, LM216T, LM216J, and U-2 OS cell lines were grown in DMEM (Thermo Fisher) with 10% calf serum and 1% penicillin/streptomycin. The GM847 cell line was grown in DMEM (Thermo Fisher) with 10% FBS and 1% penicillin/streptomycin. LM216J/T are matched lines. Cells were regularly tested for mycoplasma using the MycoAlert Plus mycoplasma detection kit (Lonza).
Plasmids, CRISPR sgRNAs, and siRNAs
Generation of cell lines stably expressing TRF1-FokI was described previously (Dilley et al. 2016). CRISPR knockout lines were generated using a two-vector system (Tarumoto et al. 2018). The sequences for siRNAs and sgRNAs are in Supplemental Table 1. Experiments were done with complete knockout or population of cells expressing sgRNAs. For the generation of knockouts, cells expressing sgRNA were plated at single-cell dilutions to obtain individual colonies. These colonies were screened for protein knockdown and insertion/deletion (indel) mutations in the gene. Six to seven complete knockout colonies were then pooled.
BrdU pull-down dot blot, TIF assay, APB assay, Western blot, C-circle assay, pulse-field gel electrophoresis, in-gel hybridization, and telomere content dot blot
Assays were performed as described previously (Dilley et al. 2016). For pKAP-1, pPLK-1, and pChk1 blots, blocking was performed using 5% BSA made in TBST (0.2% Tween diluted in Tris-buffered saline), and TBST was used for washes. The antibodies used are listed in Supplemental Table 1.
Nascent C-circle assay
BrdU pull-down of 2 µg of gDNA was performed as described previously with two major differences (Dilley et al. 2016; Verma et al. 2018). First, sonication was omitted in order to preserve C-circle structure. Instead, gDNA was digested using AluI and MboI (New England Biolabs). Second, the digested gDNA was not denatured prior to pull-down. After BrdU pull-down, the nascent DNA was cleaned with ChIP DNA Clean and Concentrator kit (Zymo) and eluted in 20 µL. For C-circle PCR, 6 µL of eluted pull-down was used per reaction. Digested gDNA input (30 ng per reaction) was run in parallel.
EdU immunofluorescence
Cells grown on coverslips were pulsed with 100 µM EdU (Invitrogen) for 2 h and 30 min before fixing with 4% paraformaldehyde (PFA) for 10 min at room temperature. After permeabilization with PBS + 0.5% Triton X-100 at 4°C, cells were washed with 0.2% Tween diluted in PBS (PBST). EdU was labeled using the Click-iT Cell Reaction Buffer kit (Thermo Fisher Scientific) as per manufacturer's instructions. Cells were then washed with PBS + 3% BSA followed by PBS. Coverslips were then processed for telomere FISH as described before (Dilley et al. 2016). Performing the EdU labeling experiment in G2-arrested cells gave a much better signal to noise ratio compared with asynchronous cells.
Immunofluorescence
Cells grown on coverslips were fixed in 4% paraformaldehyde for 10 min at room temperature and then permeabilized with PBS + 0.5% Triton X-100 for 5 min at 4°C. For RAD51, RPA, and RAD52 immunofluorescence, cells were pre-extracted with ice-cold 0.2% Triton X-100 diluted in PBS at 4°C. For RAD51 and RPA, pre-extraction was performed for 5 min; for RAD52, the duration was 2 min (Ochs et al. 2016). SLX4 and BLM immunofluorescence was performed as described previously (Wilson et al. 2013). For most immunofluorescence experiments, fixed cells were incubated overnight at 4°C in primary antibody diluted in PBST. For RAD52, the primary antibody was diluted in filtered DMEM containing 10% FBS with 0.05% sodium azide, and incubation was performed for 1 h at room temperature. After three washes with PBST, secondary antibody (Life Technologies) diluted 1:200 in PBST or in DMEM (for RAD52 immunofluorescence) was added to cells for 1 h at room temperature. After three washes with PBST, coverslips were then processed for FISH as described earlier (Dilley et al. 2016). For pChk1 immunofluorescence, 5% BSA made in TBST was used for blocking and making antibody dilutions. Additionally, washes were performed using TBST. Images were acquired using a CoolSnap Myo camera (Photometrics) connected to Nikon NIS-Elements software. Images were processed using Fiji (National Institutes of Health), and statistical analysis was performed using Graphpad Prism. The antibodies used are listed in Supplemental Table 1.
