Zhang et al. show that manipulating the splicing regulator ESRP1 to shift alternative splicing from splice isoform CD44v to CD44s leads to an induction of cancer stem cell properties.
Keywords: CD44s, alternative splicing, CSC, breast cancer, PDGFRβ/Stat3
Abstract
Although changes in alternative splicing have been observed in cancer, their functional contributions still remain largely unclear. Here we report that splice isoforms of the cancer stem cell (CSC) marker CD44 exhibit strikingly opposite functions in breast cancer. Bioinformatic annotation in patient breast cancer in The Cancer Genome Atlas (TCGA) database reveals that the CD44 standard splice isoform (CD44s) positively associates with the CSC gene signatures, whereas the CD44 variant splice isoforms (CD44v) exhibit an inverse association. We show that CD44s is the predominant isoform expressed in breast CSCs. Elimination of the CD44s isoform impairs CSC traits. Conversely, manipulating the splicing regulator ESRP1 to shift alternative splicing from CD44v to CD44s leads to an induction of CSC properties. We further demonstrate that CD44s activates the PDGFRβ/Stat3 cascade to promote CSC traits. These results reveal CD44 isoform specificity in CSC and non-CSC states and suggest that alternative splicing provides functional gene versatility that is essential for distinct cancer cell states and thus cancer phenotypes.
Metastatic breast cancers are essentially incurable, causing nearly 40,000 breast cancer patients to succumb each year in the United States. A small population of cancer cells, termed “cancer stem cells” (CSCs), contributes to tumor metastasis and relapse (Dalerba et al. 2007a; Polyak and Weinberg 2009). These CSCs possess cellular plasticity that enables them to repropagate and differentiate, allowing for switches between different cellular phenotypes. The plasticity of switchable cell states and phenotypes contributes to tumor heterogeneity and could be a critical determinant of successful metastasis and tumor recurrence. However, its underlying mechanisms have been largely unexplored.
Alternative splicing constitutes a prevalent mechanism that produces more than one protein from a single gene, greatly increasing proteome diversity. For decades, dysregulation of alternative splicing has been observed in cancer (Srebrow and Kornblihtt 2006; Venables 2006; Liu and Cheng 2013), including recent findings showing that therapeutic resistance to the RAF (V600E) inhibitor vemurafenib is caused by the appearance of aberrantly spliced B-RAF in melanoma patients (Poulikakos et al. 2011). Despite these important observations, the functional contribution of splice isoforms and alternative splicing regulation in cancer, especially in the field of CSCs, is not well understood.
The cell surface protein CD44 has been widely used as a CSC marker in breast cancer and various other types of cancers (Al-Hajj et al. 2003; Jin et al. 2006; Dalerba et al. 2007b; Fillmore and Kuperwasser 2007; Li et al. 2007; Liu et al. 2007; Prince et al. 2007). CD44 undergoes extensive alternative splicing, generating two families of isoforms: the variable exon-containing CD44 variant isoforms (CD44v) and the variable exon-absent CD44 standard isoform (CD44s). We previously reported that isoform switching from CD44v to CD44s is functionally essential for cells to undergo epithelial–mesenchymal transition (EMT) (Brown et al. 2011; Reinke et al. 2012). Shifting alternative splicing to produce different CD44 splice isoforms allows for changes of cellular phenotypes between epithelial and mesenchymal states, suggesting that alternative splicing regulates phenotypic plasticity (Brown et al. 2011; Xu et al. 2014).
While questions remain on whether CD44 serves merely as a CSC marker or also exhibits a functional role in sustaining the essential qualities of CSCs, increasing evidence has pointed to a role for CD44 in promoting cancer progression through mechanisms such as alterations of signaling cascades and enhancing CD44–extracellular matrix interactions (Xu et al. 2010; Brown et al. 2011; Su et al. 2011; Hiraga et al. 2013; Zhao et al. 2013; Pietras et al. 2014; Xu et al. 2014; Gao et al. 2015; Wang et al. 2015; Zhao et al. 2016). Interestingly, the isoform specificity of CD44 in cancer is somewhat controversial (Lopez et al. 2005; Kim et al. 2008; Brown et al. 2011; Yae et al. 2012; Hiraga et al. 2013; Zhao et al. 2016). The CD44s isoform was reported to promote tumor cell survival, invasiveness, and metastasis (Ouhtit et al. 2007; Mima et al. 2012; Hiraga et al. 2013; Zhao et al. 2016). CD44v was also reported to promote CSC activities, especially in gastric cancer, where tumor initiation ability was drastically altered in Cd44 knockout or Cd44v-expressing mice (Yoshikawa et al. 2013; Lau et al. 2014; Todaro et al. 2014; Zeilstra et al. 2014). CD44v was shown to contribute to reactive oxygen species (ROS) defense through its interaction with xCT, protecting CSCs from ROS-induced stress (Ishimoto et al. 2011). Clinically, pan-CD44 as well as both CD44s and CD44v splice isoforms have been reported as prognosis markers for various types of cancers (Lee et al. 2008; Zhou et al. 2010; Ko et al. 2011; Olsson et al. 2011; Auvinen et al. 2013; Deng et al. 2013; Jung et al. 2013; Cao et al. 2014; Ni et al. 2014; Tei et al. 2014; Yan et al. 2015). These seemingly conflicting observations highlight the complex nature of CD44 and its splice isoforms in different cancer stages and cancer types.
In this study, we show that distinct activities are associated with CD44s and CD44v. CD44s is preferentially expressed in breast CSCs and promotes CSC traits by activating the PDGFRβ and Stat3 signaling cascades. Conversely, CD44v is inversely correlated with CSC signatures. Manipulating CD44 alternative splicing shifts cells between CSC and non-CSC states. These results thus suggest that the plasticity of breast CSC phenotypes can be modulated at the level of alternative RNA splicing.
Results
CD44s and CD44v splice isoforms show distinct associations with breast cancer phenotypes and subtypes
To objectively evaluate the activities of CD44s and CD44v splice isoforms in breast cancer, we used a bioinformatics approach to extract CD44 isoform-associated gene signatures from RNA sequencing (RNA-seq) data sets of >1000 patient breast tumor specimens in The Cancer Genome Atlas (TCGA). We used the TCGA exon expression data set and developed a method to obtain the levels of CD44v and CD44s isoforms (see detailed information in the Supplemental Material). As shown in Supplemental Figure S1A, the human CD44 gene contains nine variable exons located between its nine constitutive exons. Plotting the CD44 exon expression levels revealed that the CD44 v8, v9, and v10 exons were the most abundant variable exons (Supplemental Fig. S1A), and the expression of these three variable exons was most highly correlated among the breast cancer specimens (Supplemental Fig. S1B). These results are in agreement with previous reports that most of the CD44v isoforms contain the v8 to v10 exons (Ponta et al. 1998). Thus, the average expression of exons v8, v9, and v10 was used to represent levels of CD44v. Likewise, the average of three constitutive exons (c6, c7, c8, which appeared in all CD44 transcripts) was used as a surrogate for total CD44. The levels of CD44s were then calculated by subtraction of the two values. Visualization of the TCGA RNA-seq read distribution on the CD44 gene confirmed that the calculated levels of CD44 isoforms were reliable (Supplemental Fig. S1C).
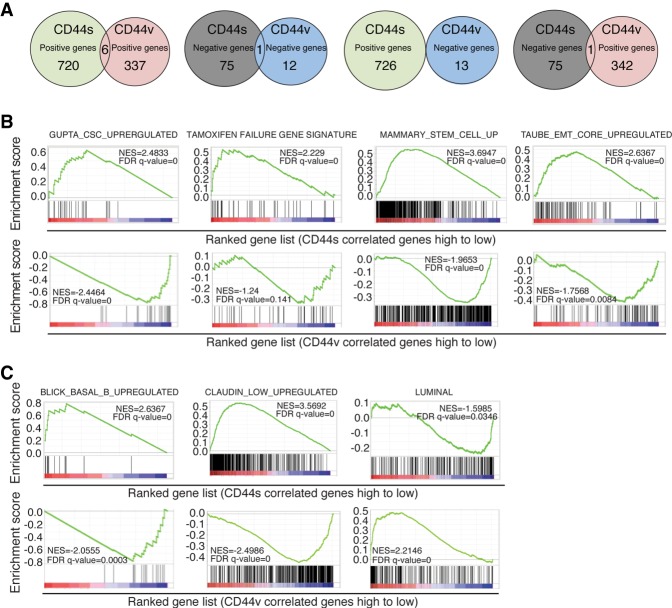
We first identified gene sets that were correlated with CD44s and CD44v, respectively. Using an absolute value of 0.3 as the cutoff for correlation coefficient, we identified 802 and 356 genes whose expression levels were correlated with CD44s and CD44v, respectively (Fig. 1A; Supplemental Table 1). The vast majority of these CD44s- and CD44v-associated genes do not overlap, suggesting that these two splice isoforms could be linked with distinct functions in breast cancer.
Figure 1.
Genome-wide TCGA analysis reveals distinct association of CD44 isoforms with breast cancer phenotypes and subtypes. (A) Venn diagram plots showing the overlapping of CD44s- and CD44v-correlated genes analyzed from the breast cancer TCGA database. CD44v levels were calculated as the average of exon expression of v8, v9, and v10. (B) Gene set enrichment analysis (GSEA) showing the enrichment of CSC, tamoxifen failure, mammary stem cell, and EMT gene signatures in the CD44s-correlated gene list and negative enrichment in the CD44v-correlated gene list. (C) GSEA showing the positive enrichment of the Basal_B gene signature and Claudin_low signature in the CD44s-correlated gene list and the Luminal gene signature in the CD44v-correlated gene list.
