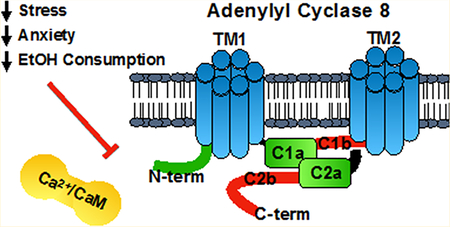
Abstract
Adenylyl cyclases (AC) catalyze the formation of cyclic AMP (cAMP) from ATP and are involved in a number of disease states, making them attractive potential drug targets. AC8, in particular, has been implicated in several neurological disorders. While development of small molecule AC inhibitors has generated some chemical leads, the lack of inhibitor specificity among AC family members has limited the identification of successful drug candidates. Therefore, finding alternative novel methods to suppress AC activity are needed. Because only AC1 and AC8 are robustly stimulated by calmodulin (CaM), we set out to explore the mechanism of disrupting the AC/CaM interaction as a way to selectively inhibit AC8. Through the development and implementation of a novel biochemical high-throughput-screening paradigm, we identified six small molecules from an FDA-approved compound library that are capable of disrupting the AC8/CaM interaction. These compounds were also shown to be able disrupt formation of this complex in cells, ultimately leading to decreased AC8 activity. Interestingly, further mechanistic analysis determined that these compounds functioned by binding to CaM and blocking its interaction with AC8. While these particular compounds could inhibit CaM interaction with both AC1 and AC8, they provide significant proof of concept for inhibition of ACs through disruption of CaM binding. These compounds, as dual AC1/AC8 inhibitors, provide important tools for probing pathological conditions where AC1/AC8 activity are enhanced, such as chronic pain and ethanol consumption. Furthermore, unlike tools such as genetic deletion, these compounds can be used in a dose-dependent fashion to determine the role of AC/CaM interactions in these pathologies.
Keywords: Adenylyl cyclase, calmodulin, high-throughput screening, protein–protein interaction, drug discovery, assay development
Graphical Abstract
■ INTRODUCTION
The adenylyl cyclase (AC) family of proteins catalyzes the formation of the secondary messenger cAMP from ATP, serving as effector enzymes for numerous G protein-coupled receptor-dependent signaling cascades. As they represent a central signaling hub, transforming extracellular stimuli into intracellular cAMP signals, ACs have become attractive targets for drug development. Membrane-bound ACs are composed of an intracellular N-terminus, two transmembrane domains connected by a cytosolic domain (C1), and another cytosolic domain (C2) at the C-terminus. Together, the C1 and C2 domains can form a soluble, catalytically active AC.1,2 Each cytosolic domain is further divided into catalytic (Ca) and regulatory (Cb) regions.
AC activity is dynamically regulated by various signaling proteins, such as Gβγ, Gαi, Gαs, protein kinases A and C, and Ca2+/calmodulin (CaM) (recently reviewed by Dessauer et al.3). Notably, CaM only stimulates the activity of group 1 ACs (AC1, 3, and 8), and robust CaM-stimulated activity is only observed for AC1 and 8.4–6 This stimulation occurs through direct interactions between CaM and intracellular AC regions, such that inhibition of these interactions can serve as a mechanism to attenuate AC-mediated cAMP signaling.4,5 Interestingly, AC1 and AC8 interact with CaM differently. As described above, the intracellular regions of AC can be roughly divided into 5 regions: the N-terminus, C1a, C1b, C2a, and C2b. While the first cytosolic regulatory (C1b) and second catalytic (C2a) domains of AC1 bind CaM, AC8’s N-terminus (Nt) and second cytosolic regulatory domain (C2b) mediate the CaM interaction.7,8 Additionally the CaM binding regions on AC8 interact with CaM differently. AC8-Nt conforms to the classical 1-5-8-14 motif and requires the C-terminal domain of CaM to be Ca2+ saturated. In contrast, AC8-C2b contains an IQ-like motif and binding requires the N-terminus of CaM to be Ca2+ bound.7,9 Since these binding regions are distinct between AC1 and AC8, there is hope that small molecule inhibitors may be able to achieve selectivity between these AC isoforms.
Inhibitors of AC1 and AC8 could be clinically useful, as mice lacking AC1 and AC8 exhibit decreased chronic inflammatory pain10 and mice lacking AC8 show decreased ethanol consumption, stress, and stress-induced anxiety.11–14 Mice lacking both AC1 and AC8 simultaneously show deficits in both long-term memory and long-term potentiation, but these deficits are less pronounced in mice lacking either AC1 or AC8 alone.15 This highlights the importance of achieving inhibitor selectivity for AC8, rather than complete inhibition of CaM-stimulated AC activity. Additionally, selective inhibition of the Bacillus anthracis AC toxin edema factor, which is also a CaM-stimulated cyclase, could be clinically useful for treatment of symptoms associated with anthrax.16
To date, efforts to identify AC inhibitors have resulted in molecules that fit into several distinct classes. One class of molecules competes with the ATP substrate for binding to the catalytic site. As this site is conserved across the AC family, achieving true isoform selectivity has proved challenging. A second class of molecules, the P-site binding inhibitors, act as transition state mimics, largely through uncompetitive/non-competitive mechanisms, and also suffer from lack of isoform selectivity. A third class of inhibitors takes advantage of the forskolin-binding site, a bona fide small molecule-binding site present on all ACs. Forskolin binding to this site results in AC activation, and as this site is highly conserved, isoform selectivity has been a major concern. For recent reviews of previously identified AC inhibitors, see Dessauer et al. and Seifert et al.3,17 Alternatively, recent work has identified at least one compound that prima facie appears to be selective for AC1 over other isoforms, providing hope that future efforts to directly modulate the activity of specific AC isoforms could prove fruitful.18 However, due to general concerns about lack of specificity across the AC family, alternative mechanisms for achieving inhibition of AC activity demand further attention. One such mechanism is the modulation of protein–protein interactions involving ACs and specifically the interaction between CaM and AC1 or AC8.
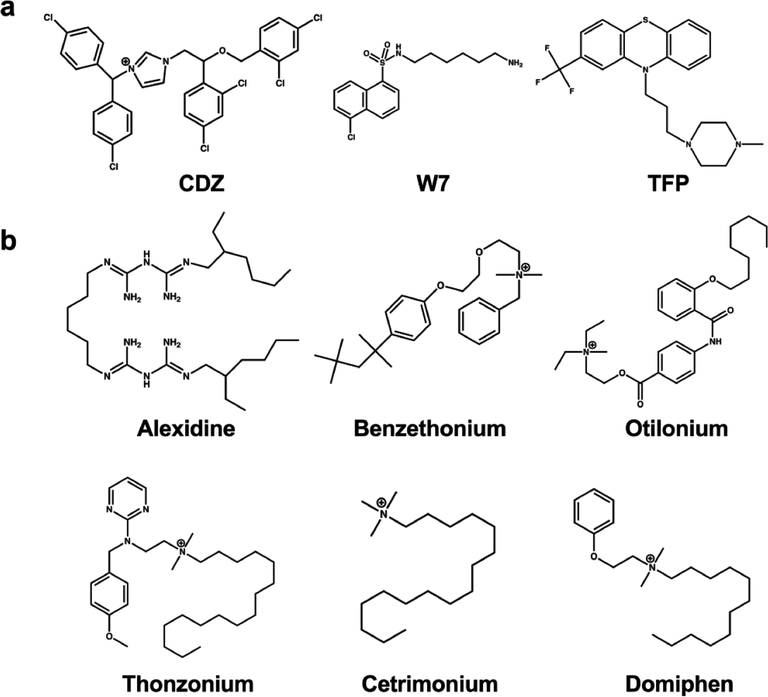
CaM is a highly evolutionarily conserved cytosolic signaling molecule that senses intracellular Ca2+ levels via its EF hand motifs. It is composed of two lobes, one at the N-terminus and one at the C-terminus, each of which contains two EF hands; these two lobes are connected by a flexible linker region. Upon Ca2+ binding, CaM undergoes conformational changes, allowing it to interact with various CaM-target proteins, including AC1 and AC8. During this conformational change, hydrophobic patches become exposed, and previous efforts have identified a number of compounds capable of binding to these regions. Structure–activity relationship studies of these molecules, which have been previously reviewed, have identified a general pharmacophore requirement of an amine located close to a hydrophobic region.19 Three previously described and well-studied CaM inhibitors are trifluoperazine (TFP), W7, and calmidazolium chloride (CDZ). TFP is a phenothiazine class antipsychotic that induces a conformational change in CaM, preventing its association with CaM-targets.20 W7, another CaM antagonist, was first identified for its ability to inhibit CaM activity and has been a useful tool compound for interrogating CaM-mediated signaling.21 CDZ was first described as an inhibitor of CaM-dependent Ca2+ transporters.22 Chemical structures of these compounds are shown in Figure 1a. Notably, all three of these CaM antagonists have been previously reported to inhibit CaM-mediated AC activity.16,23 CDZ, in particular, was the most effective of 39 tested CaM inhibitors at reducing CaM-stimulated AC1 activity.16
Figure 1.