GFP-CRISPR screen
LM216J cells were engineered to stably express Cas9 (Addgene, 108100). Individual sgRNAs for each gene were cloned into a lentiviral vector with a U6 promoter and an IRIS GFP (Addgene, 108098). Virus for sgRNAs was generated in a 96-well format by transfecting HEK293T cells in each well with sgRNA containing 1050 ng of vector, 750 ng of pPAX2, and 500 ng of pVSVG using PEI. Viruses were then transduced at an MOI of <0.6 on Cas9-expressing LM216J wild-type or LM216J RAD52 knockout cells seeded in a 96-well plate. The percent of GFP in each well was monitored using a Guava Easycyte HT instrument (Milllipore) for seven population doublings after the second day of transduction (which represented the day of maximum GFP expression). Live cells were gated using forward and side scatter before measuring GFP-positive cells. Two-thousand cells were analyzed at most time points. Data points obtained from <500 cells were excluded from the analysis.
Drugs
Doxycyline was used at a concentration of 40 ng/mL for 16 h to induce TRF1-FokI expression. Cells were treated with 10 µM 4-hydroxytamoxifen (4-OHT) (Sigma-Aldrich) for 2 h to allow nuclear translocation of TRF1-FokI. HU (98%) (Sigma) was used at 1 mM concentration for 1 h. Unless indicated otherwise, mitosis-arrested cells were isolated by shakeoff after ∼16 h of incubation with 100 ng/mL nocodazole. Nocodazole remained in the medium after shakeoff to ensure continued arrest. Alternatively, cells were arrested in G2 for ∼16 h with 10 µM RO-3306 and subsequently released into nocodazole for 20 min before shakeoff. For sustained G2 arrest, cells were kept in RO-3306.
PCNA chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously with minor changes (Tang et al. 2013). Briefly, G2-arrested cells were fixed for 1 h in 1% formaldehyde, lysed, sonicated, and subjected to immunoprecipitation with 2 µg of anti-PCNA (mouse PC10; Cell Signaling Technologies, 2586) antibody overnight at 4°C. The next day, samples were incubated with Protein G magnetic beads (Pierce) for 2 h rotating at 4°C, washed, and eluted overnight at 65°C. Eluted DNA was then cleaned and processed for dot blot analysis of telomere content.
Neutral–neutral two-dimensional (2D) gel electrophoresis
Neutral–neutral 2D gel electrophoresis was performed as described before (Nabetani and Ishikawa 2009). HinfI- and RsaI-digested DNA samples were subjected to a first-dimensional gel, which was prepared by 0.4% agarose in 1× TBE, and electrophoresized at 1 V/cm for 12 h at room temperature. Afterward, sample lanes were cut out and immersed in 1 × TBE containing 0.3 µg/mL ethidium bromide (EB; Sigma) for 30 min. The 1% agarose gel containing EB was casted to embed the sample lanes for the second-dimensional gel, which was subjected to electrophoresis at 3 V/cm for 6 h at 4°C in a cold room.