To better understand the functions of CD44 splice isoforms in breast cancer, we determined CD44 isoform-enriched gene signatures and molecular pathways by genome-wide gene set enrichment analysis (GSEA) of the TCGA data set. The CD44s and CD44v gene signatures showed striking inverse relationships (Fig. 1B; Supplemental Fig. S1D). The CD44s gene set exhibited significant positive association with signatures of CSC, tamoxifen (TAM) therapeutic failure in breast cancer patients and mammary stem cells, and EMT, while the CD44v gene set negatively correlated with all of these signatures. Conversely, CD44v positively correlated with a G1–S proliferative signature (Supplemental Fig. S1E), a property of CD44v that was reported previously (Matter et al. 2002; Cheng et al. 2006). Moreover, the opposing relationship between CD44s and CD44v was also observed when analyzing an independent Cancer Cell Line Encyclopedia (CCLE) breast cancer cell line data set (Supplemental Fig. S1F).
Analysis of the association between the CD44 isoform gene signatures and breast cancer subtypes showed that the CD44s gene set exhibited positive enrichment of more aggressive basal-like and Claudin-low signatures and negative enrichment of a less aggressive luminal signature, whereas the CD44v gene set displayed exactly the opposite enrichment relationships (Fig. 1C; Supplemental Fig. S1G). These results, along with additional opposing enrichment patterns of breast cancer phenotypes (Supplemental Fig. S1E), showed that the CD44s and CD44v isoforms correlated with different cancer cell states and phenotypes: CD44s is closely associated with breast CSC features and aggressive phenotypes, but CD44v is negatively associated with these phenotypes and is positively associated with cell proliferation.
CD44s is the predominant isoform expressed in human CSCs and mediates CSC properties
CSCs have been regarded as the source of therapeutic relapse and metastasis, and CD44 is widely used as a marker for CSCs (Al-Hajj et al. 2003; Jin et al. 2006; Dalerba et al. 2007b; Fillmore and Kuperwasser 2007; Li et al. 2007; Liu et al. 2007; Prince et al. 2007). The fact that the gene sets specifically associated with CD44s, but not CD44v, were significantly enriched in signatures of CSCs prompted us to examine the composition of CD44 isoforms in the CD44hi/CD24lo CSCs.
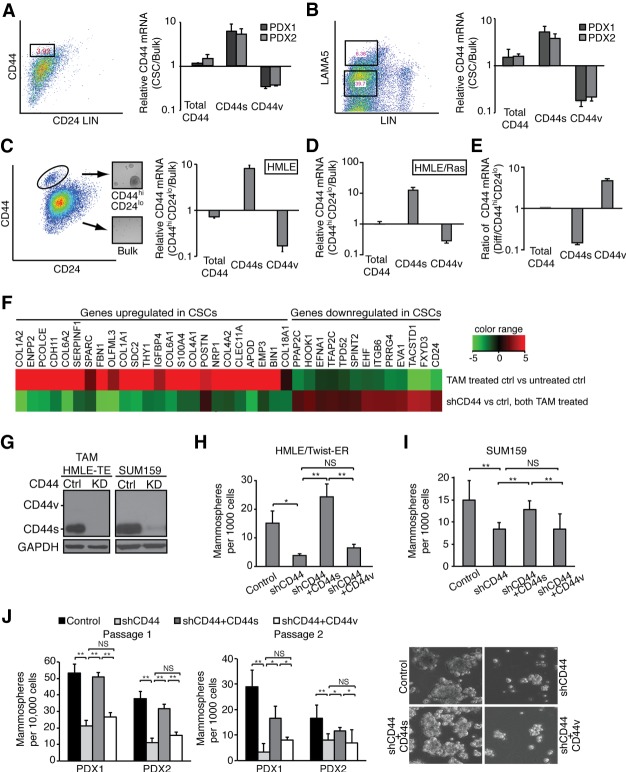
We isolated CD44hi/CD24lo CSCs from two patient-derived xenografts (PDXs) of triple-negative breast tumors. RT–PCR analysis of this population compared with the bulk population showed that CD44s was highly enriched in the CSC population, with an increase in CD44s by 5.4-fold and an accompanied decrease of CD44v by 3.3-fold (Fig. 2A). We noted that the levels of total CD44 mRNA transcripts were very similar between the CD44hi/CD24lo fraction and the bulk. The increased intensity of CD44 in the CD44hi/CD24lo fraction by FACS was due in part to the fact that the CD44 antibody recognizes CD44s better than CD44v. We also isolated CSCs from two breast PDX tumors using a different set of previously defined CSC markers, LAMA5/lineage-negative (Chang et al. 2015), and observed that the relative ratio of CD44s to CD44v in LAMA5/lineage-negative CSCs was ∼20-fold (Fig. 2B). These data demonstrate that the CD44s splice isoform is enriched in breast CSCs.
Figure 2.
CD44s is the major splice isoform expressed in CSCs and mediates CSC properties. (A) CD44 isoform expression was analyzed in CD44hiCD24lo CSCs and bulk populations that are lineage-negative isolated from two PDX tumors. Distinct primer pairs that amplify all CD44 isoforms (total CD44), CD44s, or v5 and v6 exon-containing CD44v (CD44v5/v6) were used. Ratios of CD44 isoform mRNAs in CSC and bulk populations are shown. (B) CD44 isoform expression was analyzed in LAMA5hi and LAMA5lo populations isolated from two PDX tumors. (C, left panel) The CD44hiCD24lo population was isolated from human mammary epithelial (HMLE) cells by FACS. Inset images show the mammosphere-forming ability of the CD44hiCD24lo population. (Right panel) Ratios of CD44 isoform mRNA levels analyzed in CD44hiCD24lo and bulk HMLE populations are shown. (D) Ratios of CD44 isoform mRNAs in CD44hiCD24lo and bulk HMLE/Ras populations are shown. (E) The FACS-sorted CD44hiCD24lo population of HMLE cells was grown in monolayer culture for 8 d for epithelial differentiation. Ratios of CD44 isoform mRNAs at day 8 relative to day 0 are shown. (F) TAM-treated HMLE/Twist-ER cells expressing control or CD44 shRNA were analyzed by quantitative RT–PCR (qRT–PCR) for expression of the CSC signature. (G) Immunoblot of CD44 isoform expression in TAM-treated HMLE/Twist-ER (HMLE-TE), SUM159, and their corresponding CD44 knockdown cell lines. (H–J) The effect of CD44 isoforms on mammosphere-forming ability was analyzed using breast cell lines (H,I) and PDX-derived tumor cells (J). (J) Representative images of mammospheres are shown in PDX-derived tumor cells with differential CD44 isoform expression. The numbers of mammospheres are presented. Error bars indicate SEM. n = 3. (*) P < 0.05; (**) P < 0.01.
In accordance with the above results, experimental systems of the CD44hi/CD24lo population sorted from human mammary epithelial (HMLE) cells (Fig. 2C) or their tumorigenic derivative HMLE/Ras cells (Fig. 2D) showed that CD44s was highly enriched in the CD44hi/CD24lo fraction despite the fact that CD44v was the predominant isoform in the unsorted cells (Supplemental Fig. S2A). The minuscule fraction of the CD44hi/CD24lo cells prevented us from detecting CD44 isoform expression by immunoblot analysis. However, the relative levels of CD44s to the housekeeping gene TATA-binding protein (TBP) in the CD44hi/CD24lo cells were on par with that in the HMLE/Twist cells where the CD44s protein was readily detectable (Supplemental Fig. S2B), inferring that the CD44s protein was the major isoform in the CD44hi/CD24lo cells. Furthermore, experimental differentiation of the CD44hi/CD24lo cells to epithelial cells (Supplemental Fig. S2C) by growing them in culture resulted in an isoform switch from CD44s to CD44v (Fig. 2E), demonstrating a return to expression levels similar to that of bulk HMLE cells. Since the levels of CD44 transcripts (total CD44) did not vary significantly in any comparisons (Fig. 2A–E), these results indicate that isoform expression is switched from CD44s to CD44v between CSC and non-CSC states.
To verify these results, we used an additional HMLE model in which the induction of Twist expression using a TAM-inducible Twist-ER fusion construct leads to a substantial increase in CD44hi/CD24lo cells, which are mesenchymal in nature (Mani et al. 2008). Accordingly, we found highly expressed CD44s in the TAM-treated cells, whereas the parental non-TAM-treated cells expressed CD44v (Supplemental Fig. S2D, cf. lanes 1 and 2). Moreover, these TAM-treated cells showed enriched expression of a CSC signature (Fig. 2F, top line; Gupta et al. 2009). Eliminating CD44 by shRNA in the TAM-treated cells (Supplemental Fig. S2D) broadly abolished the CSC gene signature (Fig. 2F, bottom line), suggesting a functional role for CD44 in CSC traits.
In addition to the CD44hi/CD24lo mesenchymal-like CSCs, it was reported recently that ALDH+ cells were representative of epithelial-like CSCs (Liu et al. 2014). The CD44hi/CD24lo mesenchymal-like CSCs were found to be primarily quiescent and localized at the tumor invasive front, whereas the ALDH+ epithelial-like CSCs are proliferative and located in the interior of tumors (Liu et al. 2014). We found that the epithelial HMLE cells contained 21% ALDH+ cells, whereas the mesenchymal HMLE cells induced by Twist contained 8% ALDH+ cells (Supplemental Fig. S2E,F). shRNA depletion of CD44 did not change the ALDH+ cell fraction in either cell state (Supplemental Fig. S2E,F). Thus, we focused on the effect of CD44 isoforms in the mesenchymal-like CSCs.