CaM/AC8 inhibitor structures. (a) Chemical structures of previously identified CaM inhibitors: calmidazolium chloride (CDZ), N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide (W7), and trifluoperazine (TFP). (b) Chemical structures of FDA-approved CaM/AC inhibitors identified in this study.
Due to the potential clinical utility of selective AC8 inhibitors as therapies to reduce ethanol consumption, stress, and stress-induced anxiety, and the fact that few drug discovery efforts have focused on inhibition of CaM-stimulated AC8 activity, we used high throughput screening (HTS) to identify novel small molecule inhibitors of the AC8/CaM interaction. Completion of a pilot screen of more than 1000 FDA-approved small molecules identified six compounds capable of preventing CaM association with AC8. These compounds could disrupt the CaM/AC8 association both biochemically and in cells and were capable of decreasing AC8 activity in cells, though they lacked specificity between AC8 and AC1.
■ RESULTS AND DISCUSSION
Development of a Biochemical Assay To Detect AC8–CaM Interaction.
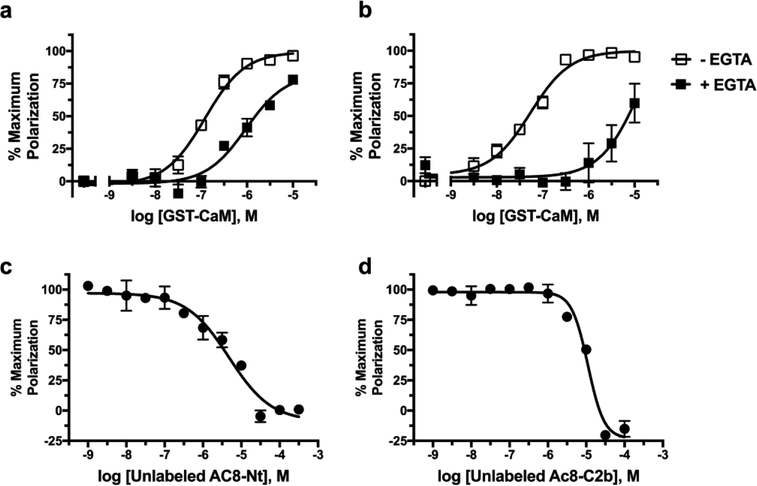
In order to identify novel AC8–CaM inhibitors, we developed two fluorescence polarization (FP) assays utilizing Cy5-labeled peptides derived from the Nt and C2b regions of AC8, as peptides derived from these areas have previously been shown to mediate Ca2+-dependent CaM binding.7,9 FP was chosen, as it is a relatively inexpensive robust methodology that is easily amenable to a HTS microplate format. FP requires a substantial size difference between the labeled “ligand” and unlabeled “receptor”, so incorporation of a GST-tagged CaM was used to further increase the size difference between CaM and the AC8 peptides. As expected, increasing concentrations of GST-CaM resulted in increased binding as measured by increased polarization for both AC8 peptides with pEC50 values of 6.92 ± 0.07 and 7.27 ± 0.07 for Nt and C2b, respectively (Figure 2a,b). Addition of the Ca2+-chelator EGTA resulted in rightward shifted concentration response curves (Figure 2a,b). Ca2+-loaded CaM in the absence of EGTA had approximately 8- and 250-fold higher affinity for AC8-Nt and AC8-C2b, respectively, than in the presence of EGTA (Figure 2a,b). This data indicates that the association of the AC8 peptides is greatly enhanced when CaM is saturated with Ca2+. Additionally, unlabeled AC8-Nt and C2b peptides are able to compete with labeled peptides, resulting in pIC50 values of 5.3 ± 0.1 and 4.95 ± 0.04, respectively (Figure 2c,d). Taken together these data help to validate this newly developed FP assay, as both AC8–CaM interactions are responsive to the amount of free Ca2+ available and can be inhibited by unlabeled AC8-derived peptides.
Figure 2.
Ca2+ dependent and competitive association of CaM with AC8 peptides. Ca2+-dependence of the GST-CaM/AC8 association examined in concentration response for 100 nM Cy5-labeled AC8-Nt (a) and AC8-C2b (b) in the presence and absence of 10 mM EGTA using FP. Inhibition of the FP signal generated using 316 nM GST-CaM and 100 nM Cy5-labeled AC8-Nt (c) or AC8-C2b (d) peptide in the presence of increasing concentrations of unlabeled AC8-Nt (c) or AC8-C2b (d). Data represent mean ± SEM from three independent experiments.
Screening of an FDA-Approved Compound Library.
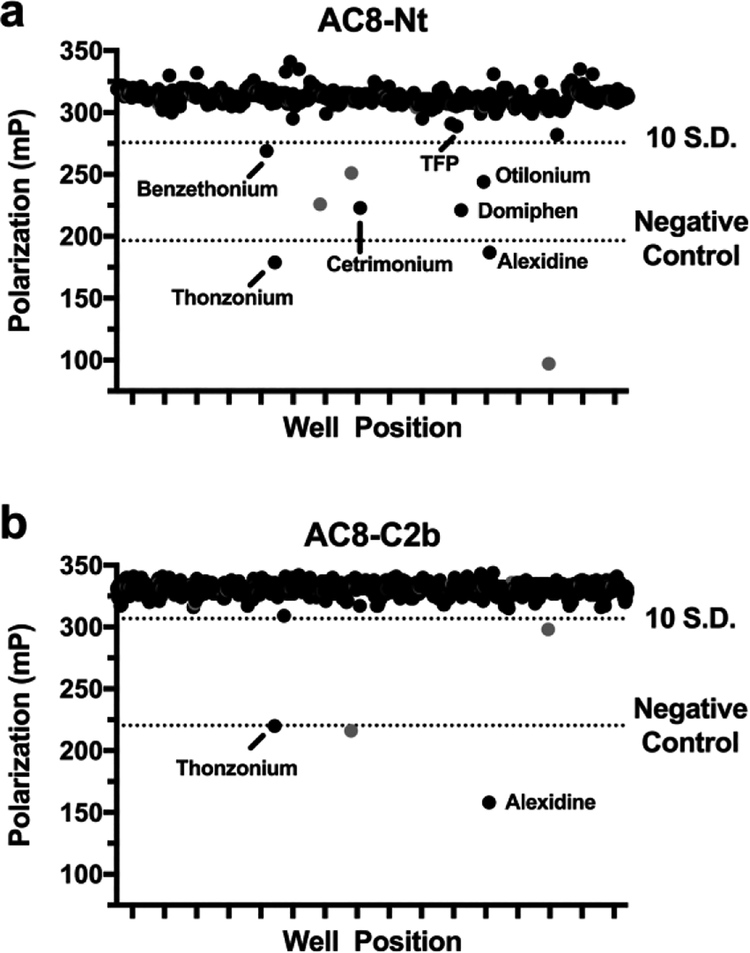
To determine assay stability and suitability for HTS, the Z′-factor was determined at various time points and concentrations of DMSO using the inclusion of 10 mM EGTA as a positive control, as earlier experiments had shown that the CaM/AC8 interaction is Ca2+ dependent (Figure 2a,b). Under every condition tested for both AC8 peptides, Z′-factor was greater than 0.5, indicating an excellent assay for HTS (Supporting Table 1, Supporting Figure 1a–d).24 Subsequently, a chemical library composed of approximately 1000 FDA-approved small molecules was screened against both CaM–AC8 peptide interactions. Screening data is presented in Figure 3 and summarized in Table 1. Throughout the screen, the Z′-factor remained well above the established threshold of 0.5, with Z′ values ranging from 0.74 to 0.83 between plates. The initial hit rates were below 1% at a cutoff of 10 standard deviations from the average negative control. This cutoff corresponds to 32% and 24% inhibition relative to EGTA control for the AC8-Nt and C2b interactions, respectively (Figure 3a,b). Though red-shifted-dyes like Cy5 are less likely to be affected by fluorescent interference than other commonly used shorter wavelength dyes,25 absorbance spectra for all initial hits were collected (Supporting Figure 2). Two compounds showed significant absorbance in the spectral regions used for the FP assay, leading to their designation as assay interfering compounds and exclusion from further work. Also excluded due to its unfavorable structure was a large cyclic peptide (caspofungin), that was a hit in both AC8 screens. Six hits were identified in this primary screen that inhibit the AC8-Nt/CaM FP assay, and two of these hits also inhibit the AC8-C2b/CaM assay. The chemical structures along with the common name of each of these compounds are presented in Figure 1b. Though the CaM inhibitor TFP was a member of the chemical library screened, it did not result in inhibition above the 10 SD threshold in either assay (Figure 3a); however, TFP, along with the CaM inhibitors W7 and CDZ, were tested in subsequent experiments, as previous reports indicated their ability to disrupt CaM-mediated AC activity.16,23
Figure 3.