Metaphase FISH and Q-FISH
Metaphase FISH and Q-FISH were performed as described previously (Perner et al. 2003; Ourliac-Garnier and Londoño-Vallejo 2017). Colcemid (Roche) was added at 100 ng/mL to the indicated cell lines at ∼70% confluency and incubated for 3 h. The cells were harvested by trypsinization, rinsed once with PBS, swelled in prewarmed 0.075 M KCl for 30 min in 37°C water bath, and then fixed overnight with freshly prepared methanol:acetic acid (3:1) at 4°C. Metaphase spread was done by dropping the cells onto slides preheated at 42°C. Slides were dried overnight; rehydrated in PBS; fixed with formaldehyde for 2 min; rinsed three times with PBS; dehydrated with 75%, 95%, and 100% ethanol; and air dried. Hybridization was performed for 2 h at room temperature in hybridization solution (70% formamide, 10 mM Tris-HCl at pH 7.2, 0.5% blocking solution [prepared by blocking reagent from Roche]) containing telomere probe (TelC-Cy3; PNA Bio) after heating for 3 min at 75°C. Afterward, slides were washed twice with buffer 1 (70% formamide, 10 mM Tris-HCl at pH 7.2) and another three times in PBST. During the second wash with PBST, chromosomes were stained with DAPI. The samples were then dehydrated again with 75%, 95%, and 100% ethanol; air-dried; mounted with VectaShield mounting medium without DAPI (Vector Laboratories); and sealed with clear nail polish. Images of 40∼50 metaphases for each sample were taken by a Nikon Eclipse 80i fluorescent microscope. Telomere signal intensity was analyzed with Fiji software.
Cell cycle analysis
Cells were collected by trypsinization from a 70%–80% confluent 10-cm dish. The pellet was suspended in 300 µL of PBS, and 700 µL of 100% chilled ethanol was added dropwise while simultaneously vortexing the cells. The cells were fixed at least overnight at −20°C. At the time of staining, cells were pelleted and washed twice with PBS. The pellet was then resuspended in 250 µL of PBS containing 250 µg/mL RNase A (Roche) and incubated for 30 min at 37°C. PBS (250 µL) containing 50 µg/mL propidium iodide (Santa Cruz Biotechnology) was then added, and cells were incubated in the dark for 10 min at room temperature. Data for at least 10,000 live cells were collected using FACSCalibur (BD Biosciences). Analysis was performed using the FlowJo software. A double-discrimination gate was used to exclude doublets, and G0/G1, S, and G2 percentages were calculated using the Watson pragmatic algorithm. Cells >4N were manually gated.
Supplementary Material
Acknowledgments
We thank J. Lukas (University of Copenhagen) for sharing the RAD52 antibody, J. Rouse (University of Dundee) for the SLX4 antibody, and J. Shi (University of Pennsylvania) for sharing reagents and providing guidance with the CRISPR screen. This work was supported by National Institutes of Health (NIH) grants GM101149 and CA17494 to R.A.G., who is also supported by funds from the Basser Center for BRCA and a V Foundation Team Convergence Award. P.V. is supported by the Ann and Sol Schreiber Mentored Investigator Award (Ovarian Cancer Research Fund Alliance). R.L.D. was supported by NIH grants T32GM007170 and T32GM008216.
Authors contribution: P.V., R.L.D., and R.A.G. designed the study. P.V., R.L.D., and T.Z. performed most of the experiments, with assistance from M.T.G. and Y.L. P.V., R.L.D., and R.A.G. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.319723.118.