Next, we performed mammosphere formation assays to examine the effect of CD44 splice isoforms on CSC traits. We used TAM-treated HMLE/Twist-ER and SUM159 cells, both of which expressed high levels of CD44s with little or no detectable CD44v (Fig. 2G). CD44 shRNA expression in these cells effectively depleted CD44s (Fig. 2G) and significantly reduced mammosphere-forming ability (Fig. 2H,I). Interestingly, re-expressing CD44s restored the mammosphere-forming ability, but re-expressing CD44v3–10, the dominant form detected in the HMLE cells, did not (Fig. 2H,I; Supplemental Fig. S2G,H). Results from serial passaging of mammospheres using breast cancer cells isolated from two independent PDX models further reveal that only the CD44s-specific splice isoform, and not CD44v, is functionally required to promote CSC properties (Fig. 2J; Supplemental Fig. S2I).
CD44s is required for CSC properties in a breast cancer mouse model
To further evaluate CD44s-mediated CSC potential, we performed in vivo tumor initiation experiments by limiting dilution. We used breast tumor cells that were derived from recurrent tumors of a HER2/NEU-inducible breast cancer mouse model (Moody et al. 2002, 2005). In this model, doxycycline-induced expression of oncogenic HER2/Neu in the mammary epithelium resulted in primary breast tumor formation. The primary tumor cells expressed high levels of CD44v but no detectable level of CD44s (Supplemental Fig. S3A, left lane). Withdrawal of doxycycline led to primary tumor regression. However, several weeks to months later, recurrent tumors developed at the primary site (Moody et al. 2005). These recurrent tumor cells predominantly expressed CD44s with no detectable expression of CD44v (Supplemental Fig. S3A, right lane) and showed enhanced CSC signatures (Supplemental Fig. S3B). We therefore used these recurrent tumor cells to examine the role of CD44s in CSCs by CD44 shRNA knockdown. Orthotopic injection of as few as 50 recurrent tumor cells was sufficient to form tumors in mice (Fig. 3A). The tumor-forming efficiency was six out of 24 mice when injecting 50 tumor cells, and this increased to 11 out of 14 mice when injecting 500 tumor cells. Based on these results, we calculated the frequency of CSCs to be roughly one out of 230 in control tumor cells. Knockdown of CD44 in these cells drastically decreased the efficiency of tumor initiation. Only two out of 23 mice formed tumors when injecting 50 CD44s-depleted cells, and three out of 14 mice that received 500 tumor cells produced tumors, resulting in a significantly reduced frequency of CSCs of one out of 987. Since CD44s is the most predominant isoform in this model system, these data indicate that CD44s is required for breast tumor initiation in vivo, a key property of CSCs.
Figure 3.
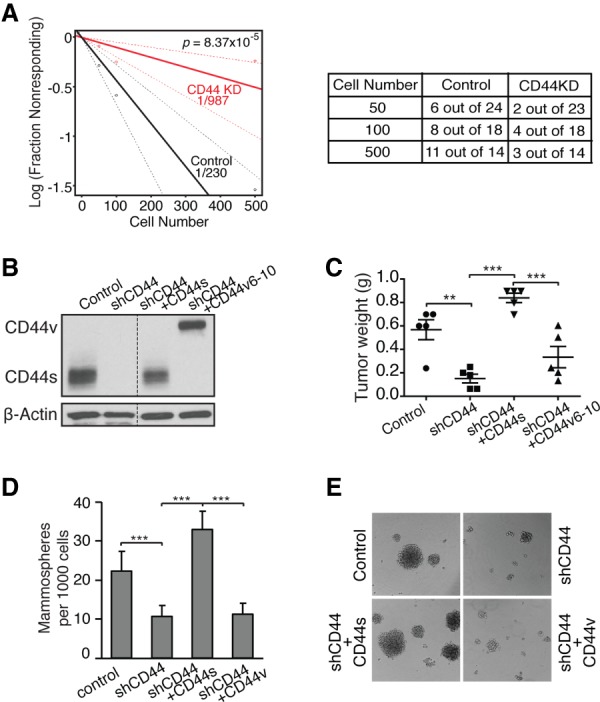
CD44s is a functional mediator of tumor-initiating cell properties in a mouse model of breast cancer progression. (A) Recurrent breast tumor cells expressing control or CD44 shRNA (CD44KD) were transplanted into mammary fat pads of FVB mice. Data are presented as a log–log plot. Frequency of CSCs is calculated by extreme limiting dilution analysis. (B) Immunoblot analysis showing reconstituted expression of CD44s and CD44v in CD44 knockdown cells that ectopically expressed CD44s and CD44v cDNA. (C) Analysis of tumor weight in the indicated mice. The CD44v6–10 reconstituted cells formed the least number of tumors; i.e., five tumors from 12 injections. Thus, the five largest tumors from each group were compared. (D) The effect of CD44s in mammosphere-forming ability was assessed in recurrent tumor cells that have had CD44 depleted and its isoforms reconstituted. (E) Representative mammosphere images are shown. Error bars in C and D indicate SEM (n = 5; C) and SD (n = 9; D). (**) P < 0.01; (***) P < 0.001.
To address the isoform specificity of CD44 in promoting recurrent tumor growth in mice, we injected recurrent tumor cells that have had CD44 knocked down and those that have had reconstituted expression of CD44s or CD44v isoforms (Fig. 3B). The CD44v6–v10 cDNA was used for CD44v because the HER2/NEU-induced primary tumor cell line from which the recurrent tumor cells were derived predominantly expressed the v6–v10-containing CD44v. While knockdown of CD44 inhibited tumor growth, re-expression of CD44s in the knockdown cells restored this tumor growth capacity, and re-expression of CD44v6–10 showed a lower extent (Fig. 3C). These results show that the CD44s splice isoform exhibit a higher activity to promote recurrent tumor growth in mice.
We further performed mammosphere-forming assays using recurrent cells with the CD44 knocked down and its isoform reconstituted. Knockdown of CD44 in the recurrent tumor cells reduces mammosphere-forming potential, a defect that was rescued by re-expressing CD44s but not CD44v (Fig. 3D,E), suggesting an essential role for the CD44s isoform in promoting CSC traits.
The splicing factor ESRP1 suppresses CD44s-mediated CSC function and inversely correlates with CSC signatures
The above results led us to postulate that regulation of alternative splicing by controlling the switchable expression patterns of CD44 isoforms may modulate the plasticity of cancer cells between CSC and non-CSC states. To identify splicing factors that may control CD44 isoform switching in CSCs, we analyzed the correlation between the expression of splicing factors and the ratio of CD44s to CD44v isoforms in the breast cancer TCGA database. While no significant positively correlated splicing factors were found, three splicing factors exhibited significant negative correlations (Fig. 4A). Among them, ESRP1 was the top hit (Fig. 4A,B), a phenomenon that was also observed when analyzing the CCLE breast cancer cell line data set (Supplemental Fig. S4A).
Figure 4.
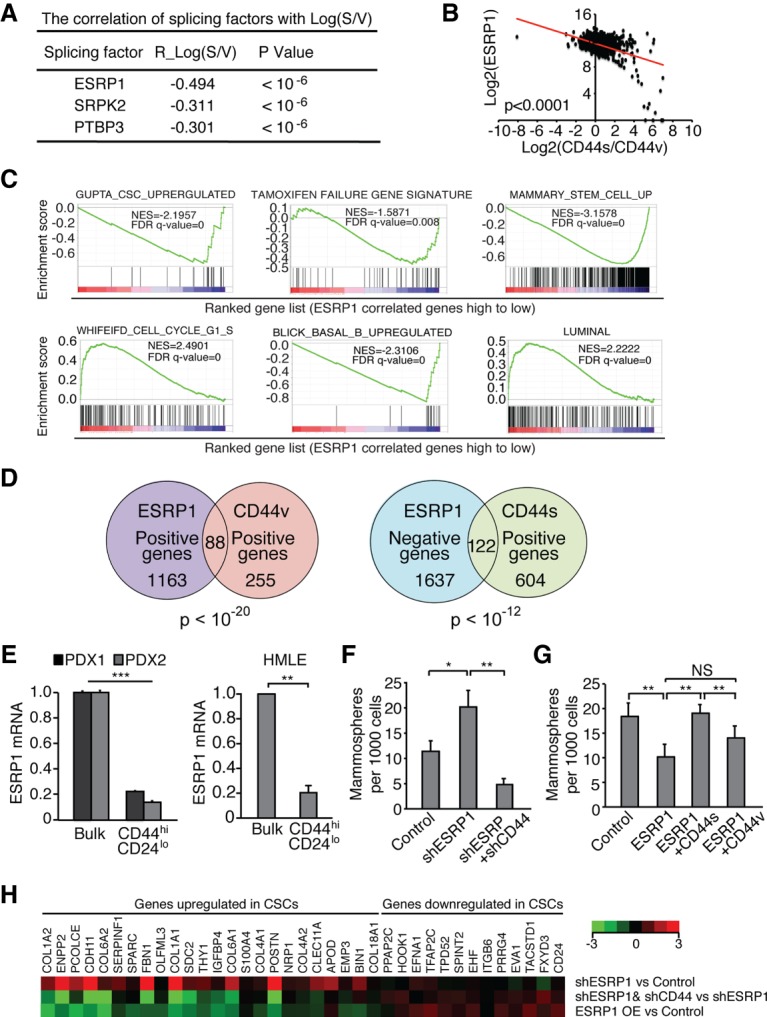
The splicing factor ESRP1 suppresses CD44s-mediated CSC function and inversely correlates with CSC signatures. (A) The correlation values between splicing factors and the ratio of CD44s to CD44v from analysis of the breast cancer TCGA data set are shown. (B) ESRP1 levels show significant negative correlation with the ratios of CD44s to CD44v. Log2 values are shown. (C) GSEA plots indicate the negative enrichment of CSC, TAM failure, mammary stem cell, and Basal_B gene signatures and positive enrichment of G1–S and Luminal A gene signatures in the ESRP1-correlated gene list. (D) Venn diagram plots showing the overlapping of ESRP1-, CD44v-, and CD44s-associated genes using the breast cancer TCGA data set. (E) qRT–PCR analysis indicates that ESRP1 mRNA levels in the CD44hiCD24lo population was reduced compared with the bulk population in PDX tumors and HMLE cells. (F,G) Mammosphere-forming ability is depicted in TGFβ-treated HMLE cells (control, shESRP1, and shESRP1 + shCD44) or SUM159 cells (control, ESRP1, ESRP1 + CD44s, and ESRP1 + CD44v) after growth in mammosphere culture for 2 wk. (H) HMLE cell lines (control, shESRP1, shESRP1 + shCD44, and ESRP1 overexpression) were analyzed by qRT–PCR for expression of the CSC signature. Error bars in E–G indicate SEM. n = 3. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
ESRP1 is an RNA-binding protein that is expressed in an epithelial cell type-specific manner (Warzecha et al. 2009). ESRP1 has been demonstrated to promote CD44v exon inclusion, resulting in production of CD44v and inhibition of CD44s (Brown et al. 2011; Reinke et al. 2012). These results are in agreement with our observations from TCGA data mining. Interestingly, previous studies have reported connections between ESRP1 and breast cancer but with conflicting conclusions: ESRP1 was shown to both promote (Yae et al. 2012) and inhibit (Goel et al. 2014) CSC traits.