CaM/AC8 FP HTS Screening Results. (a) Primary screening results of 1018 FDA-approved small molecules against CaM/AC8-Nt identified 9 compounds that inhibit the assay >10 SD (32% inhibition). Three compounds later determined to be optically interfering with assay or have unfavorable structures are shown in gray. CaM inhibitor TFP, a member of the screening library, is also highlighted. (b) Screening result for CaM/AC8-C2b identified four compounds that inhibit the assay >10 SD (24% inhibition), two of which also inhibit CaM/AC8-Nt and two of which interfere with assay. Numbers shown correspond to chemical structures in Figure 1. Screening results are summarized in Table 1.
Table 1.
Primary FP HTS Screening of CaM/AC8 Peptide Interactions
| AC8-Nt | AC8-C2b | |
|---|---|---|
| Z′- factor | 0.80–0.83 | 0.74–0.79 |
| compounds screened | 1018 | 1018 |
| initial hits (10 SD cutoff) | 9 (0.88%) | 4 (0.38%) |
| assay interfering compounds and unfavorable structures | 3 | 2 |
| hits | 6 (0.59%) | 2 (0.20%) |
Concentration Response of Primary Screen Hits.
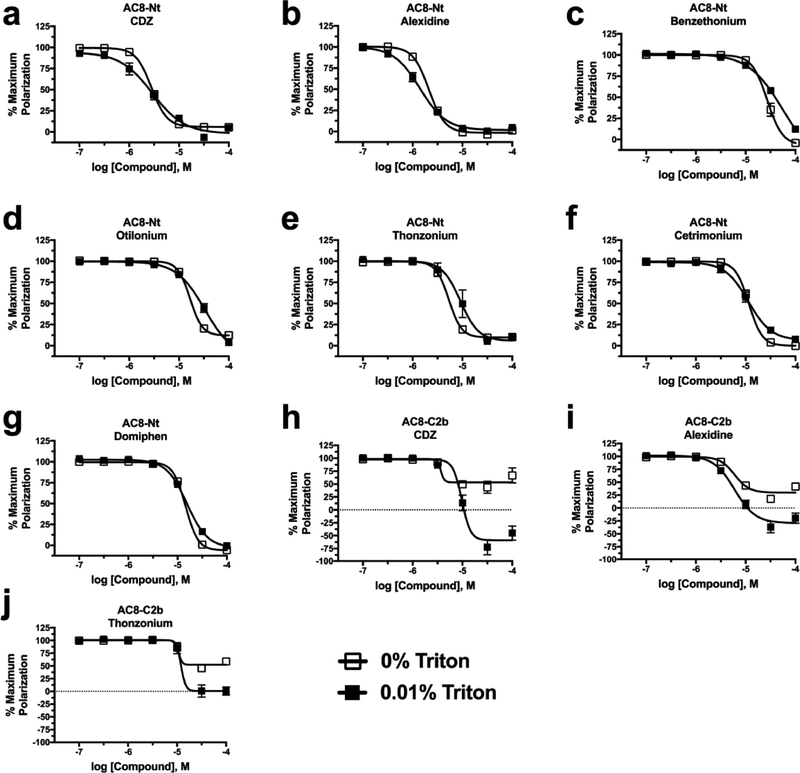
In order to confirm hits and rank-order them according to affinity, concentration–response curves were generated for all hits, along with CDZ, W7, and TFP. Interestingly, the CaM inhibitor W7 showed little to no inhibition in either assay (Supporting Figure 3a,b). TFP did not inhibit the CaM/AC8-C2b interaction and only showed appreciable inhibition against AC8-Nt at 100 μM, the highest concentration tested, resulting in a pIC50 of 4.4 ± 0.6 (Table 2, Supporting Figure 3a,b). Alternatively, CDZ exhibited complete inhibition relative to EGTA control of CaM/AC8-Nt with a pIC50 of 5.56 ± 0.01 (Figure 4a) and inhibited CaM/AC8-C2b approximately 50%, relative to EGTA control (Figure 4h). Concentration–response experiments with newly identified hit compounds resulted in pIC50 ranging from 5.67 ± 0.02 for alexidine to 4.57 ± 0.02 for benzethonium against the CaM/AC8-Nt interaction (Figure 4b,c, Table 2). The AC8-C2b interaction was inhibited approximately 50% relative to EGTA control by thonzonium (Figure 4j, Table 2) and by alexidine with a pIC50 of 5.22 ± 0.07 (Figure 4i, Table 2).
Table 2.
Concentration Response and Detergent Sensitivity of FP HTS Hits
| AC8-Nt pIC50 | AC8-C2b pIC50 | ||||
|---|---|---|---|---|---|
| cLogPa | 0% Triton | 0.01% Triton | 0% Triton | 0.01% Triton | |
| CDZ | 10.3 | 5.55 ± 0.01 | 5.54 ± 0.07 | 5.5b | 5.01 ± 0.05 |
| TFP | 4.9 | 4.4 ± 0.6 | c | ||
| alexidine | 4.3 | 5.67 ± 0.01 | 5.85 ± 0.03 | 5.23 ± 0.07 | 5.23 ± 0.06 |
| benzethonium | 6.1 | 4.57 ± 0.02 | 4.3 ± 0.2 | ||
| otilonium | 6.3 | 4.78 ± 0.01 | 4.48 ± 0.05 | ||
| thonzonium | 8.1 | 5.27 ± 0.02 | 5.04 ± 0.06 | 5.0b | 4.9b |
| cetrimonium | 6.2 | 4.94 ± 0.02 | 4.94 ± 0.03 | ||
| domiphen | 6.1 | 4.84 ± 0.02 | 4.81 ± 0.02 | ||
Retrieved from ZINC15.26
Data represent the mean of three independent experiments ± SEM.
SEM not calculated.
Not determined.
Figure 4.
Screening hits inhibit CaM/AC8 association through a detergent-insensitive mechanism. Concentration–response curve analysis of CaM/AC8-Nt (a–g) and CaM/AC8-C2b (h–j) inhibition by CaM inhibitor CDZ and indicated FDA hit in the presence and absence of 0.01% Triton X-100. (h–j) As in panel a, but using CaM/AC8-C2b assay. Data represent mean ± SEM from three independent experiments and summarized in Table 2.
FDA-Approved Hit Compounds’ Mechanism of Inhibition Is Not Detergent Sensitive.
Every compound tested resulted in concentration–response curves with steep slopes deviating greatly from −1. These values ranged from −2.7 ± 0.2 for alexidine to −3.5 ± 0.2 for otilonium (Table 2). This phenomenon is often associated with aggregation-based inhibitors.27 Additionally, every compound identified via screening has a log P larger than 3 (Table 2), another indication that aggregation could be a concern, so concentration–response curves were generated in the presence of 0.01% Triton X-100, as inclusion of a detergent has been shown to prevent aggregate formation for some compounds.28,29 All compounds identified as AC8-Nt inhibitors showed minimal sensitivity to inclusion of detergent in the buffer, indicating that the mechanism of inhibition was likely not due to aggregation (Figure 4a–g, Table 2). For the CaM/AC8-C2b interaction, the inclusion of detergent had almost no effect on the potency of alexidine as the pIC50 was nearly identical, but there was approximately 60% more inhibition in the presence of detergent (Figure 4i, Table 2). Increased inhibition of the CaM/AC8-C2b interaction in the presence of Triton X-100 was also observed for CDZ and thonzonium (Figure 4h,j). Though the exact cause of this apparent “super inhibition” remains unknown, one possibility is that the presence of detergent somehow alters the C2b peptide itself or stabilizes exposed hydrophobic patches on CaM. Interestingly, the identity of the peptide is the only variable between the AC8-Nt and C2b assays, and these results are not observed with AC8-Nt.
Development of Cell-Based CaM–AC8 Interaction Assay.