References
- Anand RP, Lovett ST, Haber JE. 2013. Break-induced DNA replication. Cold Spring Harb Perspect Biol 5: a010397 10.1101/cshperspect.a010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson FE, Baumann P, West SC. 1998. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391: 401–404. 10.1038/34937 [DOI] [PubMed] [Google Scholar]
- Bhowmick R, Minocherhomji S, Hickson ID. 2016. RAD52 facilitates mitotic DNA synthesis following replication stress. Mol Cell 64: 1117–1126. 10.1016/j.molcel.2016.10.037 [DOI] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14: 4240–4248. 10.1002/j.1460-2075.1995.tb00098.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Rondinelli B, D'Andrea AD. 2016. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26: 52–64. 10.1016/j.tcb.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR. 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11: 319–330. 10.1038/nrg2763 [DOI] [PubMed] [Google Scholar]
- Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. 2009. Spontaneous occurrence of telomeric DNA damage response in the absence of chromosome fusions. Nat Struct Mol Biol 16: 1244–1251. 10.1038/nsmb.1725 [DOI] [PubMed] [Google Scholar]
- Chen Q, Ijpma A, Greider CW. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol 21: 1819–1827. 10.1128/MCB.21.5.1819-1827.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NW, Dilley RL, Lampson MA, Greenberg RA. 2014. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell 159: 108–121. 10.1016/j.cell.2014.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Symington LS. 2016. Stressing out about RAD52. Mol Cell 64: 1017–1019. 10.1016/j.molcel.2016.11.036 [DOI] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. 2014. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343: 88–91. 10.1126/science.1243211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA. 2016. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539: 54–58. 10.1038/nature20099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR. 2009. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic Acids Res 37: 1740–1754. 10.1093/nar/gkn995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. 2011. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci 108: 686–691. 10.1073/pnas.1010959107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E, Kim Y, Lach FP, Kottemann MC, Smogorzewska A. 2013. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep 5: 207–215. 10.1016/j.celrep.2013.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S, Belotserkovskaya R, Jackson SP. 2010. DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol 190: 197–207. 10.1083/jcb.200911156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamshet K, Mazina OM, Mazin AV. 2016. Reappearance from obscurity: mammalian Rad52 in homologous recombination. Genes (Basel) 7: 63 10.3390/genes7090063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AYM, Pickett HA, Reddel RR. 2009. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol 27: 1181–1185. 10.1038/nbt.1587 [DOI] [PubMed] [Google Scholar]
- Hustedt N, Durocher D. 2017. The control of DNA repair by the cell cycle. Nat Cell Biol 19: 1–9. 10.1038/ncb3452 [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. 2001. Homologous pairing promoted by the human Rad52 protein. J Biol Chem 276: 35201–35208. 10.1074/jbc.M104938200 [DOI] [PubMed] [Google Scholar]
- Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. 2002. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell 10: 359–371. 10.1016/S1097-2765(02)00587-7 [DOI] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider CW. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok BH, Carley AC, Tchang B, Powell SN. 2013. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32: 3552–3558. 10.1038/onc.2012.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360. 10.1016/0092-8674(93)90234-H [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823. 10.1038/nature06047 [DOI] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci 93: 7131–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Wright WE, Shay JW. 2017. Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break-induced replication processes. Mol Cell Biol 37: 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. 2015. Replication stress activates DNA repair synthesis in mitosis. Nature 528: 286–290. 10.1038/nature16139 [DOI] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F. 2009. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol 29: 703–713. 10.1128/MCB.00603-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A, Yokoyama O, Ishikawa F. 2004. Localization of hRad9, hHus1, hRad1, and hRad17 and caffeine-sensitive DNA replication at the alternative lengthening of telomeres-associated promyelocytic leukemia body. J Biol Chem 279: 25849–25857. 10.1074/jbc.M312652200 [DOI] [PubMed] [Google Scholar]
- Ochs F, Somyajit K, Altmeyer M, Rask M-B, Lukas J, Lukas C. 2016. 53BP1 fosters fidelity of homology-directed DNA repair. Nat Struct Mol Biol 23: 714–721. 10.1038/nsmb.3251 [DOI] [PubMed] [Google Scholar]
- Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, Escribano-Diaz C, Durocher D. 2014. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 344: 189–193. 10.1126/science.1248024 [DOI] [PubMed] [Google Scholar]
- O'Sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G, Karlseder J. 2014. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol 21: 167–174. 10.1038/nsmb.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourliac-Garnier I, Londoño-Vallejo A. 2017. Telomere length analysis by quantitative fluorescent in situ hybridization (Q-FISH). Methods Mol Biol 1587: 29–39. 10.1007/978-1-4939-6892-3_3 [DOI] [PubMed] [Google Scholar]
- Özer Ö, Bhowmick R, Liu Y, Hickson ID. 2018. Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget 9: 15836–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe A, West SC. 2014. MUS81–EME2 promotes replication fork restart. Cell Rep 7: 1048–1055. 10.1016/j.celrep.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Brüderlein S, Hasel C, Waibel I, Holdenried A, Ciloglu N, Chopurian H, Nielsen KV, Plesch A, Högel J, et al. 2003. Quantifying telomere lengths of human individual chromosome arms by centromere-calibrated fluorescence in situ hybridization and digital imaging. Am J Pathol 163: 1751–1756. 10.1016/S0002-9440(10)63534-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzaru AM, Hom RA, Beal A, Phillips AF, Ni E, Cardozo T, Nair N, Choi J, Wuttke DS, Sfeir A, et al. 2016. Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep 15: 2170–2184. 10.1016/j.celrep.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol 18: 6423–6429. 10.1128/MCB.18.11.6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera T, Haggblom C, Cosconati S, Karlseder J. 2017. A balance between elongation and trimming regulates telomere stability in stem cells. Nat Struct Mol Biol 24: 30–39. 10.1038/nsmb.3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root H, Larsen A, Komosa M, Al-Azri F, Li R, Bazett-Jones DP, Stephen Meyn M. 2016. FANCD2 limits BLM-dependent telomere instability in the alternative lengthening of telomeres pathway. Hum Mol Genet 25: 3255–3268. 10.1093/hmg/ddw175 [DOI] [PubMed] [Google Scholar]
- Roumelioti F-M, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD, Gagos S. 2016. Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep 17: 1731–1737. 10.15252/embr.201643169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51–BRCA1/2. Cancer Cell 22: 106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103. 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33: 661–667. 10.1038/nbt.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobinoff AP, Allen JA, Neumann AA, Yang SF, Walsh ME, Henson JD, Reddel RR, Pickett HA. 2017. BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J 36: 2907–2919. 10.15252/embj.201796889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Ré C, Huber F, Padayachy L, Tardy S, Nicati NL, Barriot S, et al. 2016. Mammalian RAD52 functions in break-induced replication repair of collapsed DNA replication forks. Mol Cell 64: 1127–1134. 10.1016/j.molcel.2016.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20: 317–325. 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarumoto Y, Lu B, Somerville TDD, Huang Y-H, Milazzo JP, Wu XS, Klingbeil O, El Demerdash O, Shi J, Vakoc CR. 2018. LKB1, Salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol Cell 69: 1017–1027.e6. 10.1016/j.molcel.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Zakian VA. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19: 8083–8093. 10.1128/MCB.19.12.8083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell 6: 947–952. 10.1016/S1097-2765(05)00094-8 [DOI] [PubMed] [Google Scholar]
- Van Dyck E, Stasiak AZ, Stasiak A, West SC. 1999. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398: 728–731. 10.1038/19560 [DOI] [PubMed] [Google Scholar]
- Van Dyck E, Stasiak AZ, Stasiak A, West SC. 2001. Visualization of recombination intermediates produced by RAD52-mediated single-strand annealing. EMBO Rep 2: 905–909. 10.1093/embo-reports/kve201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J. 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561. 10.1016/j.molcel.2005.09.024 [DOI] [PubMed] [Google Scholar]
- Verma P, Greenberg RA. 2016. Noncanonical views of homology-directed DNA repair. Genes Dev 30: 1138–1154. 10.1101/gad.280545.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Dilley RL, Gyparaki MT, Greenberg RA. 2018. Direct quantitative monitoring of homology-directed DNA repair of damaged telomeres. Methods Enzymol 600: 107–134. 10.1016/bs.mie.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JSJ, Tejera AM, Castor D, Toth R, Blasco MA, Rouse J. 2013. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell Rep 4: 853–860. 10.1016/j.celrep.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HDM, Sarbajna S, Matos J, West SC. 2013. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol Cell 52: 234–247. 10.1016/j.molcel.2013.08.035 [DOI] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59: 4175–4179. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.