We analyzed ESRP1's activity in clinical breast cancer specimens in the TCGA. The ESRP1-associated gene set exhibited a significant negative correlation with signatures of CSC, TAM resistance, and mammary stem cells but showed a positive association with a proliferative signature (Fig. 4C), exactly mirroring the signatures observed from the CD44v-associated gene set (Fig. 1B; Supplemental Fig. S1C). Like CD44v (Fig. 1C), the ESRP1 gene set also displayed a positive correlation with a luminal signature and a negative correlation with a basal phenotype (Fig. 4C). These ESRP1-associated signatures were also confirmed when using the CCLE breast cancer cell line data set (Supplemental Fig. S4B) or mining published RNA-seq data in response to ESRP1 silencing (Supplemental Fig. S4C; Yang et al. 2016). Furthermore, ESRP1 positively correlated genes significantly overlapped with CD44v positively associated genes (P < 10−20), and, in contrast, ESRP1 negatively correlated genes significantly overlapped with CD44s positively correlated genes (P = 2.69 × 10−13) (Fig. 4D; Supplemental Fig. S4D).
The above patient data-derived results suggest that ESRP1 exhibits an inhibitory activity on CSCs and aggressive breast cancer phenotypes and acts in opposition to the activities of CD44s. Given the role of ESRP1 in promoting the CD44v splice isoform (Brown et al. 2011; Reinke et al. 2012), it is conceivable that ESRP1 inhibits CSC traits at least in part through its regulation of CD44 alternative splicing, which inhibits the production of CD44s. To test this hypothesis, we first examined the expression of ESRP1 in CD44hi/CD24lo CSCs where CD44s is the predominant isoform. The expression levels of ESRP1 were roughly fivefold lower in CSCs than non-CSCs in two independent PDX tumors and the HMLE cells (Fig. 4E). We then determined whether shifting isoform expression to CD44s by knocking down ESRP1 elevates mammosphere-forming activity and, furthermore, whether shRNA-mediated depletion of CD44s in the ESRP1 knockdown cells abolishes this activity. Expression of ESRP1 shRNA in HMLE cells showed accelerated isoform switching from CD44v to CD44s in response to TGFβ treatment (Supplemental Fig. S4E). These ESRP1-depleted and TGFβ-treated HMLE cells possessed enhanced mammosphere-forming ability, and, importantly, silencing CD44 in these ESRP1-depleted cells abrogated mammosphere formation (Fig. 4F). As a complementary approach, we expressed ESRP1 in SUM159 breast cancer cells and observed reduced potential for mammosphere formation. This defect was rescued by coexpression of CD44s but not CD44v (Fig. 4G). Thus, these gain- and loss-of-function experiments indicate that ESRP1 suppresses CSC traits by shifting alternative splicing to inhibit the CD44s isoform production.
Further supporting the above findings, silencing ESRP1 promoted expression of the CSC signature, and this enhancement was largely reversed when CD44s was depleted in the ESRP1 silenced cells (Fig. 4H, top two lines). In contrast, ectopic expression of ESRP1 in TGFβ-treated HMLE cells, where endogenous ESRP1 was low, greatly inhibited the CSC gene signature (Fig. 4H, bottom line). Collectively, these results reveal that the splicing factor ESRP1 inhibits CSC properties by reducing the production of the CD44s splice isoform, demonstrating that ESRP1-mediated switching of CD44 alternative splicing modulates the phenotypes between CSC and non-CSC states.
CD44s mediates CSC-like properties through the PDGFRβ/Stat3 signaling pathway
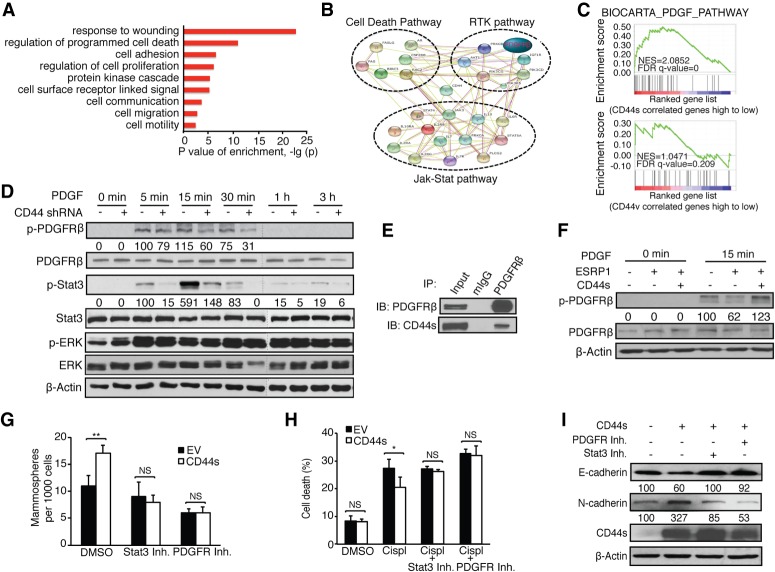
We next set out to investigate the mechanism underlying CD44s-potentiated CSC properties. Given that CD44 is a cell surface protein that interacts with receptor tyrosine kinases (RTKs) (Ponta et al. 2003; Orian-Rousseau 2015), we focused on RTK-mediated signaling pathway alterations. Gene ontology analysis showed that the CD44s gene signature was associated with functions involved in wounding, cell death regulation, and cell surface receptor-linked signal transduction (Fig. 5A; Supplemental Fig. S5A). The CD44v gene signature, on the other hand, showed association with cell morphogenesis and epithelial tube morphogenesis (Supplemental Fig. S5B). Of particular interest, Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) showed that CD44s was tightly connected to interacting networks of the cell death pathway, RTK pathways (including PDGFRβ and IGF1R), and the Jak–Stat pathway (Fig. 5B)—signal nodes that have been implicated in CSC activities (Dallas et al. 2009; Marotta et al. 2011; Tam et al. 2013). Noticeably, GSEA on RTK pathways showed that the PDGF pathway was the only cascade that exerted significant enrichment uniquely associated with the signature of CD44s but not CD44v (Fig. 5C; Supplemental Fig. S5C).
Figure 5.
CD44s mediates CSC-like properties through the PDGFRβ/Stat3 signaling pathway. (A) Gene ontology analysis of CD44s-correlated genes. (B) STRING analysis showing functional association networks of CD44s-correlated genes. (C) GSEA plots showing the enrichment of the PDGF pathway signature in the CD44s-correlated gene list. (D) The levels of p-PDGFRβ, p-Stat3, and p-ERK were examined in HMLE-Twist control and shCD44 cells using immunoblot analysis. Cells were treated with 10 ng/mL PDGF for the indicated time intervals. The relative intensity of phosphorylated to unphosphorylated signals is depicted below the images. Representative images are shown from three biological replicates. (E) 293FT cells cotransfected with CD44s and Flag-tagged PDGFRβ were subjected to immunoprecipitation (IP). (F) The levels of p-PDGFRβ were examined using immunoblot analysis in HMLE/Twist cells expressing control and ESRP1 cDNA or coexpressing ESRP1 and CD44s cDNA. Cells were treated with 10 ng/mL PDGF for 15 min. The relative intensity of p-PDGFRβ to total PDGFRβ is depicted below the images. Representative images are shown from three biological replicates. (G) Mammosphere-forming ability assay in control and CD44s-expressing Mes10A cells that were treated with 5 µM PDGFR inhibitor IV or 10 µM Stat3 inhibitor XVIII. (H) Mes10A cells expressing control and CD44s cDNA were pretreated with 5 µM PDGFR inhibitor IV or 10 µM Stat3 inhibitor XVIII for 1 h followed by treatment with 100 µM cisplatin (Cispl) for 24 h. Cell death was assessed and plotted as percent dead cells. (I) Mes10A cells expressing CD44s cDNA were treated with 10 µM Stat3 inhibitor XVIII or 5 µM PDGFR inhibitor IV for 48 h. The expression level of E-cadherin and N-cadherin was examined using immunoblot analysis. The parental Mes10A cells were used as control. Representative images are shown from three biological replicates. Error bars in G and H indicate SEM. n = 3. (*) P < 0.05; (**) P < 0.01.