Determination of whether these newly identified CaM/AC8 inhibitors are active against full length AC8 in a cellular context required the development of a cell-based CaM/AC8 interaction assay. The recently described NanoBiT bimolecular luminescence complementation assay was selected due to the minimal protein tag requirements and the ability to monitor complex formation in real-time.30 This assay involves tagging one binding partner with an 18 kDa portion of NanoLuc and the other binding partner with a 1.3 kDa NanoLuc fragment.30 When the protein–protein interaction is formed between the proteins of interest, a competent NanoLuc is formed and luminescence is observed. As there are two binding partners (AC8 and CaM) that can be tagged on either the N-or C-terminus with one of two NanoLuc segments, eight (23) potential combinations exist. For each of these combinations, the area under the curve was used to quantify the NanoBiT response following treatment with the Ca2+ mobilizers A23187 (Ca2+ carrier) or thapsigargin (SERCA inhibitor) in the presence and absence of the cell permeable Ca2+ chelator BAPTA-AM, as our earlier work had shown that the CaM/AC8 interaction was Ca2+ dependent (Supporting Figure 4a). A representative NanoBiT kinetic trace is presented in Supporting Figure 4b. The most dynamic changes in luminescence in response to Ca2+ are observable when the AC8 N-terminus is labeled with the small NanoLuc portion. Alternately, the terminus of CaM (N- or C-) tagged with the large NanoLuc fragment is less critical (Supporting Figure 4a). This is interesting given that residues in the AC8 N-terminus directly interact with CaM. Furthermore, it is possible that inclusion of the large NanoLuc portion at this location physically occludes CaM binding. For all subsequent experiments, CaM N-terminally tagged with the large NanoLuc fragment and AC8 N-terminally tagged with the small NanoLuc fragment were used. Functional activity of the NanoBiT-tagged AC8 constructs was confirmed by measuring A23187- and forskolin-stimulated cAMP accumulation in transiently transfected HEK cells (Supporting Figure 5). Overall, this is a new microplate-based assay that allows the monitoring of Ca2+-stimulated CaM/AC8 binding in cells using small protein tags.
FDA-Approved Compounds Inhibit CaM–AC8 Complex Formation in Cells.
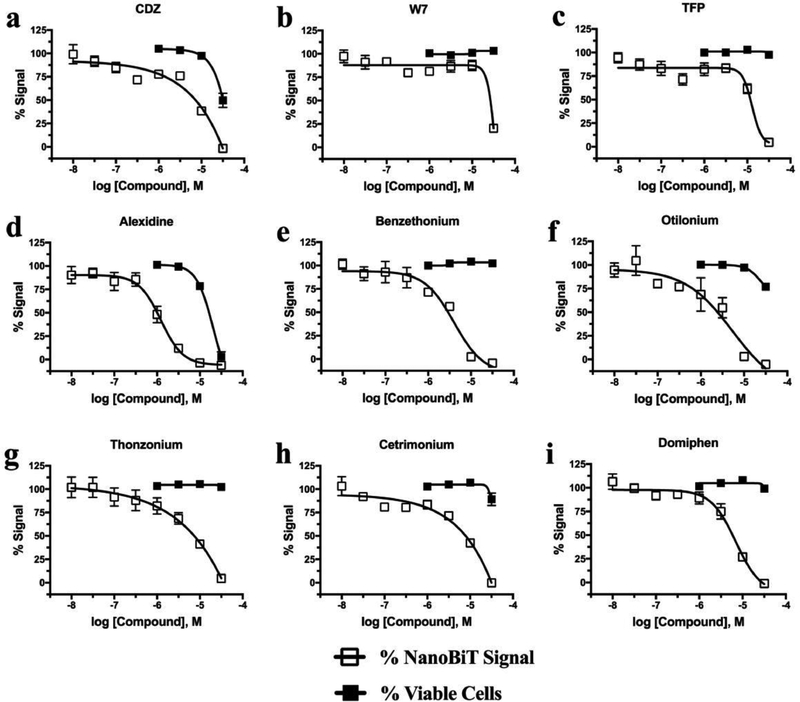
The screening hits from FP and CDZ, W7, and TFP were tested in the NanoBiT interaction paradigm in concentration–response experiments (Figure 5). Additionally, the CytoTox Fluor system was used to ensure that apparent NanoBiT inhibition was not due solely to decreased cell viability or membrane integrity, as several of the FDA hit compounds are used clinically as surfactants. CDZ remained the most potent previously identified CaM inhibitor tested, but it exhibited appreciable cell toxicity at 31.6 μM, the highest concentration tested (Figure 5a). Furthermore, there was only approximately half of an order of magnitude difference between the degree of NanoBiT inhibition and cell toxicity observed during CDZ treatment. Due to this limitation, higher inhibitor concentrations were not explored. Both W7 and TFP were more active in the cell-based complementation assay than in FP, without negatively affecting cell viability (Figure 5b,c), but were less potent than nearly all of the FDA leads. At 31.6 μM, W7 exhibited 80% ± 1% inhibition of the NanoBiT signal, whereas this same concentration showed little to no inhibition in either FP assay (Figure 5b, Supporting Figure 3a,b). Similarly, TFP at this concentration showed 95.4% ± 0.8% NanoBiT inhibition and very little FP inhibition (Figure 5c, Supporting Figure 3a,b).
Figure 5.
CaM inhibitors and FDA hits are active in cell-based NanoBiT complementation assay. (a–i) Concentration response curve of HEK-293T transfected with NanoBiT labeled CaM and AC8, pretreated for 30 min with compound indicated. Also shown is concentration–response curve of cell viability using CytoTox Fluor assay in HEK-293T cells pretreated for 30 min with compound. Viability normalized to 100% for vehicle treatment and 0% for 30 μg mL−1 digitonin control. Data represent mean ± SEM from at least three independent experiments.
The most potent FP hit, alexidine, was also found to be the most potent in the NanoBiT assay with a pIC50 of 5.91 ± 0.07, similar to that observed in the biochemical assay (Figure 5d, Table 2). Alexidine began to reduce cell viability at concentrations at or above 10 μM, though there was more than an order of magnitude difference between the NanoBiT and cell viability inhibition curves, which is larger than the difference observed for CDZ (Figure 5a,d). Benzethonium and otilonium were the next most potent hits in vitro, with pIC50s of 5.4 ± 0.1 and 5.3 ± 0.5, respectively (Figure 5e,f). Benzethonium showed no negative effect on cell viability, whereas otilonium caused a slight reduction in viability at the highest concentration tested, resulting in 77% ± 6% viability (Figure 5e,f). Thonzonium and cetrimonium inhibited the NanoBiT assay with potencies similar to that observed for CDZ (Figure 5a,g,h). Thonzonium did not adversely affect cell viability, and cetrimonium had minimal effects that only became evident at the highest concentration (89% ± 13% viability at 31.6 μM) (Figure 5g,h). Domiphen inhibited CaM/AC8 complementation with a pIC50 of 5.2 ± 0.1 with minimal cytotoxicity, as 99% ± 6% viability was observed at the highest concentrations used (Figure 5i). Taken together these data show that hits identified in the screen were in fact able to inhibit CaM/AC8 association in cells, validating both the FP assay as a discovery method and these FDA-approved small molecules as AC8/CaM inhibitors. Furthermore, though these compounds are known surfactants, only alexidine caused appreciable loss of cell viability and membrane integrity, and only at concentrations beyond that required for CaM/AC8 inhibition.
FDA-Approved Compounds Inhibit CaM-Stimulated AC8 and AC1 Activity in Vitro.
Disrupting the interaction between AC8 and calmodulin may allow selective inhibition of AC activity. Thus, we evaluated whether the compounds could inhibit CaM-stimulated AC8 activity in membrane preparations of HEK 293 cells stably expressing AC8. Since calmodulin also interacts with and stimulates AC1, we also examined the ability of these compounds to inhibit AC1 activity to assess the isoform selectivity of the compounds’ inhibitory action. All of the test compounds, including the CDZ control, inhibited both CaM-stimulated AC1 and CaM-stimulated AC8 activity. The average of the pIC50 values and % inhibition at 100 μM concentration are summarized in Table 3.
Table 3.
IC50 Values and Percentage (%) Inhibition of FP HTS Hits for AC1 and AC8 Activity in Membrane Preparationsa
| AC1 | AC8 | |||
|---|---|---|---|---|
| pIC50 | % inhibitionb |
pIC50 | % inhibitionb |
|
| CDZ | 4.87 ± 0.06 | 97 ± 3 | 5.33 ± 0.11 | 100 ± 0 |
| alexidine | 5.24 ± 0.1 | 162 ± 10 | 5.31 ± 0.06 | 106 ± 2 |
| benzethonium | 4.25 ± 0.03 | 85 ± 1 | 4.52 ± 0.07 | 100 ± 1 |
| otilonium | 4.58 ± 0.03 | 103 ± 4 | 4.62 ± 0.12 | 99 ± 1 |
| thonzonium | 4.20 ± 0.06 | 77 ± 6 | 4.41 ± 0.06 | 91 ± 1 |
| cetrimonium | 4.40 ± 0.07 | 86 ± 7 | 4.65 ± 0.18 | 102 ± 2 |
| domiphen | 4.55 ± 0.09 | 96 ± 4 | 4.76 ± 0.04 | 104 ± 1 |
Data represent the mean of at least three independent experiments ± SEM.
The % inhibition of AC1 or AC8 activity at the highest concentration tested (100 μM) relative to the control, 100 μM CDZ (100%).