To examine whether CD44s stimulates the activity of PDGFRβ, thus promoting CSC activities, we introduced CD44 shRNA in the CSC-like HMLE-Twist cells (Mani et al. 2008). Only the CD44s isoform is detectable in the HMLE-Twist cells, allowing us to assess the role of CD44s in PDGFRβ signaling (Supplemental Fig. S5D). Treatment with PDGF in control cells resulted in a burst activation of PDGFRβ, and the signal declined with time (Fig. 5D). Depletion of CD44s impaired PDGFRβ activation (Fig. 5D). Assessment of the PDGFRβ downstream effector Stat3 phosphorylation also showed that CD44s depletion inhibited Stat3 activation. Similarly, PDGFRβ downstream effector Akt phosphorylation was also impaired in the absence of CD44s (Supplemental Fig. S5E). This CD44s-dependent activation was specific, and no alterations of MAPK (ERK) phosphorylation were observed (Fig. 5D). Furthermore, reciprocal immunoprecipitation experiments showed that CD44s and PDGFRβ interact (Fig. 5E; Supplemental Fig. S5F). To determine whether the CD44–PDGFRβ interaction is CD44s isoform-specific, we ectopically expressed an HA-tagged CD44v cDNA in the HMLE/Twist cells or an HA-tagged PDGFRβ in the CD44v highly expressed HMLE cells, where endogenous levels of PDGFRβ were not detectable. In both cell conditions, no interactions were detected between CD44v and PDGFRβ (Supplemental Fig. S5G). Thus, the PDGFRβ interaction is CD44s isoform-specific.
Next, we ectopically expressed ESRP1 in HMLE-Twist cells to examine whether the ESRP1-potentiated switch from CD44s to CD44v negatively impacts PDGFRβ activity (Supplemental Fig. S5H). Forced expression of ESRP1 caused a reduction in PDGFRβ phosphorylation (Fig. 5F). Notably, this defect was corrected when CD44s was overexpressed (Fig. 5F), demonstrating that the splicing factor ESRP1 suppresses PDGFRβ signaling through shifting the splicing product away from the CD44s splice isoform.
To functionally determine whether CD44s-mediated CSC traits were dependent on its activation of the PDGFR–Stat3 pathway, we examined whether inhibiting PDGFRβ and Stat3 affects the CD44s-dependent CSC phenotypes, as gauged by mammosphere formation, survival, and EMT markers. As shown in Figure 5G, forced expression of CD44s in Mes10A cells increased mammosphere-forming ability, and this activity was abolished when cells were treated with either the PDGFR inhibitor IV or the Stat3 inhibitor XVIII. Similarly, treatment with either PDGFR or Stat3 inhibitors diminished the survival advantage in cisplatin-treated CD44s-expressing cells (Fig. 5H). These PDGFR or Stat3 inhibitor-treated cells also showed increased expression of the epithelial marker E-cadherin and reduced expression of the mesenchymal marker N-cadherin, suggesting a partial reversal of EMT marker expression (Fig. 5I). Taken together, these results demonstrate that PDGFRβ/Stat3 activation serves as an important downstream pathway mediating CD44s-dependent CSC traits.
CD44s is up-regulated in triple-negative tumors and correlates with CSC gene signature in multiple types of cancers
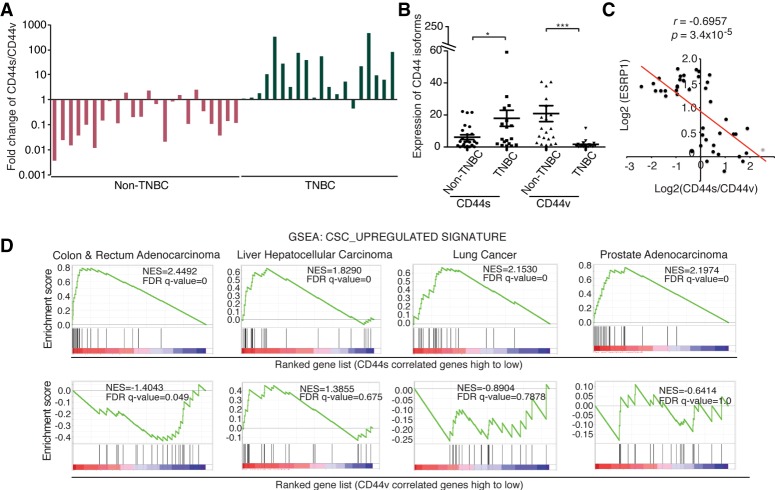
The triple-negative breast cancer (TNBC) is among the most difficult types of tumors to treat and has been shown to contain an increased number of dedifferentiated CSCs (Pece et al. 2010; Bhola et al. 2013; Ma et al. 2014; Brooks et al. 2015). To further validate our bioinformatics and experimental findings in patient tumors, we assessed the expression of CD44 isoforms and their correlation with ESRP1 as well as Stat3 target gene expression in a cohort of TNBC and non-TNBC specimens. We found that non-TNBC tumors predominantly expressed CD44v, which was reflected by the values of the CD44s to CD44v ratio that were <1 (Fig. 6A). In TNBC tumors, however, there was a shift of CD44 isoform expression toward CD44s, accompanied by a decrease in CD44v expression (Fig. 6A,B). These data support the notion that a switch in splice isoform expression toward CD44s occurs in CSC-enriched TNBC tumors. Interestingly, our data corroborate previous finding that ESRP1 expression is significantly lower in TNBC tumors than that in non-TNBC tumors using the same cohort (Goel et al. 2014).
Figure 6.
CD44s expression is elevated in TNBC and associates with a CSC signature in multiple cancer types. (A) RNA isolated from frozen clinical specimens of TNBC (n = 20) and non-TNBC (n = 24) was subjected to qRT–PCR analysis of CD44s and CD44v. Ratios of CD44s to CD44v are shown. (B) Expression levels of CD44s and CD44v in TNBC and non-TNBC samples are shown. (C) Correlation graphing reveals a significant negative correlation between ESRP1 and the ratio of CD44s to CD44v in breast tumor samples. (D) GSEA plots indicate the enrichment of the CSC gene signature in the CD44s-correlated gene list in colon and rectum adenocarcinoma, liver hepatocellular carcinoma, lung cancer, and prostate adenocarcinoma. Error bars in B indicate SEM. n = 24 non-TNBC; n = 20 TNBC. (*) P < 0.05; (***) P < 0.001.
RT–PCR analysis showed an inverse correlation between the expression of ESRP1 and the ratio of CD44s to CD44v in breast tumors that we examined (Fig. 6C), consistent with the results from the TCGA data analysis shown in Figure 4B. Moreover, evaluating expression levels of Stat3 downstream targets that are involved in metastasis (Carpenter and Lo 2014) showed that depletion of CD44s in breast cancer cells repressed the expression of these genes (Supplemental Fig. S6), supporting the notion that CD44s promotes Stat3 signaling in breast cancer patients.
Because CD44 is widely used as a CSC marker in breast cancers and various other types of cancers (Al-Hajj et al. 2003; Jin et al. 2006; Dalerba et al. 2007b; Fillmore and Kuperwasser 2007; Li et al. 2007; Liu et al. 2007; Prince et al. 2007), we evaluated whether the CD44s splice isoform is associated with CSC signatures in four additional cancer types: colon and rectum adenocarcinoma, liver hepatocellular carcinoma, lung cancer, and prostate adenocarcinoma. Our analysis revealed that the CD44s-associated gene set positively correlated with the CSC signature in all of the analyzed cancer types, whereas CD44v showed either a negative correlation or no correlation (Fig. 6D). These results imply that CD44s is associated with a CSC phenotype in many different types of cancers.
Discussion
Although CD44 is a widely used marker of CSCs, it has remained controversial whether CD44 and, in particular, any of its splice isoforms are functionally critical for CSCs. In this study, we provide evidence demonstrating that the splice isoforms CD44s and CD44v exhibit distinct activities in breast cancer. Mining of the TCGA breast cancer database revealed that CD44s positively associates with gene signatures of CSCs and therapeutic resistance, whereas CD44v negatively associates with these signatures but positively associates with cell proliferation. Profiling of CSCs from PDXs and experimental systems revealed that the CD44s splice isoform is predominantly expressed in breast CSCs.
Cancer cell heterogeneity is partially controlled by the plasticity of cancer cell states, promoting therapeutic resistance and cancer metastasis. While the change of cell states can be influenced by tumor microenvironment and signaling activation or perturbation, we argue in this study that alternative splicing provides an effective means to contribute to cell state plasticity. In addition to demonstrating that CD44s is the major splice isoform in CSCs, our results show that expression of CD44s is switched to that of CD44v when CSCs are differentiated to non-CSCs. In a similar notion, we showed previously that CD44 splicing is tightly regulated during EMT and that isoform switching from CD44v to CD44s is required for cells to undergo EMT. Given the functional contribution of CD44s in CSCs and the connection between CSCs and EMT (Mani et al. 2008; Singh and Settleman 2010), these results suggest that regulation at the level of alternative splicing can causally change the status of CSCs, which in turn contributes to distinct cancer phenotypes.
The CSCs are known to promote tumor metastasis and relapse. Using a mouse recurrent breast cancer model system and limiting dilution experiments, we demonstrated that CD44 is important for tumor initiation in vivo. We found previously that CD44s is predominantly expressed in metastatic breast cancer cells (Zhao et al. 2016). We and others have also shown that in these metastatic breast cancer cell lines, knockdown of CD44 significantly inhibits breast cancer metastasis to the bone and lungs (Hiraga et al. 2013; Zhao et al. 2016). Thus, our in vivo data here are in accordance with previous findings and support the notion that CD44s is a functional component in CSCs.
In the search for splicing regulators for CD44 isoform switching, our bioinformatics analysis of TCGA breast cancer specimens revealed the highest negative correlation between the CD44s/CD44v ratio and ESRP1 among all examined splicing factors. Importantly, manipulation of ESRP1 to shift CD44 expression to the CD44v isoform inhibited CSC phenotypes, which could be corrected by enforced expression of CD44s. The role of ESRP1 has been linked previously to breast cancer. Specifically, it was reported that ESRP1 regulates the splicing of the α6β1 integrin and that the α6Bβ1 integrin splice variant promotes the function of breast CSCs and tumor initiation (Goel et al. 2014). ESRP1 inhibits CSC properties by increasing expression of the α6Aβ1 splice variant at the expense of α6Bβ1 (Goel et al. 2014). Our results are congruent with these findings. Given that CSCs have been implicated in metastasis, a previous report that ESPR1 promotes breast cancer metastasis by a mechanism that involves ESRP1-stimulated up-regulation of CD44v (Yae et al. 2012) seems contradictory. However, an emerging consensus is that metastasis involves both epithelial and mesenchymal cells, possibly in clusters (Aceto et al. 2014; Cheung and Ewald 2016), which could explain the opposing functions of ESRP1 in the metastatic process. Indeed, the reported highly metastatic CD44v+ 4T1 cells still express a significant amount of the CD44s protein isoform (Yae et al. 2012). Interestingly, in a study that separated metastatic MCF10CA1h cells into CD44med/CD24low and CD44high/CD24low fractions, it was shown that the CD44med/CD24low cells expressed higher levels of ESRP1 and CD44v and gave rise to more metastatic nodules. However, the CD44high/CD24low cells showed decreased expression of ESRP1, expressed higher levels of CD44s, and showed higher potential for tumor initiation (Hu et al. 2017). These results are in agreement with our findings that CD44s plays an important role in tumor initiation and also suggest that the CD44v-high cells have proliferation advantage. It is conceivable that increased levels of ESRP1 in the metastatic cells help sustain an epithelial phenotype and produce the CD44v splice isoform (Warzecha et al. 2009; Brown et al. 2011), which stimulates Ras/MAPK signaling and cell proliferation (Matter et al. 2002; Cheng et al. 2006). However, loss of ESRP1 triggers an EMT and acquisition of CSC properties (Brown et al. 2011; Goel et al. 2014). Thus, the plasticity that alternative splicing provides allows cancer cells to adapt to the need to proliferate and survive in different circumstances and may enable distinct populations of cells to function in concert to facilitate processes such as tumor recurrence and metastasis. Interestingly, expression of the ESRP1 gene is epigenetically regulated (Yae et al. 2012). It would be an exciting future direction to investigate how epigenetic regulation affects ESRP1-mediated alternative splicing, resulting in changes of cancer phenotypes.
While widely used as a CSC marker, the function of CD44 in CSCs was largely unclear. A major conclusion of this study is that the CD44s splice isoform, but not CD44v, is indispensable for breast CSC activities and tumor initiation, indicating a functional requirement for CD44s in maintaining CSCs. Importantly, our results reveal a mechanism by which CD44s promotes CSC properties through activation of the PDGFRβ/Stat3 signaling pathways, connecting CD44s to a downstream signaling cascade necessary in maintaining a CSC phenotype. These results were further supported by the global GSEA of the TCGA data set, where the PDGF-stimulated signaling signature was significantly enriched in the signature of CD44s but not CD44v. As a cell surface protein, CD44 has long been proposed to act as a coreceptor for activating signaling cascades (Ponta et al. 2003), although the mechanisms underlying CD44-mediated signaling activation remain elusive. Our results revealed that CD44s interacts with PDGFRβ, suggesting that this interaction may facilitate the activation of PDGFRβ signaling. While further work will be needed to investigate the detailed mechanism of CD44s-mediated PDGFRβ/Stat3 activation, these findings are significant because of the critical role of PDGFRβ/Stat3 in CSCs (Tam et al. 2013). Several PDGFR and Stat3 inhibitors have been developed and are used primarily for the treatment of TNBC patients whose tumors have enriched CSCs and show poor prognosis. Previous studies have suggested that combination of the PDGFR inhibitor pazopanib with other kinase inhibitors produces improved efficacy in the treatment of TNBC (Gril et al. 2013; Van Swearingen et al. 2017). Similarly, targeting STAT3 by its inhibitors also resulted in a promising response against TNBC (Liu et al. 2017). Our work presented here illustrates how enrichment of CD44s expression by manipulating alternative splicing can lead to increased PDGFRβ/Stat3 activation and CSC features. Therefore, therapeutic targeting of CD44 splicing may open up a new approach for inhibiting the PDGFR/Stat3 pathway to target CSC-enriched TNBCs.
In addition to the PDGFRβ/Stat3 pathway, CD44s may also act on other signaling cascades to promote a CSC phenotype. CD44s attenuates endocytosis-mediated degradation of RTKs, including EGFR and cMet, resulting in prolonged pAkt activity that is critical for cancer cell survival and therapeutic resistance (Wang et al. 2017). Interestingly, Akt activation further feeds back to CD44s, resulting in a positive feedback loop that sustains Akt signaling (Liu and Cheng 2017). These results suggest a central role of CD44s in regulating a set of RTKs and downstream signaling that promote activities favoring CSCs. Since the most effective treatment of cancer is to eliminate the small population of CSCs in addition to the bulk tumor cells, we argue that tilting splicing patterns of CD44 to diminish the production of CD44s may be effective for eradicating CSCs.
In conclusion, we described the identification of CD44s as the specific CD44 isoform that is predominantly expressed in breast CSCs and found that CD44s promotes CSCs traits by activating the PDGFRβ/Stat3 signaling pathway. Our results provide evidence that splicing regulation shifts CSC and non-CSC states and thus cancer phenotypes. These findings stress the importance of understanding the mechanisms of RNA alternative splicing regulation in cancer. They also emphasize the need to investigate the function of different splice isoforms in the context of cancer phenotypes. The targeting of dysregulated splicing or cancer-specific isoforms presents a novel strategy for therapeutic intervention.
Materials and methods
Antibodies and reagents
For FACS analysis and isolation of CD44+/CD24− cells, the following antibodies were used: anti-CD44 conjugated to APC (BD Pharmingen, 559942) and anti-CD24 conjugated to PE (BD Pharmingen, 555428). For immunoblotting analysis, the following antibodies were used: CD44-IM7 (Santa Cruz Biotechnology, sc18849), CD44H (R&D, BBA10), p-PDGFRβ (Cell Signaling, 3124s), PDGFRβ (Cell Signaling, 3169s), p-Stat3 (Cell Signaling, 9145s), Stat3 (Cell Signaling, 4904s), p-ERK (Cell Signaling, 9101s), E-cadherin (Cell Signaling, 3195s), N-cadherin (BD Pharmingen, 610920), β-actin (Sigma, A2228), and GAPDH (Millipore, MAB374). The Aldefluor assay kit (Stem Cell Technologies) was used for the ALDH assay. PDGFR inhibitor IV (Millipore, 521233) and Stat3 inhibitor XVIII (Millipore, 573132) were used for inhibition analysis. shRNAs targeting CD44 and ESRP1 were in the pLKO.1 vector backbone, and plasmids expressing the human CD44s and CD44v3–10 cDNA and mouse CD44s and CD44v6–10 were in the pBrit-HA/Flag vector with an HA tag on its C terminus as described previously (Brown et al. 2011; Xu et al. 2014; Zhao et al. 2016).
Cell culture
All cell lines were maintained at 37°C in 5% CO2. All HMLE lines and Mes10A lines were cultured as described previously (Brown et al. 2011). Induction of EMT by treatment with TAM or TGFβ was performed as described (Brown et al. 2011). The primary and recurrent tumor cell lines derived from the HER2/neu mouse model were cultured as described (Moody et al. 2005). SUM159 cells were cultured in F12 medium supplied with 5% fetal bovine serum (FBS), 5 µg/mL insulin, 1 ng/mL hydrocortisone, and 10 mM HEPES.
Mammosphere assays
Cells were grown in low-attachment plates in serum-free MEGM supplemented with 2 ng/mL EGF, 2 ng/mL bFGF, 4 µg/mL heparin, 1% methylcellulose, and B27 supplement diluted 1:50. Either 1000 or 500 cells were plated in each well of a 96-well plate. Spheres were allowed to form for 10–14 d and then quantified.
Fresh/frozen PDX tumors were chopped with scalpel blades under sterile conditions and digested with 1 g of 2 mg/mL collagenase (Sigma, C0130) and 100 mg of 1 mg/mL hyaluronidase (Sigma, H3506) for 4–6 h at 37°C with shaking. Tissue fragments were vortexed gently every 15–30 min. The single-cell suspension was then passed through a 40-µm cell strainer, centrifuged at 2000 rpm for 5 min, and washed three times with 1× PBS. Cells were then FACS-sorted using antimouse CD45 (BD Bioscience, 553081), TER119 (eBioscience, 12-5921-81), and CD31 (eBioscience, 12-0311-81) clones to remove mouse stromal cells. The resulting tumor cells were used for functional assays.