CDZ was the most potent compound tested against AC8 with a pIC50 of 5.33 ± 0.11. Alexidine was the most potent newly identified inhibitor of AC8 activity with a pIC50 of 5.31, in accordance with the pIC50 values observed for the FP (Nt 5.67 ± 0.01, C2b 5.23 ± 0.07) and NanoBiT (5.91 ± 0.07) luminescence assays. Alexidine was also the most potent inhibitor against AC1 with a pIC50 of 5.24. Domiphen was the second most potent hit with a pIC50 of 4.76 ± 0.04, which is similar to results seen in FP (pIC50 4.84 ± 0.02) but less potent than results observed in NanoBiT (pIC50 5.2 ± 0.1). Benzethonium and otilonium exhibited approximately equal potencies against AC8 activity with pIC50s of 4.52 ± 0.07 and 4.65 ± 0.18, respectively. These results closely match those observed in the AC8 Nt FP assay where benzethonium and otilonium had pIC50s of 4.57 ± 0.02 and 4.78 ± 0.01, respectively, but both compounds were approximately half of an order of magnitude more potent in the NanoBiT cell-based complementation assay. Alexidine, otilonium, and domiphen had similar potencies on both AC1 and AC8, while benzethonium, thonzonium, and cetrimonium were slightly more potent against AC8 than AC1. Promisingly, every newly identified inhibitor except thonzonium resulted in complete inhibition of AC8 activity relative to CDZ control at the highest concentration tested (100 μM). Although selectivity was not achieved, these data indicate that disruption of the AC–CaM interaction can result in the inhibition of the cyclase activity.
Hit Compound’s Mechanism of Action Involves Direct Binding to Calmodulin.
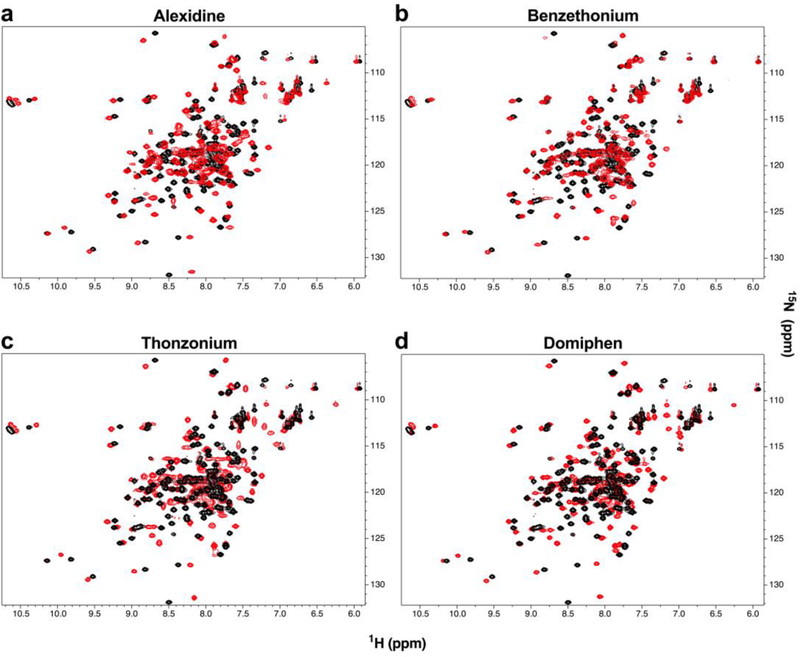
Given that each identified hit (1) contains a basic nitrogen atom (some which are positively charged) attached to large hydrophobic groups and (2) was incapable of discriminating between AC8 and AC1 in vitro, we hypothesized that these compounds mechanism of AC8/CaM inhibition involved binding directly to CaM. In order to explore this possibility, NMR was implemented to examine CaM in both the presence and absence of hit compounds. For all compounds tested a 5-fold molar excess of compound was sufficient to induce conformational changes in CaM, as evidenced by large chemical shift perturbations observed in 1H,15N HSQC spectra of 15N-labeled CaM (Figure 6a–d). Chemical shift perturbations in CaM were large and numerous enough that the accurate determination of the compound binding site was not possible within the scope of this study without numerous additional multidimensional NMR experiments.
Figure 6.
FDA hit compounds cause structural changes in CaM. (a–d) 1H,15N-HSQC of 100 μM 15N-labeled CaM in the absence (black) and presence (red) of 5-fold molar excess for the indicated compound collected at 298 K in 20 mM HEPES, pH 7.4, 100 mM KCl, 10 mM CaCl2, 10% D2O, 5% DMSO.
FDA Approved CaM Inhibitors Identified in This Study.
The most potent molecule identified in this study, alexidine, is a member of the bisbiguanide class of antiseptics that is bactericidal and has also been noted as a possible anticancer drug lead due to its ability to produce apoptosis in some in vitro and in vivo cancer models.31,32 This compound has been used in oral antiseptic solutions at concentrations ranging from 0.035% to 0.05% (600 to 860 μM), which is well above the concentrations at which inhibition was noted in this study.33,34 Alexidine is also used as a preservative in contact lens solutions at 4 ppm (approximately 7 μM), which is similar to the IC50 values observed in the assays developed here.35 Alexidine was more potent than even CDZ in both biochemical and cell-based AC8/CaM assays (Table 2, Figure 5a,d).
Benzethonium is an antiseptic and a preservative used in some ketamine formulations,36 which has recently begun to be found in other first aid products as a disinfectant and preservative. It is often found at concentrations ranging from 0.1% to 0.2% (2–4 mM), which is higher than concentrations needed here for AC/CaM inhibition.37 It has previously been reported to inhibit nicotinic acetylcholine receptors, as well as to exhibit anticancer activity, similar to alexidine.36,38
Otilonium is used to treat diarrhea due to its ability to reduce gut smooth muscle cell contractility by binding L-type Ca2+ channels, which it inhibits with a single-digit micromolar IC50 value, approximately 2.5-fold more potent than the observed AC/CaM inhibition.39,40 Additionally, otilonium has previously been identified as capable of binding CaM and inhibiting CaM-stimulated cyclic nucleotide phosphodiesterase activity.41
Thonzonium, cetrimonium, and domiphen are structurally related, as each contains a quaternary amine bound to two methyl groups and a long alkyl chain. Thonzonium is commonly used as an additive and surfactant in nasal and otic pharmaceutical formulations to increase tissue penetration of coadministered drugs. It is used at concentrations up to 0.05% (>800 μM), well above the concentrations required to inhibit AC/CaM association and activity. Additionally, it has been shown to prevent lipopolysaccharide-induced bone loss.42 Additionally, it can prevent proton transport by vacuolar ATPase pumps, as can alexidine.43 Cetrimonium is also used as surfactant in the dermal disinfectant cetrimide and may have some use as a potential anticancer therapy, as it can induce apoptosis in cancerous cells.44 Clinically relevant concentrations (approximately 5 mM in Cetavlon) are significantly higher than any condition required for AC inhibition here. Domiphen is used as an oral and topical antiseptic for infectious disease.45 Additionally, it can inhibit the HERG potassium channel with single-digit nanomolar IC50 values, which is much more potent than the AC inhibition results obtained here.46 Given the hydrophobic nature of these compounds, as indicated by high log P values (Table 2), it is unsurprising that most are used as either antiseptics or surfactants, and it is likely that their hydrophobic nature drives CaM binding.
Future efforts focused on the structure–activity relationships for these compounds have several avenues to explore. For example, analogs of alexidine, thonzonium, cetrimonium, and domiphen with varying alkyl chain lengths are worth investigation. Additionally, as alexidine is symmetrical, analogs consisting of only half of this compound deserve attention, as do other bisbiguanide compounds, like chlorhexidine. Altering the aromatic hydrophobic groups on hits benzethonium, otilonium, thonzonium, and domiphen to larger, bulkier groups or heterocyclic groups could yield interesting results. Finally, as these compounds were not selective for AC8 over AC1, the determination of the specificity of these compounds as AC/CaM inhibitors will be evaluated. CaM is able to alter the activity of numerous proteins, such as phosphodiesterases,47 phospholipases,48 NO synthases,49 and many more, and pan-CaM inhibition is not desirable.
■ CONCLUSIONS
In this study, our aim was to identify small molecules capable of specifically inhibiting AC8 by disrupting its association with CaM. Two novel assays to detect AC8/CaM binding were developed, and screening of 1018 FDA-approved small molecules identified six molecules capable of disrupting this interaction. Screening against both the Nt and C2b regions of AC8 identified several compounds that only inhibited the AC8-Nt/CaM interaction, likely due to different interactions between the peptides and CaM N-and C-terminal lobes. Subsequent work revealed that hit compounds were in fact capable of reducing CaM-stimulated AC8 activity, but they were found to inhibit AC1 with similar potencies, likely due to their mechanism of action involving direct binding to CaM. Future efforts will focus on developing a FP-based assay to detect CaM/AC1 peptide interactions, as this could provide a cheap, high throughput counterscreen method to quickly discriminate between AC8 selective and AC1/AC8 dual inhibitors. Additionally, this work validates the idea that disruption of the interaction between CaM and CaM-stimulated cyclases is a viable means to decrease cAMP formation. This mechanism could also be clinically valuable for chronic pain through inhibition of AC1 or anthrax infection through inhibition of the AC toxin edema factor.
This work provides a new platform to discover AC8/CaM inhibitors, and we speculate that screening larger, more complex chemical libraries could yield molecules capable of selectively inhibiting AC8 and not AC1. Further, as the compounds identified in this study were potent CaM inhibitors and CaM is largely evolutionarily conserved, it is possible that these molecules exert some of their clinical effects through CaM-dependent mechanisms, though further experimentation will be needed to test this hypothesis.
■ METHODS
Chemical Reagents.
Alexidine (≥98% purity), W7 (≥98% purity), CDZ (≥98% purity), TFP (≥98% purity), caspofungin (≥98% purity), and mitoxantrone (≥95% purity) were obtained from Cayman Chemical (Ann Arbor, MI). Domiphen bromide (≥97% purity) and crystal violet (≥90% purity) were obtained from Alfa Aesar (Harborhill, MA). Thonzonium bromide (≥98% purity) and otilonium bromide (≥98% purity) were obtained from MedChem Express (Monmouth Junction, NJ). Benzethonium chloride (≥97% purity) and cetrimonium bromide (≥98% purity) were obtained from TCI America (Portland, OR).
Cloning.
Human CaM protein coding sequence, residues 1–149 (Addgene plasmid no. 47598), was cloned into pET-His6-GST-TEV LIC (Addgene plasmid no. 29655) using ligation independent cloning to generate the bacterial expression vector, coding for N-terminally 6X-His-GST-tagged CaM with a TEV protease cleavage site located between the GST and CaM regions. Human CaM residues 1–149 and rat AC8 residues 1–1248 were cloned into NanoBit PPI TK/BiT MCS vectors, according to manufacturer directions using BglII/XhoI (rAC8) and NheI/XhoI (hCaM) restriction sites (Promega, Madison, WI). Sanger sequencing was used to confirm all DNA sequences (Iowa Institute of Human Genetics, Iowa City, IA).
Protein Purification.
BL21CodonPlus (DE3) - RIPL E. coli (Agilent, Santa Clara, CA) were transformed and grown while shaking at 37 °C and 300 rpm until OD600 of 2.0 was reached, at which point protein production was induced with 1.0 mM isopropyl β-d-1-thiogalactopyranoside (RPI, Mt Prospect, IL), and protein production proceeded for 16 h at 300 rpm, 18 °C. Culture was then pelleted, resuspended in 50 mL of 50 mM Tris, pH 8, 150 mM NaCl, 10 mM imidazole, supplemented with protease inhibitors, and flash frozen. Pellets were thawed, supplemented with 1 mg mL−1 chicken egg lysozyme (Sigma-Aldrich, St Louis, MO), agitated for 1 h, and subjected to two additional freeze–thaw cycles in liquid N2 to achieve cell lysis, and 100 μg of DNase (Roche, Indianapolis, IN) was added. Lysate was clarified by 100 000g centrifugation, and the resulting supernatant was purified largely as described previously.50 Briefly, lysate was loaded on NiS6FF resin using an AKTA system (GE Life Sciences, Chicago, IL) and eluted via a gradient over 20 column volumes of resuspension buffer supplemented with 400 mM imidazole. Peak fractions were then pooled, diluted ~1:6 with pH 7.5 50 mM Tris, 1 mM CaCl2 in order to reduce NaCl concentration, and subjected to hydrophobic interaction chromatography on Phenyl FF (hi sub) 16/10 column (GE Life Sciences, Chicago, IL) equilibrated with 50 mM Tris, pH 7.5, 1 mM CaCl2. GST-CaM was eluted via gradient elution over 10 column volumes using pH 7.5 50 mM Tris, 5 mM EGTA. For GST-CaM, pooled fractions (90+% pure, via SDS-PAGE and coomassie staining) were exhaustively dialyzed against 20 mM HEPES, 100 mM KCl, pH 7.4, at 4 °C to remove EGTA/Ca2+, flash frozen in liquid N2, and stored at −80 °C until needed. To produce untagged CaM, GST-CaM was incubated with 1:20 molar ratio of His-TEV to GST-CaM overnight at 4 °C during dialysis against 5 L of pH 7.4 20 mM HEPES, 100 mM KCl, 1 mM CaCl2, and subjected to further IMAC to capture cleaved His-GST and His-TEV. The resulting flow through, containing 95+% pure (SDS-PAGE, coomassie staining) cleaved CaM, was collected, concentrated to 10 mg mL−1 using a stirred concentration cell (Amicon, EMD Millipore, Billerica, MA), flash frozen in liquid N2, and stored at −80 °C until needed. 15N-labeled hCaM for NMR experiments was produced as above, with the exception that RIPL were grown on M9 minimal media supplemented with 1 g of 15NH4Cl per L of media.
Fluorescence Polarization (FP) Assay.
Peptides corresponding to human AC8 residues 30–54 and 1191–1214 (AC8-NT and AC8-C2b, respectively) containing an additional N-terminal Cys residue labeled with Cy5 were obtained (Genscript, Piscataway, NJ). FP data was collected in 384-well, black polystyrene, nonbinding plates (Corning 3575) and read on a BioTek Synergy 2 (Winooski, VT) equipped with excitation 620/40 nm and emission 680/30 nm filters and a 660 nm dichroic mirror with polarizers. Twenty microliters of assay buffer (20 mM HEPES, pH 7.4, 100 mM KCl, 50 μM CaCl2) either with or without 3× EGTA (10 mM final) or 3× final percent DMSO or indicated concentration of compound was added to each well. For Triton X-100 experiment, buffer included 3× final Triton X-100 concentration, and for competition experiments, unlabeled peptide (Genscript, Piscataway, NJ) was included at 3× concentration indicated. This was followed by addition of 20 μL of 3× concentrated GST-CaM (indicated concentration in Figure 2 or 316 nM final for all subsequent experiments). The plate was then allowed to incubate for 30 min at ambient temperature. Finally, 20 μL of Cy5-labeled peptide at 3× concentration (100 nM final) in assay buffer was added, and the plate was incubated for 2 h (or indicated time during assay stability experiments). The plate was then read, and polarization (in mP) was calculated as follows:
where P represents polarization and I represents fluorescence intensity in indicated polarity.
FP Screen of FDA-Approved Library.
For each well, 1 μL of 800 μM FDA-approved Drug Library (Selleck Chemical, Houston TX) in 100% DMSO was added to 19 μL of assay buffer in assay plate (Corning 3575) using a Hamilton MicroLab. This was followed by additions of GST-CaM and Cy5-labeled peptide, and FP was measured as described above. Optically interfering compounds were identified by performing absorbance wavelength scan every 5 nm from 350 to 700 nm on BioTek Synergy 2 (Winooski, VT).
AC8/CaM NanoBiT Assay.
HEK293T cells were cultured in DMEM (Gibco) supplemented with 10% FBS and 1% Pen/Strep at 37 °C and 5% CO2. HEK cells were seeded at 25 000 cells/well in 100 μL of growth medium in white, half area 96-well plates (Greiner Bio-one 675098) coated with poly(d-lysine) and allowed to grow for 16 h, at which time they were transfected with NanoBiT DNAs constructed above using Lipofectamine 3000 (Life Technologies, Carlsbad, CA), following manufacturer’s instructions and incubated 48 h further. On the day of the assay, growth medium was exchanged for 40 μL of HBSS supplemented with 20 mM HEPES, pH 7.4, and either 10 μL of 5× BAPTA-AM (10 μM final), 10 μL of 5× compound, or vehicle was added and incubated at 37 °C for 30 min. Next 12.5 μL of 5× NanoGlo Live Cell Substrate was added (prepared as per manufacturer instructions), and luminescence was monitored using BioTek Synergy 2 at 37 °C for 30 min to establish baseline luminescence. After baseline, 12.5 μL of 6× thapsigargin (1 μM final) (Acros Organics, Geel, Belgium), A23187 (1 μM final) (Sigma-Aldrich, St Louis, MO), or vehicle was added, and the plate was read for an additional 1.5 h. Baseline luminescence was normalized to zero, and area under curve (AUC) analysis was used to quantify thapsigargin- or A23187-induced AC8/CaM association observed over first 20–25 min.
CytoTox-Fluor Acute Cytotoxicity Assay.