Bioinformatics analysis
The TCGA breast invasive carcinoma (BRCA) exon expression data set by RNA-seq (polyA+ IlluminaHiSeq) was downloaded from the University of California at Santa Cruz (UCSC) cancer browser. This data set can be accessed through a new platform: UCSC Xena browser (https://xenabrowser.net/datapages). Bam files of CCLE breast-invasive carcinoma RNA-seq data were downloaded from the Genomic Data Commons Legacy Archive. Detailed analysis is described in the Supplemental Material.
Quantitative RT–PCR (qRT–PCR)
RNA was isolated from cells using the E.Z.N.A. Total RNA kit I (Omega Bio-tek). Reverse transcription was performed using GoScript RT and reagents (Promega). qRT–PCR was performed using GoTaq master mix (Promega) and a Roche 480 LightCycler. The qRT–PCR primers used were as follows: CD44 total (forward: 5′-GATGGAGAAAGCTCTGAGCATC-3′; reverse: 5′-TTGCTG CACAGATGGAGTTG-3′), CD44s (forward: 5′-TACTGATGAT GACGTGAGCA-3′; reverse: 5′-GAATGTGTCTTGGTCTCTG GT-3′), CD44v5/6 (forward: 5′-GTAGACAGAAATGGCACC AC-3′; reverse: 5′-CAGCTGTCCCTGTTGTCGAA-3′), ESRP1 (forward: 5′- CAGAGGCACAAACATCACAT-3′; reverse: 5′- AG AAACTGGGCTACCTCATTGG-3′), and TBP (forward: 5′GG AGAGTTCTGGGATTGTAC-3′; reverse: 5′-CTTATCCTCATG ATTA CCGCAG-3′).
Immunoblotting and immunoprecipitation
Cells were harvested in RIPA lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors and lysed for 30 min. Whole-cell lysates were centrifuged, and protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad). Equal amounts of proteins were used for immunoblotting analysis. For immunoprecipitation assay, cells were lysed in lysis buffer (20 mM Tris-HCl, 100 mM NaCl, 5 mM EDTA, 0.2% NP40, 16% glycerol, 20 mM NaF, 1 mM Na3VO4, 20 mM β-glycerophosphate) supplemented with protease inhibitor cocktails. Cell lysates were precleared with sepharose beads and incubated with the primary antibody and agitated overnight. Protein A or Protein G beads (depending on the species of the immunoprecipitation antibody) were added the next day and agitated for 4 h. Beads were washed three times with the lysis buffer and boiled for 10 min in 2× SDS sample buffer (100 mM Tris-HCl, 4% SDS, 20% glycerol, 2% β-mercaptoethanol, 0.005% Bromophenol Blue). Eluates were subjected to immunoblotting procedure.
Mice
All animal experiments were performed with approval from the Institutional Animal Care and Use Committee at Baylor College of Medicine and Northwestern University. Fifty, 100, or 500 recurrent tumor cells were injected bilaterally into the fourth mammary fat pads of 8-wk=old FVB mice.
Statistics
Two-tailed and unpaired Student's t-tests were done in Excel. The statistical significance of the differences between the means was determined appropriately with one-way ANOVA followed by Tukey's post hoc. Correlation statistical analyses were performed using the Graphpad Prism program. For all figures, P-values of <0.05 were considered significant. One asterisk denotes a P-value of <0.05, two asterisks denote a P-value of <0.01, and three asterisks denote a P-value of <0.001.
Supplementary Material
Acknowledgments
We thank Rong Zheng for help with bioinformatics analysis. This research was supported in part by grants from the US National Institutes of Health (T32CA080621) to S.L., the Brewster Foundation and the Susan G. Komen Foundation to Y.K., R01CA203439 to A.M.M., and R01 CA182467 and R01GM11 0146 to C.C. C.C. is a Cancer Prevention and Research Institute of Texas Scholar in Cancer Research (RR160009).
Author contributions: H.Z, R.L.B., and C.C. designed the study. H.Z. and R.L.B., along with the help of P.Z., performed most of the experiments. Y.W. initiated the bioinformatics analysis. H.Z., Y.W., X.L., X.D.G., and Y.K. provided bioinformatics analysis and interpretation. H.L.G. and A.M.M. provided findings from experiments using PDXs and breast cancer patient specimens. S.L., Y.D., X.H., and J.Z. provided experimental help. C.C. conceived the project. H.Z., R.L.B., A.M.M., and C.C. wrote the manuscript with inputs from coauthors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.319889.118.
References
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. 2014. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158: 1110–1122. 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. 2003. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci 100: 3983–3988. 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvinen P, Tammi R, Kosma VM, Sironen R, Soini Y, Mannermaa A, Tumelius R, Uljas E, Tammi M. 2013. Increased hyaluronan content and stromal cell CD44 associate with HER2 positivity and poor prognosis in human breast cancer. Int J Cancer 132: 531–539. 10.1002/ijc.27707 [DOI] [PubMed] [Google Scholar]
- Bhola NE, Balko JM, Dugger TC, Kuba MG, Sánchez V, Sanders M, Stanford J, Cook RS, Arteaga CL. 2013. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest 123: 1348–1358. 10.1172/JCI65416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks MD, Burness ML, Wicha MS. 2015. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell 17: 260–271. 10.1016/j.stem.2015.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. 2011. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial–mesenchymal transition and breast cancer progression. J Clin Invest 121: 1064–1074. 10.1172/JCI44540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Hu X, Zhang J, Liang P, Zhang Y. 2014. CD44+ CD324− expression and prognosis in gastric cancer patients. J Surg Oncol 110: 727–733. 10.1002/jso.23690 [DOI] [PubMed] [Google Scholar]
- Carpenter RL, Lo HW. 2014. STAT3 target genes relevant to human cancers. Cancers 6: 897–925. 10.3390/cancers6020897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Goel HL, Gao H, Pursell B, Shultz LD, Greiner DL, Ingerpuu S, Patarroyo M, Cao S, Lim E, et al. 2015. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the α6Bβ1 integrin to sustain breast cancer stem cells. Genes Dev 29: 1–6. 10.1101/gad.253682.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Yaffe MB, Sharp PA. 2006. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev 20: 1715–1720. 10.1101/gad.1430906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KJ, Ewald AJ. 2016. A collective route to metastasis: seeding by tumor cell clusters. Science 352: 167–169. 10.1126/science.aaf6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Cho RW, Clarke MF. 2007a. Cancer stem cells: models and concepts. Annu Rev Med 58: 267–284. 10.1146/annurev.med.58.062105.204854 [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. 2007b. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci 104: 10158–10163. 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G II, Samuel S, Kim MP, Lim SJ, Ellis LM. 2009. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res 69: 1951–1957. 10.1158/0008-5472.CAN-08-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Niu G, Cai L, Wei R, Zhao X. 2013. The prognostic significance of CD44V6, CDH11, and β-catenin expression in patients with osteosarcoma. Biomed Res Int 2013: 496193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore C, Kuperwasser C. 2007. Human breast cancer stem cell markers CD44 and CD24: enriching for cells with functional properties in mice or in man? Breast Cancer Res 9: 303 10.1186/bcr1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Foster R, Yang X, Feng Y, Shen JK, Mankin HJ, Hornicek FJ, Amiji MM, Duan Z. 2015. Up-regulation of CD44 in the development of metastasis, recurrence and drug resistance of ovarian cancer. Oncotarget 6: 9313–9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel HL, Gritsko T, Pursell B, Chang C, Shultz LD, Greiner DL, Norum JH, Toftgard R, Shaw LM, Mercurio AM. 2014. Regulated splicing of the α6 integrin cytoplasmic domain determines the fate of breast cancer stem cells. Cell Rep 7: 747–761. 10.1016/j.celrep.2014.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gril B, Palmieri D, Qian Y, Anwar T, Liewehr DJ, Steinberg SM, Andreu Z, Masana D, Fernandez P, Steeg PS, et al. 2013. Pazopanib inhibits the activation of PDGFRβ-expressing astrocytes in the brain metastatic microenvironment of breast cancer cells. Am J Pathol 182: 2368–2379. 10.1016/j.ajpath.2013.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. 2009. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138: 645–659. 10.1016/j.cell.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga T, Ito S, Nakamura H. 2013. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res 73: 4112–4122. 10.1158/0008-5472.CAN-12-3801 [DOI] [PubMed] [Google Scholar]
- Hu J, Li G, Zhang P, Zhuang X, Hu G. 2017. A CD44v+ subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death Dis 8: e2679 10.1038/cddis.2017.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, et al. 2011. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc− and thereby promotes tumor growth. Cancer Cell 19: 387–400. 10.1016/j.ccr.2011.01.038 [DOI] [PubMed] [Google Scholar]
- Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. 2006. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med 12: 1167–1174. 10.1038/nm1483 [DOI] [PubMed] [Google Scholar]
- Jung WY, Kang Y, Lee H, Mok YJ, Kim HK, Kim A, Kim BH. 2013. Expression of moesin and CD44 is associated with poor prognosis in gastric adenocarcinoma. Histopathology 63: 474–481. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lee YS, Choe J, Lee H, Kim YM, Jeoung D. 2008. CD44-epidermal growth factor receptor interaction mediates hyaluronic acid-promoted cell motility by activating protein kinase C signaling involving Akt, Rac1, Phox, reactive oxygen species, focal adhesion kinase, and MMP-2. J Biol Chem 283: 22513–22528. 10.1074/jbc.M708319200 [DOI] [PubMed] [Google Scholar]
- Ko YH, Won HS, Jeon EK, Hong SH, Roh SY, Hong YS, Byun JH, Jung CK, Kang JH. 2011. Prognostic significance of CD44s expression in resected non-small cell lung cancer. BMC Cancer 11: 340 10.1186/1471-2407-11-340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WM, Teng E, Chong HS, Lopez KA, Tay AY, Salto-Tellez M, Shabbir A, So JB, Chan SL. 2014. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res 74: 2630–2641. 10.1158/0008-5472.CAN-13-2309 [DOI] [PubMed] [Google Scholar]