HEK293T cells were cultured as above and plated at 25 000 cells/well in 100 μL of growth medium in black, half area 96-well plates (Greiner Bio-one 675090) coated with poly(d-lysine) and allowed to grow for 48 h. On the day of the assay, growth medium was exchanged for 25 μL of HBSS supplemented with 20 mM HEPES, pH 7.4, and either 25 μL of 2× compound or vehicle was added and incubated at 37 °C for 30 min. CytoTox-Fluor reagent (2×) was prepared according to manufacturer’s protocol (Promega, Madison, WI), and the plate was incubated in BioTek Synergy 2 at 37 °C for 30 min, after which fluorescence was measured using 485/20 and 528/20 excitation and emission filters, respectively. Toxicity was normalized to vehicle (0%) and 30 μg mL−1 digitonin (Sigma-Aldrich, St Louis, MO) (100%). For ease of data interpretation, viability was determined as the difference between 100% (0% toxicity, i.e., DMSO vehicle) and the observed % Toxicity (relative to 100% digitonin control).
AC Activity in Cellular Membranes.
Membranes from HEK293 cells stably expressing AC1 or AC8 were isolated as previously described in the presence of 1 mM EGTA.18 On the day of the assay, membranes were resuspended in membrane buffer containing 33 mM HEPES, 0.1% Tween 20, and 0.5 mM EGTA (pH 7.4). Protein concentration was measured using the Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Protein concentration was adjusted to 50 μg/mL and 0.5 μg/mL for AC1 and AC8 membranes, respectively, and 10 μL of membrane preparations was plated into a white, flat-bottom, 384-well plate (PerkinElmer, Shelton, CT). Test compounds (4×) diluted into membrane buffer without EGTA or vehicle were added to the wells, and preincubated with membranes at room temperature for 20 min. Membranes were then incubated for 45 min at room temperature with 12 μM CaM (Enzo Life Sciences) diluted in a 4× stimulation buffer containing 33 mM HEPES, 0.1% Tween 20, 10 mM MgCl2, 1 mM ATP, 4 μM GppNHp, and 2 mM IBMX in the presence of 260 μM CaCl2/10 μM free Ca2+ or 355 μM CaCl2/100 μM free Ca2+ for AC1 and AC8 membranes, respectively. Cyclic AMP accumulation was measured using Cisbio′s dynamic 2 kit (Cisbio Bioassays, Bedford, MA) according to the manufacturer’s instructions.
Cyclic AMP Accumulation in Intact Cells.
HEK293 cells were transiently transfected with the NanoBiT tagged AC8 plasmids for 48 h using Lipofectamine 2000 (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Transfected cells were then plated in 384 well plates and incubated for 1 h at 37 °C prior stimulation with 0.5 mM IBMX and 10 μM A23187 in the presence or absence of forskolin as indicated. After 1 h incubation at room temperature, Cisbio’s dynamic 2 kit reagents were added for detection of cAMP accumulation.
Nuclear Magnetic Resonance.
For all NMR samples, 100 μM 15N-CaM was incubated with or without indicated compound at 5× molar excess in the following buffer: 20 mM HEPES, pH 7.4, 100 mM KCl, 10 mM CaCl2, 10% D2O, 5% DMSO. All spectra were acquired at 298 K using a 600 MHz Varian INOVA NMR spectrometer equipped with a triple resonance gradient probe. NMR spectra were processed and analyzed using NMRPipe.51
Data Analysis.
GraphPad Prism was used to analyze all dose–response curves as well as cAMP accumulation standard curves.
Supplementary Material
■ ACKNOWLEDGMENTS
We thank members of the Roman lab past and present for helpful discussion, and Meng Wu of the University of Iowa HTS facility for assistance in chemical library formatting.
Funding
This work was supported by the UI College of Pharmacy (D.L.R., M.P.H.), Purdue University, and NIH MH101673 (M.S.V., V.J.W.).
■ ABBREVIATIONS
- AC
adenylyl cyclase
- CaM
calmodulin
- cAMP
cyclic adenosine monophosphate
- CDZ
calmidazolium chloride
- FP
fluorescence polarization
- GppNHp
5′-guanylyl imidodi-phosphate
- HTS
high-throughput screening
- IBMX
3-isobutyl-1-methylxanthine
- TFP
trifluoperazine
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneur-o.7b00349.
AC8 FP assay stability for HTS as determined by Z′ score, absorbance spectra of FDA hits to identify optically interfering compounds, concentration–response curves of W7 and TFP against AC8 FP assays, AC8/CaM NanoBiT assay development, confirmation of NanoBiT tagged AC cyclase activity, and summary of AC8 FP HTS Z′ score (PDF)
The authors declare no competing financial interest.
■ REFERENCES
- (1).Dessauer CW, Scully TT, and Gilman AG (1997) Interactions of forskolin and ATP with the cytosolic domains of mammalian adenylyl cyclase. J. Biol. Chem 272, 22272–22277. [DOI] [PubMed] [Google Scholar]
- (2).Sunahara RK, Dessauer CW, Whisnant RE, Kleuss C, and Gilman AG (1997) Interaction of Gsalpha with the cytosolic domains of mammalian adenylyl cyclase. J. Biol. Chem 272, 22265–22271. [DOI] [PubMed] [Google Scholar]
- (3).Dessauer CW, Watts VJ, Ostrom RS, Conti M, Dove S, and Seifert R (2017) International Union of Basic and Clinical Pharmacology. CI. Structures and Small Molecule Modulators of Mammalian Adenylyl Cyclases. Pharmacol. Rev 69, 93–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Tang WJ, Krupinski J, and Gilman AG (1991) Expression and characterization of calmodulin-activated (type I) adenylylcyclase. J. Biol. Chem 266, 8595–8603. [PubMed] [Google Scholar]
- (5).Cali JJ, Zwaagstra JC, Mons N, Cooper DM, and Krupinski J (1994) Type VIII adenylyl cyclase. A Ca2+/calmodulin-stimulated enzyme expressed in discrete regions of rat brain. J. Biol. Chem 269, 12190–12195. [PubMed] [Google Scholar]
- (6).Choi EJ, Xia Z, and Storm DR (1992) Stimulation of the type III olfactory adenylyl cyclase by calcium and calmodulin. Biochemistry 31, 6492–6498. [DOI] [PubMed] [Google Scholar]
- (7).Masada N, Schaks S, Jackson SE, Sinz A, and Cooper DM (2012) Distinct mechanisms of calmodulin binding and regulation of adenylyl cyclases 1 and 8. Biochemistry 51, 7917–7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Diel S, Beyermann M, Llorens JM, Wittig B, and Kleuss C (2008) Two interaction sites on mammalian adenylyl cyclase type I and II: modulation by calmodulin and G(betagamma). Biochem. J 411, 449–456. [DOI] [PubMed] [Google Scholar]
- (9).Herbst S, Masada N, Pfennig S, Ihling CH, Cooper DM, and Sinz A (2013) Structural insights into calmodulin/adenylyl cyclase 8 interaction. Anal. Bioanal. Chem 405, 9333–9342. [DOI] [PubMed] [Google Scholar]
- (10).Vadakkan KI, Wang H, Ko SW, Zastepa E, Petrovic MJ, Sluka KA, and Zhuo M (2006) Genetic reduction of chronic muscle pain in mice lacking calcium/calmodulin-stimulated adenylyl cyclases. Mol. Pain 2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Maas JW Jr., Vogt SK, Chan GC, Pineda VV, Storm DR, and Muglia LJ (2005) Calcium-stimulated adenylyl cyclases are critical modulators of neuronal ethanol sensitivity. J. Neurosci 25, 4118–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bernabucci M, and Zhuo M (2016) Calcium activated adenylyl cyclase AC8 but not AC1 is required for prolonged behavioral anxiety. Mol. Brain 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Razzoli M, Andreoli M, Maraia G, Di Francesco C, and Arban R (2010) Functional role of Calcium-stimulated adenylyl cyclase 8 in adaptations to psychological stressors in the mouse: implications for mood disorders. Neuroscience 170, 429–440. [DOI] [PubMed] [Google Scholar]
- (14).Schaefer ML, Wong ST, Wozniak DF, Muglia LM, Liauw JA, Zhuo M, Nardi A, Hartman RE, Vogt SK, Luedke CE, Storm DR, and Muglia LJ (2000) Altered stress-induced anxiety in adenylyl cyclase type VIII-deficient mice. Journal of neuroscience 20, 4809–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wong ST, Athos J, Figueroa XA, Pineda VV, Schaefer ML, Chavkin CC, Muglia LJ, and Storm DR (1999) Calcium-stimulated adenylyl cyclase activity is critical for hippocampus-dependent long-term memory and late phase LTP. Neuron 23, 787–798. [DOI] [PubMed] [Google Scholar]