- Lee SM, Lee KE, Chang HJ, Choi MY, Cho MS, Min SK, Lee HK, Mun YC, Nam EM, Seong CM, et al. 2008. Prognostic significance of CD44s expression in biliary tract cancers. Ann Surg Oncol 15: 1155–1160. 10.1245/s10434-007-9786-9 [DOI] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. 2007. Identification of pancreatic cancer stem cells. Cancer Res 67: 1030–1037. 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- Liu S, Cheng C. 2013. Alternative RNA splicing and cancer. Wiley Interdiscip Rev RNA 4: 547–566. 10.1002/wrna.1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cheng C. 2017. Akt signaling is sustained by a CD44 splice isoform-mediated positive feedback loop. Cancer Res 77: 3791–3801. 10.1158/0008-5472.CAN-16-2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. 2007. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med 356: 217–226. 10.1056/NEJMoa063994 [DOI] [PubMed] [Google Scholar]
- Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al. 2014. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep 2: 78–91. 10.1016/j.stemcr.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Huang TT, Chu PY, Huang CT, Lee CH, Wang WL, Lau KY, Tsai WC, Chao TI, Su JC, et al. 2017. The tyrosine kinase inhibitor nintedanib activates SHP-1 and induces apoptosis in triple-negative breast cancer cells. Exp Mol Med 49: e366 10.1038/emm.2017.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JI, Camenisch TD, Stevens MV, Sands BJ, McDonald J, Schroeder JA. 2005. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res 65: 6755–6763. 10.1158/0008-5472.CAN-05-0863 [DOI] [PubMed] [Google Scholar]
- Ma F, Li H, Wang H, Shi X, Fan Y, Ding X, Lin C, Zhan Q, Qian H, Xu B. 2014. Enriched CD44+/CD24− population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC). Cancer Lett 353: 153–159. 10.1016/j.canlet.2014.06.022 [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, et al. 2011. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J Clin Invest 121: 2723–2735. 10.1172/JCI44745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N, Herrlich P, König H. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695. 10.1038/nature01153 [DOI] [PubMed] [Google Scholar]
- Mima K, Okabe H, Ishimoto T, Hayashi H, Nakagawa S, Kuroki H, Watanabe M, Beppu T, Tamada M, Nagano O, et al. 2012. CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res 72: 3414–3423. 10.1158/0008-5472.CAN-12-0299 [DOI] [PubMed] [Google Scholar]
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. 2002. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2: 451–461. 10.1016/S1535-6108(02)00212-X [DOI] [PubMed] [Google Scholar]
- Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. 2005. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8: 197–209. 10.1016/j.ccr.2005.07.009 [DOI] [PubMed] [Google Scholar]
- Ni J, Cozzi PJ, Hao JL, Beretov J, Chang L, Duan W, Shigdar S, Delprado WJ, Graham PH, Bucci J, et al. 2014. CD44 variant 6 is associated with prostate cancer metastasis and chemo-/radioresistance. Prostate 74: 602–617. 10.1002/pros.22775 [DOI] [PubMed] [Google Scholar]
- Olsson E, Honeth G, Bendahl PO, Saal LH, Gruvberger-Saal S, Ringner M, Vallon-Christersson J, Jonsson G, Holm K, Lovgren K, et al. 2011. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer 11: 418 10.1186/1471-2407-11-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian-Rousseau V. 2015. CD44 acts as a signaling platform controlling tumor progression and metastasis. Front Immunol 6: 154 10.3389/fimmu.2015.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouhtit A, Abd Elmageed ZY, Abdraboh ME, Lioe TF, Raj MH. 2007. In vivo evidence for the role of CD44s in promoting breast cancer metastasis to the liver. Am J Pathol 171: 2033–2039. 10.2353/ajpath.2007.070535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. 2010. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 140: 62–73. 10.1016/j.cell.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Pietras A, Katz AM, Ekström EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EC. 2014. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell 14: 357–369. 10.1016/j.stem.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9: 265–273. 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Ponta H, Wainwright D, Herrlich P. 1998. The CD44 protein family. Int J Biochem Cell Biol 30: 299–305. 10.1016/S1357-2725(97)00152-0 [DOI] [PubMed] [Google Scholar]
- Ponta H, Sherman L, Herrlich PA. 2003. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 4: 33–45. 10.1038/nrm1004 [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480: 387–390. 10.1038/nature10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF, Ailles LE. 2007. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci 104: 973–978. 10.1073/pnas.0610117104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke LM, Xu Y, Cheng C. 2012. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial–mesenchymal transition. J Biol Chem 287: 36435–36442. 10.1074/jbc.M112.397125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Settleman J. 2010. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29: 4741–4751. 10.1038/onc.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebrow A, Kornblihtt AR. 2006. The connection between splicing and cancer. J Cell Sci 119: 2635–2641. 10.1242/jcs.03053 [DOI] [PubMed] [Google Scholar]
- Su YJ, Lai HM, Chang YW, Chen GY, Lee JL. 2011. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J 30: 3186–3199. 10.1038/emboj.2011.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W, et al. 2013. Protein kinase Cα is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 24: 347–364. 10.1016/j.ccr.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H, Miyake H, Harada K, Fujisawa M. 2014. Expression profile of CD44s, CD44v6, and CD44v10 in localized prostate cancer: effect on prognostic outcomes following radical prostatectomy. Urol Oncol 32: 694–700. 10.1016/j.urolonc.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G, et al. 2014. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 14: 342–356. 10.1016/j.stem.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Van Swearingen AED, Sambade MJ, Siegel MB, Sud S, McNeill RS, Bevill SM, Chen X, Bash RE, Mounsey L, Golitz BT, et al. 2017. Combined kinase inhibitors of MEK1/2 and either PI3K or PDGFR are efficacious in intracranial triple-negative breast cancer. Neuro Oncol 19: 1481–1493. 10.1093/neuonc/nox052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP. 2006. Unbalanced alternative splicing and its significance in cancer. Bioessays 28: 378–386. 10.1002/bies.20390 [DOI] [PubMed] [Google Scholar]
- Wang L, Yang H, Abel EV, Ney GM, Palmbos PL, Bednar F, Zhang Y, Leflein J, Waghray M, Owens S, et al. 2015. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev 29: 171–183. 10.1101/gad.253591.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang H, Liu S, Kim CK, Xu Y, Hurley LA, Nishikawa R, Nagane M, Hu B, Stegh AH, et al. 2017. Internalized CD44s splice isoform attenuates EGFR degradation by targeting Rab7A. Proc Natl Acad Sci 114: 8366–8371. 10.1073/pnas.1701289114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. 2009. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33: 591–601. 10.1016/j.molcel.2009.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Stamenkovic I, Yu Q. 2010. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res 70: 2455–2464. 10.1158/0008-5472.CAN-09-2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, et al. 2014. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev 28: 1191–1203. 10.1101/gad.241968.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, Wada T, Masuko T, Mogushi K, Tanaka H, et al. 2012. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun 3: 883 10.1038/ncomms1892 [DOI] [PubMed] [Google Scholar]
- Yan Y, Zuo X, Wei D. 2015. Concise review: emerging role of CD44 in cancer stem cells: a promising biomarker and therapeutic target. Stem Cells Transl Med 4: 1033–1043. 10.5966/sctm.2015-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Park JW, Bebee TW, Warzecha CC, Guo Y, Shang X, Xing Y, Carstens RP. 2016. Determination of a comprehensive alternative splicing regulatory network and combinatorial regulation by key factors during the epithelial-to-mesenchymal transition. Mol Cell Biol 36: 1704–1719. 10.1128/MCB.00019-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Tsuchihashi K, Ishimoto T, Yae T, Motohara T, Sugihara E, Onishi N, Masuko T, Yoshizawa K, Kawashiri S, et al. 2013. xCT inhibition depletes CD44v-expressing tumor cells that are resistant to EGFR-targeted therapy in head and neck squamous cell carcinoma. Cancer Res 73: 1855–1866. 10.1158/0008-5472.CAN-12-3609-T [DOI] [PubMed] [Google Scholar]
- Zeilstra J, Joosten SP, van Andel H, Tolg C, Berns A, Snoek M, van de Wetering M, Spaargaren M, Clevers H, Pals ST. 2014. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene 33: 665–670. 10.1038/onc.2012.611 [DOI] [PubMed] [Google Scholar]
- Zhao P, Damerow MS, Stern P, Liu AH, Sweet-Cordero A, Siziopikou K, Neilson JR, Sharp PA, Cheng C. 2013. CD44 promotes Kras-dependent lung adenocarcinoma. Oncogene 32: 5186–5190. 10.1038/onc.2012.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Xu Y, Wei Y, Qiu Q, Chew TL, Kang Y, Cheng C. 2016. The CD44s splice isoform is a central mediator for invadopodia activity. J Cell Sci 129: 1355–1365. 10.1242/jcs.171959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jiang Y, Yan T, Di G, Shen Z, Shao Z, Lu J. 2010. The prognostic role of cancer stem cells in breast cancer: a meta-analysis of published literatures. Breast Cancer Res Treat 122: 795–801. 10.1007/s10549-010-0999-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.