- (16).Lubker C, and Seifert R (2015) Effects of 39 Compounds on Calmodulin-Regulated Adenylyl Cyclases AC1 and Bacillus anthracis Edema Factor. PLoS One 10, e0124017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Seifert R, Lushington GH, Mou TC, Gille A, and Sprang SR (2012) Inhibitors of membranous adenylyl cyclases. Trends Pharmacol. Sci 33, 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Brust TF, Alongkronrusmee D, Soto-Velasquez M, Baldwin TA, Ye Z, Dai M, Dessauer CW, van Rijn RM, and Watts VJ (2017) Identification of a selective small-molecule inhibitor of type 1 adenylyl cyclase activity with analgesic properties. Sci. Signaling 10, eaah5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Weiss B, Prozialeck WC, and Wallace TL (1982) Interaction of drugs with calmodulin. Biochemical, pharmacological and clinical implications. Biochem. Pharmacol 31, 2217–2226. [DOI] [PubMed] [Google Scholar]
- (20).Vandonselaar M, Hickie RA, Quail JW, and Delbaere LT (1994) Trifluoperazine-induced conformational change in Ca(2+)-calmodulin. Nat. Struct. Biol 1, 795–801. [DOI] [PubMed] [Google Scholar]
- (21).Caulfield MP, Robbins J, Sim JA, Brown DA, Mac Neil S, and Blackburn GM (1991) The naphthalenesulphonamide calmodulin antagonist W7 and its 5-iodo-1-C8 analogue inhibit potassium and calcium currents in NG108–15 neuroblastoma x glioma cells in a manner possibly unrelated to their antagonism of calmodulin. Neurosci. Lett 125, 57–61. [DOI] [PubMed] [Google Scholar]
- (22).Gietzen K, Wuthrich A, and Bader H (1981) R 24571: a new powerful inhibitor of red blood cell Ca++-transport ATPase and of calmodulin-regulated functions. Biochem. Biophys. Res. Commun 101, 418–425. [DOI] [PubMed] [Google Scholar]
- (23).Ahlijanian MK, and Cooper DM (1987) Antagonism of calmodulin-stimulated adenylate cyclase by trifluoperazine, calm-idazolium and W-7 in rat cerebellar membranes. J. Pharmacol Exp Ther 241, 407–414. [PubMed] [Google Scholar]
- (24).Zhang JH, Chung TD, and Oldenburg KR (1999) A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screening 4, 67–73. [DOI] [PubMed] [Google Scholar]
- (25).Turek-Etienne TC, Small EC, Soh SC, Xin TA, Gaitonde PV, Barrabee EB, Hart RF, and Bryant RW (2003) Evaluation of fluorescent compound interference in 4 fluorescence polarization assays: 2 kinases, 1 protease, and 1 phosphatase. J. Biomol. Screening 8, 176–184. [DOI] [PubMed] [Google Scholar]
- (26).Sterling T, and Irwin JJ (2015) ZINC 15 - Ligand Discovery for Everyone. J. Chem. Inf. Model 55, 2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Feng BY, Simeonov A, Jadhav A, Babaoglu K, Inglese J, Shoichet BK, and Austin CP (2007) A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem 50, 2385–2390. [DOI] [PubMed] [Google Scholar]
- (28).Irwin JJ, Duan D, Torosyan H, Doak AK, Ziebart KT, Sterling T, Tumanian G, and Shoichet BK (2015) An Aggregation Advisor for Ligand Discovery. J. Med. Chem 58, 7076–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ryan AJ, Gray NM, Lowe PN, and Chung CW (2003) Effect of detergent on ″promiscuous″ inhibitors. J. Med. Chem 46, 3448–3451. [DOI] [PubMed] [Google Scholar]
- (30).Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP, and Wood KV (2016) NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol 11, 400–408. [DOI] [PubMed] [Google Scholar]
- (31).Yip KW, Ito E, Mao X, Au PY, Hedley DW, Mocanu JD, Bastianutto C, Schimmer A, and Liu FF (2006) Potential use of alexidine dihydrochloride as an apoptosis-promoting anticancer agent. Mol. Cancer Ther 5, 2234–2240. [DOI] [PubMed] [Google Scholar]
- (32).McDonnell G, and Russell AD (1999) Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 12, 147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Spolsky VW, and Forsythe AB (1977) Effects of alexidine.2HCL mouthwash on plaque and gingivitis after six months. J. Dent. Res 56, 1349–1358. [DOI] [PubMed] [Google Scholar]
- (34).Lobene RR, and Soparkar PM (1973) The effect of an alexidine mouthwash on human plaque and gingivitis. J. Am. Dent. Assoc., JADA 87, 848–851. [DOI] [PubMed] [Google Scholar]
- (35).Elder BL, Bullock JD, Warwar RE, Khamis HJ, and Khalaf SZ (2012) Pan-antimicrobial failure of alexidine as a contact lens disinfectant when heated in Bausch & Lomb plastic containers: implications for the worldwide Fusarium keratitis epidemic of 2004 to 2006. Eye Contact Lens 38, 222–226. [DOI] [PubMed] [Google Scholar]
- (36).Coates KM, and Flood P (2001) Ketamine and its preservative, benzethonium chloride, both inhibit human recombinant alpha7 and alpha4beta2 neuronal nicotinic acetylcholine receptors in Xenopus oocytes. Br. J. Pharmacol 134, 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Dao H Jr., Fricker C, and Nedorost ST (2012) Sensitization prevalence for benzalkonium chloride and benzethonium chloride. Dermatitis 23, 162–166. [DOI] [PubMed] [Google Scholar]
- (38).Yip KW, Mao X, Au PY, Hedley DW, Chow S, Dalili S, Mocanu JD, Bastianutto C, Schimmer A, and Liu FF (2006) Benzethonium chloride: a novel anticancer agent identified by using a cell-based small-molecule screen. Clin. Cancer Res 12, 5557–5569. [DOI] [PubMed] [Google Scholar]
- (39).Lee KJ (2015) Pharmacologic Agents for Chronic Diarrhea. Intest Res. 13, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Strege PR, Evangelista S, Lyford GL, Sarr MG, and Farrugia G (2004) Otilonium bromide inhibits calcium entry through L-type calcium channels in human intestinal smooth muscle. Neurogastroenterol. Motil 16, 167–173. [DOI] [PubMed] [Google Scholar]
- (41).Ronca-Testoni S, Hrelia S, Hakim G, and Rossi CA (1985) Interaction of smooth muscle relaxant drugs with calmodulin and cyclic nucleotide phosphodiesterase. Experientia 41, 75–76. [DOI] [PubMed] [Google Scholar]
- (42).Zhu X, Gao JJ, Landao-Bassonga E, Pavlos NJ, Qin A, Steer JH, Zheng MH, Dong Y, and Cheng TS (2016) Thonzonium bromide inhibits RANKL-induced osteoclast formation and bone resorption in vitro and prevents LPS-induced bone loss in vivo. Biochem. Pharmacol 104, 118–130. [DOI] [PubMed] [Google Scholar]
- (43).Chan CY, Prudom C, Raines SM, Charkhzarrin S, Melman SD, De Haro LP, Allen C, Lee SA, Sklar LA, and Parra KJ (2012) Inhibitors of V-ATPase proton transport reveal uncoupling functions of tether linking cytosolic and membrane domains of V0 subunit a (Vph1p). J. Biol. Chem 287, 10236–10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Ito E, Yip KW, Katz D, Fonseca SB, Hedley DW, Chow S, Xu GW, Wood TE, Bastianutto C, Schimmer AD, Kelley SO, and Liu FF (2009) Potential use of cetrimonium bromide as an apoptosis-promoting anticancer agent for head and neck cancer. Mol. Pharmacol 76, 969–983. [DOI] [PubMed] [Google Scholar]
- (45).Scaglione F, Scarpazza G, Marchi E, Biella G, and Fraschini F (1983) Evaluation of domifen bromide in the treatment of acute infectious oral diseases. Int. J. Clin Pharmacol Res 3, 261–264. [PubMed] [Google Scholar]
- (46).Long Y, Chen W, Lin Z, Sun H, Xia M, Zheng W, and Li Z (2014) Inhibition of HERG potassium channels by domiphen bromide and didecyl dimethylammonium bromide. Eur. J. Pharmacol 737, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lin YM, Liu YP, and Cheung WY (1974) Cyclic 3′:5′- nucleotide phosphodiesterase. Purification, characterization, and active form of the protein activator from bovine brain. J. Biol. Chem 249, 4943–4954. [PubMed] [Google Scholar]
- (48).Sidhu RS, Clough RR, and Bhullar RP (2005) Regulation of phospholipase C-delta1 through direct interactions with the small GTPase Ral and calmodulin. J. Biol. Chem 280, 21933–21941. [DOI] [PubMed] [Google Scholar]
- (49).Forstermann U, Pollock JS, Schmidt HH, Heller M, and Murad F (1991) Calmodulin-dependent endothelium-derived relaxing factor/nitric oxide synthase activity is present in the particulate and cytosolic fractions of bovine aortic endothelial cells. Proc. Natl. Acad. Sci. U. S. A 88, 1788–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).McCluskey AJ, Poon GM, and Gariepy J (2007) A rapid and universal tandem-purification strategy for recombinant proteins. Protein Sci. 16, 2726–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.